Summary
Extensive metabolic heterogeneity in breast cancers has limited the deployment of metabolic therapies. To enable patient stratification, we studied the metabolic landscape in breast cancers (∼3000 patients combined) and identified three subtypes with increasing degrees of metabolic deregulation. Subtype M1 was found to be dependent on bile-acid biosynthesis, whereas M2 showed reliance on methionine pathway, and M3 engaged fatty-acid, nucleotide, and glucose metabolism. The extent of metabolic alterations correlated strongly with tumor aggressiveness and patient outcome. This pattern was reproducible in independent datasets and using in vivo tumor metabolite data. Using machine-learning, we identified robust and generalizable signatures of metabolic subtypes in tumors and cell lines. Experimental inhibition of metabolic pathways in cell lines representing metabolic subtypes revealed subtype-specific sensitivity, therapeutically relevant drugs, and promising combination therapies. Taken together, metabolic stratification of breast cancers can thus aid in predicting patient outcome and designing precision therapies.
Subject areas: Medical informatics, computational bioinformatics, cancer
Graphical abstract

Highlights
-
•
Discovered 3 metabolic subtypes with graded deregulation
-
•
M1 subtype not represented by any of the 50 cell line models analyzed
-
•
Bile acid biosynthesis is associated with less aggressive tumors and better prognosis
-
•
TP53 mutation and MYC amplification are predictive of metabolic deregulation
Medical informatics; Computational bioinformatics; Cancer
Introduction
Cancer cells undergo reprogramming of metabolic pathways to satisfy their metabolic demands.1 In recent years, accumulating evidence points to the role of metabolic reprogramming beyond providing energy and biomass such as influencing gene regulation,2 epigenetic regulation,3 DNA repair,4 cell cycle,5 apoptosis,6 angiogenesis,7 metastasis,8 inflammation9 and immune evasion.10 Cancer metabolism has also been implicated in chemo- and radioresistance.11,12 Consequently, cancer metabolism is perceived as a therapeutic hotspot.13 However, clinical targeting of this hallmark to get desirable results has been challenging.14,15 This in part is due to our limited understanding of the complexity and dynamics of metabolic pathways in cancer. Most of the studies exploring cancer metabolism focus on a particular metabolic perturbation. However, metabolic reprogramming is much more complicated, as it may involve the derangement of numerous pathways synergistically promoting tumor growth. Moreover, tumor metabolism exhibits flexibility and plasticity in relation to genetic heterogeneity, tissue biology and tumor microenvironment.16 Taken together, a full spectrum of metabolic perturbations needs to be elucidated and a context-specific stratification of tumors is warranted for better understanding of metabolic heterogeneity and to design coherent treatments.
Multiomics analysis of patient tumors provides insights into molecular differences, disease markers, and biological pathways, and is thus a powerful tool for precision medicine.17 In our previous work, using multiomics data, we discovered antagonistic roles of epigenetic readers CBX2 and CBX7 in breast cancer metabolism.18 Genomic and transcriptomic analysis of pan-cancer samples provided useful insights into metabolic perturbations in cancer.19,20,21,22 Studies applying pathway-based approaches on diverse cancer types provided a richer comparative understanding on how metabolism differs across different cancers.23,24 However, pathway-based analysis of metabolism across multiple cancers provides a “zoomed-out” view of the metabolic landscape within a cancer type. It is crucial to analyze a “zoomed-in” view of metabolism that exists within a cancer type to be able to understand the depth of heterogeneity and identify metabolic subtypes. With regards to breast cancer, studies have shown the existence of metabolic heterogeneity,25,26 however, a pathway-based global metabolic view of breast cancer and the clinical relevance of metabolic subtypes remains poorly understood.
Here, we use a systems approach to stratify patients based on their metabolic types, validate these patterns in independent cohorts, study the subtype genomic alterations, biological pathways, and clinical parameters, and identify likely treatment responses in a context-specific manner.
Results
Pathway-based analysis of breast cancer metabolism reveals three distinct metabolic subtypes
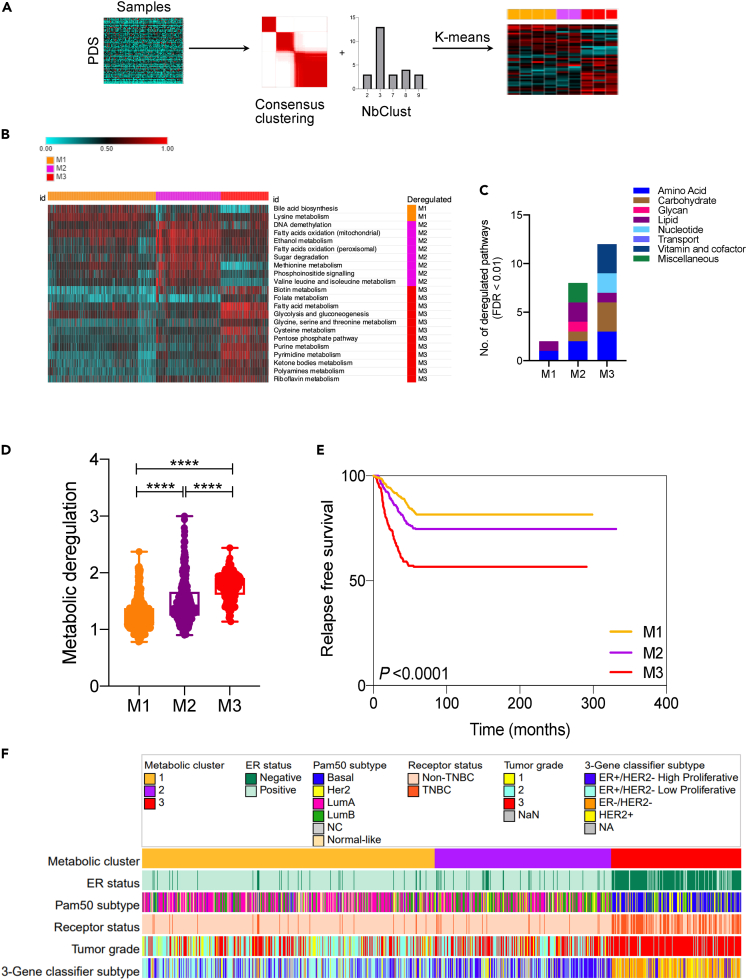
To investigate the metabolic landscape of breast tumors, we interrogated the METABRIC dataset of 1987 tumors (5 samples removed for missing expression values) and 144 normal samples. Gene sets were generated using 1519 metabolic genes from Recon1, with each set representing one of the 90 metabolic pathways.27,28 Using the gene sets’ expression data as input into Pathifier, we calculated pathway deregulation scores (PDS) for the 90 pathways in each tumor and normal sample in the discovery and validation cohorts. Pathifer is a powerful tool that converts gene-level information into pathway-level information and assigns a deregulation score based on the extent of deviation of a pathway from normal samples. Based on the PDS of 90 pathways, normal and tumor samples showed discriminating metabolic profile with much higher degree of metabolic deregulation observed in tumor samples (Figures S1A–S1C).
Moreover, the metabolic deregulation in tumors showed a high degree of heterogeneity compared to normal samples (Figure S1D), suggesting the existence of metabolic subtypes in breast tumors. Hence, we used a 2-fold approach employing consensus clustering and NbClust, to identify the number of metabolic clusters in tumor samples based on supplied PDS values (Figure 1A). Both approaches concurred strongly to suggest the number of clusters to be three, in both discovery and validation sets (Figures S2A and S2B). Then, using k-means clustering (k = 3), samples were clustered based on PDS and top deregulated pathways unique to each cluster were identified (Figure 1B). For instance, deregulated bile acid metabolism was found to be associated with the M1 subtype whereas methionine and pyrimidine metabolism were associated with M2 and M3 subtypes, respectively, along with other pathways (FDR<0.01). Overall, M3 exhibited deregulation in the highest number of pathways whereas M1 showed the least number of pathways deregulated (Figure 1C). Merging the pathways to super pathways highlighted the deregulation of lipid metabolism pathways in M2, compared to nucleotide and carbohydrate metabolism pathways in M3 (Figure 1C). Next, to measure the extent of metabolic deregulation in each cluster, Euclidean distance was calculated. As shown in Figure 1D, Euclidean distance consistently increased from M1 to M3, suggesting M1 and M3 as least and highest metabolically deregulated clusters, respectively. M2, however, exhibited somewhat intermediate deregulation.
Figure 1.
Identification of three metabolic subtypes in breast cancer
(A) Flow chart depicting the analysis pipeline. First, pathway deregulation score (PDS) was calculated for 90 metabolic pathways in each tumor sample using the Pathifier algorithm. The PDS matrix generated was then subjected to consensus clustering and NbClust to identify the best number of clusters (k). Finally, k-means clustering was used to cluster the PDS matrix using k = 3 as identified by 2-fold approach.
(B) Heatmap showing top pathways deregulated in each cluster compared to the remaining two (FDR<0.01 and PDS difference ≥0.1). PDS value varies from 0 (lowest) to 1 (highest).
(C) Bar chart showing super pathway category of deregulated pathways in M1, M2 and M3.
(D) Box and whiskers plot representing Euclidean distance as a measure of metabolic deregulation.
(E) Relapse free survival (RFS) of three metabolic subtypes with most metabolically deregulated subtype M3 showing worst and least deregulated subtype M1 best RFS.
(F) Heatmap showing the variation in key clinicopathological parameters across three metabolic subtypes. p values were calculated using Kruskal-Wallis test represented as ∗∗∗∗p < 0.0001. For the K-M survival curve, p value was calculated by log rank test.
To understand the clinical significance of differential metabolic deregulation in breast cancer, survival, and other clinicopathological indicators were examined. Notably, the extent of metabolic deregulation correlated strongly with patient outcome with M3 showing worst and M1 best prognosis (Figure 1E). Moreover, M3 subtype tumor samples were of higher grade compared to other two subtypes (Figure 1E). Interestingly, 3-gene classifiers based on ER/HER2 status and proliferation index (one of the clinicopathological indicator of METABRIC samples, Figure S4) differentiated M2 as being high proliferative compared to M1, albeit both subtypes being predominantly ER-positive (Figure 1F). Results from the clinicopathological analysis for both cohorts are provided as Figures S3 and S4. All results were reproducible in the validation set (Figure S2). Additionally, we queried the TCGA dataset, and all key findings were reproduced (Figure S5). In summary, these results demonstrate the existence of three major metabolic subtypes in breast cancer with varying degree of metabolic perturbations, patient outcome, and other clinicopathological indicators.
Genomic characterization of metabolic subtypes
Amplifications and mutations in key genes are associated with metabolic reprogramming in cancer.29 For instance, MYC amplification and TP53 mutations have been linked with the rewiring of key metabolic pathways in cancer.30,31 For the identification of mutations related with each subtype, we focused on forty driver mutations reported in the METABRIC dataset32 and their association with clusters were tested using the Chi-square test. Contingency tables showing the frequency of top 5 mutations in each cluster are displayed in Figure 2A. TP53 mutation is predominant in M3 (80% mutated samples) followed by M2 (28%) and M1 (15%). On the contrary, the PIK3CA mutation showed the opposite trend with highest in M1 (47% mutated samples), by M2 (39%) and M3 (24%) (Figure 2A). GATA3, MAP3K1 and CDH1 showed a mutation trend such as PIK3CA, that is, decreasing frequency from M1 to M3.
Figure 2.
Genomic characterization of metabolic clusters in breast cancer
(A) The Chi-Square test was applied to mutation data for forty genes in METABRIC to test for associations with the metabolic clusters. The top five genes based on the resulting p values are shown in the contingency tables. Pearson’s correlation between these genes’ mutations and metabolic deregulation is shown in the heatmap.
(B) The Kruskal-Wallis test was applied to CNV data for 1568 (frequency cut-off 10%) genes to test for associations with the metabolic clusters. The top 5 genes based on the resulting p values are shown via boxplots. Pearson’s correlation between these genes’ CNV and metabolic deregulation is shown in the heatmap.
To understand the relation between driver mutations and metabolic subtypes, we performed a correlation analysis of top five driver mutations with all metabolic pathways. Heatmaps revealed strong correlations of TP53 mutation with nucleotide metabolism (positive) and bile acid metabolism (negative) (chi-square test; p < 0.05) (Figure 2A). The correlation behavior of PIK3CA mutation was less pronounced and opposite to that of TP53.
To study the copy number variations (CNVs) associated with metabolic clusters, cancer genes (OncoKB) with CNVs in 10% or more samples were selected and subjected to Kruskal-Wallis testing. All top 5 genes, based on resulting p values, were amplified to the highest degree in M3, followed by M2 and with least amplification in M1 (Kruskal-Wallis test; p < 0.05) (Figure 2B). The difference in copy number gains for all 5 genes is much less in M2/M3 compared to M1/M3 and M1/M2 scenario, consistent with metabolic deregulation (Figure 1). The correlation of CNVs with all metabolic pathways exhibited a graded pattern with highest correlations with pyrimidine metabolism and other M3 deregulated pathways followed by M2 and M1 pathways (Figure 2B). As noted in the heatmap, bile acid and methionine metabolism were found to be negatively correlated with gene amplification (Figure 2B). Combined with mutation data, the M3 subtype demonstrated the strongest correlation between genomic alterations and metabolism.
Metabolic subtypes and their association with cancer hallmarks
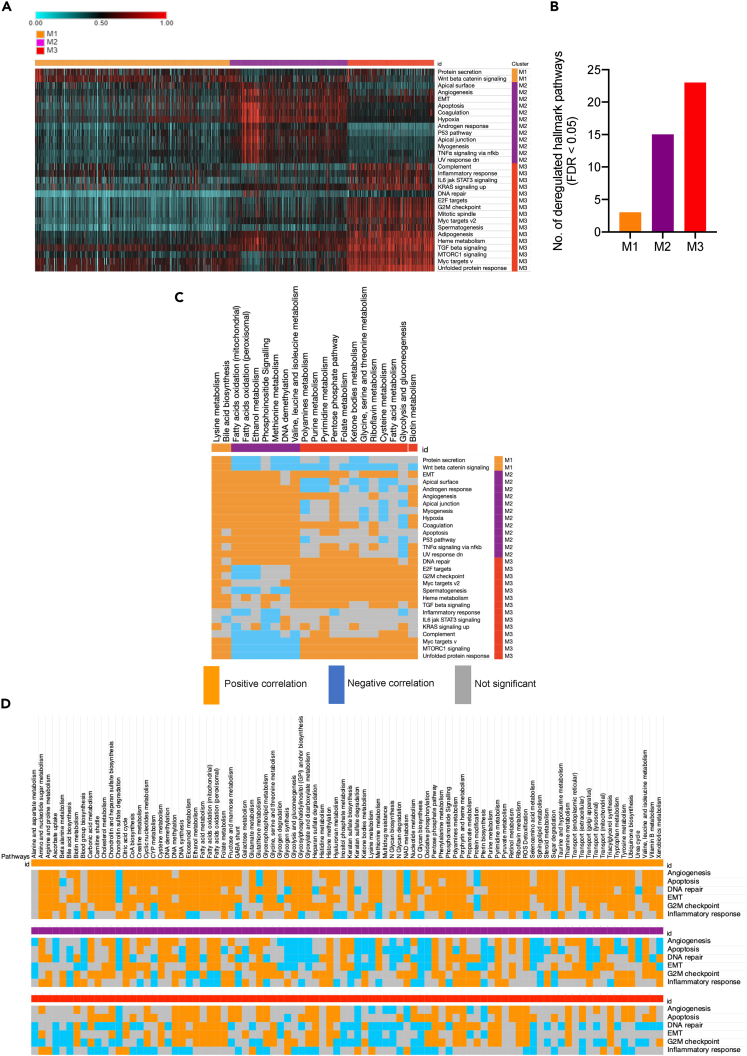
To assess the biological relevance of the metabolic clusters, we calculated the PDS of 50 cancer hallmark pathways (from MSigDB) in each sample and analyzed their deregulation. As shown in the Figure 3A heatmap, such as metabolic deregulation, deregulation of hallmark pathways increased from M1 (lowest) to M3 (highest), FDR < 0.05 (Figures 3A and 3B). Out of six major cancer hallmarks, none was found to be deregulated in M1, consistent with this cluster’s pattern of least metabolic deregulation and better patient outcome (Figures 1D and 1E). However, three hallmarks (apoptosis, EMT, and angiogenesis) were deregulated in M2 and another three (inflammatory response, G2M checkpoint, and DNA repair) were found to be deregulated in M3. Metabolic pathways deregulated in each subtype were positively correlated with each cluster’s hallmark pathways (Figure 3C). For instance, mTORC1 signaling was found to be positively correlated with M3 metabolic pathways but negatively correlated with M2 metabolic pathways. However, the inflammatory response pathway did not show any positive correlation with M3 or other the two clusters for that matter.
Figure 3.
Metabolic subtypes and associated cancer hallmarks
(A) Heatmap showing PDS of cancer hallmark pathways (MSigDB); only the top hallmarks pathways deregulated in each cluster compared to remaining two (FDR < 0.01 and PDS difference ≥ 0.1) are shown.
(B) Bar chart showing number of significantly deregulated pathways in M1, M2 and M3.
(C) The heatmap showing directional correlation between top metabolic and hallmark pathways in each cluster (p < 0.05).
(D) The correlation of all 90 metabolic pathways with six major cancer hallmarks pathways in each metabolic subtype. Only significant correlations are shown (p < 0.05).
Next, to understand whether the correlation between metabolic and hallmark pathways is cluster-specific, we studied the association of all metabolic pathways (in all three clusters) with six cancer hallmarks. Interestingly, a differential correlation of metabolic pathways with six cancer hallmarks was observed in three metabolic subtypes (Figure 3C). In other words, correlation between metabolism and hallmark pathways was found to be dependent on the cluster type. For example, the glycolysis pathway showed a positive correlation with all six hallmarks in M1, but a predominantly negative correlation in M2 and was relatively insignificant in M3. On the contrary, pyrimidine metabolism showed a consistent positive correlation with all six hallmarks in all three clusters. Of note, the M1 subtype showed an overall positive correlation between metabolic pathways and cancer hallmarks. Overall, these results highlight that the relationship between metabolic and biological pathways is contextual and may change depending on the metabolic subtype of breast tumor.
Machine-learning analysis of the clusters reveals the predictive potential of metabolism
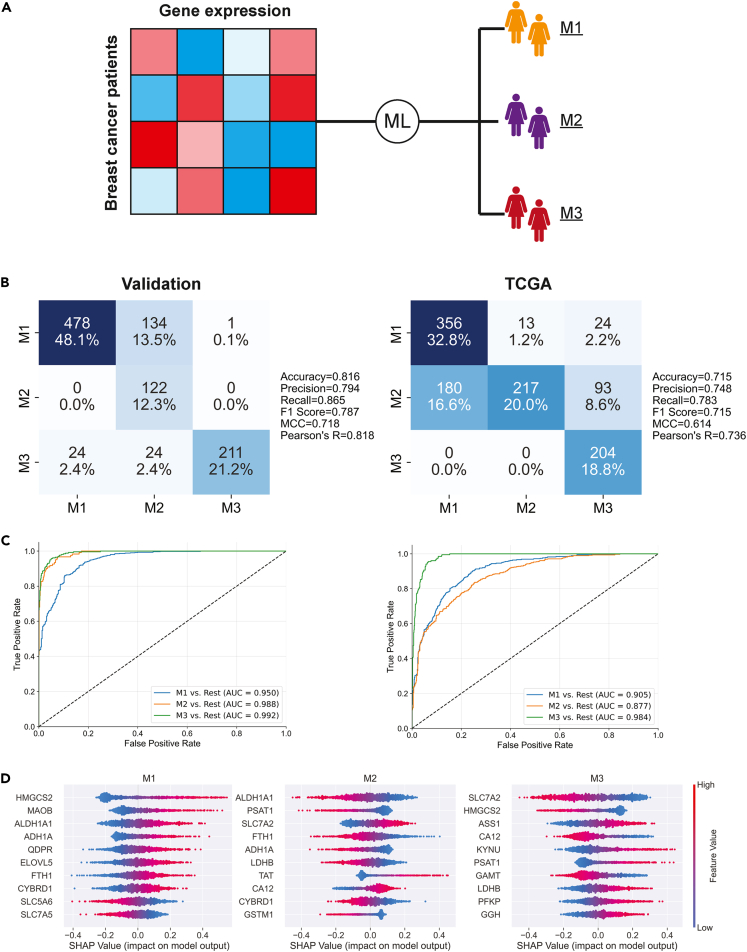
After applying the unsupervised clustering to the METABRIC and TCGA datasets, we set out to show that the clusters could be recaptured using supervised machine-learning methods. The goal was to use metabolic transcriptomics data to predict the cluster labels (Figure 4A). Transcriptomics data from the discovery cohort was used to train the initial ridge regression model. This included 993 samples and 1330 metabolic genes. Cross-validation was used to test the model’s robustness, in addition to tuning the regularization hyperparameter, alpha. Aggregation of the 5-fold cross-validation predictions indicated that the model was performing well (accuracy = 0.913, Matthew’s correlation coefficient (MCC) = 0.863). A final model was then trained on the entire discovery dataset, using the tuned value of alpha from the best-performing cross-validation fold. We found that the model tended to perform better with stronger regularization. This was expected due to the large number of input features (genes), as the regularization parameter prevents the model from over-fitting by reducing the coefficients of highly correlated and non-contributing features.
Figure 4.
Supervised machine-learning analysis of the metabolic clusters
(A) Gene expression samples from the discovery dataset were labeled with the unsupervised metabolic cluster results and used to train a ridge regression model.
(B and C) Confusion matrices and ROC curves showing the results of the ridge regression model’s classification of the validation and TCGA samples.
(D) Shapley values indicate which genes were most important for the ridge model’s classification of each cluster.
The fully trained model was used to make predictions on the validation and TCGA datasets, where it also showed good performance (MCC > 0.614, AUC > 0.877 in both datasets) (Figures 4B and 4C). The strong performance in TCGA data, which used a different transcriptomics methodology than the METABRIC training data, suggests that the model is able to learn robust and generalizable signatures for each cluster. The ROC curves were generated using the one-vs-rest method, where the classification of each cluster versus the other two is measured. The confusion matrices and ROC curves indicate that the model had the most difficulty separating patients in the M1 and M2 clusters. This was expected, as these clusters are metabolically similar relative to M3.
Shapley values were generated for the validation samples, using the discovery-trained model as the input for the SHAP package in Python.33 SHAP summary plots were created for each class and were used to determine which genes are most positively and negatively associated with the clusters, in regards to our model (Figure 4D). Expression of HMGCS2 (steroid metabolism) was positively associated with M1 and negatively with M3. Meanwhile, the model heavily relied on ALDH1A1 (ethanol metabolism) to differentiate between the M1 (lower expression) and M2 (higher expression) classes. Lower expression of SLC7A2 was the largest driving factor for the model’s prediction of the M3 cluster.
Metabolomics data is consistent with gene expression-based pathway analysis
To investigate whether gene expression-based pathway analysis of metabolic subtypes is consistent with metabolite levels in tumors, we used metabolomics data of 67 breast tumors from Terunuma et al.34 First, the using machine-learning approach described in Figure 4, we predicted and labeled tumors as M1/M2/M3. Thereafter, we compared 344 annotated metabolites (representing 63 pathways) across three subtypes. A differential metabolite abundance was observed in the three clusters, as shown in the Figure 5A heatmap (top 5 pathways indicated, FDR < 0.05). A similar pattern of perturbation at the metabolite level was observed, with M3 showing maximal increase in metabolite levels, followed by M2 and M1 (Figure 5B).
Figure 5.
Validation of metabolic subtypes using metabolite data
(A) Heatmap showing difference in metabolite levels (FDR < 0.05) across three metabolic subtypes; top 5 pathways are indicated. Metabolomics data of breast tumors from Terunuma et al.34
(B) Survival and metabolic deregulation (Euclidean distance) based on metabolite data.
(C–G) Boxplots showing the differences in metabolites of fatty acid C), pyrimidine D), glycolysis E), pentose phosphate pathway F) and bile acid metabolism G) in M1, M2 and M3. Boxplots represent minimum, maximum and median. p values were calculated using Kruskal-Wallis test represented as represented as ∗p < 0.03, ∗∗p < 0.0021, ∗∗∗p < 0.0002, ∗∗∗∗p < 0.0001.
Patient outcome across the three clusters suggested the worst prognosis for M3 and best for M1, consistent with the transcriptomics-based survival analysis (compare Figures 1E and 5B). The log rank ptrend for survival curves was not significant though (ptrend = 0.15), possibly due to relatively lower sample size. Furthermore, we compared the abundance of metabolites representing the top 5 pathways that differ across three subtypes. Interestingly, metabolites of fatty acid (Figure 5C), pyrimidine (Figure 5D), glycolysis (Figure 5E), and pentose sugars (Figure 5F) metabolism showed highest abundance in M3, whereas bile acid metabolites (Figure 5G) were upregulated in M1. To summarize, the metabolite-level results were consistent with data obtained from the gene expression-based pathway analysis, demonstrating the robustness of identified metabolic subtypes in breast cancer.
Metabolic subtypes show differential sensitivity to drugs
Next, we were interested in understanding whether cell line characteristics such as doubling time and drug sensitivity are affected by metabolic subtypes. To this end, we first predicted and labeled cell lines as M1 or M2 or M3, using the same machine learning pipeline as described above. Expression data of cell lines for prediction purposes was taken from Heiser et al.35 We trained an additional ridge model for making predictions on the unlabeled cell lines using the entire METABRIC dataset, with the intention of using all available training data in order to ensure that the cell line cluster predictions were robust. Surprisingly, out of 56 cell lines, no cell line was predicted to represent M1 metabolic subtype. To rule out algorithmic bias, we repeated the prediction using prediction analysis of microarrays (PAMR), a nearest shrunken centroid classifier,36 and obtained the same results with no cell line predicted for M1 metabolic subtype. A possible explanation could be difficulty in establishing cell lines from less aggressive tumors.37
The doubling time of M3 cell lines was found to be significantly lower than M2 cell lines (p < 0.0021), in agreement with the higher metabolic deregulation in M3 compared to M2 (Figure 6A). Furthermore, cell lines representing the M2 and M3 clusters differed significantly in their response to metabolic inhibitors. For example, M3 cell lines were found to be more sensitive to inhibitors of pyrimidine (leflunomide), fatty acid (C75), and glycolysis (CAP232), in accordance with higher deregulation of these three metabolic pathways in M3 tumor samples (Figure 6A). On the other hand, M2 cell lines were found to be more sensitive to methionine deprivation compared to M3 cells, corresponding with higher deregulation of methionine metabolism in M2 compared to M3 subtype tumors (Figure 6B). To evaluate if metabolic inhibition could improve the effect of chemotherapeutic drugs, a combination of leflunomide (pyrimidine inhibitor) or C75 (fatty acid synthesis inhibitor) with doxorubicin was used. Importantly, up to 35% improved the inhibition of cell growth by doxorubicin was observed in two different M3 group cell lines (Figure 6C).
Figure 6.
Metabolic subtypes and sensitivity to drugs
(A) Boxplot showing M2 and M3 comparison of doubling time, sensitivity to leflunomide (pyrimidine synthesis inhibitor), C75 (fatty acid synthesis inhibitor) and CAP232 (glycolysis inhibitor).
(B) Methionine dependency tested on four cell lines collectively representing M2 and M3 subtype.
(C) Cell viability analysis in presence of 10 μg/mL C75, 100 μmol/L leflunomide (LFD) and 1 μmol/L doxorubicin for 24 h in M3 representative cell lines MDA-MB-231 and MDA-MB-468. For combination treatments, cells were either pretreated with C75 or leflunomide for 12 h before incubation with doxorubicin for another 48 h. DMSO was used as vehicle control.
(D) Drug sensitivities heatmap of cell lines representing M2 and M3 subtype based on data from Heiser et al.35 Vertical red and purple bars indicate top drugs (p < 0.01) to which M3 and M2 cell lines, respectively, are most sensitive. Gray triangles indicate the direction of decreasing (inverted) or increasing (upright) drug sensitivities.
(E) Heatmap showing difference in sensitivities of M2 and M3 type cell lines to repurposing drugs (PRISM). Top 10 drug sensitivities are shown (FDR < 0.05). Viability after 24 h brigatinib treatment compared experimentally in T47D (M2) and MDA-MB-231 (M3) cell lines. Boxplots represent minimum, maximum and median. Error bars represent SEM. p values were calculated using Kruskal-Wallis or Mann-Whitney test or ANOVA and represented as ∗p < 0.03, ∗∗p < 0.0021, ∗∗∗p < 0.0002, ∗∗∗∗p < 0.0001.
To further assess the clinical utility of metabolic subtypes, sensitivity data against 77 therapeutic compounds (including FDA-approved drugs) tested on 45 different breast cancer cell lines from Heiser et al.35 was extracted. As presented in Figure 6D, we identified seven drugs against M2 and four drugs against M3 with highest sensitivities (FDR < 0.05). Moreover, we evaluated the PRISM database, which records sensitivities of large numbers of cancer cell lines against > 4500 compounds, to identify repurposing drugs.38 Brigatinib and arecaidine ester were the most effective inhibitors of M3 and M2 cell lines, respectively (Figure 6E; FDR < 0.05). Upon experimental testing, the M3 cell line MDA-MB-231 was found to be substantially more sensitive compared to M2 cell line T47D which showed negligible response to brigatinib dosage (p < 0.0002).
Discussion
Since the discovery of deranged metabolism in cancer cells by Otto Warburg in the 1920s, mounting evidence has been generated that underlines the biological significance of metabolic transformation in neoplastic progression. It is now well-recognized that metabolic rewiring confers a variety of benefits including the promotion of key cancer hallmarks.39 Furthermore, metabolic capacities drive clonal selection during tumor evolution.40 Consequently, metabolic reprogramming is considered a hallmark of therapeutic relevance.13 However, such as genetic heterogeneity, metabolic heterogeneity could be a challenge in the clinical management of cancer. Therefore, a systematic stratification of patients based on metabolic subtype is required for tailoring treatments targeting metabolic liabilities of cancer. Accordingly, the present study was designed to investigate the metabolic landscape of breast cancer and to identify metabolic subtypes of clinical relevance.
Analysis of transcriptomics-based deregulation scoring of 90 metabolic pathways provided the first line of evidence that within breast cancer three metabolic subtypes exists (Figure 1; Figure S1). The use of Euclidean distance metrics provided evidence for the extent of metabolic deregulation, and a clear pattern of least (M1), intermediate (M2), and most deregulated (M3) subtypes emerged. Of note, both M1 and M2 samples are largely ER positive, yet their metabolism was found to be different with M2 showing greater deregulation of pathways than M1 (Figures 1B–1D; Figure S2). Interestingly, METABRIC 3-gene classifier’s high proliferative samples populated significantly more in M2 compared to M1 (Figure 1). This endorses the relation of cancer metabolism with cell proliferation and rightly so because cancer metabolism provides for the biosynthetic and energetic demands of growing tumor.1 The higher bile acid deregulation in the M1 subtype is consistent with reports where the accumulation of bile acids in breast tumors improves clinical outcome.41 Moreover, the deregulation of methionine metabolism in M2 is in agreement with the study by Toker and colleagues where they demonstrate that oncogenic PIK3CA creates methionine dependency in breast cancer.42 We also observed deregulated ethanol metabolism in M2, which was in concurrence with work documenting the role of ethanol in the promotion of proliferation of ER positive M2-type cell lines such as MCF7 and ZR75.1.43 In short, these consistencies not only highlight the reliability of results, but also underscore the diverse metabolic dependencies in breast cancer. Moreover, the variation in survival and clinical parameters among the three clusters emphasize how metabolic subtype is informative about both tumor characteristics and patient outcome (Figures 1E and 1F; Figures S1F–S1G). The reproducibility of gene expression-based pathway analysis (Figure 1) at metabolite levels in patients with breast cancer (Figure 5) conveys the robustness of metabolic subtypes. It also demonstrates that metabolic gene expression reflects well on patient tumor metabolism.
Genomic characterization revealed how oncogenic drivers vary across metabolic clusters. For instance, mutations in TP53 and PIK3CA behaved contrastingly with regards to their correlation with metabolic pathways (Figure 2A). Likewise, MYC amplification correlated positively with M3 pathways, such as pyrimidine metabolism, but negatively with bile acid and methionine metabolism, pathways deregulated in M1 and M2 clusters, respectively (Figure 2B). Unlike mutations, the correlation patterns of PDS across the top 5 amplified genes was well graded, which is possibly due to a similar pattern of increase in copy number and metabolic deregulation from M1 to M3 (Figures 1D and 2B). Overall, genomic analysis of metabolic clusters revealed that genetic heterogeneity may translate into metabolic heterogeneity, as different oncogenic drivers may result in different metabolic dependencies.16
The deregulation pattern of hallmark pathways in the three clusters was quite similar to the pattern of metabolic alterations observed, suggesting a systemic deregulation due to the distinct biology of each cluster (Figures 3A and 3B). However, the positive correlation patterns between uniquely deregulated metabolic and hallmark pathways in each subtype indicate an interdependence between metabolism and other biological processes in a cluster-specific manner (Figure 3C). Significantly, the direction of correlation between metabolic and hallmark pathways appears to be dependent on cluster type, suggesting a heterogeneous role of breast cancer metabolism in relation to the biology of each subtype (Figure 4D).
Big data analysis using machine learning is a powerful approach for stratifying patients for precision medicine.44 Our supervised machine learning model showed strong and robust accuracy in both the METABRIC validation and TCGA datasets. Although the model performance decreased when evaluated on the TCGA dataset, this drop in accuracy could be attributed to differences in measurement platform (microarray vs. RNA-seq) between METABRIC and TCGA. Further, most machine learning models tend to underperform beyond the data they were trained on.45 Hence considering these issues, our model’s high AUC of 0.87 in TCGA is quite remarkable. This model can be used to classify new samples into these three clinically relevant subtypes. It also allowed us to classify model cell lines representing metabolic subtypes to enable targeted drug discovery.
Metabolic reprogramming is perceived as a promising candidate for anti-cancer therapeutics.46 However, heterogeneity in metabolism needs to be characterized in a subtype-specific manner to be able to exploit the metabolic liabilities successfully. In line with this view, we attempted to identify drugs to which cell line model of each metabolic subtype is responsive. Cell line panels are considered an excellent tool for the preclinical testing of drugs to efficiently identify responsive subtypes for guiding clinical trials.35 Drugs such as gefitinib, trastuzumab, lapatinib, imatinib have been identified using cell line panels.47,48,49,50 The M2 and M3 subtypes showing maximal sensitivity to rapamycin (mTOR inhibitor) and GSK461364 (Polo-like kinase 1 inhibitor), respectively, is a clinically relevant observation suggesting that M2-and M3-type patients could benefit from these drugs (Figure 6D). Incidentally, Polo-like kinase 1 (PLK1) inhibitor is over-expressed in aggressive M3-like breast cancers.51 Additionally, our identification of the M3 subtype sensitivity to brigatinib is an interesting observation with potential clinical implications (Figure 6E). Brigatinib is an FDA approved drug used for the treatment of patients with metastatic anaplastic lymphoma kinase (ALK)-positive non-small cell lung cancer (NSCLC).
In conclusion, the metabolism of breast cancer exhibits significant heterogeneity and three metabolic subtypes exist, each with a unique fingerprint of metabolic deregulation. Further, the metabolic make-up of each subtype is informative about patient outcome and other clinically relevant parameters including sensitivity to anti-cancer drugs. Finally, the results presented suggest targeting metabolic behavior in a personalized manner could be a way forward in breast cancer therapy.
Limitations of the study
Our study does have certain limitations. The transcriptomics samples in METABRIC were collected from the UK and Canadian tumor banks with a large majority of patients with European ancestry, and this data may not be representative of other ethnic and racial groups. Furthermore, we have extrapolated tumor metabolic activity using transcriptomics and metabolomic data. Gene expression and metabolite levels do not strongly correlate with metabolic pathway flux. Moreover, given the dynamic nature of metabolism, metabolomics-based subtypes may not be as consistent as gene expression-based subtypes across datasets. Tumor metabolism may also interconvert between different metabolic subtypes. More complex computational models built using temporal patient samples can be applied in the future to dissect these dynamic nonlinear effects.52 Finally, we have validated promising vulnerabilities for each subtype using cell line models; while cell lines have been a great tool in cancer drug discovery, sensitivities observed in vitro may not necessarily reflect in vivo tumor sensitivity to a drug.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Brigatinib | MedChemExpress | Cat#HY-12857 |
| C75 | Sigma-Aldrich | Cat#C5490 |
| Doxorubicin | Sigma-Aldrich | Cat#D5220 |
| Fetal Bovine Serum | Gibco | Cat#16000044 |
| Leflunomide | Sigma-Aldrich | Cat#L5025 |
| Penicillin/Streptomycin | Gibco | Cat#15-140-122 |
| Methionine deficient media | Gibco | Cat#A1451701 |
| Critical commercial assays | ||
| CCK8 kit | Abcam | Cat#ab228554 |
| Deposited data | ||
| Code availability | This paper | https://github.com/kirksmi/BreastCancerClustering |
| METABRIC microarray data | European Genome-Phenome Archive |
https://ega-archive.org accession number: EGAD00010000210, EGAD00010000211, and EGAD00010000212. |
| METABRIC copy-number and mutation data | cbioportal | https://www.cbioportal.org |
| TCGA mRNA-seq and clinical data | UCSC Xena | http://xena.ucsc.edu |
| Drug sensitivity data | ArrayExpress E-MTAB-181 | https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-181 |
| Metabolomics data | Terunuma et al. (GSE37751) | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3871244/ |
| Experimental models: Cell lines | ||
| MDA-MB-231 | ATCC | ATCC HTB-26 |
| MDA-MB-468 | ATCC | ATCC HTB-132 |
| MCF7 | ATCC | ATCC HTB-22 |
| T47D | ATCC | ATCC HTB-133 |
| Software and algorithms | ||
| Consensus clustering | Gene pattern version 2.0, GSEA | http://www.broadinstitute.org/gsea/ |
| GraphPad Prism v8 | GraphPad prism software | https://www.graphpad.com/features |
| MSigDB | The Broad Institute | http://software.broadinstitute.org/gsea/msigdb/index.jsp |
| Morpheus | Broad institute | https://software.broadinstitute.org/morpheus/ |
| Pathifier | Bioconductor package | https://www.bioconductor.org/packages/release/bioc/html/pathifier.html |
| Nbclust | CRAN | https://cran.r-project.org/web/packages/NbClust/index.html |
| PRISM | Broad institute | https://depmap.org/repurposing |
| Python version | Python Software Foundation | https://www.python.org |
| SHAP | Python | https://shap.readthedocs.io/en/latest/ |
Resource availability
Lead contact
Mohammad Askandar Iqbal (dr.askandar@gmu.ac.ae).
Materials availability
All data are provided in figures and in the supplementary materials. No materials were generated in this study.
Method details
Pathway analysis, clustering, metabolic and clinical analysis of patients with breast cancer
From the European-Genome Archive (EGA), we obtained transcriptomics and clinical data (N = 1992 tumor, 144 normal) of the METABRIC dataset. Details about the METABRIC patient population, consents, approvals, tissue collection and sample processing are provided in the publication by Curtis et al.53 Tumor samples were randomly split into discovery (N = 993) and validation (N = 994) cohort using RAND function in excel. We used 1500 metabolic genes from Recon127 to represent 90 metabolic pathways.28 TCGA-BRCA data of 1104 tumor and 114 normal sample data were obtained from UCSC Xena. Pathifier was used to calculate pathway deregulation score (PDS) of metabolic and MSigDB hallmark pathways for each tumor sample. Pathifier calculates deregulation scores based on supplied expression data of gene sets and PDS values are normalized between 0 and 1.54 Consensus clustering and NbClust were used to identify the number of clusters in breast tumor samples based on the PDS of 90 pathways in each sample. Consensus clustering is a robust method that classifies samples into clusters and provide metrics to evaluate their stability along with a heatmap for visualization.55 NbClust uses 30 indices to determine the number of clusters in a dataset and the number supported by the maximum number of indices is usually selected.56 Once the cluster number (k = 3) was determined by consensus and NbClust, k-means clustering was performed and heatmaps were prepared as described here.18 For the validation of subtypes at the metabolite level, metabolomics, transcriptomics, and survival data of 67 patients with breast cancer were obtained from Terunuma et al.34 Kaplan–Meier curves were prepared for survival analysis; p values were calculated using the log rank test in GraphPad software.
Genomic analysis
Copy number and mutation data of the METABRIC dataset were taken from cBioportal. For copy number, genes with more than 10% frequency in tumors were subjected to Kruskal-Wallis test to select the ones with significant copy number changes in metabolic clusters. For mutation analysis, 40 driver mutation genes in METABRIC32 were studied across metabolic subtypes and chi-square tests were used to find out significant associations.
Machine learning analysis
A ridge regression classification model was trained using the Scikit-Learn package in Python. This algorithm was well-suited for our scenario, in which there were many features (metabolic genes), some of which are likely to be highly correlated. Ridge regression incorporates L2 regularization, which adds a penalty to the loss function and helps prevent overfitting. This method helps deal with multi-collinearity and drives the coefficients of less important features to zero, making it easier to determine which genes are most associated with the clusters. First, a model was trained on the discovery subset of the METABRIC data, which we referred to as the discovery dataset (994 samples x 1330 metabolic genes). We first used 5-fold cross-validation on this training data to evaluate the model’s robustness before training a final model on the entire dataset. The final model’s performance was measured by predicting on the METABRIC-validation (993 samples) and TCGA (1087 samples) datasets. For the ensuing cell line drug sensitivity analysis, we trained an additional model using the entire METABRIC dataset, to increase model's robustness. The TCGA data was not included due to known differences in the transcriptomics distributions.
For determining which metabolic genes were most important in the ridge regression model we used the SHAP (SHapley Additive exPlanation) package in Python to generate Shapley values.33 Based on cooperative game theory, Shapley values are essentially calculated by iteratively adding and removing features from a machine-learning model and using the average change in the model outcome as a measure of the feature’s contribution. This information provided insight into whether the ridge model was more likely to predict the M1, M2 or M3 class as the expression of each gene either increases or decreases.
Drug sensitivity analysis
Drug sensitivity data of breast cancer cell lines against metabolic inhibitors C75, leflunomide and CAP232 were taken from Genomics of Drug Sensitivity in Cancer (GDSC). Doubling time and drug data in Figure 6D were obtained from Heiser et al.35; Z-scores of -log10(GI 50) values were used for heatmap. Repurposing drug sensitivity data (Figure 6E) were obtained from the PRISM resource.38
Cell culture experiments
MDA-MB-231, MDA-MB-468, T47D and MCF7 breast cancer cell lines were procured from the American Type Culture Collection (ATCC), authenticated through STR profiling and maintained as described.57 Briefly, cell lines were either grown in DMEM or RPMI media (Gibco, ThermoFisher Scientific Inc., Waltham, MA, USA) supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco). C75, leflunomide and doxorubicin were purchased from Sigma-Aldrich (St. Louis, MO, USA) and dissolved in DMSO to prepare stocks of recommended concentrations and stored at −80°C until further use. For brigatinib treatment: T47D and MDA-MB-231 cells were treated with indicated concentrations of brigatinib (MedChemExpress, NJ, USA) for 24 h. For methionine deprivation: MDA-MB-231, MDA-MB468, T47D and MCF7 cells were seeded in 24 or 96 well plates and after 24 h spent media was removed and replaced with methionine deficient media and incubated for additional 24 h. Viability measurements were done using CCK8 assay (Abcam, Cambridge, UK).
Statistical analysis
All experiments were performed in three independent replicates to calculate mean and its standard error (SEM). Unless otherwise noted, Kruskal-Wallis or Mann-Whitney tests were used to calculate significance and Benjamini-Hochberg procedure was used to calculate false discovery rate (FDR). p < 0.05 was considered statistically significant and denoted as ∗p < 0.03, ∗∗p < 0.0021, ∗∗∗p < 0.0002, ∗∗∗∗p < 0.0001.
Acknowledgments
Internal research grant by Gulf Medical University (M.A.I.) and startup funds from the University of Michigan (S.C.). Funders had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Author contributions
MAI conceived and designed the study. MAI, SS, KS, PS, and BK participated in data acquisition. MAI, SS, KS, PS, BK, SCh and SC participated in the data analysis and interpretation. MAI, SS, and KS prepared figures. MAI wrote the article with contributions from KS and SC. MAI and SC participated in study supervision, critical reviewing of article and decision to submit final version. All authors contributed to article revision.
Declaration of interests
SC has served as a consultant for Axcella Health and Tempus. The authors declare that they have no other competing interests.
Published: September 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108059.
Contributor Information
Mohammad A. Iqbal, Email: dr.askandar@gmu.ac.ae.
Sriram Chandrasekaran, Email: csriram@umich.edu.
Supplemental Information
Data and code availability
-
•
No new datasets were generated during the current study.
-
•
Codes used are made available at https://github.com/kirksmi/BreastCancerClustering.
-
•
Information and requests for resources and any additional data should be directed to and will be fulfilled by the lead contact.
References
- 1.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enzo E., Santinon G., Pocaterra A., Aragona M., Bresolin S., Forcato M., Grifoni D., Pession A., Zanconato F., Guzzo G., et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015;34:1349–1370. doi: 10.15252/embj.201490379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J.V., Carrer A., Shah S., Snyder N.W., Wei S., Venneti S., Worth A.J., Yuan Z.F., Lim H.W., Liu S., et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turgeon M.O., Perry N.J.S., Poulogiannis G. DNA Damage, Repair, and Cancer Metabolism. Front. Oncol. 2018;8:15. doi: 10.3389/fonc.2018.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalucka J., Missiaen R., Georgiadou M., Schoors S., Lange C., De Bock K., Dewerchin M., Carmeliet P. Metabolic control of the cell cycle. Cell Cycle. 2015;14:3379–3388. doi: 10.1080/15384101.2015.1090068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuura K., Canfield K., Feng W., Kurokawa M. Metabolic Regulation of Apoptosis in Cancer. Int. Rev. Cell Mol. Biol. 2016;327:43–87. doi: 10.1016/bs.ircmb.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiménez-Valerio G., Casanovas O. Angiogenesis and Metabolism: Entwined for Therapy Resistance. Trends Cancer. 2017;3:10–18. doi: 10.1016/j.trecan.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Nokin M.J., Bellier J., Durieux F., Peulen O., Rademaker G., Gabriel M., Monseur C., Charloteaux B., Verbeke L., van Laere S., et al. Methylglyoxal, a glycolysis metabolite, triggers metastasis through MEK/ERK/SMAD1 pathway activation in breast cancer. Breast Cancer Res. 2019;21:11. doi: 10.1186/s13058-018-1095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaughan R.A., Garcia-Smith R., Dorsey J., Griffith J.K., Bisoffi M., Trujillo K.A. Tumor necrosis factor alpha induces Warburg-like metabolism and is reversed by anti-inflammatory curcumin in breast epithelial cells. Int. J. Cancer. 2013;133:2504–2510. doi: 10.1002/ijc.28264. [DOI] [PubMed] [Google Scholar]
- 10.Lim S.O., Li C.W., Xia W., Lee H.H., Chang S.S., Shen J., Hsu J.L., Raftery D., Djukovic D., Gu H., et al. EGFR Signaling Enhances Aerobic Glycolysis in Triple-Negative Breast Cancer Cells to Promote Tumor Growth and Immune Escape. Cancer Res. 2016;76:1284–1296. doi: 10.1158/0008-5472.CAN-15-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao F., Ming J., Zhou Y., Fan L. Inhibition of Glut1 by WZB117 sensitizes radioresistant breast cancer cells to irradiation. Cancer Chemother. Pharmacol. 2016;77:963–972. doi: 10.1007/s00280-016-3007-9. [DOI] [PubMed] [Google Scholar]
- 12.Li Q., Qin T., Bi Z., Hong H., Ding L., Chen J., Wu W., Lin X., Fu W., Zheng F., et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat. Commun. 2020;11:1456. doi: 10.1038/s41467-020-15308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vander Heiden M.G. Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 14.Papaldo P., Lopez M., Cortesi E., Cammilluzzi E., Antimi M., Terzoli E., Lepidini G., Vici P., Barone C., Ferretti G., et al. Addition of either lonidamine or granulocyte colony-stimulating factor does not improve survival in early breast cancer patients treated with high-dose epirubicin and cyclophosphamide. J. Clin. Oncol. 2003;21:3462–3468. doi: 10.1200/JCO.2003.03.034. [DOI] [PubMed] [Google Scholar]
- 15.Long G.V., Dummer R., Hamid O., Gajewski T.F., Caglevic C., Dalle S., Arance A., Carlino M.S., Grob J.J., Kim T.M., et al. Epacadostat plus pembrolizumab versus placebo plus pembrolizumab in patients with unresectable or metastatic melanoma (ECHO-301/KEYNOTE-252): a phase 3, randomised, double-blind study. Lancet Oncol. 2019;20:1083–1097. doi: 10.1016/S1470-2045(19)30274-8. [DOI] [PubMed] [Google Scholar]
- 16.Kim J., DeBerardinis R.J. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab. 2019;30:434–446. doi: 10.1016/j.cmet.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akhoundova D., Rubin M.A. Clinical application of advanced multi-omics tumor profiling: Shaping precision oncology of the future. Cancer Cell. 2022;40:920–938. doi: 10.1016/j.ccell.2022.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Iqbal M.A., Siddiqui S., Ur Rehman A., Siddiqui F.A., Singh P., Kumar B., Saluja D. Multiomics integrative analysis reveals antagonistic roles of CBX2 and CBX7 in metabolic reprogramming of breast cancer. Mol. Oncol. 2021;15:1450–1465. doi: 10.1002/1878-0261.12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu J., Locasale J.W., Bielas J.H., O'Sullivan J., Sheahan K., Cantley L.C., Vander Heiden M.G., Vitkup D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 2013;31:522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsson R., Jain M., Madhusudhan N., Sheppard N.G., Strittmatter L., Kampf C., Huang J., Asplund A., Mootha V.K. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haider S., McIntyre A., van Stiphout R.G.P.M., Winchester L.M., Wigfield S., Harris A.L., Buffa F.M. Genomic alterations underlie a pan-cancer metabolic shift associated with tumour hypoxia. Genome Biol. 2016;17:140. doi: 10.1186/s13059-016-0999-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oruganty K., Campit S.E., Mamde S., Lyssiotis C.A., Chandrasekaran S. Common biochemical properties of metabolic genes recurrently dysregulated in tumors. Cancer Metab. 2020;8:5. doi: 10.1186/s40170-020-0211-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng X., Chen Z., Farshidfar F., Xu X., Lorenzi P.L., Wang Y., Cheng F., Tan L., Mojumdar K., Du D., et al. Molecular Characterization and Clinical Relevance of Metabolic Expression Subtypes in Human Cancers. Cell Rep. 2018;23:255–269.e4. doi: 10.1016/j.celrep.2018.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosario S.R., Long M.D., Affronti H.C., Rowsam A.M., Eng K.H., Smiraglia D.J. Pan-cancer analysis of transcriptional metabolic dysregulation using The Cancer Genome Atlas. Nat. Commun. 2018;9:5330. doi: 10.1038/s41467-018-07232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haukaas T.H., Euceda L.R., Giskeødegård G.F., Lamichhane S., Krohn M., Jernström S., Aure M.R., Lingjærde O.C., Schlichting E., Garred Ø., et al. Metabolic clusters of breast cancer in relation to gene- and protein expression subtypes. Cancer Metab. 2016;4:12. doi: 10.1186/s40170-016-0152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y., Du S., Liu J., Huang W., Liu W., Zhang M., Li N., Wang R., Wu J., Chen W., et al. Diagnosis and prognosis of breast cancer by high-performance serum metabolic fingerprints. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2122245119. e2122245119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duarte N.C., Becker S.A., Jamshidi N., Thiele I., Mo M.L., Vo T.D., Srivas R., Palsson B.Ø. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaude E., Frezza C. Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nat. Commun. 2016;7:13041. doi: 10.1038/ncomms13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N.C., Nakano K., Bartrons R., Gottlieb E., Vousden K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh A.L., Walton Z.E., Altman B.J., Stine Z.E., Dang C.V. MYC and metabolism on the path to cancer. Semin. Cell Dev. Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereira B., Chin S.F., Rueda O.M., Vollan H.K.M., Provenzano E., Bardwell H.A., Pugh M., Jones L., Russell R., Sammut S.J., et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott L. 2020. SHAP Documentation (Release Latest Septemebr 12, 2020)https://shap.readthedocs.io/en/latest/index.html [Google Scholar]
- 34.Terunuma A., Putluri N., Mishra P., Mathé E.A., Dorsey T.H., Yi M., Wallace T.A., Issaq H.J., Zhou M., Killian J.K., et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J. Clin. Invest. 2014;124:398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heiser L.M., Sadanandam A., Kuo W.L., Benz S.C., Goldstein T.C., Ng S., Gibb W.J., Wang N.J., Ziyad S., Tong F., et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. USA. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tibshirani R., Hastie T., Narasimhan B., Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masters J.R. Human cancer cell lines: fact and fantasy. Nat. Rev. Mol. Cell Biol. 2000;1:233–236. doi: 10.1038/35043102. [DOI] [PubMed] [Google Scholar]
- 38.Corsello S.M., Nagari R.T., Spangler R.D., Rossen J., Kocak M., Bryan J.G., Humeidi R., Peck D., Wu X., Tang A.A., et al. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer. 2020;1:235–248. doi: 10.1038/s43018-019-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroemer G., Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Yun J., Rago C., Cheong I., Pagliarini R., Angenendt P., Rajagopalan H., Schmidt K., Willson J.K.V., Markowitz S., Zhou S., et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang W., Putluri V., Ambati C.R., Dorsey T.H., Putluri N., Ambs S. Liver- and Microbiome-derived Bile Acids Accumulate in Human Breast Tumors and Inhibit Growth and Improve Patient Survival. Clin. Cancer Res. 2019;25:5972–5983. doi: 10.1158/1078-0432.CCR-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lien E.C., Ghisolfi L., Geck R.C., Asara J.M., Toker A. Oncogenic PI3K promotes methionine dependency in breast cancer cells through the cystine-glutamate antiporter xCT. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aao6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singletary K.W., Frey R.S., Yan W. Effect of ethanol on proliferation and estrogen receptor-alpha expression in human breast cancer cells. Cancer Lett. 2001;165:131–137. doi: 10.1016/s0304-3835(01)00419-0. [DOI] [PubMed] [Google Scholar]
- 44.Cammarota G., Ianiro G., Ahern A., Carbone C., Temko A., Claesson M.J., Gasbarrini A., Tortora G. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 2020;17:635–648. doi: 10.1038/s41575-020-0327-3. [DOI] [PubMed] [Google Scholar]
- 45.Sung J., Wang Y., Chandrasekaran S., Witten D.M., Price N.D. Molecular signatures from omics data: from chaos to consensus. Biotechnol. J. 2012;7:946–957. doi: 10.1002/biot.201100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galluzzi L., Kepp O., Vander Heiden M.G., Kroemer G. Metabolic targets for cancer therapy. Nat. Rev. Drug Discov. 2013;12:829–846. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 47.Paez J.G., Jänne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 48.Scappini B., Gatto S., Onida F., Ricci C., Divoky V., Wierda W.G., Andreeff M., Dong L., Hayes K., Verstovsek S., et al. Changes associated with the development of resistance to imatinib (STI571) in two leukemia cell lines expressing p210 Bcr/Abl protein. Cancer. 2004;100:1459–1471. doi: 10.1002/cncr.20131. [DOI] [PubMed] [Google Scholar]
- 49.Konecny G.E., Pegram M.D., Venkatesan N., Finn R., Yang G., Rahmeh M., Untch M., Rusnak D.W., Spehar G., Mullin R.J., et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 50.Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salama M.E., Khairy D.A. Polo-Like Kinase 1(PLK1) Immunohistochemical Expression in Triple Negative Breast Carcinoma: A Probable Therapeutic Target. Asian Pac. J. Cancer Prev. 2021;22:3921–3925. doi: 10.31557/APJCP.2021.22.12.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung C.H., Lin D.W., Eames A., Chandrasekaran S. Next-Generation Genome-Scale Metabolic Modeling through Integration of Regulatory Mechanisms. Metabolites. 2021;11 doi: 10.3390/metabo11090606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drier Y., Sheffer M., Domany E. Pathway-based personalized analysis of cancer. Proc. Natl. Acad. Sci. USA. 2013;110:6388–6393. doi: 10.1073/pnas.1219651110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monti S., Tamayo P., Mesirov J., Golub T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Mach. Learn. 2003;52:91–118. doi: 10.1023/A:1023949509487. [DOI] [Google Scholar]
- 56.Charrad M., Ghazzali N., Boiteau V., Niknafs A. NbClust: An R Package for Determining the Relevant Number of Clusters in a Data Set. J. Stat. Softw. 2014;61:1–36. doi: 10.18637/jss.v061.i06. [DOI] [Google Scholar]
- 57.Iqbal M.A., Chattopadhyay S., Siddiqui F.A., Ur Rehman A., Siddiqui S., Prakasam G., Khan A., Sultana S., Bamezai R.N. Silibinin induces metabolic crisis in triple-negative breast cancer cells by modulating EGFR-MYC-TXNIP axis: potential therapeutic implications. FEBS J. 2021;288:471–485. doi: 10.1111/febs.15353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
No new datasets were generated during the current study.
-
•
Codes used are made available at https://github.com/kirksmi/BreastCancerClustering.
-
•
Information and requests for resources and any additional data should be directed to and will be fulfilled by the lead contact.