Abstract
Background:
A prostate-specific antigen density (PSAd) cutoff of 0.15 ng/ml/cc is a commonly recommended threshold to identify patients with negative prostate magnetic resonance imaging (MRI) who should proceed to a prostate biopsy. We were unable to find any study that explicitly examined the properties of this threshold compared with others.
Objective:
To investigate whether the 0.15 cutoff is justified for selecting patients at risk of harboring high-grade cancer (Gleason score ≥3 + 4) despite negative MRI.
Design, setting, and participants:
A cohort of 8974 prostate biopsies provided by the Prostate Biopsy Collaborative Group (PBCG) was included in the study.
Outcome measurements and statistical analysis:
Locally weighted scatterplot smoothing was used to investigate whether there was a change in the risk of high-grade cancer around this value. We examined whether the use of this cutoff in patients with negative MRI corresponds to a reasonable threshold probability for a biopsy (defined as a 10% risk of high-grade disease). To do so, we applied the negative likelihood ratio of MRI, calculated from eight studies on prostate MRI, to the risk curve derived from the PBCG.
Results and limitations:
There was no discontinuity in the risk of high-grade prostate cancer at a PSAd cutoff of 0.15. This cutoff corresponded to a probability of high-grade disease ranging from 2.6% to 10%, depending on MRI accuracy. Using 10% as threshold probability, the corresponding PSAd cutoff varied between 0.15 and 0.38, with the threshold increasing for greater MRI accuracy. Possible limitations include difference between studies on MRI and the use of ultrasound to measure prostate volume.
Conclusions:
The 0.15 cutoff to recommend prostate biopsies in patients with negative MRI is justified only under an extreme scenario of poor MRI properties. We recommend a value of at least ≥0.20. Our results suggest the need for future studies to look at how to best identify patients who need prostate biopsies despite negative MRI, likely by using individualized risk prediction.
Patient summary:
In this study, we investigated whether the commonly used prostate-specific antigen density cutoff of 0.15 is justified to identify patients with negative magnetic resonance imaging (MRI) who should proceed to a prostate biopsy. We found that this cutoff is appropriate only in case of very poor MRI quality, and a higher cutoff (≥ 0.20) should be used for the average MRI.
Keywords: Prostatic cancer, Magnetic resonance imaging, Prostate-specific antigen density, Prostate biopsy, Clinical decision-making
1. Introduction
It is known that men with negative magnetic resonance imaging (MRI) have a low risk of high-grade prostate cancer (hgPCa—defined as any Gleason score of ≥3 + 4), leading to clinical recommendations to avoid a biopsy. However, while the probability of a missed hgPCa is low (5–5%) in one recent systematic review [1]), it is not negligible, leading to a search for secondary markers.
There has been particular interest in prostate-specific antigen (PSA) density (PSAd) as this can be calculated from information already available to the urologist. Many papers suggest a PSAd cutoff of 0.15 ng/ml/cc to identify patients with negative MRI who nonetheless require a prostate biopsy [2–5]. The same cutoff is also explicitly mentioned in the most recent European Association of Urology (EAU) guideline for exactly this purpose [6]. However, the origin and rationale for this cut-point are unclear. We conducted an extensive review of the literature and were unable to find any papers in the literature that explicitly examined the properties of the 0.15 threshold compared with other possible thresholds following negative MRI (score ≤2). The 0.15 cutoff is best explained as a holdover from the pre-MRI era. The earliest references to 0.15 we could find were two papers from the early 1990s, with 95 [7] and 142 [8] patients, respectively, and the cut-point derived from an analysis of the receiver operating curve. In other words, the most used PSAd cut-point for patients undergoing MRI seems be derived from analyses of patients who could not have received MRI, and furthermore, it has not been evaluated extensively for its accuracy in this context. Critically, no study has explicitly examined the diagnostic properties of the 0.15 threshold compared with the complete range of PSAd values in the negative MRI setting.
The aim of this study was to investigate whether it is justified to use a PSAd of 0.15 to recommend a biopsy in men with negative MRI. We hypothesized two possible justifications for the use of this cutoff. First, there could be a sudden change in the risk of hgPCa around the PSAd 0.15 cutoff. Second, the use of this cutoff in patients with negative MRI might correspond to a threshold probability that is reasonable for the biopsy decision. To test this second hypothesis, we first calculated the risk of hgPCa for each level of PSAd in patients who underwent a systematic prostate biopsy without MRI. Then, to estimate the risk of hgPCa on biopsy following negative MRI, we applied a multiplication factor reflecting that patients with negative MRI have a reduced risk.
2. Patients and methods
After obtaining institutional review board approval, we used data from one of the major studies on prostate cancer diagnosis—the Prostate Biopsy Collaborative Group (PBCG)—to estimate the risk of hgPCa associated with PSAd values in patients who underwent a systematic prostate biopsy without MRI. The PBCG was a consortium of 11 different North American and European institutions created to build a prediction tool for hgPCa in men being considered for a biopsy [9]. Both biopsy-naïve and previously biopsied patients were included. From this database, we selected patients who underwent a systematic prostate biopsy and who had complete clinical information on age, race, family history, previous biopsy history, PSA, prostate volume, and biopsy results. We excluded patients who had MRI and those with very high PSA levels (≥50 ng/ml) because we considered these patients to be at a high risk of hgPCa regardless of MRI results. This resulted in a final population of 8974 biopsies taken in ten different centers (Cleveland Clinic, Hamburg, Mayo Clinic, San Raffaele, Zurich, Memorial Sloan Kettering Cancer Center, Durham Veterans Affairs, San Juan Veterans Affairs, Sunnybrook, and UT Health). Overall, 62% of the final cohort data were collected prospectively.
The definition of clinically significant prostate cancer is not uniform in literature. For this reason, we selected, as the outcome of the present study, the risk of hgPCa defined as any Gleason score of ≥3 + 4 (International Society of Urological Pathology grade group ≥2). The PSAd 0.15 cutoff could be justified if there was an important change in the risk of hgPCa around this value. To test this hypothesis, we plotted the risk of hgPCa against PSAd using locally weighted scatterplot smoothing.
Another justification for the 0.15 ng/ml/cc as a cutoff could be that, in patients with negative MRI, this value corresponded to a probability of hgPCa that would be close to a reasonable threshold. We chose 10% on the basis that few urologists would want to biopsy more than ten patients to find one hgPCa [10]. The risk of hgPCa in patients with negative MRI is lower than that of a population biopsied without MRI. To obtain the risk of hgPCa on biopsy following negative MRI for each PSAd value, we applied a multiplication factor to the risk derived from the PBCG. This multiplication factor is the negative likelihood ratio of MRI and reflects the diagnostic value of MRI as a test: a negative likelihood ratio close to 1 means that risk is not much different in patients with a negative versus a positive test; a negative likelihood ratio close to zero means that a test is highly accurate, such that patients with a negative test are at a very low risk of having the disease.
To derive the negative likelihood ratio for MRI, we reviewed a number of articles, including those in the 2021 EAU guidelines [6] and selected those where a five-point scoring system was used to evaluate the MRI, patients were biopsied irrespective of MRI results, prostate biopsy was used as the reference standard, and the study provided either sensitivity and specificity of prostate MRI or data sufficient for the negative likelihood ratio to be calculated. Using this approach, we selected seven original articles (those by Ahmed et al [11], Boesen et al [5], Distler et al [3], Hansen et al [12], Mannaerts et al [13], Rouvière et al [14], and van der Leest et al [15]) and one Cochrane review [16]. Study characteristics are summarized in Table 1. For all papers, we were able to extract results for a score of ≥3 and a Gleason score ≥3 + 4 as positive MRI and positive biopsy cutoffs. We then calculated the odds of hgPCa at each level of PSAd by converting the risk obtained from PBCG using locally weighted scatterplot smoothing. The odds were multiplied by the negative likelihood ratio of MRI and converted back to probabilities (see the Supplementary material for further details). We conducted analyses to evaluate how the PSAd cutoff varies with different patient risk factors. This analysis serves a dual role of examining how different population prevalences of hgPCa would influence our findings. We repeated the analyses but by plotting the posterior probability of hgPCa for patients with versus without each risk factor for prostate cancer (digital rectal examination [DRE] results, family history for prostate cancer, African ancestry, and prior biopsy history). Finally, as some urologists use a score of ≥4 to define positive MRI, we repeated all the analyses using this cutoff. Statistical analyses were performed using R version 4.0.2 statistical software.
Table 1 –
Selected studies about prostate MRIFirst author (year)
| First author (year) | Study design | Biopsy history | Other patients’ selection criteria | MRI scoring system |
|---|---|---|---|---|
| Ahmed et al (2017) [11] | Prospective | Biopsy naïve | Patients with suspicion of prostate cancer on the basis of PSA increase, suspect DRE, and/or family history. PSA <15 ng/ml cT ≤ cT2 | Likert |
| Boesen et al (2019) [5] | Retrospective; analysis of prospective database | Biopsy naïve | PSA <20 ng/ml cT < cT3 | PI-RADS v.2 (adapted for bpMRI) |
| Distler et al (2017) [3] | Retrospective; analysis of prospective database | Biopsy naïve or previous negative biopsy | Patients with suspicion of prostate cancer PSA >4 ng/ml and/or suspicious DRE | PI-RADS v.1 |
| Drost et al (2019) a [16] | Cochrane review | Biopsy naïve or previous negative biopsy | Not reported | Likert or PI-RADS |
| Hansen et al (2018) [12] | Retrospective | Biopsy naïve | Patients with suspicion of prostate cancer on the basis of PSA increase, suspect DRE, and/or family history PSA ≤30 ng/ml | PI-RADS v.1 or v.2 |
| Mannaerts et al (2018) [13] | Prospective | Biopsy naïve | Patients with suspicion of prostate cancer on the basis of PSA of ≥3 ng/ml and/or abnormal DRE. MRI was generally omitted for men with high PSA level of >20 ng/ml in one of the two participant centers | PI-RADS v.2 |
| Rouvière et al (2019) [14] | Prospective | Biopsy naïve | Patients with suspicion of prostate cancer on the basis of PSA increase, suspect DRE, and/or family history SA <20 ng/ml cT < cT3 | Likert |
| van der | Prospective | Biopsy | Patients with suspicion of prostate | PI-RADS |
| Leest et al (2019) [15] | naïve | Cancer PSA >3 ng/ml | v.2 |
bpMRI = biparametric MRI; cT = clinical T stage; DRE = digital rectal examination; MRI = magnetic resonance imaging; PI-RADS = Prostate Imaging Reporting and Data System; PSA = prostate-specific antigen.
We used the results for MRI as an index test and systematic biopsy as the reference standard.
3. Results
Cohort characteristics are summarized in Table 2. In 2402 patients (27%), hgPCa was found. The median PSAd was 0.13 ng/ml/cc (interquartile range: 0.09–0.21), comparable with the values reported by the selected studies about MRI (Table 3).
Table 2 –
Characteristics of 8974 patients receiving prostate biopsy
| Characteristic | N = 8974 a |
|---|---|
| Age at biopsy (yr) | 64 (59, 69) |
| Black or African American | 1964 (22%) |
| Positive family history | 1461 (16%) |
| Prior negative biopsy | 2361 (26%) |
| Abnormal DRE | 2477 (28%) |
| Prostate volume at TRUS (cc) | 43 (30, 60) |
| PSA (ng/ml) | 5.9 (4.3, 8.5) |
| PSA density (ng/ml/cc) | 0.13 (0.09, |
| 0.21) | |
| Total number of biopsy cores (N = 8345) | 12 (12, 12) |
| Biopsy total Gleason score | |
| Negative | 4916 (55%) |
| 6 | 1656 (18%) |
| 7 | 1780 (20%) |
| 8 | 337 (3.8%) |
| 9 | 263 (2.9%) |
| 10 | 22 (0.2%) |
DRE = digital rectal examination; IQR = interquartile range; PSA = prostate-specific antigen; TRUS = transrectal ultrasound.
Values are given as median (IQR) or n (%).
Table 3 –
Population characteristics and derived MRI accuracies of the selected studies about MRI
| First author (year) | Population number | PSA (ng/ml) a | PSAd (ng/ml/cc) a | Positive MRI b | Positive biopsy b | Sensitivity | Specificity | Negative likelihood ratio |
|---|---|---|---|---|---|---|---|---|
| Ahmed et al (2017) [11] | 576 | 7.1 (2.9) c | NR | 418 (72) | 308 (53) | 0.88 | 0.45 | 0.27 |
| Boesen et al (2019) [5] | 808 | 6.3 (5.1–8.7) | 0.10 (0.07–0.13) | 508 (63) | 283 (35) | 0.92 | 0.53 | 0.14 |
| Distler et al (2017) [3] | 1040 | 7.2 (5.3–10.4) | 0.16 (0.1–0.24) | 696 (67) | 451 (43) | 0.84 | 0.46 | 0.34 |
| Drost et al (2019) [16] | 3091 | NR | NR | NR | NR (29) | 0.91 | 0.37 | 0.24 |
| Hansen et al (2018) [12] | 807 | 6.5 (4.9–8.8) | 0.15 (0.10–0.22) | 571 (71) | 392 (49) | 0.88 | 0.45 | 0.27 |
| Mannaerts et al (2018) [13] | 200 | 6.4 (5.1–9.1) | 0.13 (0.10–0.22) | 104 (52) | 67 (33) | 0.83 | 0.64 | 0.26 |
| Rouvière et al (2019) [14] | 251 | 6.5 (5.6–9.6) | NR | 198 (79) | 94 (37) | 0.94 | 0.3 | 0.21 |
| van der Leest et al (2019) [15] | 626 | 6.4 (4.6–8.2) | 0.11 (0.08–0.18) | 317 (51) | 190 (30) | 0.95 | 0.68 | 0.08 |
IQR = interquartile range; MRI = magnetic resonance imaging; NR = not reported; PSA = prostate-specific antigen; PSAd = prostate-specific anstigen density; SD = standard deviation.
Median (IQR).
n (%).
Mean (SD).
We first examined whether there is a discontinuity in the risk of hgPCa around a PSAd value of 0.15. The results are shown in Figure 1, where we see that increasing PSAd values were associated with increased probabilities of hgPCa, but there was no obvious increase in slope at any PSAd level.
Fig. 1 –
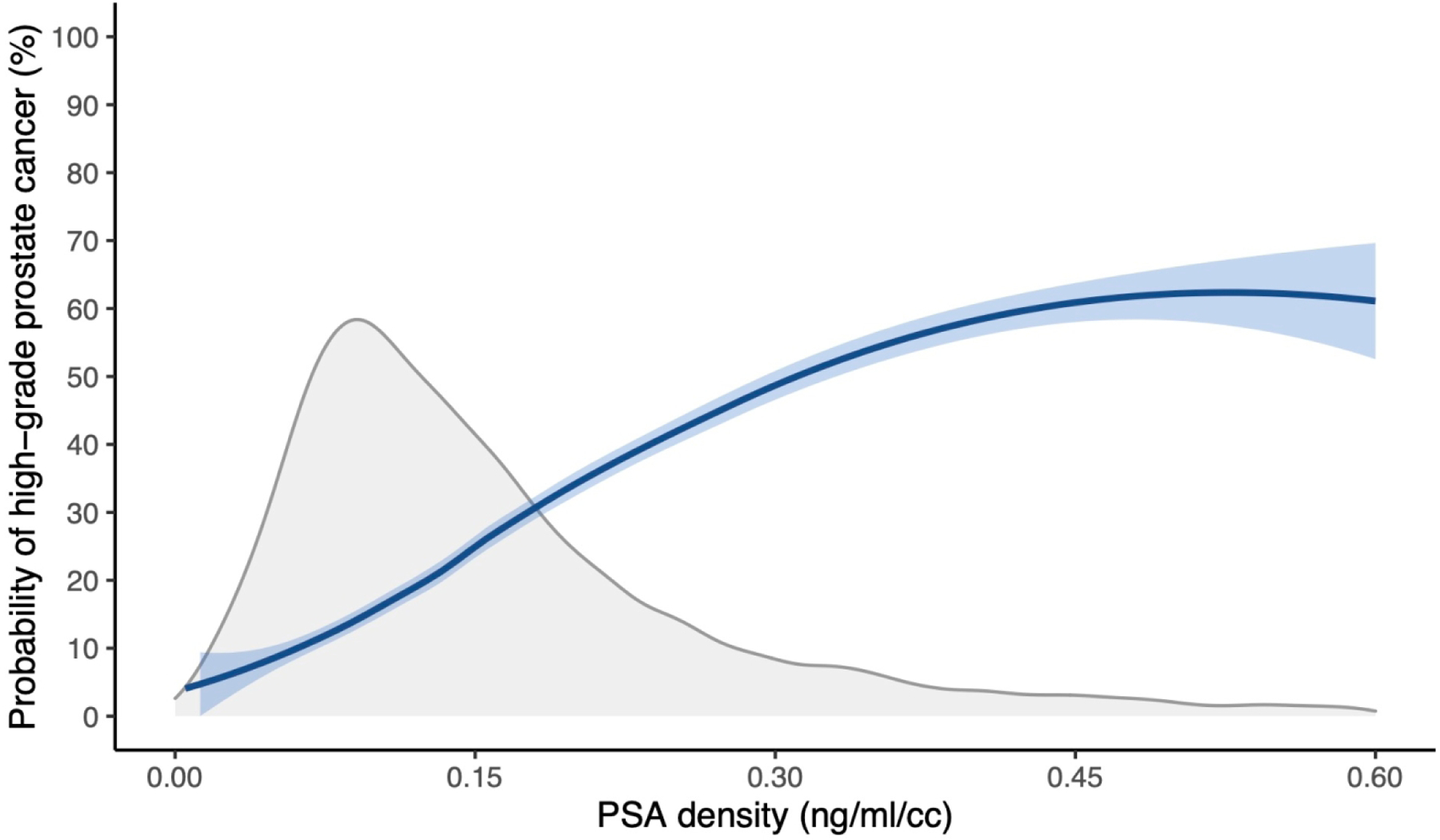
Probability of high-grade prostate cancer by PSAd. The gray curve represents the population distribution of PSAd. PSA = prostate-specific antigen; PSAd = prostate-specific antigen density.
We then examined the risk of hgPCa due to PSAd values in a patient with negative MRI to determine whether the use of 0.15 ng/ml/cc as a cutoff corresponds to a threshold probability that is reasonable for the biopsy decision. Population characteristics and derived MRI accuracy across the selected papers on prostate MRI are given in Table 3. Based on the published results, prostate MRI accuracy varied greatly between articles (sensitivity 0.83–0.95 and specificity 0.30–0.68). This resulted in negative likelihood ratios that ranged from 0.08 to 0.34. Figure 2 shows the probability of harboring hgPCa (negative MRI and PSAd value) for the extremes of the MRI likelihood ratio estimates and for the meta-analytic estimate of a negative likelihood ratio from the Cochrane review. Using a PSAd cutoff of 0.15, the corresponding threshold probabilities of hgPCa varied between 2.6% and 10%. Setting a threshold probability of 10% for hgPCa (indicated by the dotted red line in Fig. 2), PSAd cutoffs ranged from 0.15 to 0.38 for the studies. Therefore, the 0.15 cutoff is justified only where MRI accuracy is the worst reported in the literature. For the median of MRI accuracy (a negative likelihood ratio of 0.25), the threshold probability of 10% corresponded to a PSAd cutoff of 0.18.
Fig. 2 –
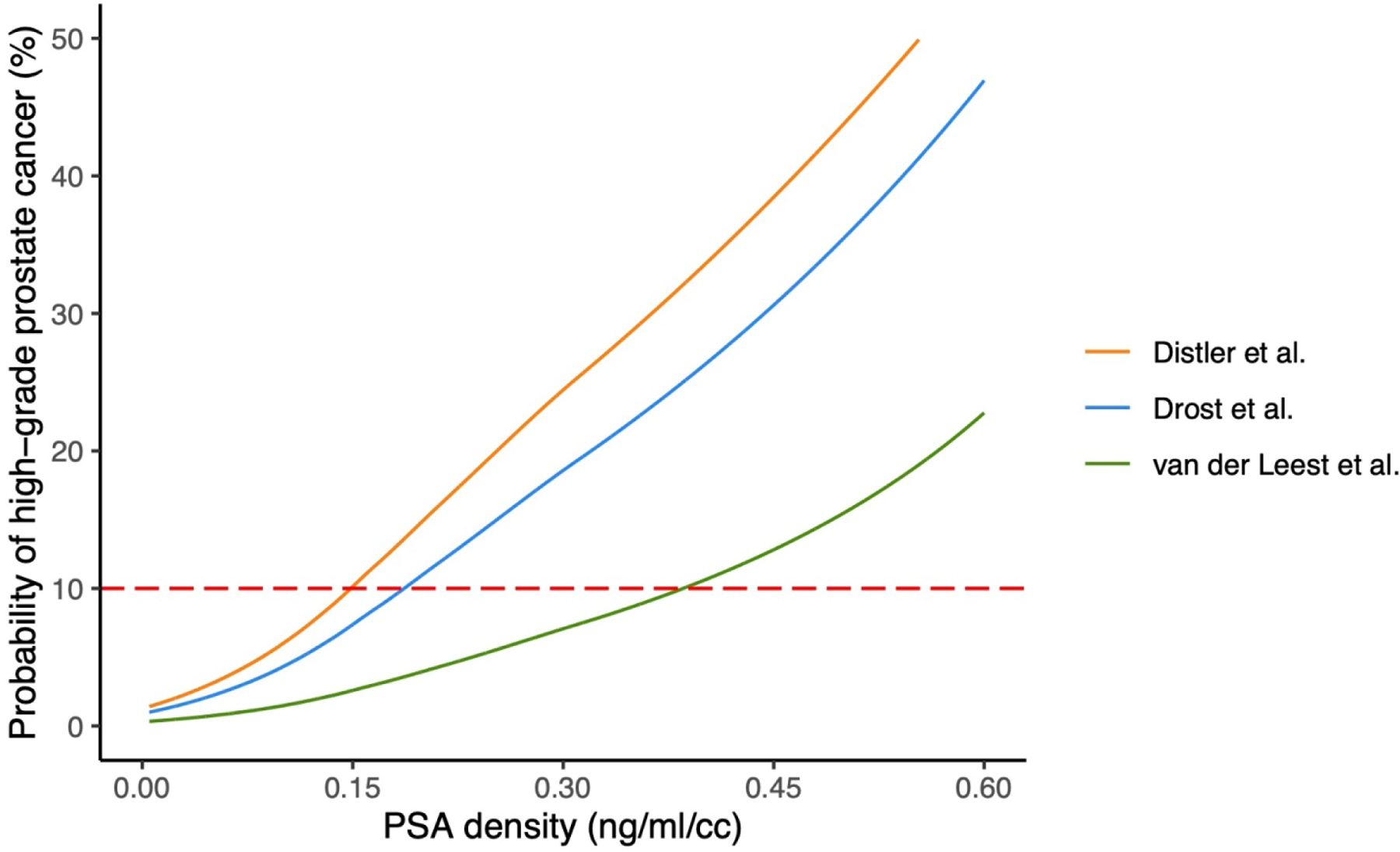
Predicted probability of high-grade prostate cancer by PSAd and different MRI accuracies from Distler et al (lowest estimate of MRI accuracy) [3], Drost et al (meta-analytic estimate including older studies) [16], and van der Leest et al (high estimate of MRI accuracy) [15]. The dotted red line indicates a threshold probability of 10% for high-grade prostate cancer. MRI = magnetic resonance imaging; PSAd = prostate-specific antigen density.
We then evaluated how the probability of hgPCa corresponding to 0.15 ng/ml/cc varies due to patient characteristics (Supplementary Fig. 1–5 and Supplementary Table 1). As expected, in patients with a risk factor, and in particular, a positive DRE, the probability of hgPCa corresponding to a PSAd of 0.15 was higher than that in the general population. For instance, the probability of hgPCa at 0.15 ng/ml/cc ranged between 6% and 21% for patients with a positive DRE versus between 2% and 8% for patients with a negative DRE.
Finally, we repeated our analyses using a score of ≥4 as a positive MRI cutoff. Only some of the selected studies provided data to calculate the negative likelihood ratio for MRI using this cutoff. Supplementary Table 2 reports the derived MRI accuracy across the selected papers using this cutoff. Supplementary Figure 6 shows the probability of harboring hgPCa for the extremes of the MRI likelihood ratio estimate when using a score of ≥4 to define positive MRI. As expected, the shapes of the curves are very similar, but they are shifted to the left. Supplementary Figure 7 shows the probability of harboring hgPCa for the MRI negative likelihood ratio derived from articles using Prostate Imaging Reporting and Data System version 2 (PI-RADS v.2). Comparing this with Figure 2, we can observe that the curves are shifted to the right, and so even a higher PSAd cutoff should be used.
4. Discussion
We found that a PSAd cutoff of 0.15 does not correspond to any discontinuity in the risk of hgPCa. It corresponds to a reasonable threshold probability of hgPCa following negative MRI only where the accuracy of MRI is at the extreme low end of estimates reported in the literature. Moreover, we found that the probability of hgPCa corresponding to this cutoff is affected by patient characteristics such as DRE and prior biopsy.
Our results are in apparent contrast with several other articles that, irrespective of different estimates for MRI accuracy or patient risk factors, recommended the use of the 0.15 ng/ml/cc cutoff in patients with negative MRI [2–5]. However, these articles prespecified the cutoff as 0.15 rather than conducting analyses to compare different possible thresholds. For instance, Oishi et al reported that merely a PSAd of ≥0.15 had a statistically significant association with the risk of hgPCa, something that would likely have also been true for cutoffs of 0.10, 0.30, or, for that matter, 0.2673 ng/ml/cc. It is illustrative that the results of these studies were quite different despite using the same threshold. Indeed, Distler et al [3] found that obtaining a biopsy in patients with negative MRI and PSAd ≥0.15 increased the detection of hgPCa in 13% versus only in 4% in the report of Boesen et al [5].
The use of a single cutoff leads to different results because, as suggested by our data, the risk of harboring hgPCa after negative MRI strongly depends on the properties of the MRI test, which differs among studies. The high variability of MRI accuracy has been reported in several articles [1,16–19], and it should, indeed, be expected given variation in technique and interobserver variability of radiologists [20].
Our data suggest that the 0.15 ng/ml/cc cutoff is appropriate only in the case of very low MRI accuracy. For the average MRI, a higher cutoff should be used—at least 0.20, assuming that MRI accuracy has improved over the years: in our study, the negative likelihood ratio was 0.25 versus 0.14 for all selected articles on MRI versus the most recent ones, respectively. A cutoff of ≥0.35 could instead be considered in centers that are very confident about their MRI accuracy. A similar cutoff was proposed by few previous studies [21,22], but these articles, as well as the majority of other papers on this topic, did not evaluate the entire range of PSAd vales. For instance, Hansen et al [21] suggested the use of a PSAd cutoff of 0.20 in patients with negative or equivocal MRI. However, they arbitrarily categorized their patients in three groups based on PSAd (≤0.10, 0.10–0.20, and >0.20), and they found that a PSAd of ≤0.20 was associated with low detection of hgPCa in patients with MRI scores of 1–3.
To the best of our knowledge, only one article has evaluated the complete PSAd range rather than using investigator-specified cut-points [23]. This study had two main limitations. First, it was relatively small, with hgPCa found in only 33 cases. Second, the statistical method used is questionable. The authors reported sensitivity and specificity rather than risk, and used a method than introduced a nonmonotonic relationship between diagnostic performance and diagnostic threshold.
Our sensitivity analyses show that the risk of hgPCa corresponding to a PSAd value in a patient with negative MRI is also affected by patient characteristics such as DRE and prior biopsy. A clearly superior alternative to using a fixed PSAd cutoff is to use an individualized risk prediction tool to estimate the risk of hgPCa for each patient combining patient information (eg, age, race, prior negative biopsy, and DRE), markers (eg, PSA and free-to-total PSA ratio), and MRI characteristics (eg, PI-RADS score and PSA volume). Prediction models have widely been used in prostate cancer for many years [9,24], but do not appear to have been developed for use in the setting of negative MRI.
A potential limitation of our study was that the two original articles represented in Figure 2 (articles by van der Leest et al [15] and Distler et al [3]) used different populations and scoring systems. Van der Leest et al [15] used PI-RADS v.2 and biopsy-naïve men only, whereas Distler et al [3] used PI-RADS v.1 and both biopsy-naïve men and men with a prior negative biopsy.
Nevertheless, the risk of hgPCa varied greatly even if we compared two studies that evaluated the same population (biopsy-naïve men) and used the same scoring system (PI-RADS v.2). For example, the corresponding threshold probabilities of hgPCa at a PSAd of 0.15 were 2.6% and 8% using data form van der Leest et al [15] and Mannaerts et al [13], respectively. Similar results were obtained by setting a threshold probability of 10% for hgPCa. The corresponding PSAd cutoffs ranged from 0.15 to 0.38 for all selected studies, and from 0.18 to 0.38 using data from van der Leest et al [15] and Mannaerts et al [13].
Another possible limitation of our study is that in the subpopulation of PBCG that we used for our analysis, prostate volume was measured by transrectal ultrasound (TRUS) rather than by MRI. However, although MRI volume estimation is more accurate, the change of our results due to the use of a difference measurement method is unlikely. Indeed, the use of MRI cannot introduce a discontinuity to Figure 1. Moreover, TRUS would have to systematically underestimate MRI volume by about 20–40% in order to have a PSAd of 0.15 correspond to a risk of hgPCa of 10% for the average MRI. Nevertheless, further studies should investigate how the PSAd cutoff may change with different volume measurement methods in this setting.
Finally, the method used may be a potential limitation. Indeed, to obtain the risk of hgPCa on biopsy following negative MRI for each PSAd value, we did not analyze a population of patients who underwent a prostate biopsy following negative MRI, but we applied the negative likelihood ratio of MRI to the risk curve of hgPCa by PSAd derived from a population of patients who underwent a systematic prostate biopsy without MRI. This approach, considering the retrospective nature of the current study, allowed us to avoid a potential selection bias of those patients undergoing a prostate biopsy for a high pretest probability of hgPCa. Ideally, our results should be confirmed in a prospective study in which all patients will undergo a prostate biopsy after negative MRI, although such a study might raise some ethical concerns.
5. Conclusions
We demonstrated that the use of a PSAd cutoff of 0.15 ng/ml/cc to recommend a prostate biopsy to patients with negative MRI is justified only in the case of very low MRI accuracy. For the average MRI, a higher cutoff of at least 0.20 is indicated. Furthermore, our results point to the need for future studies exploring how to best identify which patients need a prostate biopsy following negative MRI, most likely by using individualized risk prediction incorporating a variety of predictors in addition to PSAd.
Supplementary Material
Financial disclosures:
Francesco Pellegrino certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Andrew Vickers is named on a patent for a statistical method to detect prostate cancer that has been commercialized by OPKO Health as the 4Kscore. Andrew Vickers receives royalties from sales of the test and has stock options in OPKO Health.
Funding/Support and role of the sponsor:
This work was supported in part by the National Institutes of Health/National Cancer Institute (NIH/NCI) with a Cancer Center Support Grant to Memorial Sloan Kettering Cancer Center (P30 CA008748), a SPORE grant in Prostate Cancer to Dr.
H. Scher (P50-CA92629), and the Sidney Kimmel Center for Prostate and Urologic Cancers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
For the average magnetic resonance imaging (MRI), a prostate-specific antigen density cutoff of ≥0.20 ng/ml/cc should be used to recommend a prostate biopsy to patients with negative MRI. Future studies should look at how to best identify these patients, likely by using individualized risk prediction.
References
- [1].Sathianathen NJ, Omer A, Harriss E, et al. Negative predictive value of multiparametric magnetic resonance imaging in the detection of clinically significant prostate cancer in the Prostate Imaging Reporting and Data System era: a systematic review and meta-analysis. Eur Urol 2020;78:402–14. [DOI] [PubMed] [Google Scholar]
- [2].Oishi M, Shin T, Ohe C, et al. Which patients with negative magnetic resonance imaging can safely avoid biopsy for prostate cancer? J Urol 2019;201:268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Distler FA, Radtke JP, Bonekamp D, et al. The value of PSA density in combination with PI-RADS™ for the accuracy of prostate cancer prediction. J Urol 2017;198:575–82. [DOI] [PubMed] [Google Scholar]
- [4].Washino S, Okochi T, Saito K, et al. Combination of Prostate Imaging Reporting and Data System (PI-RADS) score and prostate-specific antigen (PSA) density predicts biopsy outcome in prostate biopsy naïve patients. BJU Int 2017;119:225–33. [DOI] [PubMed] [Google Scholar]
- [5].Boesen L, Nørgaard N, Løgager V, et al. Prebiopsy biparametric magnetic resonance imaging combined with prostate-specific antigen density in detecting and ruling out Gleason 7–10 prostate cancer in biopsy-naïve men. Eur Urol Oncol 2019;2:311–9. [DOI] [PubMed] [Google Scholar]
- [6].Mottet N, Cornford P, van den Bergh RCN, et al. EAU-EANM-ESTRO-ESUR-ISUP-SIOG guidelines: prostate cancer. European Association of Urology; 2021. [DOI] [PubMed] [Google Scholar]
- [7].di Donna A, Bazzocchi M, Guerra UP, et al. [The role of the absolute value and “density” of the prostate-specific antigen estimated echographically in the selection of patients to undergo a biopsy in suspected prostatic carcinoma. A comparison between PSA, palpation and echography in 95 patients undergoing echo-guided endorectal prostatic biopsy.] Radiol Med 1993;85:84–9. [PubMed] [Google Scholar]
- [8].Bazinet M, Meshref AW, Trudel C, et al. Prospective evaluation of prostate-specific antigen density and systematic biopsies for early detection of prostatic carcinoma. Urology 1994;43:44–51. [DOI] [PubMed] [Google Scholar]
- [9].Ankerst DP, Straubinger J, Selig K, et al. A contemporary prostate biopsy risk calculator based on multiple heterogeneous cohorts. Eur Urol 2018;74:197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Vickers AJ, van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ 2016; 352:i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ahmed HU, El-Shater Bosaily A, Brown LC, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet 2017;389:815–22. [DOI] [PubMed] [Google Scholar]
- [12].Hansen NL, Barrett T, Kesch C, et al. Multicentre evaluation of magnetic resonance imaging supported transperineal prostate biopsy in biopsy-naïve men with suspicion of prostate cancer. BJU Int 2018;122:40–9. [DOI] [PubMed] [Google Scholar]
- [13].Mannaerts CK, Gayet M, Verbeek JF, et al. Prostate cancer risk assessment in biopsy-naïve patients: the Rotterdam Prostate Cancer Risk Calculator in multiparametric magnetic resonance imaging-transrectal ultrasound (TRUS) fusion biopsy and systematic TRUS biopsy. Eur Urol Oncol 2018;1:109–17. [DOI] [PubMed] [Google Scholar]
- [14].Rouvière O, Puech P, Renard-Penna R, et al. Use of prostate systematic and targeted biopsy on the basis of multiparametric MRI in biopsy-naive patients (MRI-FIRST): a prospective, multicentre, paired diagnostic study. Lancet Oncol 2019;20:100–9. [DOI] [PubMed] [Google Scholar]
- [15].van der Leest M, Cornel E, Israël B, et al. Head-to-head comparison of transrectal ultrasound-guided prostate biopsy versus multiparametric prostate resonance imaging with subsequent magnetic resonance-guided biopsy in biopsy-naïve men with elevated prostate-specific antigen: a large prospective multicenter clinical study. Eur Urol 2019;75:570–8. [DOI] [PubMed] [Google Scholar]
- [16].Drost FH, Osses DF, Nieboer D, et al. Prostate MRI, with or without MRI-targeted biopsy, and systematic biopsy for detecting prostate cancer. Cochrane Database Syst Rev 2019;4:CD012663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mazzone E, Stabile A, Pellegrino F, et al. Positive predictive value of Prostate Imaging Reporting and Data System version 2 for the detection of clinically significant prostate cancer: a systematic review and meta-analysis. Eur Urol Oncol 2021;4:697–713. [DOI] [PubMed] [Google Scholar]
- [18].Barkovich EJ, Shankar PR, Westphalen AC. A systematic review of the existing Prostate Imaging Reporting and Data System version 2 (PI-RADSv2) literature and subset meta-analysis of PI-RADSv2 categories stratified by Gleason scores. Am J Roentgenol 2019;212:847–54. [DOI] [PubMed] [Google Scholar]
- [19].Westphalen AC, McCulloch CE, Anaokar JM, et al. Variability of the positive predictive value of PI-RADS for prostate MRI across 26 centers: experience of the Society of Abdominal Radiology Prostate Cancer Disease-focused Panel. Radiology 2020;296:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stabile A, Giganti F, Kasivisvanathan V, et al. Factors influencing variability in the performance of multiparametric magnetic resonance imaging in detecting clinically significant prostate cancer: a systematic literature review. Eur Urol Oncol 2020;3:145–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hansen NL, Barrett T, Koo B, et al. The influence of prostate-specific antigen density on positive and negative predictive values of multiparametric magnetic resonance imaging to detect Gleason score 7–10 prostate cancer in a repeat biopsy setting. BJU Int 2017;119:724–30. [DOI] [PubMed] [Google Scholar]
- [22].Falagario UG, Jambor I, Lantz A, et al. Combined use of prostate-specific antigen density and magnetic resonance imaging for prostate biopsy decision planning: a retrospective multi-institutional study using the Prostate Magnetic Resonance Imaging Outcome Database (PROMOD). Eur Urol Oncol 2021;4:971–9. [DOI] [PubMed] [Google Scholar]
- [23].Mortezavi A, Eklund M, Bergman M, Kjosavik SR, Discacciati A, Nordström T. Association between PSA density and prostate cancer in men without significant MRI lesions. BJU Int 2020;125:763–4. [DOI] [PubMed] [Google Scholar]
- [24].Alberts AR, Roobol MJ, Verbeek JFM, et al. Prediction of high-grade prostate cancer following multiparametric magnetic resonance imaging: improving the Rotterdam European Randomized Study of Screening for Prostate Cancer Risk Calculators. Eur Urol 2019;75:310–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


