Abstract
Riboswitches are cis-regulatory RNA elements that regulate gene expression in response to ligand binding through the coordinated action of a ligand-binding aptamer domain (AD) and a downstream expression platform (EP). Previous studies of transcriptional riboswitches have uncovered diverse examples that utilize structural intermediates that compete with the AD and EP folds to mediate the switching mechanism on the timescale of transcription. Here we investigate whether similar intermediates are important for riboswitches that control translation by studying the Escherichia coli thiB thiamin pyrophosphate (TPP) riboswitch. Using cellular gene expression assays, we first confirmed that the riboswitch acts at the level of translational regulation. Deletion mutagenesis showed the importance of the AD–EP linker sequence for riboswitch function. Sequence complementarity between the linker region and the AD P1 stem suggested the possibility of an intermediate nascent RNA structure called the antisequestering stem that could mediate the thiB switching mechanism. Experimentally informed secondary structure models of the thiB folding pathway generated from chemical probing of nascent thiB structures in stalled transcription elongation complexes confirmed the presence of the antisequestering stem, and showed it may form cotranscriptionally. Additional mutational analysis showed that mutations to the antisequestering stem break or bias thiB function according to whether the antisequestering stem or P1 is favored. This work provides an important example of intermediate structures that compete with AD and EP folds to implement riboswitch mechanisms.
Keywords: RNA folding, riboswitches, SHAPE-Seq, translation regulation
INTRODUCTION
Riboswitches are RNA elements that control gene expression in response to a ligand (Garst et al. 2011; Breaker 2012). Riboswitches achieve their function through the coordinated action of two domains: the aptamer domain (AD) that binds a particular ligand and the expression platform (EP) that converts ligand-binding information into a gene regulatory outcome. Much is known about AD structures that can bind ligands such as metabolites, ions, amino acids, and other small molecules (Roth and Breaker 2009; Serganov and Patel 2012; McCown et al. 2017; Ray et al. 2018). There is also a great deal known about the different classes of EPs that allow riboswitches to control multiple aspects of gene expression. For example, EPs can control transcriptional termination by forming a Rho-independent terminator hairpin or exposing a Rho-termination factor binding site (Wickiser et al. 2005; Ray-Soni et al. 2016); translation initiation through structures that occlude or expose a ribosome binding site (RBS) (Nou and Kadner 2000); RNA degradation by controlling exposure to RNase E sites (Winkler et al. 2002; Caron et al. 2012); and even the formation of alternative splice sites (Cheah et al. 2007). While AD sequences and structures are highly conserved (McCown et al. 2017), EP sequences are much more diverse. In addition, there are many examples of riboswitch sequences where the same AD can be found next to different classes of EPs (Lemay et al. 2011; Baker et al. 2012; Li and Breaker 2013; Kim et al. 2015), making it a major challenge to understand how ligand binding to the AD results in the EP enacting a regulatory decision.
Recent studies aimed at understanding riboswitch mechanisms have focused on transcriptional riboswitches, which must make their regulatory decision before the RNA polymerase finishes transcribing the riboswitch through forming either a terminator or antiterminator (Watters et al. 2016; Hua et al. 2020). Because of the mechanistic time constraint, transcriptional riboswitch processes are enacted by nascent RNA folds that form in a cotranscriptional folding regime, a regime which has been shown to be important for proper riboswitch function (Wickiser et al. 2005; Frieda and Block 2012; Perdrizet et al. 2012; Hua et al. 2020; Chauvier et al. 2021; Scull et al. 2021). Several studies of diverse transcriptional riboswitches have shown that these function through an internal strand-displacement mechanism, a kinetically efficient process by which RNA folds can rapidly change conformations (Supplemental Fig. S1; LeCuyer and Crothers 1994; Hong and Šulc 2019). Specifically, for the ZTP (Strobel et al. 2019; Hua et al. 2020), fluoride (Watters et al. 2016), and I-A type 2′ dG-sensing riboswitch (Helmling et al. 2017) riboswitches, it has been shown that EPs form through strand displacing apo-AD structures, while AD–ligand interactions block strand displacement to implement the genetic switch.
While nascent RNA folding has been shown to be important for transcriptional riboswitches, its importance is less clear for translational riboswitches (Scull et al. 2021). Previously, translational riboswitches, in which the EP fold sequesters the RBS to block translation initiation, were shown to operate post-transcriptionally through a two-state mechanism in which ligand binding tilts the structural equilibrium toward either the AD or EP fold (Smith et al. 2010; Liberman et al. 2015; Polaski et al. 2016). However, complex folding mechanisms can exist for translational riboswitches. For example, studies on the Vibrio vulnificus add adenine riboswitch showed that it operates through a three-state mechanism where the AD structure competes both with the EP-folded structure and an alternative ligand-binding incompetent structure (Reining et al. 2013). Intriguingly, the coupling of transcription and translation in bacteria (Wang et al. 2020; Blaha and Wade 2022) makes it possible that some translational riboswitches could operate cotranscriptionally to navigate a complex folding regime where strand displacement could be important (Reining et al. 2013; Bushhouse et al. 2022).
Here, we aimed to investigate the mechanism of translational riboswitches through studying the thiB thiamin pyrophosphate (TPP) translational riboswitch from Escherichia coli (Winkler et al. 2002). TPP is the only known riboswitch class represented in all domains of life, with the second highest number of unique riboswitches identified (Cheah et al. 2007; Li and Breaker 2013; Moldovan et al. 2018; Antunes et al. 2019), and diverse EPs that can control transcription, translation, degradation, and splicing all with the same highly conserved AD (Cheah et al. 2007; Li and Breaker 2013; Chauvier et al. 2017; Bushhouse et al. 2022). The TPP AD is a well-conserved three-way junction, consisting of a long-range P1 hairpin with two branched hairpins that comprise the ligand-binding pocket (Fig. 1A; Supplemental Fig. S2; Serganov et al. 2006). Notably, the P1 helix makes no direct contacts with the TPP ligand, but rather is stabilized by long-range ligand-binding interactions between the two branched helices. Currently, there are three identified E. coli TPP riboswitches that use RBS and start codon sequestration to down-regulate downstream operon gene expression: thiM, thiC, and thiB (Winkler et al. 2002). Recent studies have revealed dual control mechanisms in the thiM and thiC riboswitches that use NusG-dependent RNA polymerase pausing, Rho-dependent termination, and RBS sequestration to perform their regulation (Bastet et al. 2017; Chauvier et al. 2017, 2019). We, therefore, chose to study the E. coli thiB TPP translational riboswitch. While the E. coli thiB sequence contains identified RNA polymerase pause sites and suggested Rho-dependent termination in the downstream gene body, the mechanism this riboswitch uses to down-regulate gene expression is still unclear (Bastet et al. 2017; Chauvier et al. 2017). In addition, the thiB aptamer has been found to fold most efficiently in a cotranscriptional folding regime (Lang et al. 2007; Haller et al. 2013; Chauvier et al. 2021), suggesting that nascent RNA structures may play a role in the switching mechanism.
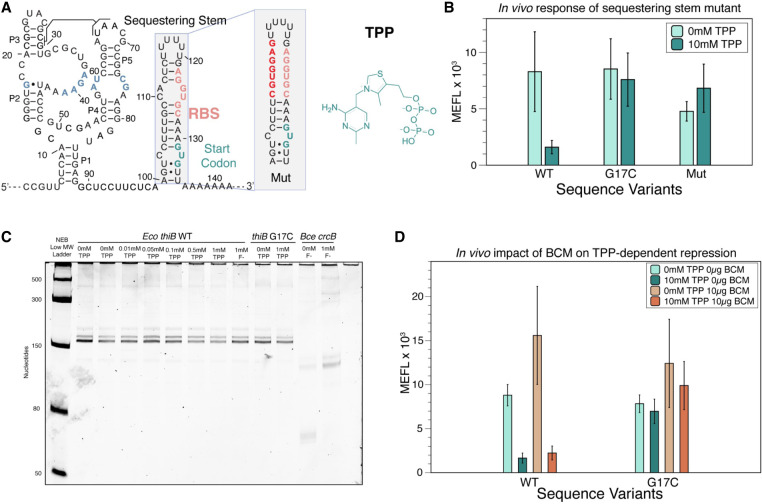
FIGURE 1.
The thiB E. coli TPP riboswitch represses translation through a sequestering stem that includes the RBS. (A) Proposed secondary structure of the ligand-bound E. coli thiB TPP riboswitch based on the published crystal structure (Serganov et al. 2006), Rfam sequence alignment (Kalvari et al. 2017), and the sequestering stem secondary structure modeled using RNAstructure (Reuter and Mathews 2010). Ligand-binding nucleotides within the ligand-binding pocket are colored blue. The RBS and the start codon for the naturally encoded downstream thiB gene are colored in salmon and teal, respectively. The chemical structure of TPP is drawn next to the RNA, and a known tertiary interaction between the P2 and P3 stems is highlighted. Inset shows secondary structure model of a mutant sequestering stem designed to weaken base-pairing with the RBS. Mutated nucleotides in red. Secondary structures determined by using RNAstructure version 6.4 (Reuter and Mathews 2010). (B) Wild-type (WT) and mutant thiB riboswitch-regulated super folder green fluorescent protein (sfGFP) expression in E. coli cells measured by flow cytometry. Units shown are molecules of equivalent fluorescein (MEFL) determined by flow cytometry. (C) Single-round in vitro E. coli RNA polymerase transcription assay measuring transcription products on a 10% urea poly-acrylamide gel. The Bce crcB F− riboswitch, used as a positive control, terminates in the absence of F− at an expected length of 82 nt, and antiterminates in the presence of F− at an expected length of 124 nt. The expected length of the full thiB transcription product is 173 nt. (D) WT and mutant thiB riboswitch-regulated sfGFP expression in E. coli cells in the presence and absence of the Rho-inhibitor bicyclomycin (BCM) measured by flow cytometry. Bar graphs in B and D represent mean values across three biological replicates, each performed in technical triplicate for nine total data points (n = 9). Error bars represent the standard deviation from the mean. Data in C are n = 2 representative gels (Supplemental Fig. S7).
We first confirmed that the E. coli thiB TPP riboswitch regulates at the level of translation using cellular gene expression assays. Using a targeted mutagenesis approach, we next investigated deletions to the linker region connecting the AD and EP, and discovered a key sequence element in this region that was essential for function. Further exploration revealed that this sequence contained a complementary sequence to the P1 stem, indicating this sequence could base pair with the P1 stem to form alternative intermediate structures during the riboswitch folding pathway. Cotranscriptional and equilibrium-refolded structure probing on intermediate lengths of the thiB RNA, followed by experimentally informed computational structure modeling, showed that the linker region can form into an antisequestering stem that competes with P1 formation as well as full EP formation, suggesting that this intermediate structure is involved in implementing the switching mechanism. We next performed detailed site-directed mutagenesis experiments focusing on this linker region by designing sequence changes to the P1 stem to favor or disfavor the formation of the predicted antisequestering stem, and found that the function of the riboswitch changed to favor the ON or OFF state according to the direction of bias. Additional mutagenesis experiments that lengthened or shortened the P1 stem or the antisequestering stem showed similar biasing effects in gene expression according to which RNA structure was favored. Overall, this investigation uncovered that the E. coli thiB switching mechanism is mediated by an antisequestering stem that forms in between the AD and EP, providing further evidence of riboswitches that utilize transient intermediate structures to implement genetic decisions.
RESULTS
The Eco thiB TPP riboswitch down-regulates translation through a sequestering stem that occludes the ribosome binding site
We first sought to develop an assay to characterize E. coli thiB riboswitch gene regulation in response to TPP in E. coli cells. The thiB TPP riboswitch is predicted to function similarly to the thiM TPP riboswitch by occluding the RBS in the presence of ligand (Rodionov et al. 2002). A secondary structure model of the thiB riboswitch was generated from the consensus TPP riboswitch aptamer structure (Kalvari et al. 2017) and the predicted fold of the EP using RNAstructure (Fig. 1A; Reuter and Mathews 2010). Based on this knowledge, we designed and constructed a plasmid consisting of a constitutive σ70 E. coli RNA polymerase promoter followed by the E. coli thiB TPP riboswitch sequence and a sfGFP coding sequence (Supplemental Fig. S3). The riboswitch coding sequence was designed to start at the transcription start site (Vogel et al. 2003) for the thiBPQ operon through the first 12 nt of the thiB gene, with the downstream coding sequence included based on the predicted location of the sequestering stem that is thought to fold to occlude the RBS in the riboswitch OFF state (Fig. 1A; Rodionov et al. 2002).
We next used this expression construct to optimize an E. coli flow cytometry assay to characterize sfGFP fluorescence as regulated by the thiB TPP riboswitch (Supplemental Fig. S4). An important part of this assay is a ligand unresponsive control that should produce constitutive fluorescence independent of TPP. To generate this mutant, we used the crystal structure of the well-studied E. coli thiM TPP riboswitch (Serganov et al. 2006), along with mutagenesis data (Ontiveros-Palacios et al. 2007), to generate a series of point mutations predicted to interfere with ligand binding but not overall aptamer structure (Supplemental Fig. S5). Constructs were transformed in E. coli TG1 chemically competent cells, grown overnight in LB media, and then subcultured for 6 h in thiamine hydrochloride deficient M9 media in the absence or presence of indicated concentrations of TPP. While the WT sequence showed repression in the presence of TPP, all mutants tested showed a broken “ON” phenotype (high fluorescence independent of ligand) with fluorescence levels similar to or above the WT sequence (Supplemental Fig. S5B), indicating that the WT repression was due to ligand interactions with the riboswitch and not TPP-dependent toxicity. We chose to use mutant G17C for additional experiments as it showed the most similar sfGFP expression to the WT thiB riboswitch with no ligand.
We next performed a dose–response curve comparing the WT and G17C mutant riboswitches to determine the optimum concentration of TPP for further experiments, which revealed that 10 mM TPP provided distinguishable repression between these two constructs (Supplemental Fig. S6). We next tested the importance of EP structural context on thiB repression. We hypothesized the formation of the predicted EP hairpin blocked ribosome binding to the RBS, therefore repressing gene expression. To test this hypothesis, we created a mutation to the EP that was designed to weaken base-pairing with the RBS in this sequestering stem (Fig. 1A). Fluorescence characterization of this mutant resulted in a broken response to TPP, indicating that base-pairing context with the RBS within the proposed sequestering stem is important for the thiB repression mechanism (Fig. 1B).
To further confirm that thiB regulates at the level of translation, we tested whether this RBS sequestering stem could also function as an intrinsic transcriptional terminator by completing single-round in vitro transcription at increasing concentrations of TPP. In this assay, we observed only full-length transcription products with increased concentrations of TPP, indicating that the riboswitch does not function through transcriptional termination in these conditions (Fig. 1C; Supplemental Fig. S7). Previous studies have shown thiC and thiM E. coli TPP riboswitches repress gene expression through dual control mechanisms that use both an RBS sequestering hairpin and Rho-dependent transcriptional termination (Bastet et al. 2017; Chauvier et al. 2017). To test whether this could also be true for thiB, we tested whether the Rho-inhibitor BCM altered the riboswitch's repression due to TPP. Characterization of the WT riboswitch sequence in cultures where BCM was added in the subculture M9 media at 10 µg/mL, showed a similar repression in the presence of TPP, suggesting that Rho is not important for riboswitch function (Fig. 1D).
Taken together, these results demonstrate that the thiB TPP riboswitch regulates at the level of translation in our experimental context, and that base-pairing interactions with the RBS within a putative sequestering stem are important for this regulation.
Nucleotides in the linker region are essential for thiB gene regulation
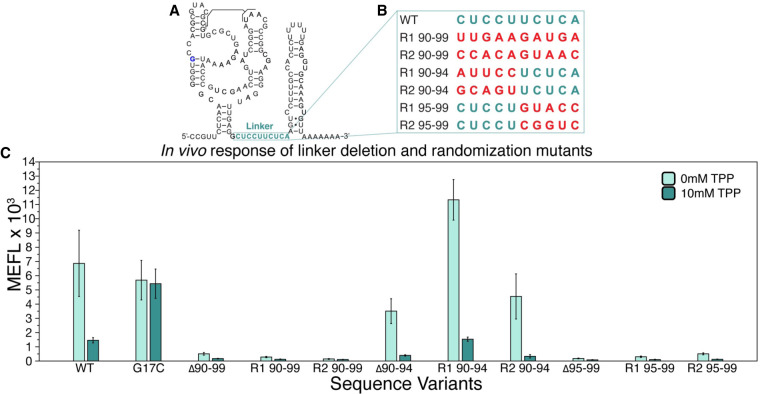
Previous work has shown that sequences directly after a P1 helix can play a role in riboswitch mechanisms through the formation of intermediate structures that compete with EP folding to enact the switch (Cheng et al. 2022). Consequently, we next sought to determine whether this sequence could play a role in the thiB switching mechanism. We were specifically interested in investigating nucleotides 90–99, a nonconserved sequence region that is predicted to form a single-stranded “linker” between the aptamer and the EP sequestering stem in the thiB OFF state (Fig. 2A). While this 10 nt sequence does not have sequence complementarity with nucleotides in the model of the sequestering stem, we sought to investigate whether this region, or portions of this region, are essential for thiB function. To address this question, we performed functional mutagenesis of this region by deleting or randomizing portions of the sequence (Fig. 2B). Upon deletion or randomization of the entire 10 nt sequence, we observed total loss of sfGFP expression in the presence or absence of TPP, indicating that these mutant riboswitches could possibly be trapped in the RBS sequestering state (Fig. 2C). We next investigated deleting or mutating portions of the linker region. Deleting or randomizing the 5′ half (nts 90–94) resulted in mutants that demonstrated repression in the presence of TPP similar to the WT sequence (Fig. 2C), indicating that this sequence was not essential for riboswitch function. However, deletion or randomization of the 3′ half (nts 95–99) showed the broken OFF phenotype (low fluorescence independent of ligand), indicating that this region is essential for riboswitch function. Taken together, these results demonstrate that the linker plays an important role in thiB regulation.
FIGURE 2.
A specific portion of the linker region in the E. coli thiB TPP riboswitch is essential for TPP-dependent repression. (A) Secondary structure model of the thiB E. coli TPP riboswitch after Figure 1A, with linker sequence (nts 90–99) outlined in teal. (B) A table of sequence randomizations (R) tested within the linker sequence, labeled according to which nucleotides were randomized. (C) Flow cytometry assay of plasmid sfGFP expression regulated by the ThiB riboswitch sequence variants. X-axis labels indicate the riboswitch sequence variant tested ([WT] wild-type, [G17C] ligand unresponsive mutant, [Δ] deletion). Bar graphs represent mean values across three biological replicates, each performed in technical triplicate for nine total data points (n = 9). Error bars represent the standard deviation from the mean.
Cotranscriptional and equilibrium-refolded SHAPE-Seq of thiB intermediates confirms the presence of an antisequestering stem formation
The above results demonstrated the importance of the 3′ half of the linker region for thiB function. A closer analysis of this sequence revealed that these nucleotides (95–99) are complementary to the 3′ side of P1 (nts 85–89), and thus potentially able to form an alternative intermediate hairpin together that, if extended through ligand-binding portions of the aptamer sequence, would prevent the EP from forming and thus act as an “antisequestering” stem. In addition, this linker region sequence is identical to the 5′ side of the P1 stem (nts 5–9), suggesting that the P1 helix and this antisequestering helix could be mutually exclusively base-paired regions within the riboswitch. Combined, these observations suggest the hypothesis that the ligand-dependent formation of such an antisequestering hairpin could be a core feature of the thiB switching mechanism.
To test this hypothesis, we generated experimentally informed models of nascent thiB RNA structures using a combination of cotranscriptional Selective 2′-Hydroxyl Acylation Analyzed by Primer Extension sequencing (cotranscriptional SHAPE-Seq) (Watters et al. 2016) and the computational algorithm Reconstructing RNA Dynamics from Data (R2D2) (Yu et al. 2021). Cotranscriptional SHAPE-Seq combines in vitro transcription, transcriptional road blocking, and SHAPE probing to collect RNA structure chemical probing data from RNAs folded in stalled transcription elongation complexes (Watters et al. 2016). To perform cotranscriptional SHAPE-Seq, we performed PCR using biotinylated dNTPs using the TPP riboswitch expression construct as a template (Strobel et al. 2017). In vitro transcription from this template pool in the presence of streptavidin generates a randomly distributed mixture of all intermediate length TPP nascent RNAs within stalled E. coli RNA polymerase transcription complexes, which we chemically probed with benzoyl cyanide (BzCN) (Watters et al. 2016). Extracting these RNAs, performing reverse transcription, and formatting and sequencing the resulting cDNAs yielded a matrix of SHAPE-Seq reactivities for each transcript length (TL), which we generated using 0 mM or 1 mM TPP in the transcription conditions (Supplemental Fig. S8). We also performed a similar experiment in which nascent RNAs were generated in the same way, but were extracted and equilibrium refolded without RNAP before probing to study the difference between cotranscriptional and equilibrium folding regimes (Supplemental Fig. S9).
Chemical probing reactivities were then fed into the R2D2 algorithm to generate experimentally informed computational models of nascent RNA structures. R2D2 uses a sample-and-select approach to generate models of families of RNA structures that are maximally consistent with the experimental data by first using folding algorithms to stochastically sample hundreds of thousands of possible RNA folds from a given sequence, and then using experimental data to select from those possibilities the ones that are most consistent with the data. Running R2D2 hundreds of times for a given SHAPE-Seq data set will generate populations of structures at each nascent TL that are consistent with the data. These populations are conveniently visualized as RNAbow plots (Aalberts and Jannen 2013), where an arc between two nucleotides indicates they are paired in a particular structure, the thickness of the arcs represents the proportion of structures in the population that have that base pair, and the color corresponds to a particular structural unit. For convenience, the populations are also converted into a consensus structure representing the average structural state of each base pair over the population.
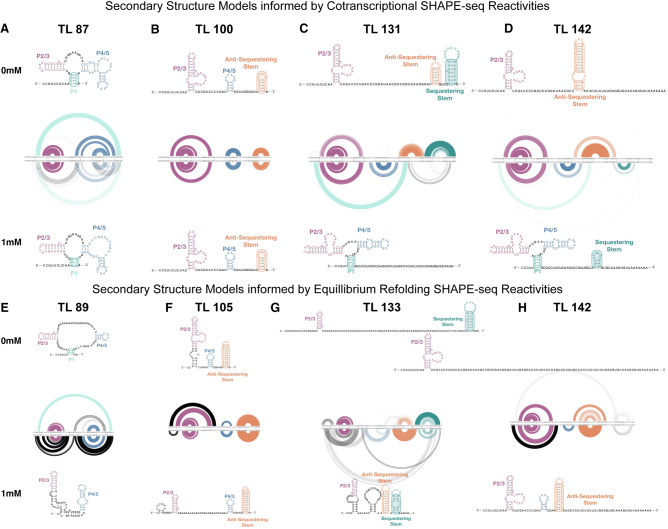
An R2D2 analysis of cotranscriptional SHAPE-Seq reactivity data from 0 and 1 mM TPP conditions showed that early TLs form a secondary structure similar to the aptamer (Fig. 3A). Specifically, these structures contain portions of P1, P2/P3, and P4/5, but like our previous R2D2 analysis of the yxjA purine riboswitch folding pathway (Cheng et al. 2022), lack noncanonical interactions and pseudoknots present in the three-dimensional aptamer since R2D2 only models at these pathways secondary structure level (Yu et al. 2021). We also note that the R2D2 P1 stem prediction contained differences in the 5′ paired nucleotides from the crystal structure, possibly due to difficulties in sampling correctly paired P1 structures. Interestingly at TL 100, both in the presence and absence of TPP, the P1 hairpin was not selected and instead a nascent structure corresponding to the antisequestering stem was found to form in both ligand conditions (Fig. 3B). However, by TL 131, the folding pathways were found to diverge: in the absence of TPP, the P4/5 hairpin was not present, while a portion of the antisequestering stem remained, allowing a portion of the sequestering stem containing the RBS to form; in the presence of TPP, the P1 hairpin reformed, with the antisequestering stem no longer present. At full length (TL 142), in the absence of TPP, the EP was found to fold into an extended antisequestering stem corresponding to a riboswitch ON state, while in the presence of TPP, the TPP aptamer was predicted to form alongside a portion of the sequestering stem corresponding to a riboswitch OFF state. Similar models were obtained with a replicate cotranscriptional SHAPE-Seq data set, although the presence of the P1 hairpin in the presence of 1 mM TPP could not be detected past TL 89 nt (Supplemental Figs. S10, S11).
FIGURE 3.
Experimentally informed models of the cotranscriptional and equilibrium-refolded RNA folding pathways for E. coli thiB riboswitch in the presence and absence of TPP. R2D2 models of secondary structure intermediate in the folding pathway. One hundred secondary structure models were generated from the experimental data for each intermediate RNA length. Models are depicted as RNA bow plots (0 mM TPP top, 1 mM TPP bottom), where arcs between nucleotides represent base pairs in a particular structure, arc thickness is proportional to the number of times that base pair is present in the one hundred structure set, and arcs are color coded according to portions of the riboswitch models. Secondary structure depictions are the consensus structure over the 100 iterations. Models and consensus structures from cotranscriptional SHAPE-Seq data sets at TLs (A) 87 nt, (B) 100 nt, (C) 131 nt, (D) 142 nt, and from equilibrium-refolded SHAPE-Seq data sets at TL (E) 89 nt, (F) 105 nt, (G) 133 nt, (H) 142 nt. Representative lengths for cotranscriptional and refolded conditions were chosen to be similar but do not exactly match due to differences in read coverage (see Materials and Methods).
R2D2 analysis of equilibrium-refolded SHAPE-Seq data generated in the presence and absence of TPP showed structural models that differed from those generated with cotranscriptional SHAPE-Seq data (Fig. 3E–H; Supplemental Fig. S10). For one replicate data set, at the full-length transcript (length 142), R2D2 equilibrium models predicted the presence of the antisequestering stem in a large fraction of the structural subpopulations independent of ligand concentration, suggesting that the RNA folding pathway is less likely to bifurcate into different structures under these folding conditions (Fig. 3H). Similar conclusions hold for a replicate equilibrium refolding data set, though with the antisequestering stem present alongside a portion of the sequestering stem in the 1 mM TPP condition, making it difficult to conclude if this is a functional OFF riboswitch state (Supplemental Figs. S10, S11).
Notably, the experimentally informed secondary structures modeled by R2D2 both with and without the TPP ligand did not accurately represent the known ligand-bound secondary structure of the AD in either folding condition. This is an expected limitation of using R2D2, since the underlying computational algorithms used to sample possible structures are not able to incorporate known features of the TPP AD including noncanonical RNA base pairs, pseudoknots, and other sophisticated RNA–ligand interactions (Spasic 2018; Ermolenko 2021; Casadio 2022). As we have observed before in our study of the yxjA purine riboswitch folding pathway (Cheng et al. 2022), despite inaccuracies in AD modeling, cotranscriptional SHAPE-Seq and R2D2 can still provide important models for EP intermediate folding pathways at the secondary structural level which can be tested and corroborated through functional experiments.
Overall, these data-driven structural models provide evidence for the occurrence of an intermediate antisequestering structure in the E. coli thiB folding pathway, that the switching mechanism facilitated by this intermediate structure may be able to occur both co- and post-transcriptionally, and are inconclusive as to which folding regime would allow most efficient switching.
Linker region mutations show the antisequestering stem is important for thiB gene regulation
The above results suggest a model in which the TPP riboswitch bifurcates its folding pathway to form an antisequestering stem ON state in the absence of TPP, and a sequestering stem OFF state in the presence of TPP. A closer inspection of the structural models of the antisequestering stem shows that a portion of its structure overlaps with the P1 stem, suggesting that the P1 helix and the antisequestering helix could be mutually exclusively base-paired regions within the riboswitch. Notably, such internal competing structures have been identified in other riboswitches that appear to leverage strand displacement in their switching mechanisms (Strobel et al. 2019; Cheng et al. 2022).
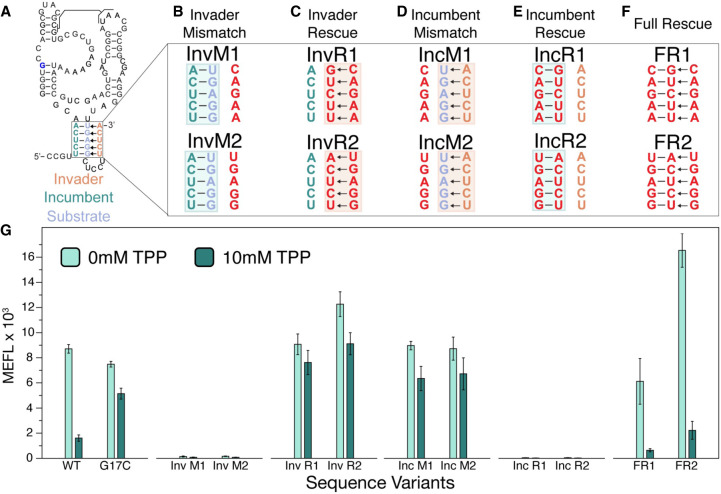
Strand displacement is a process in which nucleotide strands base-pair at the cost of displacing another previously bound nucleotide strand (Supplemental Fig. S1; Hong and Šulc 2019). Specifically, an internal helix, composed of an “incumbent” strand:“substrate” strand duplex, can be broken apart when an “invader” strand forms the competing invader:substrate duplex. Within the thiB system, this nomenclature can be used to label the 5′ side of the P1 (nts 5–9) as the incumbent strand, the 3′ side of P1 (nts 85–89) as the substrate strand, and nucleotides 95–99 of the linker region as the invading strand (Fig. 4A). Using this framework, we hypothesized that mutations to these strands would bias riboswitch function according to which structure they favored: mutations that favored incumbent:substrate pairing would favor the formation of P1 and the sequestering stem and bias riboswitch function to the OFF state, while mutations that favored substrate:invader pairing would favor the formation of the antisequestering stem and favor the riboswitch ON state.
FIGURE 4.
Complementary base pairs within P1 and within the antisequestering stem are required for E. coli thiB TPP riboswitch TPP-dependent repression. (A) Secondary structure model of the thiB E. coli TPP riboswitch after Figure 1A including the proposed top portion of the antisequestering stem drawn as invading the P1 helix. Invader, incumbent and substrate strands are labeled according to a mechanism in which the incumbent:substrate duplex forms the P1 stem of the aptamer and the substrate:invader duplex forms the top of the antisequestering stem. Arrows indicate a potential mechanism where the invader displaces the incumbent during antisequestering stem folding. (B–F) Mutant nucleotides are depicted in red, while WT nucleotides are teal (incumbent), orange (invader), and blue (substrate). Mutations were designed to break the invasion of the antisequestering stem (B, invader mismatch, InvM), rescue the invasion and break P1 (C, invader rescue, InvR), break P1 and allow invasion by the antisequestering stem (D, incumbent mismatch, IncM), rescue P1 and break the invasion (E, incumbent rescue, IncR), and rescue all base-pairing interactions with sequences different than the WT sequence (F, full rescue, FR). (G) Flow cytometry assay of plasmid sfGFP expression regulated by the thiB riboswitch sequence variants. X-axis labels indicate the riboswitch sequence variant tested ([WT] wild-type, [G17C] ligand unresponsive mutant). Bar graphs represent mean values across three biological replicates, each performed in technical triplicate for nine total data points (n = 9). Error bars represent the standard deviation from the mean.
To test these hypotheses, we constructed sets of mismatch mutants designed to break and rescue specific substrate:incumbent and substrate:invader base pairs and characterized their gene expression. As predicted, when the incumbent:substrate interaction (P1 stem) was favored, the riboswitch function was broken in the OFF state (Fig. 4B,E,G). In contrast, when the substrate:invader interaction (antisequestering stem) was favored, the riboswitch was broken in the ON state (Fig. 4C,D,G). Finally, a full rescue of both incumbent:substrate and substrate:invader interactions, such that the mutually exclusive hairpin structures are both possible, fully rescued the ability of the thiB sequence to regulate expression in response to TPP, although with varying dynamic range than the WT sequence (Fig. 4F,G).
Taken together, these results support the importance of pairing between the 5′ and 3′ sides of P1 and the 3′ side of P1 with the linker to form an antisequestering hairpin for riboswitch switching.
Lengthening or shortening the P1 stem or the antisequestering stem bias TPP riboswitch regulation
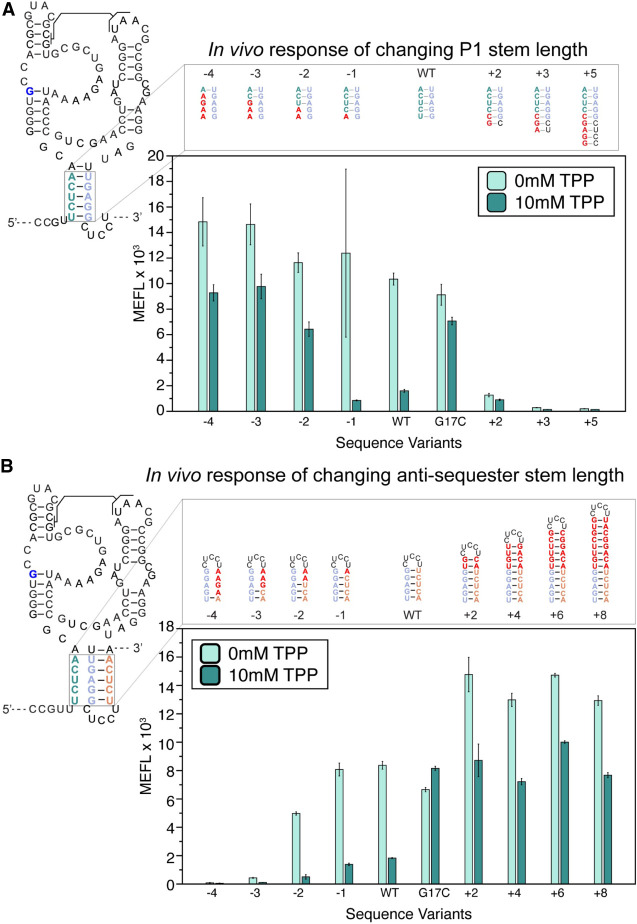
The above results demonstrate the importance of overlapping sequence complementarity between P1 and the antisequestering stem for proper riboswitch function. Based on this data, we predicted that manipulating the length of either helix could impact riboswitch function by biasing the fold to favor either P1 or the antisequestering stem. Specifically, we hypothesized that strengthening P1 by adding base pairs, or shortening the antisequestering stem with mismatches, could inhibit antisequestering hairpin formation and bias the riboswitch to the OFF state even in the presence of ligand. Conversely, promoting the formation of the antisequestering stem—either through P1 weakening mutations or antisequestering stem strengthening mutations—could bias the riboswitch in the ON state.
To test these hypotheses, we lengthened the P1 stem by adding complementary nucleotides to the 5′ side which could base-pair with the loop of the antisequestering stem (Fig. 5A). These mutants expressed low amount of sfGFP both with and without TPP, confirming that strengthening the P1 helix with more base pairs favored the OFF pathway (Fig. 5A). We next shortened the P1 helix by making 5′ side mismatch mutations (Fig. 5A), which resulted in mutants that repressed in the presence of TPP, but with overall higher sfGFP expression both in the presence and absence of TPP. To demonstrate that this effect was independent of sequence, we designed a second set of P1 shortening mutants, which had the same impact on sfGFP expression and repression due to TPP (Supplemental Fig. S12). This further supported our hypothesis that weakening the P1 stem biases the folding pathway toward the ON state (Fig. 5A).
FIGURE 5.
Changing the length of P1 and the antisequestering stem have the opposite effects on E. coli thiB TPP riboswitch function. (A) P1 stem mutants that lengthen and shorten the P1 stem. Lengthening mutants change nucleotides in the leader sequence before the riboswitch to extend P1. Mismatches on the 5′ end shorten the P1 helix from the bottom. Secondary structure depictions of the mutations are depicted above flow cytometry assay data of plasmid sfGFP expression regulated by the thiB riboswitch P1 sequence variants. X-axis labels indicate the riboswitch sequence variant tested ([WT] wild-type, [G17C] ligand unresponsive mutant). (B) Sequence mutants for the antisequestering stem. Mutants to the left of the WT hairpin are mismatch mutations which shorten the antisequestering helix, while mutants to the right extend the helix. Secondary structure depictions and flow cytometry assay data as in (A). X-axis labels indicate the riboswitch sequence variant tested ([WT] wild-type, [G17C] ligand unresponsive mutant). Bar graphs represent mean values across one biological replicate, each performed in technical triplicate for three total data points (n = 3). Error bars represent the standard deviation from the mean.
Next, we designed mutations to weaken and strengthen the antisequestering stem, testing our hypothesis that antisequestering stem length has a direct relationship with thiB TPP riboswitch expression (Fig. 5B). As predicted, mismatch mutations to the antisequestering stem 3′ side, which shortens the antisequestering stem by three or more base pairs, result in always OFF phenotypes where sfGFP expression is low both in the presence and absence of TPP. We next designed a set of mutants which added base pairs to the top of the antisequestering helix two base pairs at a time. As predicted, these antisequestering stem lengthening mutants resulted in higher overall sfGFP expression than the WT riboswitch. However, interestingly these antisequestering stem lengthening mutants were able to repress in the presence of TPP, and increasing the length of the antisequestering stem beyond four base pairs did not increase the sfGFP expression above the two base pair addition. This suggests there is a limit to the impact antisequestering stem lengthening may have on riboswitch expression. Similar to our previous results, we created a second set of antisequestering stem lengthening and shortening mutants to test overall sequence bias, and the second set of mutants had similar results (Supplemental Fig. S13). Overall, these results matched the behavior of the corresponding P1 mutations, demonstrating the direct relationship between antisequestering stem length and thiB TPP riboswitch expression.
Taken together, these results show that the riboswitch regulatory decision can be biased in opposite directions through mutations that favor the formation of P1 or the antisequestering stem, pointing to the central role these structures play in determining riboswitch fate.
DISCUSSION
In this work, we present evidence that the E. coli thiB TPP riboswitch mechanism regulates translation via an intermediate “antisequestering” RNA structure that competes with the folding of the AD and the RBS sequestering EP stem.
Using cellular gene expression assays, we were able to first verify that thiB acts at the level of translational regulation (Fig. 1). Mutagenesis of the thiB linker region that connects the AD to the EP revealed a sequence element that when deleted resulted in riboswitches that were broken OFF, indicating it is important for the switching mechanism (Fig. 2). Experimentally informed secondary structure models of both cotranscriptionally and equilibrium-refolded thiB intermediates revealed that once the linker sequence is available, the antisequestering stem can form and compete with the formation of the aptamer P1 stem. In addition, these models showed a bifurcation between the antisequestering stem and the sequestering stem at full length that favors the sequestering stem in the presence of TPP (Fig. 3). Characterization of riboswitch mutants that were designed to favor the formation of P1 or this antisequestering stem, either through point mutations (Fig. 4) or lengthening/shortening mutations (Fig. 5), biased the riboswitch toward the OFF or ON gene expression states, respectively, indicating both the importance of the antisequestering stem for the switching mechanism, as well as the balance needed between the stabilities of the P1 stem and the antisequestering stem for proper riboswitch function.
As noted above, our approach of using cotranscriptional SHAPE-Seq and R2D2 to capture and model folds of intermediate riboswitch structures is limited to modeling secondary structures that can be computationally sampled using existing folding algorithms, precluding accurate modeling of riboswitch ADs that utilize intricate structures to form ligand-binding pockets. However, as also noted above, this approach can be used to capture and model EP states, and demonstrate for example the ligand-dependence of different intermediate EP folds (Fig. 3). This feature allowed us to discover new aspects of the TPP riboswitch mechanism, for example by designing mutants to test the dependence of gene expression on intermediate EP structures, and demonstrate the importance of proper antisequestering stem formation for TPP riboswitch function (Figs. 4,5).
Interestingly, our finding of the importance of an intermediate RNA structure in the thiB mechanism is remarkably similar to studies of other OFF riboswitches. Specifically, in the case of the Bacillus subtilis yxjA transcriptional OFF purine riboswitch, we found the presence of an intermediate central helix that competes with the aptamer P1 stem and the EP to implement ligand-dependent transcriptional termination (Cheng et al. 2022). In this way, the central helix acts to flip the logic of the purine riboswitch architecture—in riboswitches without this central helix such as the pbuE riboswitch, the lack of a central helix allows direct competition between the folding of the aptamer and the transcriptional terminator EP which results in a ligand-dependent ON switch. We recently showed that the addition of a synthetic sequence to the Clostridium beijerinkii ZTP transcriptional riboswitch, designed to create an intermediate that competes with both the aptamer and the EP, can also flip the logic of this switch from ON to OFF (Bushhouse and Lucks 2023). In addition, the presence of an intermediate structure necessary for proper switching in thiB is similar to the mechanism for the V. vulnificus add riboswitch (Reining et al. 2013). The finding that a similar competition exists in the thiB translational OFF riboswitch points to a potential generality of this internal RNA structure competition mechanism that could be at play in riboswitches that utilize diverse EPs.
While it is clear from our results that the antisequestering stem is a critical intermediate necessary for the thiB mechanism, less clear is whether the mechanism must operate in a cotranscriptional folding regime. The observation of the antisequestering stem and the ligand-dependent bifurcation toward folds that contain the sequestering stem in cotranscriptional folding conditions (Fig. 3A–D; Supplemental Fig. S12A–D) support that the mechanism could operate in a cotranscriptional folding regime. This would be consistent with results from a recent cotranscriptional smFRET study, which found transcription to be essential for the thiB riboswitch to sense ligand, and that antisequestering hairpin formation defines the end of the ligand sensing transcription window (Chauvier et al. 2021). We also note that our mutagenesis results are consistent with a cotranscriptional strand-displacement mechanism like we found for the yxjA purine transcriptional riboswitch (Cheng et al. 2022). However, in one of the experimental replicates, the data-driven equilibrium-refolded models of the riboswitch folding intermediates suggested a similar ligand-dependent bifurcation could happen. More work is needed to resolve the impact of the RNA folding regime on the thiB mechanism.
This work could also shed light onto the evolution of the riboswitch EP sequence. While aptamer sequences are highly conserved, EP sequences are known to show a high degree of variability (Moldovan et al. 2018). As seen in this work, changes to EP sequence can bias the riboswitch toward the ON or OFF state, indicating that EP sequence variation could be an evolutionary mechanism to tune the riboswitch function (Bushhouse and Lucks 2023).
Overall, this work continues to connect our understanding of the folding mechanisms of transcriptional and translational riboswitches showing, in the case of the E. coli thiB TPP riboswitch, that similar intermediate RNA structures can implement the switching mechanism used by diverse riboswitches. As such, this study provides inspiration for further investigation of how nascent RNA intermediates influence other RNA regulatory mechanisms (Bushhouse et al. 2022).
MATERIALS AND METHODS
Strains, primers, and media
All plasmids were derived from the p15a plasmid backbone with chloramphenicol resistance. Plasmid sequences are listed in Supplemental Data File S1 and were created either using Gibson Assembly or inverse PCR (iPCR). Some strains were deposited in Addgene with accession numbers listed in Supplemental Data File S1. All strains were grown in Difco LB broth for cloning and purification. Gibson assembly was used to add the E. coli thiB WT sequence, including 2 nt of the thiB promoter sequence, downstream from the E. coli sigma 70 consensus promoter (annotated as J23119 in the registry of standard parts, Supplemental Data Table S1) followed by the sfGFP sequence. In order to create different mutants using iPCR, primers designed to add mutant sequences were ordered from Integrated DNA Technologies (IDT). Then, 200 µL PCR reactions were mixed with 1–10 ng/µL of template DNA, 200 µM dNTPs, 1× Phusion Buffer, 100 nM each primer and 0.25 µL of Phusion Polymerase (2000 U/mL; NEB). PCR products were run on a 1% agarose gel to confirm the desired length. Dpn1 (NEB) was used to digest template DNA and PCR Cleanup (Qiagen) was used to purify PCR products. Digested and purified PCR products were then phosphorylated and ligated simultaneously using T4 PNK Enzyme (NEB), T4 DNA Ligase (NEB), and 10× T4 DNA Ligase Buffer by incubating at room temperature for 1 h. Ligation products were then transformed into NEBTurbo competent cells and plated on chloramphenicol LB agar plates and incubated at 37°C overnight. The next day, isolated colonies were inoculated in 4 mL LB media cultures, mini-prepped and then sequence confirmed using Sanger Sequencing (Quintara Biosciences).
Flow cytometry data collection of sfGFP fluorescence regulated by riboswitches
For each experiment, the indicated constructs were transformed into E. coli strain TG1 (F′ [traD36 proAB lacIqZ ΔM15] supE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM)5(rK - mK -). Plasmids were transformed into chemically competent E. coli cells, plated on Difco LB agar plates containing 34 µg/mL chloramphenicol, and incubated at 37°C for 16–18 h. Plates were then placed at room temperature for 6–8 h. Three colonies per plasmid were inoculated in 200 µL of LB chloramphenicol (34 µg/mL) and incubated overnight at 37°C. Following this, subcultures were created by adding 4 µL of the overnight culture to 196 µL of prewarmed M9 minimal media lacking thiamin hydrochloride (M9 Salts, 0.4% glycerol, 0.2% casamino acids, 2 mM MgSO4, 0.1 mM CaCl2) with the indicated growth conditions. TPP at the indicated concentration was added to prewarmed M9 media from solid powder directly before the experiment. Each culture block was covered with a Breath-Easier sealing membrane (Sigma-Aldrich) and incubated at 37°C while shaking at 1000 rpm (Vortemp Shaking Incubator) for 6 h. After 6 h of subculture incubation, cells were diluted 1:100 in phosphate buffered saline (PBS) and kanamycin (50 µg/mL). A BD Accuri C6 Plus Flow Cytometer fitted with a high-throughput sampler was then used to measure the sfGFP fluorescence for each sample. E. coli events were collected using the FSC-A threshold set to 2000. SfGFP fluorescence was measured by collecting the FITC signal from the FL1-A channel. Fifty-thousand events were captured for each sample.
Flow cytometry data analysis
Flow cytometry data analysis was performed using FlowJo (v10.4.1). Cells were density-gated using an ellipsoid gate around the FSC-A versus SSC-A plot, and the same gate was used for all samples prior to calculating the geometric mean of fluorescence in the FITC-A channel. Arbitrary fluorescent values from the flow cytometer were converted to MEFL by measuring the fluorescence from Spherotech Rainbow 8-peak calibration beads (cat# RCP305A). A calibration curve was then created by calculating the linear regression between the arbitrary relative fluorescent values from the flow cytometer and the known MEFL provided by the manufacturer. The geometric mean of the FITC channel peaks after gating for E. coli cells was converted to MEFL by multiplying by the slope of the calibration curve and adding the y-intercept. Supplemental Figure S4 contains representative flow cytometry data from the measurement and calibration procedure.
Single-round in vitro transcription of linear templates with E. coli RNA polymerase
Linear double-stranded DNA templates were prepared by 500 µL PCR reactions using 50 µL 10× Thermo Pol buffer (NEB), 10 µL dNTP (10 mM) (NEB), 2.5 µL forward template primer (100 µM) and reverse template primer (100 µM), 2.5 µL mini-prepped plasmid, and 2.5 µL of Taq DNA Polymerase. The PCR reactions were then thermocycled at 95°C for 3 min, then 24 cycles of 95°C for 30 sec, 57°C for 1 min, and 68°C for 1 min and 30 sec, and final extension at 68°C for 5 min. The PCR products were purified by ethanol precipitation, and 1% agarose gel was purified using the QIAquick Gel Extraction Kit (Qiagen). Single-round E. coli RNA polymerase in vitro transcription was then performed by mixing reactions on ice consisting of 100 nM DNA template, 20 mM Tris pH 8.0, 1 µM EDTA pH 8.0, 1 mM DTT, 50 mM KCl, 1 mM each NTP, 0.2 µg BSA, two units of E. coli RNA Polymerase Holoenzyme (NEB), and the indicated concentration of ligand. We found that above 1 mM, TPP was toxic to in vitro transcription. Each reaction was then incubated at 37°C for 10 min, then transcription was initiated by adding 2.5 µL of 100 mM MgCl2 and 0.1 mg/mL Rifampicin. After 30 sec, the reaction was stopped with 75 µL of TRIzol RNA Isolation Reagents (Thermo Fisher Scientific). TRIzol extraction was then completed as directed to purify nucleic acids from the reaction. Extracted nucleic acids were resuspended in 43 µL sterile, RNase-free water, 5 µL Turbo DNase 10× Buffer, and 2 µL of Turbo DNase. Samples were then incubated at 37°C for 60 min, and then RNA was purified with TRIzol extraction as described previously. Purified RNA was resuspended in 10 µL of sterile, RNase-free water and then mixed with 10 µL of RNA loading dye (7 M Urea, 0.01% xylene cyanol and bromophenol blue). Then, samples and ssRNA LR ladder (NEB) were run on a 10% Urea PAGE at 16 W for 90 min. The resulting PAGE was stained with SYBR Gold (Thermo Fisher Scientific) for 5 min and imaged using a ChemiDoc Imaging System (Bio-Rad). Raw PAGE images are shown in Supplemental Figure S15. Template sequences and primers used to create templates are listed in Supplemental Data Table S1.
Biotinylated DNA template preparation for in vitro transcription structural probing
Biotinylated DNA templates were prepared as described above with modifications described in Strobel et al. (2017) to include biotinylated dNTPs. Briefly, 500 µL PCR reactions were set up as above, and PRC performed using Vent-Exo DNA polymerase (NEB) and biotinylated nucleotides spiked in at approximately a 1:140 molar ratio corresponding to the sequence of the template, as described previously. Templates were then ethanol precipitated and purified from 1% agarose gels as described above.
Single-round in vitro transcription assay with SHAPE modification
Cotranscriptional SHAPE-Seq probing of thiB intermediates was performed as in Strobel et al. (2017). Transcription reactions in the presence or absence of 1 mM TPP were prepared using biotinylated DNA templates according to “Single-round in vitro transcription of linear templates with E. coli RNA Polymerase” above with several modifications: doubling the transcription reaction volume to 50 µL, adding 5 µL of 95 µM streptavidin (Promega) after a 7.5 min incubation at 37°C, then allowing another 7.5 min incubation at 37°C before adding the transcription start solution (5 µL of 100 mM MgCl2 and 0.1 mg/mL Rifampicin). Thirty seconds after the transcription start, chemical probing was performed by splitting the sample into 25 µL aliquots and mixing with 2.78 µL of 400 mM BzCN (Pfaltz & Bauer) dissolved in anhydrous DMSO ([+] sample) or anhydrous DMSO ([−] sample) for ∼2 sec (Mortimer and Weeks 2007). Nucleic acid was purified through a TRIzol extraction and isopropanol purification as described above. Afterward, the DNA was removed through a DNase digestion for 30 min at 37°C, followed by another TRIzol extraction and isopropanol purification. RNA was resuspended in 10 µL of 10% DMSO prior to “RNA linker ligation” (below).
Equilibrium-refolded SHAPE modification
Equilibrium-refolded SHAPE-Seq probing of thiB intermediates was performed as in Strobel et al. (2017). RNA was prepared alongside the RNA for “Single-round in vitro transcription assay with SHAPE modification” (above), except SHAPE probing with BzCN was not performed. Purified RNAs from these reactions were resuspended in 25 µL of water, heated at 95°C for 2 min, snapped cooled on ice for 1 min, then 25 µL of 2× folding buffer (20 mM Tris pH 8.0, 1 µM EDTA pH 8.0, 1 mM DTT, 50 mM KCl, 1 mM each NTP, 0.2 µg BSA, 10 mM MgCl2) was added, including a final concentration of 1 mM of TPP for the “with ligand” condition. The RNA was allowed to fold at 37°C for 20 min. Afterward, 25 µL of samples were added to tubes with either 2.78 µL of 400 mM of BzCN and 2.78 µL of anhydrous DMSO. The samples went through another round of TRIzol extraction and isopropanol purification. RNA was resuspended in 10 µL of 10% DMSO prior to “RNA linker ligation” (below).
RNA linker 5′-adenylation
The 3′ RNA linker used to ligate a reverse transcription site for SHAPE-Seq libraries was 5′ adenylated using the Mth Ligase (NEB). A 225 µL Mth ligase reaction was mixed containing 5 µM Phosphorylated RNA Linker (Supplemental Table S1), 100 µM ATP, 1× NEB 5′ adenylation buffer, and 0.5 µM Mth RNA Ligase and incubated at 65°C for 1 h. Then the reaction was purified by TRIzol extraction and resuspended in sterile, nuclease-free water. The resulting adenylated RNA linker was then quantified by NanoDrop.
RNA linker ligation
The linker ligation reaction was carried out in 20 µL reactions with 10 µL RNA sample in 10% DMSO, 0.5 µL SUPERase-IN (20 U/µL; Thermo Fisher Scientific, cat# AM2696), 6 µL of 50% PEG 8000, 2 µL of 10× T4 RNA Ligase Buffer, 1 µL of 2 µM 5′-adenylated RNA linker, and 0.5 µL of T4 RNA ligase, truncated KQ (200 U/µL; NEB, Cat# M0373L). The reactions were mixed well and incubated overnight (>10 h) at 25°C.
Reverse transcription
Following linker ligation, RNA samples were precipitated with 15 µL 3 M NaOAc, 1 µL glycoblue, and 450 µL 100% EtOH stored at −80°C for 30 min, centrifuged at 4°C for 30 min, washed once with 400 µL of 70% EtOH (v/v), and air dried to remove residual EtOH. A total of 9.09 nM of reverse transcription primer (Supplemental Table S1) was added to samples and heated to 95°C for 2 min, and then primer annealing was carried out at 65°C for 5 min. The samples were cooled to 45°C while 9 µL of SSIII reverse transcription master mix (per reaction: 4 µL of 5× FS buffer, 1 µL of 10 mM dNTPs, 1 µL of 0.1 M DTT, 2.5 µL nuclease-free water, and 0.5 µL SSIII) was added to all samples. The samples were then held at 45°C for 1 min, incubated at 52°C for 25 min, then 65°C for 5 min, and cooled to 4°C. The RNA was hydrolyzed by adding 1 µL of 4 M NaOH and incubated at 95°C for 5 min, then cooled again to 4°C. The reaction was partially neutralized by the addition of 2 µL of 1 M HCl.
Adapter ligation
Sequencing adapter ligation followed the Structure-Seq 2 protocol (Ritchey et al. 2017). Following reverse transcription, DNA samples were precipitated with 69 µL 100% EtOH stored at −80°C for 30 min, centrifuged at 4°C for 30 min, washed once with 400 µL of 70% EtOH (v/v), and air dried to remove residual EtOH. Samples were resuspended in 7 or 10 µL of RNase-free water. To each reaction, 0.5 µL of 100 µM of dumbbell ligation primer (Supplemental Table S1) was added and incubated at 95°C for 2 min. The samples were cooled to 25°C for 3 min. A total of 17.5 µL of T4 DNA Ligase Master Mix (per reaction: 2.5 µL 10× T4 DNA Ligase Buffer, 2.5 µL of 5 M Betaine, 10 µL of 50% PEG 8000, 2.5 µL T4 DNA Ligase [400,000 U/mL NEB, Cat# M0202S]) was then added to all samples. The samples were incubated at 16°C for 6 h, 30°C for 6 h, and then cooled to 4°C.
Illumina sequencing library preparation
Following adapter ligation, DNA was precipitated with 2.8 µL of 3 M NaOAc, 1 µL of glycoblue, and 84 µL of 100% EtOH stored at −80°C for 30 min, centrifuged at 4°C for 30 min, washed once with 400 µL of 70% EtOH (v/v), and air dried to remove residual EtOH. The sample was further purified using Agencourt AmpureXP beads following the manufacturer's protocol. Samples were extracted from the beads in 20 µL of TE Buffer. A total of 3 µL of sample was added to a PCR tube and 0.25 µL of 100 µM of Illumina TruSeq Indexing Primer was added (Supplemental Table S1). Then, 46.75 µL of library master mix (per reaction: 33 µL of nuclease-free water, 10 µL of 5× Phusion Buffer, 0.5 µL of 10 mM dNTPs, 2.5 µL), 0.1 µM Selection Primer (Supplemental Table S1), and 0.5 µL of Phusion Polymerase (2000 U/mL; NEB, Cat# M0530S) was added. For the selection primer, the positive channel primer was used to barcode the samples reacted with BzCN while the negative channel primer was used to barcode control samples. Amplification was performed with a primer annealing at 65°C for 30 sec, polymerase extension at 72°C for 30 sec. After 15 cycles, 0.25 µL of 100 µM primer PE_F (Supplemental Table S1) was added at 4°C and the amplification was continued for 10 additional cycles. The samples were cooled to 4°C for at least 5 min before 0.25 µL of ExoI was added. Samples were incubated at 37°C for 30 min to degrade excess ssDNA and then the ExoI was heat inactivated by incubating the samples at 80°C for 20 min. The libraries were purified again with Agencourt AmpureXP beads and extracted in 20 µL of TE Buffer. DNA concentration and purity were measured on a NanoDrop. Additionally, samples were analyzed for quality on a 8% nondenaturing TBE PAGE gel stained with Gel Red.
High-throughput sequencing was performed on the Illumina NextSeq 500 by NUSeq Core with mid-output 2 × 75 bp output.
SHAPE-Seq data analysis and generating secondary structure models
SHAPE-Seq reactivity analysis and secondary structure models are detailed in Supplemental Note S1. SHAPE-Seq reactivities were calculated from FASTQ files using Spats v2.0.5 and run using Python 2.7 through Anaconda 2.4. SHAPE-Seq informed secondary structures were generated by Reconstructing RNA Dynamics from Data (R2D2), which is detailed in the methods of Yu et al. (2021). Predicted structures were visualized as RNAbow plots and consensus secondary structures using RNAbow software (Aalberts and Jannen 2013) and VARNA, version 3.9 (Darty et al. 2009), respectively.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Adam Silverman and Luyi Cheng for experimental contributions to this study not included in this manuscript; Angela Yu for many helpful conversations about cotranscriptional RNA folding and strand displacement; and Alexis Reyes for careful feedback on manuscript drafts and figures. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (1R01GM130901 to J.B.L.) as well as Northwestern University's Office of Undergraduate Research (1275ACADYR2119473 and 782WCASSUM1915810 to R.S.) and the Chemistry of Life Processes Institute.
Author contributions: K.E.B. and J.B.L. conceived the projects. The data was curated, analyzed and checked by K.E.B. and J.B.L. K.E.B. and J.B.L. devised the methods as well as administered and validated the project. Flow cytometry experiments were completed by K.E.B. and R.S. Single-round in vitro transcription was completed by K.E.B. and L.M.H. Manuscript was written and edited by K.E.B. and J.B.L. Drafts were revised and edited by K.E.B., L.M.H., R.S., and J.B.L.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079427.122.
REFERENCES
- Aalberts DP, Jannen WK. 2013. Visualizing RNA base-pairing probabilities with RNAbow diagrams. RNA 19: 475–478. 10.1261/rna.033365.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes D, Jorge NAN, Garcia de Souza Costa M, Passetti F, Caffarena ER. 2019. Unraveling RNA dynamical behavior of TPP riboswitches: a comparison between Escherichia coli and Arabidopsis thaliana. Sci Rep 9: 4197. 10.1038/s41598-019-40875-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. 2012. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335: 233–235. 10.1126/science.1215063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastet L, Chauvier A, Singh N, Lussier A, Lamontagne A-M, Prévost K, Masśe E, Wade JT, Lafontaine DA. 2017. Translational control and Rho-dependent transcription termination are intimately linked in riboswitch regulation. Nucleic Acids Res 45: 7474–7486. 10.1093/nar/gkx434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha GM, Wade JT. 2022. Transcription-translation coupling in bacteria. Annu Rev Genet 56: 187–205. 10.1146/annurev-genet-072220-033342 [DOI] [PubMed] [Google Scholar]
- Breaker RR. 2012. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol 4: a003566. 10.1101/cshperspect.a003566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushhouse DZ, Lucks JB. 2023. Tuning strand displacement kinetics enables programmable ZTP riboswitch dynamic range in vivo. Nucleic Acids Res 51: 2891–2903. 10.1093/nar/gkad110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushhouse DZ, Choi EK, Hertz LM, Lucks JB. 2022. How does RNA fold dynamically? J Mol Biol 34: 167665. 10.1016/j.jmb.2022.167665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron M-P, Bastet L, Lussier A, Simoneau-Roy M, Massé E, Lafontaine DA. 2012. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc Natl Acad Sci 109: E3444–E3453. 10.1073/pnas.1214024109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio R, Mathews DH, Sternberg MJE. 2022. Computational resources for molecular biology 2022. J Mol Biol 434: 167625. 10.1016/j.jmb.2022.167625 [DOI] [PubMed] [Google Scholar]
- Chauvier A, Picard-Jean F, Berger-Dancause J-C, Bastet L, Naghdi MR, Dubé A, Turcotte P, Perreault J, Lafontaine DA. 2017. Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation. Nat Commun 8: 13892. 10.1038/ncomms13892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvier A, Nadon JF, Grondin JP, Lamontagne AM, Lafontaine DA. 2019. Role of a hairpin-stabilized pause in the Escherichia coli thiC riboswitch function. RNA Biol 16: 1066–1073. 10.1080/15476286.2019.1616354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvier A, St-Pierre P, Nadon J-F, Hien EDM, Pérez-González C, Eschbach SH, Lamontagne A-M, Penedo JC, Lafontaine DA. 2021. Monitoring RNA dynamics in native transcriptional complexes. Proc Natl Acad Sci 118: 2106564118. 10.1073/pnas.2106564118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. 2007. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 447: 497–500. 10.1038/nature05769 [DOI] [PubMed] [Google Scholar]
- Cheng L, White EN, Brandt NL, Yu AM, Chen AA, Lucks JB. 2022. Cotranscriptional RNA strand exchange underlies the gene regulation mechanism in a purine-sensing transcriptional riboswitch. Nucleic Acids Res 50: 12001–12018. 10.1093/nar/gkac102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. 2009. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25: 1974–1975. 10.1093/bioinformatics/btp250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Mathews DH. 2021. Making ends meet: new functions of mRNA secondary structure. Wiley Interdiscip Rev RNA 12: e1611. 10.1002/wrna.1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieda KL, Block SM. 2012. Direct observation of cotranscriptional folding in an adenine riboswitch. Science (New York, NY) 338: 397–400. 10.1126/science.1225722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Edwards AL, Batey RT. 2011. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol 3: a003533. 10.1101/cshperspect.a003533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller A, Altman RB, Soulière MF, Blanchard SC, Micura R. 2013. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc Natl Acad Sci 110: 4188–4193. 10.1073/pnas.1218062110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmling C, Wacker A, Wolfinger MT, Hofacker IL, Hengesbach M, Fü B, Schwalbe H. 2017. NMR structural profiling of transcriptional intermediates reveals riboswitch regulation by metastable RNA conformations. J Am Chem Soc 139: 2647–2656. 10.1021/jacs.6b10429 [DOI] [PubMed] [Google Scholar]
- Hong F, Šulc P. 2019. An emergent understanding of strand displacement in RNA biology. J Struct Biol 207: 241–249. 10.1016/j.jsb.2019.06.005 [DOI] [PubMed] [Google Scholar]
- Hua B, Jones CP, Mitra J, Murray PJ, Rosenthal R, Ferré-D'Amaré AR, Ha T. 2020. Real-time monitoring of single ZTP riboswitches reveals a complex and kinetically controlled decision landscape. Nat Commun 11: 4531. 10.1038/s41467-020-18283-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI. 2017. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46: D335–D342. 10.1093/nar/gkx1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PB, Nelson JW, Breaker RR. 2015. An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism HHS public access. Mol Cell 57: 317–328. 10.1016/j.molcel.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K, Rieder R, Micura R. 2007. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res 35: 5370–5378. 10.1093/nar/gkm580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCuyer KA, Crothers DM. 1994. Kinetics of an RNA conformational switch. Proc Natl Acad Sci 91: 3373–3377. 10.1073/pnas.91.8.3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J-F, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Massé E, Lafontaine DA. 2011. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet 7: e1001278. 10.1371/journal.pgen.1001278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Breaker RR. 2013. Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing. Nucleic Acids Res 41: 3022–3031. 10.1093/nar/gkt057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman JA, Suddala KC, Aytenfisu A, Chan D, Belashov IA, Salim M, Mathews DH, Spitale RC, Walter NG, Wedekind JE. 2015. Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics. Proc Natl Acad Sci 112: E3485–E3494. 10.1073/pnas.1503955112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. 2017. Riboswitch diversity and distribution. RNA 23: 995–1011. 10.1261/rna.061234.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan MA, Petrova SA, Gelfand MS. 2018. Comparative genomic analysis of fungal TPP-riboswitches. Fungal Genet Biol 114: 34–41. 10.1016/j.fgb.2018.03.004 [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. 2007. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc 129: 4144–4145. 10.1021/ja0704028 [DOI] [PubMed] [Google Scholar]
- Nou X, Kadner RJ. 2000. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci 97: 7190–7195. 10.1073/pnas.130013897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontiveros-Palacios N, Smith AM, Grundy FJ, Soberon M, Henkin TM, Miranda-Ríos J. 2007. Molecular basis of gene regulation by the THI-box riboswitch. Mol Microbiol 67: 793–803. 10.1111/j.1365-2958.2007.06088.x [DOI] [PubMed] [Google Scholar]
- Perdrizet GA, Artsimovitch I, Furman R, Sosnick TR, Pan T. 2012. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc Natl Acad Sci 109: 3323–3328. 10.1073/pnas.1113086109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaski JT, Holmstrom ED, Nesbitt DJ, Batey RT. 2016. Mechanistic insights into cofactor-dependent coupling of RNA folding and mRNA transcription/translation by a cobalamin riboswitch. Cell Rep 15: 1100–1110. 10.1016/j.celrep.2016.03.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Chauvier A, Walter NG. 2018. Kinetics coming into focus: single-molecule microscopy of riboswitch dynamics. RNA Biol 16: 1077–1085. 10.1080/15476286.2018.1536594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Soni A, Bellecourt MJ, Landick R. 2016. Mechanisms of bacterial transcription termination: all good things must end. Annu Rev Biochem 85: 319–347. 10.1146/annurev-biochem-060815-014844 [DOI] [PubMed] [Google Scholar]
- Reining A, Nozinovic S, Schlepckow K, Buhr F, Fürtig B, Schwalbe H. 2013. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature 499: 355–359. 10.1038/nature12378 [DOI] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchey LE, Su Z, Tang Y, Tack DC, Assmann SM, Bevilacqua PC. 2017. Structure-seq2: sensitive and accurate genome-wide profiling of RNA structure in vivo. Nucleic Acids Res 45: e135. 10.1093/nar/gkx533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2002. Comparative genomics of thiamin biosynthesis in procaryotes new genes and regulatory mechanisms. J Biol Chem 277: 48949–48959. 10.1074/jbc.M208965200 [DOI] [PubMed] [Google Scholar]
- Roth A, Breaker RR. 2009. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78: 305–334. 10.1146/annurev.biochem.78.070507.135656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scull CE, Dandpat SS, Romero RA, Walter NG. 2021. Transcriptional riboswitches integrate timescales for bacterial gene expression control. Front Mol Biosci 7: 607158. 10.3389/fmolb.2020.607158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. 2012. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu Rev Biophys 41: 343–370. 10.1146/annurev-biophys-101211-113224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. 2006. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 441: 1167–1171. 10.1038/nature04740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Fuchs RT, Grundy FJ, Henkin TM. 2010. The SAM-responsive SMK box is a reversible riboswitch. Mol Microbiol 78: 1393–1402. 10.1111/j.1365-2958.2010.07410.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spasic A, Assmann SM, Bevilacqua PC, Mathews DH. 2018. Modeling RNA secondary structure folding ensembles using SHAPE mapping data. Nucleic Acids Res 46: 314–323. 10.1093/nar/gkx1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel EJ, Watters KE, Nedialkov Y, Artsimovitch I, Lucks JB. 2017. Distributed biotin-streptavidin transcription roadblocks for mapping cotranscriptional RNA folding. Nucleic Acids Res 45: e109. 10.1093/nar/gkx233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel EJ, Cheng L, Berman KE, Carlson PD, Lucks JB. 2019. A ligand-gated strand displacement mechanism for ZTP riboswitch transcription control. Nat Chem Biol 15: 1067–1076. 10.1038/s41589-019-0382-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jäger JG, Hüttenhofer A, Wagner EGH. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res 31: 6435–6443. 10.1093/nar/gkg867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Molodtsov V, Firlar E, Kaelber JT, Blaha G, Su M, Ebright RH. 2020. Structural basis of transcription-translation coupling. Science 369: 1359–1365. 10.1126/science.abb5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Strobel EJ, Yu AM, Lis JT, Lucks JB. 2016. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat Struct Mol Biol 23: 1124–1131. 10.1038/nsmb.3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. 2005. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell 18: 49–60. 10.1016/j.molcel.2005.02.032 [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956. 10.1038/nature01145 [DOI] [PubMed] [Google Scholar]
- Yu AM, Gasper PM, Cheng L, Lai LB, Kaur S, Gopalan V, Chen AA, Lucks JB. 2021. Computationally reconstructing cotranscriptional RNA folding from experimental data reveals rearrangement of non-native folding intermediates. Mol Cell 81: 870–883.e10. 10.1016/j.molcel.2020.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]