Summary
Background
Deep learning has shown potential in various scientific domains but faces challenges when applied to complex, high-dimensional multi-omics data. Alzheimer's Disease (AD) is a neurodegenerative disorder that lacks targeted therapeutic options. This study introduces the Circular-Sliding Window Association Test (c-SWAT) to improve the classification accuracy in predicting AD using serum-based metabolomics data, specifically lipidomics.
Methods
The c-SWAT methodology builds upon the existing Sliding Window Association Test (SWAT) and utilizes a three-step approach: feature correlation analysis, feature selection, and classification. Data from 997 participants from the Alzheimer's Disease Neuroimaging Initiative (ADNI) served as the basis for model training and validation. Feature correlations were analyzed using Weighted Gene Co-expression Network Analysis (WGCNA), and Convolutional Neural Networks (CNN) were employed for feature selection. Random Forest was used for the final classification.
Findings
The application of c-SWAT resulted in a classification accuracy of up to 80.8% and an AUC of 0.808 for distinguishing AD from cognitively normal older adults. This marks a 9.4% improvement in accuracy and a 0.169 increase in AUC compared to methods without c-SWAT. These results were statistically significant, with a p-value of 1.04 × 10ˆ-4. The approach also identified key lipids associated with AD, such as Cer(d16:1/22:0) and PI(37:6).
Interpretation
Our results indicate that c-SWAT is effective in improving classification accuracy and in identifying potential lipid biomarkers for AD. These identified lipids offer new avenues for understanding AD and warrant further investigation.
Funding
The specific funding of this article is provided in the acknowledgements section.
Keywords: Deep learning, Machine learning, Alzheimer's disease, Metabolomics, Lipidomics
Research in context.
Evidence before this study
A PubMed search up to August 7, 2023, using the terms “deep learning” “machine learning” "Alzheimer's Disease” “metabolomics” and “lipidomics” yielded studies that mainly applied traditional machine learning algorithms and neuroimaging techniques for diagnosing and understanding Alzheimer's Disease (AD). While there has been interest in applying deep learning to metabolomics and multi-omics data, these efforts often overlook the importance of feature correlations, which could potentially improve the accuracy of predictive models. Additionally, deep learning models struggle with high-dimensional, complex data sets like those commonly found in multi-omics research, particularly when data are sparse.
Added value of this study
We introduced a three-step deep learning method, the circular-Sliding Window Association Test (c-SWAT), specifically designed to account for correlated features and improve model performance. The method was applied to serum-based lipidomics data from 997 individuals to predict AD. c-SWAT achieved an accuracy of up to 80.8% and an Area Under the Curve (AUC) of 0.808, outperforming models not using c-SWAT by 9.4% in accuracy and 0.169 in AUC. Moreover, c-SWAT identified key lipids correlated with AD, including Cer(d16:1/22:0), PI(37:6), CE (20:4) [+OH], and LPE (20:4) [sn1], which have biological implications related to neural cell membranes, microglial signaling, cholesterol storage, and glycerophospholipid metabolism. These lipids showed a 1.27%–3.23% increase in AD samples compared to cognitively normal older adults (CN).
Implications of all the available evidence
The c-SWAT approach offers a notable advancement in the application of deep learning to multi-omics data, particularly for complex diseases like AD. The method not only improves predictive accuracy but also offers insights into potential biomarkers crucial for understanding AD pathophysiology. The identified lipids offer new possibilities for targeted research and therapeutic interventions. Furthermore, the approach has the potential to be adapted to other types of biological data and diseases, contributing to the development of personalized medicine for a range of conditions.
Introduction
Deep learning is able to recognize complex patterns in input data by weighting key features during backpropagation.1, 2, 3 Because deep learning can handle large volumes of data without feature selection procedures,4 researchers have been using it to uncover new biological phenomena,5,6 develop new drugs,7 and find important clues to help delineate diseases.8,9 Deep learning excels when applied to data with complex structures such as images10 and language.11 However, its application to tabular data,12 which often has a smaller sample size and a more straightforward feature set, has not consistently shown effectiveness. In light of the widespread use of tabular data in genomic and multi-omics studies,13 it is imperative to refine deep learning applications for this type of data. Therefore, we developed the circular- Sliding Window Association Test (c-SWAT), an advanced variant of SWAT,14 initially designed for genetic data and adapted to accommodate the specificities of multi-omics data. Our c-SWAT method enhances the efficiency of the deep learning algorithm in classifying data by incorporating correlated feature groups within each flexible-sized window. This study reports on the application of this method to lipidomic data, a subfield of metabolomics, to detect Alzheimer's disease, as well as identify metabolites associated with the disease.
Alzheimer's disease (AD) is a neurodegenerative disorder leading to conditions such as dementia, personality changes, impairment of judgment and speech, and memory loss.15,16 In the preclinical phase of AD, patients' function declines steadily for more than 10 years.17 However, no clinical evidence enables a diagnosis of AD, and there are no targeted pharmacological interventions or prophylactic interventions to treat AD.18,19 A further understanding of the underlying cause of AD is therefore essential to identifying new treatment targets. Neurofibrillary tangles and amyloid plaques are two typical AD pathological lesions.20 When the disease advances, there are also changes in the structure of the brain, glucose metabolism, and measurements of biomarkers in circulating blood.21,22 In recent years, metabolomics technologies have been used to identify a number of disease-specific biomarkers, enabling insights into the pathophysiology of diseases like AD, type 2 diabetes, and cancer.23, 24, 25, 26, 27, 28, 29, 30, 31
Metabolites, products of biological cascades that include DNA, transcripts, and proteins, may have beneficial properties which may help identify AD biomarkers. In this paper, we present a three-step deep learning approach for detecting AD using lipidomic data, a specific aspect of the broader metabolomics data.
We used serum-based metabolomics data from 997 participants from the Alzheimer's Disease Neuroimaging Initiative (ADNI). c-SWAT, an extension of SWAT, calculates the phenotype influence score (PIS), which represents the association between the metabolites and the phenotype of interest, based on the correlation between these metabolites using deep learning. Using weighted gene co-expression network analysis (WGCNA),32 the correlation between features was calculated and feature groups were created. WGCNA is utilized in our study to uncover highly correlated clusters of features among metabolites. We then implement a flexible windowing approach, choosing the most influential features within each feature group and thereby defining the structure for input into the Convolutional Neural Networks (CNN). The purpose of this process is not simply to filter the data, but rather to determine the structured input, based on feature correlations, for our deep learning model. The structure required by CNN is maintained in this approach, with dense layers becoming more appropriate due to the derived correlations from the WGCNA and SWAT processes. Subsequently, CNN is employed to classify AD from cognitively normal older adults (CN).
Methods
Data acquisition
The Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort served as the source for all study participants. This cohort included 382 cognitively normal individuals (CN), 212 individuals with early mild cognitive impairment (EMCI), 254 with late mild cognitive impairment (LMCI), and 149 with Alzheimer's Disease (AD). For the EMCI, LMCI, and AD groups, only those with confirmed amyloid-β positivity were included. Overall, our investigation involved a total of 997 fasted individuals. The metabolomics data used in this study was obtained from the ADNI database (http://adni.loni.usc.edu/). The ADNI, launched in 2003 as a public-private partnership and led by Principal Investigator Michael W. Weiner, MD, has been primarily concerned with testing whether the progression of AD can be effectively measured using serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment.
Extension of SWAT to c-SWAT
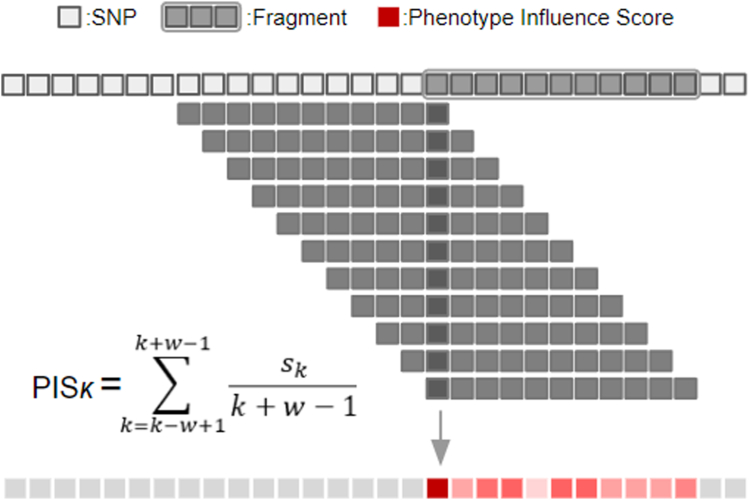
We developed an innovative method named c-SWAT, a variation on the original Sliding Window Association Test (SWAT)14 approach we previously developed. The original SWAT was proposed as a three-step, deep learning-based procedure to identify informative single nucleotide polymorphisms (SNPs) and create classification models for phenotypes of interest. This process, as shown in Fig. 1, was initiated by dividing the whole genome into optimally sized, non-overlapping fragments, upon which deep learning algorithms were employed to select phenotype-associated fragments containing relevant SNPs. SWAT then computed a PIS for each SNP within a fragment. When the kth SNP is Sk, the PIS in SWAT is calculated using the following formula:
Fig. 1.
The figure illustrates the original Sliding Window Association Test (SWAT) in Genome-Wide Association Studies (GWAS). SWAT begins by partitioning the entire genome into smaller, nonoverlapping fragments. For every fragment, SWAT employs a sliding window technique in conjunction with a Convolutional Neural Network (CNN) to compute a phenotype influence score (PIS) for each Single Nucleotide Polymorphism (SNP). This computation considers ‘w’, the number of SNPs in a fragment, and ‘Sk’, the position of each SNP. By distinguishing SNPs with significant PIS values, SWAT efficiently identifies phenotype-associated genetic variants.
In this process, the fragment length is denoted as ‘w’. A window of length ‘w-1’ is initially positioned at the first SNP of the fragment. The window then slides one SNP at a time until it reaches the end of the fragment. This SWAT procedure is implemented across all selected fragments, leading to the calculation of a PIS for each SNP involved. This score reflected the association between the SNP and the phenotype of interest, aiding in the identification of the most informative SNPs. Notably, in a real-world application to Alzheimer's disease, this deep learning approach using SWAT succeeded in identifying significant genetic loci for the disease and achieved a higher classification accuracy than existing machine learning methods. While SWAT proved effective for high-dimensional data, it is not as well-suited for the tabular data formats frequently found in multi-omics studies, which often have a smaller set of features. These data types inherently constrain the range and number of features that can be analyzed effectively.
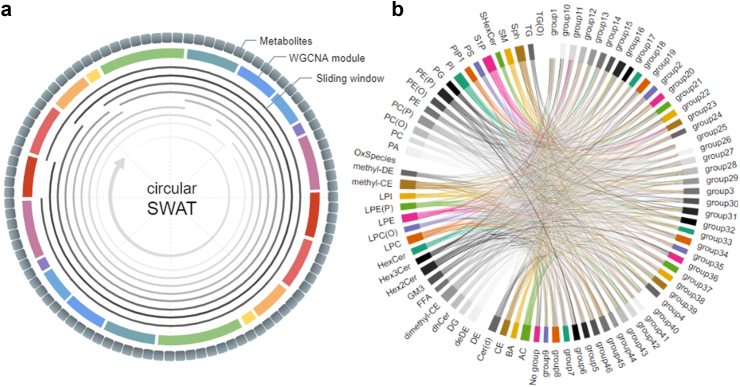
To improve upon these limitations, c-SWAT was developed. Fig. 2 illustrates an overview of c-SWAT, designed to enhance the learning effect by grouping related input features. While SWAT demonstrates high efficiency with complex, unfiltered data, it may encounter limitations with tabular data possessing fewer features, mainly due to constraints in acquiring a sufficient number of windows for PIS computation. To address this, c-SWAT computes the PIS for each group, as demonstrated in Fig. 2a, distinguishing itself from SWAT by constructing a circular window that links the start and end of all features. This window size is uniquely tailored for each group. As seen in Fig. 2b, we applied the WGCNA to determine these feature groups. Each group, represented by a circle in the figure, encompasses correlated metabolite features. Variable-sized sliding windows captured their collective properties, and based on these arrangements and the class of each lipid, a PIS for each metabolite is calculated. These PIS values then subsequently inform the classification of AD.
Fig. 2.
Overall structure of the c-SWAT. The phenotype influence score for the feature groups was calculated as shown in (a). Sliding windows of varying sizes overlap all feature groups except one to perform the classification prediction, thereby determining the importance of the excluded group. WGCNA was used to determine the group as shown in (b). Based on these results and the lipid classes, PIS for each metabolite was calculated and used to classify AD.
Deep learning for phenotype influence score (PIS) calculation in c-SWAT
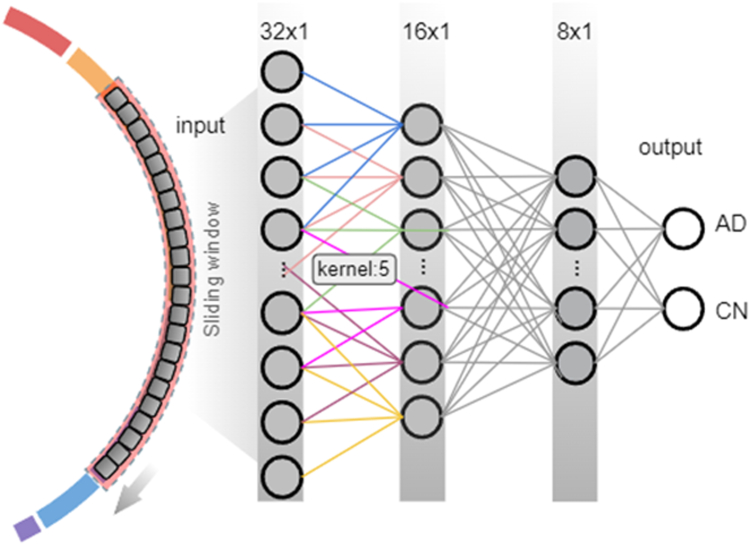
In the stages of c-SWAT that required the calculation of PIS, we employed CNN as our base classifier. As illustrated in Fig. 3, the CNN consists of a convolutional layer containing 64 filters with a kernel size of 5, applying ReLU33 activation function, followed by a global max pooling layer. Subsequent to the pooling, four dense layers with units of the corresponding number of features, 32, 16, and 8 respectively, are implemented, all utilizing ReLU activation functions. The final output layer is tailored for binary AD classification, containing 2 units and employing a softmax activation function. We utilized Adam (preprint34) as the optimizer within the context of deep learning. Each input value was initially rescaled using the log(x) transformation. The robustness of our model was confirmed using 5-fold cross-validation.
Fig. 3.
An overview of how a deep learning approach was implemented in steps 1 and 3. Our model utilizes three main hidden layers, with the number of nodes in these layers optimized from 32 down to 8 using a grid search approach. The classification between AD and CN was performed with top-ranked features from each group using the CNN algorithm, and the performance was assessed by a 5-fold cross-validation.
Step1: grouping correlated metabolites
To detect network modules in metabolomics data, we used the WGCNA R package, which utilizes hierarchical clustering and dynamic tree cut algorithm (preprint34). We used the biweight mid-correlation method to calculate the correlation between lipids with a soft-thresholding power of 7. The minimum number of lipids within modules was set to 5. The levels of lipids in a module were represented by the module eigen-lipid value (ME), which is defined as the first principal component of the lipid matrix of the corresponding module.
Step2: calculating phenotype influence score based on deep learning
The importance of each feature group was determined using a modified k-fold Cross-Validation approach. In this approach, each feature group was left out once, and the model was trained on the remaining feature groups. The performance was then averaged across the k folds to provide an estimate of the feature group's importance. The error for this method can be represented by the following formula:
Here, n represents the total number of feature modules, yi denotes the actual value of the ith data, and refers to the predicted value obtained by feeding the ith data into the model trained using all other data, and k’ serves as the index for each individual fold in k-fold Cross-Validation. j is the index for the feature group, defined either through WGCNA or though lipid classes. The PIS is the value obtained by applying the above formula to the feature groups defined by WGCNA, and to the pre-defined lipid classes. Specifically, represents the CV-based module impact for the ith feature module defined using WGCNA, and represents the CV-based module impact for the ith feature module divided according to the group subclasses given in ADNI data. In addition to the CV approach, Random Forest's feature importance, denoted as , was employed to further assess the relevance of each feature in the model. This process can be represented as follows:
Step3: classifying Alzheimer's disease (AD) from cognitively normal older adults (CN)
After computing the PIS in the second step, we applied the Random Forest algorithm for AD-CN classification. This process was conducted using 10-fold cross-validation with stratification, utilizing the features with the highest PIS from each feature group. Our Random Forest model consists of 100 decision trees. The final prediction is obtained by aggregating the results from individual trees using majority voting. To evaluate the performance of our model, we used both average accuracy and average AUC as primary metrics, computed over the 10 validation folds.
Ethics
For the ADNI data used in this study, all participants provided written informed consent approved by the institutional review board of each participating institution.
Statistics
To assess the effectiveness of the c-SWAT approach, the model's performance was compared against a baseline where an equal number of features were randomly selected. This choice of comparison was made to evaluate the impact of feature selection by c-SWAT in a controlled setting. The classification accuracy and the Area Under the Receiver Operating Characteristic (ROC) Curve (AUC) were primary metrics, calculated using a 10-fold cross-validation approach. Additionally, the Precision-Recall (PR) curve was employed to provide a comprehensive assessment of the model's performance. A p-value of approximately 1.04 × 10−4 was obtained, confirming the statistical significance of the improvement in classification accuracy. The sample size for the study was determined by the availability of multi-omics data for each participant in the Alzheimer's Disease Neuroimaging Initiative (ADNI) database, totaling 997 participants. No further inclusion or exclusion criteria were applied. Randomization was applied in the selection of features for the baseline comparison model. All data used are publicly available and there are no restrictions on their availability.
Role of funders
The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Results
In our c-SWAT approach, we used serum metabolomics data from 997 ADNI participants. WGCNA identified 48 network modules (feature groups) from 781 metabolites. The modified CV approach identified the influential factors of each lipid within their respective feature groups, which enabled us to assess each metabolite's impact on classification. Utilizing a CNN model with a 5-fold cross validation method, we calculated the PIS for each metabolite. Finally, metabolites with higher PIS values were used to construct a robust AD classification model.
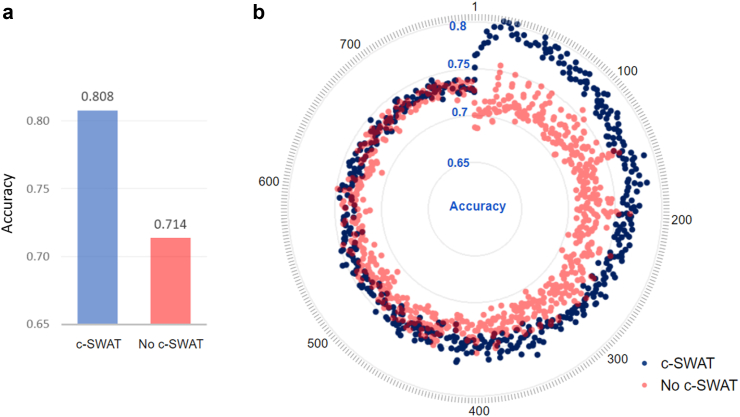
The overall result of a 10-fold cross-validation is shown in Fig. 4a. With c-SWAT, the Random Forest model could classify AD from CN with a highest accuracy of 0.808 when using 22 features, compared to an accuracy of 0.714 when the same number of features were randomly applied without using PIS. The difference in accuracy between the two methods was found to be statistically significant with a p-value of approximately 1.04 × 10−4. Fig. 4b shows the accuracies for AD/CN classification, using subsets containing the top 1 to 781 features based on PIS. A blue dot represents a testing accuracy on a number of feature sets after applying c-SWAT, and a red dot represents a testing accuracy without c-SWAT. When features with high PIS rankings were applied, the classification accuracy improved, and as the number of features increased, the differences between classifiers narrowed.
Fig. 4.
Visualization of AD/CN classification results. (a) Bar graph on the y-axis representing the average accuracy of a 10-fold cross validation. With c-SWAT, the Random Forest model could classify AD from CN with a highest accuracy of 0.807 when using 22 features, compared to an accuracy of 0.714 when the same number of features were randomly applied without using PIS. (b) The y-axis presents the accuracy for AD/CN classification in each subset, both with and without the implementation of c-SWAT, considering subsets ranging from the top 1 to 781 features. An outer circle represents the number of metabolite features utilized. Blue dots indicate classification accuracy when incorporating the results of PIS with c-SWAT, while red dots represent cases without the application of c-SWAT.
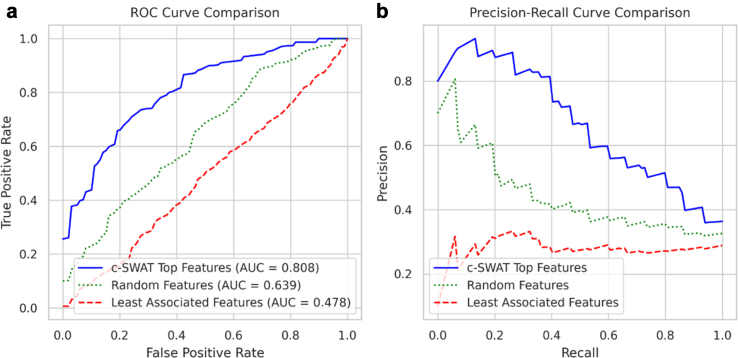
Fig. 5a displays the Receiver Operating Characteristic (ROC) curve for the classification capability using three sets of features: the top features selected by c-SWAT, randomly selected features, and the least associated features determined by c-SWAT. Utilizing a 10-fold cross-validation, the highest average Area Under the Curve (AUC) achieved with the top features was 0.808 when using 22 features. For a similar set of 22 features chosen randomly, the AUC was 0.639. In stark contrast, the AUC for the same number of least associated features was significantly lower at 0.478. Fig. 5b further explores the Precision-Recall (PR) curve for these different sets of features. The top features selected by c-SWAT consistently showed higher precision and recall values when compared to both randomly selected and least associated features. This underscores the predictive proficiency of the top features in AD classification.
Fig. 5.
Performance comparison between top features, randomly selected features, and least associated features in AD classification. (a) The Receiver Operating Characteristic (ROC) curve illustrates the classification capability using the top features selected by c-SWAT, randomly selected features, and the least associated features determined by c-SWAT. The highest average Area Under the Curve (AUC) from a 10-fold cross-validation for the top features reached 0.808 with 22 features. When randomly selecting the same number of 22 features, the AUC was 0.639. In comparison, the AUC for the same number of least associated features was significantly lower at 0.478. (b) The Precision-Recall (PR) curve showcases the predictive performance of these feature sets, including the top features selected by c-SWAT, randomly selected features, and the least associated features determined by c-SWAT. The top features consistently exhibited higher precision and recall values relative to the randomly selected and least associated features, underscoring their enhanced predictive proficiency in AD classification.
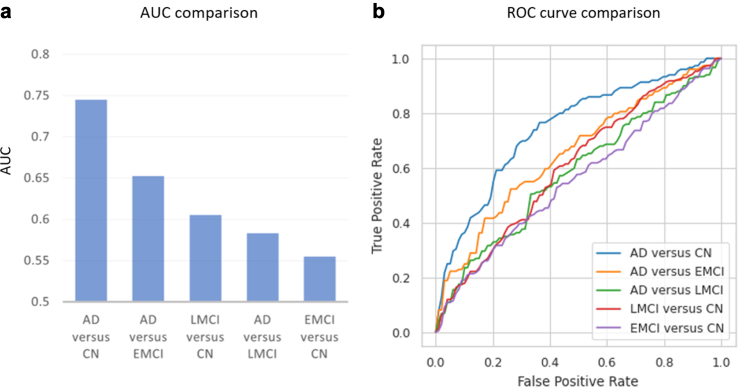
We also conducted a comprehensive analysis on the classification accuracy across distinct stages of Alzheimer's disease, focusing on the top features selected by c-SWAT, as they showed superior performance. By applying these top features to each Alzheimer's disease stage (Alzheimer's Disease [AD], Late Mild Cognitive Impairment [LMCI], Early Mild Cognitive Impairment [EMCI], and Cognitively Normal [CN]), we developed classification models. Fig. 6a presents the AUC values for various Alzheimer's disease stages, and Fig. 6b showcases the ROC curves for different disease stage comparisons. Our analysis revealed that the classification of AD from CN had the highest accuracy. Following the AD vs CN comparison in accuracy ranking, the next best classifications were AD vs ENCI, then LMCI vs CN, followed by AD vs LMCI, and lastly EMCI vs CN.
Fig. 6.
Classification accuracy across Alzheimer's Disease stages. (a) Bar chart illustrating the area under the curve (AUC) values for the classification among different Alzheimer's disease stages. (b) ROC curves representing classification performance for various disease stage comparisons, with each curve displaying the relationship between the true positive rate and false positive rate.
Using PIS obtained through c-SWAT, we identified highly AD-associated lipids. In Table 1, the highest scoring lipids are listed, with Cer(d16:1/22:0), PI(37:6), CE (20:4) [+OH], and LPE (20:4) [sn1] being the top-ranking lipid classes.
Table 1.
Lipids with the highest PIS scores.
| Identifier | Full name | Category | Class | PIS |
|---|---|---|---|---|
| CER.D16.1.22.0 | Cer (d16:1/22:0) | sphingolipids | ceramide | 0.158 |
| PI.37.6 | PI (37:6) | glycerophospholipids | phosphatidylinositol | 0.157 |
| CE.20.4....OH | CE (20:4) [+OH] | Neutral/Other | oxidised lipids | 0.155 |
| LPE.20.4...SN1 | LPE (20:4) [sn1] | glycerophospholipids | Lysophosphatidylethanolamine | 0.144 |
| PE.P.19.0.20.4...A | PE (P-19:0/20:4) (a) | glycerophospholipids | alkenylphosphatidylethanolamine | 0.142 |
| PE.P.17.0.20.4...A | PE (P-17:0/20:4) (a) | glycerophospholipids | alkenylphosphatidylethanolamine | 0.139 |
| PC.O.16.0.20.3 | PC (O-16:0/20:3) | glycerophospholipids | alkylphosphatidylcholine | 0.133 |
| CER.D18.1.20.0 | Cer (d18:1/20:0) | sphingolipids | ceramide | 0.126 |
| CER.D19.1.20.0 | Cer (d19:1/20:0) | sphingolipids | ceramide | 0.118 |
| PI.34.1 | PI (34:1) | glycerophospholipids | phosphatidylinositol | 0.115 |
| PE.P.15.0.20.4...B | PE (P-15:0/20:4) (b) | glycerophospholipids | alkenylphosphatidylethanolamine | 0.114 |
| PE.P.16.0.20.5 | PE (P-16:0/20:5) | glycerophospholipids | alkenylphosphatidylethanolamine | 0.110 |
| CER.D18.1.24.1 | Cer (d18:1/24:1) | sphingolipids | ceramide | 0.109 |
| LPE.20.4...SN2 | LPE (20:4) [sn2] | glycerophospholipids | Lysophosphatidylethanolamine | 0.101 |
| PE.P.18.0.22.5...N6 | PE (P-18:0/22:5) (n6) | glycerophospholipids | alkenylphosphatidylethanolamine | 0.101 |
| PC.P.16.0.20.5 | PC (P-16:0/20:5) | glycerophospholipids | alkenylphosphatidylcholine | 0.100 |
| LPE.18.2...SN1 | LPE (18:2) [sn1] | glycerophospholipids | Lysophosphatidylethanolamine | 0.097 |
| DE.20.4 | DE (20:4) | Neutral/Other | dehydrocholesteryl ester | 0.097 |
Using the phenotype influence score obtained through c-SWAT, highly AD-associated lipids were identified.
Discussion
In this study, we developed and evaluated a deep learning-based approach to select phenotype-related features and construct an AD classification model using feature correlations. This approach was applied to serum-based metabolomics data related to AD. From our analysis, we discerned specific features with a strong correlation to AD. Using these features enabled more accurate classification and played a crucial role in enhancing the accuracy of classification across various Alzheimer's Disease stages.
Our findings highlight the effectiveness of our method in isolating metabolites that correlate with AD (Table 1). By analyzing these associations, we can better understand the role of lipidomic data, a subset of metabolomics, in the broader context of AD's pathophysiology.
Based on our results, Cer(d16:1/22:0), known as N-docosanoyl-hexadecasphing-4-enine, emerged as one of the metabolites with elevated PIS values. This ceramide is notably found in cell membranes, especially in peripheral nerve cells and the central nervous system. Its roles in fundamental cellular processes like cell division, differentiation, and cell death are significant. Changes in levels of ceramides like Cer(d16:1/22:0) have been observed in neurodegenerative diseases like AD.35,36 They might be involved in AD-related pathways, including inflammation in the brain, neuronal damage and death, and the formation of amyloid-beta plaques.37,38 PI(37:6), a member of the phosphatidylinositols family, has been reported to be integral to microglial signaling pathways. These phosphatidylinositols, recognized as essential secondary messenger lipids, are notably affected by amyloid-β protein deposits in AD.39 Originating from various phosphatidylinositol kinases, the significance of their phosphorylation state in their functional roles has been emphasized. CE (20:4) is a cholesteryl ester involved in cholesterol movement and storage. Recent studies found that certain cholesteryl esters, including CE (20:4), have decreased levels in Alzheimer's disease patients. This observation hints at a possible connection between these esters and the disease, though the exact role is still being explored.40 LPE (20:4) [sn1] is a component in the Glycerophospholipid metabolism pathway and interacts with specific enzymes. Studies suggest that variations in such lipids could be associated with brain health and conditions like AD, with peroxisomal function potentially being a key factor.41 In AD samples, the levels of key metabolites—Cer(d16:1/22:0), PI(37:6), CE (20:4) [+OH], and LPE (20:4) [sn1]—were found to be 1.27%, 3.23%, 1.54%, and 1.49% higher, respectively, compared to CN samples. deDE (18:2) and deDE (20:4) from the Dehydrodesmosteryl ester class showed high associations. While these compounds showed a strong correlation with AD, the potential effects of medications, such as donepezil, on these associations should be taken into account. Further research and interpretation are essential to confirm this correlation.
In instances where the dataset may not be sufficiently extensive for exhaustive learning, our approach, c-SWAT, has proven adept at confronting these limitations effectively. This efficacy can be largely attributed to our pre-processing stage, where we leverage feature correlations to tactically arrange our data. Our approach is engineered to diminish the weight of less pertinent features during the training phase, with an objective of filtering out unnecessary noise and thereby minimizing the risk of overfitting. Essentially, this phase plays a critical role in safeguarding our model against the potential negative influence of extraneous features on its performance. This element of our work, in particular, demonstrates the potential of c-SWAT to operate effectively even under less than ideal conditions, thus underscoring its value in the pursuit of improving deep learning methodologies.
Conclusion
We developed a deep learning-based approach, c-SWAT, which uses the strength of feature correlations to maximize the predictive capabilities of tabular datasets. A key feature of c-SWAT is its ability to handle correlations inherent in biological data and incorporate them into the learning process, contributing to improved predictive performance. In the research area of AD, a neurodegenerative disorder marked by progressive cognitive impairment, c-SWAT appear promising for identifying potentially AD-related lipids and building predictive models for AD. It's important to note that while our results using c-SWAT appear promising, these findings are needed to be validated using large independent data sets. Furthermore, c-SWAT's ability to efficiently identify meaningful patterns from complex biological data, especially when the dataset may not be large enough for exhaustive learning, underscores the potential of this method. In the future, this method will be applied to large-scale metabolite datasets to further refine its predictive capabilities. Quantitative endophenotypes will also be leveraged to evaluate the early stages of AD, including mild cognitive impairment (MCI), providing insights into the preclinical phase of the disease. Thus, by utilizing the advantages of deep learning and innovative feature correlation methods, c-SWAT offers a significant opportunity to enhance disease prediction and biomarker discovery in AD and beyond.
Contributors
TJ contributed to the conception and design of the study, analysis of data, and drafting of the manuscript. JP contributed to analysis of data and drafting of the manuscript. PB contributed to drafting of the manuscript. KH contributed to the acquisition of data and drafting of the manuscript. TW contributed to the acquisition of data and drafting of the manuscript. MA contributed to the data preprocessing and drafting of the manuscript. PJM contributed to the acquisition of data and drafting of the manuscript. CG contributed to drafting of the manuscript. RKD contributed to the conception and design of the study, acquisition of data, and drafting of the manuscript. AJS contributed to the conception and design of the study, acquisition of data, and drafting of the manuscript. KN contributed to the conception and design of the study and drafting of the manuscript. TJ, AJS and KN accessed and verified the data. All authors read and approved the final version of the manuscript. All authors had access to the underlying data, which is also available to the scientific community through the ADNI website.
Data sharing statement
The ADNI data used in this study were obtained from the ADNI database (https://adni.loni.usc.edu). The source code for executing the c-SWAT approach, as described in this paper, is openly available. The code can be accessed at https://github.com/taehojo/c-SWAT.
Declaration of interests
Dr. Saykin receives support from multiple NIH grants (P30 AG010133, P30 AG072976, R01 AG019771, R01 AG057739, U19 AG024904, R01 LM013463, R01 AG068193, T32 AG071444, U01 AG068057, U01 AG072177, and U19 AG074879). He has also received support from Avid Radiopharmaceuticals, a subsidiary of Eli Lilly (in kind contribution of PET tracer precursor); Siemens Medical Solutions USA, Inc. (Dementia Advisory Board); NIH NHLBI (MESA Observational Study Monitoring Board); Eisai (Scientific Advisory Board); NIH/NIA: External Advisory Committees, Multiple NIH-funded centers/programs; and Springer-Nature Publishing (Editorial Office Support as Editor-in-Chief, Brain Imaging and Behavior). Dr. Kaddurah-Daouk receives support from multiple NIH grants (3U01AG061359, 1RF1AG059093, 1RF1AG058942, 5U19AG063744, 3U19AG063744-04S1, 1R01AG069901, 3U01AG061359-05S1). She is an inventor on a series of patents related to metabolomics signatures in neuropsychiatric diseases. Dr. Kaddurah-Daouk holds equity and stock in Metabolon, Inc., and PsyProtix, which were not involved in this study. Matthias Arnold is coinventor (through Duke University/Helmholtz Zentrum München) on patents on applications of metabolomics in diseases of the central nervous system. Matthias Arnold also holds equity in Chymia LLC and IP in PsyProtix and Atai that is unrelated to this work. The other authors declare no conflict of interest.
Acknowledgements
Funding for ADMC (Alzheimer's Disease Metabolomics Consortium, led by Dr R.K.-D. at Duke University) was provided by the National Institute on Aging grants 1U19AG063744, 1R01AG069901-01A1, U01AG061357, P30AG10161, P30AG72975, R01AG15819, R01AG17917, U01AG46152, U01AG61356, RF1AG058942, RF1AG059093, and U01AG061359, a component of the Accelerating Medicines Partnership for AD (AMP-AD) Target Discovery and Preclinical Validation Project (https://www.nia.nih.gov/research/dn/amp-ad-target-discovery-and-preclinical-validation-project) and the National Institute on Aging, a component of the M2OVE-AD Consortium (Molecular Mechanisms of the Vascular Etiology of AD—Consortium https://www.nia.nih.gov/news/decoding-molecular-ties-between-vascular-disease-and-alzheimers).
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Additional support for data analysis was provided in part by grants: AARG 22-974053, P30 AG010133, P30 AG072976, R01 AG019771, R01 AG057739, U01 AG024904, R01 LM013463, R01 AG068193, T32 AG071444, U01 AG068057, U01 AG072177, R01 LM012535, R01 AG069901 and R03 AG063250.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104820.
Contributor Information
Andrew J. Saykin, Email: asaykin@iupui.edu.
Kwangsik Nho, Email: knho@iupui.edu.
Alzheimer's Disease Metabolomics Consortium (ADMC):
Rima Kaddurah-Daouk, Alexandra Kueider-Paisley, P. Murali Doraiswamy, Colette Blach, Arthur Moseley, Will Thompson, Lisa St John-Williams, Siamak Mahmoudiandehkhordi, Jessica Tenenbaum, Kathleen Welsh-Balmer, Brenda Plassman, Andrew J. Saykin, Kwangsik Nho, Shannon L. Risacher, Gabi Kastenmüller, Matthias Arnold, Xianlin Han, Rebecca Baillie, Rob Knight, Pieter Dorrestein, James Brewer, Emeran Mayer, Jennifer Labus, Pierre Baldi, Arpana Gupta, Oliver Fiehn, Dinesh Barupal, Peter Meikle, Sarkis Mazmanian, Dan Rader, Mitchel Kling, Leslie Shaw, John Trojanowski, Cornelia van Duijin, Alejo Nevado-Holgado, David Bennett, Ranga Krishnan, Ali Keshavarzian, Robin Vogt, Arfan Ikram, Thomas Hankemeier, Ines Thiele, Nathan Price, Cory Funk, Priyanka Baloni, Wei Jia, David Wishart, Roberta Brinton, Rui Chang, Lindsay Farrer, Rhoda Au, Wendy Qiu, Peter Würtz, Therese Koal, Lara Mangravite, Jan Krumsiek, Karsten Suhre, John Newman, Herman Moreno, Tatania Foroud, Frank Sacks, and Janet Jansson
Alzheimer's Disease Neuroimaging Initiative (ADNI):
Michael W. Weiner, Paul Aisen, Ronald Petersen, Clifford R. Jack, Jr., William Jagust, John Q. Trojanowki, Arthur W. Toga, Laurel Beckett, Robert C. Green, Andrew J. Saykin, John C. Morris, Richard J. Perrin, Leslie M. Shaw, Zaven Khachaturian, Maria Carrillo, William Potter, Lisa Barnes, Marie Bernard, Hector Gonzalez, Carole Ho, John K. Hsiao, Jonathan Jackson, Eliezer Masliah, Donna Masterman, Ozioma Okonkwo, Richard Perrin, Laurie Ryan, Nina Silverberg, Adam Fleisher, Diana Truran Sacrey, Juliet Fockler, Cat Conti, Dallas Veitch, John Neuhaus, Chengshi Jin, Rachel Nosheny, Miriam Ashford, Derek Flenniken, Adrienne Kormos, Tom Montine, Michael Rafii, Rema Raman, Gustavo Jimenez, Michael Donohue, Devon Gessert, Jennifer Salazar, Caileigh Zimmerman, Yuliana Cabrera, Sarah Walter, Garrett Miller, Godfrey Coker, Taylor Clanton, Lindsey Hergesheimer, Stephanie Smith, Olusegun Adegoke, Payam Mahboubi, Shelley Moore, Jeremy Pizzola, Elizabeth Shaffer, Brittany Sloan, Danielle Harvey, Arvin Forghanian-Arani, Bret Borowski, Chad Ward, Christopher Schwarz, David Jones, Jeff Gunter, Kejal Kantarci, Matthew Senjem, Prashanthi Vemuri, Robert Reid, Nick C. Fox, Ian Malone, Paul Thompson, Sophia I. Thomopoulos, Talia M. Nir, Neda Jahanshad, Charles DeCarli, Alexander Knaack, Evan Fletcher, Duygu Tosun-Turgut, Stephanie Rossi Chen, Mark Choe, Karen Crawford, Paul A. Yushkevich, Sandhitsu Das, Robert A. Koeppe, Eric M. Reiman, Kewei Chen, Chet Mathis, Susan Landau, Nigel J. Cairns, Erin Householder, Erin Franklin, Haley Bernhardt, Lisa Taylor-Reinwald, Magdalena Korecka, Michal Figurski, Scott Neu, Kwangsik Nho, Shannon L. Risacher, Liana G. Apostolova, Li Shen, Tatiana M. Foroud, Kelly Nudelman, Kelley Faber, Kristi Wilmes, Leon Thal, Lisa C. Silbert, Betty Lind, Rachel Crissey, Jeffrey A. Kaye, Raina Carter, Sara Dolen, Joseph Quinn, Lon S. Schneider, Sonia Pawluczyk, Mauricio Becerra, Liberty Teodoro, Karen Dagerman, Bryan M. Spann, James Brewer, Helen Vanderswag, Jaimie Ziolkowski, Judith L. Heidebrink, Lisa Zbizek-Nulph, Joanne L. Lord, Sara S. Mason, Colleen S. Albers, David Knopman, Kris Johnson, Javier Villanueva-Meyer, Valory Pavlik, Nathaniel Pacini, Ashley Lamb, Joseph S. Kass, Rachelle S. Doody, Victoria Shibley, Munir Chowdhury, Susan Rountree, Mimi Dang, Yaakov Stern, Lawrence S. Honig, Akiva Mintz, Beau Ances, David Winkfield, Maria Carroll, Georgia Stobbs-Cucchi, Angela Oliver, Mary L. Creech, Mark A. Mintun, Stacy Schneider, David Geldmacher, Marissa Natelson Love, Randall Griffith, David Clark, John Brockington, Daniel Marson, Hillel Grossman, Martin A. Goldstein, Jonathan Greenberg, Effie Mitsis, Raj C. Shah, Melissa Lamar, Patricia Samuels, Ranjan Duara, Maria T. Greig-Custo, Rosemarie Rodriguez, Marilyn Albert, Chiadi Onyike, Leonie Farrington, Scott Rudow, Rottislav Brichko, Stephanie Kielb, Amanda Smith, Balebail Ashok Raj, Kristin Fargher, Martin Sadowski, Thomas Wisniewski, Melanie Shulman, Arline Faustin, Julia Rao, Karen M. Castro, Anaztasia Ulysse, Shannon Chen, Mohammed O. Sheikh, Jamika Singleton-Garvin, P. Murali Doraiswamy, JeffreyR. Petrella, Olga James, Terence Z. Wong, Salvador Borges-Neto, Jason H. Karlawish, David A. Wolk, Sanjeev Vaishnavi, Christopher M. Clark, Steven E. Arnold, Charles D. Smith, Gregory A. Jicha, Flavius D. Raslau, Oscar L. Lopez, MaryAnn Oakley, Donna M. Simpson, Anton P. Porsteinsson, Kim Martin, Nancy Kowalski, Melanie Keltz, Bonnie S. Goldstein, Kelly M. Makino, M. Saleem Ismail, Connie Brand, Gaby Thai, Aimee Pierce, Beatriz Yanez, Elizabeth Sosa, Megan Witbracht, Brendan Kelley, Trung Nguyen, Kyle Womack, Dana Mathews, Mary Quiceno, Allan I. Levey, James J. Lah, Ihab Hajjar, Janet S. Cellar, Jeffrey M. Burns, Russell H. Swerdlow, William M. Brooks, Daniel H.S. Silverman, Sarah Kremen, Liana Apostolova, Kathleen Tingus, Po H. Lu, George Bartzokis, Ellen Woo, Edmond Teng, Neill R. Graff-Radford, Francine Parfitt, Kim Poki-Walker, Martin R. Farlow, Ann Marie Hake, Brandy R. Matthews, Jared R. Brosch, Scott Herring, Christopher H. van, Adam P. Mecca, Susan P. Good, Martha G. MacAvoy, Richard E. Carson, Pradeep Varma, Howard Chertkow, Susan Vaitekunis, Chris Hosein, Sandra Black, Bojana Stefanovic, Chris (Chinthaka) Heyn, Ging-Yuek Robin Hsiung, Ellen Kim, Benita Mudge, Vesna Sossi, Howard Feldman, Michele Assaly, Elizabeth Finger, Stephen Pasternak, Irina Rachinsky, Andrew Kertesz, Dick Drost, John Rogers, Ian Grant, Brittanie Muse, Emily Rogalski, Jordan Robson, M.-Marsel Mesulam, Diana Kerwin, Chuang-Kuo Wu, Nancy Johnson, Kristine Lipowski, Sandra Weintraub, Borna Bonakdarpour, Nunzio Pomara, Raymundo Hernando, Antero Sarrael, Howard J. Rosen, Bruce L. Miller, David Perry, Raymond Scott Turner, Kathleen Johnson, Brigid Reynolds, Kelly MCCann, Jessica Poe, Reisa A. Sperling, Keith A. Johnson, Gad A. Marshall, Jerome Yesavage, Joy L. Taylor, Steven Chao, Jaila Coleman, Jessica D. White, Barton Lane, Allyson Rosen, Jared Tinklenberg, Christine M. Belden, Alireza Atri, Kelly A. Clark, Edward Zamrini, Marwan Sabbagh, Ronald Killiany, Robert Stern, Jesse Mez, Neil Kowall, Andrew E. Budson, Thomas O. Obisesan, Oyonumo E. Ntekim, Saba Wolday, Javed I. Khan, Evaristus Nwulia, Sheeba Nadarajah, Alan Lerner, Paula Ogrocki, Curtis Tatsuoka, Parianne Fatica, Pauline Maillard, John Olichney, Owen Carmichael, Vernice Bates, Horacio Capote, Michelle Rainka, Michael Borrie, T.-Y. Lee, Dr Rob Bartha, Sterling Johnson, Sanjay Asthana, Cynthia M. Carlsson, Allison Perrin, Anna Burke, Douglas W. Scharre, Maria Kataki, Rawan Tarawneh, David Hart, Earl A. Zimmerman, Dzintra Celmins, Delwyn D. Miller, Laura L. BolesPonto, Karen Ekstam Smith, Hristina Koleva, Hyungsub Shim, Ki Won Nam, Susan K. Schultz, Jeff D. Williamson, Suzanne Craft, Jo Cleveland, Mia Yang, Kaycee M. Sink, Brian R. Ott, Jonathan Drake, Geoffrey Tremont, Lori A. Daiello, Jonathan D. Drake, Aaron Ritter, Charles Bernick, Donna Munic, Abigail O'Connelll, Jacobo Mintzer, Arthur Wiliams, Joseph Masdeu, Jiong Shi, Angelica Garcia, Paul Newhouse, Steven Potkin, Stephen Salloway, Paul Malloy, Stephen Correia, Smita Kittur, Godfrey D. Pearlson, Karen Blank, Karen Anderson, Laura A. Flashman, Marc Seltzer, Mary L. Hynes, Robert B. Santulli, Norman Relkin, Gloria Chiang, Athena Lee, Michael Lin, Lisa Ravdin, Ron Petersen, Thomas Neylan, Jordan Grafman, Sarah Danowski, Catherine Nguyen-Barrera, Jacqueline Hayes, Shannon Finley, Matthew Bernstein, Matt Senjem, Norm Foster, Sungeun Kim, Ajay Sood, Kimberly S. Blanchard, Debra Fleischman, Konstantinos Arfanakis, Daniel Varon, Maria T. Greig, Jeffrey R. Petrella, Bonnie Goldstein, Kimberly S. Martin, Christopher Reist, Carl Sadowsky, Walter Martinez, Teresa Villena, Howard Rosen, Gad Marshall, Elaine R. Peskind, Eric C. Petrie, Gail Li, Scott Mackin, Gustavo Jimenez-Maggiora, Erin Drake, Mike Donohue, Craig Nelson, David Bickford, Meryl Butters, Michelle Zmuda, Denise Reyes, Kelley M. Faber, Kelly N. Nudelman, Yiu Ho Au, Kelly Scherer, Daniel Catalinotto, Samuel Stark, Elise Ong, and Dariella Fernandez
Appendix A. Supplementary data
References
- 1.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 2.Jo T. Gilbut; 2019. Deep-learning for everyone. [Google Scholar]
- 3.Schmidhuber J. Deep learning in neural networks: an overview. Neural Network. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Bengio Y., LeCun Y. Scaling learning algorithms towards AI. Large-scale kernel Machines. 2007;34(5):1–41. [Google Scholar]
- 5.Jo T., Hou J., Eickholt J., Cheng J. Improving protein fold recognition by deep learning networks. Sci Rep. 2015;5(1):1–11. doi: 10.1038/srep17573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jumper J., Evans R., Pritzel A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciloglu F.U., Caliskan A., Saridag A.M., et al. Drug-resistant Staphylococcus aureus bacteria detection by combining surface-enhanced Raman spectroscopy (SERS) and deep learning techniques. Sci Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-97882-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jo T., Nho K., Saykin A.J. Deep learning in Alzheimer's disease: diagnostic classification and prognostic prediction using neuroimaging data. Front Aging Neurosci. 2019;11:220. doi: 10.3389/fnagi.2019.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jo T., Nho K., Risacher S.L., Saykin A.J. Deep learning detection of informative features in tau PET for Alzheimer's disease classification. BMC Bioinf. 2020;21(21):1–13. doi: 10.1186/s12859-020-03848-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krizhevsky A., Sutskever I., Hinton G.E. Advances in neural information processing systems. 2012. Imagenet classification with deep convolutional neural networks. [Google Scholar]
- 11.Hinton G., Deng L., Yu D., et al. Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups. IEEE Signal Process Mag. 2012;29(6):82–97. [Google Scholar]
- 12.Shwartz-Ziv R., Armon A. Tabular data: deep learning is not all you need. Inf Fusion. 2022;81:84–90. [Google Scholar]
- 13.Chervitz S.A., Deutsch E.W., Field D., et al. Data standards for Omics data: the basis of data sharing and reuse. Methods Mol Biol. 2011;719:31–69. doi: 10.1007/978-1-61779-027-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jo T., Nho K., Bice P., Saykin A.J., Initiative AsDN Deep learning-based identification of genetic variants: application to Alzheimer's disease classification. Briefings Bioinf. 2022;23(2):bbac022. doi: 10.1093/bib/bbac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Strooper B., Karran E. The cellular phase of Alzheimer's disease. Cell. 2016;164(4):603–615. doi: 10.1016/j.cell.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 16.Terracciano A., Sutin A.R. Personality and Alzheimer's disease: an integrative review. Personal Disord. 2019;10(1):4. doi: 10.1037/per0000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Younes L., Albert M., Moghekar A., Soldan A., Pettigrew C., Miller M.I. Identifying changepoints in biomarkers during the preclinical phase of Alzheimer's disease. Front Aging Neurosci. 2019;11:74. doi: 10.3389/fnagi.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J., Howard R.S., Schneider L.S. The current landscape of prevention trials in dementia. Neurotherapeutics. 2022;19:1–20. doi: 10.1007/s13311-022-01236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Bokhoven P., de Wilde A., Vermunt L., et al. The Alzheimer's disease drug development landscape. Alzheimer's Res Ther. 2021;13(1):1–9. doi: 10.1186/s13195-021-00927-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanseeuw B.J., Betensky R.A., Jacobs H.I., et al. Association of amyloid and tau with cognition in preclinical Alzheimer disease: a longitudinal study. JAMA Neurol. 2019;76(8):915–924. doi: 10.1001/jamaneurol.2019.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sweeney M.D., Sagare A.P., Zlokovic B.V. Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neurol. 2018;14(3):133–150. doi: 10.1038/nrneurol.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butterfield D.A., Halliwell B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat Rev Neurosci. 2019;20(3):148–160. doi: 10.1038/s41583-019-0132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michalkova L., Hornik S., Sýkora J., Habartova L., Setnicka V., Bunganic B. Early detection of pancreatic cancer in type 2 diabetes mellitus patients based on 1H NMR metabolomics. J Proteome Res. 2021;20(3):1744–1753. doi: 10.1021/acs.jproteome.0c00990. [DOI] [PubMed] [Google Scholar]
- 24.Liang L., Sun F., Wang H., Hu Z. Metabolomics, metabolic flux analysis and cancer pharmacology. Pharmacol Ther. 2021;224 doi: 10.1016/j.pharmthera.2021.107827. [DOI] [PubMed] [Google Scholar]
- 25.Escobar M.Q., de Moraes Pontes J.G., Tasic L. Metabolomics in degenerative brain diseases. Brain Res. 2021;1773 doi: 10.1016/j.brainres.2021.147704. [DOI] [PubMed] [Google Scholar]
- 26.Hone-Blanchet A., Bohsali A., Krishnamurthy L.C., et al. Relationships between frontal metabolites and Alzheimer's disease biomarkers in cognitively normal older adults. Neurobiol Aging. 2022;109:22–30. doi: 10.1016/j.neurobiolaging.2021.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marksteiner J., Blasko I., Kemmler G., Koal T., Humpel C. Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer's disease. Metabolomics. 2018;14(1):1–10. doi: 10.1007/s11306-017-1297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peña-Bautista C., Roca M., Hervás D., et al. Plasma metabolomics in early Alzheimer's disease patients diagnosed with amyloid biomarker. J Proteonomics. 2019;200:144–152. doi: 10.1016/j.jprot.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Toledo J.B., Arnold M., Kastenmueller G., et al. Metabolic network failures in Alzheimer's disease: a biochemical road map. Alzheimer's Dementia. 2017;13(9):965–984. doi: 10.1016/j.jalz.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nho K., Kueider-Paisley A., Arnold M., et al. Serum metabolites associated with brain amyloid beta deposition, cognition and dementia progression. Brain Commun. 2021;3(3):fcab139. doi: 10.1093/braincomms/fcab139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varma V.R., Oommen A.M., Varma S., et al. Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: a targeted metabolomics study. PLoS Med. 2018;15(1) doi: 10.1371/journal.pmed.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9(1):1–13. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair V., Hinton G.E. Proceedings of the 27th international conference on machine learning. ICML-10; 2010. Rectified linear units improve restricted Boltzmann machines. [Google Scholar]
- 34.Kingma D.P., Ba J. Adam: a method for stochastic optimization. arXiv. 2014 Preprint. arXiv:14126980. [Google Scholar]
- 35.Chai J.-F., Raichur S., Khor I.W., et al. Associations with metabolites in Chinese suggest new metabolic roles in Alzheimer's and Parkinson's diseases. Hum Mol Genet. 2020;29(2):189–201. doi: 10.1093/hmg/ddz246. [DOI] [PubMed] [Google Scholar]
- 36.Haughey N.J., Bandaru V.V., Bae M., Mattson M.P. Roles for dysfunctional sphingolipid metabolism in Alzheimer's disease neuropathogenesis. Biochim Biophys Acta. 2010;1801(8):878–886. doi: 10.1016/j.bbalip.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He X., Huang Y., Li B., Gong C.-X., Schuchman E.H. Deregulation of sphingolipid metabolism in Alzheimer's disease. Neurobiol Aging. 2010;31(3):398–408. doi: 10.1016/j.neurobiolaging.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimm M.O., Grimm H.S., Pätzold A.J., et al. Regulation of cholesterol and sphingomyelin metabolism by amyloid-β and presenilin. Nat Cell Biol. 2005;7(11):1118–1123. doi: 10.1038/ncb1313. [DOI] [PubMed] [Google Scholar]
- 39.Desale S.E., Chinnathambi S. Phosphoinositides signaling modulates microglial actin remodeling and phagocytosis in Alzheimer's disease. Cell Commun Signal. 2021;19(1):1–12. doi: 10.1186/s12964-021-00715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Proitsi P., Kim M., Whiley L., et al. Plasma lipidomics analysis finds long chain cholesteryl esters to be associated with Alzheimer's disease. Transl Psychiatry. 2015;5(1):e494. doi: 10.1038/tp.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood P.L. Lipidomics of Alzheimer's disease: current status. Alzheimer's Res Ther. 2012;4(1):1–10. doi: 10.1186/alzrt103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.