Summary
Ongoing HIV transmission is a public health priority in Indonesia. We developed a new multiassay algorithm (MAA) to identify recent HIV infection. The MAA is a sequential decision tree based on multiple biomarkers, starting with CD4+ T cells >200/μL, followed by plasma viral load (pVL) > 1,000 copies/ml, avidity index (AI) < 0 · 7, and pol ambiguity <0 · 47%. Plasma from 140 HIV-infected adults from 19 hospitals across Indonesia (January 2018 – June 2020) was studied, consisting of a training set (N = 60) of longstanding infection (>12-month) and a test set (N = 80) of newly diagnosed (≤1-month) antiretroviral (ARV) drug naive individuals. Ten of eighty (12 · 5%) newly diagnosed individuals were classified as recent infections. Drug resistance mutations (DRMs) against reverse transcriptase inhibitors were identified in two individuals: one infected with HIV subtype C (K219Q, V179T) and the other with CRF01_AE (V179D). Ongoing HIV transmission, including infections with DRMs, is substantial in Indonesia.
Subject areas: Health sciences, Medicine, Computer science
Graphical abstract

Highlights
-
•
First report of a feasible, scalable assay to identify recent HIV infection in Indonesia
-
•
The assay also identifies transmitted drug resistance mutations in recent HIV infections
-
•
The assay identified 10/80 (12.5%) newly diagnosed individuals as recent HIV infections
-
•
2/10 (20%) of recent HIV infections contained transmitted drug resistance mutations
Health sciences; Medicine; Computer science
Introduction
The HIV epidemic is an urgent public health challenge in Indonesia. Overall HIV prevalence is estimated at 0 · 4% (560,000) and may exceed 17 · 9% in key populations, with approximately 27,000 individuals diagnosed with HIV infection in 2021.1 The accuracy of these estimates is uncertain, however, as only an estimated 66% individuals know their diagnosis and there are disparities of surveillance across the >15,000 islands in the country.1 Even less is known regarding HIV incidence, particularly the proportion of recently infected individuals (<12-month after seroconversion) among the newly diagnosed people living with HIV (PLWH). Reliable methods are essential to determine the proportion of recent HIV infection and identify other factors associated with ongoing events in HIV epidemic, including transmitted drug resistance mutations (tDRMs).
The “gold standard” method to identify recent HIV infection consists of measuring seroconversion rates in prospective longitudinal studies of seronegative individuals.2 The “gold standard” method provides accurate measures of recent infection, but is impractical in large populations, especially those spread across large geographic areas. Results comparable to the “gold standard” approach have been obtained in cross-sectional studies using multiassay algorithms (MAAs), which combine measurements of HIV biomarkers to differentiate between recent and longstanding infections. MAAs have demonstrated to be feasible alternatives to conducting a prospective longitudinal cohort study and are used by various countries to estimate incidence.3,4,5,6,7,8
The backbone of many MAAs is a high throughput serology assay to measure antibody (Ab) avidity for HIV antigens (Ags), typically adapting commercial enzymatic immunoassay (EIA) kits to quantify the strength of Ag – Ab binding (avidity) over time after seroconversion.6,8,9 Less mature antibodies found during the early period of infection are generally less avid and are more prone to dissociate in stringent buffer conditions. Recent infection is estimated using avidity index (AI) value, which is the ratio of Ag – Ab binding in stringent and non-stringent buffer conditions.4,6,9 Compared to other recency assays based on Ab titer or specific IgG proportion, avidity-based assays are less affected by dysfunctional immune responses due to advanced stage of disease or consumption of ARV drugs, leading to the lowest false recent rate (FRR).10 The threshold of an AI assay is ideally chosen for the most accurate classification of recent infection in an evaluation panel by minimizing the FRR; i.e., an avidity assay that can yield FRR <2%,2,11 as well as reducing misclassification of recent infections as longstanding ones (false long term rate; FLTR).9
To improve estimation of recent HIV infection in an MAA, additional tests with other biomarkers are used to minimize the FRR and FLTR. A notable biomarker that can distinguish between stages of HIV infection is the proportion of mixed, or ambiguous, bases in the viral genome from population-based DNA sequencing. In general, infection with HIV proceeds with the expansion of a single or very few infecting virions; yielding a genetically uniform population.12 Over time after infection, HIV populations diversify by accumulating mutations in the HIV genome. Electropherograms of population-based sequencing from recently infected individuals contain few or no mixed bases, while those containing more mixed bases are likely to be from long-standing infection.12,13 Quantification of ambiguous bases has successfully identified recent HIV infection, including in CRF01_AE, with proportion of ambiguous bases 0 · 45–0 · 5% representing limited variability in the pol gene of HIV from infection within 12 months12,13,14
In addition to Ab avidity and sequence ambiguity, other biomarkers of HIV infection are useful to prevent mis-classification of recent infection. CD4+ T cells and plasma viral load (pVL) are the most practical and widely used biomarkers for routine monitoring of disease progression. For the use in MAA, CD4+ T cell count <200 cells/μL and pVL <400 or <1,000 copies/ml suggest immunocompromised and virally suppressed stages of chronic infection among treated individuals.2,3,15,16,17,18,19
Minimizing the FRR and FLTR by employing multiple biomarkers of HIV infection in an MAA increases the chance to identify recent HIV infection more accurately. The ultimate evidence of recency remains seroconversion date and in circumstances where documented seroconversion is not available, assessment of medical records and clinical manifestation at the time samples are collected can avoid misclassification.20,21
Taken together, biomarkers of HIV infection have been used to estimate the proportion of recent HIV infection in various countries. Here, we describe a sequential decision tree MAA to identify recent HIV infection among newly diagnosed ARV naive PLWH in Indonesia. The MAA combines Ab avidity, ambiguity of HIV pol DNA sequence, CD4+ T cell count, and pVL; and was employed to provide the first survey of recent HIV infection in Indonesia.
Results
Study participants
From 4,329 “INA – PROACTIVE” participants (Figure S1), we included 60 individuals with longstanding infection and 80 newly diagnosed individuals (Figure 1, Table 1). Newly diagnosed individuals were all ARV-naïve, younger, and more frequently male with HIV risk factor MSM compared to the long-standing group (Table 1). Most individuals with long-standing infection (55/60) were undergoing antiretroviral therapy (ART), but the majority were not virologically suppressed (<1,000 copies/ml) at the time of sampling. CD4+ T cell numbers were comparable and pVL differs significantly in the two groups.
Figure 1.
Flowchart of participant and specimen selection
Long-standing individuals made a training set to develop the serology AI EIA that has never been applied to the Indonesian population. Newly diagnosed individuals were the testing set of this study, where recent infection was identified using MAA.
Table 1.
Characteristics of study participants
| Long-standing |
Newly diagnosed |
p | |
|---|---|---|---|
| (N = 60) | (N = 80) | ||
| Sex at birtha | <0 · 01 | ||
| Female | 23 (38 · 3%) | 14 (17 · 5%) | |
| Male | 37 (61 · 7%) | 66 (82 · 5%) | |
| Age, year, median (IQR) | 33 (29, 45) | 28 (25, 33) | <0 · 01 |
| Risk factorsa | <0 · 01 | ||
| Unprotected sex | 40 (66 · 67%) | 32 (40%) | |
| MSM | 17 (28 · 33%) | 44 (55%) | |
| Other | 3 (5%) | 4 (5%) | |
| Diagnosis, months, median (IQR) | 32 (22, 48) | 0 · 2 (0 · 025, 0 · 5) | <0 · 01 |
| ARTa | <0 · 01 | ||
| Yes | 55 (91 · 67%) | 0 (0%) | |
| No | 5 (8 · 33%) | 80 (100%) | |
| ART, months, median (IQR) | 31 (16, 43) | (-0 · 3) ((-0 · 58), (−0 · 10)) | <0 · 01 |
| CD4+ T cell count (cells/μL), median (IQR) | 141 (67, 228) | 181 (51, 314) | 0 · 35 |
| CD4+ T cell count (cells/μL), distribution | |||
| >500 | 1 (1 · 67%) | 7 (8 · 75%) | |
| 350–499 | 6 (10%) | 9 (11 · 25%) | |
| 200–349 | 14 (23 · 33%) | 19 (23 · 75%) | |
| <200 | 39 (65%) | 45 (56 · 25%) | |
| log10 pVL (copies/ml), median (IQR) | 4 · 9 (3 · 7–5 · 4) | 5 · 5 (4 · 8–5 · 8) | <0 · 01 |
| pVL (copies/ml) distribution | |||
| >1,000,000 | 1 (1 · 67%) | 17 (21 · 25%) | |
| >100,000–1,000,000 | 22 (36 · 67%) | 38 (47 · 5%) | |
| >10,000–100,000 | 19 (31 · 67%) | 20 (25%) | |
| >1,000–10,000 | 4 (6 · 67%) | 4 (5%) | |
| ≤1,000 | 14 (23 · 33%) | 1 (1 · 25%) |
Minus values represent timepoint after blood drawn.
MSM: men having sex with men; ART: antiretroviral drug therapy; pVL: plasma viral load; IQR: interquartile range.
Categorical variables.
Avidity index enzymatic immunoassay (AI EIA)
The distribution of AI values in chronically infected individuals (long-standing group) was left (negative) skewed (Shapiro-Wilk p < 0 · 001, W = 0 · 60), with median (Q1, Q3) = 0 · 98 (0 · 90, 0 · 99) (Table S5). As expected, the majority (75%) of samples had high AI values (>0 · 9, Figure 2). This is consistent with observations of increased avidity along with disease progression. Previous AI EIA studies of HIV-1 subtype B and other subtypes have used an AI cut-off of 0 · 7 to discriminate recent from long-standing infection. Seven long-standing infections had AI <0 · 7 in this study. As such, the FRR of the 2-well AI EIA was 11 · 7% (7/60). Among newly diagnosed group (N = 80), AI values were also left skewed (Shapiro-Wilk p < 0 · 001, W = 0 · 79), with median (IQR) = 0 · 92 (0 · 55, 0 · 98) (Table S6). The left skewness of AI, with most individuals had AI ≥0 · 7, implies a large proportion of newly diagnosed individuals had long-standing infection. Triplicate measure revealed the assay was reproducible (Tables S5 and S6). We observed <10% coefficient of variation (CoV) of OD for each sample, indicating that differences in AI among all individuals measured in this study were due to variation in immune response and not technical errors.
Figure 2.
Distribution of AI values in long-standing and newly diagnosed individuals
In individuals with long-standing infection (N = 60), 53 (88 · 33%) have AI ≥0 · 7, and were classified as not recent, while seven (11 · 67%) have AI <0 · 7 that is the characteristics of recent infection. Among newly diagnosed individuals (N = 80), 51 (63 · 75%) have AI ≥0 · 7, suggesting long-standing infection that was just recently diagnosed, while 29 (36 · 25%) have AI <0 · 7. The newly diagnosed individuals having AI <0 · 7 were classified as potentially recent and subjected to ambiguity analysis.
HIV pol DNA sequence ambiguity
The order of screening in this MAA was designed to reduce FLTR early in the sequence of the algorithm and to optimize overall resource allocation. Decreases in CD4+ T cells to ≤200 cells/μl within the first year of HIV infection are rare, yielding a low FLTR (estimated at 0.2%)22,23,24,25,26,27,28,29; as such, the CD4+ T cell count was placed first in the MAA sequence. pVL >1,000 copies/ml excludes individuals undergoing antiretroviral therapy,30 and was ordered second. AI was positioned third; as such, HIV sequence analysis to determine nucleotide ambiguity of HIV-1 pol sequence was used to resolve whether newly diagnosed individuals having AI <0 · 7 (N = 28) were recent infection (Figure 3). HIV plasma RNA from 27 individuals were successfully sequenced; HIV from one subject (2–1507, pVL 3,290 copies/ml), did not successfully amplify. The proportion of ambiguous bases in the HIV pol sequence ranged 0 · 00–0 · 58%. Ambiguity <0 · 47% was observed in 23 individuals. Four individuals had discordant results between serology and molecular assays (AI <0 · 7 characterizing recent infection and HIV pol ambiguity ≥0 · 47% characterizing chronic infection). Detailed outcomes on HIV pol sequence are presented in Table S7.
Figure 3.
DNA sequencing strategy of the HIV pol gene
The sequencing strategy covers the HIV-1 pol gene that consists of protease (PR) amino acids 1–99, reverse transcriptase (RT) amino acids 1–560, and integrase (IN) amino acids 1–288. NFLG cDNA was synthesized in two separate reactions, each using the GSPs Pan-HIV 1F or Pan-HIV 4R. Pre-sequencing amplification was done using both cDNAs as templates in nested amplification. The target region was initially amplified in two large fragments, using primer set 1 that covers HXB2 1031–5308 (4278-bp) and primer set 2 that covers HXB2 2030–4604 (2575-bp). Amplicon 1 was re-amplified into 4 overlapping fragments: F0 (1031–2682), F1 (2030–3817), F2 (2519–4604), and F3 (2984–5308) whereas amplicon 2 was re-amplified into 3 overlapping fragments: FA (2519–3817), FB (2030–3410), and FC (2984–4604). All fragments were sequenced using several overlapping primers, 8 primers each for F0 – F3 and 6 primers each for FA – FC (Table S3).
Multiassay algorithm (MAA) to identify recently infected individuals
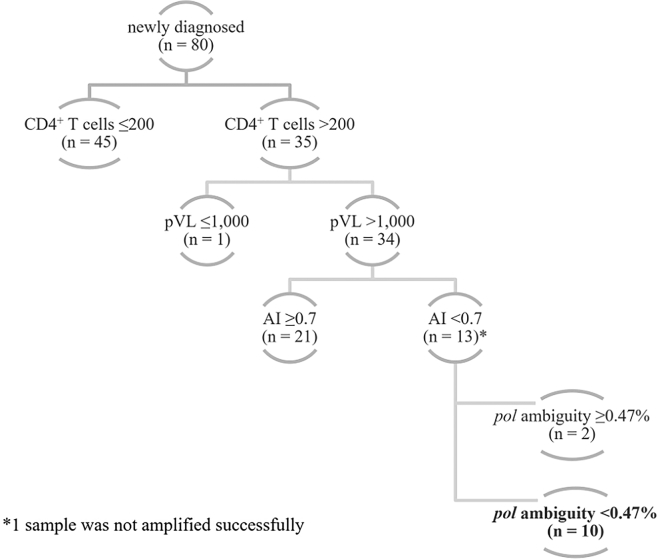
The outcomes of serology AI EIA and molecular HIV pol ambiguity were combined with CD4+ T cell count and pVL to build a stepwise multiassay decision tree (Figure 4). The algorithm screens first for CD4+ T cells >200 per μL, followed by pVL >1,000 copies/ml, AI <0 · 7 and HIV pol ambiguity <0 · 47%. This approach resulted in 10/80 (12 · 5%) newly diagnosed individuals presenting concordant immunological and virological evidence of recent HIV infection, or seroconversion within 12 months.
Figure 4.
MAA classifying recent infection
The MAA considers recently infected individuals in a hierarchical selection starting from CD4+ T cells >200 per μL, pVL >1,000 copies/ml, AI <0 · 7, and HIV pol ambiguity <0 · 47%. In 80 newly diagnosed, ARV naive, adult PLWH, 45 (56 · 25%) had CD4+ T cells ≤200 per μL and 35 (43 · 75%) had CD4+ T cells >200 per μL. Almost all individuals with CD4+ T cells >200 per μL had pVL >1,000 copies/ml (N = 34). Further differentiation based on the AI values resulted in a larger proportion had the long-standing AI ≥0 · 7 (N = 21). The remaining 13 individuals having CD4+ T cells >200 per μL and pVL >1,000 copies/ml and AI <0 · 7 were differentiated based on the ambiguity of HIV pol sequence. In the end, 10 individuals presented concordant evidence of recent HIV infection in the MAA (CD4+ T cells >200 per μL, pVL >1,000 copies/ml, AI <0 · 7, and HIV pol ambiguity <0 · 47%).
The identification was complemented with detailed assessment on clinical information (Table 2). The clinical characteristics in individuals 2–0511, 2–1515, 2–1707, 2–1710, 2–1902 (clinical stage I) and 2–1708 and 2–0302 (clinical stage II) was consistent with the presumptively asymptomatic/mild symptomatic condition of one-year period after seroconversion. The majority had CD4+ T cells ≥350 cells/μL, except 2–1708 and 2–1902 (299 and 246 cells/μL). HBV infection, which was observed in individual 2–1708, shares modes of transmission with HIV. While it is possible that the individual was infected with both viruses at the same time in the past (long-standing infection), the biomarker evidence of HIV infection suggests a recent infection: the relatively weak Ab – Ag binding, shown by AI <0 · 7 can be a marker of immature Ab affinity in the early period of infection or the loss of avidity in severely immunocompromised individuals, but the genomic sequence of viral population was relatively uniform, as shown by the proportion of ambiguous bases 0 · 12 (<0 · 47%), which is consistent with the small number of mixed bases in HIV pol sequence from a one-year period after seroconversion. Taken together, the clinical information of these individuals agrees with the categorization of recent HIV infection by MAA.
Table 2.
Clinical information of newly diagnosed individuals with concordant biomarker evidence of recent HIV infection
| No | ID | Sex | Age (years) | pVL (copies/ml) | CD4 (cells/μL) | AI | Ambiguity (%) | Subtype | DRMs | Relevant Clinical Information | Stage | Transmission |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2–0302 | M | 24 | 29,000 | 422 | 0 · 63 | 0 · 14 | CRF01_AE | NONE | NONE | 2 | MSM |
| 2 | 2–0507 | M | 33 | 80,900 | 229 | 0 · 68 | 0 · 06 | CRF01_AE | NONE | Oral candidiasis | 3 | unprotected heterosexual |
| 3 | 2–0511 | M | 25 | 16,300 | 506 | 0 · 69 | F1: 0 · 06 | CRF01_AE | NONE | NONE | 1 | unprotected heterosexual |
| F2: 0 · 00 | ||||||||||||
| 4 | 2–0903 | F | 35 | 3,480,000 | 262 | 0 · 08 | 0 · 18 | CRF01_AE | NONE | Pulmonary TB | 3 | unprotected heterosexual |
| 5 | 2–1515 | M | 25 | 21,300 | 683 | 0 · 42 | 0 · 03 | CRF01_AE | NONE | NONE | 1 | unprotected heterosexual |
| 6 | 2–1517 | F | 41 | 294,000 | 292 | 0 · 16 | 0 · 41 | CRF01_AE | NNRTI: V179D | TB lymphadenitis | 4 | unprotected heterosexual |
| 7 | 2–1707 | M | 26 | 97,400 | 399 | 0 · 28 | 0 · 12 | C | NRTI: K219Q | NONE | 1 | MSM |
| NNRTI: V179T | ||||||||||||
| 8 | 2–1708 | M | 30 | 130,000 | 299 | 0 · 6 | 0 · 12 | CRF01_AE | NONE | Chronic Hep-B | 2 | MSM |
| 9 | 2–1710 | F | 25 | 3,010 | 586 | 0 · 2 | 0 · 00 | CRF01_AE | NONE | NONE | 1 | unprotected heterosexual |
| 10 | 2–1902 | M | 19 | 1,600,000 | 246 | 0 · 09 | 0 · 00 | CRF01_AE | NONE | NONE | 1 | MSM |
ID: subject identification; ART: antiretroviral drug therapy; pVL: plasma viral load; CD4: cluster of differentiation 4; AI: ambiguity index; DRMs: drug resistance mutations; CRF: circulating recombinant form; sex M: male; sex F: female; MSM: men having sex with men; TB: infection by Mycobacterium tuberculosis; Hep-B: infection by hepatitis B virus (HBV), Stage: WHO’s clinical staging.
The MAA outcome of 2–0507 (clinical stage III) revealed biomarker evidence of recent HIV infection similar with the seven individuals described above, while having a relatively low CD4+ T cell count (229 cells/μL) and oral candidiasis, suggesting advanced HIV infection. However, this individual had a negative HIV test eight months prior to diagnosis, which corresponds to a period of seroconversion shorter than 12 months. Individuals 2–0903 (stage III: pulmonary TB) and 2–1517 (stage IV: non-pulmonary TB lymphadenitis) had a relatively severe disease manifestation and were co-infected with Mycobacterium tuberculosis. Further evaluation of case report forms revealed that 2–0903 was diagnosed with HIV and TB at the same time, indicated by treatment for TB directly after diagnosis and initiation of ART one month after (Table 2), whereas 2–1517 had a documented HIV negative test result 19 months prior to diagnosis.
HIV subtypes and DRMs
The HIV pol gene sequences from 27 newly diagnosed individuals identified five subtypes of HIV: CRF01_AE (N = 20), CRF02_AG (N = 1), B (N = 1), C (N = 2), CRF52_01B (N = 1), and CRF01_AE/B recombinant forms (N = 2). DRMs in RT gene were observed in four individuals V179T (2–1011, HIV C), V179D (2–1517, CRF01_AE), K103N and G190A (2–1905, CRF02_AG) and K219Q and V179T (2–1707, HIV C). K219Q is an NRTI-associated mutation, whereas K103N, G190A, and V179D/T are NNRTI-associated mutations. No DRMs were observed in PR and IN genes.
Discussion
This is the first report that investigates recent HIV infection among newly diagnosed individuals in Indonesia using samples across broad geographic and demographic backgrounds. We designed the MAA with a future implementation in mind, using CD4+ T cell count and pVL, the two widely available HIV biomarker tests in Indonesia as initial MAA steps. Studies estimating recent HIV infection within and across risk groups and geographic regions are essential to identify areas with higher activity of ongoing transmission. In addition, as Indonesia continues to expand ART, identifying transmitted drug resistance in recently infected individuals will provide critical information that will preserve future ARV choices. This MAA combines clinical, immunologic, and virologic characteristics, to identify recent HIV infection, similar to its application in various regions such as in North America, Southeast Asia, and Africa.3,5,7,8,16
We evaluated samples from newly diagnosed (≤1 month) ARV naive adult PLWH. Our MAA utilizes decision tree strategy, beginning with differentiation based on CD4+ T cell count. Statistical analyses and public health recommendations, including from the European CDC and WHO,2,15,31 have supported exclusion of individuals with AIDS and/or CD4+ T cells ≤200 cells/μL as a conservative approach to identify individuals with recent HIV infection, as progressive loss of CD4 cells to 200 cells/μL may take up to eight years after infection.32 Decreases of CD4+ T cell count to ≤200 cells/μL within the first year of HIV infection are rare, although it may occur in those accompanied by opportunistic infections.20,21 Prior longitudinal studies of HIV infection revealed a low frequency (0.2%) of individuals with CD4 ≤200 cells/μL within the first year of HIV infection,.22,23,24,25,26,27,28,29 For an MAA, reducing the FLTR at the beginning is critical and early placement of CD4+ T cell count 200 cells/μL effectively excludes most (99.8%) of chronic infections, while less likely to eliminate true recent infection.
CD4+ T cell count is the most universally available measure from all newly diagnosed individuals and for monitoring disease progression in PLWH in Indonesia. Placing CD4+ T cell count at the top of the algorithm makes the MAA feasible and cost effective for use in places where resource allocations are limited.33,34 Similar approaches were also employed in previous studies where recency is evaluated in the context of health care settings..17,34,35,36,37
The second criterion of this MAA is exclusion of individuals having pVL ≤1,000 copies/ml. pVL is another generic biomarker to monitor disease progression in HIV-infected individuals that has also been used as a screening variable in an MAA.3,7,8,16,19,38,39,40,41 Individuals who are recently infected typically have the relatively high HIV RNA levels with ARV naive individuals who are in the 12 months after seroconversion maintain a setpoint level, approximately within the log10 3–4 copies/ml. Circulating RNA >1,000 copies/ml from two consecutive pVL tests in three months with full adherence following the 1st test or two consecutive >400 copies/ml VL measurements within 0.5–2 years from the initiation of ART (baseline) and still using the similar class of drugs are known as the limitation for virological failure (VF) among treated individuals30,42,43; and pVL 400 or 1,000 cp/ml have been used as a threshold to differentiate between recent and chronic infection in several MAAs to omit long-standing infection with ARV suppression.
Following the initial differentiation by CD4+ T cell count and pVL, we employed serologic and population sequencing assays to further evaluate recent HIV infection in individuals who fulfilled the preliminary CD4+ T cell count and pVL screening. Avidity immunoassays have been the backbone of many MAAs, but can be affected by false recency from individuals initiating ART during primary HIV infection (PHI),44,45,46 in individuals undergoing ART with suppressed pVL,9,47 or in individuals who have AIDS.48,49,50 We initiated the serology assay by investigating the performance of 2-well AI EIA (GenScreen Ultra HIV Ag-Ab Kit [Bio-Rad, Marnes-la-Coquette, France]) in known chronically infected individuals. The kit has well described performance characteristics (sensitivity and specificity) with robust signal/cut-off ratios with the most commonly recognized HIV-1 subtypes,51 and was prequalified for use in HIV diagnosis by the WHO.52 It has been validated for avidity analysis among ARV naive samples in an evaluation panel containing CRF01_AE, with AI 0 · 7 as the optimized threshold9; with performance comparable to the Genetic Systems HIV-1/HIV-2 Plus O EIA [Bio-Rad, Marnes-la-Coquette, France] that have been established for avidity-based recency test.53
In this study, most long-standing samples (53/60; 88 · 3%) (Figure 2) had AIs exceeding the 0 · 7 threshold,9 while the remaining (7/60; 11.7%) had AI values lower than the threshold, yielding FRR of 11 · 7% (7/60). The FRR of our 2-well AI EIA from long-standing samples is higher than that observed in the avidity analysis of long-standing samples47,54; but the potential reasons for the false recent outcomes were identified, which are similar with prior reports.44,45,46,48,49,50,54 Low AI mimicking avidity in a recently infected person can occur in long-standing infection with advanced immunodeficiency,48,49,50 marked by high pVL and low CD4+ T cell count, as seen in subjects 1–0201, 1–1502, and 1–1602 who presented with pVL of 88,000; 783,000; 1,220,000 copies/ml, and low CD4+ T cells counts (91, 68, 7 cells/μL; Table S5). In addition, low AI values in individuals with long-standing infection who were treated early following infection have been reported44,45,46 and was likely the explanation for subjects 1–0501, 1–1301, 1–1402, and 1–1901; who initiated therapy shortly after diagnosis (Table S5).
The inherent false recency of AI EIA is a compelling reason to use multidisciplinary approach for identification of recent HIV infection in an MAA. Inclusion of genomic diversity has been shown to reduce the FRR of an MAA containing avidity assay.55 Proportion of ambiguous bases is positioned as the final differentiating variable in this MAA, thereby reserving the costliest and labor–intensive parameter for the fewest samples.16 In doing so, we did not determine the HIV pol sequences for all samples and have not formally insured the degree of sequence ambiguity that correlates with duration of infection. However, the utility of using proportion of ambiguous bases in the pol gene to determine the duration of infection in CRF01_AE, the most common subtype in Indonesia, has been evaluated by Anderson et al. (2013) and Meixenberger et al. (2019).13,14 HIV infection is typically initiated with a single virus and subsequent accumulation of substitutions in HIV genome of untreated individuals, following approximately 3 X 10−5 errors per bp per replication cycle,56,57 is useful to differentiate between recent and long-standing infection, measured as an increased proportion of ambiguous bases in the HIV pol gene sequence.12,13,14 The 0 · 47% threshold in this study is somewhat more stringent than 0 · 5% proportion used in other studies.12,13,14,16 Using the 0 · 47% threshold, we identified ten newly diagnosed, ARV naive, adult PLWH who had biomarker evidence of recent infection.
MAAs containing combinations of AI, pVL, and CD4+ T cell count have successfully estimated recent infection in diverse geographic regions, including in developing countries such as Thailand and Kenya.7,8 In our study, the ten individuals were young, mostly male (7:3), with HIV transmission risk of unprotected sex and MSM (Table 2). Nearly all individuals were infected with HIV CRF01_AE, except one male individual from Banda Aceh (2–1707) who was infected with HIV subtype C from MSM risk and had K219Q (nucleoside reverse transcriptase inhibitor (NRTI) resistance) and V179T (non-nucleoside reverse transcriptase inhibitor (NNRTI) resistance) mutations. The DRMs are consistent with the first-line ARV drug regimen available in Indonesia at the time the samples were collected (2018–2020), typically the combination of tenofovir (TDF) + lamivudine (3TC) and efavirenz (EFV).
MAAs provide estimates of recent infection based on multiple biomarker testing outcomes and have a certain degree of false recent or false long-term rates and, therefore, require cautious interpretation. Our MAA employed samples from Indonesia where many newly diagnosed individuals present late in infection and often accompanied with disease symptoms.58,59 CD4+ lymphopenia was observed in three individuals (2–1708, 2–1902, and 2–0507) and is not uncommon in PHI, as have been described in individuals with OIs20,21 or in individuals infected by the highly virulent variant.60 Two PLWH with biomarker evidence of recent HIV infection (2–0903 and 2–1517) had active TB infection. In Indonesia, co-infection of TB and HIV is not uncommon, with the epidemic of TB (incidence = 759 · 1 per 100,0000 individuals) exceeds that of HIV infection.61 TB-HIV coinfection may result in impairment of immune responses, including affinity maturation of HIV-specific antibody, that eventually leads to false recent MAA outcome.62,63 Alternatively, they may represent latent TB that was activated by immunocompromised conditions of acute HIV infection, which explains the biomarker evidence of recent HIV infection.20,21 Overall, the clinical information and the biomedical evidence from MAA were in agreement that ten individuals fulfilled the prerequisites defining HIV infection in the period of 12 months after seroconversion, or recent infection.
The presence of DRMs was readily detected among newly diagnosed ARV naive PLWH, in recent and non-recent HIV infection. We observed four HIV carrying RT-inhibitor DRMs from 27 newly diagnosed individuals (14 · 8%) (Table S7). The prevalence of RT inhibitor DRMs in this study is somewhat higher than that reported at the global level, where NNRTI-associated tDRMs was 4 · 5% and NRTI’s was 4%.64 With regards to subtype distribution, the circulating subtypes are genetically diverse and consistent with prior characterization of circulating HIV subtypes in Indonesia based on partial PR and RT sequencing for DRMs genotyping, with CRF01_AE as the most common subtype; other less prevalence subtypes were B, CRF02_AG, C, and recombinant forms between CRF01_AE and other subtypes/CRFs.65,66 As the current sample is relatively limited, a larger sample size representing the countrywide survey is required to accurately determine the prevalence of tDRMs and HIV subtype distribution. Nevertheless, the finding of DRMs in four of 27 newly diagnosed ARV naive individuals suggests transmitted drug resistance is a public health concern as ART continues to be expanded in Indonesia.
The use of a conservative MAA to determine recent HIV infection, identified that 10 of 80 (12 · 5%) newly diagnosed individuals from 19 hospitals in 16 large cities in Indonesia, sampled in 1 · 5 years (January 2018 – June 2020), were recently infected. HIV infection in Indonesia was first reported in 1987 in Bali and has spread to rural and urban areas across the archipelago ever since, concentrated within key populations.67 The early epidemic was largely driven by male injection drug users but then transitioned to unprotected sexual transmission beginning approximately 2007,68,69 with commercial sex workers currently comprising the largest proportion within key populations and the highest per-key population prevalence is among MSM.1 Notably, the increasing proportion of women acquiring HIV infection has resulted in increased mother-to-child-transmission (MTCT) rate of the country, which was nearly three times the global average in 2020.70 ART was introduced in 1997,71 and strong efforts are underway to expand therapy to key populations. Understanding the proportion of recent HIV infection, including transmission of drug resistant HIV, will improve our understanding of HIV epidemic in Indonesia.
Limitations of the study
This study has limitations. First, the overall sample size (long-standing = 60, newly diagnosed = 80) remains small for a country’s population of more than 270 million. Next, the serology 2-well AI EIA and the molecular assay quantification of ambiguous bases has not been compared with an evaluation panel consisting of well-characterized pre- and post-seroconversion specimens from Indonesian population; as such seroconversion panels of Indonesian samples area are not yet available. The GenScreen Ultra has been optimized to differentiate the avidity of anti-HIV Ab between recent and longstanding infections in multiple HIV-1 subtypes,9 including CRF01_AE, with sensitivity and specificity comparable to other commercial EIA kits that have been similarly adapted for avidity testing. It would be ideal to analyze samples using other EIA kits to minimize the FRR and FLTR, and as direct comparison to evaluate the kit’s performance for this sample set.72,73 As availability of such kits is rare in Indonesia and generating the HIV pol sequences is costly; establishing the performance characteristics of the GenScreen Ultra as well as analyzing the proportion of ambiguous bases from existing CRF01_AE evaluation panels could become an option.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Diethylamine | Merck Millipore | Cat# 803010 |
| Invitrogen™ PBS Tablets | Thermo Fisher Scientific | Cat# 003002 |
| Applied Biosystems™ Anode Buffer Container (ABC) | Thermo Fisher Scientific | Cat# 4393927 |
| Applied Biosystems™ Cathode Buffer Container (CBC) | Thermo Fisher Scientific | Cat# 4408256 |
| Applied Biosystems™ POP-7™ Polymer for 3500 | Thermo Fisher Scientific | Cat# 4393708 |
| Invitrogen™ BlueJuice™ Gel Loading Buffer (10X) | Thermo Fisher Scientific | Cat# 10816105 |
| Invitrogen™ 100 bp DNA Ladder | Thermo Fisher Scientific | Cat# 15628019 |
| Invitrogen™ SYBR™ Safe DNA Gel Stain | Thermo Fisher Scientific | Cat# S33102 |
| Invitrogen™ Nuclease-Free Water | Thermo Fisher Scientific | Cat# AM9937 |
| Agarose, LE, Analytical Grade | Promega | Cat# V3121 |
| TBE Buffer, 10X, Molecular Biology Grade | Promega | Cat# V4251 |
| Sigma-Aldrich Ethyl alcohol, Pure | Merck | Cat# E7023-500ML |
| Critical commercial assays | ||
| GenScreen™ Ultra HIV Ag-Ab | Bio-Rad | Cat# 72388 |
| QIAamp Viral RNA Mini Kit | Qiagen | Cat# 52906 |
| Invitrogen™ SuperScript™ III First-Strand Synthesis System | Thermo Fisher Scientific | Cat# 18080051 |
| Invitrogen™ Platinum™ SuperFi™ DNA Polymerase | Thermo Fisher Scientific | Cat# 12351010 |
| Applied Biosystems™ ExoSAP-IT™ Express PCR Product Cleanup Reagent | Thermo Fisher Scientific | Cat# 75001.4X.1.ML |
| Invitrogen™ SuperScript™ III Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18080093 |
| Invitrogen™ Ribonuclease H | Thermo Fisher Scientific | Cat# 18021014 |
| Invitrogen™ dNTP Mix (10 mM ea) | Thermo Fisher Scientific | Cat# 18427013 |
| Applied Biosystems™ BigDye v3.1 Cycle Sequencing Kit | Thermo Fisher Scientific | Cat# 4337455 |
| Applied Biosystems™ BigDye XTerminator™ Purification Kit | Thermo Fisher Scientific | Cat# 4376486 |
| Deposited data | ||
| Raw and analyzed data | This paper | https://data.mendeley.com/datasets/ktg27krty7/1 |
| HIV-1 pol DNA sequence | This paper | Genbank: OR139650 – OR139679 |
| Oligonucleotides | ||
| Primer: PanHIV-1_1F Forward: AGCCYGGGAGCTCTCTG | Gall et al.74 | N/A |
| Primer: PanHIV-1_4R Reverse: CTTWTATGCAGCWTCTGAGGG | Gall et al.74 | N/A |
| Primer: PanHIV-1_2F Forward: GGGAAGTGAYATAGCWGGAAC | Gall et al.74 | N/A |
| Primer: PanHIV-1_2R Reverse: CTGCCATCTGTTTTCCATARTC | Gall et al.74 | N/A |
| Primer: KVL84 Reverse: TCCTGTATGCARACCCCAATATG | Van Laethem et al.75 | N/A |
| Primer: Rty Reverse: GTGTCTCATTGTTTATACTAGG | Yabar et al.76 | N/A |
| Primer: MAW26 Forward: TTGGAAATGTGGAAAGGAAGGAC | Yabar et al.76 | N/A |
| Primer: 1243 Reverse: ACTAAGGGAGGGGTATTGACAAACTC | Yabar et al.76 | N/A |
| Primer: Rta Forward: GTTGACTCAGATTGGTTGCAC | Yabar et al.76 | N/A |
| Primer: Rtb Forward: CCTAGTATAAACAATGAGACAC | Yabar et al.76 | N/A |
| Primer: MAW70 Reverse: TAATCCCTGCGTAAATCTGACTTGCCCA | Yabar et al.76 | N/A |
| Software and algorithms | ||
| Applied Biosystems 3500/3500xL Genetic Analyzer System | Thermo Fisher Scientific | Cat# 4405673; RRID:SCR_021901 |
| Sequencher v5.4.6 | Genecodes, | RRID:SCR_001528 |
| GraphPad Prism v9.3.0 | GraphPad | RRID:SCR_002798 |
| jpHMM (jumping profile Hidden Markov Model) | Schultz et al., 200977 | http://jphmm.gobics.de/ |
| RIP (Recombinant Identification Program) | Siepel et al., 199578 | https://www.hiv.lanl.gov/content/sequence/RIP/RIP.html |
| HIVdb Program: Sequence Analysis | Rhee et al., 200379 | https://hivdb.stanford.edu/hivdb/by-sequences/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Frank Maldarelli (Fmalli@mail.nih.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Study participants and samples
Blood plasma collected from individuals participating in the multicentre study “HIV Infection and Risk Related Coinfections/Comorbidities in Indonesia” (INA-PROACTIVE; ClinicalTrials.gov Identifier: NCT03663920) conducted by the Indonesia Research Network on Infectious Disease (INA-RESPOND)80 were used. INA-PROACTIVE enrolled 4,329 HIV outpatients from 19 hospital sites across Indonesia from January 2018 to June 2020 (Figure S1).81 Subjects were diagnosed as HIV positive by the standard of care (SoC) in Indonesia. On enrollment in INA-PROACTIVE, confirmation of HIV diagnosis was done using a rapid diagnostic test (RDT) SD Bioline HIV/Syphilis Duo [Standard Diagnostics, Inc., Suwon, South Korea]. For those unable to provide evidence of HIV diagnosis, re-affirmation was done using other RDT brands. Determine HIV-1/2 Ag/Ab Combo [Alere, Waltham, USA] was used to resolve discordant diagnoses.81 All participants provided written informed consent for the collection of clinical data and specimens for the subsequent analyses, as well as to have their medical records, including WHO clinical staging,82 recorded in the case record forms (CRFs). CD4+ T cell count and pVL were measured at blood drawn. CD4+ T cells was counted using the point-of-care (POC) PIMA CD4 Analyser [Abbott, Scarborough, USA] and pVL was measured using the Xpert HIV-1 Viral Load [Cepheid, Sunnyvale, USA], with plasma was later stored at −80°C for subsequent serology and molecular tests. This is a cross-sectional study that used two groups of “INA – PROACTIVE” participants, a training set of individuals with long-standing (chronic) HIV infection, and a test set of newly diagnosed ARV naive individuals (Figure 1).
Ethics
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of The Faculty of Medicine, Universitas Indonesia – Cipto Mangunkusumo Hospital (Number KET-1499/UN2.F1/ETIK/PPM.00.02/2020 dated 28 December 2020). Written informed consent were obtained from all participants.81
Role of the funding source
The funders have no role in study design, data collection, data analyses, interpretation and writing of the manuscript or decision to publish it. Study, writing, and review of the manuscript was completed by the authors.
Method details
2-Well avidity index enzymatic immunoassay (AI EIA)
The 2-well AI EIA was done using the GenScreen Ultra HIV Ag-Ab Kit [Bio-Rad, Marnes-la-Coquette, France] that detects antibody against the gp160 of HIV-1/2 and 0 · 1 M diethylamine (DEA) as the dissociation buffer.4,9 Frozen plasma (−80°C) was thawed to 4°C and diluted 1:10 in 1X PBS [Thermo Fisher Scientific, Carlsbad, USA]. Diluted plasma (75 μl) was added into both the control and dissociation wells in an EIA plate and incubated for antibody binding (4°C, 1 hour). To dissociate the binding of non-avid antibodies, 100-ul 0·1M DEA [Merck, Darmstadt, Germany] was added to the dissociation wells and incubated at 37°C for 1-hour. In parallel, 100-ul 1X PBS [Thermo Fisher Scientific, Carlsbad, USA] was added to the control wells. EIA was carried out according to the manufacturer’s instructions83 and optical density (OD) measured using a Multi Skan Go microplate spectrophotometer and SkanIt Software 6.0.1 [Thermo Fisher Scientific, Carlsbad, USA] and had expected performance characteristics (Table S4). AI was calculated as [(OD of the dissociation wells)/(OD of control wells)].4,9 Plasma from a subject was tested in triplicate and the averaged AI values were analysed. This approach classifies samples with AI <0 · 7 as potential recent infection cases,9 which were further subjected to quantification of ambiguous bases proportion in HIV pol gene.
HIV pol DNA sequencing
HIV pol gene sequence was generated using an in-house Sanger sequencing protocol described in Figure 3. The sequencing strategy allows to retrieve a continuous pol sequence that covers a region within the HIV genome that is generally analysed for DRMs.75,76,84 Briefly, HIV RNA was extracted from 140-μl of patient’s plasma using a guanidium-based spin column protocol [Qiagen, Hilden, Germany] after concentrating viral particles by centrifuging 500-μl plasma in 20,000g, 4°C for 1 hour. Near full length genomic (NFLG) cDNA was synthesized using Superscript III First Strand cDNA Synthesis Kit [Thermo Fisher Scientific, Carlsbad, USA] and gene-specific primers (GSPs) Pan-HIV 1F and Pan-HIV 4R that anneal to the 5′-LTR and 3′-LTR, respectively.74 Pre-sequencing amplification was done using Platinum SuperFi™ DNA Polymerase [Thermo Fisher Scientific, Carlsbad, USA] using a low cycle number (25X) to minimize random mutations introduced by repeated copying of DNA polymerase. The 1st PCR mixture was a 25 μl volume of 1X SuperFi Buffer, 0 · 2 mM dNTP Mix, 0 · 08 μM of each primer, 0 · 02 U Platinum SuperFi DNA Polymerase, and template (2 · 5 μL of each 1F- and 4R-cDNA). PCR conditions consisted of 98°C 30 seconds initial denaturation, 25 cycles of 98°C 10 seconds (denaturation), 55°C 15 seconds (annealing), 72°C 5 minutes (extension), and a final extension of 72°C for 5 minutes for primer set 1. For primer set 2, the annealing temperature was 58°C. Nested PCR mixture and reaction were similar with the 1st PCR, only differing in the template (2 · 5 μL of 1st PCR product) and extension time (2 minutes). The primers are listed in Tables S1–S3. PCR products were visualized in 1 · 5% agarose gel electrophoresis and successful amplicons were purified using ExoSAP-IT Express [Thermo Fisher Scientific, Carlsbad, USA], then cycle-sequenced using the BigDye v3.1 Cycle Sequencing Kit [Thermo Fisher Scientific, Carlsbad, USA]. Cycle sequencing product was purified using the BigDye Xterminator Cycle Sequencing Kit [Thermo Fisher Scientific, Carlsbad, USA], then electrophoresed using FastSeq50_POP7_Assay protocol in an Applied Biosystems 3500/3500xL Genetic Analyzer (GA) System (RRID:SCR_021901) [Thermo Fisher Scientific, Carlsbad, USA]. Fluorescence signals from the capillary electrophoresis were converted into nucleotide base sequences by automated base calling in the 3500 Series Data Collection Software 3.3. This in-house sequencing procedure is the strategy for HIV genotypic resistance testing (GRT) for INA-RESPOND. Performance characteristics are maintained by strict quality control (QC), including yearly “HIV Viral Load Survey (HV2/HIVG)” [College of American Pathologist, Northfield, USA] since 2019.
Analysis of ambiguous bases in pol sequence
The electropherogram outcome of Sanger DNA sequencing was reviewed for analysis of base ambiguity using Sequencher (RRID:SCR_001528) v5.4.6 [Genecodes, Ann Arbor, USA]. Mixed bases were determined from a consensus. Multiple bases in a position were scored as mixed/ambiguous when they appeared in more than one sequencing read, and the minor peak height was at least 20% of that of the first base and not due to heightened background. The proportion of ambiguous bases (%) was calculated as the number of mixed bases divided by the total sequence length: [(number of mixed base positions)/(total sequence length)] X 100%.
Identification of drug resistance mutations (DRMs) and HIV subtypes
The final consensus HIV pol sequence (fasta) was submitted to the Stanford University HIV Drug Resistant Database (https://hivdb.stanford.edu/hivdb/by-sequences/) to determine DRMs and HIV subtype. If subtype identification indicated a recombinant form, the recombination junction was confirmed by jumping profile HMM (http://jphmm.gobics.de/submission_hiv.html) and recombination identification program (https://www.hiv.lanl.gov/content/sequence/RIP/RIP.html).
Multiassay algorithm (MAA) to identify recently infected individuals
Clinical information of patients, including past and current illnesses (PCIs), transmission risks, and the WHO’s clinical staging of AIDS were retrieved from the CRFs. AI EIA was done to all newly diagnosed individuals, and ambiguity analysis was done to those having AI <0 · 7. Recently infected individuals were inferred using a decision tree with hierarchical screening starting from samples with CD4+ T cell count >200 cells/μL, pVL >1,000 copies/ml, AI values < 0 · 7, and HIV pol ambiguity threshold <0 · 47%.2,9,13,15,16 Individuals conforming to the hierarchical classification criteria or presented the recent infection profile based on the biomarker testing outcomes were verified using their clinical information.
Quantification and statistical analysis
To set up the 2-well AI EIA (GenScreen™ Ultra HIV Ag-Ab Kit [Bio-Rad, Marnes-la-Coquette, France]), 30 random samples of individuals fulfilling inclusion criteria of longstanding infection were tested in a pilot to estimate the standard deviation (SD) of the AI values. The SD value was applied in Cochran’s formula to estimate the sample size necessary to achieve 95% confidence (5% absolute error) of the 2-well AI EIA. A minimum of 53 longstanding individuals was required and we evaluated 60 (Figure 1; Table 1). To ensure the consistency of AI values, the coefficient of variation (CoV) of OD in untreated (1X PBS) and treated (0·1M DEA) wells were determined from triplicate measures from each subject, with only triplicate measures having CoV <10% proceeded for AI calculation. The Shapiro-Wilk test was done for measures of participant characteristics and AI values from longstanding and newly diagnosed individuals to identify whether values are normally distributed. Depending on the data distribution, means values of participants characteristics and AI values were compared using either the parametric Student’s t test for normally distributed data or the nonparametric Mann-Whitney test for data that were not normally distributed. Categorical variables were analysed using contingency analysis Fisher’s exact (2X2) or Chi-square (2X3) test. The level of significance was set at p < 0 · 05. Analysis was performed in GraphPad Prism (RRID:SCR_002798) software (v9.3.0, GraphPad, San Diego, USA).
Acknowledgments
We thank the patients who participated in this study and the site study team. This work has been funded in whole or in part with the Ministry of Health Indonesia and the Intramural Research Program, National Institutes of Health; National Cancer Institute, Center for Cancer Research, National Institutes of Health under Contract No ZIA BC 010819. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. This work was completed in partial fulfilment for the doctorate degree from Universitas Indonesia (W.N.W) and presented at the 36th Annual Meeting of the Japanese Society for AIDS (18–20 November 2022). We thank the INA – RESPOND’s INA – PROACTIVE study and the network reference laboratory personnels for their support.
Funding: Ministry of Health Indonesia; the Intramural Research Program, National Institutes of Health; National Cancer Institute, Center for Cancer Research, National Institutes of Health, under Contract No ZIA BC 010819.
Author contributions
Conceptualization, W.N.W., E.Y., P.P.S., and F.M.; methodology, W.N.W., K.H., Z.G., and F.M.; software, W.N.W.; validation, D.A., D.L., and H.K.; formal analysis, W.N.W., E.Y., H.K., C.Y., and F.M.; investigation, W.N.W. and F.M.; resources, T.P.M., R.W., A.T.A., S.A., R.E., A.A.A.Y.G., C.H., A.R.I., J.K., V.K.M., M.B.R.N., A.N., R.R., A.R., M.R., D.Y.S., J.S., M.A.U.S., W.E.S., J.T., F.L.T., and A.N.; data curation, N.H.S.; writing—original draft preparation, W.N.W.; writing—review and editing, W.N.W., E.Y., D.A., Z.G., K.H., C-Y.L., H.K., and F.M.; visualization, W.N.W. and F.M.; supervision, P.P.S.; project administration, D.A.; funding acquisition, M.K. and F.M. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no conflict of interest.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research. We avoided “helicopter science” practices by including the participating local contributors from the region where we conducted the research as authors on the paper.
Published: September 21, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107986.
Supplemental information
Data and code availability
-
•
The HIV-1 pol DNA sequence data have been deposited to the GenBank with Accession Number GenBank: OR139650 – OR139679. All data presented in this manuscript are also available online as supplementary material that can be downloaded at https://data.mendeley.com/datasets/ktg27krty7/1. All datasets generated for this study are available from the corresponding author upon reasonable request.
-
•
Sequences generated in this study have been deposited to GenBank. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Joint_United_Nations_Programme_on_HIV/AIDS_(UNAIDS) 2021. UNAIDS Data 2021. [Google Scholar]
- 2.World_Health_Organization_(WHO) 2011. When and How to Use Assays for Recent Infection to Estimate HIV Incidence at a Population Level. [Google Scholar]
- 3.Laeyendecker O., Brookmeyer R., Cousins M.M., Mullis C.E., Konikoff J., Donnell D., Celum C., Buchbinder S.P., Seage G.R., 3rd, Kirk G.D., et al. HIV Incidence Determination in the United States: A Multiassay Approach. J. Infect. Dis. 2013;207:232–239. doi: 10.1093/infdis/jis659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huik K., Soodla P., Pauskar M., Owen S.M., Luo W., Murphy G., Jõgeda E.L., Kallas E., Rajasaar H., Avi R., et al. The concordance of the limiting antigen and the Bio-Rad avidity assays in persons from Estonia infected mainly with HIV-1 CRF06_cpx. PLoS One. 2019;14 doi: 10.1371/journal.pone.0217048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karatzas-Delgado E.F., Ruiz-González V., García-Cisneros S., Olamendi-Portugal M.L., Herrera-Ortiz A., López-Gatell H., González-Rodríguez A., Sánchez-Alemán M.A. Evaluation of an HIV recent infection testing algorithm withserological assays among men who have sex with men in Mexico. J. Infect. Public Health. 2020;13:509–513. doi: 10.1016/j.jiph.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Suligoi B., Massi M., Galli C., Sciandra M., Di Sora F., Pezzotti P., Recchia O., Montella F., Sinicco A., Rezza G. Identifying recent HIV infections using the avidity index and an automated enzyme immunoassay. J. Acquir. Immune Defic. Syndr. 2003;32:424–428. doi: 10.1097/00126334-200304010-00012. [DOI] [PubMed] [Google Scholar]
- 7.Otecko N., Inzaule S., Odhiambo C., Otieno G., Opollo V., Morwabe A., Were K., Ndiege K., Otieno F., Kim A.A., Zeh C. Viral and Host Characteristics of Recent and Established HIV-1 Infections in Kisumu based on a Multiassay Approach. Sci. Rep. 2016;6:37964. doi: 10.1038/srep37964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant-McAuley W., Klock E., Laeyendecker O., Piwowar-Manning E., Wilson E., Clarke W., Breaud A., Moore A., Ayles H., Kosloff B., et al. Evaluation of multi-assay algorithms for identifying individuals with recent HIV infection: HPTN 071 (PopART) PLoS One. 2021;16:e0258644. doi: 10.1371/journal.pone.0258644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauser A., Heiden M.a.d., Meixenberger K., Han O., Fiedler S., Hanke K., Koppe U., Hofmann A., Bremer V., Bartmeyer B., et al. Evaluation of a BioRad Avidity assay for identification of recent HIV-1 infections using dried serum or plasma spots. J. Virol. Methods. 2019;266:114–120. doi: 10.1016/j.jviromet.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Dobbs T., Kennedy S., Pau C.-P., McDougal J.S., Parekh B.S. Performance Characteristics of the Immunoglobulin G-Capture BED-Enzyme Immunoassay, an Assay To Detect Recent Human Immunodeficiency Virus Type 1 Seroconversion. J. Clin. Microbiol. 2004;42:2623–2628. doi: 10.1128/JCM.42.6.2623-2628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauser A., Santos-Hoevener C., Meixenberger K., Zimmermann R., Somogyi S., Fiedler S., Hofmann A., Bartmeyer B., Jansen K., Hamouda O., et al. Improved Testing of Recent HIV-1 Infections with the BioRad Avidity Assay Compared to the Limiting Antigen Avidity Assay and BED Capture Enzyme Immunoassay: Evaluation Using Reference Sample Panels from the German Seroconverter Cohort. PLoS One. 2014;9:e98038. doi: 10.1371/journal.pone.0098038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouyos R.D., von Wyl V., Yerly S., Böni J., Rieder P., Joos B., Taffé P., Shah C., Bürgisser P., Klimkait T., et al. Ambiguous Nucleotide Calls From Population based Sequencing of HIV-1 are a Marker for Viral Diversity and the Age of Infection. Clin. Infect. Dis. 2011;52:532–539. doi: 10.1093/cid/ciq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson E., Shao W., Bontell I., Cham F., Cuong D.D., Wondwossen A., Morris L., Hunt G., Sönnerborg A., Bertagnolio S., et al. Evaluation of Sequence Ambiguities of the HIV-1 pol gene as a Method to Identify Recent HIV-1 Infection in Transmitted Drug Resistance Surveys. Infect. Genet. Evol. 2013;18:125–131. doi: 10.1016/j.meegid.2013.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meixenberger K., Hauser A., Jansen K., Yousef K.P., Fiedler S., von Kleist M., Norley S., Somogyi S., Hamouda O., Bannert N., et al. Assessment of Ambiguous Base Calls in HIV-1 pol Population Sequences as a Biomarker for Identification of Recent Infections in HIV-1 Incidence Studies. J. Clin. Microbiol. 2014;52:2977–2983. doi: 10.1128/JCM.03289-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European_Centre_for_Disease_Prevention_and_Control . European Centre for Disease Prevention and Control (ECDC); 2013. Monitoring Recently Acquired HIV Infections in the European Context. [Google Scholar]
- 16.Cousins M.M., Konikoff J., Sabin D., Khaki L., Longosz A.F., Laeyendecker O., Celum C., Buchbinder S.P., Seage G.R., 3rd, Kirk G.D., et al. A Comparison of Two Measures of HIV Diversity in Multi-Assay Algorithms for HIV Incidence Estimation. PLoS One. 2014;9:e101043. doi: 10.1371/journal.pone.0101043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann A., Hauser A., Zimmermann R., Santos-Hövener C., Bätzing-Feigenbaum J., Wildner S., Kücherer C., Bannert N., Hamouda O., Bremer V., Bartmeyer B. Surveillance of recent HIV infections among newly diagnosed HIV cases in Germany between 2008 and 2014. BMC Infect. Dis. 2017;17:484. doi: 10.1186/s12879-017-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eshleman S.H., Hughes J.P., Laeyendecker O., Wang J., Brookmeyer R., Johnson-Lewis L., Mullis C.E., Hackett J., Jr., Vallari A.S., Justman J., Hodder S. Use of a Multifaceted Approach to Analyze HIV Incidence in a Cohort Study of Women in the United States: HIV Prevention Trials Network 064 Study. J. Infect. Dis. 2013;207:223–231. doi: 10.1093/infdis/jis658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laeyendecker O., Piwowar-Manning E., Fiamma A., Kulich M., Donnell D., Bassuk D., Mullis C.E., Chin C., Swanson P., Hackett J., Jr., et al. Estimation of HIV Incidence in a Large, Community-Based, Randomized Clinical Trial: NIMH Project Accept (HIV Prevention Trials Network 043) PLoS One. 2013;8:e68349. doi: 10.1371/journal.pone.0068349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo S., James C.W., Telford G. Unusual Presentations of Primary Human Immunodeficiency Virus Infection. AIDS Patient Care STDS. 2002;16:251–254. doi: 10.1089/10872910260066688. [DOI] [PubMed] [Google Scholar]
- 21.Subedee A., Kimmel W., Donato A.A. Never too early for the opportunists. BMJ Case Rep. 2013;2013 doi: 10.1136/bcr-2013-009612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.AIDS-Hemophilia_French_Study_Group Natural History of Primary Infection With LAV in Multitransfused Patients. Blood. 1986;68:89–94. [PubMed] [Google Scholar]
- 23.Daul C.B., deShazo R.D., Andes W.A. Human immunodeficiency virus infection in hemophiliac patients: A three-year prospective evaluation. Am. J. Med. 1988;84:801–809. doi: 10.1016/0002-9343(88)90056-3. [DOI] [PubMed] [Google Scholar]
- 24.Robb M.L., Eller L.A., Kibuuka H., Rono K., Maganga L., Nitayaphan S., Kroon E., Sawe F.K., Sinei S., Sriplienchan S., et al. Prospective Study of Acute HIV-1 Infection in Adults in East Africa and Thailand. N. Engl. J. Med. 2016;374:2120–2130. doi: 10.1056/NEJMoa1508952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen C., Lindhardt B.O., Jensen B.L., Lauritzen E., Gerstoft J., Dickmeiss E., Gaub J., Scheibel E., Karlsmark T. Clinical course of primary HIV infection: consequences for subsequent course of infection. BMJ. 1989;299:154–157. doi: 10.1136/bmj.299.6692.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margolick J.B., Donnenberg A.D., Muñoz A., Park L.P., Bauer K.D., Giorgi J.V., Ferbas J., Saah A.J. Changes in T and Non-T Lymphocyte Subsets Following Seroconversion to HIV-1 Stable CD3+ and Declining CD3- Populations Suggest Regulatory Responses Linked to Loss of CD4 Lymphocytes. J. Acquir. Immune Defic. Syndr. 1993;6:153–161. [PubMed] [Google Scholar]
- 27.Lindbäck S., Broström C., Karlsson A., Gaines H. Does symptomatic primary HIV-1 infection accelerate progression to CDC stage IV disease, CD4 count below 200 x 106/L, AIDS, and death from AIDS? BMJ. 1994;309:1535–1537. doi: 10.1136/bmj.309.6968.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De_Wolf F., Roos M., Lange J.M.A., Houweling J.T.M., Coutinho R.A., Van_Der_Noordja J., Schellekens P.T., Goudsmit J. Decline in CD4+ Cell Numbers Reflects Increase in HIV-1 Replication. AIDS Res. Hum. Retrovir. 2009;4 doi: 10.1089/aid.1988.4.433. [DOI] [PubMed] [Google Scholar]
- 29.Nkone P., Loubser S., Quinn T.C., Redd A.D., Laeyendecker O., Tiemessen C.T., Mayaphi S.H. Evaluation of the HIV-1 Polymerase Gene Sequence Diversity for Prediction of Recent HIV-1 Infections Using Shannon Entropy Analysis. Viruses. 2022;14 doi: 10.3390/v14071587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkin N., Gao F., Grebe E., Cutrell A., Das M., Donnell D., Duerr A., Glidden D.V., Hughes J.P., Murray J., et al. Facilitating Next-Generation Pre-Exposure Prophylaxis Clinical Trials Using HIV Recent Infection Assays: A Consensus Statement from the Forum HIV Prevention Trial Design Project. CLINICAL PHARMACOLOGY & THERAPEUTICS. 2023 doi: 10.1002/cpt.2830. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 31.Fellows I.E., Hladik W., Eaton J.W., Voetsch A.C., Parekh B.S., Shiraishi R.W. Improving Biomarker-based HIV Incidence Estimation in the Treatment Era. Epidemiology. 2023;34:353–364. doi: 10.1097/EDE.0000000000001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodi S., Phillips A., Touloumi G., Geskus R., Meyer L., Thiébaut R., Pantazis N., Amo J.d., Johnson A.M., Babiker A., et al. Time From Human Immunodeficiency Virus Seroconversion to Reaching CD4+ Cell Count Thresholds <200, <350, and <500 Cells/mm3: Assessment of Need Following Changes in Treatment Guidelines. Clin. Infect. Dis. 2011;53:817–825. doi: 10.1093/cid/cir494. [DOI] [PubMed] [Google Scholar]
- 33.Chauhan C.K., Lakshmi P.V.M., Sagar V., Sharma A., Arora S.K., Kumar R. Immunological markers for identifying recent HIV infection in North-West India. Indian J. Med. Res. 2020;152:227–233. doi: 10.4103/ijmr.IJMR_2007_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Q., Wang Y., Liu J., Duan X., Chen M., Yang J., Yang T., Yang S., Guan P., Jiang Y., et al. Identifying major drivers of incident HIV infection using recent infection testing algorithms (RITAs) to precisely inform targeted prevention. Int. J. Infect. Dis. 2020;101:131–137. doi: 10.1016/j.ijid.2020.09.1421. [DOI] [PubMed] [Google Scholar]
- 35.Slurink I., van_Aar F., Parkkali S., Heijman T., Gotz H., Kampman K., Weert Y.v., Benthem B.v., Laar T.v.d., Coul E.O.d., for_the_RITA_surveillance_network_of_sexual_health_centres Recently acquired HIV infections and associated factors among men who have sex with men diagnosed at Dutch sexual health centres. Int. J. STD AIDS. 2021;0:1–11. doi: 10.1177/09564624211008071. [DOI] [PubMed] [Google Scholar]
- 36.Mastro T.D., Kim A.A., Hallett T., Rehle T., Welte A., Laeyendecker O., Oluoch T., Garcia-Calleja J.M. Estimating HIV Incidence in Populations Using Tests for Recent Infection: Issues, Challenges and the Way Forward. J. HIV AIDS Surveill. Epidemiol. 2010;2:1–14. [PMC free article] [PubMed] [Google Scholar]
- 37.Widgren K., Skar H., Berglund T., Kling A.-M., Tegnell A., Albert J. Delayed HIV diagnosis common in Sweden, 2003 – 2010. Scand. J. Infect. Dis. 2014;46:862–867. doi: 10.3109/00365548.2014.953575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brookmeyer R., Konikoff J., Laeyendecker O., Eshleman S.H. Estimation of HIV Incidence Using Multiple Biomarkers. Am. J. Epidemiol. 2013;177:264–272. doi: 10.1093/aje/kws436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laeyendecker O., Kulich M., Donnell D., Komárek A., Omelka M., Mullis C.E., Szekeres G., Piwowar-Manning E., Fiamma A., Gray R.H., et al. Development of Methods for Cross-Sectional HIV Incidence Estimation in a Large, Community Randomized Trial. PLoS One. 2013;8:e78818. doi: 10.1371/journal.pone.0078818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konikoff J., Brookmeyer R., Longosz A.F., Cousins M.M., Celum C., Buchbinder S.P., Seage G.R., 3rd, Kirk G.D., Moore R.D., Mehta S.H., et al. Performance of a Limiting-Antigen Avidity Enzyme Immunoassay for Cross-Sectional Estimation of HIV Incidence in the United States. PLoS One. 2013;8:e82772. doi: 10.1371/journal.pone.0082772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laeyendecker O., Konikoff J., Morrison D.E., Brookmeyer R., Wang J., Celum C., Morrison C.S., Abdool Karim Q., Pettifor A.E., Eshleman S.H. Identification and validation of a multi-assay algorithm for cross-sectional HIV incidence estimation in populations with subtype C infection. J. Int. AIDS Soc. 2018;21 doi: 10.1002/jia2.25082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice B., Wit M.d., Hargreaves J., all_members_of_the_MeSH_Working_Group . MeSH Consortium; 2019. The Feasibility and Utility of HIV Recent Infection Testing in a Range of Routine Service-Provision Contexts. [Google Scholar]
- 43.ICAP’s_Clinical_and_Training_Unit . 2016. Standard Operating Procedures on Viral Load Monitoring for ICAP Clinical Staff and Healthcare Workers Version 1. [Google Scholar]
- 44.Le Guillou H., Le Meur A., Bourdon S., Riou M., Loison J., Fialaire P., Chennebault J.M., Kouyoumdjian S., Payan C. Antibody avidity: use for the diagnosis of HIV early infection [article in French] Ann. Biol. Clin. 2001;59:41–47. [PubMed] [Google Scholar]
- 45.Selleri M., Orchi N., Zaniratti M.S., Bellagamba R., Corpolongo A., Angeletti C., Ippolito G., Capobianchi M.R., Girardi E. Effective Highly Active Antiretroviral Therapy in Patients With Primary HIV-1 Infection Prevents the Evolution of the Avidity of HIV-1–Specific Antibodies. J. Acquir. Immune Defic. Syndr. 2007;46:145–150. doi: 10.1097/QAI.0b013e318120039b. [DOI] [PubMed] [Google Scholar]
- 46.Re M.C., Schiavone P., Bon I., Vitone F., De Crignis E., Biagetti C., Gibellini D. Incomplete IgG response to HIV-1 proteins and low avidity levels in recently converted HIV patients treated with early antiretroviral therapy. Int. J. Infect. Dis. 2010;14:e1008–e1012. doi: 10.1016/j.ijid.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 47.Laeyendecker O., Brookmeyer R., Mullis C.E., Donnell D., Lingappa J., Celum C., Baeten J.M., Campbell M.S., Essex M., de Bruyn G., et al. Specificity of Four Laboratory Approaches for Cross-Sectional HIV Incidence Determination: Analysis of Samples from Adults with Known Nonrecent HIV Infection from Five African Countries. AIDS Res. Hum. Retrovir. 2012;28:1177–1183. doi: 10.1089/aid.2011.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sciascia C., Palomba E., Gay V., Tovo P.A. Anti-HIV-1 antibody avidity is correlated with clinical status in infected children. Pediatr. AIDS HIV Infect. 1996;7:14–19. [PubMed] [Google Scholar]
- 49.Chargelegue D., Stanley C.M., O'Toole C.M., Colvin B.T., Steward M.W. The affinity of IgG antibodies to gag p24 and p17 in HIV-1-infected patients correlates with disease progression. Clin. Exp. Immunol. 1995;99:175–181. doi: 10.1111/j.1365-2249.1995.tb05529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curtis K.A., Kennedy M.S., Owen S.M. Longitudinal Analysis of HIV-1-Specific Antibody Responses. AIDS Res. Hum. Retrovir. 2014;30:1099–1105. doi: 10.1089/aid.2014.0105. [DOI] [PubMed] [Google Scholar]
- 51.Ly T.D., Martin L., Daghfal D., Sandridge A., West D., Bristow R., Chalouas L., Qiu X., Lou S.C., Hunt J.C., et al. Seven Human Immunodeficiency Virus (HIV) Antigen- Antibody Combination Assays: Evaluation of HIV Seroconversion Sensitivity and Subtype Detection. J. Clin. Microbiol. 2001;39:3122–3128. doi: 10.1128/JCM.39.9.3122-3128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.World_Health_Organization . 2023. WHO List of Prequalified in Vitro Diagnostic Products. [Google Scholar]
- 53.Masciotra S., Dobbs T., Candal D., Hanson D., Delaney K., Rudolph D., Charurat M., Harrigan R., McDougal S., Owen M. 2010. Antibody Avidity-Based Assay for Identifying Recent HIV-1 Infections Based on Genetic Systems TM 1/2 Plus O EIA. . 17th Conference on Retroviruses and Opportunistic Infections. [Google Scholar]
- 54.Longosz A.F., Mehta S.H., Kirk G.D., Margolick J.B., Brown J., Quinn T.C., Eshleman S.H., Laeyendecker O. Incorrect Identification of Recent HIV Infection in Adults in the United States Using a Limiting-Antigen Avidity Assay. AIDS. 2014;28:1227–1232. doi: 10.1097/QAD.0000000000000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moyo S., Vandormael A., Wilkinson E., Engelbrecht S., Gaseitsiwe S., Kotokwe K.P., Musonda R., Tanser F., Essex M., Novitsky V., de Oliveira T. Analysis of Viral Diversity in Relation to the Recency of HIV-1C Infection in Botswana. PLoS One. 2016;11:e0160649. doi: 10.1371/journal.pone.0160649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao F., Chen Y., Levy D.N., Conway J.A., Kepler T.B., Hui H. Unselected Mutations in the Human Immunodeficiency Virus Type 1 Genome Are Mostly Nonsynonymous and Often Deleterious. J. Virol. 2004;78:2426–2433. doi: 10.1128/JVI.78.5.2426-2433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roberts J.D., Bebenek K., Kunkel T.A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 58.Jeong S.J., Italiano C., Chaiwarith R., Ng O.T., Vanar S., Jiamsakul A., Saphonn V., Nguyen K.V., Kiertiburanakul S., Lee M.P., et al. Late Presentation into Care of HIV Disease and Its Associated Factors in Asia: Results of TAHOD. AIDS Res. Hum. Retrovir. 2016;32:255–261. doi: 10.1089/AID.2015.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiertiburanakul S., Boettiger D., Lee M.P., Omar S.F., Tanuma J., Ng O.T., Durier N., Phanuphak P., Ditangco R., Chaiwarith R., et al. Trends of CD4 cell count levels at the initiation of antiretroviral therapy over time and factors associated with late initiation of antiretroviral therapy among Asian HIV-positive patients. J. Int. AIDS Soc. 2014;17:18804. doi: 10.7448/IAS.17.1.18804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wymant C., Bezemer D., Blanquart F., Ferretti L., Gall A., Hall M., Golubchik T., Bakker M., Ong S.H., Zhao L., et al. A highly virulent variant of HIV-1 circulating in the Netherlands. Science. 2022;375:540–545. doi: 10.1126/science.abk1688. [DOI] [PubMed] [Google Scholar]
- 61.Noviyani A., Nopsopon T., Pongpirul K. Variation of tuberculosis prevalence across diagnostic approaches and geographical areas of Indonesia. PLoS One. 2021;16 doi: 10.1371/journal.pone.0258809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chetty S., Govender P., Zupkosky J., Pillay M., Ghebremichael M., Moosa M.-Y.S., Ndung’u T., Porichis F., Kasprowicz V.O. Co-Infection with Mycobacterium tuberculosis Impairs HIV-Specific CD8+ and CD4+ T Cell Functionality. PLoS One. 2015;10:e0118654. doi: 10.1371/journal.pone.0118654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Streeck H., D’Souza M.P., Littman D.R., Crotty S. Harnessing CD4+ T cell responses in HIV vaccine development. Nat. Med. 2013;19:143–149. doi: 10.1038/nm.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baxter J.D., Dunn D., White E., Sharma S., Geretti A.M., Kozal M.J., Johnson M.A., Jacoby S., Llibre J.M., Lundgren J., International Network for Strategic Initiatives in Global HIV Trials INSIGHT START Study Group Global HIV-1 transmitted drug resistance in the INSIGHT Strategic Timing of AntiRetroviral Treatment trial. HIV Med. 2015;16 Suppl 1:77–87. doi: 10.1111/hiv.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.seq-info@lanl.gov . 2022. Geographic Distribution of Subtyped Sequences in the HIV Database.https://www.hiv.lanl.gov/components/sequence/HIV/geo/geo.html [Google Scholar]
- 66.Fischer W., Giorgi E.E., Chakraborty S., Nguyen K., Bhattacharya T., Theiler J., Goloboff P.A., Yoon H., Abfalterer W., Foley B.T., et al. HIV-1 and SARS-CoV-2: Patterns in the evolution of two pandemic pathogens. Cell Host Microbe. 2021;29:1093–1110. doi: 10.1016/j.chom.2021.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mboi N. Cases in Global Health Delivery; 2011. HIV/AIDS in Indonesia: Building a Coordinated National Response. [Google Scholar]
- 68.Rahmalia A., Wisaksana R., Meijerink H., Indrati A.R., Alisjahbana B., Roeleveld N., van der Ven A.J.A.M., Laga M., van Crevel R. Women with HIV in Indonesia: are they bridging a concentrated epidemic to the wider community? BMC Res. Notes. 2015;8:757. doi: 10.1186/s13104-015-1748-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iskandar S., Basar D., Hidayat T., Siregar I.M.P., Pinxten L., van Crevel R., Van der Ven A.J.A.M., De Jong C.A.J. High risk behavior for HIV transmission among former injecting drug users: a survey from Indonesia. BMC Publ. Health. 2010;10:472. doi: 10.1186/1471-2458-10-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.United_Nations_Children's_Fund_(UNICEF) 2022. HIV/AIDS Indicator: Mother-To-Child HIV Transmission Rate UNAIDS 2021 Estimates.http://aidsinfo.unaids.org/ [Google Scholar]
- 71.Communication_and_Public_Service_Ministry_of_Health_Republic_of_Indonesia . 2019. Rekam Jejak ARV Pertama di Indonesia Sampai Menurunkan Angka Kematian ODHA.https://sehatnegeriku.kemkes.go.id/baca/umum/20191127/2232393/rekam-jejak-arv-pertama-indonesia-sampai-menurunkan-angka-kematian-odha/ [Google Scholar]
- 72.Kassanjee R., Pilcher C.D., Keating S.M., Facente S.N., McKinney E., Price M.A., Martin J.N., Little S., Hecht F.M., Kallas E.G., et al. Independent assessment of candidate HIV incidence assays on specimens in the CEPHIA repository. AIDS. 2014;28:2439–2449. doi: 10.1097/QAD.0000000000000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murphy G., Pilcher C.D., Keating S.M., Kassanjee R., Facente S.N., Welte A., Grebe E., Marson K., Busch M.P., Dailey P., et al. Moving towards a reliable HIV incidence test – current status, resources available, future directions and challenges ahead. Epidemiol. Infect. 2017;145:925–941. doi: 10.1017/S0950268816002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gall A., Ferns B., Morris C., Watson S., Cotten M., Robinson M., Berry N., Pillay D., Kellam P. Universal Amplification, Next-Generation Sequencing, and Assembly of HIV-1 Genomes. J. Clin. Microbiol. 2012;50:3838–3844. doi: 10.1128/JCM.01516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Laethem K., Schrooten Y., Covens K., Dekeersmaeker N., De Munter P., Van Wijngaerden E., Van Ranst M., Vandamme A.-M. A genotypic assay for the amplification and sequencing of integrase from diverse HIV-1 group M subtypes. J. Virol. Methods. 2008;153:176–181. doi: 10.1016/j.jviromet.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 76.Yabar C.A., Acuña M., Gazzo C., Salinas G., Cárdenas F., Valverde A., Romero S. New Subtypes and Genetic Recombination in HIV Type 1-Infecting Patients with Highly Active Antiretroviral Therapy in Peru (2008–2010) AIDS Res. Hum. Retrovir. 2012;28:1712–1722. doi: 10.1089/aid.2012.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schultz A.K., Zhang M., Bulla I., Leitner T., Korber B., Morgenstern B., Stanke M. jpHMM: improving the reliability of recombination prediction in HIV-1. Nucleic Acids Res. 2009;37:W647–W651. doi: 10.1093/nar/gkp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Siepel A.C., Halpern A.L., Macken C., Korber B.T. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res. Hum. Retroviruses. 1995;11:1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 79.Rhee S.Y., Gonzales M.J., Kantor R., Betts B.J., Ravela J., Shafer R.W. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karyana M., Kosasih H., Samaan G., Tjitra E., Aman A.T., Alisjahbana B., Fatmawati, Gasem M.H., Gasem M.H., Arif M., et al. INA-RESPOND: a multi-centre clinical research network in Indonesia. Health Res. Pol. Syst. 2015;13:34. doi: 10.1186/s12961-015-0024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.ClinicalTrials.gov[Internet] 2018. HIV Infection and Risk Related Coinfections/Comorbidities in Indonesia (INAPROACTIVE) [Google Scholar]
- 82.World_Health_Organization . Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection: Recommendations for a Public Health Approach. 2nd edition. World Health Organization; 2016. ANNEX 10: WHO clinical staging of HIV disease in adults, adolescents and children. [PubMed] [Google Scholar]
- 83.Bio-Rad . 2010. Genscreen™ Ultra HIV Ag-Ab: Screening Kit for the Detection of HIV P24 Antigen and Antibodies to HIV-1 and HIV-2 in Human Serum/Plasma by Enzyme Immunoassay. [Google Scholar]
- 84.Saravanan S., Vidya M., Balakrishnan P., Kumarasamy N., Solomon S.S., Solomon S., Kantor R., Katzenstein D., Ramratnam B., Mayer K.H. Evaluation of two human immunodeficiency virus-1 genotyping systems: ViroSeq™ 2.0 and an in-house method. J. Virol. Methods. 2009;159:211–216. doi: 10.1016/j.jviromet.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The HIV-1 pol DNA sequence data have been deposited to the GenBank with Accession Number GenBank: OR139650 – OR139679. All data presented in this manuscript are also available online as supplementary material that can be downloaded at https://data.mendeley.com/datasets/ktg27krty7/1. All datasets generated for this study are available from the corresponding author upon reasonable request.
-
•
Sequences generated in this study have been deposited to GenBank. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.