Abstract
Integration of the human immunodeficiency virus type 1 (HIV-1) cDNA is a required step for viral replication. Integrase, the virus-encoded enzyme important for integration, has not yet been exploited as a target for clinically useful inhibitors. Here we report on the identification of new polyhydroxylated aromatic inhibitors of integrase including ellagic acid, purpurogallin, 4,8,12-trioxatricornan, and hypericin, the last of which is known to inhibit viral replication. These compounds and others were characterized in assays with subviral preintegration complexes (PICs) isolated from HIV-1-infected cells. Hypericin was found to inhibit PIC assays, while the other compounds tested were inactive. Counterscreening of these and other integrase inhibitors against additional DNA-modifying enzymes revealed that none of the polyhydroxylated aromatic compounds are active against enzymes that do not require metals (methylases, a pox virus topoisomerase). However, all were cross-reactive with metal-requiring enzymes (restriction enzymes, a reverse transcriptase), implicating metal atoms in the inhibitory mechanism. In mechanistic studies, we localized binding of some inhibitors to the catalytic domain of integrase by assaying competition of binding by labeled nucleotides. These findings help elucidate the mechanism of action of the polyhydroxylated aromatic inhibitors and provide practical guidance for further inhibitor development.
Early during infection of a sensitive cell, the RNA genome of human immunodeficiency virus (HIV) type 1 (HIV-1) is reverse transcribed to yield a double-stranded cDNA copy, and that DNA copy is then integrated into a chromosome of the host (for reviews, see references 4, 14, 19, 29, 38, 58, and 63). Retroviruses encode a protein, named integrase, that carries out the initial DNA breaking and joining reactions involved in integration. The integrase enzyme is a potentially attractive target for antiretroviral agents, since it is known to be required for HIV replication (1, 18, 66, 70, 71). Although many inhibitors that are active against purified recombinant integrase protein have been described, none have yet proven to be useful clinically (2, 12, 13, 22, 23, 25, 28, 33, 34, 48, 51–53, 55, 59–62, 64, 65, 73).
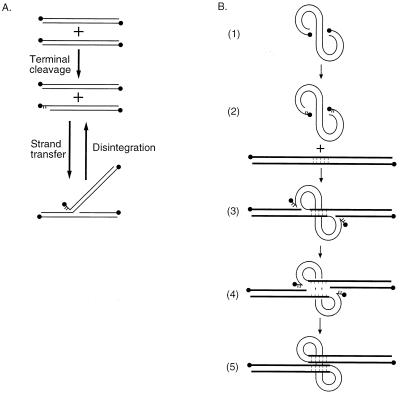
Several in vitro assays are available for assessing the function of integrase inhibitors. Integrase protein purified after overexpression in insect cells or Escherichia coli can carry out DNA cleavage and joining reactions that mimic normal integration in several respects. Integrase protein can remove two nucleotides from the 3′ end of a model viral cDNA end (6, 9, 21, 35, 47, 68), probably to prepare a defined substrate for joining (57), and can then join the recessed 3′ end to a 5′ phosphate in a target DNA (11, 21, 46) (Fig. 1A). Integrase can also catalyze a reversal of this reaction, playfully named “disintegration” (17). However, reactions catalyzed by purified HIV-1 integrase mainly model reactions at one viral cDNA end only, whereas integration in vivo involves the coordinated joining of both cDNA ends (Fig. 1B).
FIG. 1.
DNA cutting and joining reactions involved in cDNA integration. (A) Reactions of purified integrase. The lines represent double-stranded oligonucleotides that match in sequence a cDNA end (LTR). DNA 5′ ends are shown as balls; unpaired bases are shown as short lines. Integrase first cleaves the LTR to remove two nucleotides (middle) and then attaches the exposed 3′ hydroxyl to the 5′ end of a break in the target DNA (bottom). The reversal of this reaction is named “disintegration.” (B) Reactions mediating integration in vivo. The viral cDNA is shown as the curved line; target DNA is shown as the straight line. Integrase first removes two nucleotides from each 3′ cDNA end (parts 1 and 2). The recessed 3′ ends are then joined to protruding 5′ ends of breaks made in the target DNA (part 3), yielding the integration intermediate labeled “II.” The target DNA between the points of joining then comes unpaired, leaving gaps at each host-virus DNA junction (part 4). These gaps are then repaired (part 5) to yield the integrated provirus. Reactions with preintegration complexes recapitulate parts 1 to 4 but do not support the final repair step (part 5).
More authentic integration reactions can be carried out in vitro by using preintegration complexes (PICs) (5) isolated from HIV-1-infected cells (26, 30). To prepare PICs, human T cells are infected with HIV-1 and are then permeabilized 4 to 6 h after infection. The resulting extracts contain the viral cDNA in association with integrase (31), further viral proteins (7, 36, 57), and also at least one cellular protein (27). Reactions with PICs in vitro yield products with the structures expected for coupled integration (Fig. 1B). Importantly in this context, reactions with PICs are much less sensitive to small-molecule inhibitors than reactions with purified integrase (28, 39). Thus, PIC reactions represent attractive secondary screens for assessing the promise of integrase inhibitors.
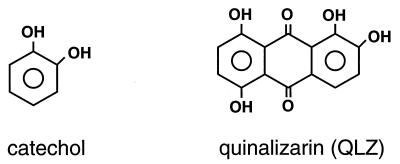
Many reported inhibitors of purified integrase share a common characteristic, a benzene ring with cis-hydroxyls (catechol; Fig. 2) (8). We have previously reported on studies of one group of compounds containing this structure, the polyhydroxylated anthraquinones such as quinalizarin (Fig. 2). These compounds are of particular interest, since they are active against PICs and have been reported to be active against retroviruses in cell culture, although as discussed below, it is not clear that these compounds are active against integrase itself in cell culture (28, 67).
FIG. 2.
Structures of catechol and QLZ.
Here we describe the characterization of additional polyhydroxylated aromatic compounds as integrase inhibitors and studies of their mechanisms of action. A series of new anthraquinones and related quinones yielded new inhibitors, as did a family of compounds containing the catechol moiety embedded in larger structures. We also found that hypericin, a previously described polyhydroxylated aromatic inhibitor of viral replication, was in fact an inhibitor of integration in vitro. We used assays with both purified integrase and PICs to better understand the promise of different inhibitors. We also present the results of counterscreening of several integrase inhibitors against a battery of DNA-modifying enzymes; these results emphasize the role of metal atoms in inhibition. In mechanistic studies, we find that the polyhydroxylated anthraquinone and several other compounds compete with a compound that binds the integrase catalytic domain, placing the likely site of action of these compounds on the catalytic domain.
MATERIALS AND METHODS
Materials.
Unless indicated otherwise, test compounds were obtained from Sigma. Anthrarobin (Aldrich), alizarin red S (ARS; Aldrich), quercetagetin (QTN; Indofine Chemicals), and myricetin (Indofine Chemicals) were obtained elsewhere. l-Chicoric acid (LCH) was a gift of Manfred Reinecke (Texas Christian University, Fort Worth, Tex.).
Integration assays.
Integrase protein and deletion derivatives were purified as described previously (10). Assays for inhibition of reactions with purified integrase were performed as described elsewhere (9, 14, 28). Preparation of PICs was carried out as described previously (30). Tests of inhibition were performed as described previously (28).
Counterscreening.
Assays of inhibition of EcoRT and PvuII (New England Biolabs, Beverly, Mass.) were carried out by incubating 1 U of enzyme with several concentrations of inhibitor for 5 min in 20 μl of the manufacturer’s recommended buffer (containing 10 mM MgCl2) and then adding 0.2 μg of pUC19 reaction substrate. Reactions were stopped after 5 min by the addition of loading dye containing excess EDTA. The products were separated by electrophoresis and were visualized by staining with ethidium bromide. The reactions were quantitated with a Speedlight Gel Documentation System and IP Lab Gel software.
Reactions with EcoRT methylase (New England Biolabs) and SssI methylase (also known as CpG methylase; New England Biolabs) were carried out in 50 mM NaCl–50 mM Tris HCl (pH 8.0)–10 mM EDTA–80 μM S-adenosylmethionine. Compounds were incubated with methylase (0.8 U) in the buffer described above for 5 min at room temperature, and then pUC19 DNA substrate was added and the reaction mixtures were incubated for 15 min at 37°C. Reactions were stopped by phenol extraction, and the reaction mixtures were chilled to 4°C. DNAs were recovered by ethanol precipitation, resuspended in restriction digestion buffer, and cleaved with EcoRI (to test for methylation by EcoRI methylase) or AatII (to test for methylation by CpG methylase). The reaction products were analyzed by electrophoresis as described above. In some cases inhibition was seen with concentrations of inhibitors greater than 50 μM, thereby arguing against various possible systematic errors in assaying the methylases.
Assays of molluscum contagiosum virus (MCV) topoisomerase were carried out as described previously (44). Briefly, purified enzyme was incubated for 5 min with test compounds in buffer containing 10 mM EDTA, 20 mM Tris (pH 8), 200 mM potassium glutamate, 1 mM dithiothreitol, and 0.1% Nonidet P-40, and then the assays were started by the addition of 0.2 μg of pUC19 DNA substrate. Relaxation of DNA substrates was monitored by gel electrophoresis and quantitated as described above.
Assays of Moloney murine leukemia virus (MoMuLV) reverse transcriptase (RT) were carried out essentially as described previously (37). The reaction mixtures contained 2.5 U of RT (New England Biolabs), 50 mM Tris (pH 7.9), 75 mM KCl, 2 mM dithiothreitol, 25 μg of poly (rA)-oligo(dT) template (Pharmacia) per ml, 5 mM MgCl2 or MnCl2, 0.05% Nonidet P-40, and 2.3 mM [3H]TTP. RT was preincubated in the reaction mixture with the compound to be tested for 5 min at room temperature and was then incubated for 1 h at 37°C. Reactions were stopped by spotting the reaction mixtures onto filter paper disks (Whatman DE81). The dried filters were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and then in 95% ethanol, dried, and counted by liquid scintillation counting in the presence of EcoLume scintillation cocktail (ICN).
Nucleotide binding to integrase and drug competition.
Assays of binding of oxidized [α-32P]ATP to integrase were carried out exactly as described previously (50). To test for competition by integrase inhibitors, the compounds were added to integrase, and the mixture was preincubated for 5 min at room temperature prior to the addition of oxidized ATP.
Synthesis of emodin-9-anthrone (DZ-III-77).
A solution of emodin 1,3-dimethyl ether (200 mg, 0.67 mmol) in glacial acetic acid (20 ml) and HI (47% aqueous, 10 ml) was refluxed for 3 h. The solution was cooled at 3°C overnight, and the product crystallized as pale yellow plates. The solid was collected, washed with water, and air dried (121 mg; 71%). An analytical sample was obtained via recrystallization from acetone; melting point (mp) 255 to 256°C, dec. (literature mp 255°C, dec.); 1H nuclear magnetic resonance (NMR) (300 HMz, dimethyl sulfoxide [DMSO]-d6) δ 2.33 (s, 3H), 4.32 (s, 2H), 6.24 (d, 1H, J = 2.3 Hz), 6.43 (d, 1H, J = 2.3 Hz), 6.70 (bs, 1H), 6.80 (bs, 1H), 10.84 (s, 1H), 12.34 (s, 1H), 12.39 (s, 1H); electron ionization-mass spectrometry (EIMS) m/e 256 (100), 241 (17), 227 (10), 213 (14); Fourier transform infrared (FTIR) (Nujol) 3378, 3312, 1625, 1598, 1277, 1242, 1157 cm−1. Exact mass calculated for C15H12O4: 256.0736. Found: 256.0748.
Synthesis of emodic acid N,N-diethylamide-9-anthrone (DZ-III-126).
A solution of emodic acid N,N-diethylamide (130 mg, 0.37 mmol) was dissolved in glacial acetic acid (20 ml) by refluxing under nitrogen. A suspension of stannous chloride (3.5 g) in concentrated HCl (8.75 ml) was carefully added and the solution was stirred at reflux for 2 min. The solution was cooled to 60°C, diluted with water (50 ml), and cooled at 3°C. The light tan solid was collected by vacuum filtration, washed with water, and air dried. Recrystallization from acetone afforded pale yellow needles (85 mg, 67%); mp 233 to 235°C (dec) with darkening at 221 to 222°C. 1H NMR (300 MHz, acetone-d6) δ 1.10–1.26 (m, 6H), 3.31 (m, 2H), 3.52 (m, 2H), 4.43 (s, 2H), 6.35 (d, 1H, J = 2.2 Hz), 6.56 (m, 1H), 6.79 (bs, 1H), 6.93 (bs, 1H), 12.39 (s, 1H), 12.45 (s, 1H); EIMS m/e 341 (84), 324 (19), 269 (100), 242 (54), 213 (32), 184 (15), 155 (10), 139 (28), 128 (22); infrared (Nujol) 1627, 1594, 1286, 1161 cm−1. Exact mass calculated for C19H19NO5: 341.1263. Found: 341.1279. Analysis calculated for C19H19NO5 · 1/2 H2O: C, 65.13; H, 5.75. Found: C, 65.15; H, 5.23.
Synthesis of bromoquinalizarin.
Bromoquinalizarin was prepared as follows. A 500-ml, two-neck, round-bottom flask was charged with quinalizarin (QLZ; 2.02 g, 7.4 mmol) and dimethylformamide (100 ml) and cooled to 0°C. A solution of N-bromosuccinimide in dimethylformamide (2.64 g, 14.8 mmol in 110 ml) was added dropwise to the cooled reaction mixture. After the addition, the reaction mixture was warmed to room temperature and was stirred overnight. The solvent was then removed by distillation at reduced pressure to leave a red precipitate. The solid was triturated with chloroform to remove the residual succinimide. After trituration, the red solid was characterized as bromoquinalizarin (1.7 g, 65% yield). mp 280 to 280°C (dec); 1H NMR (DMSO-d6) d ppm 7.84 (s, 1H), 7.43 (d, 2H, 9 Hz), 7.39 (d, 2H, 9 Hz); 13C NMR (DMSO-d6) d ppm 189.9, 184.0, 156.9, 156.5, 150.3, 150.1, 129.6, 128.8, 123.6, 123.4, 116.4, 115.0, 112.4, 112.0. EIMS (m+) = 351.
RESULTS
Assays of inhibition of purified HIV-1 integrase.
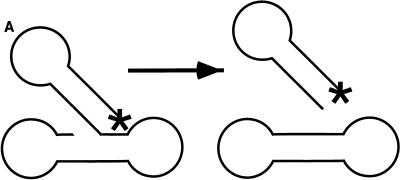
HIV-1 integrase protein was purified after overexpression in E. coli and was assayed with oligonucleotide substrates that model the viral long terminal repeat (LTR) and target DNA. A particularly convenient assay monitors disintegration (Fig. 1A) (17). If integrase protein is provided with a Y-shaped model substrate resembling the in vitro integration product, integrase can catalyze the release of the viral DNA analog and reclosure of the target DNA. One useful version of these substrates has the DNA ends joined by loops, permitting particularly easy synthesis and analysis of the reaction products (“dumbbell disintegration”) (15, 28). An example of the use of this substrate for inhibitor screening is shown in Fig. 3B. Incubation of this substrate with purified integrase results in the release of an LTR-like duplex and resealing of the model target DNA. When a 32P label is placed in the DNA at the 5′ end (Fig. 3A), the disintegration product can be visualized by the appearance of a single band of lower molecular weight (16).
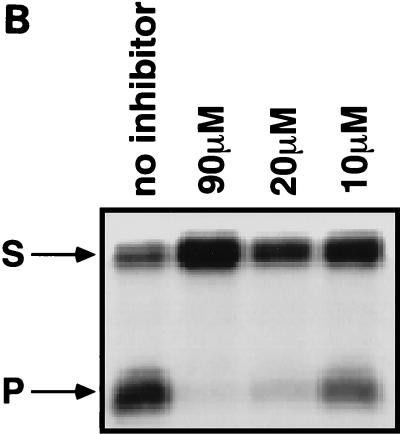
FIG. 3.
Tests of integrase inhibitors by dumbbell disintegration. (A) Diagram of the dumbbell disintegration reaction. The asterisk marks the position of a 5′ 32P label. (B) Inhibition of dumbbell disintegration by hypericin. The concentrations of hypericin are indicated above the lanes. S, substrate; P, product.
This dumbbell disintegration reaction was used as a first screen in testing candidate inhibitors. Previous studies have indicated that the disintegration reaction is relatively permissive compared with other integrase assays. The disintegration reaction was used here as a primary screen in an effort to select for particularly potent inhibitors. To determine the concentrations required for 50% inhibition (IC50s), different concentrations of small molecules were added to test reactions, and the yield of product was quantitated with a PhosphorImager instrument. IC50s were derived from these measurements as described previously (28).
Assays of inhibition of PICs.
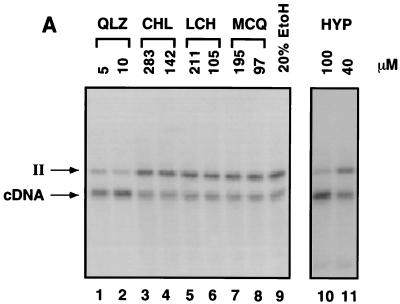
As discussed above, in vitro assays based on PICs isolated from HIV-1-infected cells more closely resemble integration in vivo than do assays with purified integrase (see reference 28 and 29 for a discussion). To characterize the inhibitors, PICs containing HIV cDNA were incubated with a naked DNA target in vitro. The reaction products were then deproteinized and assayed on Southern blots probed with labeled HIV LTR sequences. The compounds to be tested for inhibition were titrated into the reaction mixtures, and inhibition was quantitated by determining the level of reduction in the appearance of the integration product (an example is shown in Fig. 4A) (28).
FIG. 4.
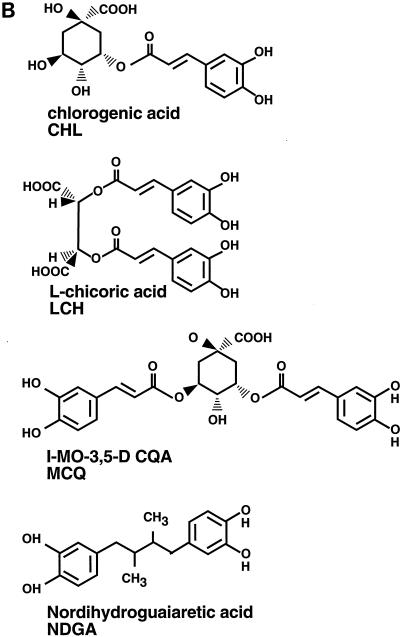
Tests of integrase inhibitors in PIC assays and the structures of LCH and derivatives. (A) Assay of inhibitors of PIC reactions. Shown is a Southern blot analysis of reaction products probed with labeled LTR sequences. Bands corresponding to the unreacted cDNA and integration intermediate (II; part 4 in Fig. 1) are marked beside the autoradiogram. Lanes 1 to 12, inhibitors at the indicated concentrations; lane 10, 20% ethanol (EtOH), the solvent used to dissolve CHL. HYP, hypericin. (B) Structures of LCH and its chemical relatives.
Test of hypericin.
The polyhydroxylated aromatic compound hypericin, isolated from the medicinal herb St. John’s wort (Hypericum perforatum), has previously been reported to inhibit the replication of HIV and other viruses (49, 56). Hypericin was tested for its ability to inhibit dumbbell disintegration and was found to be active, with an IC50 of 15 μM (Fig. 3B). Hypericin also displayed inhibitory activity against PICs, with an IC50 of about 100 μM (Fig. 4A, lanes 10 and 11).
Tests of new polyhydroxylated aromatic compounds.
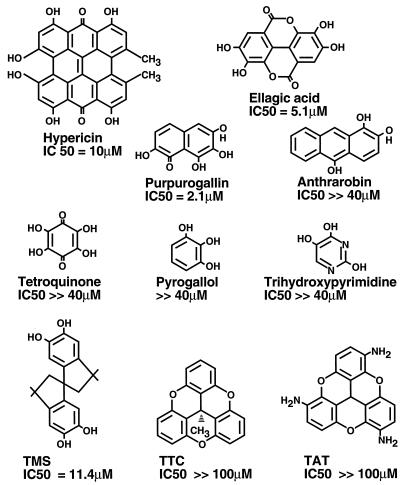
Since many polyhydroxylated aromatic compounds were found to be integrase inhibitors, further compounds of this class were tested for their inhibitory activities. The compounds were titrated into mixtures containing purified integrase, and the mixtures were incubated for 5 min at room temperature. Dumbbell disintegration substrates were then added to start the reaction. Compounds were analyzed for inhibitory activity initially at 40 μM. Those compounds showing any inhibitory activity at this concentration were then characterized further to determine the IC50. The structures of the compounds studied and the measured IC50s are shown in Fig. 5. The IC50s of the single-ring compounds tetroquinone, pyrogallol, and trihydroxypyrimidine were well above 40 μM, as was the IC50 of catechol itself. Of the multiring compounds tested, purpurogallin, ellagic acid (ELA), and 3,3,3′,3′-tetramethyl-1,1′-spirobis(indan)-5,5′,6,6′-tetraol (TMS) were active, while the parasiticide anthrarobin was inactive. Two compounds related in structure to TMS—4,8,12-trioxatricornan and 2,6,10-triamino-4,8,12-trioxatricornan—were also inactive. These data emphasize the importance of residues in addition to the catechol moiety in potentiating the inhibition of integrase.
FIG. 5.
Structures of polyhydroxylated aromatic compounds and some chemical relatives with IC50s for inhibition of dumbbell disintegration. The measured IC50 is given beneath each structure. TTC, 4,8,12-trioxatricornan; TAT, 2,6,10-triamino-4,8,12-trioxatricornan.
The active compounds described above were tested against the PICs isolated from infected cells. None were detectably inhibitory at 40 μM (data not shown).
Test of LCH and derivatives against PICs.
Previously, LCH and several derivatives were found to inhibit purified integrase in vitro and viral replication in vivo (64, 65), raising the possibility that these compounds inhibit integrase in vivo. LCH, the inactive relative chlorogenic acid (CHL), and the active related molecule 1-MO-3,5-DCQA (MCQ) were tested for their inhibitory activities against PICs (Fig. 4A and B). None of these compounds displayed detectable inhibitory activity, even at concentrations of 100 μM (Fig. 4A, lanes 4 to 10). QLZ was tested in parallel as a control and was shown to be inhibitory, as reported previously (28) (Fig. 4A, lane 3).
A further chemical relative of LCH, nordihydroguaiaretic acid (NDGA; Fig. 4B), was also tested. This compound is composed of two catechol moieties, as in LCH and MCQ, but NDGA is tethered by a 2,3-dimethylbutane moiety. NDGA displayed no inhibitory activity against the terminal cleavage or strand transfer activities of purified integrase (data not shown). Evidently, features in addition to the catechol rings are important for inhibition in this series of compounds.
Effects of ring substituents on inhibition by anthraquinones.
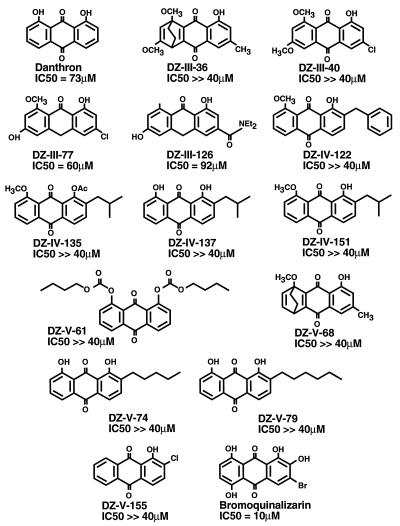
To explore the potential of the anthraquinone nucleus, 14 further anthraquinones and two related anthrones (72) were tested for their abilities to inhibit dumbbell disintegration (Fig. 6). The compounds were initially tested at 40 μM. Those showing detectable inhibition at this concentration were tested to determine the IC50. The IC50s of danthron, DZ-III-77, and DZ-III-126 were between 40 and 70 μM, while the other compounds tested were inactive. These data emphasize the importance of the cis-hydroxyls present in the previously reported compounds.
FIG. 6.
Anthraquinones and anthrones tested for inhibition of dumbbell disintegration. IC50s are shown beneath each structure.
All of the compounds described above were also tested against PICs and were found to be inactive at a concentration of 40 μM (data not shown).
A new derivative of QLZ containing a bromine atom was synthesized in an effort to improve its inhibitory activity. Bromoquinalizarin did inhibit dumbbell disintegration, but the IC50 was higher than that of QLZ itself (10 versus 2 μM) (data not shown) (28).
Tests of function of integrase inhibitors against other DNA-modifying enzymes.
Many previous reports of integrase inhibitors have not presented data on the specificity of inhibition from counterscreening experiments. To investigate specificity more carefully, seven representative polyhydroxylated aromatic integrase inhibitors were tested against a panel of six DNA-modifying enzymes (Table 1). In addition to representatives of the compounds mentioned above (QLZ, TMS, LCH, and ELA), three other previously described inhibitors were also tested: aurintricarboxylic acid (ATC) (23), QTN (a flavone) (34), and ARS (an anthraquinone) (28).
TABLE 1.
IC50s for inhibition of DNA-modifying enzymes by polyhydroxylated aromatic inhibitors of integrasea
| Enzyme | IC50 (μM)
|
||||||
|---|---|---|---|---|---|---|---|
| QLZ | ATC | TMS | QTN | LCH | ELA | ARS | |
| HIV-1 integrase | 4 | 0.5 | 12 | 1 | 0.13 | 5 | 3 |
| MoMLV RT (Mn) | 0.11 ± 0.06 | 0.45 ± 0.3 | >50 | 0.23 ± 0.15 | 0.58 ± 0.24 | 0.68 ± 0.45 | 0.14 ± 0.09 |
| MoMLV RT (Mg) | 2.9 ± 0.8 | 3.8 ± 1 | >50 | 1.2 ± 0.9 | 8.3 ± 6.3 | 1.7 ± 1.9 | 0.75 ± 0.08 |
| PvuII | 15 ± 9 | >50 | >50 | 7 ± 6 | >50 | 48 ± 22 | 22 ± 2 |
| EcoRI | 6 ± 4 | >50 | 13 ± 2 | 2 ± 0.1 | >50 | 2 ± 1 | 16 ± 1 |
| MCV topoisomerase | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| EcoRI methylase | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| CpG methylase | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
Enzymes that either required metal cofactors (MoMLV RT and the restriction enzymes PvuII and EcoRI) or functioned in their absence (MCV topoisomerase, EcoRI methylase, or CpG methylase) were selected. Integrase inhibitors were titrated into reaction mixtures containing each enzyme, and reactions were started by the addition of substrate. Inhibition of product formation was quantitated for each compound and was expressed as an IC50 (Table 1).
All compounds tested were found to inhibit at least some of the metal-utilizing enzymes. TMS was most specific, inhibiting EcoRI but not the other enzymes. QLZ was least specific, inhibiting all the metal-requiring enzymes tested. Strikingly, none of the compounds were active against enzymes that did not require metal cofactors at less than 50 μM, highlighting the likely role of metal atoms in inhibition. Note that the inhibitors are present in the test reaction mixtures at concentrations much lower than those of the metal ions themselves, ruling out the trivial possibility that the added inhibitors are simply sequestering the required metal cofactor.
The role of the particular metal atom bound was probed by comparing the IC50s for MoMLV RT in the presence of Mg2+ or Mn2+. In all cases the compounds inhibited MoMLV RT more effectively in the presence of Mn2+. More potent inhibition of HIV integrase itself by polyhydroxylated aromatic compounds in the presence of Mn2+ than in the presence of Mg2+ has also been reported (40).
Binding of QLZ and other inhibitors to integrase measured by competition of nucleotide binding.
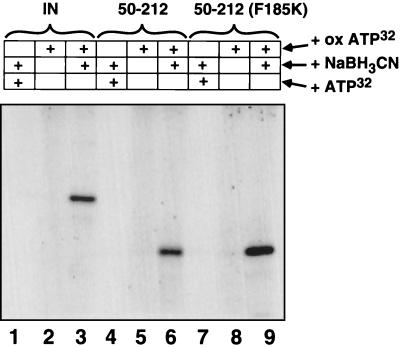
Previously, it was reported that labeled nucleotides can bind to HIV-1 integrase (50, 51, 54), an observation that forms the basis for localizing the binding site of integrase inhibitors. Purified HIV-1 integrase can be incubated with oxidized [α-32P]ATP and then reduced with NaBH3CN to form a covalent complex, likely involving cross-linking of ATP to a lysine residue on the protein. Such reaction mixtures can then be separated by electrophoresis on sodium dodecyl sulfate-polyacrylamide gels and visualized by autoradiography (Fig. 7, lane 3). The formation of the labeled complex was dependent on the addition of [α-32P]ATP and the reducing agent NaBH3CN (Figure 7, lanes 1 and 2). To localize the site of binding, oxidized ATP was incubated with just the catalytic domain fragment of integrase (residues 50 to 212) and was subsequently reduced. Two catalytic domain fragments were compared: one containing the wild-type sequence (Fig. 7, lanes 4 to 6) and one containing a mutation (F185K) that was reported to improve solubility (45). Both catalytic domain fragments bound oxidized ATP with an efficiency comparable to that of full-length integrase (Fig. 7, lanes 3, 6, and 9). These data indicate that ATP associates primarily with the catalytic domain.
FIG. 7.
Nucleotide binding to the catalytic domain of integrase. Integrase (IN) protein (lanes 1 to 3), the catalytic domain only (integrase residues 50 to 212; lanes 4 to 6), and the catalytic domain containing the F185K mutant (lanes 7 to 9) were incubated with [α-32P]ATP and NaBH3CN (lanes 1, 4, and 7), oxidized ATP (ox ATP) (lanes 2, 5, and 8), or oxidized ATP and NaBH3CN (lanes 3, 6, and 9). In each case, Coomassie brilliant blue staining confirmed that the labeled band had the mobility of the integrase derivative studied (data not shown).
To investigate whether QLZ or other inhibitors bound to sites overlapping the ATP-binding site on the catalytic domain, integrase was preincubated with inhibitors and then oxidized [α-32P]ATP was added. Several other integrase inhibitors, in addition to those described above, were tested: curcumin, isolated from the spice turmeric (55); myricetin and quercetin, two flavones (34); doxorubicin, a topoisomerase inhibitor (28); genistein, an isoflavone; and dihydroxynaphthaquinone, a naphthaquinone (28, 33).
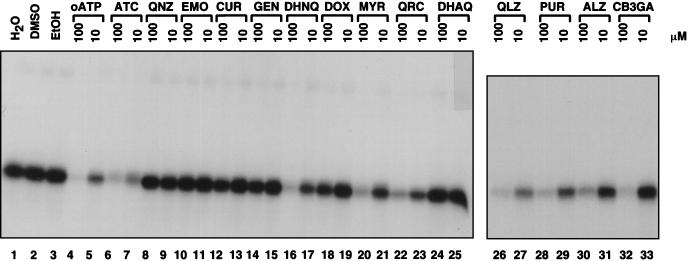
The nucleotide binding activity detected in this assay (Fig. 8, lane 1) was not changed by the addition of 10% DMSO or 10% ethanol, solvents used to resuspend some of the inhibitors (Fig. 8, lanes 1 to 3). Preincubation with 100 μM unlabeled oxidized ATP prevented subsequent binding of labeled oxidized ATP, while preincubation with 10 μM ATP blocked binding only partially (Fig. 8, lanes 4 and 5). Several of the anthraquinones (QLZ, purpurin, alizarin and CB3GA) inhibited cross-linking to oxidized ATP (Fig. 8, lanes 26 to 33). Several other compounds also inhibited binding of oxidized ATP (ATC [Fig. 8, lanes 7 and 8], dihydroxynaphthaquinone [Fig. 8, lanes 16 and 17], myrcetin [Fig. 8, lanes 20 and 21], and quercetin [Fig. 8, lanes 22 and 23]). However, not all of the polyhydroxylated aromatic compounds inhibited the binding of oxidized ATP, indicating that the binding sites of these compounds may not be the same in all cases. These data taken together support a model in which some of the polyhydroxylated anthraquinones and the other inhibitors of binding inhibit integrase by binding to the catalytic domain. Alternatives are possible, however, since indirect effects of ATP binding on inhibitor binding are not ruled out. Some of these compounds may be good candidates for cocrystallization trials with the integrase catalytic domain.
FIG. 8.
Analysis of the binding site of integrase inhibitors by competition with nucleotide binding. In each case integrase was incubated with the indicated compound and then oxidized [α-32P]ATP was added and cross-linked by the subsequent addition of NaBH3CN. Reaction products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by autoradiography. Compound codes: EtOH, ethanol; oATO, oxidized ATP; ATC, aurintricarboxylic acid; QNZ, quinizarin; EMO, emodin; CUR, curcumin; GEN, genistein; DHNQ, dihydroxynaphthaquinone; DOX, doxorubicin; MYR, myricetin; QRC, quercetin; DHAQ, 1,8-dihydroxyanthraquinone; QLZ, quinalizarin; PUR, purpurin; ALZ, alizarin. CB3GA is an anthraquinone.
To control for the possibility that the added compounds were interacting with the oxidized ATP directly, compounds were incubated with oxidized [α-32P]ATP and the reaction products were analyzed by thin-layer chromatography. No new labeled forms were detected, arguing against the idea that the drugs reacted with the oxidized nucleotide itself (data not shown).
DISCUSSION
Here we describe new polyhydroxylated aromatic inhibitors of the HIV-1 integrase protein and report on studies of their mechanisms of action. We previously found that certain polyhydroxylated anthraquinones are of particular interest, since they inhibited both purified integrase protein and PICs (28), so in the present study we analyzed new anthraquinones and related polyhydroxylated phenolic molecules for their inhibitory activities. We also report on the counterscreening of molecules of this class against six other DNA-modifying enzymes and studies of binding to integrase. These data help clarify the nature of the active pharmacophore and its mechanism of action and also provide practical guidance for further screening of inhibitors.
Structure of the active pharmacophore.
No anthraquinones lacking the catechol motif were as active as those containing this motif, highlighting the importance of the catechol structure. However, simply embedding the catechol moiety in an aromatic backbone is not sufficient to generate an inhibitor, as indicated by the lack of inhibition by anthrarobin (Fig. 5). The paired carbonyls in the central ring of the anthraquinones do not both appear to be strictly necessary, since two anthrones (containing a single carbonyl) were at least somewhat inhibitory. Thus, one functionality sufficient for the inhibition of purified integrase by the anthraquinone-related catechol inhibitors appears to be a naphthaquinone nucleus with cis-hydroxyls on one ring and a carbonyl on the adjacent ring. Two other groups have also concluded that substituents in addition to the catechol moiety are important for inhibition by related compounds (8, 34, 48, 62).
However, as described below it may be that not all of the polyhydroxylated aromatic inhibitors act by the same mechanism, raising the possibility that multiple pharmacophores may be contributing to the inhibition. Potentially consistent with this idea are recent studies with several different pharmacophore models to identify new integrase inhibitors in small-molecule databases (42, 62). Also consistent with the idea that multiple pharmacophores are involved, LCH and hypericin do not contain the dihydroxynaphthaquinone motif described above.
Mechanism of inhibition by the polyhydroxylated anthraquinones.
Data presented here clarify a previously unresolved issue, that of whether the polyhydroxylated anthraquinone inhibitors act on the integrase protein or on substrate DNA. Anthraquinones may have some potential to intercalate into DNA due to their planar shape, although they lack the net positive charge characteristic of many strong intercalators, and the results of gel shift assays for intercalation were negative (unpublished data) (33). The data from competition with binding of nucleotides indicate that the polyhydroxylated anthraquinones can bind to integrase in the absence of DNA, supporting the inference that integrase binding is at least part of the inhibitory mechanism. Recently, zidovudine has been cross-linked to integrase and the binding site was mapped, and a possible site for the binding of oxidized ATP and the drugs with which it competes was localized (24). For some of the other polyhydroxylated aromatic inhibitors, binding to the catalytic domain was not detected by competition with nucleotide binding. These compounds may bind to sites that do not overlap the ATP-binding site or that instead bind to DNA.
Previous work has suggested that many of the aromatic polyhydroxylated inhibitors may act by blocking binding of integrase to DNA (40). This observation potentially explains the lack of activity against PICs, in which integrase and cDNA are stably prebound (3, 5, 26, 27, 30). However, the finding that QLZ and related compounds are active against PICs indicates that, at least for these compounds, blocking of binding is not the only mechanism of inhibition.
Some of the polyhydroxylated aromatic integrase inhibitors can potentially chelate metal atoms, and the data from counterscreening experiments highlight the likely role of metal binding in inhibition. Three of the six enzymes screened, EcoRI methylase, CpG methylase, and MCV topoisomerase (44), do not require metals to function, while the other three, MoMLV RT, PvuII, and EcoRI, do require metals. Strikingly, none of the inhibitors tested were active at the concentrations tested against enzymes that did not require metal cofactors, while all inhibitors were cross-reactive with one or more of the metal-containing enzymes. Further support for the idea that metals are involved in inhibition comes from the observation that the IC50 for inhibition of MoMuLV RT differs depending on the metal cofactor, with inhibition being more efficient in the presence of Mn2+ than in the presence of Mg2+. It was similarly reported for HIV integrase that the actions of many of the polyhydroxylated aromatic inhibitors were more potent in the presence of Mn2+ than in the presence of Mg2+ (34, 40). Interestingly, the catalytic domain of integrase, the site of action of the QLZ and certain other inhibitors, also binds to metals. This raises the possibility that these drugs may form a ternary complex with enzymes and metals. In any case it seems likely that the inhibitors act as metal complexes.
Polyhydroxylated anthraquinones have been reported previously to be active against HIV in vivo (41, 67), but the issue of whether this effect is due to inhibition of integrase itself in vivo remains unresolved. QLZ at concentrations close to or below the concentrations necessary for viral inhibition is toxic to cells, depending on the assay (41, 65a, 67). Furthermore, QLZ at 5 μM can inhibit the growth of yeast cells when QLZ is added to the growth medium (44a). Certain polyhydroxylated aromatic compounds have been reported to be able to cross-link proteins when added to cells, and a specific chemical mechanism has been proposed (69). Taken together, these data raise concerns about the toxic effects of polyhydroxylated anthraquinones in vivo.
Mechanism of inhibition by LCH.
LCH and related compounds are of particular interest since they are potent inhibitors of purified integrase and are active against virus in vivo (64, 65). The suggestion that LCH and the related molecule MCQ are active against integrase in vivo, however, is complicated by the finding that LCH also inhibits interactions between the HIV envelope protein and the cellular ligand CD4 (cited in reference 65). Here we report that LCH and MCQ are not active against PICs, findings that do not strengthen the idea that these compounds are active against integrase in vivo. However, it may be possible that compounds that cannot inhibit PICs might still inhibit integration by acting earlier, during the assembly of PICs following the completion of reverse transcription but prior to the assembly of integrase with the cDNA. It is also possible that LCH acts in vivo against other targets, disrupting env-CD4 interactions or reverse transcription. A further possibility is that the in vivo effect on HIV is indirect, mediated in part by toxic effects on cells. Consistent with this idea, LCH is cross-reactive with other metal-requiring enzymes (although antiviral activity has been reported at nontoxic concentrations [64, 65]). The analysis of LCH-resistant derivatives of HIV may resolve some of these issues.
Mechanism of inhibition by hypericin.
Hypericin has been reported to show anti-HIV activity in cell culture (49, 56), and the findings presented here of action against purified integrase and PICs raise the possibility that integrase may be a target protein in vivo. In the presence of light, hypericin can produce singlet oxygen and also superoxide and hypericinium ion. Possibly, these species mediate inhibition (although for another view, see reference 32).
The IC50 of hypericin for the blocking of HIV-mediated fusion in cell cultures is in the high nanomolar range, and the IC50 for the blocking of HIV-induced syncytium formation is about 1 μM (depending on the assay and the time points compared) (49). However, values for inhibition of integrase or PICs are higher (IC50s, 10 to 100 μM). Although the assays differ in important respects, these differences in IC50s raise the possibility that integration is not the major target in vivo. Hypericin may also be active against membranes, since hypericin inhibits the replication of several enveloped viruses, some of which do not encode integrases, but not the replication of nonenveloped viruses (20, 43, 56). The isolation of HIV mutants insensitive to hypericin could clarify whether this antiviral agent is active against integrase in vivo.
Practical lessons for identifying integrase inhibitors.
This work emphasizes the usefulness of counterscreens for integrase inhibitors based on metal-requiring DNA-modifying enzymes (see also reference 48). Counterscreens against enzymes that do not require metals have been reported (61), but on the basis of the data reported here, such studies risk concluding incorrectly that the studied inhibitors are specific for integrase. The observation that different inhibitors are cross-reactive with different metal-utilizing enzymes also emphasizes the importance of studying multiple enzymes in secondary screens.
The promise of the polyhydroxylated aromatic inhibitors of integrase is unclear. Several lines of data argue against the utility of these compounds. The observed cross-reactivity with metal-requiring enzymes is potentially consistent with observations of toxic effects on cells. The tight binding of QLZ to PICs (data not shown) raises the possibility that the compound binds to PICs covalently, as do studies of the cross-linking of proteins by some compounds of this class. However, in support of the utility of the polyhydroxylated aromatic inhibitors, covalent binding of QLZ to PICs would have to be somewhat specific given the observation that the IC50 is not altered when high concentrations of cellular proteins are present in test reaction mixtures (28). Furthermore, cells can clearly tolerate the presence of catechol-containing compounds. For example, the catechol-containing catecholamines are an important group of neurotransmitters. Perhaps the catechol-related pharmacophore will be a useful inhibitor substituent when it is combined with other chemical groups that improve the specificity for integrase.
ACKNOWLEDGMENTS
We thank members of the laboratory of F. D. Bushman for suggestions and helpful discussions.
This work was supported by grants GM56553 and AI34786 to F.D.B. F.D.B. is a scholar of the Leukemia Society of America.
ADDENDUM IN PROOF
A mutant of HIV-1 has been isolated that displays reduced sensitivity to LCH (P. G. King and W. E. Robinson, Jr., J. Virol., in press). Sequencing revealed a change in the integrase coding region. A virus in which wild-type integrase was replaced with the mutant integrase also displayed reduced sensitivity. These data strengthen the view that LCH acts at least in part against integrase in vivo.
REFERENCES
- 1.Adachi A, Ono N, Sakai H, Ogawa K, Shibata R, Kiyomasu T, Masuike H, Ueda S. Generation and characterization of the human immunodeficiency virus type 1 mutants. Arch Virol. 1991;117:45–58. doi: 10.1007/BF01310491. [DOI] [PubMed] [Google Scholar]
- 2.Billich A, Schauer M, Frank S, Rosenwirth B, Billich S. HIV-1 integrase: high-level production and screening assay for the endonucleolytic activity. Antivir Chem Chemother. 1992;3:113–119. [Google Scholar]
- 3.Bowerman B, Brown P O, Bishop J M, Varmus H E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O. Integration of retroviral DNA. Curr Top Microbiol Immunol. 1990;157:19–48. doi: 10.1007/978-3-642-75218-6_2. [DOI] [PubMed] [Google Scholar]
- 5.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 6.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent complex and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley G W, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke T R, Fesen M R, Mazumder A, Wang J, Carothers A M, Grunberger D, Driscoll J, Kohn K, Pommier Y. Hydroxylated aromatic inhibitors of HIV-1 integrase. J Med Chem. 1995;38:4171–4178. doi: 10.1021/jm00021a006. [DOI] [PubMed] [Google Scholar]
- 9.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bushman F D, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushman F D, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 12.Carteau S, Mouscadet J F, Goulaouic H, Subra F, Auclair C. Effect of topoisomerase inhibitors on the in vitro HIV DNA integration reaction. Biochem Biophys Res Commun. 1993;192:1409–1414. doi: 10.1006/bbrc.1993.1573. [DOI] [PubMed] [Google Scholar]
- 13.Carteau S, Mouscadet J F, Goulaouic H, Subra F, Auclair C. Inhibitory effect of the polyanionic drug suramin on the in vitro HIV DNA integration reaction. Arch Biochem Biophys. 1993;305:606–610. doi: 10.1006/abbi.1993.1468. [DOI] [PubMed] [Google Scholar]
- 14.Chow S A. In vitro assays for activities of retroviral integrase. Methods. 1997;12:306–317. doi: 10.1006/meth.1997.0484. [DOI] [PubMed] [Google Scholar]
- 15.Chow S A, Brown P O. Juxtaposition of two viral DNA ends in a bimolecular disintegration reaction mediated by multimers of human immunodeficiency virus type 1 or murine leukemia virus integrase. J Virol. 1994;68:7869–7878. doi: 10.1128/jvi.68.12.7869-7878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow S A, Brown P O. Substrate features important for recognition and catalysis by human immunodeficiency virus type 1 integrase identified by using novel DNA substrates. J Virol. 1994;68:3896–3907. doi: 10.1128/jvi.68.6.3896-3907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 18.Clavel F, Hoggan M D, Willey R L, Strebel K, Martin M, Repaske R. Genetic recombination of human immunodeficiency virus. J Virol. 1989;63:1455–1459. doi: 10.1128/jvi.63.3.1455-1459.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley R M, editors. Virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1767–1848. [Google Scholar]
- 20.Cohen P A, Hudson J B, Towers G H. Antiviral activities of anthraquinones, bianthrones and hypericin derivatives from lichens. Experientia. 1996;52:180–183. doi: 10.1007/BF01923366. [DOI] [PubMed] [Google Scholar]
- 21.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 22.Cushman M, Golegiewski W M, Pommier Y, Mazumder A, Reymen D, De Clercq E, Graham L, Rice W G. Cosalane analogues with enhanced potencies as inhibitors of HIV-1 protease and integrase. J Med Chem. 1995;38:443–452. doi: 10.1021/jm00003a007. [DOI] [PubMed] [Google Scholar]
- 23.Cushman M, Sherman P. Inhibition of HIV-1 integration protein by aurintricarboxylic acid monomers, monomer analogs, and polymer fractions. Biochem Biophys Res Commun. 1992;185:85–90. doi: 10.1016/s0006-291x(05)80958-1. [DOI] [PubMed] [Google Scholar]
- 24.Drake R R, Neamati N, Hong H, Pilon A A, Sunthanker P, Hume S D, Milne G W, Pommier Y. Identification of a nucleotide binding site in HIV-1 integrase. Proc Natl Acad Sci USA. 1998;95:4170–4175. doi: 10.1073/pnas.95.8.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eich E, Pertz H, Kaloga M, Schulz J, Fesen M R, Mazumder A, Pommier Y. (−)-Arctigenin as a lead structure for inhibitors of human immunodeficiency virus type-1 integrase. J Med Chem. 1996;39:89–95. doi: 10.1021/jm950387u. [DOI] [PubMed] [Google Scholar]
- 26.Ellison V H, Abrams H, Roe T, Lifson J, Brown P O. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farnet C, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 28.Farnet C, Lipford R, Wang B, Bushman F D. Differential inhibition of HIV-1 preintegration complexes and purified integrase protein by small molecules. Proc Natl Acad Sci USA. 1996;93:9742–9747. doi: 10.1073/pnas.93.18.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farnet, C. M., and F. D. Bushman. 1996. HIV cDNA integration: molecular biology and inhibitor development. AIDS 10(Suppl. A):3–11. [PubMed]
- 30.Farnet C M, Haseltine W A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farnet C M, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fehr M J, Carpenter S L, Petrich J W. The role of oxygen in the photoinduced antiviral activity of hypericin. Bioorg Med Chem Lett. 1994;4:1339–1344. [Google Scholar]
- 33.Fesen M R, Kohn K W, Leteurtre F, Pommier Y. Inhibitors of human immunodeficiency virus integrase. Proc Natl Acad Sci USA. 1993;90:2399–2403. doi: 10.1073/pnas.90.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fesen M R, Pommier Y, Leteurtre F, Hiroguchi S, Yung J, Kohn K W. Inhibition of HIV-1 integrase by flavones, caffeic acid phenethyl ester (CAPE) and related compounds. Biochem Pharmacol. 1994;48:595–608. doi: 10.1016/0006-2952(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 35.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 36.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;17:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 37.Goff S, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goff S P. Genetics of retroviral integration. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 39.Hansen M, Bushman F D. HIV-2 preintegration complexes: activities in vitro and response to inhibitors. J Virol. 1997;71:3351–3356. doi: 10.1128/jvi.71.4.3351-3356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hazuda D J, Felock P J, Hastings J C, Pramanik B, Wolfe A L. Differential divalent cation requirements uncouple the assembly and catalytic reactions of human immunodeficiency virus type 1 integrase. J Virol. 1997;71:7005–7011. doi: 10.1128/jvi.71.9.7005-7011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higuchi H, Mori K, Kato A, Ohkuma T, Endo T, Kaji H, Kaji A. Antiretroviral activities of anthraquinones and their inhibitory effects on reverse transcriptase. Antivir Res. 1991;15:205–216. doi: 10.1016/0166-3542(91)90067-2. [DOI] [PubMed] [Google Scholar]
- 42.Hong H, Neamati N, Wang S, Nicklaus M C, Mazumder A, Zhao H, Burke T R, Pommier Y, Milne G W A. Discovery of HIV-1 integrase inhibitors by pharmacophore searching. J Med Chem. 1997;40:930–936. doi: 10.1021/jm960754h. [DOI] [PubMed] [Google Scholar]
- 43.Hudson J B, Lopez-Bazzocchi I, Towers G H. Antiviral activities of hypericin. Antivir Res. 1991;15:101–112. doi: 10.1016/0166-3542(91)90028-p. [DOI] [PubMed] [Google Scholar]
- 44.Hwang Y, Wang B, Bushman F D. Molluscum contagiosum virus topoisomerase: purification, activities, and response to inhibitors. J Virol. 1998;72:3401–3406. doi: 10.1128/jvi.72.4.3401-3406.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Jamison, G., and F. D. Bushman. Unpublished data.
- 45.Jenkins T M, Hickman A B, Dyda F, Ghirlando R, Davies D R, Craigie R. Catalytic domain of human immunodeficiency virus type 1 integrase: identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc Natl Acad Sci USA. 1995;92:6057–6061. doi: 10.1073/pnas.92.13.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 47.Katzman M, Katz R A, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaFemina R L, Graham P L, LeGrow K, Hastings J C, Wolfe A, Young S D, Emini E A, Hazuda D J. Inhibition of human immunodeficiency virus integrase by bis-catechols. Antimicrob Agents Chemother. 1995;39:320–324. doi: 10.1128/aac.39.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lenard J, Rabson A, Vanderoef R. Photodynamic inactivation of infectivity of human immunodeficiency virus and other enveloped viruses using hypericin and rose bengal: inhibition of fusion and syncytia formation. Proc Natl Acad Sci USA. 1993;90:158–162. doi: 10.1073/pnas.90.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipford J R, Worland S T, Farnet C M. Nucleotide binding by the HIV-1 integrase protein in vitro. J Acquired Immune Defic Syndr. 1994;7:1215–1223. [PubMed] [Google Scholar]
- 51.Mazumder A, Cooney D, Agbaria R, Gupta M, Pommier Y. Inhibition of human immunodeficiency virus type 1 integrase by 3′-azido-3′-deoxythymidylate. Proc Natl Acad Sci USA. 1994;91:5771–5775. doi: 10.1073/pnas.91.13.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazumder A, Gazit A, Levitzki A, Nicklaus M, Yung J, Kohlhagen G, Pommier Y. Effects of tyrphostins, protein kinase inhibitors, on human immunodeficiency virus type 1 integrase. Biochemistry. 1995;34:15111–15122. doi: 10.1021/bi00046a018. [DOI] [PubMed] [Google Scholar]
- 53.Mazumder A, Gupta M, Perrin D M, Sigman D S, Rabinovitz M, Pommier Y. Inhibition of human immunodeficiency virus type 1 integrase by a hydrophobic cation: the phenanthroline-cuprous complex. AIDS Res Hum Retroviruses. 1995;11:115–125. doi: 10.1089/aid.1995.11.115. [DOI] [PubMed] [Google Scholar]
- 54.Mazumder A, Neamati N, Sommadossi J-P, Gosselin G, Schinazi R F, Imbach J-L, Pommier Y. Effects of nucleotide analogs on human immunodeficiency virus type-1 integrase. Mol Pharmacol. 1996;49:621–628. [PubMed] [Google Scholar]
- 55.Mazumder A, Raghavan K, Weinstein J, Kohn K W, Pommier Y. Inhibition of human immunodeficiency virus type-1 integrase by curcumin. Biochem Pharmacol. 1995;49:1165–1170. doi: 10.1016/0006-2952(95)98514-a. [DOI] [PubMed] [Google Scholar]
- 56.Meruelo D, Lavie G, Lavie D. Therapeutic agents with dramatic antiretroviral activity and little toxicity at effective doses: aromatic polycyclic diones hypericin and pseudohypericin. Proc Natl Acad Sci USA. 1988;85:5230–5234. doi: 10.1073/pnas.85.14.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mizuuchi K. Polynucleotidyl transfer reactions in transpositional DNA recombination. J Biol Chem. 1992;287:21273–21276. [PubMed] [Google Scholar]
- 59.Mouscadet J F, Carteau S, Goulaouic H, Subra F, Auclair C. Triplex-mediated inhibition of HIV DNA integration in vitro. J Biol Chem. 1994;269:21635–21638. [PubMed] [Google Scholar]
- 60.Mouscadet J F, Ketterle C, Goulaouic H, Carteau S, Subra F, Le Bret M, Auclair C. Triple helix formation with short oligonucleotide-intercalator conjugates matching the HIV-1 U3 LTR end sequence. Biochemistry. 1994;33:4187–4196. doi: 10.1021/bi00180a011. [DOI] [PubMed] [Google Scholar]
- 61.Neamati N, Hong H, Mazumder A, Wang S, Sunder S, Nicklaus M C, Milne G W A, Proksa B, Pommier Y. Depsides and depsidones as inhibitors of HIV-1 integrase: discovery of novel inhibitors through 3D database searching. J Med Chem. 1997;40:942–951. doi: 10.1021/jm960759e. [DOI] [PubMed] [Google Scholar]
- 62.Nicklaus M C, Neamati N, Hong H, Mazumder A, Sunder S, Chen J, Milne G W A, Pommier Y. HIV-1 integrase pharmacophore: discovery of inhibitors through three-dimensional database searching. J Med Chem. 1997;40:920–929. doi: 10.1021/jm960596u. [DOI] [PubMed] [Google Scholar]
- 63.Plasterk R H A. The HIV integrase catalytic core. Struct Biol. 1995;2:87–90. doi: 10.1038/nsb0295-87. [DOI] [PubMed] [Google Scholar]
- 64.Robinson W E, Cordeiro M, Abdel-Malek S, Jia Q, Chow S A, Reinecke M G, Mitchell W M. Dicaffeoylquinic acid inhibitors of human immunodeficiency virus integrase: inhibition of the core catalytic domain of human immunodeficiency virus integrase. Mol Pharmacol. 1996;50:846–855. [PubMed] [Google Scholar]
- 65.Robinson W E, Reinecke M G, Abdel-Malek S, Jia Q, Chow S A. Inhibitors of HIV-1 replication that inhibit HIV integrase. Proc Natl Acad Sci USA. 1996;93:6326–6331. doi: 10.1073/pnas.93.13.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65a.Robinson, W. E., Jr. Unpublished data.
- 66.Sakai H, Kawamura M, Sakuragi J-I, Sakuragi S, Shibata R, Ishimoto A, Ono N, Ueda S, Adachi A. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J Virol. 1993;67:1169–1174. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schinazi R F, Chu C K, Babu J R, Oswald B J, Saalmann V, Cannon D L, Eriksson B F H, Nasr M. Anthraquinones as a new class of antiviral agents against human immunodeficiency virus. Antivir Res. 1990;13:265–272. doi: 10.1016/0166-3542(90)90071-e. [DOI] [PubMed] [Google Scholar]
- 68.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stanwell C, Ye B, Yuspa S H, Burke T R. Cell protein cross-linking by erbstatin and related compounds. Biochem Pharmacol. 1996;52:475–480. doi: 10.1016/0006-2952(96)00250-x. [DOI] [PubMed] [Google Scholar]
- 70.Stevenson M, Haggerty S, Lamonica C A, Meier C M, Welch S-K, Wasiak A J. Integration is not necessary for expression of human immunodeficiency virus type 1 protein products. J Virol. 1990;64:2421–2425. doi: 10.1128/jvi.64.5.2421-2425.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stevenson M, Stanwick T L, Dempsey M P, Lamonica C A. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zembower D E, Kam C-M, Powers J C, Zalkow L H. Novel anthraquinone inhibitors of human leukocyte elastase and cathepsin. J Med Chem. 1992;35:1597–1605. doi: 10.1021/jm00087a014. [DOI] [PubMed] [Google Scholar]
- 73.Zhao H, Neamati N, Hong H, Mazumder A, Wang S, Sunder S, Milne G W A, Pommier Y, Burke T R. Coumarin-based inhibitors of HIV integrase. J Med Chem. 1997;40:242–249. doi: 10.1021/jm960450v. [DOI] [PubMed] [Google Scholar]