Abstract
Congenital defects in the pancreas can cause severe health issues such as pancreatic cancer and diabetes which require lifelong treatment. Regenerating healthy pancreatic cells to replace malfunctioning cells has been considered a promising cure for pancreatic diseases including birth defects. However, such therapies are currently unavailable in the clinic. The developmental gene regulatory network underlying pancreatic development must be reactivated for in vivo regeneration and recapitulated in vitro for cell replacement therapy. Thus, understanding the mechanisms driving pancreatic development will pave the way for regenerative therapies. Pancreatic progenitor cells are the precursors of all pancreatic cells which use epigenetic changes to control gene expression during differentiation to generate all of the distinct pancreatic cell types. Epigenetic changes involving DNA methylation and histone modifications can be controlled by non-coding RNAs (ncRNAs). Indeed, increasing evidence suggests that ncRNAs are indispensable for proper organogenesis. Here, we summarize recent insight into the role of ncRNAs in the epigenetic regulation of pancreatic development. We further discuss how disruptions in ncRNA biogenesis and expression lead to developmental defects and diseases. This review summarizes in vivo data from animal models and in vitro studies using stem cell differentiation as a model for pancreatic development.
Keywords: pancreatic development, pancreatic birth defects, ncRNA, miRNA, lncRNA, epigenetic regulation
Introduction
The pancreas has diverse and essential roles in nutrition and metabolism. The pancreas is composed of endocrine and exocrine cells. The endocrine pancreas consists of five types of cells that form distinct clusters called islets of Langerhans and secrete hormones required to regulate blood glucose and energy homeostasis. The exocrine pancreas contains acinar and ductal cells. Acinar cells synthesize and secret digestive zymogens, and the ductal cells constitute the duct system that funnels zymogens into the duodenum where they are activated to digest food (Longnecker et al., 2018; Mahadevan, 2019). The endocrine and exocrine cells are generated from the same pancreatic progenitor cells (PPCs) during development. Disruptions in PPC proliferation and differentiation lead to pancreatic birth defects including pancreatic agenesis, pancreas divisum, annular pancreas, and some types of diabetes mellitus (Lorberbaum et al., 2020). Diabetes is a chronic disease that requires life-long treatment. Over time, abnormal blood glucose leads to complications such as heart disease, stroke, kidney disease, loss of visual function, and peripheral nerve damage (Nathan, 1993). More than 11% of the population in the United States have diabetes mellitus (National Diabetes Statistics Report | Diabetes | CDC, n.d.), costing an estimated $327 billion annually (American Diabetes Association, 2018). In newborns, β-cell differentiation defects lead to insufficient insulin production and elevated blood glucose, which can ultimately develop into diabetes mellitus. Increasing functional β-cell mass is the optimal cure for diabetes patients and two goals have been set forward, (1) activating endogenous β-cell proliferation or transdifferentiation from other pancreatic cell types such as acinar cells and (2) transplanting exogenous β-cells such as β-cells differentiated from stem cells in vitro. However, the field is still trying to figure out how to achieve the first goal in animal models and we are still far from translating these findings to the clinic. There are ongoing clinical trials that transplant stem cell-derived islets into patients (Ramzy et al., 2021; Shapiro et al., 2021), however, these trials are still in their infancy so the outcomes still need to be determined and they use costly devices which makes them unaffordable to some patients. Birth defects in the pancreas also lead to inefficient transportation of zymogens which can be activated prematurely inside the pancreas instead of the duodenum, causing acinar cell damage and inflammation in the pancreas termed pancreatitis. Acinar cells have plasticity to de-differentiate into ductal-like cells after injury through a process called acinar-to-ductal metaplasia (ADM). However, for pancreatic regeneration to occur, the ADM must be resolved by the re-differentiation of metaplastic cells back into acinar cells. Otherwise, persistent ADM and pancreatic inflammation can result in neoplastic precancerous lesions which may develop into pancreatic cancer (Stanger & Hebrok, 2013). The five-year survival rate for pancreatic cancer is less than 11% which is the lowest among all types of cancer (Siegel et al., 2022). Indeed, increasing acinar cell re-differentiation after ADM can significantly decrease the risk of pancreatic cancer (Storz, 2017). Even though regenerative medicine is a promising, potentially permanent therapy for patients with pancreatic birth defects, there are no effective therapies currently available. Most patients adapt their lifestyles to delay disease progression and manage symptoms. Understanding how the pancreas develops is critical for identifying the cause of pancreatic birth defects. Since the underlying mechanisms controlling pancreatic development are reactivated during pancreatic regeneration to replace damaged cells, it is also critical to understand the gene regulatory network driving development to determine targetable pathways for future regenerative therapies.
As mentioned, the pancreatic endocrine and exocrine cells are derived from the same progenitor population with the same genetic material. Epigenetic modifications change chromatin accessibility and structure to alter gene expression and cellular identity without changing the DNA sequence, and their effects on gene expression are inherited during cell division, thereby pushing PPCs to adopt different cell fates (Goldberg et al. 2007; Bernstein and Allis 2005). Chromatin is composed of DNA and histones (Akhtar et al., 2000). DNA methylation and histone modifications modulate the opening and closing of chromatin and thereby control the accessibility of chromatin to transcriptional machinery (Bannister et al. 2001; Cosgrove et al. 2004; Hebbes et al. 1988; Hall et al. 2002; Keshet et al. 1985). Non-coding RNA (ncRNA) is also considered to be an epigenetic modifier (Penny et al. 1996; Bernstein and Allis 2005; Mohammad et al. 2012). Non-coding RNAs are functional RNA molecules that do not translate into proteins and it has been estimated that ncRNAs take up more than 90% of the transcriptome in eukaryotic cells (Palazzo & Lee, 2015). NcRNAs can be classified based on their lengths into long ncRNAs (longer than 200bp) and small ncRNAs (shorter than 200bp) such as PIWI-interacting RNAs (piRNAs) and microRNAs (miRNAs) (Bhat et al., 2020). Both long and short ncRNAs have been shown to control epigenetic status via multiple mechanisms including directing DNA methylation, recruiting chromatin remodelers and histone modifiers, and acting as a scaffold for transcriptional regulators. Not only do ncRNAs epigenetically regulate normal biological processes such as differentiation and development (Bhat et al. 2020), but also are involved in the pathology of human diseases such as cancer and diabetes (Hajjari & Salavaty, 2015; Kondo et al., 2017; Kumar et al., 2020; Lardenoije et al., 2015; Zarzour et al., 2019). Thus, targeting ncRNAs may be a novel strategy for manipulating epigenetic status and gene expression for regenerative medicine.
Animal models are the most widely used tools to study pancreatic development and disease (Lorberbaum et al., 2020). Additionally, the differentiation potential of stem cells has emerged as a powerful in vitro tool for modeling pancreatic development (Rezania et al. 2013; Huang et al. 2021; Trott et al. 2017; Lee and Chung 2011; Rezania et al. 2014; Rezania et al. 2012; Kroon et al. 2008). Here we summarize results from in vivo and in vitro models on the epigenetic regulation of pancreatic development by ncRNAs and how disruptions in ncRNA biogenesis and function contribute to pancreatic birth defects.
Overview of pancreas development
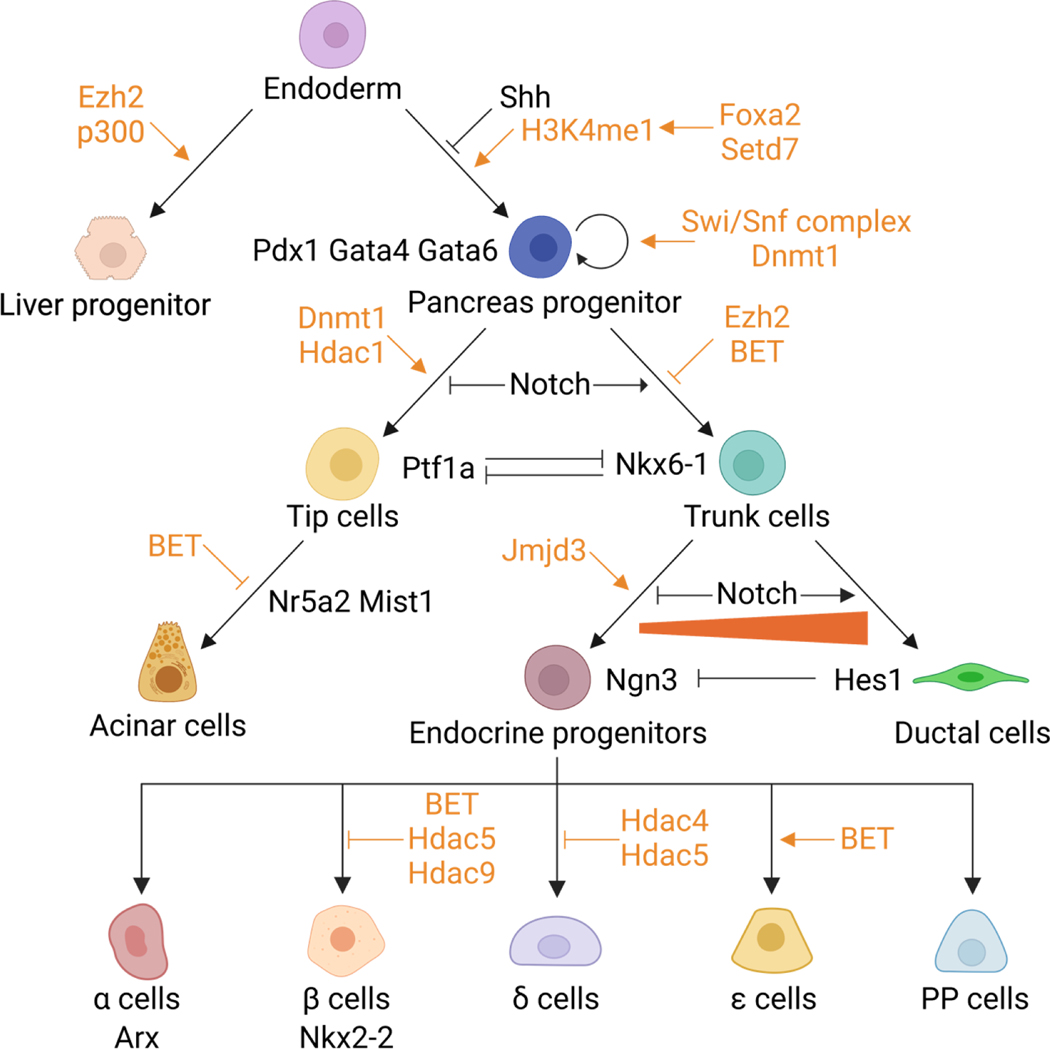
The development of the pancreas is similar between humans and rodents. In this section, pancreatic development in mice is used as an example (Figure 1). After gastrulation, the primary germ layers (endoderm, mesoderm, and ectoderm) are established. The endoderm is a flat layer of epithelial cells adjacent to the mesoderm (Wells & Melton, 1999). A crescent-shaped fold forms at both the anterior (anterior intestinal portal, AIP) and posterior end (caudal intestinal portal, CIP) of the endoderm. From embryonic (E) day E8.0 to E9.0 in mouse embryos (E17 to E21 in humans), the AIP and CIP elongate and migrate towards each other until they meet at the yolk stalk where they connect to the yolk sac and form the primitive gut tube (Spence et al., 2011). Concurrently, the gut tube is patterned along the anterior-posterior axis with different transcription factors expressed in each region, namely the foregut, midgut, and hindgut. The dorsal endoderm is in contact with the notochord between E8.5 and E8.75 in mice, and by E9.0, the two paired dorsal aortae fuse between the notochord and gut epithelium. The aorta and notochord are both associated with the formation of the pancreatic primordia (Lammert et al. 2001; Kim et al. 1997). Sonic hedgehog (Shh) is expressed along the gut tube except in the pancreatic domain. The notochord secrets factors including activin-βB and FGF2 to repress Shh and induce the expression of Pdx1 which is the earliest transcription factor expressed in the developing pancreas (Hebrok et al., 1998). On the ventral side of the gut tube, a bipotential precursor population is capable of activating both pancreatic and hepatic programs. The default fate of these bipotential progenitors is to express pancreatic genes. However, FGF signals from the cardiogenic mesoderm (Deutsch et al., 2001) and BMP signals from the septum transversum mesenchyme (Rossi et al., 2001) divert those cells to the hepatic track.
Figure 1.
Illustration of pancreatic development. Transcription factors and signaling pathways regulating pancreatic development are labeled in black. DNA methylation and histone modifications are labeled in orange.
Once the pancreatic domain is specified along the gut tube, additional transcription factors required for pancreatic development are induced including Ptf1a, Sox9, Nkx6–1, and Hnf6 (Jørgensen et al., 2007). The mesenchyme expands to envelop the pancreatic epithelium and separate it from the dorsal aorta. Signals from the pancreatic mesenchyme are essential for expanding the pancreatic progenitor pool (Golosow & Grobstein, 1962; Landsman et al., 2011). The mesenchymal FGF10 promotes the expression of Sox9 in the pancreatic progenitors, and Sox9 maintains the expression of FGF receptor 2b (Fgfr2) which transduces the mesenchymal FGF signal into PPCs. Sox9, Fgfr2, and Fgf10 form a feed-forward loop to maintain the pancreatic identity and promote the proliferation of PPCs (Bhushan et al., 2001; Norgaard et al., 2003; Seymour et al., 2012). From E8.5 to E10.5 in mice (E20 to E30 in humans), PPCs are induced on the dorsal and ventral side of the gut tube, proliferate and form two pancreatic buds, and convert from a “predifferentiatied” to “protodifferentiated” state where low levels of pancreas-specific proteins are expressed. This process is referred to as the primary regulatory transition (Pictet et al., 1972; Rutter et al., 1968).
Shortly after the primary transition, the PPCs remain as undifferentiated multipotent stem cells that are tightly packed within pancreatic buds. After E10.5, the pancreatic epithelium expands rapidly and branches into two domains, the tip and trunk. This expansion is initiated by transcriptional cross-repression between Ptf1a and Nkx6–1 (Schaffer et al., 2010). The tip cells expressing Ptf1a will develop into acinar cells while the trunk cells marked by Nkx6–1 will form the bipotent progenitors which give rise to ductal and endocrine cells. Notch inhibits Ptf1a activity and acinar differentiation while promoting the expansion of trunk cells (Afelik et al., 2012; Esni et al., 2004). As the PPCs adopt a tip or trunk fate, the secondary regulatory transition begins when the “protodifferentiated” cells undergo cytodifferentiation and upregulate the synthesis of pancreatic proteins (Pictet et al., 1972). By E12.5 in mice (E42 in humans), the gut tube rotates, and the dorsal and ventral pancreatic buds fuse into an interconnected organ. In tip cells, Ptf1a forms a transcriptional activating complex with Rbp-jl to activate the expression of key acinar genes like Nr5a2 and Mist1. Together, these genes maintain the acinar identity and drive the expansion of acinar cells (Masui et al. 2008; Thompson et al. 2012; Masui et al. 2010; Masui et al. 2007). Within the trunk domain, some bipotent progenitor cells activate the expression of Ngn3, which initiates endocrine differentiation (Gu et al., 2002). Ngn3 induces epithelial-to-mesenchymal transition (EMT) in endocrine cells and promotes delamination out of the epithelium (Gouzi et al., 2011). The Ngn3 positive cells differentiate into all five types of endocrine cells within the islet of Langerhans: α-cells (produce glucagon), β-cells (produce insulin), δ-cells (produce somatostatin), PP-cells (produce pancreatic polypeptide), and ε-cells (produce ghrelin). The Notch signaling pathway controls the activation of Ngn3 in the bipotent progenitors. Intermediate Notch activity promotes Sox9 expression, which induces Ngn3 cell-autonomously. However, high Notch activity also activates the expression of Hes1, an Ngn3 repressor (Lee et al. 2001). Bipotent progenitors that do not express Ngn3 will adopt a ductal fate and eventually develop into the branching ductal system (Delous et al. 2012; Shih et al. 2012).
Outside of mice, other systems such as flies, fish, frogs and birds have been used to study pancreatic development. Invertebrate species such as D. melanogaster do not have pancreatic islets, however, they do have cells producing insulin- and glucagon-like peptides. In flies, such cells are present in midgut muscle and pars intercerebralis of the brain (Ikeya et al., 2002; Veenstra et al., 2008). Basal chordates such as Hagfish represent the most primitive vertebrates with a pancreas-like structure containing insulin and somatostatin-producing cells (Heller, 2010). Over the course of vertebrate evolution, insulin-producing cells along and with more pancreatic hormone-producing cells became more organized into distinct structures resembling islets (Madsen, 2007). The structure of the pancreas in fish, frogs, and birds are similar to mammals with distinct localization of endocrine cells in islets, but they differ in the composition and position of different hormone-producing cells (Heller, 2010). The gene regulatory network governing pancreatic development in vertebrates is also conserved. Pdx1 expression can drive pancreas formation from nonpancreatic regions across multiple species (Afelik et al., 2006; Grapin‑Botton et al., 2001; Yee et al., 2001). Inhibiting Ptf1a in zebrafish converts acinar cells to insulin-producing cells, suggesting Ptf1a is also required for maintaining acinar identity in zebrafish (Hesselson et al., 2011). Ectopic expression of Ngn3 in chicken embryos is sufficient to differentiate endodermal cells into endocrine cells expressing glucagon and somatostatin (Grapin‑Botton et al., 2001). The conserved gene regulatory network across species allows us to take advantage of different model organisms to study pancreatic organogenesis.
Compared to animal models, the availability of human embryonic and fetal tissue to study pancreas development is limited. Current knowledge from animal models has guided the development of protocols for differentiating human pluripotent stem cells (hPSCs) into pancreatic cells. There are multiple protocols, but in general, they involve using Activin and Wnt agonists to induce definitive endoderm formation from hPSCs, retinoic acid and FGF to pattern the posterior foregut cells, and Shh and BMP inhibitors to promote pancreatic progenitor cells induction (Pagliuca et al. 2014; Rezania et al. 2013; Rezania et al. 2014; Rezania et al. 2012). Despite the success in differentiating hPSCs into pancreatic cells, there are drawbacks limiting the capacity of those differentiated cells. The cells generated from hPSCs are still immature compared to somatic cells. The differentiation is conducted and cultured in 2D and thus, cannot model the interaction with surrounding tissues. In addition, the efficiency of generating exocrine cells remains low compared to endocrine cells (Gaertner et al., 2019). Further optimizations are needed to better model human development in vitro and serve as a source of cells for regenerative therapies.
Models for pancreatic birth defects
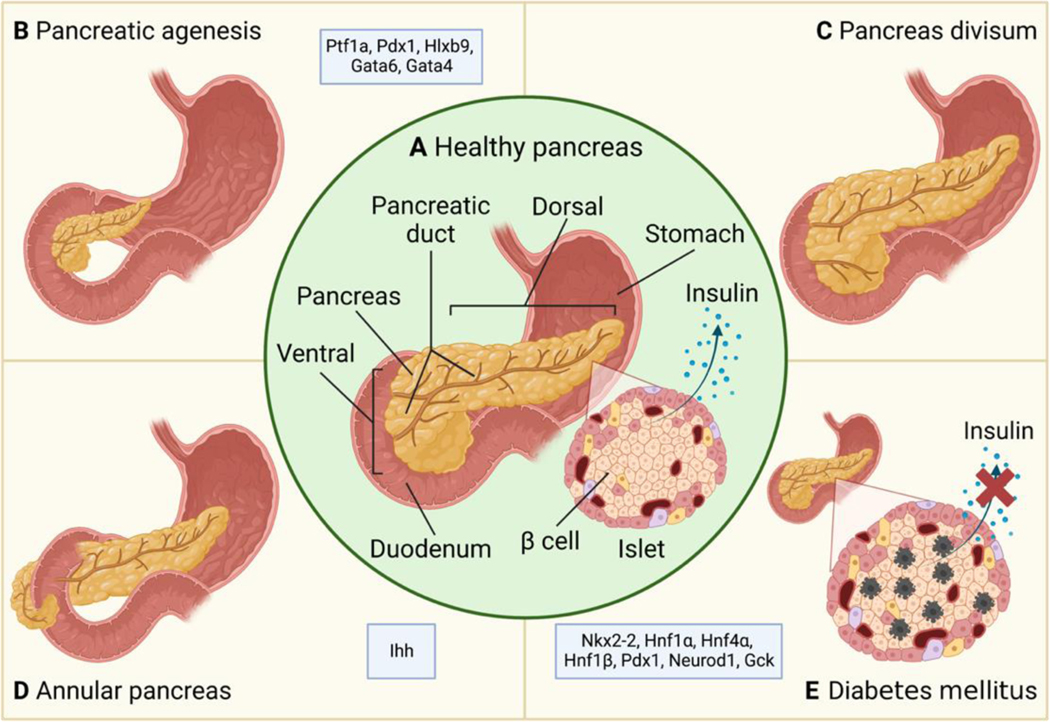
Human and rodent pancreas development is similar in that they both undergo primary and secondary transition and share many conserved transcription factors that govern these processes. In addition, the mature pancreas structure in rodents (Figure 2A) also resembles the mature human pancreas. Thus, rodent models have been a very powerful tool to understand pancreas development. In this section, we summarize different types of human pancreatic birth defects and how they can be recapitulated in animal models.
Figure 2.
Illustration of birth defects in the pancreas. (A) In a healthy pancreas, the ducts in the dorsal and ventral pancreas fuse to form a single duct, and the β-cells in the islet of Langerhans synthesize and secret insulin. (B) Agenesis of the pancreas with partially developed pancreas. (C) Failure in the fusion between dorsal and ventral ducts leads to pancreas divisum. (D) Formation of the annular pancreas. Instead of rotating with the gut tube and fusing with the dorsal pancreas, the ventral pancreas encircles the duodenum. (E) Insulin secretion is decreased due to β-cell loss in diabetes. Factors known to contribute to each birth defect are labeled in each panel.
Pancreatic agenesis
Pancreatic agenesis is a rare congenital disease in which all or part of the pancreas fails to form (Figure 2B). Patients can be asymptomatic or develop pancreatic diseases like hyperglycemia and pancreatitis. Ptf1a or Pdx1 knockout in mice leads to pancreatic agenesis, hyperglycemia, and perinatal death (Krapp et al., 1998; Offield et al., 1996; Sellick et al., 2004). Persistent expression of Hlxb9 also results in agenesis of the pancreas (H. Li & Edlund, 2001). One genomic study in 27 patients identified GATA6 haploinsufficiency as a cause of pancreatic agenesis (Allen et al., 2011). The role of GATA6 in human pancreatic development was later confirmed using in vitro stem cell differentiation which found that loss of GATA6 prevented endoderm differentiation and β-cell formation (Shi et al., 2017; Tiyaboonchai et al., 2017). In mice, pancreas-specific deletion of Gata6 did not significantly impact pancreatic formation. Only when another GATA family member, Gata4, was deleted together with Gata6, did the mice develop pancreatic agenesis and hyperglycemia and die shortly after birth (Carrasco et al., 2012; Xuan et al., 2012). Xuan et al. deleted Gata6 and Gata4 together in the gut endoderm using Foxa3-Cre at E8.5 and found that the PPCs were still induced in the KO embryos, but PPC proliferation and branching morphogenesis were arrested by E11.5, thereby contributing to agenesis in the pancreas.
Pancreas divisum
During pancreatic development, the ventral pancreas rotates with the gut tube and fuses with the dorsal pancreas, and the ducts in the ventral and dorsal pancreas also fuse together and form the main pancreatic duct. Pancreas divisum (Figure 2C) occurs when the ventral and dorsal ducts fail to fuse together. It is the most common congenital anomaly in the pancreas and is found in 4–14% of the population (Nijs et al., 2005). The pancreatic duct system drains and funnels the pancreatic enzymes into the duodenum through the papilla which is the opening of the duct into the duodenum. In embryos, the majority of the gland drains from the dorsal duct into the minor papilla (opening of the accessory pancreatic duct) and the ventral duct drains the minority of the pancreas into the major papilla (opening of the main pancreatic duct). In adults, the situation is reversed. If the fusion of dorsal and ventral fails, the dorsal duct will still drain the majority of the pancreas. This can lead to functional stenosis in the ducts causing some patients to develop recurrent pancreatitis which increases the risk for pancreatic cancer (Chalazonitis et al., 2008; Kuzel et al., 2017).
Annular pancreas
The annular pancreas (Figure 2D) is a rare birth defect that occurs when rotation of the ventral pancreas has failed or is incomplete, causing pancreatic tissue to encircle the duodenum (Borghei et al., 2013; Etienne et al., 2012). Newborns with annular pancreas may not feed well, spit up more often, and develop pancreatitis and duodenal obstruction. Pancreatitis usually occurs in the annulus of the pancreas since the pancreatic secretions cannot pass through the annular duct, resulting in the accumulation of secretions behind the obstruction and increasing the susceptibility of the annular pancreas to damage (Aleem & Shah, 2022; Itoh et al., 1989). Only one example of annular pancreas is found in mice, which is caused by the inactivation of Indian hedgehog (Ihh) (Hebrok et al., 2000).
Diabetes mellitus
Multiple forms of congenital diabetes (Figure 2E) can occur in newborns and children due to defects in β-cell differentiation and function, and they are often caused by genetic mutations and environmental insults during in utero stages of development. Neonatal diabetes mellitus (NDM) patients present with severe hyperglycemia at birth. Flanagan et al. analyzed mutations in key transcription factors involved in pancreatic development using samples from 37 NDM patients and identified mutations in NKX2–2 as an etiological gene for NDM (Flanagan et al., 2014). Consistent with that, mice lacking Nkx2–2 develop severe hyperglycemia and die shortly after birth (Sussel et al., 1998). Maturity onset diabetes of the young (MODY) is a familial monogenic disease where patients harbor a single mutation that disrupts insulin production (Fajans et al., 2001). MODY is classified based on its genetic causes, including mutations in Hnf1α, Hnf4α, Hnf1β, Gck, Pdx1, and Neurod1 (Dickens et al., 2019; Fajans et al., 2001; Horikawa et al., 1997; Pihoker et al., 2013). Patients inheriting these mutations usually develop MODY and are diagnosed with diabetes under the age of 25 (McDonald & Ellard, 2013). Disruptions in these genes during animal development and hPSC differentiation also recapitulate the MODY phenotypes (De Vas et al. 2015; Niborski et al. 2021; Servitja et al. 2009; Low et al. 2021; Cardenas-Diaz et al. 2019; Shih et al. 2002; Ng et al. 2019; Song et al. 2020; Bali et al. 1995; van Bürck et al. 2010; Sachdeva et al. 2009; Bohuslavova et al. 2021; Hermann et al. 2022).
Disruption of epigenetic regulation can cause developmental defects in the pancreas
Epigenetic regulation of pancreatic development and diseases, especially DNA methylation and histone modification, has been reviewed in detail elsewhere (Arnes & Sussel, 2015; Golson & Kaestner, 2017; Kaimala et al., 2022; Quilichini & Haumaitre, 2015). Here we briefly summarize how these two epigenetic mechanisms contribute to pancreatic birth defects (Figure 1).
Histone modifications
Histones are basic proteins that provide structural support for chromosomes. DNA wraps around the histone octamers which consist of H2A, H2B, H3, and H4 dimers. The histone H1 is a linker that connects DNA to the octamers. The N-terminal tails of histones can undergo post-translational modifications which affect their interaction with DNA, chromatin structure and the accessibility of DNA to transcriptional machinery. The most well-known types of histone modifications include acetylation, methylation, phosphorylation, and ubiquitylation. Other types of modifications such as deamination, GlcNAcylation, ADP ribosylation, sumoylation, and isomerization have also been discovered (Andrew J Bannister & Kouzarides, 2011). Histone modifications are added (writers) and removed (erasers) by histone-modifying enzymes. Histone modification readers contains effector domains that recognize and bind to specific modifications and mediate downstream functional outcomes such as transcriptional activation and inactivation. For example, histone acetylation, which usually activates gene expression, is added by histone acetyltransferases (HATs) and removed by histone deacetylases (HDACs), and is recognized by bromodomain.
In embryonic stem cells (ESCs), the promoters of developmental genes are enriched with bivalent chromatin domains which contain both repressive (H3K27me3) and active (H3K4me3) marks. These bivalent chromatin domains keep developmental genes poised for expression (Bernstein et al. 2006). In response to developmental cues, the repressive H3K27me3 marks are removed by demethylases Kdm6a and Kdm6b which allows rapid activation of lineage-specific genes. The demethylases drive definitive endoderm differentiation by activating Eomes and regulating the Wnt signaling pathway components, Wnt3 and Dkk1 (Jiang et al., 2013; Kartikasari et al., 2013). The undifferentiated endoderm is pre-patterned to express pancreatic genes by default. However, in a subset of cells, the H3K27 methyltransferase Ezh2 and histone acetyltransferase p300 promote hepatic instead of pancreatic fate by suppressing Pdx1 and activating liver genes (Figure 1) (Xu et al., 2011). Decreases in heterochromatin marked by H3K9me3 are also required for liver gene expression (Nicetto et al., 2019). The pioneer factor Foxa2 binds to compact nucleosomal DNA and establishes a primed enhancer state by interacting with H2A.Z, a histone variant that is involved in nucleosome depletion, deposit H3K4me1, and recruit Gata6 during endodermal and pancreatic differentiation (Lee et al. 2019; Li et al. 2012). In Xenopus embryos, knocking out the methyltransferase that catalyzes H3K4me1, Setd7, eliminates the specification of PPCs (Kofent et al., 2016). Once PPCs are induced, Pdx1 engages with the Swi/Snf chromatin remodeling complex to promote proliferation (Spaeth et al., 2019). The Swi/Snf complex is an ATP-dependent chromatin remodeling complex with multiple subunits that can hydrolyze ATP to relocate the nucleosomes to adjacent DNA (sliding) or even evict nucleosomes from the chromatin. Brg1 and Brm are two ATPase subunits in the complex. Losing Brg1 and Brm during development leads to pancreatic agenesis due to reduced proliferation of PPCs while deleting both units in the adult results in fasting hyperglycemia, glucose intolerance, and reduced serum insulin levels (Spaeth et al., 2019). Histone modifications also regulate the differentiation of PPCs. Acinar cell differentiation is abolished when HDACs are inhibited or deleted, while endocrine differentiation is enhanced (Haumaitre et al., 2008; Lenoir et al., 2011; Noël et al., 2008; Zhou et al., 2011). During endocrine differentiation, the repressive mark H3K27me3 on endocrine-specific genes is lost both in vitro and in vivo (Xie et al., 2013; Xu et al., 2014). Consistent with that finding, deleting Ezh2 during development increased Ngn3 expression and β-cell number. Indeed, Ezh2 heterozygous animals showed enhanced glucose tolerance. However, Ezh2 knockout animals developed diabetes due to aberrant Ink4a/Arf induction which impaired β-cell function (H. Chen et al., 2009; Xu et al., 2014). H3K27me3 demethylase is also required for endocrine differentiation as deleting Jmjd3 leads to impaired endocrine progenitor formation (Yu et al., 2018). Inhibiting the Bromodomain extra-terminal (BET) proteins, the reader of acetylated histones, increases the number of Ngn3+ endocrine progenitors and Ghrl+ ε-cells, but it leads to incomplete development of both β-cells and acinar cells as shown by decreased expression of mature cell markers (Huijbregts et al., 2019).
DNA methylation
In addition to histones, DNA molecules themselves are often methylated. The methyl group is added to the C5 position of the cytosine by DNA methyltransferases to form 5-methylcytosine (5-mC). Interestingly, no enzymes have been found that cleave the methyl group directly off the 5-mC. Instead, DNA methylation can be diluted during cell division (passive demethylation), or the methyl group on the 5-mC can be oxidized by ten-eleven translocation (TET) enzymes to form 5-hydroxymethylcytosine (5hmC). The TET enzymes further oxidize 5hmC into 5-carboxylcytosine (5caC) and 5-formylcytosine (5fC), both of which can be recognized by base excision repair system and revert back to unmethylated state (active demethylation) (Liyanage et al., 2014; Moore et al., 2013). DNA methylation represses gene expression by inhibiting the binding of transcription factors to DNA and recruiting HDAC and chromatin remodelers.
Dynamic DNA methylation is required during development. Georgia et al. found that one of the DNA methyltransferases, Dnmt1, methylates the p53 locus in PPCs to inhibit apoptosis. Knocking out Dnmt1 decreases the PPC number and leads to pancreatic agenesis, and the decrease in PPC number in Dnmt1 KO embryos can be rescued by p53 haploinsufficiency (Georgia et al., 2013). In addition, two studies showed that Dnmt1 was indispensable for acinar cell differentiation as the knockdown of Dnmt1 in embryos inhibited acinar cell formation. They also showed that the regulation of acinar differentiation by Dnmt1 is dependent on the carboxyterminal domain which catalyzes cytosine methylation (Anderson et al., 2009; Rai et al., 2006). In the endocrine lineage, Dnmt1 controls the differentiation from endocrine progenitor cells to α- or β-cells. In β-cells, the promoter of Arx (α-cell specific gene) is methylated by Dnmt1. Inhibiting Dnmt1 during pancreatic development leads to hypomethylation of Arx and promotes α-cell formation at the expense of β-cells (Dhawan et al. 2011; Liu et al. 2019). The repression of Arx in β-cells is also mediated by Dnmt3a which forms a repressive complex with Nkx2–2, a gene critical for β-cell differentiation (Doyle & Sussel, 2007). DNA demethylation is also required for proper differentiation. Li et al. deleted TET enzymes in hPSCs and found that pancreatic progenitor and β-cell formation were significantly inhibited. TET depletion disrupted the demethylation of FOXA2 binding sites which leads to decreased binding of other pancreatic transcription factors such as GATA6 and PDX1 (Li et al. 2022). However, there are still many open questions about how TET enzymes regulate demethylation during pancreatic development, particularly the role of TET enzymes in vivo is currently limited. For example, what is the role of TET enzymes in activating the expression of endoderm genes in ESCs and what genes are TET enzymes targeting during pancreas organogenesis? Since DNA methylation is also enriched in heterochromatin which is removed during liver specification of bipotential endoderm progenitors, do TET enzymes control the pancreatic versus liver fate decisions?
Non-coding RNAs
NcRNAs are RNA molecules that are not translated into proteins. The coding exons of protein-coding genes only account for less than 2% of the genome (Venter et al., 2001), and more than 90% of the RNA in the transcriptome of mammalian cells are ncRNAs (Palazzo & Lee, 2015). An increasing amount of data has highlighted that ncRNAs, like protein-coding genes, play an important role in regulating normal biological processes, and misregulation of ncRNA expression and function lead to birth defects (Esteller, 2011). Based on their function, ncRNAs can be categorized into housekeeping ncRNAs and regulatory ncRNAs. Housekeeping ncRNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) and are the key components of cellular structure and metabolism. Regulatory RNAs can be classified by their lengths into small ncRNAs (shorter than 200bp) and long ncRNAs (lncRNA, longer than 200bp). The small RNAs include micro RNAs (miRNAs), piwi RNAs (piRNAs), small interfering RNAs (siRNAs), small nucleolar RNAs (snoRNAs), and small nuclear RNAs (snRNAs). LncRNAs can also be further categorized based on their genomic location and orientation, such as intronic, enhancer, intergenic, promoter, antisense, sense, and bidirectional lncRNAs (Dahariya et al., 2019).
NcRNAs regulate every step of gene expression. The piRNAs and lncRNAs are most well-known to regulate transcription in the nucleus whereas miRNAs canonically silence transcripts in the cytoplasm. The role of ncRNAs in β-cell function has been reviewed elsewhere (Dumortier & Van Obberghen, 2012; Font‑Cunill et al., 2018; Guay et al., 2012; Ozcan, 2014; Singer et al., 2015). In the following sections, we focus on how different classes of ncRNAs modulate the epigenetic status of the genome during pancreatic development, and discuss how disruptions in ncRNA expression and function can lead to pancreatic birth defects (Table 1).
Table 1.
ncRNAs involved in pancreatic diseases
| ncRNA | Targets/Mechanism | Related phenotypes/diseases | Reference |
|---|---|---|---|
| LINC00261 | Recruit SMAD2/3 to Foxa2 promoter and promote Foxa2 expression | Κnockdown of LINC00261 decreases endoderm differentiation | (Gaertner et al., 2020; Jiang et al., 2015; Swarr et al., 2019) |
| DIGIT | Upregulate GSC in trans | Depletion of DIGIT inhibits endoderm differentiation | (Daneshvar et al., 2016) |
| PLUTO | Regulate the interaction of Pdx1 enhancer clusters and promoter | PLUTO is downregulated in patients with impaired glucose tolerance and T2D | (Akerman et al., 2017) |
| HI-LNC25 | Regulate GLIS3 | Knocking down HI-LNC25 decreased GLIS3, which contributes to diabetes | (Morán et al., 2012) |
| H19 | Form a complex with MBD1 and deposit repressive histone modifications on target genes | H19 regulates β-cell proliferation during postnatal development and insulin resistance | (Monnier et al., 2013; Sanchez‑Parra et al., 2018) |
| βlinc1 | Regulate the expression of endocrine genes in cis | Mice lacking βlinc1 show reduced β-cell number during embryo stages and develop glucose intolerance | (Arnes et al., 2016) |
| miR-26a | TET enzymes | MiR-26a overexpression promotes β-cell number | (Fu et al., 2013) |
| miR-29 | YY1, Dnmt3a, Dnmt3b | MiR-29 suppresses insulin secretion | (Wang et al. 2008; Fabbri et al. 2007; Garzon et al. 2009) |
| miR-9 | Sirt1, Baf53a | MiR-9 decreases insulin exocytosis | (Ramachandran et al. 2011; Yoo et al. 2011; Yoo et al. 2009; Abernathy et al. 2017; Saunders et al. 2010) |
| miR-124 | Baf53a | MiR-124 increases Ca2+ concentration in β-cells | (Yoo et al. 2011; Yoo et al. 2009; Abernathy et al. 2017) |
| miR-375 | Cadm1, Pdk1, Mtpn, Gephyrin, Ywhaz | Diabetes | (Poy et al. 2009; Kloosterman et al. 2007; Lahmy et al. 2014; Tattikota et al. 2014; Tattikota et al. 2013; Poy et al. 2004; El Ouaamari et al. 2008) |
| miR-7 | Snca, Cspa, Pfn2, Basp1, Pax6, Mapkap1, p70S6K, Mknk1/2, eIF4E | Diabetes | (Nieto et al. 2012; Latreille et al. 2014; Wang et al. 2013; Kredo-Russo et al. 2012) |
| miR-30 | Vimentin, Snail1, NeuroD1, Map4k4, Rfx6 | Diabetes | (Tang et al. 2009; Joglekar et al. 2009; Zhao et al. 2012; Kim et al. 2013; Liao et al. 2013) |
| miR-21 | Pdcd4 | Diabetes, pancreatitis, pancreatic cancer | (Dixit et al. 2016; Ruan et al. 2011; Assmann et al. 2017; Khan et al. 2015; Rachagani et al. 2015) |
| miR-24 | Men1, Sox6, Hnf1α, Neurod1, | Diabetes | (Vijayaraghavan et al. 2014; Zhu et al. 2013; Melkman-Zehavi et al. 2011) |
Long non-coding RNA
LncRNAs have diverse biological functions, including epigenetic modifications (Rinn et al. 2007; Dinger et al. 2008; Pandey et al. 2008; Pontier and Gribnau 2011), transcription (Wang et al. 2008; Feng et al. 2006; Akerman et al. 2017), post-transcriptional modifications (Beltran et al., 2008), and nuclear trafficking (Willingham et al., 2005). LncRNAs play an essential role in pancreatic differentiation and development, and lncRNA expression is also misregulated in pancreatic diseases such as T2D (Mirzadeh Azad et al. 2021; Morán et al. 2012; Font-Cunill et al. 2018). Though it has been established that lncRNAs regulate β-cell function, the role of lncRNAs in pancreatic development is just starting to be appreciated.
Using hPSC differentiation, Jiang et al. characterized the expression of lncRNA during in vitro differentiation via transcriptome analysis (Jiang et al., 2015). They identified LINC00261 as a definitive endoderm-associated lncRNA that promotes FOXA2 expression by recruiting SMAD2/3 to the FOXA2 promoter. Deleting LINC00261 via short hairpin RNA (shRNA) decreased FOXA2 expression and endoderm differentiation efficiency (Jiang et al., 2015). The positive regulation of FOXA2 via LINC00261 also exists during lung epithelial homeostasis and regeneration (Swarr et al., 2019). However, Gaertner et al. recently found that knocking out LINC00261 via CRISPR/Cas9 impaired the differentiation of β-cells but did not decrease FOXA2 expression. They also suggested that LINC00261 regulated pancreatic differentiation via trans-regulatory mechanisms since the nearby FOXA2 and other proximal genes were not affected by Linc00261 deletion, and affected genes were distributed throughout the genome (Gaertner et al., 2020). The authors in the latter study suggested that the discrepancy between the two studies might be due to different methods of differentiation and gene manipulation. Another lncRNA that regulates definitive endoderm during hPSC differentiation is DIGIT. DIGIT expression is induced by Activin signaling during definitive endoderm differentiation, and DIGIT upregulates the expression of a gene involved in gastrulation and establishing body axis, Goosecoid (GSC), in trans to promote differentiation (Daneshvar et al., 2016). LncRNAs also regulate chromatin architecture to modulate the expression of key pancreatic transcription factors. The promoter of lncRNA PLUTO is located ~3kb upstream of PDX1. Akerman et al. used chromosome conformation capture sequencing to evaluate the interactions among different DNA sequences and showed that PLUTO promoted the contact between the PDX1 promoter and distal enhancers to regulate PDX1 expression (Akerman et al., 2017). Using human β-cells, Moran et al. showed that knocking down a β-cell-enriched lncRNA HI-LNC25 led to the downregulation of GLIS3 which is mutated in NDM (Morán et al., 2012; Senée et al., 2006). H19 is a lncRNA that forms a complex with methyl-CpG-binding domain protein 1 (MBD1). The H19 lncRNA-MBD1 complex interacts with histone lysine methyltransferases and deposits repressive histone marks on the targeted genes (Monnier et al., 2013). H19 is downregulated in the pancreas during postnatal development. Silencing of H19 in newborn rats decreased β-cell expansion. MiRNA let-7 and AKT phosphorylation are involved in promoting β-cell proliferation by H19 (Sanchez‑Parra et al., 2018). The lncRNA βlinc1 is an endocrine-specific lncRNA. Knocking out βlinc1 in mice leads to decreased endocrine differentiation in embryos and impaired glucose tolerance in adults. The genes regulated by βlinc1 are related to endocrine differentiation and β-cell function such as Pax6, Mafb, and Nkx2.2. Interestingly, those genes are significantly enriched around βlinc1 on the same chromosome, suggesting that βlinc has a cis-regulatory role during endocrine differentiation such as organizing the chromosome structure near these endocrine genes (Arnes et al., 2016). Of note, it is not known how lncRNAs regulate exocrine cell differentiation. LncRNA expression during pancreatic development has not been fully profiled either. More studies are needed to understand how lncRNAs contribute to the epigenetic regulation of pancreatic development.
MicroRNA
Biogenesis and function of microRNA
MiRNAs are the most studied class of ncRNAs. They are small ncRNAs between 20–22 nucleotides, and canonically silence gene expression by binding to the three prime untranslated region (3’ UTR) of the targeted mRNA, although targeting the 5’ UTR or coding sequence is possible. More than 60% of human protein-coding genes are regulated by miRNAs (Friedman et al., 2009). Each protein-coding gene, including the epigenetic regulators discussed above, can be targeted by multiple miRNAs, and each miRNA can target multiple genes (Peter, 2010), thus miRNAs and mRNAs form a complex gene regulatory network.
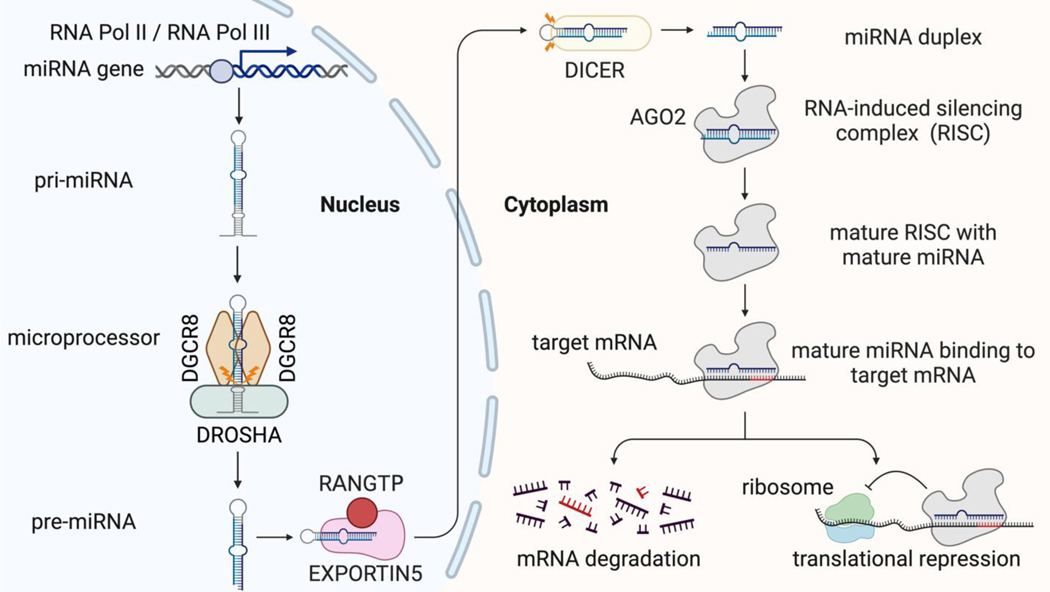
Canonically, miRNA biogenesis starts with transcription by RNA polymerase II (Lee et al. 2004) or III (Borchert et al., 2006). The transcription product, primary miRNA (pri-miRNA), folds back on itself and forms a hairpin structure. The microprocessor is a complex composed of one molecule of DROSHA and two molecules of DGCR8 (Nguyen et al., 2015). The microprocessor then cuts the hairpins at the stem and releases a ~70 nucleotides stem-loop structure termed the pre-miRNA (Lee et al. 2002; Lee et al. 2003). Then, the pre-miRNAs are transported from the nucleus to the cytoplasm by Exportin 5 and RAN-GTP (Lund et al., 2004; Yi et al., 2003). In the cytoplasm, Dicer cuts the terminal loop off the pre-miRNA to generate the miRNA duplex (Zhang et al. 2004). Once the miRNA duplex is released, it is loaded into the functional module in the gene silencing pathway termed RNA-induced silencing complex (RISC), in which the Argonaute family (e.g. Ago2) protein expels one strand of the miRNA duplex, while the remaining strand remains in RISC to form the mature RISC complex (Schwarz et al. 2003; Liu et al. 2004; Rivas et al. 2005; Suzuki et al. 2015). The mature miRNA then directs RISC to mRNA by pairing its seed sequence to the 3’ UTR of the mRNA, and gene silencing occurs through translational repression or mRNA degradation (Huntzinger & Izaurralde, 2011; Jonas & Izaurralde, 2015). Most miRNAs are generated and regulate gene expression via a canonical pathway (Figure 3); however, other non-canonical pathways have also been discovered and summarized by other reviews (Bartel, 2018; O’Brien et al., 2018; Stavast & Erkeland, 2019).
Figure 3.
The biogenesis and function of canonical miRNAs. The RNA Pol II or Pol III transcribes the miRNA gene to pri-miRNA, which then is processed by the microprocessor to form pre-miRNA. The pre-miRNA is exported to the cytoplasm by EXPORTIN5. In the cytoplasm, the pre-miRNA is cleaved by DICER to produce the miRNA duplex. The duplex is loaded onto AGO2. Only one strand will form the mature RISC with AGO2, and the other strand is ejected. The mature RISC will bind to target mRNA and silence gene expression by mRNA degradation or translational repression.
Alternations in microRNA biogenesis can cause birth defects in the pancreas
MiRNAs play an indispensable role during pancreatic development, and disruptions in miRNA biogenesis lead to severe developmental defects in the embryos. The loss of Dicer in mice results in early lethality during development (Bernstein et al. 2003), and loss of Dicer in zebrafish leads to developmental arrest around day 10 (Wienholds et al., 2003). Using a Dicer1-hypomorphic mouse model (Fukasawa et al., 2006), Kalis et al. found that histology of the pancreas showed abnormal structures in adults. Starting at four weeks of age, the Dicer1-hypomorphic pancreas showed decreased islet size, multi-nucleated cells in the duct expressing insulin and glucagon, and obscure boundaries between islets and ducts. However, glucose metabolism was not affected in the Dicer-hypomorphic animals (Morita et al., 2009).
Using another Dicer1 conditional knockout (KO) model in which the second RNase III domain in Dicer1 is knocked out in the developing gut tube by Foxa3-Cre, Prévot et al showed that Dicer1 KO repressed acinar differentiation and induced hepatic gene expression starting from E15.5, suggesting that miRNAs are required for repressing hepatic genes and promoting pancreatic differentiation in endoderm. They also knocked out Dicer1 specifically in the acinar cells and found that KO mice failed to thrive by post-natal day 15 due to intestinal malabsorption (Prévot et al., 2013). The biogenesis of let-7b and miR-495 was inhibited upon Dicer1 deletion, which led to upregulation of Hnf6 and hepatic genes. Lynn et al used Pdx1-Cre to delete Dicer1 in all pancreatic cells and showed that Dicer1 KO pancreata displayed agenesis and differentiation defects in all pancreatic lineages. KO embryos could survive until birth but died by post-natal day 3. They also found that the defects in the endocrine lineage were due to increased Notch activity and Hes1 expression which suppressed the expression of Ngn3 (Lynn et al., 2007).
When Dicer1 was depleted in endocrine progenitors using Ngn3-Cre, the mice developed hyperglycemia during neonatal development, and KO islet cells showed a significant increase in apoptosis (Kanji et al., 2013). Interestingly, Dicer1 KO islets upregulated neuronal genes after birth, suggesting that miRNAs are critical in suppressing neuronal gene expression during the maturation of endocrine lineages (Kanji et al., 2013).
Two studies deleted Dicer1 in β-cells conditionally using the rat insulin promoter (RIP)-Cre. They found that Dicer1 KO mice developed glucose intolerance at a young age (less than two months), progressive hyperglycemia, and diabetes starting from 5 weeks of age. KO pancreata had fewer β-cells and smaller islets. Ultrastructural analysis revealed that KO β-cells contained less insulin granules (Kalis et al., 2011). In the study by Mandelbaum et al, the authors labeled KO β-cells using lineage tracing and found that KO β-cells were progressively depleted during post-natal development (Mandelbaum et al., 2012).
Another key enzyme in the miRNA biogenesis pathway, Ago2, has also been investigated in the context of diabetes. Ago2 is required for insulin release from β-cells and β-cell proliferation during insulin resistance by promoting the biogenesis of miR-375, which is a β-cell enriched miRNA known to control β-cell differentiation, proliferation, and secretion (Tattikota et al. 2014; Tattikota et al. 2013; Poy et al. 2009; El Ouaamari et al. 2008; Zhang et al. 2013; Eliasson 2017; Kloosterman et al. 2007). However, unlike Dicer1, the role of Ago2 during pancreatic development is unknown.
Individual miRNAs involved in pancreatic birth defects
MiRNAs targeting epigenetic modifiers
miR-26a
MiR-26a targets ten-eleven translocation (TET) enzymes and thymine DNA glycosylase (TDG) which catalyze the oxidation of methylated DNA (Fu et al., 2013). MiR-26a overexpression in mice leads to the downregulation of TETs and increases the number of endocrine cells (Fu et al., 2013). The authors also used an in vitro assay and showed that miR-26a overexpression enhanced endocrine differentiation of pancreatic progenitor cells. Interestingly, knocking down TETs in stem cells downregulates pluripotency factors including Oct4, Sox2, and Nanog, while increasing primitive endoderm genes like Gata6 and Gata4 (Ito et al., 2010). These data suggest that miR-26a could modulate DNA methylation during multiple time points of pancreatic development.
miR-29
MiR-29 regulates lipid metabolism and glucose metabolism during insulin resistance (He et al., 2007; Kurtz et al., 2014). Overexpression of miR-29 decreases glucose-stimulated insulin secretion (GSIS) (Bagge et al., 2012) and one target of miR-29 during GSIS is Mct1 which is a plasma membrane monocarboxylate transporter. (Pullen et al., 2011). During myogenesis, miR-29 has been shown to suppress YY1 which is a member of the polycomb group which represses transcription via chromatin modifications (Wang et al. 2008). In leukemia and lung cancer, miR-29 has been shown to regulate DNA methylation via targeting DNA methyltransferases, Dnmt3a and Dnmt3b (Fabbri et al., 2007; Garzon et al., 2009). The mechanisms regulating miR-29 expression during pancreatic development require further investigation.
miR-9
Joglekar et al. profiled the miRNA expression in developing human pancreatic islets between 8 to 37 weeks of pregnancy and found that miR-9 was expressed during endocrine development (Joglekar et al. 2009). During hPSC differentiation, miR-9 expression is the highest during the formation of primitive gut tube and posterior foregut and is decreased when cells acquire pancreatic identity (Fogel et al., 2015). MiR-9 suppresses insulin release by decreasing the expression of Onecut2 which activates Granuphilin/Slp4 to promote insulin release (Plaisance et al., 2006). Sirt1, a deacetylase of histones and transcription factors, is also a target of miR-9. During spontaneous differentiation of mouse ESCs, miR-9 is upregulated and suppresses Sirt1 expression (Saunders et al., 2010). During glucose-stimulated insulin secretion, miR-9 expression is also increased and the Sirt1 protein level is downregulated (Ramachandran et al., 2011). These data indicate that miR-9 can regulate histone acetylation via targeting Sirt1 in both embryos and adults.
miR-124
The expression of miR-124a is increased in the pancreas from E14.5 to E18.5 (Baroukh et al., 2007). MiR-124a expression is also upregulated by glucose in a pancreatic β-cell line (MIN6 cells) and during differentiation from pancreatic islet-derived mesenchymal stem cells to β-cells (Coskun et al., 2018; Tang et al., 2009). MiR-124a targets two KATP channel components (Kir6.2 and Sur1) that are involved in sensitizing β-cells to Ca2+ (Baroukh et al., 2007). In a study by Lovis et al., the authors found that overexpression of miR-124a altered insulin secretion by regulating the expression of Snap25, Rab3a, synapsin1a, Rab27a, and Noc2 (Lovis et al., 2008). MiR-124a has also been found to target a plasma membrane monocarboxylate transporter, Mct1, to modulate glucose concentration and insulin secretion in β-cells (Pullen et al., 2011). Hes1, a negative regulator of Ngn3, was also reported to be targeted by miR-124a during neuronal differentiation (Wang et al. 2010). During development, miR-124a targets Foxa2 which is the pioneer factor for pancreatic differentiation as mentioned above. MiR-124 and miR-9 have been shown to target chromatin remodeling complex subunit Baf53a and facilitate the exchange of Baf53a for Baf53b during neuronal development (Yoo et al. 2011; Yoo et al. 2009). In fact, miR-124 and miR-9 induce chromatin remodeling which enables neuronal reprogramming in human fibroblasts (Abernathy et al., 2017). The epigenetic functions of miR-124 and miR-9 during neuronal development suggest that they could modulate epigenetic regulations during pancreatic development as well.
MiRNAs well studied in pancreatic diseases
miR-375
MiR-375 is one of the most highly expressed miRNAs in both the developing and mature pancreas (Lynn et al., 2007). During differentiation of hPSCs, the expression pattern of miR-375 is similar to human fetal pancreas development (Correa‑Medina et al., 2009; Wei et al., 2013). In adults, miR-375 is expressed in β-cells and acinar cells (Avnit‑Sagi et al., 2009; Bravo-Egana et al., 2008; Dixit et al., 2016; van de Bunt et al., 2013).
MiR-375 plays an important role in regulating β-cell proliferation and function. Knocking down miR-375 in zebrafish embryos leads to morphological changes in islets such as endocrine cells not being clustered, but found in a scattered distribution (Kloosterman et al., 2007). Mice lacking miR-375 are viable; however, they have decreased β-cell mass and increased α-cell mass as early as 3 weeks of age, leading to hyperglycemia at 4 weeks and ultimately diabetes (Poy et al., 2009). Consistent with the role of miR-375 in vivo, overexpression of miR-375 in human pluripotent stem cells during spontaneous differentiation promotes the formation of glucose-responsive insulin-producing cells (Lahmy et al., 2014). Upon insulin resistance, β-cells proliferate to compensate for the increased demand for insulin. MiR-375 promotes compensatory expansion along with Ago2 by targeting Cadm1, a growth suppressor (Poy et al., 2009; Tattikota et al., 2014). One study by Zhang et al. showed that miR-375 also inhibits pancreatic progenitor proliferation by targeting YAP1 which promotes transcription of genes involved in cellular proliferation. (Zhang et al. 2013). Several studies found that miR-375 reduces insulin production and secretion by targeting Pdk1 (Pdk1 is a kinase that is involved in phosphoinositide 3-kinase pathway and mediates cellular response to insulin), Mtpn (myotrophin is a cytoplasmic protein that promotes exocytosis of insulin granules), Gephyrin and Ywhaz (both are regulators of insulin release) (El Ouaamari et al., 2008; Poy et al., 2004; Tattikota et al., 2013).
MiR-375 is upregulated in animal models for T1D as well as in human patients (Assmann et al., 2017; Erener et al., 2013), thus it has been considered as a marker for β-cell death (Erener et al., 2013). Apart from its role in endocrine regulation, it is also downregulated in pancreatitis (Wang et al. 2016) and pancreatic cancer (Bloomston et al., 2007; Szafranska et al., 2007), suggesting that miR-375 can contribute to exocrine diseases as well.
miR-7
Similar to miR-375, miR-7 is highly expressed during pancreatic development and in adult β-cells (Wei et al., 2013). In mice, miR-7 expression is upregulated from E13.5 to E14.5, and then decreases at E18.5 (Nieto et al., 2012). In humans, miR-7 is induced at 9 weeks of gestational age (wGA) and reaches the highest levels between 14 to 18 wGA (Correa-Medina et al. 2009; Joglekar et al. 2009). However, unlike miR-375 which is expressed in both endocrine and exocrine cells, miR-7 appears to be predominantly enriched in the islet (Bravo-Egana et al. 2008; Joglekar et al. 2009).
Knocking down miR-7a and miR-7b simultaneously by morpholinos in mice at E10.5 leads to increased β-cell death and decreased insulin production, and newborn mice develop glucose intolerance (Nieto et al., 2012). Interestingly, another study by Latreille et al deleted miR-7a2 using RIP-Cre and glucose tolerance in adult mice increased, whereas mice overexpressing miR-7a in β-cells developed diabetes. This study also revealed that miR-7a2 targeted the SNARE complex which regulates insulin granule fusion with the plasma membrane, thereby limiting insulin secretion (Latreille et al., 2014). The discrepancy between these two studies could be due to the time points when miR-7 was being silenced or deleted, potential compensatory function of other miR-7 members, and the method of gene silencing or deletion used.
MiR-7 has also been reported to target a key regulator of endocrine differentiation, Pax6 (Dames et al., 2010; Heller et al., 2005; Sander et al., 1997; St‑Onge et al., 1997). Knocking down miR-7 in E12.5 pancreatic explants leads to upregulation of Pax6 expression and α- and β-cell formation (Kredo‑Russo et al., 2012). In adults, miR-7 limits β-cell proliferation by targeting multiple components of mTOR signaling pathway such as Mapkap1, p70S6K, Mknk1/2, and eIF4E (Wang et al. 2013). These data suggest that miR-7 family members have diverse roles in β-cell differentiation and function.
miR-30
MiR-30 family members target mesenchymal genes such as Vimentin and Snail1 to maintain an epithelial identity during endocrine cell development (Joglekar, Patil, et al., 2009). However, miR-30 family members have distinct roles in pancreatic diseases. MiR-30a targets Beta2/NeuroD (NeuroD1) to inhibit insulin secretion (Kim et al. 2013). Indeed, suppressing miR-30a increases insulin secretion and prevents β-cell dysfunction in diabetic mice (Kim et al. 2013). In contrast, miR-30d promotes insulin secretion via targeting Map4k4 (Tang et al., 2009; Zhao et al., 2012). MiR-30d also targets Rfx6, a gene that when mutated can cause neonatal diabetes (Liao et al., 2013; Smith et al., 2010).
miR-21
Using T1D murine models, Ruan et al. found that miR-21 expression was activated by NF-κB, and increased miR-21 downregulated tumor suppressor Pdcd4, which prevented β-cell death and disease progression (Ruan et al., 2011). In addition to being upregulated in T1D patients, miR-21 has also been shown to be misregulated in pancreatitis and pancreatic cancer (Assmann et al., 2017; Dixit et al., 2016; Khan et al., 2015; Rachagani et al., 2015), suggesting that miR-21 has multiple roles in pancreatic diseases.
miR-24
MiR-24 has been shown to regulate the expression of a tumor suppressor, Men1. Decreased Men1 expression promotes β-cell proliferation (Vijayaraghavan et al., 2014). Melkman-Zehavi et al. reported that miR-24 targeted Sox6 to increase the level of insulin mRNA (Melkman‑Zehavi et al., 2011). In a study by Zhu et al., two MODY genes, Hnf1α and Neurod1, were found to be direct targets of miR-24 (Zhu et al., 2013). However, the role of miR-24 in the pathogenesis of MODY requires further investigation.
Other types of ncRNAs
PiRNAs are short single-stranded RNAs. PiRNAs form a complex with PIWI proteins to induce a repressive chromatin state of transposable elements by driving DNA methylation (Watanabe et al., 2018). Silencing transposable elements is crucial in maintaining genome stability and integrity. In germline stem cells, piRNAs ensure the fidelity of genetic material for the next generation (Teixeira et al., 2017). One study by Henaoui et al. revealed that piRNAs were differentially expressed during postnatal development and were misregulated in T2D. They also found that overexpressing T2D-related piRNAs in normal islets impaired GSIS (Henaoui et al., 2017). These data suggest that piRNAs might regulate the maturation and function of β-cells. The role of other types of ncRNAs such as snoRNA and snRNA in pancreatic development is unknown.
Conclusion
Newborns with pancreatic birth defects experience problems such as obstruction of the duodenum, nutritional malabsorption, hyperglycemia, and pancreatitis. These problems impede postnatal growth and development and predispose the patient to severe pancreatic diseases like diabetes mellitus and pancreatic cancer later in life. Activating the developmental gene regulatory network is necessary to regenerate pancreatic tissue and represents a major area of focus for treating impairments during pancreatic development. Thus, improving our understanding of developmental regulation will advance our ability to treat and cure patients with birth defects. All pancreatic cells are derived from the same progenitor population. The PPCs acquire different epigenetic modifications on chromatin, thus allowing the expression of different genes and adopting different cell fates. While histone modifications and DNA methylation are mediated by specific enzymes encoded by protein-coding sequences, the non-coding RNAs, which are transcribed from non-coding sequences, make up 98% of the genome and have diverse functions in addition to the epigenetic regulation of gene expression. Understanding ncRNA biology will significantly expand our knowledge of gene regulatory mechanisms. MiRNAs can control the expression of epigenetic modifiers post-transcriptionally via a canonical pathway. Intriguingly, some data showed that miRNAs can also direct the RNA-induced transcriptional silencing complex (RITS), which includes histone deacetylases, DNA methyltransferases, and histone methyltransferases, to target DNA sequences in the nucleus to trigger chromatin structure alterations and gene transcriptional silencing. Additionally, evidence suggests that miRNAs can mediate transcriptional gene activation. For example, miR-373 has been shown to bind to the promoter of Cdh1 and Csdc2 to induce gene expression (Place et al., 2008). LncRNAs can modulate gene expression by recruiting chromatin modifiers, acting as a scaffold, and altering chromatin structure. The exact epigenetic regulatory mechanisms used by ncRNAs are gene and cell-type dependent. In this review, we summarized how epigenetic misregulation by ncRNAs can cause pancreatic birth defects, but we are just beginning to appreciate the complex interplay of ncRNAs and epigenetic regulation. In summary, expanding our knowledge of how ncRNAs regulate pancreatic development will help uncover disease mechanisms and develop therapeutics for treating birth defects.
Acknowledgments
This research was supported by funds from Andrew McDonough B+ Foundation and the NIH (R01-HD099252 and R01- HD098131). R.J.P. is a CPRIT Scholar in Cancer Research (RR150106) and a V Scholar in Cancer Research (V Foundation #V2017-017). The authors would like to thank all the lab members, Dr. Aaron R Cox, and Dr. Jennifer M Bailey for their critical feedback on the manuscript. All figures are created with BioRender.com.
Bibliography
- Abernathy DG, Kim WK, McCoy MJ, Lake AM, Ouwenga R, Lee SW, Xing X, Li D, Lee HJ, Heuckeroth RO, Dougherty JD, Wang T, & Yoo AS (2017). MicroRNAs Induce a Permissive Chromatin Environment that Enables Neuronal Subtype-Specific Reprogramming of Adult Human Fibroblasts. Cell Stem Cell, 21(3), 332–348.e9. 10.1016/j.stem.2017.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afelik S, Chen Y, & Pieler T. (2006). Combined ectopic expression of Pdx1 and Ptf1a/p48 results in the stable conversion of posterior endoderm into endocrine and exocrine pancreatic tissue. Genes & Development, 20(11), 1441–1446. 10.1101/gad.378706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afelik S, Qu X, Hasrouni E, Bukys MA, Deering T, Nieuwoudt S, Rogers W, Macdonald RJ, & Jensen J. (2012). Notch-mediated patterning and cell fate allocation of pancreatic progenitor cells. Development, 139(10), 1744–1753. 10.1242/dev.075804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman I, Tu Z, Beucher A, Rolando DMY, Sauty-Colace C, Benazra M, Nakic N, Yang J, Wang H, Pasquali L, Moran I, Garcia-Hurtado J, Castro N, Gonzalez-Franco R, Stewart AF, Bonner C, Piemonti L, Berney T, Groop L, … Ferrer J. (2017). Human Pancreatic β Cell lncRNAs Control Cell-Specific Regulatory Networks. Cell Metabolism, 25(2), 400–411. 10.1016/j.cmet.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Zink D, & Becker PB (2000). Chromodomains are protein-RNA interaction modules. Nature, 407(6802), 405–409. 10.1038/35030169 [DOI] [PubMed] [Google Scholar]
- Aleem A, & Shah H. (2022). Annular Pancreas. In StatPearls. StatPearls Publishing. [PubMed] [Google Scholar]
- Allen HL, Flanagan SE, Shaw-Smith C, De Franco E, Akerman I, Caswell R, International Pancreatic Agenesis Consortium, Ferrer J, Hattersley AT, & Ellard S. (2011). GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nature Genetics, 44(1), 20–22. 10.1038/ng.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Diabetes Association. (2018). Economic costs of diabetes in the U.S. in 2017. Diabetes Care, 41(5), 917–928. 10.2337/dci18-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Bosch JA, Goll MG, Hesselson D, Dong PDS, Shin D, Chi NC, Shin CH, Schlegel A, Halpern M, & Stainier DYR (2009). Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Developmental Biology, 334(1), 213–223. 10.1016/j.ydbio.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnes L, Akerman I, Balderes DA, Ferrer J, & Sussel L. (2016). βlinc1 encodes a long noncoding RNA that regulates islet β-cell formation and function. Genes & Development, 30(5), 502–507. 10.1101/gad.273821.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnes L, & Sussel L. (2015). Epigenetic modifications and long noncoding RNAs influence pancreas development and function. Trends in Genetics, 31(6), 290–299. 10.1016/j.tig.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann TS, Recamonde-Mendoza M, De Souza BM, & Crispim D. (2017). MicroRNA expression profiles and type 1 diabetes mellitus: systematic review and bioinformatic analysis. Endocrine Connections, 6(8), 773–790. 10.1530/EC-17-0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avnit-Sagi T, Kantorovich L, Kredo-Russo S, Hornstein E, & Walker MD (2009). The promoter of the pri-miR-375 gene directs expression selectively to the endocrine pancreas. Plos One, 4(4), e5033. 10.1371/journal.pone.0005033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagge A, Clausen TR, Larsen S, Ladefoged M, Rosenstierne MW, Larsen L, Vang O, Nielsen JH, & Dalgaard LT (2012). MicroRNA-29a is up-regulated in beta-cells by glucose and decreases glucose-stimulated insulin secretion. Biochemical and Biophysical Research Communications, 426(2), 266–272. 10.1016/j.bbrc.2012.08.082 [DOI] [PubMed] [Google Scholar]
- Bali D, Svetlanov A, Lee HW, Fusco-DeMane D, Leiser M, Li B, Barzilai N, Surana M, Hou H, & Fleischer N. (1995). Animal model for maturity-onset diabetes of the young generated by disruption of the mouse glucokinase gene. The Journal of Biological Chemistry, 270(37), 21464–21467. 10.1074/jbc.270.37.21464 [DOI] [PubMed] [Google Scholar]
- Bannister, Andrew J, & Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Research, 21(3), 381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, & Kouzarides T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410(6824), 120–124. 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, Rutter GA, & Van Obberghen E. (2007). MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. The Journal of Biological Chemistry, 282(27), 19575–19588. 10.1074/jbc.M611841200 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell, 173(1), 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Puig I, Peña C, García JM, Alvarez AB, Peña R, Bonilla F, & de Herreros AG (2008). A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes & Development, 22(6), 756–769. 10.1101/gad.455708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, & Lander ES (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell, 125(2), 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Bernstein E, & Allis CD (2005). RNA meets chromatin. Genes & Development, 19(14), 1635–1655. 10.1101/gad.1324305 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, & Hannon GJ (2003). Dicer is essential for mouse development. Nature Genetics, 35(3), 215–217. 10.1038/ng1253 [DOI] [PubMed] [Google Scholar]
- Bhat AA, Younes SN, Raza SS, Zarif L, Nisar S, Ahmed I, Mir R, Kumar S, Sharawat SK, Hashem S, Elfaki I, Kulinski M, Kuttikrishnan S, Prabhu KS, Khan AQ, Yadav SK, El-Rifai W, Zargar MA, Zayed H, … Uddin S. (2020). Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance. Molecular Cancer, 19(1), 57. 10.1186/s12943-020-01175-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan A, Itoh N, Kato S, Thiery JP, Czernichow P, Bellusci S, & Scharfmann R. (2001). Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development, 128(24), 5109–5117. 10.1242/dev.128.24.5109 [DOI] [PubMed] [Google Scholar]
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu C-G, Bhatt D, Taccioli C, & Croce CM (2007). MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. The Journal of the American Medical Association, 297(17), 1901–1908. 10.1001/jama.297.17.1901 [DOI] [PubMed] [Google Scholar]
- Bohuslavova R, Smolik O, Malfatti J, Berkova Z, Novakova Z, Saudek F, & Pavlinkova G. (2021). NEUROD1 is required for the early α and β endocrine differentiation in the pancreas. International Journal of Molecular Sciences, 22(13). 10.3390/ijms22136713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, & Davidson BL (2006). RNA polymerase III transcribes human microRNAs. Nature Structural & Molecular Biology, 13(12), 1097–1101. 10.1038/nsmb1167 [DOI] [PubMed] [Google Scholar]
- Borghei P, Sokhandon F, Shirkhoda A, & Morgan DE (2013). Anomalies, anatomic variants, and sources of diagnostic pitfalls in pancreatic imaging. Radiology, 266(1), 28–36. 10.1148/radiol.12112469 [DOI] [PubMed] [Google Scholar]
- Bravo-Egana V, Rosero S, Molano RD, Pileggi A, Ricordi C, Domínguez-Bendala J, & Pastori RL (2008). Quantitative differential expression analysis reveals miR-7 as major islet microRNA. Biochemical and Biophysical Research Communications, 366(4), 922–926. 10.1016/j.bbrc.2007.12.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas-Diaz FL, Osorio-Quintero C, Diaz-Miranda MA, Kishore S, Leavens K, Jobaliya C, Stanescu D, Ortiz-Gonzalez X, Yoon C, Chen CS, Haliyur R, Brissova M, Powers AC, French DL, & Gadue P. (2019). Modeling Monogenic Diabetes using Human ESCs Reveals Developmental and Metabolic Deficiencies Caused by Mutations in HNF1A. Cell Stem Cell, 25(2), 273–289.e5. 10.1016/j.stem.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco M, Delgado I, Soria B, Martín F, & Rojas A. (2012). GATA4 and GATA6 control mouse pancreas organogenesis. The Journal of Clinical Investigation, 122(10), 3504–3515. 10.1172/JCI63240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalazonitis NA, Lachanis BS, Laspas F, Ptohis N, Tsimitselis G, & Tzovara J. (2008). Pancreas divisum: magnetic resonance cholangiopancreatography findings. Singapore Medical Journal, 49(11), 951–954; quiz 955. [PubMed] [Google Scholar]
- Chen H, Gu X, Su I, Bottino R, Contreras JL, Tarakhovsky A, & Kim SK (2009). Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes & Development, 23(8), 975–985. 10.1101/gad.1742509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wang Y, Wang C, Hu J-F, & Li W. (2020). Lncrna functions as a new emerging epigenetic factor in determining the fate of stem cells. Frontiers in Genetics, 11, 277. 10.3389/fgene.2020.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Medina M, Bravo-Egana V, Rosero S, Ricordi C, Edlund H, Diez J, & Pastori RL (2009). MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expression Patterns, 9(4), 193–199. 10.1016/j.gep.2008.12.003 [DOI] [PubMed] [Google Scholar]
- Cosgrove MS, Boeke JD, & Wolberger C. (2004). Regulated nucleosome mobility and the histone code. Nature Structural & Molecular Biology, 11(11), 1037–1043. 10.1038/nsmb851 [DOI] [PubMed] [Google Scholar]
- Coskun E, Ercin M, & Gezginci-Oktayoglu S. (2018). The Role of Epigenetic Regulation and Pluripotency-Related MicroRNAs in Differentiation of Pancreatic Stem Cells to Beta Cells. Journal of Cellular Biochemistry, 119(1), 455–467. 10.1002/jcb.26203 [DOI] [PubMed] [Google Scholar]
- Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, & Gutti RK (2019). Long non-coding RNA: Classification, biogenesis and functions in blood cells. Molecular Immunology, 112, 82–92. 10.1016/j.molimm.2019.04.011 [DOI] [PubMed] [Google Scholar]
- Dames P, Puff R, Weise M, Parhofer KG, Göke B, Götz M, Graw J, Favor J, & Lechner A. (2010). Relative roles of the different Pax6 domains for pancreatic alpha cell development. BMC Developmental Biology, 10, 39. 10.1186/1471-213X-10-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar K, Pondick JV, Kim B-M, Zhou C, York SR, Macklin JA, Abualteen A, Tan B, Sigova AA, Marcho C, Tremblay KD, Mager J, Choi MY, & Mullen AC (2016). DIGIT Is a Conserved Long Noncoding RNA that Regulates GSC Expression to Control Definitive Endoderm Differentiation of Embryonic Stem Cells. Cell Reports, 17(2), 353–365. 10.1016/j.celrep.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L, Ma TP, Farber SA, Moens CB, & Stainier DYR (2012). Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genetics, 8(6), e1002754. 10.1371/journal.pgen.1002754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vas MG, Kopp JL, Heliot C, Sander M, Cereghini S, & Haumaitre C. (2015). Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Development, 142(5), 871–882. 10.1242/dev.110759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch G, Jung J, Zheng M, Lóra J, & Zaret KS (2001). A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development, 128(6), 871–881. 10.1242/dev.128.6.871 [DOI] [PubMed] [Google Scholar]
- Dhawan S, Georgia S, Tschen S-I, Fan G, & Bhushan A. (2011). Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx. Developmental Cell, 20(4), 419–429. 10.1016/j.devcel.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens LT, Letourneau LR, Sanyoura M, Greeley SAW, Philipson LH, & Naylor RN (2019). Management and pregnancy outcomes of women with GCK-MODY enrolled in the US Monogenic Diabetes Registry. Acta Diabetologica, 56(4), 405–411. 10.1007/s00592-018-1267-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, & Mattick JS (2008). Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Research, 18(9), 1433–1445. 10.1101/gr.078378.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit AK, Sarver AE, Yuan Z, George J, Barlass U, Cheema H, Sareen A, Banerjee S, Dudeja V, Dawra R, Subramanian S, & Saluja AK (2016). Comprehensive analysis of microRNA signature of mouse pancreatic acini: overexpression of miR-21–3p in acute pancreatitis. American Journal of Physiology. Gastrointestinal and Liver Physiology, 311(5), G974–G980. 10.1152/ajpgi.00191.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MJ, & Sussel L. (2007). Nkx2.2 regulates beta-cell function in the mature islet. Diabetes, 56(8), 1999–2007. 10.2337/db06-1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier O, & Van Obberghen E. (2012). MicroRNAs in pancreas development. Diabetes, Obesity & Metabolism, 14 Suppl 3, 22–28. 10.1111/j.1463-1326.2012.01656.x [DOI] [PubMed] [Google Scholar]
- Eliasson L. (2017). The small RNA miR-375 - a pancreatic islet abundant miRNA with multiple roles in endocrine beta cell function. Molecular and Cellular Endocrinology, 456, 95–101. 10.1016/j.mce.2017.02.043 [DOI] [PubMed] [Google Scholar]
- El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, & van Obberghen E. (2008). miR-375 targets 3’-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes, 57(10), 2708–2717. 10.2337/db07-1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Mojibian M, Fox JK, Denroche HC, & Kieffer TJ (2013). Circulating miR-375 as a biomarker of β-cell death and diabetes in mice. Endocrinology, 154(2), 603–608. 10.1210/en.2012-1744 [DOI] [PubMed] [Google Scholar]
- Esni F, Ghosh B, Biankin AV, Lin JW, Albert MA, Yu X, MacDonald RJ, Civin CI, Real FX, Pack MA, Ball DW, & Leach SD (2004). Notch inhibits Ptf1 function and acinar cell differentiation in developing mouse and zebrafish pancreas. Development, 131(17), 4213–4224. 10.1242/dev.01280 [DOI] [PubMed] [Google Scholar]
- Esteller M. (2011). Non-coding RNAs in human disease. Nature Reviews. Genetics, 12(12), 861–874. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Etienne D, John A, Menias CO, Ward R, Tubbs RS, & Loukas M. (2012). Annular pancreas: a review of its molecular embryology, genetic basis and clinical considerations. Annals of Anatomy = Anatomischer Anzeiger : Official Organ of the Anatomische Gesellschaft, 194(5), 422–428. 10.1016/j.aanat.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, & Croce CM (2007). MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proceedings of the National Academy of Sciences of the United States of America, 104(40), 15805–15810. 10.1073/pnas.0707628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajans SS, Bell GI, & Polonsky KS (2001). Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. The New England Journal of Medicine, 345(13), 971–980. 10.1056/NEJMra002168 [DOI] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, & Kohtz JD (2006). The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes & Development, 20(11), 1470–1484. 10.1101/gad.1416106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan SE, De Franco E, Lango Allen H, Zerah M, Abdul-Rasoul MM, Edge JA, Stewart H, Alamiri E, Hussain K, Wallis S, de Vries L, Rubio-Cabezas O, Houghton JAL, Edghill EL, Patch A-M, Ellard S, & Hattersley AT (2014). Analysis of transcription factors key for mouse pancreatic development establishes NKX2–2 and MNX1 mutations as causes of neonatal diabetes in man. Cell Metabolism, 19(1), 146–154. 10.1016/j.cmet.2013.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel GB, Kai ZS, Zargar S, Hinton A, Jones GA, Wong AS, Ficici SG, Lopez AD, & King CC (2015). MicroRNA dynamics during human embryonic stem cell differentiation to pancreatic endoderm. Gene, 574(2), 359–370. 10.1016/j.gene.2015.08.027 [DOI] [PubMed] [Google Scholar]
- Font-Cunill B, Arnes L, Ferrer J, Sussel L, & Beucher A. (2018). Long Non-coding RNAs as Local Regulators of Pancreatic Islet Transcription Factor Genes. Frontiers in Genetics, 9, 524. 10.3389/fgene.2018.00524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK-H, Burge CB, & Bartel DP (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Research, 19(1), 92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa M, Morita S, Kimura M, Horii T, Ochiya T, & Hatada I. (2006). Genomic imprinting in Dicer1-hypomorphic mice. Cytogenetic and Genome Research, 113(1–4), 138–143. 10.1159/000090825 [DOI] [PubMed] [Google Scholar]
- Fu X, Jin L, Wang X, Luo A, Hu J, Zheng X, Tsark WM, Riggs AD, Ku HT, & Huang W. (2013). MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proceedings of the National Academy of Sciences of the United States of America, 110(44), 17892–17897. 10.1073/pnas.1317397110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B, Carrano AC, & Sander M. (2019). Human stem cell models: lessons for pancreatic development and disease. Genes & Development, 33(21–22), 1475–1490. 10.1101/gad.331397.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B, van Heesch S, Schneider-Lunitz V, Schulz JF, Witte F, Blachut S, Nguyen S, Wong R, Matta I, Hübner N, & Sander M. (2020). A human ESC-based screen identifies a role for the translated lncRNA LINC00261 in pancreatic endocrine differentiation. ELife, 9. 10.7554/eLife.58659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CEA, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, … Marcucci G. (2009). MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood, 113(25), 6411–6418. 10.1182/blood-2008-07-170589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgia S, Kanji M, & Bhushan A. (2013). DNMT1 represses p53 to maintain progenitor cell survival during pancreatic organogenesis. Genes & Development, 27(4), 372–377. 10.1101/gad.207001.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, & Bernstein E. (2007). Epigenetics: a landscape takes shape. Cell, 128(4), 635–638. 10.1016/j.cell.2007.02.006 [DOI] [PubMed] [Google Scholar]
- Golosow N, & Grobstein C. (1962). Epitheliomesenchymal interaction in pancreatic morphogenesis. Developmental Biology, 4, 242–255. 10.1016/0012-1606(62)90042-8 [DOI] [PubMed] [Google Scholar]
- Golson ML, & Kaestner KH (2017). Epigenetics in formation, function, and failure of the endocrine pancreas. Molecular Metabolism, 6(9), 1066–1076. 10.1016/j.molmet.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzi M, Kim YH, Katsumoto K, Johansson K, & Grapin-Botton A. (2011). Neurogenin3 initiates stepwise delamination of differentiating endocrine cells during pancreas development. Developmental Dynamics, 240(3), 589–604. 10.1002/dvdy.22544 [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Majithia AR, & Melton DA (2001). Key events of pancreas formation are triggered in gut endoderm by ectopic expression of pancreatic regulatory genes. Genes & Development, 15(4), 444–454. 10.1101/gad.846001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guay C, Jacovetti C, Nesca V, Motterle A, Tugay K, & Regazzi R. (2012). Emerging roles of non-coding RNAs in pancreatic β-cell function and dysfunction. Diabetes, Obesity & Metabolism, 14 Suppl 3, 12–21. 10.1111/j.1463-1326.2012.01654.x [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, & Melton DA (2002). Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development, 129(10), 2447–2457. 10.1242/dev.129.10.2447 [DOI] [PubMed] [Google Scholar]
- Hajjari M, & Salavaty A. (2015). HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biology & Medicine, 12(1), 1–9. 10.7497/j.issn.2095-3941.2015.0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K-I, Ayoub N, Cohen A, & Grewal SIS (2002). Establishment and maintenance of a heterochromatin domain. Science, 297(5590), 2232–2237. 10.1126/science.1076466 [DOI] [PubMed] [Google Scholar]
- Haumaitre C, Lenoir O, & Scharfmann R. (2008). Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Molecular and Cellular Biology, 28(20), 6373–6383. 10.1128/MCB.00413-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes TR, Thorne AW, & Crane-Robinson C. (1988). A direct link between core histone acetylation and transcriptionally active chromatin. The EMBO Journal, 7(5), 1395–1402. 10.1002/j.1460-2075.1988.tb02956.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, & Melton DA (1998). Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes & Development, 12(11), 1705–1713. 10.1101/gad.12.11.1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, St Jacques B, McMahon AP, & Melton DA (2000). Regulation of pancreas development by hedgehog signaling. Development, 127(22), 4905–4913. 10.1242/dev.127.22.4905 [DOI] [PubMed] [Google Scholar]
- Heller RS, Jenny M, Collombat P, Mansouri A, Tomasetto C, Madsen OD, Mellitzer G, Gradwohl G, & Serup P. (2005). Genetic determinants of pancreatic epsilon-cell development. Developmental Biology, 286(1), 217–224. 10.1016/j.ydbio.2005.06.041 [DOI] [PubMed] [Google Scholar]
- Heller RS (2010). The comparative anatomy of islets. Advances in Experimental Medicine and Biology, 654, 21–37. 10.1007/978-90-481-3271-3_2 [DOI] [PubMed] [Google Scholar]
- Henaoui IS, Jacovetti C, Guerra Mollet I, Guay C, Sobel J, Eliasson L, & Regazzi R. (2017). PIWI-interacting RNAs as novel regulators of pancreatic beta cell function. Diabetologia, 60(10), 1977–1986. 10.1007/s00125-017-4368-2 [DOI] [PubMed] [Google Scholar]
- He A, Zhu L, Gupta N, Chang Y, & Fang F. (2007). Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Molecular Endocrinology, 21(11), 2785–2794. 10.1210/me.2007-0167 [DOI] [PubMed] [Google Scholar]
- Hermann FM, Kjærgaard MF, Tian C, Tiemann U, Jackson A, Olsen LR, Kraft M, Carlsson P-O, Elfving IM, Kettunen JLT, Tuomi T, Novak I, & Semb H. (2022). An insulin hypersecretion phenotype precedes pancreatic β cell failure in MODY3 patient-specific cells. Cell Stem Cell. 10.1016/j.stem.2022.12.001 [DOI] [PubMed] [Google Scholar]
- Hesselson D, Anderson RM, & Stainier DYR (2011). Suppression of Ptf1a activity induces acinar-to-endocrine conversion. Current Biology, 21(8), 712–717. 10.1016/j.cub.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, & Moazed D. (2015). RNA-mediated epigenetic regulation of gene expression. Nature Reviews. Genetics, 16(2), 71–84. 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn BN, Lindner T, Yamagata K, Ogata M, Tomonaga O, Kuroki H, Kasahara T, Iwamoto Y, & Bell GI (1997). Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nature Genetics, 17(4), 384–385. 10.1038/ng1297-384 [DOI] [PubMed] [Google Scholar]
- Huang L, Desai R, Conrad DN, Leite NC, Akshinthala D, Lim CM, Gonzalez R, Muthuswamy LB, Gartner Z, & Muthuswamy SK (2021). Commitment and oncogene-induced plasticity of human stem cell-derived pancreatic acinar and ductal organoids. Cell Stem Cell, 28(6), 1090–1104.e6. 10.1016/j.stem.2021.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbregts L, Petersen MBK, Berthault C, Hansson M, Aiello V, Rachdi L, Grapin-Botton A, Honore C, & Scharfmann R. (2019). Bromodomain and extra terminal protein inhibitors promote pancreatic endocrine cell fate. Diabetes, 68(4), 761–773. 10.2337/db18-0224 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, & Izaurralde E. (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature Reviews. Genetics, 12(2), 99–110. 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, & Hafen E. (2002). Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Current Biology, 12(15), 1293–1300. 10.1016/s0960-9822(02)01043-6 [DOI] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, & Zhang Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature, 466(7310), 1129–1133. 10.1038/nature09303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Hada T, Terano A, Itai Y, & Harada T. (1989). Pancreatitis in the annulus of annular pancreas demonstrated by the combined use of computed tomography and endoscopic retrograde cholangiopancreatography. The American Journal of Gastroenterology, 84(8), 961–964. [PubMed] [Google Scholar]
- Jiang W, Liu Y, Liu R, Zhang K, & Zhang Y. (2015). The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Reports, 11(1), 137–148. 10.1016/j.celrep.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang J, & Zhang Y. (2013). Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Research, 23(1), 122–130. 10.1038/cr.2012.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar MV, Joglekar VM, & Hardikar AA (2009). Expression of islet-specific microRNAs during human pancreatic development. Gene Expression Patterns, 9(2), 109–113. 10.1016/j.gep.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Joglekar MV, Patil D, Joglekar VM, Rao GV, Reddy DN, Mitnala S, Shouche Y, & Hardikar AA (2009). The miR-30 family microRNAs confer epithelial phenotype to human pancreatic cells. Islets, 1(2), 137–147. 10.4161/isl.1.2.9578 [DOI] [PubMed] [Google Scholar]
- Jonas S, & Izaurralde E. (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nature Reviews. Genetics, 16(7), 421–433. 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- Jørgensen MC, Ahnfelt-Rønne J, Hald J, Madsen OD, Serup P, & Hecksher-Sørensen J. (2007). An illustrated review of early pancreas development in the mouse. Endocrine Reviews, 28(6), 685–705. 10.1210/er.2007-0016 [DOI] [PubMed] [Google Scholar]
- Kaimala S, Kumar CA, Allouh MZ, Ansari SA, & Emerald BS (2022). Epigenetic modifications in pancreas development, diabetes, and therapeutics. Medicinal Research Reviews, 42(3), 1343–1371. 10.1002/med.21878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalis M, Bolmeson C, Esguerra JLS, Gupta S, Edlund A, Tormo-Badia N, Speidel D, Holmberg D, Mayans S, Khoo NKS, Wendt A, Eliasson L, & Cilio CM (2011). Beta-cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. Plos One, 6(12), e29166. 10.1371/journal.pone.0029166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanji MS, Martin MG, & Bhushan A. (2013). Dicer1 is required to repress neuronal fate during endocrine cell maturation. Diabetes, 62(5), 1602–1611. 10.2337/db12-0841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartikasari AER, Zhou JX, Kanji MS, Chan DN, Sinha A, Grapin-Botton A, Magnuson MA, Lowry WE, & Bhushan A. (2013). The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. The EMBO Journal, 32(10), 1393–1408. 10.1038/emboj.2013.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I, Yisraeli J, & Cedar H. (1985). Effect of regional DNA methylation on gene expression. Proceedings of the National Academy of Sciences of the United States of America, 82(9), 2560–2564. 10.1073/pnas.82.9.2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Zubair H, Srivastava SK, Singh S, & Singh AP (2015). Insights into the Role of microRNAs in Pancreatic Cancer Pathogenesis: Potential for Diagnosis, Prognosis, and Therapy. Advances in Experimental Medicine and Biology, 889, 71–87. 10.1007/978-3-319-23730-5_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, You YH, Jung S, Suh-Kim H, Lee IK, Cho JH, & Yoon KH (2013). miRNA-30a-5p-mediated silencing of Beta2/NeuroD expression is an important initial event of glucotoxicity-induced beta cell dysfunction in rodent models. Diabetologia, 56(4), 847–855. 10.1007/s00125-012-2812-x [DOI] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, & Melton DA (1997). Notochord to endoderm signaling is required for pancreas development. Development, 124(21), 4243–4252. 10.1242/dev.124.21.4243 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, & Plasterk RHA (2007). Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biology, 5(8), e203. 10.1371/journal.pbio.0050203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofent J, Zhang J, & Spagnoli FM (2016). The histone methyltransferase Setd7 promotes pancreatic progenitor identity. Development, 143(19), 3573–3581. 10.1242/dev.136226 [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shinjo K, & Katsushima K. (2017). Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Science, 108(10), 1927–1933. 10.1111/cas.13342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Knöfler M, Ledermann B, Bürki K, Berney C, Zoerkler N, Hagenbüchle O, & Wellauer PK (1998). The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes & Development, 12(23), 3752–3763. 10.1101/gad.12.23.3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredo-Russo S, Mandelbaum AD, Ness A, Alon I, Lennox KA, Behlke MA, & Hornstein E. (2012). Pancreas-enriched miRNA refines endocrine cell differentiation. Development, 139(16), 3021–3031. 10.1242/dev.080127 [DOI] [PubMed] [Google Scholar]
- Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, Agulnick AD, D’Amour KA, Carpenter MK, & Baetge EE (2008). Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nature Biotechnology, 26(4), 443–452. 10.1038/nbt1393 [DOI] [PubMed] [Google Scholar]
- Kumar S, Gonzalez EA, Rameshwar P, & Etchegaray J-P (2020). Non-Coding RNAs as Mediators of Epigenetic Changes in Malignancies. Cancers, 12(12). 10.3390/cancers12123657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz CL, Peck BCE, Fannin EE, Beysen C, Miao J, Landstreet SR, Ding S, Turaga V, Lund PK, Turner S, Biddinger SB, Vickers KC, & Sethupathy P. (2014). MicroRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes, 63(9), 3141–3148. 10.2337/db13-1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzel AR, Lodhi MU, & Rahim M. (2017). Pancreatic divisum: an unusual cause of chronic pancreatitis in a young patient. Cureus, 9(11), e1856. 10.7759/cureus.1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy R, Soleimani M, Sanati MH, Behmanesh M, Kouhkan F, & Mobarra N. (2014). MiRNA-375 promotes beta pancreatic differentiation in human induced pluripotent stem (hiPS) cells. Molecular Biology Reports, 41(4), 2055–2066. 10.1007/s11033-014-3054-4 [DOI] [PubMed] [Google Scholar]
- Lammert E, Cleaver O, & Melton D. (2001). Induction of pancreatic differentiation by signals from blood vessels. Science, 294(5542), 564–567. 10.1126/science.1064344 [DOI] [PubMed] [Google Scholar]
- Landsman L, Nijagal A, Whitchurch TJ, Vanderlaan RL, Zimmer WE, Mackenzie TC, & Hebrok M. (2011). Pancreatic mesenchyme regulates epithelial organogenesis throughout development. PLoS Biology, 9(9), e1001143. 10.1371/journal.pbio.1001143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardenoije R, Iatrou A, Kenis G, Kompotis K, Steinbusch HWM, Mastroeni D, Coleman P, Lemere CA, Hof PR, van den Hove DLA, & Rutten BPF (2015). The epigenetics of aging and neurodegeneration. Progress in Neurobiology, 131, 21–64. 10.1016/j.pneurobio.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latreille M, Hausser J, Stützer I, Zhang Q, Hastoy B, Gargani S, Kerr-Conte J, Pattou F, Zavolan M, Esguerra JLS, Eliasson L, Rülicke T, Rorsman P, & Stoffel M. (2014). MicroRNA-7a regulates pancreatic β cell function. The Journal of Clinical Investigation, 124(6), 2722–2735. 10.1172/JCI73066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, & Chung HM (2011). Differentiation into Endoderm Lineage: Pancreatic differentiation from Embryonic Stem Cells. International Journal of Stem Cells, 4(1), 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Smith SB, Watada H, Lin J, Scheel D, Wang J, Mirmira RG, & German MS (2001). Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes, 50(5), 928–936. 10.2337/diabetes.50.5.928 [DOI] [PubMed] [Google Scholar]
- Lee K, Cho H, Rickert RW, Li QV, Pulecio J, Leslie CS, & Huangfu D. (2019). FOXA2 Is Required for Enhancer Priming during Pancreatic Differentiation. Cell Reports, 28(2), 382–393.e7. 10.1016/j.celrep.2019.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, & Kim VN (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature, 425(6956), 415–419. 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee J-T, Kim S, & Kim VN (2002). MicroRNA maturation: stepwise processing and subcellular localization. The EMBO Journal, 21(17), 4663–4670. 10.1093/emboj/cdf476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom K-H, Lee S, Baek SH, & Kim VN (2004). MicroRNA genes are transcribed by RNA polymerase II. The EMBO Journal, 23(20), 4051–4060. 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir O, Flosseau K, Ma FX, Blondeau B, Mai A, Bassel-Duby R, Ravassard P, Olson EN, Haumaitre C, & Scharfmann R. (2011). Specific control of pancreatic endocrine β- and δ-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9. Diabetes, 60(11), 2861–2871. 10.2337/db11-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Xue H, Wang Y-C, Nazor KL, Guo S, Trivedi N, Peterson SE, Liu Y, Loring JF, & Laurent LC (2013). Matched miRNA and mRNA signatures from an hESC-based in vitro model of pancreatic differentiation reveal novel regulatory interactions. Journal of Cell Science, 126(Pt 17), 3848–3861. 10.1242/jcs.123570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Jidong, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J-J, Hammond SM, Joshua-Tor L, & Hannon GJ (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science, 305(5689), 1437–1441. 10.1126/science.1102513 [DOI] [PubMed] [Google Scholar]
- Liu Jing, Banerjee A, Herring CA, Attalla J, Hu R, Xu Y, Shao Q, Simmons AJ, Dadi PK, Wang S, Jacobson DA, Liu B, Hodges E, Lau KS, & Gu G. (2019). Neurog3-Independent Methylation Is the Earliest Detectable Mark Distinguishing Pancreatic Progenitor Identity. Developmental Cell, 48(1), 49–63.e7. 10.1016/j.devcel.2018.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage VRB, Jarmasz JS, Murugeshan N, Del Bigio MR, Rastegar M, & Davie JR (2014). DNA modifications: function and applications in normal and disease States. Biology, 3(4), 670–723. 10.3390/biology3040670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, & Edlund H. (2001). Persistent expression of Hlxb9 in the pancreatic epithelium impairs pancreatic development. Developmental Biology, 240(1), 247–253. 10.1006/dbio.2001.0440 [DOI] [PubMed] [Google Scholar]
- Li J, Wu X, Ke J, Lee M, Lan Q, Li J, Yu J, Huang Y, Sun D-Q, & Xie R. (2022). TET1 dioxygenase is required for FOXA2-associated chromatin remodeling in pancreatic beta-cell differentiation. Nature Communications, 13(1), 3907. 10.1038/s41467-022-31611-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, & Kaestner KH (2012). Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell, 151(7), 1608–1616. 10.1016/j.cell.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker DS, Gorelick F, & Thompson ED (2018). Anatomy, histology, and fine structure of the pancreas. In Beger HG, Warshaw AL, Hruban RH, Büchler MW, Lerch MM, Neoptolemos JP, Shimosegawa T, Whitcomb DC, & Groβ C. (Eds.), The pancreas: an integrated textbook of basic science, medicine, and surgery (pp. 10–23). John Wiley & Sons, Ltd. 10.1002/9781119188421.ch2 [DOI] [Google Scholar]
- Lorberbaum DS, Docherty FM, & Sussel L. (2020). Animal models of pancreas development, developmental disorders, and disease. Advances in Experimental Medicine and Biology, 1236, 65–85. 10.1007/978-981-15-2389-2_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovis P, Gattesco S, & Regazzi R. (2008). Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biological Chemistry, 389(3), 305–312. 10.1515/BC.2008.026 [DOI] [PubMed] [Google Scholar]
- Low BSJ, Lim CS, Ding SSL, Tan YS, Ng NHJ, Krishnan VG, Ang SF, Neo CWY, Verma CS, Hoon S, Lim SC, Tai ES, & Teo AKK (2021). Decreased GLUT2 and glucose uptake contribute to insulin secretion defects in MODY3/HNF1A hiPSC-derived mutant β cells. Nature Communications, 12(1), 3133. 10.1038/s41467-021-22843-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE, & Kutay U. (2004). Nuclear export of microRNA precursors. Science, 303(5654), 95–98. 10.1126/science.1090599 [DOI] [PubMed] [Google Scholar]
- Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, & German MS (2007). MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes, 56(12), 2938–2945. 10.2337/db07-0175 [DOI] [PubMed] [Google Scholar]
- Madsen OD (2007). Pancreas phylogeny and ontogeny in relation to a “pancreatic stem cell”. Comptes Rendus Biologies, 330(6–7), 534–537. 10.1016/j.crvi.2007.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan V. (2019). Anatomy of the pancreas and spleen. Surgery (Oxford), 37(6), 297–301. 10.1016/j.mpsur.2019.04.008 [DOI] [Google Scholar]
- Mandelbaum AD, Melkman-Zehavi T, Oren R, Kredo-Russo S, Nir T, Dor Y, & Hornstein E. (2012). Dysregulation of Dicer1 in beta cells impairs islet architecture and glucose metabolism. Experimental Diabetes Research, 2012, 470302. 10.1155/2012/470302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui T, Long Q, Beres TM, Magnuson MA, & MacDonald RJ (2007). Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes & Development, 21(20), 2629–2643. 10.1101/gad.1575207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui T, Swift GH, Deering T, Shen C, Coats WS, Long Q, Elsässer H-P, Magnuson MA, & MacDonald RJ (2010). Replacement of Rbpj with Rbpjl in the PTF1 complex controls the final maturation of pancreatic acinar cells. Gastroenterology, 139(1), 270–280. 10.1053/j.gastro.2010.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui T, Swift GH, Hale MA, Meredith DM, Johnson JE, & Macdonald RJ (2008). Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Molecular and Cellular Biology, 28(17), 5458–5468. 10.1128/MCB.00549-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald TJ, & Ellard S. (2013). Maturity onset diabetes of the young: identification and diagnosis. Annals of Clinical Biochemistry, 50(Pt 5), 403–415. 10.1177/0004563213483458 [DOI] [PubMed] [Google Scholar]
- Melkman-Zehavi T, Oren R, Kredo-Russo S, Shapira T, Mandelbaum AD, Rivkin N, Nir T, Lennox KA, Behlke MA, Dor Y, & Hornstein E. (2011). miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. The EMBO Journal, 30(5), 835–845. 10.1038/emboj.2010.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Azad F, Polignano IL, Proserpio V, & Oliviero S. (2021). Long noncoding rnas in human stemness and differentiation. Trends in Cell Biology, 31(7), 542–555. 10.1016/j.tcb.2021.02.002 [DOI] [PubMed] [Google Scholar]
- Mohammad F, Pandey GK, Mondal T, Enroth S, Redrup L, Gyllensten U, & Kanduri C. (2012). Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development, 139(15), 2792–2803. 10.1242/dev.079566 [DOI] [PubMed] [Google Scholar]
- Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, & Dandolo L. (2013). H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proceedings of the National Academy of Sciences of the United States of America, 110(51), 20693–20698. 10.1073/pnas.1310201110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T, & Fan G. (2013). DNA Methylation and Its Basic Function. Neuropsychopharmacology, 38(1), 23–38. 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakić N, García-Hurtado J, Rodríguez-Seguí S, Pasquali L, Sauty-Colace C, Beucher A, Scharfmann R, van Arensbergen J, Johnson PR, Berry A, Lee C, Harkins T, … Ferrer J. (2012). Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metabolism, 16(4), 435–448. 10.1016/j.cmet.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Hara A, Kojima I, Horii T, Kimura M, Kitamura T, Ochiya T, Nakanishi K, Matoba R, Matsubara K, & Hatada I. (2009). Dicer is required for maintaining adult pancreas. Plos One, 4(1), e4212. 10.1371/journal.pone.0004212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DM (1993). Long-term complications of diabetes mellitus. The New England Journal of Medicine, 328(23), 1676–1685. 10.1056/NEJM199306103282306 [DOI] [PubMed] [Google Scholar]
- National Diabetes Statistics Report| Diabetes | CDC. (n.d.). Retrieved November 25, 2022, from https://www.cdc.gov/diabetes/data/statistics-report/index.html
- Ng NHJ, Jasmen JB, Lim CS, Lau HH, Krishnan VG, Kadiwala J, Kulkarni RN, Ræder H, Vallier L, Hoon S, & Teo AKK (2019). HNF4A Haploinsufficiency in MODY1 Abrogates Liver and Pancreas Differentiation from Patient-Derived Induced Pluripotent Stem Cells. IScience, 16, 192–205. 10.1016/j.isci.2019.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TA, Jo MH, Choi Y-G, Park J, Kwon SC, Hohng S, Kim VN, & Woo J-S (2015). Functional anatomy of the human microprocessor. Cell, 161(6), 1374–1387. 10.1016/j.cell.2015.05.010 [DOI] [PubMed] [Google Scholar]
- Niborski LL, Paces-Fessy M, Ricci P, Bourgeois A, Magalhães P, Kuzma-Kuzniarska M, Lesaulnier C, Reczko M, Declercq E, Zürbig P, Doucet A, Umbhauer M, & Cereghini S. (2021). Hnf1b haploinsufficiency differentially affects developmental target genes in a new renal cysts and diabetes mouse model. Disease Models & Mechanisms, 14(5). 10.1242/dmm.047498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicetto D, Donahue G, Jain T, Peng T, Sidoli S, Sheng L, Montavon T, Becker JS, Grindheim JM, Blahnik K, Garcia BA, Tan K, Bonasio R, Jenuwein T, & Zaret KS (2019). H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science, 363(6424), 294–297. 10.1126/science.aau0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Hevia P, Garcia E, Klein D, Alvarez-Cubela S, Bravo-Egana V, Rosero S, Damaris Molano R, Vargas N, Ricordi C, Pileggi A, Diez J, Domínguez-Bendala J, & Pastori RL (2012). Antisense miR-7 impairs insulin expression in developing pancreas and in cultured pancreatic buds. Cell Transplantation, 21(8), 1761–1774. 10.3727/096368911X612521 [DOI] [PubMed] [Google Scholar]
- Nijs E, Callahan MJ, & Taylor GA (2005). Disorders of the pediatric pancreas: imaging features. Pediatric Radiology, 35(4), 358–373; quiz 457. 10.1007/s00247-004-1326-1 [DOI] [PubMed] [Google Scholar]
- Noël ES, Casal-Sueiro A, Busch-Nentwich E, Verkade H, Dong PDS, Stemple DL, & Ober EA (2008). Organ-specific requirements for Hdac1 in liver and pancreas formation. Developmental Biology, 322(2), 237–250. 10.1016/j.ydbio.2008.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgaard GA, Jensen JN, & Jensen J. (2003). FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development. Developmental Biology, 264(2), 323–338. 10.1016/j.ydbio.2003.08.013 [DOI] [PubMed] [Google Scholar]
- O’Brien J, Hayder H, Zayed Y, & Peng C. (2018). Overview of microrna biogenesis, mechanisms of actions, and circulation. Frontiers in Endocrinology, 9, 402. 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, & Wright CV (1996). PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development, 122(3), 983–995. 10.1242/dev.122.3.983 [DOI] [PubMed] [Google Scholar]
- Ozcan S. (2014). Minireview: microRNA function in pancreatic β cells. Molecular Endocrinology, 28(12), 1922–1933. 10.1210/me.2014-1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, & Melton DA (2014). Generation of functional human pancreatic β cells in vitro. Cell, 159(2), 428–439. 10.1016/j.cell.2014.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, & Lee ES (2015). Non-coding RNA: what is functional and what is junk? Frontiers in Genetics, 6, 2. 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, & Kanduri C. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Molecular Cell, 32(2), 232–246. 10.1016/j.molcel.2008.08.022 [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, & Brockdorff N. (1996). Requirement for Xist in X chromosome inactivation. Nature, 379(6561), 131–137. 10.1038/379131a0 [DOI] [PubMed] [Google Scholar]
- Peter ME (2010). Targeting of mRNAs by multiple miRNAs: the next step. Oncogene, 29(15), 2161–2164. 10.1038/onc.2010.59 [DOI] [PubMed] [Google Scholar]
- Pictet RL, Clark WR, Williams RH, & Rutter WJ (1972). An ultrastructural analysis of the developing embryonic pancreas. Developmental Biology, 29(4), 436–467. 10.1016/0012-1606(72)90083-8 [DOI] [PubMed] [Google Scholar]
- Pihoker C, Gilliam LK, Ellard S, Dabelea D, Davis C, Dolan LM, Greenbaum CJ, Imperatore G, Lawrence JM, Marcovina SM, Mayer-Davis E, Rodriguez BL, Steck AK, Williams DE, Hattersley AT, & SEARCH for Diabetes in Youth Study Group. (2013). Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the SEARCH for Diabetes in Youth. The Journal of Clinical Endocrinology and Metabolism, 98(10), 4055–4062. 10.1210/jc.2013-1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li L-C, Pookot D, Noonan EJ, & Dahiya R. (2008). MicroRNA-373 induces expression of genes with complementary promoter sequences. Proceedings of the National Academy of Sciences of the United States of America, 105(5), 1608–1613. 10.1073/pnas.0707594105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, & Regazzi R. (2006). MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. The Journal of Biological Chemistry, 281(37), 26932–26942. 10.1074/jbc.M601225200 [DOI] [PubMed] [Google Scholar]
- Pontier DB, & Gribnau J. (2011). Xist regulation and function explored. Human Genetics, 130(2), 223–236. 10.1007/s00439-011-1008-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, & Stoffel M. (2004). A pancreatic islet-specific microRNA regulates insulin secretion. Nature, 432(7014), 226–230. 10.1038/nature03076 [DOI] [PubMed] [Google Scholar]
- Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, & Stoffel M. (2009). miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proceedings of the National Academy of Sciences of the United States of America, 106(14), 5813–5818. 10.1073/pnas.0810550106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévot P-P, Augereau C, Simion A, Van den Steen G, Dauguet N, Lemaigre FP, & Jacquemin P. (2013). Let-7b and miR-495 stimulate differentiation and prevent metaplasia of pancreatic acinar cells by repressing HNF6. Gastroenterology, 145(3), 668–78.e3. 10.1053/j.gastro.2013.05.016 [DOI] [PubMed] [Google Scholar]
- Pullen TJ, da Silva Xavier G, Kelsey G, & Rutter GA (2011). miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Molecular and Cellular Biology, 31(15), 3182–3194. 10.1128/MCB.01433-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini E, & Haumaitre C. (2015). Implication of epigenetics in pancreas development and disease. Best Practice & Research. Clinical Endocrinology & Metabolism, 29(6), 883–898. 10.1016/j.beem.2015.10.010 [DOI] [PubMed] [Google Scholar]
- Qureshi IA, & Mehler MF (2012). Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nature Reviews. Neuroscience, 13(8), 528–541. 10.1038/nrn3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachagani S, Macha MA, Menning MS, Dey P, Pai P, Smith LM, Mo Y-Y, & Batra SK (2015). Changes in microRNA (miRNA) expression during pancreatic cancer development and progression in a genetically engineered KrasG12D;Pdx1-Cre mouse (KC) model. Oncotarget, 6(37), 40295–40309. 10.18632/oncotarget.5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, Cairns BR, & Jones DA (2006). Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Molecular and Cellular Biology, 26(19), 7077–7085. 10.1128/MCB.00312-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran D, Roy U, Garg S, Ghosh S, Pathak S, & Kolthur-Seetharam U. (2011). Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic β-islets. The FEBS Journal, 278(7), 1167–1174. 10.1111/j.1742-4658.2011.08042.x [DOI] [PubMed] [Google Scholar]
- Ramzy A, Thompson DM, Ward-Hartstonge KA, Ivison S, Cook L, Garcia RV, Loyal J, Kim PTW, Warnock GL, Levings MK, & Kieffer TJ (2021). Implanted pluripotent stem-cell-derived pancreatic endoderm cells secrete glucose-responsive C-peptide in patients with type 1 diabetes. Cell Stem Cell, 28(12), 2047–2061.e5. 10.1016/j.stem.2021.10.003 [DOI] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, Yang YHC, Johnson JD, & Kieffer TJ (2014). Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nature Biotechnology, 32(11), 1121–1133. 10.1038/nbt.3033 [DOI] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Riedel MJ, Mojibian M, Asadi A, Xu J, Gauvin R, Narayan K, Karanu F, O’Neil JJ, Ao Z, Warnock GL, & Kieffer TJ (2012). Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes, 61(8), 2016–2029. 10.2337/db11-1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Xu J, Narayan K, Fox JK, O’Neil JJ, & Kieffer TJ (2013). Enrichment of human embryonic stem cell-derived NKX6.1-expressing pancreatic progenitor cells accelerates the maturation of insulin-secreting cells in vivo. Stem Cells, 31(11), 2432–2442. 10.1002/stem.1489 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, & Chang HY (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell, 129(7), 1311–1323. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song J-J, Aragon JP, Liu J, Hannon GJ, & Joshua-Tor L. (2005). Purified Argonaute2 and an siRNA form recombinant human RISC. Nature Structural & Molecular Biology, 12(4), 340–349. 10.1038/nsmb918 [DOI] [PubMed] [Google Scholar]
- Rossi JM, Dunn NR, Hogan BL, & Zaret KS (2001). Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes & Development, 15(15), 1998–2009. 10.1101/gad.904601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Wang T, Kameswaran V, Wei Q, Johnson DS, Matschinsky F, Shi W, & Chen YH (2011). The microRNA-21-PDCD4 axis prevents type 1 diabetes by blocking pancreatic beta cell death. Proceedings of the National Academy of Sciences of the United States of America, 108(29), 12030–12035. 10.1073/pnas.1101450108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter WJ, Kemp JD, Bradshaw WS, Clark WR, Ronzio RA, & Sanders TG (1968). Regulation of specific protein synthesis in cytodifferentiation. Journal of Cellular Physiology, 72(2), Suppl 1:1–18. 10.1002/jcp.1040720403 [DOI] [PubMed] [Google Scholar]
- Sachdeva MM, Claiborn KC, Khoo C, Yang J, Groff DN, Mirmira RG, & Stoffers DA (2009). Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proceedings of the National Academy of Sciences of the United States of America, 106(45), 19090–19095. 10.1073/pnas.0904849106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Parra C, Jacovetti C, Dumortier O, Lee K, Peyot M-L, Guay C, Prentki M, Laybutt DR, Van Obberghen E, & Regazzi R. (2018). Contribution of the Long Noncoding RNA H19 to β-Cell Mass Expansion in Neonatal and Adult Rodents. Diabetes, 67(11), 2254–2267. 10.2337/db18-0201 [DOI] [PubMed] [Google Scholar]
- Sander M, Neubüser A, Kalamaras J, Ee HC, Martin GR, & German MS (1997). Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes & Development, 11(13), 1662–1673. 10.1101/gad.11.13.1662 [DOI] [PubMed] [Google Scholar]
- Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, Willenbring H, & Verdin E. (2010). miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging, 2(7), 415–431. 10.18632/aging.100176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AE, Freude KK, Nelson SB, & Sander M. (2010). Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Developmental Cell, 18(6), 1022–1029. 10.1016/j.devcel.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, & Zamore PD (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell, 115(2), 199–208. 10.1016/s0092-8674(03)00759-1 [DOI] [PubMed] [Google Scholar]
- Sellick GS, Barker KT, Stolte-Dijkstra I, Fleischmann C, Coleman RJ, Garrett C, Gloyn AL, Edghill EL, Hattersley AT, Wellauer PK, Goodwin G, & Houlston RS (2004). Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nature Genetics, 36(12), 1301–1305. 10.1038/ng1475 [DOI] [PubMed] [Google Scholar]
- Senée V, Chelala C, Duchatelet S, Feng D, Blanc H, Cossec J-C, Charon C, Nicolino M, Boileau P, Cavener DR, Bougnères P, Taha D, & Julier C. (2006). Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nature Genetics, 38(6), 682–687. 10.1038/ng1802 [DOI] [PubMed] [Google Scholar]
- Servitja J-M, Pignatelli M, Maestro MA, Cardalda C, Boj SF, Lozano J, Blanco E, Lafuente A, McCarthy MI, Sumoy L, Guigó R, & Ferrer J. (2009). Hnf1alpha (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Molecular and Cellular Biology, 29(11), 2945–2959. 10.1128/MCB.01389-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour PA, Shih HP, Patel NA, Freude KK, Xie R, Lim CJ, & Sander M. (2012). A Sox9/Fgf feed-forward loop maintains pancreatic organ identity. Development, 139(18), 3363–3372. 10.1242/dev.078733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro AMJ, Thompson D, Donner TW, Bellin MD, Hsueh W, Pettus J, Wilensky J, Daniels M, Wang RM, Brandon EP, Jaiman MS, Kroon EJ, D’Amour KA, & Foyt HL (2021). Insulin expression and C-peptide in type 1 diabetes subjects implanted with stem cell-derived pancreatic endoderm cells in an encapsulation device. Cell Reports. Medicine, 2(12), 100466. 10.1016/j.xcrm.2021.100466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih DQ, Heimesaat M, Kuwajima S, Stein R, Wright CVE, & Stoffel M. (2002). Profound defects in pancreatic beta-cell function in mice with combined heterozygous mutations in Pdx-1, Hnf-1alpha, and Hnf-3beta. Proceedings of the National Academy of Sciences of the United States of America, 99(6), 3818–3823. 10.1073/pnas.062605899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Kopp JL, Sandhu M, Dubois CL, Seymour PA, Grapin-Botton A, & Sander M. (2012). A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development, 139(14), 2488–2499. 10.1242/dev.078634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z-D, Lee K, Yang D, Amin S, Verma N, Li QV, Zhu Z, Soh C-L, Kumar R, Evans T, Chen S, & Huangfu D. (2017). Genome Editing in hPSCs Reveals GATA6 Haploinsufficiency and a Genetic Interaction with GATA4 in Human Pancreatic Development. Cell Stem Cell, 20(5), 675–688.e6. 10.1016/j.stem.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Fuchs HE, & Jemal A. (2022). Cancer statistics, 2022. CA: A Cancer Journal for Clinicians, 72(1), 7–33. 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- Singer RA, Arnes L, & Sussel L. (2015). Noncoding RNAs in β cell biology. Current Opinion in Endocrinology, Diabetes, and Obesity, 22(2), 77–85. 10.1097/MED.0000000000000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, Qu H-Q, Taleb N, Kishimoto NY, Scheel DW, Lu Y, Patch A-M, Grabs R, Wang J, Lynn FC, Miyatsuka T, Mitchell J, Seerke R, Désir J, Vanden Eijnden S, Abramowicz M, Kacet N, Weill J, Renard M-E, … German MS (2010). Rfx6 directs islet formation and insulin production in mice and humans. Nature, 463(7282), 775–780. 10.1038/nature08748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Sui T, Zhang Y, Wang Y, Chen M, Deng J, Chai Z, Lai L, & Li Z. (2020). Genetic deletion of a short fragment of glucokinase in rabbit by CRISPR/Cas9 leading to hyperglycemia and other typical features seen in MODY-2. Cellular and Molecular Life Sciences, 77(16), 3265–3277. 10.1007/s00018-019-03354-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaeth JM, Liu J-H, Peters D, Guo M, Osipovich AB, Mohammadi F, Roy N, Bhushan A, Magnuson MA, Hebrok M, Wright CVE, & Stein R. (2019). The Pdx1-Bound Swi/Snf Chromatin Remodeling Complex Regulates Pancreatic Progenitor Cell Proliferation and Mature Islet β-Cell Function. Diabetes, 68(9), 1806–1818. 10.2337/db19-0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Lauf R, & Shroyer NF (2011). Vertebrate intestinal endoderm development. Developmental Dynamics, 240(3), 501–520. 10.1002/dvdy.22540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, & Gruss P. (1997). Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature, 387(6631), 406–409. 10.1038/387406a0 [DOI] [PubMed] [Google Scholar]
- Stanger BZ, & Hebrok M. (2013). Control of cell identity in pancreas development and regeneration. Gastroenterology, 144(6), 1170–1179. 10.1053/j.gastro.2013.01.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavast CJ, & Erkeland SJ (2019). The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells, 8(11). 10.3390/cells8111465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz P. (2017). Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nature Reviews. Gastroenterology & Hepatology, 14(5), 296–304. 10.1038/nrgastro.2017.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O’Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, & German MS (1998). Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development, 125(12), 2213–2221. 10.1242/dev.125.12.2213 [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Katsura A, Yasuda T, Ueno T, Mano H, Sugimoto K, & Miyazono K. (2015). Small-RNA asymmetry is directly driven by mammalian Argonautes. Nature Structural & Molecular Biology, 22(7), 512–521. 10.1038/nsmb.3050 [DOI] [PubMed] [Google Scholar]
- Swarr DT, Herriges M, Li S, Morley M, Fernandes S, Sridharan A, Zhou S, Garcia BA, Stewart K, & Morrisey EE (2019). The long noncoding RNA Falcor regulates Foxa2 expression to maintain lung epithelial homeostasis and promote regeneration. Genes & Development, 33(11–12), 656–668. 10.1101/gad.320523.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, & Hahn SA (2007). MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene, 26(30), 4442–4452. 10.1038/sj.onc.1210228 [DOI] [PubMed] [Google Scholar]
- Tang X, Muniappan L, Tang G, & Ozcan S. (2009). Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA (New York), 15(2), 287–293. 10.1261/rna.1211209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattikota SG, Rathjen T, McAnulty SJ, Wessels H-H, Akerman I, van de Bunt M, Hausser J, Esguerra JLS, Musahl A, Pandey AK, You X, Chen W, Herrera PL, Johnson PR, O’Carroll D, Eliasson L, Zavolan M, Gloyn AL, Ferrer J, … Poy MN (2014). Argonaute2 mediates compensatory expansion of the pancreatic β cell. Cell Metabolism, 19(1), 122–134. 10.1016/j.cmet.2013.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattikota SG, Sury MD, Rathjen T, Wessels H-H, Pandey AK, You X, Becker C, Chen W, Selbach M, & Poy MN (2013). Argonaute2 regulates the pancreatic β-cell secretome. Molecular & Cellular Proteomics, 12(5), 1214–1225. 10.1074/mcp.M112.024786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Okuniewska M, Malone CD, Coux R-X, Rio DC, & Lehmann R. (2017). piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature, 552(7684), 268–272. 10.1038/nature25018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson N, Gésina E, Scheinert P, Bucher P, & Grapin-Botton A. (2012). RNA profiling and chromatin immunoprecipitation-sequencing reveal that PTF1a stabilizes pancreas progenitor identity via the control of MNX1/HLXB9 and a network of other transcription factors. Molecular and Cellular Biology, 32(6), 1189–1199. 10.1128/MCB.06318-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyaboonchai A, Cardenas-Diaz FL, Ying L, Maguire JA, Sim X, Jobaliya C, Gagne AL, Kishore S, Stanescu DE, Hughes N, De Leon DD, French DL, & Gadue P. (2017). GATA6 Plays an Important Role in the Induction of Human Definitive Endoderm, Development of the Pancreas, and Functionality of Pancreatic β Cells. Stem Cell Reports, 8(3), 589–604. 10.1016/j.stemcr.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott J, Tan EK, Ong S, Titmarsh DM, Denil SLIJ, Giam M, Wong CK, Wang J, Shboul M, Eio M, Cooper-White J, Cool SM, Rancati G, Stanton LW, Reversade B, & Dunn NR (2017). Long-Term Culture of Self-renewing Pancreatic Progenitors Derived from Human Pluripotent Stem Cells. Stem Cell Reports, 8(6), 1675–1688. 10.1016/j.stemcr.2017.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bürck L, Blutke A, Kautz S, Rathkolb B, Klaften M, Wagner S, Kemter E, Hrabé de Angelis M, Wolf E, Aigner B, Wanke R, & Herbach N. (2010). Phenotypic and pathomorphological characteristics of a novel mutant mouse model for maturity-onset diabetes of the young type 2 (MODY 2). American Journal of Physiology. Endocrinology and Metabolism, 298(3), E512–23. 10.1152/ajpendo.00465.2009 [DOI] [PubMed] [Google Scholar]
- van de Bunt M, Gaulton KJ, Parts L, Moran I, Johnson PR, Lindgren CM, Ferrer J, Gloyn AL, & McCarthy MI (2013). The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis. Plos One, 8(1), e55272. 10.1371/journal.pone.0055272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra JA, Agricola H-J, & Sellami A. (2008). Regulatory peptides in fruit fly midgut. Cell and Tissue Research, 334(3), 499–516. 10.1007/s00441-008-0708-3 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, … et al. (2001). The sequence of the human genome. Science, 291(5507), 1304–1351. 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan J, Maggi EC, & Crabtree JS (2014). miR-24 regulates menin in the endocrine pancreas. American Journal of Physiology. Endocrinology and Metabolism, 307(1), E84–92. 10.1152/ajpendo.00542.2013 [DOI] [PubMed] [Google Scholar]
- Wang C, Yao N, Lu C-L, Li D, & Ma X. (2010). Mouse microRNA-124 regulates the expression of Hes1 in P19 cells. Frontiers in Bioscience (Elite Edition), 2(1), 127–132. 10.2741/e74 [DOI] [PubMed] [Google Scholar]
- Wang D, Zhu Z-M, Tu Y-L, Dou C-Q, Xu Y, Tan X-L, Han M-M, Yang Z-J, Jin X, Zhang B, Cai S, & Liu Z-W (2016). Identfication of key miRNAs in pancreatitis using bioinformatics analysis of microarray data. Molecular Medicine Reports, 14(6), 5451–5460. 10.3892/mmr.2016.5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, & Guttridge DC (2008). NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell, 14(5), 369–381. 10.1016/j.ccr.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, & Kurokawa R. (2008). Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature, 454(7200), 126–130. 10.1038/nature06992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu J, Liu C, Naji A, & Stoffers DA (2013). MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells. Diabetes, 62(3), 887–895. 10.2337/db12-0451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Cui X, Yuan Z, Qi H, & Lin H. (2018). MIWI2 targets RNAs transcribed from piRNA-dependent regions to drive DNA methylation in mouse prospermatogonia. The EMBO Journal, 37(18). 10.15252/embj.201695329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R, Yang J, Liu G-Q, Gao M-J, Hou W-F, Zhang L, Gao H-W, Liu Y, Chen G-A, & Hong T-P (2013). Dynamic expression of microRNAs during the differentiation of human embryonic stem cells into insulin-producing cells. Gene, 518(2), 246–255. 10.1016/j.gene.2013.01.038 [DOI] [PubMed] [Google Scholar]
- Wells JM, & Melton DA (1999). Vertebrate endoderm development. Annual Review of Cell and Developmental Biology, 15, 393–410. 10.1146/annurev.cellbio.15.1.393 [DOI] [PubMed] [Google Scholar]
- Wienholds E, Koudijs MJ, van Eeden FJM, Cuppen E, & Plasterk RHA (2003). The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nature Genetics, 35(3), 217–218. 10.1038/ng1251 [DOI] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, & Schultz PG (2005). A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science, 309(5740), 1570–1573. 10.1126/science.1115901 [DOI] [PubMed] [Google Scholar]
- Xie R, Everett LJ, Lim H-W, Patel NA, Schug J, Kroon E, Kelly OG, Wang A, D’Amour KA, Robins AJ, Won K-J, Kaestner KH, & Sander M. (2013). Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell, 12(2), 224–237. 10.1016/j.stem.2012.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan S, Borok MJ, Decker KJ, Battle MA, Duncan SA, Hale MA, Macdonald RJ, & Sussel L. (2012). Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. The Journal of Clinical Investigation, 122(10), 3516–3528. 10.1172/JCI63352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C-R, Cole PA, Meyers DJ, Kormish J, Dent S, & Zaret KS (2011). Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science, 332(6032), 963–966. 10.1126/science.1202845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C-R, Li L-C, Donahue G, Ying L, Zhang Y-W, Gadue P, & Zaret KS (2014). Dynamics of genomic H3K27me3 domains and role of EZH2 during pancreatic endocrine specification. The EMBO Journal, 33(19), 2157–2170. 10.15252/embj.201488671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee NS, Yusuff S, & Pack M. (2001). Zebrafish pdx1 morphant displays defects in pancreas development and digestive organ chirality, and potentially identifies a multipotent pancreas progenitor cell. Genesis, 30(3), 137–140. 10.1002/gene.1049 [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, & Cullen BR (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Development, 17(24), 3011–3016. 10.1101/gad.1158803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, & Crabtree GR (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature, 460(7255), 642–646. 10.1038/nature08139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, & Crabtree GR (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature, 476(7359), 228–231. 10.1038/nature10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X-X, Qiu W-L, Yang L, Li L-C, Zhang Y-W, & Xu C-R (2018). Dynamics of chromatin marks and the role of JMJD3 during pancreatic endocrine cell fate commitment. Development, 145(6). 10.1242/dev.163162 [DOI] [PubMed] [Google Scholar]
- Zarzour A, Kim HW, & Weintraub NL (2019). Epigenetic regulation of vascular diseases. Arteriosclerosis, Thrombosis, and Vascular Biology, 39(6), 984–990. 10.1161/ATVBAHA.119.312193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, & Filipowicz W. (2004). Single processing center models for human Dicer and bacterial RNase III. Cell, 118(1), 57–68. 10.1016/j.cell.2004.06.017 [DOI] [PubMed] [Google Scholar]
- Zhang Z-W, Men T, Feng R-C, Li Y-C, Zhou D, & Teng C-B (2013). miR-375 inhibits proliferation of mouse pancreatic progenitor cells by targeting YAP1. Cellular Physiology and Biochemistry, 32(6), 1808–1817. 10.1159/000356614 [DOI] [PubMed] [Google Scholar]
- Zhao X, Mohan R, Özcan S, & Tang X. (2012). MicroRNA-30d induces insulin transcription factor MafA and insulin production by targeting mitogen-activated protein 4 kinase 4 (MAP4K4) in pancreatic β-cells. The Journal of Biological Chemistry, 287(37), 31155–31164. 10.1074/jbc.M112.362632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Liang I-C, & Yee NS (2011). Histone deacetylase 1 is required for exocrine pancreatic epithelial proliferation in development and cancer. Cancer Biology & Therapy, 11(7), 659–670. 10.4161/cbt.11.7.14720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, You W, Wang H, Li Y, Qiao N, Shi Y, Zhang C, Bleich D, & Han X. (2013). MicroRNA-24/MODY gene regulatory pathway mediates pancreatic β-cell dysfunction. Diabetes, 62(9), 3194–3206. 10.2337/db13-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]