Abstract
Horizontal gene transfer (HGT) has been considered the most important pathway to introduce antibiotic resistance genes (ARGs), which seriously threatens human health and biological security. The presence of ARGs in the aquatic environment and their effect on the intestinal micro-ecosystem of aquatic animals can occur easily. To investigate the HGT potential and rule of exogenous ARGs in the intestinal flora, a visual conjugative model was developed, including the donor of dual-fluorescent bacterium and the recipient of Xenopus tropicalis intestinal microbiome. Some common pollutants of oxytetracycline (OTC) and three heavy metals (Zn, Cu and Pb) were selected as the stressor. The multi-techniques of flow cytometry (FCM), scanning electron microscopy (SEM), atomic force microscopy (AFM), single-cell Raman spectroscopy with sorting (SCRSS) and indicator analysis were used in this study. The results showed that ARG transfer could occur more easily under stressors. Moreover, the conjugation efficiency mainly depended on the viability of the intestinal bacteria. The mechanisms of OTC and heavy metal stressing conjugation included the upregulation of ompC, traJ, traG and the downregulation of korA gene. Moreover, the enzymatic activities of SOD, CAT, GSH-PX increased and the bacterial surface appearance also changed. The predominant recipient was identified as Citrobacter freundi by SCRSS, in which the abundance and quantity of ARG after conjugation were higher than those before. Therefore, since the diversity of potential recipients in the intestine are very high, the migration of invasive ARGs in the microbiome should be given more attention to prevent its potential risks to public health.
Subject terms: Microbiome, Environmental microbiology
Introduction
Antibiotic resistance could be selected by antibiotics released into the environment at low (subinhibitory) concentrations [1] and antibiotic resistance genes (ARGs) from the environmental resistome have been detected in human commensals and pathogens [2]. HGT is the main pathway to mediate the spread of genes encoding antibiotic resistance among microbial hosts [3]. Moreover, as a significant HGT mechanism, conjugation can disseminate ARGs by transferring mobile genetic elements, including plasmids, integrons and transposons [4–7]. The horizontal spread of plasmids usually carries one or more resistance genes and deserves more extensive attention, especially in high-density flora such as the intestinal environment of humans and animals [8]. The long-term alteration of the normal healthy gut microbiota and horizontal transfer of resistance genes could result in a reservoir of organisms with a multidrug-resistant gene pool [9]. Among different environments, the human intestine-associated microbiota has 25 times more likelihood of having horizontal gene transfer [10]. Meanwhile, the intestine can serve as a reservoir for a variety of ARGs, such as β-lactam resistance and quinolone resistance [11], in which the chromosomal ancestral source of qnrB is theorized to be Citrobacter [12]. Therefore, it should be noted that the intestines of aquatic animals would be a hot site for HGT to occur.
Some factors, including heavy metals and antibacterial biocides, can co-select for antibiotic-resistant strains via cross-resistance (that is, via the same mechanism) or co-resistance (that is, via genetically linked mechanisms) [13–15]. For example, E. xiangfangensis from the intestine showed resistance not only to β-lactam, quinolone, aminoglycoside and sulfonamide antibiotics but also to an unusually high number of plasmid-based heavy metal resistance gene clusters, such as copper, mercury and cadmium [16]. Gram-negative bacteria increased significantly after the addition of heavy metals and were more prone to receiving resistant plasmids from donors [17, 18]. It was shown that OTC served as a selective pressure to increase the proportion of tet-bacteria in the red seabream (Pagrus major) intestine, where tet (B) and tet (M) abundance increases immediately [19]. The presence of OTC promotes the spread and proliferation of ARG and transforms the microbial community structure [20]. The concentration of heavy metals may promote bacterial antibiotic resistance and thereby increase the prevalence of ARGs across aquatic environments [21]. For instance, Cu could accelerate the propagation of MRGs and ARGs via intI1 in the horizontal gene transmission process in urban rivers [22]. In duck/fish polyculture ponds, levels of Zn and Cu were significantly correlated with numerous ARG types, in which sul2, floR, and tetM were identified as potential ARG indicators [23]. The observed increase in the abundance of antibiotic resistance in aquatic ecosystems is likely due to the co-selection of Pb, Zn and Co [24]. A diversity and high abundance of ARGs were observed to be disseminated via water source to aquaculture pond water and existed in the reared organism [25], while the intestine was a hotspot environment for receiving ARGs. Therefore, there is a great need to investigate the effects of ARBs and ARGs on the intestinal microbiome and evaluate the potential for horizontal migration of ARGs in the aquatic environment with the combined presence of heavy metals and antibiotics.
In this study, a conjugative system consisting of both multiple-resistant bacteria and a potential intestinal microbiome was established to study the conjugative transfer capability and rule of four ARGs carried by the plasmid, in which ARGs of the donor were harbored on the environmentally relevant conjugative plasmid RP4. Some common aquatic pollutants, namely, three heavy metals (Zn, Cu, Pb) and one antibiotic (OTC), were selected as stressor. Tests including cell viability differences, plasmid validation, mRNA expression levels, bacterial reactive oxygen system enzyme index and cell membrane variation were conducted to reveal the underlying mechanisms. The results aid in comprehending the mechanism for the enrichment of the ARG level of each stressor and reveal the ARG dissemination approaches between bacteria during conjugation.
Materials and methods
Bacterial strains
Dual fluorescent E. coli K12 (chromosome labeled with red fluorescent protein mCherry) carrying the RP4 plasmid (labeled with green fluorescent protein sfgfp) was selected as the donor, while indigenous bacteria from intestinal samples of Xenopus tropicalis were selected as the recipient. Since the mCherry gene expresses red fluorescence and the sfgfp gene expresses green fluorescence, E. coli K12 will exhibit dual-fluorescence (red and green). When the plasmid RP4 of E. coli K12 was transferred to other bacteria via HGT, the recipient would exhibit green fluorescence, while the donor itself, which had lost the plasmid, would express only red fluorescence in chromosome. The culture conditions and acquisition method of the recipient are shown in Text S1.
Mobile plasmid-mediated conjugative transfer and plasmid verification
To test the effect of three heavy metals (Zn, Cu, Pb) and the antibiotic OTC on ARG transfer to indigenous intestinal microbes, mating conjugation was established. The conjugative system was conducted in the LB-based solution and consisted of both donor and recipient bacteria at cell densities of 108 CFU/mL. In this case, 4 mL of the donors and recipients were mixed, followed by immediately adding 12 mL of different stress solutions (containing OTC, Zn, Cu, Pb, Zn + OTC, Cu + OTC, Pb + OTC groups) subsequently. Treatment with equal volume sterile water addition was used as a control. The mixed cultures were harvested for subsequent experimental assays after an 8 h incubation at 37 °C [26].
The donors, recipients and transconjugants in mating systems were isolated on LB plates using a sterile inoculation loop after recognizing the fluorescence signal under the inverted fluorescence microscopy (Carl Zeiss, Germany). The plasmids of transconjugants were extracted using the E.Z.N.A. BAC/PAC DNA Isolation Kit (D2156-01, Omega, USA). The specific traG gene of plasmid RP4 was amplified by PCR (Eppendorf, Germany) [17] and the amplicons were observed using 1% agarose gel electrophoresis. PCR primers and conditions are described in Text S2 and Supplementary Table S1.
Analysis of conjugative transfer frequency with the aid of FCM
The mixed culture was harvested by centrifuging for 8 min at 8000 rpm and washing twice with PBS (pH 7.2–7.4). The acquired cell pellet was resuspended in PBS and filtered with a 70 μm cell sieve and the OD600 of the final cell suspension was set to 0.5. The frequency of conjugative transfer was assessed by FCM (Flowsight, Merck, USA). The number of bacteria collected by FCM was set to 10,000. The following formula was used to calculate the conjugative transfer frequency (f) [27]: f = (Ng–Nd)/(Nt–Nr), where Ng is the number of bacteria carrying green fluorescent (donors and transconjugants); Nd is the number of bacteria carrying both red and green fluorescent (donors with plasmid RP4); Nt is the total number of collected bacteria, Nr is the number of bacteria carrying red fluorescent (donors with and without plasmid RP4).
Measurement of viable bacteria counting
Verify the stressor response induced by exogenous heavy metals and OTC over time by examining the absorbance and cell viability of donors and recipients. The Cell Counting Kit-8 (CCK-8) (APE × BIO, USA) assay was applied to assess the activity of donors and recipients. The OD450 value of each well was measured at 0, 2, 4, 6 and 8 h with the Varioskan LUX Multifunctional microplate reader (VLBL0TD2, Thermo Fisher, USA). Calculation methods of cell viability are shown in Text S3.
Evaluation of mRNA expression for conjugative transfer
The mechanism of various stressors affecting HGT can be explained by quantifying HGT-related mRNA. Firstly, the donor, recipient and mating systems were treated by control and experimental groups for 8 h for the RNA extractions. The bacterial cell pellets were collected by centrifugation at 8000 rpm, 4 °C for 8 min. Total RNA was extracted from pellets using Bacterial RNA Kit (R6950, Omega, USA). Secondly, extracted RNA was transcribed to cDNA with ReverTra Ace qPCR RT Kit (FSQ-101, TOYOBO, Japan) by PCR. Thirdly, the expression of the target genes was accurately quantified by qPCR (Bio-Rad, Singapore). Conditions of PCR reverse transcription reaction and QPCR reaction are described in Text S4 and Text S5. The HGT-related mRNA genes include porin genes (ompA, ompC, ompF), oxidative stress genes (rpoS), conjugative genes (trbBp), regulatory plasmid transfer and replication genes (trfAp, traJ), global regulator genes (korA, korB, trbA) and coupling genes (traG). The 16S rRNA gene was used as an internal control. The primers used in this study are provided in Table S2.
Determination of the bacterial reactive oxygen system index
ROS is an important mechanism by which HGT occurs in bacteria and the activity of ROS-related enzymes is usually positively correlated with their concentration [28, 29]. Catalase (CAT) assay kit (A007-1-1), superoxide dismutase (SOD) assay kit (A001-3) and glutathione peroxidase (GSH-PX) assay kit (A005-1) were used to evaluate the bacterial reactive oxygen system index in conjugative solution. The activity of the indicators was measured according to the manufacturer’s instructions. All kits were purchased from Nanjing Jiancheng Institute of Biological Engineering.
Effect of stressor on the cell membrane surface
Bacteria in the conjugatiive solution were prepared according to the steps in Text S6 and then observed by SEM (Tescan, Lyra 3 xmu). The conjugative solutions were adhered to fresh cut mica and then dried for more than 2 h [30], after which they were transferred to the sample stage of AFM (Dimension Fast-Scan, Bruker, USA) for bacterial imaging and images were acquired. Fields of vision were selected for observation in each sample based on the random distribution of bacteria and represented the final condition with the dominant images.
Cultivable and nonculturable transconjugant identification and qPCR
The cultivable transconjugants were separated from the total conjugative solution using plate streaking. Both cultivable and nonculturable transconjugants were separated with SCRSS (PRECI SCS-P300, HOOKE, Instruments Ltd.). SCRSS method for isolating single cells is described in Text S7. The transconjugant isolated from SCRSS were used to obtain amplified DNA with REPLI-g Single Cell kit (150345, QIAGEN, Germany) and the products were PCR amplified using bacterial primers 341F (5′- AGAGTTTGATCCTGGCTCAG-3′) and 806R (5′- TACGACTTAACCCCAATCGC-3′). The PCR products were then sent to Comate Bioscience Co.,Ltd. for sequencing. Then the obtained 16S rRNA gene sequences were put into BLAST for homology comparison.
Relative abundance of resistance genes was examined to explore the resistance evolution of recipients after receiving plasmids. Selection of tetM, zntA, pcoD and pbrT as indicator genes for OTC, Zn, Cu and Pb, respectively [19, 23, 31]. Gene tetA carried by the plasmid RP4 was selected to verify the entry of plasmid RP4 into the recipient [32]. Since intI1 are associated with HMRGs and ARGs gene cassette and sul1 is frequently associated with integrons [33, 34], intI1 and sul1 were selected to assess the migration potential and resistance risk of the recipient after receiving plasmid RP4. The qPCR tracked tetA, tetM, zntA, pcoD, pbrT, sul1 and intI1 in the recipients and transconjugants. The relative abundance of all measured target genes was calculated using 16S rRNA as an internal reference gene control. DNA was extracted from the collected bacteria using a bacterial DNA kit (D3350, Omega, USA) according to the manufacturer’s instructions. All the qPCR procedures and primers are described in detail in Text S4 and Table S3 in SI.
Correlation tests and statistical analysis
All experiments were conducted in biological triplicate. Transfer frequency data were derived from a total of nine data from three tests per parallel sample and expressed as mean ± standard deviation (SD). All data were analyzed with SPSS 27.0 (SPSS, Chicago, USA). The results were analyzed by Independent-sample t test methods. The p values less than 0.05 were considered to be statistically significant. Further, Pearson correlation was applied to calculate correlation coefficient, which was significant if p value was less than 0.05. Data visualization was performed using Origin 2021 (OriginLab Co., MA, USA).
Results
Stressor affected foreign ARG transfer to the intestinal microbiome
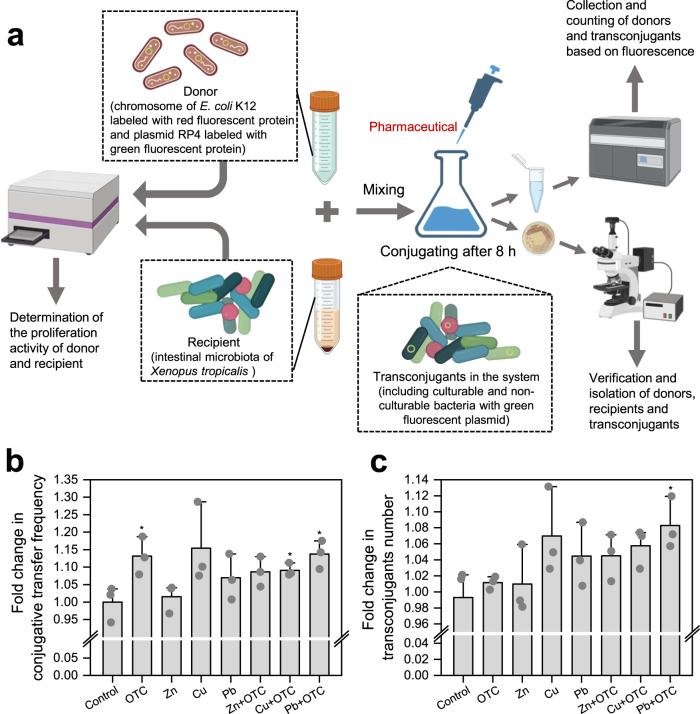
To evaluate the single and combined effects of these four stressors on conjugation at low concentrations, E. coli K12 with the RP4 plasmid was selected as the donor. Meanwhile, some indigenous microbiome from the intestine of Xenopus tropicalis were the recipients (Fig. 1a). Based on some studies, Zn, Cu and Pb exhibited the greatest conjugative transfer ability at concentrations of 50, 5 and 100 μg/L in freshwater microcosms, respectively [27]. Therefore, these concentrations were chosen to further explore whether HGT among animal intestinal microbiome has the same ability. The 100 μg/L concentration of OTC was chosen because it was found to strongly promote intI1 abundance in our previous study [35]. To verify the transfer of the RP4 plasmid, transconjugants were isolated on plates containing antibiotics (ampicillin, kanamycin and tetracycline) and green fluorescence verification was performed. Moreover, the plasmids in transconjugants were identified as the same as those in the donor by PCR amplification and gel electrophoresis, while no plasmid was detected in the recipient (Supplementary Fig. S1).
Fig. 1. Schematic depicting experimental design and conjugation results of ARGs induced by OTC and heavy metals.
a Schematic experimental design of the conjugation. b Fold change of conjugation frequency of RP4 plasmid in E. coli K12 under OTC and heavy metal. c Fold change of transconjugant number under antibiotic and heavy metal. Significant differences between individual exposure groups and the control were tested with the independent-sample t test: *(p < 0.05).
OTC and heavy metals might increase the conjugative transfer of ARGs by increasing the transconjugant quantity. The frequency under OTC, Zn, Cu and Pb stressors were up to 1-fold higher than that of the control group, where the OTC, Cu + OTC and Pb + OTC groups were significant (p = 0.022–0.045, Fig. 1b). Among the single heavy metals, Cu had the maximum transfer frequency [(2.0869 ± 0.0096) × 10−1 transconjugants per recipient cell], where the number of transconjugants was 1.0493–1.1314-fold higher than the control and the fold change in conjugation frequency was 1.0721–1.2826-fold higher, followed by Pb and Zn in that order (p = 0.10–0.71, Fig. 1c). In addition, the combined effects of any one of the three heavy metals and OTC were effective in increasing the number of transconjugants. The conjugative transfer frequency increased with Pb + OTC [(2.0614 ± 0.0078) × 10−1 transconjugants per recipient cell] and was up to 1.0943–1.1752-fold higher than that of the control group (p = 0.048). Therefore, the combined effect of low concentrations of Pb and OTC was more likely to promote the risk of HGT of ARGs in the environment. It should be noted that Xenopus tropicalis were not exposed to any OTC or heavy metals during their development from embryonic periods to adults, so their indigenous microbiome in the intestine should have low resistance. In this conjugative system, the coupling of low concentrations of OTC and heavy metals was enhanced by constructing intestine-potential bacteria in the environmentally relevant model compared to the control. Collectively, we concluded that OTC and Zn, Cu, Pb at environmental concentrations increased the conjugative transfer of ARGs.
Relationship between intestinal microbiome activity and conjugative transfer efficiency
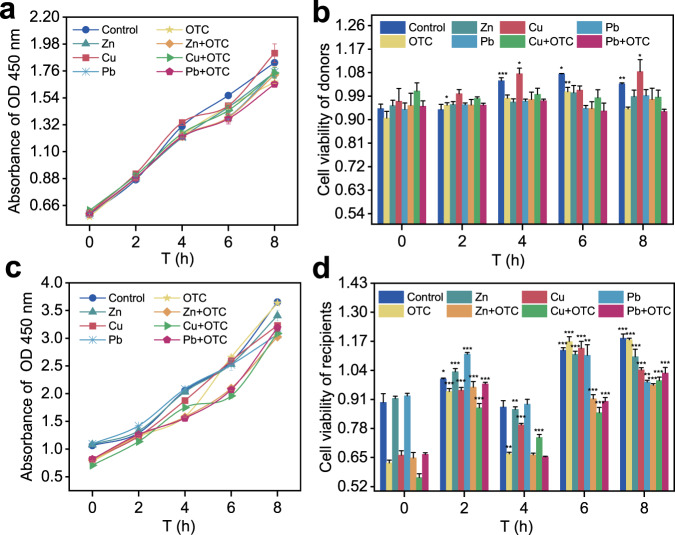
For the donor, the absorbance of OD450 progressively increased in all groups from 0 h to 4 h (Fig. 2a), but there was no significant difference in donor cell viability within the group for most of times (Fig. 2b), indicating that the survival of the donor was not stressor dependent. For the recipient, since the intestinal microbiome was not resistant to OTC and heavy metals, the recipient had some inhibition when exposed to OTC and heavy metals at 0 h, except for the Zn and Pb group (Fig. 2c). However, the absorbance of OD450 of the recipient were higher than those of the donor regardless of the time, indicating that their cellular activity was higher than that of the donor. The horizontal transfer of ARGs between bacteria is affected by cell activity [28]. For cell viability, the cell viability of all groups was significantly correlated for most of the time, except for 4 h (p = 0.00000031–0.042, Fig. 2d). Cell viability recovered after 2 h, with the control, OTC, Zn, Zn + OTC and Pb + OTC groups significantly increasing (p = 0.00000031–0.00069) at all times selected for the assay (except for 4 h), while the Cu and Cu + OTC groups significantly increased at all times (p = 0.0000050–0.00032). This shows that the cell viability of the recipient was more susceptible to the perturbation of OTC and heavy metals. At the 8 h, it was shown that the cell viability of the recipient was lower in all experimental groups than in the control group but generally restored. In addition, the conjugation frequency had a positive correlation with the number of transconjugants (p < 0.05, Fig. S2), while the cell viability rate of recipients had a negative correlation with the number of transconjugants (p < 0.05). Therefore, an increase in the frequency of conjugation may depend on a moderate decrease in the activity of the recipient bacteria at a certain concentration.
Fig. 2. Effects of OTC and heavy metals on the cell viability of donors and recipients.
a Absorbance of OD 450 nm of donors. b Cell viability of donors. c Absorbance of OD 450 nm of recipients. d Cell viability of recipients. Significant differences between individual exposure groups and the control (0 h) were tested with the independent-sample t test: *(p < 0.05), **(p < 0.01) and **(p < 0.001).
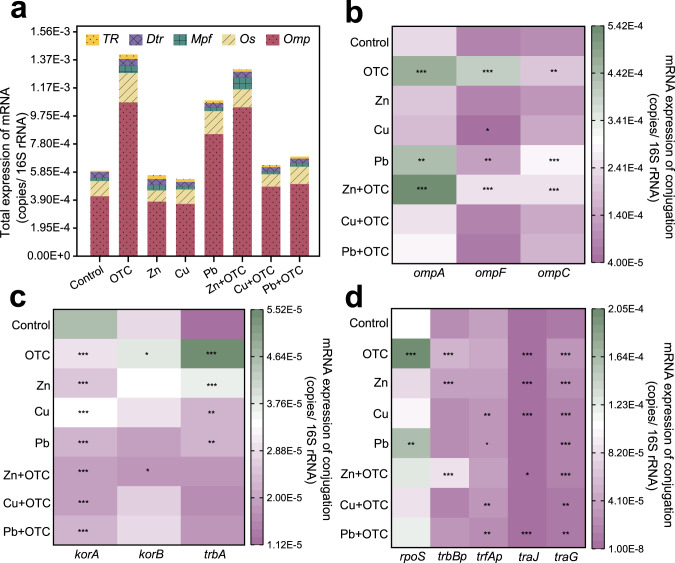
Stressor changed the mRNA expression of related genes in conjugative systems
Assays on the mRNA expression of donors and recipients revealed that that the types and abundance of mRNA expression factors were higher in the donors than in the recipients (Supplementary Fig. S3a). All selected mRNA-related genes were detected in the donor, while trbBp, trfAp, traJ, korB and trbA were not detected in the recipient. The abundances of donors were 18.3797–4659.5069-fold higher than recipients, with the most obvious being korA, ompA, traG and ompF gene. The principal component analysis showed that only Cu had a relatively large effect on multiple resistant donors (Supplementary Fig. S3b), but for potential recipients, all OTC and heavy metals had a perturbing effect on the recipient (Supplementary Fig. S3c). Core genes and proteins related to cell membrane structure and function showed significant changes with exposure to OTC and heavy metal. For example, the abundance of facilitating regulation-related mRNA expression was significantly increased (up to 0.8256–1.3639-fold) in the OTC, Pb and Zn + OTC groups (Fig. 3a). For the conjugative system, OTC and Zn + OTC showed significantly increased expression of ompA, ompF and ompC genes (p < 0.001), with ompF increasing 3.9176-fold and 2.0113-fold, respectively (Fig. 3b). The mRNA expression level of the global control system gene korA was significantly reduced under all stressor (p < 0.001) (Fig. 3c). Compared with the control group, the expression levels of korA were reduced in the Zn + OTC, Cu + OTC and Pb + OTC groups, decreased by 59.1326%, 58.0043%, 51.9323%. For other system genes, OTC significantly increased the trbBp and traJ genes by 1.2892- and 1.6328-fold, respectively (Fig. 3d). However, for ARGs, OTC and heavy metals had a significant promotion effect (p < 0.001) on the horizontal migration of all ARGs (with traG as the indicator). These had the potential to drive the horizontal migration of plasmid-mediated ARGs in the intestinal microbiome.
Fig. 3. Effects of mRNA expression in different exposures.
a Total expression of mRNA. b mRNA expression of conjugation of Omp system genes. c mRNA expression of conjugated GR system genes. d mRNA expression of conjugation about other system genes. Significant differences (c, d) between individual exposure groups and the control were tested with the independent-sample t test: *(p < 0.05), **(p < 0.01) and **(p < 0.001).
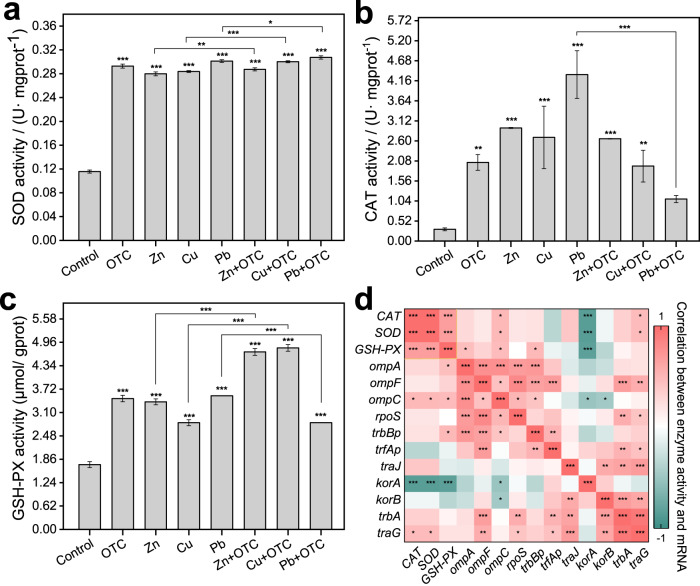
Stressor promoted both ROS-enzyme expression and conjugative transfer efficiency
SOD generation in the conjugations was extremely significantly correlated with the stressor of OTC and heavy metals (p < 0.001) (Fig. 4a) and the SOD levels increased from 0.1156 U· mgprot−1 to 0.2928, 0.2798, 0.2837 and 0.3012 U· mgprot−1 after 8 h of exposure to OTC, Zn, Cu and Pb, respectively. Moreover, no obvious difference was found in SOD generated by the conjugations between the single heavy metal group and its combined stress group with OTC. There was no significant correlation between SOD and frequency in the exposure to stressors alone group (på 0.05, Supplementary Fig. S4). However, a positive significant correlation was observed in the OTC combined with Zn/ Cu group (p < 0.001), suggesting that Zn or Cu combined with OTC could promote HGT by increasing SOD content. Both OTC and heavy metals significantly affected CAT generation in the donor and recipient during conjugation (p < 0.001) (Fig. 4b), but the fold changes in CAT levels caused by OTC and heavy metals ranged from 5.6907-fold to 13.1625-fold and the combined effects of heavy metals and OTC were even less than those of the corresponding heavy metals alone, especially Pb, with the largest reduction of 2.9621-fold. In addition, the CAT generation in the conjugation depended on the presence of the stressor more than the kind of stressor. The stressor likewise increased GSH-PX generation in the conjugation significantly (p < 0.001). GSH-PX generation increased in all stress groups, with 2.0187, 1.9673, 1.6454 and 2.0625-fold in the OTC, Zn, Cu and Pb stress groups compared with the control, respectively (Fig. 4c). Only GSH-PX, both donors and recipients during the conjugation, significantly increased with simultaneous exposure to OTC in the Zn or Cu stress group, with fold increases of 1.7381 and 1.7991-fold, respectively. Therefore, all single stressor could improve the generation of bacterial catabolic enzymes, but the combined stressor effect of antibiotics and heavy metals was different from the enzyme kind. Here, nonspecific effect, cooperative inhibition and stress-specific effect were found for the generation of SOD, CAT and GSH-PX, respectively.
Fig. 4. Effects of antibiotics and heavy metals on reactive oxygen species in mating systems of each group.
a SOD activity. b CAT activity. c GSH-PX activity. d Correlation between enzyme activity and mRNA in conjugations. Significant differences (a–c) between exposure groups and the control group were tested with an independent-sample t test and significant correlations (d) between assay factors were analyzed with Pearson correlation analysis: *(p < 0.05), **(p < 0.01) and ***(p < 0.001).
To further verify whether the production of ROS-related enzyme activity was correlated with mRNA expression, correlation analysis was performed between enzyme activity expression and mRNA expression among all groups. There was a highly significant correlation among CAT, SOD and GSH-PX (p < 0.001) (Fig. 4d). The global control system gene korA had a highly significant negative correlation with all three enzyme activities (p < 0.001). In addition, the outer membrane protein system gene ompC and transfer regulation gene traG were significantly correlated with both CAT and SOD (p < 0.05). GSH-PX also had a significant correlation with ompA, ompC and trbBp (p < 0.05). The stressor-induced increase in ROS-related enzyme activity indicated the enhanced permeability of the cell membrane, as did the increase in the outer membrane protein system genes.
Stressor altered the characteristics of the cell surface
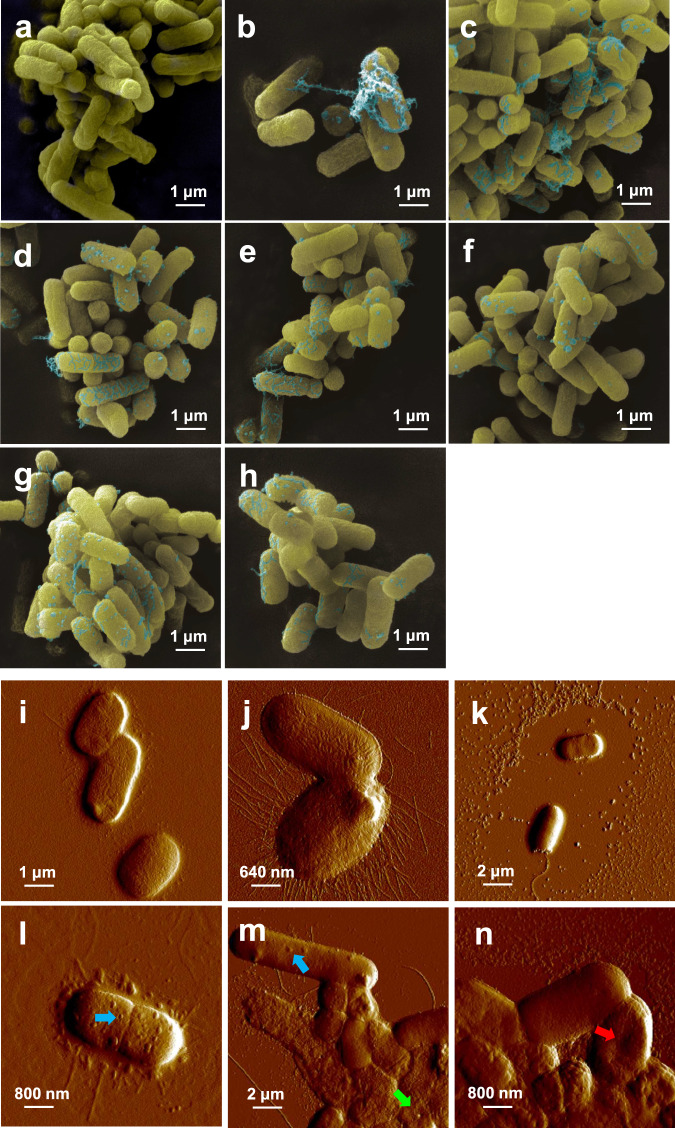
The SEM image of bacteria during the conjugation process showed that the cell membrane surface appeared to be heavily laden with globular and filamentous secretions in the presence of stressor (Fig. 5a–h) and some secretions connected the bacteria to each other, which could potentially increase the conjugation of the donor and recipient. Moreover, AFM was used to further investigate the more subtle changes on the cell membrane surface; the cell surface of both donor and recipient in the absence of stressor was intact, while the donor was smooth with no flagella, as well as a small amount of surrounding secretion (Fig. 5i). The recipient was rougher (Fig. 5j) with abundant ciliary or flagella. In the control group with no stressor, both the donor and recipient cells remained intact, while the secretion on the donors increased (Fig. 5k). Additionally, donor and recipient were covered and drawn close. In contrast, with exposure to OTC and heavy metals, granular protrusions (Fig. 5l), small pores (Fig. 5m), or a state of surface invagination (Fig. 5n) were observed on or in the bacterial cell membrane of all stress groups, which indicated that OTC and heavy metals altered the characteristics of the cell membrane. Thus, the closer cell contacts and membrane damage detected here align with the changes in membrane permeability and the correspondingly higher levels of gene transfer detected in the presence of OTC and heavy metals. These results further explain the conjugative transfer of ARGs enhanced as stressed by OTC, Zn, Cu and Pb.
Fig. 5. Effects of antibiotics and heavy metals on the characteristics of the cell membrane surface in conjugation solution.
a SEM imaging of the control group. b SEM imaging in the OTC group. c SEM imaging in the Zn group. d SEM imaging of the Cu group. e SEM imaging of the Pb group. f SEM imaging of the Zn + OTC group. g SEM imaging of the Cu + OTC group. h SEM imaging of the Pb + OTC group. i Cell membrane of the donor (AFM image). j Cell membrane of the recipient (with abundant fimbriae, AFM image). k Full state of the cell membrane in the control group (AFM image). l Granular protrusions appear on the surface of the cell membrane (blue arrow, AFM image). m Small pores appear on the surface of the cell membrane (green arrow, AFM image). n Invagination of the cell membrane surface (red arrow, AFM image).
Variation in transconjugant resistance with the obtainment of plasmid
The abundance of ARGs in the transconjugants varied more significantly than that in the recipient (Fig. 6a). The abundance of the tetA gene in the recipient increased obviously after receiving the plasmid from the donor. Interestingly, the horizontal migration of ARGs not only transferred their own resistance to the recipient but also contributed to the increase in the abundance of other ARGs, such as sul1 and intI1 genes. It was very clear that no sulfonamide antibiotics were added and the sul1 and intI1 genes were confirmed to be undetected in the plasmid (Fig. S4a, b). For the sul1 gene, expression upregulation was extremely obvious by 1002.9081–2252.7065-fold in the transconjugants and each heavy metal combined with OTC increased the relative abundance of the sul1 gene in both the recipients and transconjugants. Similarly, the relative abundance of intI1 increased more obviously in the transconjugants than in the recipients of all stress groups, which supported the occurrence of ARG HGT by applying more mobile elements. Moreover, the correlation of gene expression in recipients and transconjugants showed that the occurrence of transfer (represented by traG) was significantly correlated (p < 0.05) with the expression of the resistance genes pcoD and sul1, while the pcoD gene was also significantly correlated (p < 0.05) with the outer membrane protein system gene ompF (Fig. 6b, c). All 26 strains isolated by plate streaking were identified as Citrobacter freundii. The bacteria with only green fluorescence were sorted and identified by SCRSS (Fig. 6d, e). The results showed that 45 of the 50 strains were identified as Citrobacter freundii by bacterial identification library. The remaining five unknown strains were identified through sequencing as Aeromonas sp., Geobacillus sp. and Escherichia coli. But only Citrobacter freundii was culturable and the others were non-culturable.
Fig. 6. Transconjugant sorting and ARG analysis.
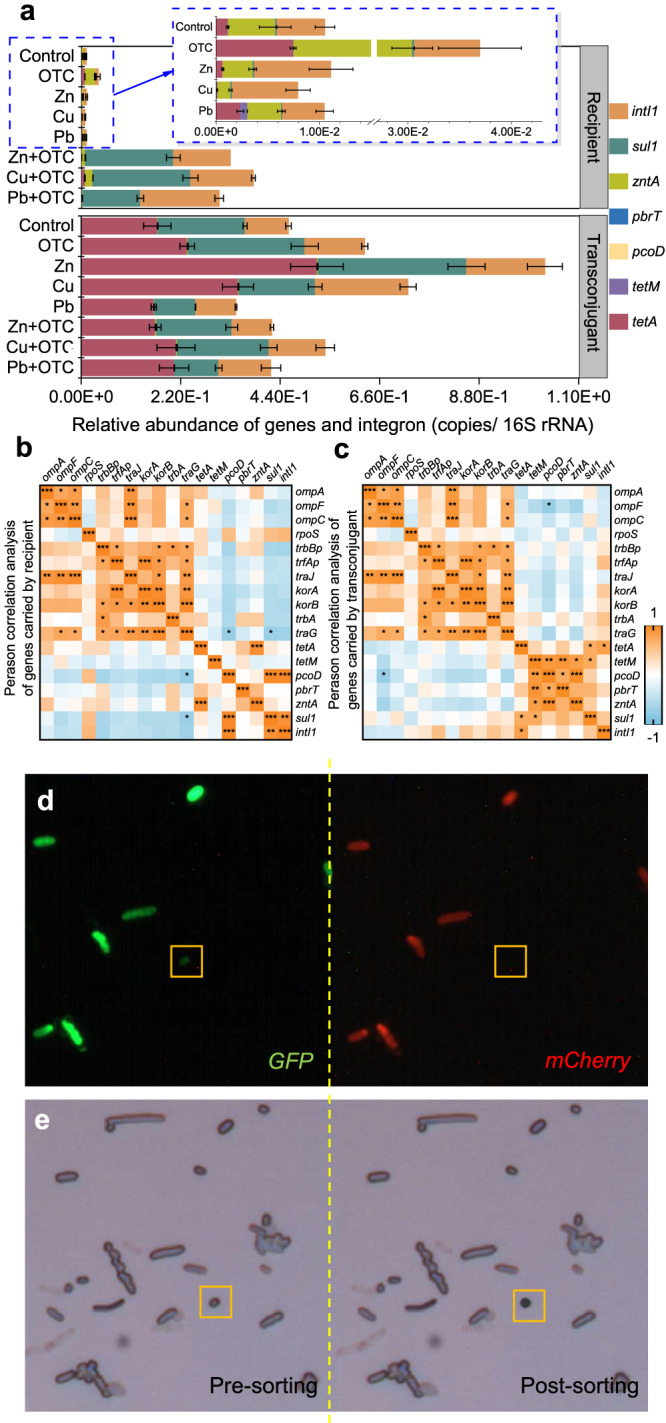
a Relative abundance of genes and integron in transconjugants and recipients. b Correlation analysis of genes carried by recipient. c Correlation analysis of genes carried by transconjugant. d Confocal fluorescence image of microbiota through GFP (green) and mCherry (red) models was collected and imaged. e Bright-field views before and after cell sorting. Significant correlations (b, c) between each gene were analyzed with Pearson correlation analysis: *(p < 0.05), **(p < 0.01) and ***(p < 0.001).
Discussion
The influence of antibiotics and heavy metals on intestinal resistance in aquatic organisms has received increasing attention in recent years [19, 36–38]. The diffusion of resistance genes can also be promoted by heavy metals, which is related to the high bacterial sensitivity to metal pollution [39]. Here, it was found that stressor at low concentrations could promote exogenous ARG transfer to the intestinal microbiome, both the fold changes in transconjugant number and conjugative transfer were enhanced and the horizontal migration potential of the invaded ARGs depended on the activity of the recipients. Considering that low-dose antibiotics could promote the spread of ARGs by pili capture and communication responses [40] and that heavy metals could co-select ARGs in the same plasmid of ARBs [41], the effects caused by these stressor on the transfer of ARGs have warranted more attention. Moreover, conjugative transfer could be affected by the bacterial stressor response and vitality [42]. Since bacterial cells often suffer a fitness cost after conjugative plasmids’ entry, transconjugants replicate slower than plasmid-free cells [43]. DNA entering the recipient as single-stranded DNA during plasmid transfer can activate the SOS response halting cell replication [44, 45]. Moderate inhibition of recipient activity by OTC resulting in decreased amounts may also be an important factor in the increase of conjugative frequency. Although the number of viable donors was nearly stable through the process of conjugation, the recipient number fluctuated, indicating that the nonresistant intestinal microbiome was continuously adapting to the stress environment, as evidenced by time.
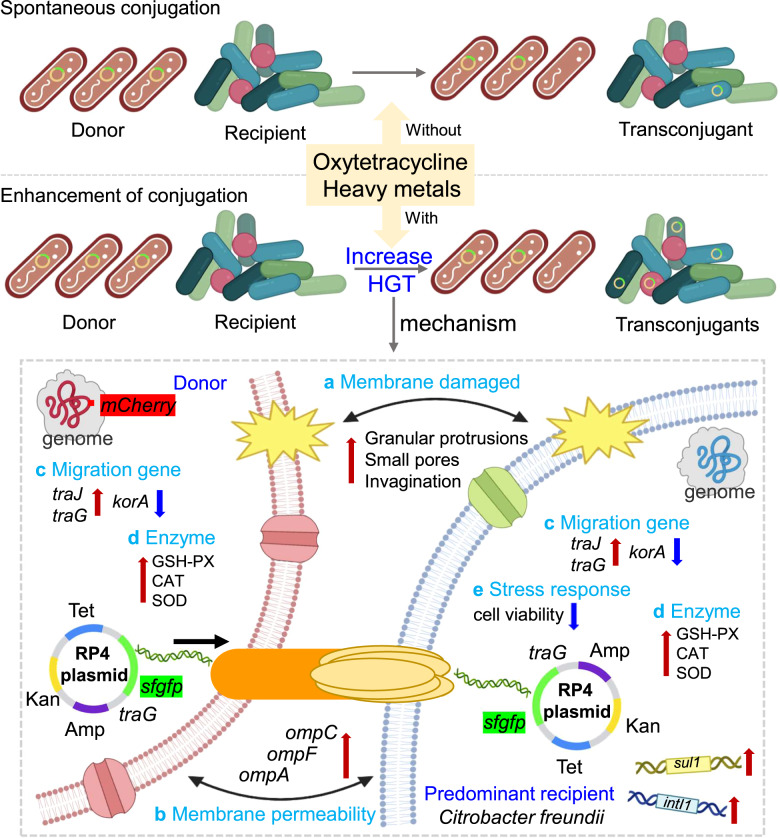
Multiple mechanisms are involved in the HGT of intestinal microbes in the host (Fig. 7), including altered expression of mRNA, ROS enzymes and cell membranes. The outer membrane protein (Omp) carried by IncF plasmid in E. coli K12 and Enterobacteriaceae family has been shown to have mating pair stabilization, which plays important roles in concert with pore-forming proteins and membrane transport, increasing membrane permeability [46–48]. The donor cooperates with different outer membrane receptors in the recipients through TraN to mediate mating pair stabilization and efficient DNA transfer [47]. However, TraN is present in IncF plasmids and not in the RP4 system [47, 49]. The significantly upregulated expression of Omp genes indicates that OTC and heavy metals might regulate IncF plasmids system in bacteria to increase porin expression and pore formation, allowing a donor pilus to attach to and access the recipient, thus facilitating cell-to-cell contact [48, 50], which contributed to the plasmid RP4 transfer. Additionally, upward expression of ompC, traJ, traG and downward expression of korA contributed to the development of HGT. SOD is the primary biological tool that microorganisms use to degrade superoxide within the cell membrane and the environment [51]. SOD activity in microorganisms has a strong correlation with the survival of bacteria in antibiotics and is protective against antibiotic toxicity [52]. Bacteria employed Zn or Cu as the catalytic metal to form SOD to reduce the toxic effects of OTC in the conjugative system [53], helping the plasmid transfer. Moreover, the increased expression levels of enzymes CAT and GSH-PX promoted the resistance of intestinal bacteria to the damage caused by harmful metal ions and then effectively protected the donors and acceptors from ROS damage, thus enhancing the uptake potential of conjugal plasmids [29]. After exposure to OTC and heavy metals, the cell membranes of bacteria in all conjugations showed different degrees of changes, with most of them showing small pores on the surface, which might be related to the migration of plasmids into and out of the cell through its membrane, facilitating plasmid DNA passing through the bacterial membrane and driving the resistance genes spread from cell to cell among bacteria [27].
Fig. 7. A model depicting the mechanisms underlying the RP4 plasmid-mediated conjugation promoted by OTC and heavy metals.
a Membrane damaged. b Improved outer membrane protein gene. c Migration gene regulation. d Increased intracellular ROS-enzyme production. e Inhibited recipient activity.
Citrobacter freundii was one of the predominant recipients (i.e., ARB) in the intestine, which could be isolated successfully in all groups. In terms of environmental risks arising from ARGs, ARB was at greater risk than recipient (i.e., non-ARB) owing to the variety and abundance of ARGs, in which sul1 and intI1 were the most sensitive genes under the combined effect of antibiotics and heavy metals. Bacteria stimulated by heavy metals were more likely to survive in an antibiotic environment [54]. Heavy metals and antibiotics showed a synergistic effect on intestinal ARGs and the coregulated resistance induced by heavy metals might aggravate the enrichment of ARGs [55]. However, the recipients in this study did not have target resistance, thus suggesting that Citrobacter freundii in the intestine is susceptible to environmental ARBs, antibiotics and heavy metals and easily develops into multidrug-resistant bacteria. It has been proven that when bacteria adapt to one plasmid, they become generally permissive to plasmid carriage [56]. In addition, there was an abundance of nonculturable bacteria in the intestinal microbiome, of which Aeromonas could also be dominant as transconjugants. Aeromonas in wildlife has the potential to be an indicator of antimicrobial resistance in aquatic environments [57]. Antibiotic stressor transmitted ARGs between bacteria via plasmid conjugation, which finally contributed to the spread of ARGs (e.g., tetC) in the aqueous environment [58], but the identification of susceptible bacterial pairs will help to explore further mechanisms and block technology for ARG conjugation.
In this study, in vitro conjugative systems were established at a laboratory scale, which cannot accurately reflect the real effects caused in vivo. Therefore, future studies on animals in vivo in real environments should be conducted to evaluate the risk of horizontal transfer of ARBs and ARGs in aquatic organisms under stressor (e.g., heavy metals and antibiotics). Considering the colonization of ARBs in vivo, the distribution of ARBs in the intestine also needs to be assessed, as well as the ecological analysis of HGT from in vitro to in vivo.
Supplementary information
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 42277260, 41977340) and the Guangdong Basic and Applied Basic Research Foundation (No. 2019A1515110937). The authors acknowledge the help of the Guangdong University of Technology analysis and test center.
Author contributions
YX conceived and supervised the project. XL conceived and designed the experiments. XL, CZ, RH, SL, HP, XZ and LH performed the experiments. XL analyzed data and wrote the manuscript.
Data availability
All data was available in the main text or Supplementary Information.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01514-w.
References
- 1.Sanchez-Cid C, Guironnet A, Keuschnig C, Wiest L, Vulliet E, Vogel TM. Gentamicin at sub-inhibitory concentrations selects for antibiotic resistance in the environment. ISME Commun. 2022;2:29. doi: 10.1038/s43705-022-00101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu YG, Zhao Y, Li B, Huang CL, Zhang SY, Yu S, et al. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat Microbiol. 2017;2:16270. doi: 10.1038/nmicrobiol.2016.270. [DOI] [PubMed] [Google Scholar]
- 3.Yao Y, Maddamsetti R, Weiss A, Ha Y, Wang T, Wang S, et al. Intra- and interpopulation transposition of mobile genetic elements driven by antibiotic selection. Nat Ecol Evol. 2022;6:555. doi: 10.1038/s41559-022-01705-2. [DOI] [PubMed] [Google Scholar]
- 4.Aminov RI. Horizontal gene exchange in environmental microbiota. Front Microbiol. 2011;2:158. doi: 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huddleston JR. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist. 2014;7:167. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lerminiaux NA, Cameron A. Horizontal transfer of antibiotic resistance genes in clinical environments. Can J Microbiol. 2019;65:34. doi: 10.1139/cjm-2018-0275. [DOI] [PubMed] [Google Scholar]
- 7.Yu Z, Wang Y, Lu J, Bond PL, Guo J. Nonnutritive sweeteners can promote the dissemination of antibiotic resistance through conjugative gene transfer. ISME J. 2021;15:2117. doi: 10.1038/s41396-021-00909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding C, Yang D, Ma J, Jin M, Shen Z, Shi D, et al. Effects of free antibiotic resistance genes in the environment on intestinal microecology of mice. Ecotoxicol Environ Saf. 2020;204:111119. doi: 10.1016/j.ecoenv.2020.111119. [DOI] [PubMed] [Google Scholar]
- 9.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar RD. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 11.Anthony WE, Burnham CD, Dantas G, Kwon JH. The gut microbiome as a reservoir for antimicrobial resistance. J Infect Dis. 2021;223:S209. doi: 10.1093/infdis/jiaa497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacoby GA, Griffin CM, Hooper DC. Citrobacter spp. as a source of qnrB alleles. Antimicrob Agents Chemother. 2011;55:4979. doi: 10.1128/AAC.05187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson D, Flach CF. Antibiotic resistance in the environment. Nat Rev Microbiol. 2022;20:257. doi: 10.1038/s41579-021-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pal C, Asiani K, Arya S, Rensing C, Stekel DJ, Larsson D, et al. Metal resistance and its association with antibiotic resistance. Adv Micro Physiol. 2017;70:261. doi: 10.1016/bs.ampbs.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Wales AD, Davies RH. Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics (Basel) 2015;4:567. doi: 10.3390/antibiotics4040567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Y, Doijad S, Falgenhauer J, Schmiedel J, Imirzalioglu C, Chakraborty T. Co-occurrence of dual carbapenemases KPC-2 and OXA-48 with the mobile colistin resistance gene mcr-9.1 in Enterobacter xiangfangensis. Front Cell Infect Microbiol. 2022;12:960892. doi: 10.3389/fcimb.2022.960892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu J, Yang D, Jin M, Liu W, Zhao X, Li C, et al. Aquatic animals promote antibiotic resistance gene dissemination in water via conjugation: role of different regions within the zebra fish intestinal tract, and impact on fish intestinal microbiota. Mol Ecol. 2017;26:5318. doi: 10.1111/mec.14255. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Li B, Wu B, Lin H, Jiang L, Qiu Y. How heavy metal stress promotes dissemination of antibiotic resistance genes in the activated sludge process. J Hazard Mater. 2022;437:129279. doi: 10.1016/j.jhazmat.2022.129279. [DOI] [PubMed] [Google Scholar]
- 19.Obayashi Y, Kadoya A, Kataoka N, Kanda K, Bak SM, Iwata H, et al. Tetracycline resistance gene profiles in red seabream (Pagrus major) intestine and rearing water after oxytetracycline administration. Front Microbiol. 2020;11:1764. doi: 10.3389/fmicb.2020.01764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YL, Li J, Hu Z, Li JL, Lu H. Oxytetracycline stress stimulates antibiotic resistance gene proliferation and quorum sensing response of marine anammox bacteria in seawater-based wastewater treatment. Chem Eng J. 2022;447:137539.
- 21.Komijani M, Shamabadi NS, Shahin K, Eghbalpour F, Tahsili MR, Bahram M. Heavy metal pollution promotes antibiotic resistance potential in the aquatic environment. Environ Pollut. 2021;274:116569. doi: 10.1016/j.envpol.2021.116569. [DOI] [PubMed] [Google Scholar]
- 22.Gupta S, Graham DW, Sreekrishnan TR, Ahammad SZ. Effects of heavy metals pollution on the co-selection of metal and antibiotic resistance in urban rivers in UK and India. Environ Pollut. 2022;306:119326. doi: 10.1016/j.envpol.2022.119326. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Q, Wang MZ, Zhong XX, Liu P, Xie XY, Wangxiao JY, et al. Dissemination of resistance genes in duck/fish polyculture ponds in Guangdong Province: correlations between Cu and Zn and antibiotic resistance genes. Environ Sci Pollut Res. 2019;26:8182. doi: 10.1007/s11356-018-04065-2. [DOI] [PubMed] [Google Scholar]
- 24.Komijani M, Eghbalpour F, Lari E, Shaykh-Baygloo N. Developing erythromycin resistance gene by heavy metals, Pb, Zn, and Co, in aquatic ecosystems. Sci Rep. 2022;12:20797. doi: 10.1038/s41598-022-25272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su HC, Liu S, Hu XJ, Xu XR, Xu WJ, Xu Y, et al. Occurrence and temporal variation of antibiotic resistance genes (ARGs) in shrimp aquaculture: ARGs dissemination from farming source to reared organisms. Sci Total Environ. 2017;607:357. doi: 10.1016/j.scitotenv.2017.07.040. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Gu AZ, He M, Li D, Chen J. Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ Sci Technol. 2017;51:570. doi: 10.1021/acs.est.6b03132. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q, Liu L, Hou Z, Wang L, Ma D, Yang G, et al. Heavy metal copper accelerates the conjugative transfer of antibiotic resistance genes in freshwater microcosms. Sci Total Environ. 2020;717:137055. doi: 10.1016/j.scitotenv.2020.137055. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Zhang G. Detection and various environmental factors of antibiotic resistance gene horizontal transfer. Environ Res. 2022;212:113267. doi: 10.1016/j.envres.2022.113267. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Lu J, Zhang S, Li J, Mao L, Yuan Z, et al. Non-antibiotic pharmaceuticals promote the transmission of multidrug resistance plasmids through intra- and intergenera conjugation. ISME J. 2021;15:2493. doi: 10.1038/s41396-021-00945-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Q, Wu H, Cai P, Fein JB, Chen W. Atomic force microscopy measurements of bacterial adhesion and biofilm formation onto clay-sized particles. Sci Rep. 2015;5:16857. doi: 10.1038/srep16857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Liu Q, Qi X, Li Y, Zhou G, Dai M, et al. Evolution and resistance of a microbial community exposed to Pb(II) wastewater. Sci Total Environ. 2019;694:133722. doi: 10.1016/j.scitotenv.2019.133722. [DOI] [PubMed] [Google Scholar]
- 32.Yu Z, Rabiee H, Guo J. Synergistic effect of sulfidated nano zerovalent iron and persulfate on inactivating antibiotic resistant bacteria and antibiotic resistance genes. Water Res. 2021;198:117141. doi: 10.1016/j.watres.2021.117141. [DOI] [PubMed] [Google Scholar]
- 33.Di Cesare A, Eckert EM, D’Urso S, Bertoni R, Gillan DC, Wattiez R, et al. Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res. 2016;94:208. doi: 10.1016/j.watres.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 34.Lin H, Sun WC, Jin DF, Yu QG, Yang YY, Zhang ZL, et al. Effect of composting on the conjugative transmission of sulfonamide resistance and sulfonamide-resistant bacterial population. J Clean Prod. 2021;285:125483. [Google Scholar]
- 35.Lin X, Xu Y, Han R, Luo W, Zheng L. Migration of antibiotic resistance genes and evolution of flora structure in the Xenopus tropicalis intestinal tract with combined exposure to roxithromycin and oxytetracycline. Sci Total Environ. 2022;820:153176. doi: 10.1016/j.scitotenv.2022.153176. [DOI] [PubMed] [Google Scholar]
- 36.Sun S, Korheina D, Fu H, Ge X. Chronic exposure to dietary antibiotics affects intestinal health and antibiotic resistance gene abundance in oriental river prawn (Macrobrachium nipponense), and provokes human health risk. Sci Total Environ. 2020;720:137478. doi: 10.1016/j.scitotenv.2020.137478. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, Lu G, Sun Y, Yan Z, Dang T, Liu J. Metagenomic analysis explores the interaction of aged microplastics and roxithromycin on gut microbiota and antibiotic resistance genes of Carassius auratus. J Hazard Mater. 2022;425:127773. doi: 10.1016/j.jhazmat.2021.127773. [DOI] [PubMed] [Google Scholar]
- 38.Zhou B, Wang C, Zhao Q, Wang Y, Huo M, Wang J, et al. Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms. J Hazard Mater. 2016;320:10. doi: 10.1016/j.jhazmat.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Wu T, Zhang Y, Wang B, Chen C, Cheng Z, Li Y, et al. Antibiotic resistance genes in Chishui River, a tributary of the Yangtze River, China: Occurrence, seasonal variation and its relationships with antibiotics, heavy metals and microbial communities. Sci Total Environ. 2022;846:157472. doi: 10.1016/j.scitotenv.2022.157472. [DOI] [PubMed] [Google Scholar]
- 40.Tang T, Chen Y, Du Y, Yao B, Liu M. Effects of functional modules and bacterial clusters response on transmission performance of antibiotic resistance genes under antibiotic stress during anaerobic digestion of livestock wastewater. J Hazard Mater. 2023;441:129870. doi: 10.1016/j.jhazmat.2022.129870. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y, Xu J, Mao D, Luo Y. Effect of the selective pressure of sub-lethal level of heavy metals on the fate and distribution of ARGs in the catchment scale. Environ Pollut. 2017;220:900. doi: 10.1016/j.envpol.2016.10.074. [DOI] [PubMed] [Google Scholar]
- 42.Ji H, Cai Y, Wang Z, Li G, An T. Sub-lethal photocatalysis promotes horizontal transfer of antibiotic resistance genes by conjugation and transformability. Water Res. 2022;221:118808. doi: 10.1016/j.watres.2022.118808. [DOI] [PubMed] [Google Scholar]
- 43.Rebelo JS, Domingues C, Nogueira T, Dionisio F. Plasmids increase the competitive ability of plasmid-bearing cells even when transconjugants are poor donors, as shown by computer simulations. Microorganisms. 2023;11:1238. doi: 10.3390/microorganisms11051238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bojer MS, Frees D, Ingmer H. SosA in Staphylococci: an addition to the paradigm of membrane-localized, SOS-induced cell division inhibition in bacteria. Curr Genet. 2020;66:495. doi: 10.1007/s00294-019-01052-z. [DOI] [PubMed] [Google Scholar]
- 45.Jones C, Holland IB. Role of the SulB (FtsZ) protein in division inhibition during the SOS response in Escherichia coli: FtsZ stabilizes the inhibitor SulA in maxicells. Proc Natl Acad Sci USA. 1985;82:6045. doi: 10.1073/pnas.82.18.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Mao D, Luo Y. Ionic liquid facilitates the conjugative transfer of antibiotic resistance genes mediated by plasmid RP4. Environ Sci Technol. 2015;49:8731. doi: 10.1021/acs.est.5b01129. [DOI] [PubMed] [Google Scholar]
- 47.Low WW, Wong J, Beltran LC, Seddon C, David S, Kwong HS, et al. Mating pair stabilization mediates bacterial conjugation species specificity. Nat Microbiol. 2022;7:1016. doi: 10.1038/s41564-022-01146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Achouak W, Heulin T, Pages JM. Multiple facets of bacterial porins. Fems Microbiol Lett. 2001;199:1. doi: 10.1111/j.1574-6968.2001.tb10642.x. [DOI] [PubMed] [Google Scholar]
- 49.Samuels AL, Lanka E, Davies JE. Conjugative junctions in RP4-mediated mating of Escherichia coli. J Bacteriol. 2000;182:2709. doi: 10.1128/jb.182.10.2709-2715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Achtman M, Morelli G, Schwuchow S. Cell-cell interactions in conjugating Escherichia coli: role of F pili and fate of mating aggregates. J Bacteriol. 1978;135:1053. doi: 10.1128/jb.135.3.1053-1061.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sutherland KM, Ward LM, Colombero CR, Johnston DT. Inter-domain horizontal gene transfer of nickel-binding superoxide dismutase. Geobiology. 2021;19:450. doi: 10.1111/gbi.12448. [DOI] [PubMed] [Google Scholar]
- 52.Martins D, McKay G, Sampathkumar G, Khakimova M, English AM, Nguyen D. Superoxide dismutase activity confers (p)ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2018;115:9797. doi: 10.1073/pnas.1804525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lynch M, Kuramitsu H. Expression and role of superoxide dismutases (SOD) in pathogenic bacteria. Microbes Infect. 2000;2:1245. doi: 10.1016/s1286-4579(00)01278-8. [DOI] [PubMed] [Google Scholar]
- 54.Reddy S, Kaur K, Barathe P, Shriram V, Govarthanan M, Kumar V. Antimicrobial resistance in urban river ecosystems. Microbiol Res. 2022;263:127135. doi: 10.1016/j.micres.2022.127135. [DOI] [PubMed] [Google Scholar]
- 55.Ding J, An XL, Lassen SB, Wang HT, Zhu D, Ke X. Heavy metal-induced co-selection of antibiotic resistance genes in the gut microbiota of collembolans. Sci Total Environ. 2019;683:210. doi: 10.1016/j.scitotenv.2019.05.302. [DOI] [PubMed] [Google Scholar]
- 56.Loftie-Eaton W, Bashford K, Quinn H, Dong K, Millstein J, Hunter S, et al. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat Ecol Evol. 2017;1:1354. doi: 10.1038/s41559-017-0243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grilo ML, Sousa-Santos C, Robalo J, Oliveira M. The potential of Aeromonas spp. from wildlife as antimicrobial resistance indicators in aquatic environments. Ecol Indic. 2020;115:106396. [Google Scholar]
- 58.Shi Y, Zhang Y, Wu X, Zhang H, Yang M, Tian Z. Potential dissemination mechanism of the tetC gene in Aeromonas media from the aerobic biofilm reactor under oxytetracycline stresses. J Environ Sci. 2021;105:90. doi: 10.1016/j.jes.2020.12.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data was available in the main text or Supplementary Information.