Abstract
Exon-skipping therapy mediated by antisense oligonucleotides is expected to provide a therapeutic option for Duchenne muscular dystrophy. Antisense oligonucleotides for exon skipping reported so far target a single continuous sequence in or around the target exon. In the present study, we investigated antisense oligonucleotides for exon 44 skipping (applicable to approximately 6% of all Duchenne muscular dystrophy patients) to improve activity by using a novel antisense oligonucleotide design incorporating two connected sequences. Phosphorodiamidate morpholino oligomers targeting two separate sequences in exon 44 were created to target two splicing regulators in exon 44 simultaneously, and their exon 44 skipping was measured. NS-089/NCNP-02 showed the highest skipping activity among the oligomers. NS-089/NCNP-02 also induced exon 44 skipping and dystrophin protein expression in cells from a Duchenne muscular dystrophy patient to whom exon 44 skipping is applicable. We also assessed the in vivo activity of NS-089/NCNP-02 by intravenous administration to cynomolgus monkeys. NS-089/NCNP-02 induced exon 44 skipping in skeletal and cardiac muscle of cynomolgus monkeys. In conclusion, NS-089/NCNP-02, an antisense oligonucleotide with a novel connected-sequence design, showed highly efficient exon skipping both in vitro and in vivo.
Keywords: MT: Oligonucleotides: Therapies and Applications, Duchenne muscular dystrophy, dystrophin, exon 44, exon skipping, antisense therapeutics, morpholino, NS-089/NCNP-02, dual-targeting, antisense oligonucleotide, splice modulation
Graphical abstract

Takeda and colleagues developed NS-089/NCNP-02, pioneering dual-targeting antisense oligonucleotides using phosphorodiamidate morpholino oligomers. NS-089/NCNP-02 selectively targets two integral splicing regulators in exon 44 of the DMD gene. Notably, it induced highly efficient exon 44 skipping and dystrophin expression in patient-derived cells from Duchenne muscular dystrophy amenable to exon 44 skipping.
Introduction
Duchenne muscular dystrophy (DMD) is an X-chromosome-linked progressive hereditary muscle disease caused by mutations in the DMD gene, which codes for dystrophin, a protein expressed underneath the muscle-cell membrane. According to estimates from newborn screening,1 DMD affects 1 in 3,500 newborn boys and is the most severe and common form of muscular dystrophy. Mutations in the DMD gene can lead to either the severe DMD or the milder Becker muscular dystrophy (BMD), depending on whether the translational reading frame is lost or maintained, respectively.2 DMD is mainly caused by frameshift mutations in the DMD gene, which result in a lack of dystrophin. BMD, in contrast, is mostly caused by in-frame mutations, which result in a reduced amount of dystrophin or a reduction in its molecular size.
Exon skipping is a therapeutic approach that uses antisense oligonucleotides (ASOs) to modulate pre-mRNA splicing, thereby correcting the translational reading frame and resulting in an internally truncated but partially functional protein. Eteplirsen (brand name, Exondys 51) is the first approved antisense drug for DMD in the United States, and it provides a treatment option for DMD patients amenable to exon 51 skipping (approximately 14% of all DMD patients).3 Golodirsen (brand name, Vyondys 53), approved in the United States, and viltolarsen (brand name, Viltepso), approved in Japan and the United States, are phosphorodiamidate morpholino oligomers (PMOs) for the treatment of DMD patients who are amenable to exon 53 skipping (approximately 8% of all DMD patients).4,5 Casimersen (brand name, Amondys 45), designed to skip exon 45, was approved in the United States.6
Exon 44 skipping mainly applies to patients with deletions in the DMD gene consisting of exon 45 or exons 45–54 and many different deletions ending at exon 43, although other exon-skipping patterns are also possible with these deletions (e.g., exon 55 skipping with deletion of exons 45–54). Exon 44 skipping is applicable to approximately 6% of DMD patients.7 An investigator-initiated phase I/II study of NS-089/NCNP-02 designed to skip exon 44 demonstrated an increase in dystrophin protein expression and suggested maintenance of motor function or a trend in its improvement.8 In this study, we describe the screening of ASOs based on a novel sequence design created by combining two targeting sequences within a single ASO. We report the development of NS-089/NCNP-02 and present a non-clinical pharmacology study of NS-089/NCNP-02 in human rhabdomyosarcoma (RD) cells, DMD patient-derived cells, and cynomolgus monkeys.
Results
Screening of ASOs targeting two separate sequences in exon 44
ASOs 20–30 nt in length targeting two separate sequences in exon 44 were investigated. Sequence screening involved three steps as described below (see Figure 1) and was evaluated based on exon 44 skipping activity in RD cells.
Figure 1.
Overview of strategy for screening sequences for exon 44 skipping
First screening: 2′-O-methyl phosphorothioate oligonucleotides targeting sequences within exon 44 were synthesized as an overlapping series of twenty-six 22-mer oligomers to provide coverage of positions 1 to 147 of this 148-nt exon in 5-nt increments from the 3′ to the 5′ end. Second screening: ten 26-mer PMOs, each consisting of two directly connected 13-mer targeting sequences, one selected from the region of positions 11–33 and the other from the region of positions 61–113 of exon 44, were synthesized, and their exon 44 skipping activity was measured. Third screening: to optimize the sequence, 18- to 30-mer PMOs consisting of two connected targeting sequences of positions 21–33, 61–73, and 91–103 were synthesized, and their exon 44 skipping activity was measured.
For the first screening, 2′-O-methyl phosphorothioate oligonucleotides targeting easily available sequences within exon 44 were synthesized as an overlapping series of twenty-six 22-mer oligomers, similar in length to viltolarsen, an ASO that shows high exon 53 skipping efficiency. These oligomers provided coverage of positions 1 to 147 of this 148-nt exon in 5-nt increments from the 3′ to the 5′ end (Table S1). The highest exon 44 skipping efficiency was observed for oligomers targeting sequences from positions 11–52 and 61–122 of the exon (Figure S1). For the second screening, ten 26-mer PMOs were synthesized and their exon 44 skipping activity was measured. Longer PMOs were not used because they are difficult to synthesize in reasonable yield. Each 26-mer consisted of two directly connected 13-mer targeting sequences, one selected from positions 11–33 and the other from positions 61–113 of exon 44. All 10 PMOs showed skipping activity (data not shown), and those consisting of the three combinations of two directly connected sequences from the group of sequences corresponding to positions 21–33, 61–73, and 91–103 showed high skipping activity (Figure S2 and Table S2). For the third screening, to optimize the sequence, 18- to 30-mer PMOs consisting of two connected targeting sequences corresponding to the region of the sequences selected for the second screening were synthesized, and their exon 44 skipping activity was measured (Table S3). The PMOs all showed high skipping activity (data not shown). The skipping activity of the 26-mer PMOs consisting of sequences around 21–33 and 91–103 was measured, and the highest activity was shown by a 26-mer PMO consisting of sequences from positions 19–31 and 89–101 of exon 44 with a mean half maximal effective concentration (EC50) value of 0.90 μmol/L (Figure S3 and Table S3). Interchanging the 19–31 and 89–101 sequences resulted in decreased activity (Figure S3 and Table S3). When the skipping activity of shortened PMOs consisting of sequences from positions around 61–73 and 91–103 was measured, the highest activity was shown by a 24-mer PMO consisting of sequences from positions 64–75 and 92–103 of exon 44 (Figure S3 and Table S3). Since the activities of the 26-mer and the 24-mer were similar, the 24-mer, NS-089/NCNP-02, was selected for further study. The exon-skipping efficiency of NS-089/NCNP-02 measured in RD cells showed a dose-dependent increase with an EC50 value of 0.83 μmol/L (95% confidence interval, 0.71–0.98 μmol/L; Figure 2).
Figure 2.
Exon 44 skipping activity of NS-089/NCNP-02 in RD cells
NS-089/NCNP-02 at concentrations of 0.1, 0.3, 1, 3, 10, and 30 μmol/L was transfected into RD cells with Nucleofector, and total RNA was extracted after 3 days. Exon skipping was measured by RT-PCR, and the PCR products were analyzed by 2100 Bioanalyzer. Each point and bar shows the mean ± standard deviation (3 wells per concentration).
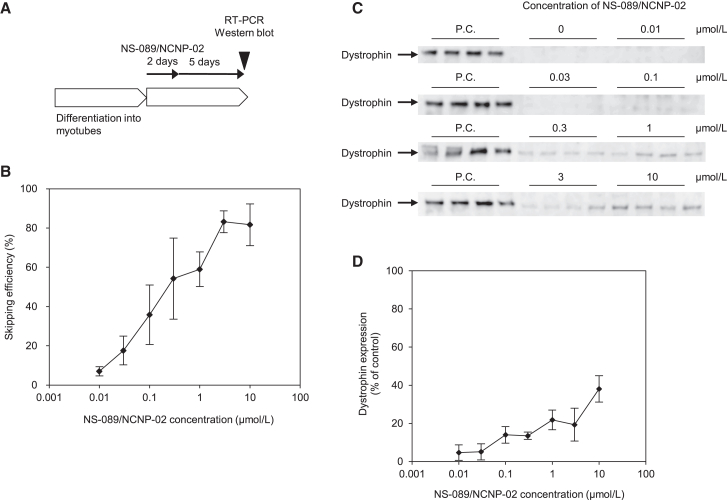
Exon 44 skipping activity of NS-089/NCNP-02 and NS-089/NCNP-02-induced expression of dystrophin protein in cells derived from patients with DMD
We next measured the exon 44 skipping activity of NS-089/NCNP-02 and the NS-089/NCNP-02-induced expression of dystrophin protein in cells derived from a DMD patient with deletion of exon 45. Patient fibroblasts were transduced with the human MYOD gene to induce differentiation into myotubes. The myotubes were then treated with NS-089/NCNP-02 at final concentrations of 0.01, 0.03, 0.1, 0.3, 1, 3, and 10 μmol/L by gymnotic transfection for 2 days, after which the differentiation medium was replaced with medium containing no NS-089/NCNP-02. The skipping activity and the expression of dystrophin protein were measured by RT-PCR and western blotting 7 days after the beginning of treatment (Figure 3A). NS-089/NCNP-02 induced exon 44 skipping in the cells with an EC50 value of 0.33 μmol/L (95% confidence interval, 0.22–0.51 μmol/L; Figures 3B and S3). When NS-089/NCNP-02 was transfected into the cells at concentrations of 0.01, 0.03, 0.1, 0.3, 1, 3, and 10 μmol/L, the expression of dystrophin protein was 4.7%, 5.1%, 14.0%, 13.5%, 21.8%, 19.3%, and 38.0%, respectively, of the expression of dystrophin protein in a cell lysate of myotubes differentiated from normal human fibroblasts by transduction with MYOD (Figures 3C and 3D).
Figure 3.
Exon 44 skipping activity and dystrophin protein expression 7 days after a 2-day treatment with NS-089/NCNP-02 of cells derived from a DMD patient with deletion of exon 45
(A) Schedule of PMO transfection for RT-PCR and western blotting. (B) Exon 44 skipping activity and (C) western blot analysis of dystrophin protein expression 7 days after the start of a 2-day treatment with NS-089/NCNP-02 by gymnotic transfection of cells derived from a DMD patient with deletion of exon 45. (B) Each point shows the mean ± standard deviation (four wells per concentration). (C) The positive control (P.C.) was a cell lysate of myotubes differentiated from normal human fibroblasts by transduction with MYOD. Cell lysates from DMD patient cells and normal human cells (3 μg protein per well) were loaded. (D) Densitometric analysis of the western blots relative to each P.C. on the same membrane. Each point shows the mean ± standard deviation (four wells per concentration).
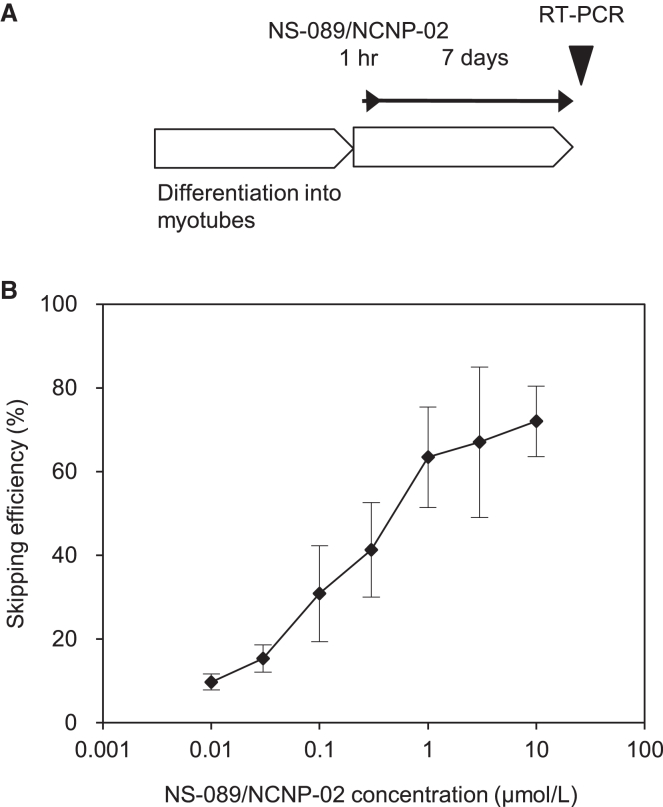
Exon 44 skipping after transfection for a short incubation time
In the clinical setting, PMOs for DMD exon skipping are administered by weekly intravenous infusion, since a PMO is eliminated from the blood with an elimination half-life of approximately 2 h after intravenous administration.9 As cells typically exhibit rapid uptake of PMOs in vitro (less than 1 h), we wanted to confirm that our PMO exhibited activity within this short period. Patient fibroblasts were transduced with the human MYOD gene to induce differentiation into myotubes. The myotubes were then treated with NS-089/NCNP-02 for 1 h at final concentrations of 0.01, 0.03, 0.1, 0.3, 1, 3, and 10 μmol/L. The exon 44 skipping activity was measured by RT-PCR 7 days after the beginning of treatment (Figure 4A). NS-089/NCNP-02 induced exon 44 skipping with an EC50 value of 0.63 μmol/L (95% confidence interval, 0.40–0.98 μmol/L; Figure 4B).
Figure 4.
Exon 44 skipping activity 7 days after starting a 1-h treatment with NS-089/NCNP-02 of cells derived from a DMD patient with deletion of exon 45
(A) Schedule of PMO transfection for RT-PCR. (B) Exon 44 skipping activity 7 days after starting a 1-h treatment with NS-089/NCNP-02 without transfection reagent of cells derived from a DMD patient with deletion of exon 45. Each point shows the mean ± standard deviation (four wells per concentration).
Exon 44 skipping in cynomolgus monkeys
Finally, we assessed the in vivo activity of NS-089/NCNP-02 in cynomolgus monkeys. Although there is a single base difference between the target sequence of NS-089/NCNP-02 in cynomolgus monkeys and humans, we confirmed that NS-089/NCNP-02 showed exon 44 skipping activity in cultured monkey cells (Figure S4). The EC50 value of skipping efficiency in cultured monkey cells was 673 μmol/L. Hence, an approximately 800-fold disparity in skipping efficiency exists between DMD patient cells and cultured monkey cells. The skipping efficiency in cultured monkey cells was about half that of a PMO that perfectly matches the monkey sequence (Figure S5). When NS-089/NCNP-02 was administered intravenously to cynomolgus monkeys once weekly for 13 weeks (Figure 5A), and the exon 44 skipping activity was measured in skeletal muscle (the right gastrocnemius muscle) the day after the last administration, the skipping efficiencies were 0.0%, 0.6%, 2.1%, and 9.5% at doses of 0, 200, 600, and 2,000 mg/kg, which are significantly different from that of the control group (0 mg/kg) for all dose groups (Figure 5B). The corresponding skipping efficiencies in cardiac muscle (the left ventricular apex) were 0.0%, 0.3%, 0.4%, and 1.4%, and those of the 600 and 2,000 mg/kg dose groups are significantly different from that of the control group (0 mg/kg) (Figure 5C). In addition, the exon 44 skipping activity was measured in skeletal muscle at the end of an 8-week recovery period (Figure 6A). The skipping efficiencies were 0.0%, 3.1%, and 16.2% at doses of 0, 600, and 2,000 mg/kg, and that of the 2,000 mg/kg dose group is significantly different from that of the control group (0 mg/kg) (Figure 6B). The corresponding skipping efficiencies in cardiac muscle were 0.0%, 0.0%, and 0.8%, which are not significantly different from that of the control group (0 mg/kg) (Figure 6C).
Figure 5.
Exon 44 skipping activity in skeletal and cardiac muscle of cynomolgus monkeys at the end of a 13-week treatment period
(A) Schedule of PMO injection and RT-PCR. (B and C) Total RNA was extracted from (B) skeletal muscle or (C) cardiac muscle after a 13-week NS-089/NCNP-02 treatment. Exon skipping was measured by RT-PCR, and the PCR products were analyzed by a 2100 Bioanalyzer. Exon-skipping efficiency was measured by RT-PCR. Each bar represents the mean and standard deviation (five animals). Shirley-Williams multiple comparison tests (one-sided) versus 0 mg/kg (saline) group. n.s., no significant difference; ∗p < 0.005; ∗∗∗p < 0.0005.
Figure 6.
Exon 44 skipping activity in skeletal and cardiac muscle of cynomolgus monkeys at the end of an 8-week recovery period
(A) Schedule of PMO injection and RT-PCR. (B and C) Total RNA was extracted from (B) skeletal muscle or (C) cardiac muscle after a 13-week NS-089/NCNP-02 treatment and an 8-week recovery period. Exon skipping was measured by RT-PCR, and the PCR products were analyzed by a 2100 Bioanalyzer. Each bar represents the mean and standard deviation (two animals). Shirley-Williams multiple comparison tests (one-sided) versus 0 mg/kg (saline) group. n.s., no significant difference.
Discussion
In this study, PMOs based on a novel design involving directly connected sequences targeting different parts of the exon were screened, and NS-089/NCNP-02 was found to have the highest exon 44 skipping activity. Antisense sequences with high exon 44 skipping activity are already known.10,11,12,13,14,15,16,17,18 For exons 10, 20, 34, and 54, more than one ASO was required for efficient exon skipping in a study on myoblasts.19 Moreover, when a series of PMOs was screened by intramuscular injection into mdx mice with deletion of exon 52 (mdx52 mice), the highest splicing efficiency was generated by a two-oligonucleotide cocktail targeting both the 5′ and 3′ splice sites of exon 51.20 The approach of combining two antisense oligonucleotides separated by a stretch of 10 uridine nucleotides into a single ASO was also investigated and resulted in improved exon-skipping activity.21 In addition, a U7 vector carrying a double antisense insert covering both the 5′ and 3′ splice sites induces efficient skipping of exon 51 in the human DMD deletion 48–50 and exon 23 in mouse cells.22,23 However, no previous report has shown that a single-strand ASO containing directly connected sequences targeting two or more sites in the same exon exhibits skipping activity. We have now found that efficient exon 44 skipping activity was shown by 18- to 30-mer single-strand ASOs that target two separate sequences within exon 44. Positions 11–52 and 61–122 of exon 44 were identified as high skipping regions in the first screening. Since the binding of the splicing factors involved in exon skipping depends on the pre-mRNA sequence, the effect of chemical modification of ASOs on skipping activity is thought to be small; therefore, the relatively easily synthesized 2′-O-methyl phosphorothioate oligonucleotides were used in the first screening. PMOs were used for optimization from the second screening. Combinations of positions 21–33 and 61–73 or 91–103 were selected from positions 11–52 and 61–122, respectively, and a combination of positions 61–73 and 91–103, both selected from positions 61–122, showed high skipping activity. This novel type of ASO with two connected sequences expands the possibilities for creating highly active new sequences.
In contrast to the substantial exon 44 skipping activity shown by NS-089/NCNP-02, little activity was shown by a mixture of two separate 12-mer PMOs with precisely the same targeting sequences as NS-089/NCNP-02 (data not shown). Yet this may not be an appropriate comparison because of the known inefficient binding of 12-mer PMO sequences. Unfortunately, we cannot include the comparison of the published sequence +61 + 84, which would have been a better comparison than the 12-mer PMO. Secondary structure prediction suggests that NS-089/NCNP-02 may bind efficiently to two targets on exon 44 that do not form stem structures (Figure S7). The result obtained with NS-089/NCNP-02 may support our idea that a single strand can simultaneously bind to two separate sites in the same exon and induce exon skipping. Interchanging the sequences 19–31 and 89–101 resulted in decreased activity. This may be because the order of the pre-mRNA target sequences is reversed, so the pre-mRNA needs to be twisted for the PMO to bind.
NS-089/NCNP-02 showed exon 44 skipping activity and promoted dystrophin protein expression in MYOD-converted fibroblasts from a patient with deletion of DMD exon 45. The EC50 value for the exon 44 skipping activity 5 days after a 2-day treatment with NS-089/NCNP-02 was 0.33 μmol/L. The EC50 value for the exon 44 skipping activity 7 days after a 1-h treatment with NS-089/NCNP-02 was 0.63 μmol/L. Thus, exon 44 skipping activity was sustained for 7 days even when the contact time with the cells was shortened to 1 h. From these results, even though PMOs are cleared from the blood quickly after intravenous administration, NS-089/NCNP-02 can be expected to have an appreciable therapeutic effect. In the phase II study of viltolarsen in the United States/Canada, dystrophin protein expression and exon 53 skipping activity were seen after treatment with viltolarsen, and preliminary results of timed function tests suggest clinical improvement in DMD boys.24 In a preclinical study, the EC50 value for the exon 53 skipping activity 5 days after a 2-day treatment with viltolarsen delivered with the transfection reagent Endo-Porter was 0.82 μmol/L in MYOD-converted fibroblasts from a DMD patient with deletion of exons 45–52.25 Since PMO was found to be active without a transfection reagent in DMD patient cells, no transfection reagent was used in the NS-089/NCNP-02 study. The EC50 value for the exon 44 skipping activity 5 days after a 2-day treatment with NS-089/NCNP-02 without a transfection reagent was 0.33 μmol/L. The skipping efficiency was 31.9% 1 week after a 1-h treatment with viltolarsen at 10 μmol/L with Endo-Porter25 and 72.0% 1 week after a 1-h treatment with NS-089/NCNP-02 at the same concentration without a transfection reagent. An investigator-initiated phase I/II study of NS-089/NCNP-02 showed an increase in dystrophin protein expression on western blots to an average of 10.27% of normal at a dose of 40 mg/kg and 15.79% of normal at a dose of 80 mg/kg.8 The clinical results of dystrophin protein expression induced by NS-089/NCNP-02 and viltolarsen correlate with the in vitro results.9,24
By exon 44 skipping, a dystrophin protein with deletion of the parts corresponding to exons 35–44, 44–45, or 44–54 would be expected to be expressed in DMD patients with deletion of exons 35–43, 45, or 45–54, respectively.26 The shorter dystrophin protein induced by exon 44 skipping therapies is expected to be partially functional because deletion of exons 44–45 has only been reported three times, which suggests an ascertainment bias attributable to a very mild phenotype.27 Additionally, there are three reports of asymptomatic cases with deletion of exons 35–44, 38–44, and 41–44.28,29,30
To investigate the activity of NS-089/NCNP-02 in vivo, exon 44 skipping activity was measured in skeletal and cardiac muscle of cynomolgus monkeys. The in vitro results for a PMO whose sequence perfectly matches that of the cynomolgus monkey target sequence shows that the single-nucleotide difference between the target sequence of NS-089/NCNP-02 in cynomolgus monkeys and humans significantly reduces the skipping efficiency by 50% and contributes to the likely underestimation of the skipping efficiency in the in vivo experiments. PMOs more efficiently enter fibers involved in active muscle regeneration, such as those in DMD patients, than fibers of normal skeletal muscle.31 Even though exon 44 skipping is of low efficiency in the normal skeletal muscle of cynomolgus monkeys, exon 44 skipping is expected to be highly efficient in the skeletal muscle of DMD patients. Although PMO delivery to cardiomyocytes is challenging,32,33 some exon 44 skipping was induced by NS-089/NCNP-02 in the cardiac muscle of cynomolgus monkeys. ASO uptake after systemic delivery differs in different muscles, and the exon-skipping activity also varies by skeletal muscle.20 In the present study, exon skipping in skeletal muscle was measured only in the gastrocnemius muscle. Exon 44 skipping activity in the skeletal muscle of cynomolgus monkeys at the end of an 8-week recovery period at the dose of 2,000 mg/kg was higher than that at the end of a 13-week treatment period. Dystrophin rescue induced by PMO-based exon skipping occurs in a sporadic patchy pattern with high geographic variability across muscle sections.34 We speculate that, in our study, the higher activity seen in the muscle section taken at the end of the recovery period compared with the end of the 13-week treatment period resulted from the fact that the gastrocnemius muscle was sampled, allowing a patchy pattern of exon skipping to manifest itself in the results. Such a sampling effect could only be eliminated by measuring the skipping activity after homogenizing the whole gastrocnemius muscle.
In conclusion, we describe the activity of NS-089/NCNP-02, a PMO for exon 44 skipping, based on a novel design involving linked sequences targeting two sites in the same exon. NS-089/NCNP-02 induced exon 44 skipping and dystrophin protein expression in cells derived from a patient with DMD amenable to exon 44 skipping and in the skeletal and cardiac muscle of cynomolgus monkeys.
Materials and methods
Antisense oligomers
The 2′-O-Methyl phosphorothioate oligonucleotides were purchased from Japan Bio Services Co., Ltd (Saitama, Japan). PMOs were synthesized by Nippon Shinyaku Co., Ltd.
Cells
RD cells were obtained from the Health Science Research Resources Bank and cultured under 5% CO2 at 37°C in Eagle’s minimum essential medium (Sigma-Aldrich, St. Louis, MO, USA) containing 10% fetal bovine serum. Fibroblasts from a DMD patient with deletion of exon 45 of the DMD gene (GM05112) were obtained from the Coriell Institute for Medical Research. After transfection of a retroviral vector for coexpressing human MYOD and ZsGreen1, ZsGreen1-positive cells were sorted by fluorescence-activated cell sorting and induced to differentiate into myotubes as previously described.25
Transfection of PMOs into cells
PMOs were dissolved in distilled water and transfected into RD cells using the Amaxa Cell Line Nucleofector Kit L and a Nucleofector II electroporation device (Lonza, Basel, Switzerland) with program T-030 or into cells from a DMD patient without a transfection reagent.
Reverse transcriptase polymerase chain reaction
Total RNA was extracted from RD cells or cells from a DMD patient, and RT-PCR was performed with 400 ng (RD cells) or 50 ng (DMD cells) of total RNA using a forward primer (5′-GCTCAGGTCGGATTGACATT-3′) and a reverse primer (5′-GGGCAACTCTTCCACCAGTA-3′) designed to be complementary to exons 43 and 47 as previously described.25 The RT-PCR program for RD cells was as follows: reverse transcription at 50°C for 30 min and heat denaturation at 95°C for 15 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min; followed by a final extension at 72°C for 10 min. The RT-PCR program for DMD patient-derived cells was as follows: reverse transcription at 50°C for 30 min and heat denaturation at 95°C for 15 min; 35 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min; followed by a final extension at 72°C for 7 min. From approximately 15-mg pieces of tissue obtained from 2-year-old male cynomolgus monkeys in a 13-week intermittent intravenous dose toxicity study of NS-089/NCNP-02 followed by an 8-week recovery period (study approved by the Institutional Animal Care and Use Committee of Nippon Shinyaku Co., Ltd; approval no. 17062701), total RNA was extracted using TissueLyser II (Qiagen, Valencia, CA, USA) and NucleoSpin RNA (Macherey-Nagel, Düren, Germany). RNA concentrations were determined from the absorbance at 260 nm using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RT-PCR was performed with 200 ng of extracted total RNA using a Qiagen OneStep RT-PCR Kit (Qiagen). The primers used were a forward primer (Hokkaido System Science, Sapporo, Japan) designed for exon 43 (5′-GCTCAGGTCGGATTGACATT-3′) and a reverse primer (Hokkaido System Science) designed for exon 47 (5′-GGGCAACTCTTCCACCAGTA-3′) to exclude exon 44. RT-PCR was performed with a Takara Thermal Cycler Dice Touch (Takara Bio, Kusatsu, Japan). The RT-PCR program was as follows: reverse transcription at 50°C for 30 min, heat denaturation at 95°C for 15 min and 35 cycles consisting of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min. The PCR reaction products were analyzed using a 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). The skipping efficiency was determined from the molarity of the PCR products by the following expression: (PCR reaction products without exon 44) × 100/[(PCR reaction products without exon 44) + (PCR reaction products with exon 44)].
Western blotting
We prepared the lysates of cells from a DMD patient and normal human cells, and used the BCA assay to quantify total protein concentration in the protein lysate. This enabled them to load an equal amount of protein into each gel well without additional manipulations, such as gel staining to visualize total proteins. Western blotting was performed on cell lysates (3 μg protein per well) as previously described.25 The positive control consisted of normal dystrophin (427 kDa) from a cell lysate of myotubes differentiated from normal human fibroblasts (TIG-119) for 14 days. The amount of dystrophin without the segment corresponding to exons 44–45 (414 kDa) was determined using an ImageQuant LAS 4000 mini imaging analyzer (FujiFilm, Tokyo, Japan). Protein levels (dystrophin expression) were determined as follows: (dystrophin expression level after exon 44 skipping [414 kDa]) × 100/(mean of dystrophin expression level of normal control [427 kDa] on the same membrane).
Statistical analysis
All analyses were performed using SAS software (ver. 9.3; SAS Institute, Cary, NC, USA) and EXSUS (ver. 8.0.0; CAC Exicare, Tokyo, Japan). The skipping efficiencies obtained were analyzed by nonlinear regression using a two-parameter logistic model to calculate EC50 values.
Data and code availability
The authors have verified that the data supporting the findings of this study are accessible within the article and its supplementary information.
Acknowledgments
The authors wish to thank Dr. Gerald E. Smyth for his assistance in preparing the manuscript. The work was performed in Kodaira, Tokyo, and Tsukuba, Ibaraki, Japan. This study was funded by Nippon Shinyaku Co., Ltd., an Intramural Research Grant (Grant number 28-6) from the National Center of Neurology and Psychiatry and the Japan Agency for Medical Research and Development (Grant number 18lm0203066).
Author contributions
N.W., Y.T., T.N., S.M., and T.S. performed the experiments; N.M., T.N., K.T., Y.A., and S.T. coordinated and supervised the project; and N.W. wrote the manuscript.
Declaration of interests
NCNP and Nippon Shinyaku Co., Ltd, are jointly developing NS-089/NCNP-02 for the treatment of DMD.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.102034.
Supplemental information
References
- 1.Emery A.E. Population frequencies of inherited neuromuscular diseases--a world survey. Neuromuscul. Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 2.Koenig M., Beggs A.H., Moyer M., Scherpf S., Heindrich K., Bettecken T., Meng G., Müller C.R., Lindlöf M., Kaariainen H., et al. The Molecular Basis for Duchenne versus Becker Muscular Dystrophy: Correlation of Severity with Type of Deletion. Am. J. Hum. Genet. 1989;45:498–506. [PMC free article] [PubMed] [Google Scholar]
- 3.Rodrigues M., Yokota T. An Overview of Recent Advances and Clinical Applications of Exon Skipping and Splice Modulation for Muscular Dystrophy and Various Genetic Diseases. Methods Mol. Biol. 2018;1828:31–55. doi: 10.1007/978-1-4939-8651-4_2. [DOI] [PubMed] [Google Scholar]
- 4.Heo Y.A. Golodirsen: First Approval. Drugs. 2020;80:329–333. doi: 10.1007/s40265-020-01267-2. [DOI] [PubMed] [Google Scholar]
- 5.Dhillon S. Viltolarsen: First Approval. Drugs. 2020;80:1027–1031. doi: 10.1007/s40265-020-01339-3. [DOI] [PubMed] [Google Scholar]
- 6.Shirley M. Casimersen: First Approval. Drugs. 2021;81:875–879. doi: 10.1007/s40265-021-01512-2. [DOI] [PubMed] [Google Scholar]
- 7.Aartsma-Rus A., Fokkema I., Verschuuren J., Ginjaar I., Van Deutekom J., Van Ommen G.J., Den Dunnen J.T. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum. Mutat. 2009;30:293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- 8.https://www.ncnp.go.jp/topics/2022/20220317e.html
- 9.Komaki H., Takeshima Y., Matsumura T., Ozasa S., Funato M., Takeshita E., Iwata Y., Yajima H., Egawa Y., Toramoto T., et al. Viltolarsen in Japanese Duchenne muscular dystrophy patients: A phase 1/2 study. Ann. Clin. Transl. Neurol. 2020;7:2393–2408. doi: 10.1002/acn3.51235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aartsma-Rus A., Bremmer-Bout M., Janson A.A.M., Den Dunnen J.T., Van Ommen G.J.B., Van Deutekom J.C.T. Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromuscul. Disord. 2002;12:S71–S77. doi: 10.1016/s0960-8966(02)00086-x. [DOI] [PubMed] [Google Scholar]
- 11.Matsuo M., Takeshima Y., Koizumi M. 2004. ENA nucleic acid drugs modifying splicing in mRNA precursor. International patent WO/2004/048570. [Google Scholar]
- 12.Wilton S.D., Fletcher S., McClorey G. 2006. Antisense oligonucleotides for inducing exon skipping and methods of use thereof. International patent WO/2006/000057. [Google Scholar]
- 13.Platenburg G.J., de Kimpe J.J., van Deutekom J.C.T., van Ommen G.-J.B., Aartsma-Rus A. 2009. Method for efficient exon (44) skipping in Duchenne muscular dystrophy and associated means. International patent WO/2009/139630. [Google Scholar]
- 14.Sazani P., Kole R. 2010. Multiple exon skipping compositions for DMD. International patent WO/2010/048586. [Google Scholar]
- 15.Popplewell L.J., Trollet C., Dickson G., Graham I.R. Design of phosphorodiamidate morpholino oligomers (PMOs) for the induction of exon skipping of the human DMD gene. Mol. Ther. 2009;17:554–561. doi: 10.1038/mt.2008.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilton S., Fletcher S., Adams A., Meloni P. 2011. Antisense molecules and methods for treating pathologies. International patent WO/2011/057350. [Google Scholar]
- 17.Echigoya Y., Yokota T. Restoring Dystrophin Expression with Exon 44 and 53 Skipping in the DMD Gene in Immortalized Myotubes. Methods Mol. Biol. 2023;2587:125–139. doi: 10.1007/978-1-0716-2772-3_7. [DOI] [PubMed] [Google Scholar]
- 18.Echigoya Y., Mouly V., Garcia L., Yokota T., Duddy W. In Silico Screening Based on Predictive Algorithms as a Design Tool for Exon Skipping Oligonucleotides in Duchenne Muscular Dystrophy. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilton S.D., Fall A.M., Harding P.L., McClorey G., Coleman C., Fletcher S. Antisense oligonucleotide-induced exon skipping across the human dystrophin gene transcript. Mol. Ther. 2007;15:1288–1296. doi: 10.1038/sj.mt.6300095. [DOI] [PubMed] [Google Scholar]
- 20.Aoki Y., Nakamura A., Yokota T., Saito T., Okazawa H., Nagata T., Takeda S. In-frame dystrophin following exon 51-skipping improves muscle pathology and function in the exon 52-deficient mdx mouse. Mol. Ther. 2010;18:1995–2005. doi: 10.1038/mt.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Vliet L., de Winter C.L., van Deutekom J.C.T., van Ommen G.J.B., Aartsma-Rus A. Assessment of the feasibility of exon 45-55 multiexon skipping for duchenne muscular dystrophy. BMC Med. Genet. 2008;9:105–107. doi: 10.1186/1471-2350-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Angelis F.G., Sthandier O., Berarducci B., Toso S., Galluzzi G., Ricci E., Cossu G., Bozzoni I. Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Delta 48-50 DMD cells. Proc. Natl. Acad. Sci. USA. 2002;99:9456–9461. doi: 10.1073/pnas.142302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brun C., Suter D., Pauli C., Dunant P., Lochmüller H., Burgunder J.M., Schümperli D., Weis J. U7 snRNAs induce correction of mutated dystrophin pre-mRNA by exon skipping. Cell. Mol. Life Sci. 2003;60:557–566. doi: 10.1007/s000180300047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemens P.R., Rao V.K., Connolly A.M., Harper A.D., Mah J.K., Smith E.C., McDonald C.M., Zaidman C.M., Morgenroth L.P., Osaki H., et al. Safety, Tolerability, and Efficacy of Viltolarsen in Boys With Duchenne Muscular Dystrophy Amenable to Exon 53 Skipping: A Phase 2 Randomized Clinical Trial. JAMA Neurol. 2020;77:982–991. doi: 10.1001/jamaneurol.2020.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe N., Nagata T., Satou Y., Masuda S., Saito T., Kitagawa H., Komaki H., Takagaki K., Takeda S. NS-065/NCNP-01: An Antisense Oligonucleotide for Potential Treatment of Exon 53 Skipping in Duchenne Muscular Dystrophy. Mol. Ther. Nucleic Acids. 2018;13:442–449. doi: 10.1016/j.omtn.2018.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Deutekom J.C., Bremmer-Bout M., Janson A.A., Ginjaar I.B., Baas F., Den Dunnen J.T., van Ommen G.J. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum. Mol. Genet. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- 27.Findlay A.R., Wein N., Kaminoh Y., Taylor L.E., Dunn D.M., Mendell J.R., King W.M., Pestronk A., Florence J.M., Mathews K.D., et al. Clinical phenotypes as predictors of the outcome of skipping around DMD exon 45. Ann. Neurol. 2015;77:668–674. doi: 10.1002/ana.24365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beggs A.H., Hoffman E.P., Snyder J.R., Arahata K., Specht L., Shapiro F., Angelini C., Sugita H., Kunkel L.M. Exploring the Molecular Basis for Variability among Patients with Becker Muscular Dystrophy: Dystrophin Gene and Protein Studies. Am. J. Hum. Genet. 1991;49:54–67. [PMC free article] [PubMed] [Google Scholar]
- 29.Zatz M., Pavanello R.D.C.M., Lourenço N.C.V., Cerqueira A., Lazar M., Vainzof M. Assessing pathogenicity for novel mutation/sequence variants: the value of healthy older individuals. NeuroMolecular Med. 2012;14:281–284. doi: 10.1007/s12017-012-8186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comi G.P., Prelle A., Bresolin N., Moggio M., Bardoni A., Gallanti A., Vita G., Toscano A., Ferro M.T., Bordoni A., et al. Clinical variability in Becker muscular dystrophy. Genetic, biochemical and immunohistochemical correlates. Brain. 1994;117(Pt 1):1–14. doi: 10.1093/brain/117.1.1-a. [DOI] [PubMed] [Google Scholar]
- 31.Aoki Y., Nagata T., Yokota T., Nakamura A., Wood M.J.A., Partridge T., Takeda S. Highly efficient in vivo delivery of PMO into regenerating myotubes and rescue in laminin-α2 chain-null congenital muscular dystrophy mice. Hum. Mol. Genet. 2013;22:4914–4928. doi: 10.1093/hmg/ddt341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alter J., Lou F., Rabinowitz A., Yin H., Rosenfeld J., Wilton S.D., Partridge T.A., Lu Q.L. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat. Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- 33.Wu B., Lu P., Benrashid E., Malik S., Ashar J., Doran T.J., Lu Q.L. Dose-dependent restoration of dystrophin expression in cardiac muscle of dystrophic mice by systemically delivered morpholino. Gene Ther. 2010;17:132–140. doi: 10.1038/gt.2009.120. [DOI] [PubMed] [Google Scholar]
- 34.Vila M.C., Klimek M.B., Novak J.S., Rayavarapu S., Uaesoontrachoon K., Boehler J.F., Fiorillo A.A., Hogarth M.W., Zhang A., Shaughnessy C., et al. Elusive sources of variability of dystrophin rescue by exon skipping. Skeletal Muscle. 2015;5:44. doi: 10.1186/s13395-015-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors have verified that the data supporting the findings of this study are accessible within the article and its supplementary information.