Lung alveoli are lined by an exquisitely thin monolayer of cells adapted for gas exchange. Guild et al. elucidate a specialized regenerative program by which this delicate, simple epithelium, continuously co-executing essential gas exchange and capillary barrier functions, is endowed with resiliency to acute injury.
Abstract
With each breath, oxygen diffuses across remarkably thin alveolar type I (AT1) cells into underlying capillaries. Interspersed cuboidal AT2 cells produce surfactant and act as stem cells. Even transient disruption of this delicate barrier can promote capillary leak. Here, we selectively ablated AT1 cells, which uncovered rapid AT2 cell flattening with near-continuous barrier preservation, culminating in AT1 differentiation. Proliferation subsequently restored depleted AT2 cells in two phases, mitosis of binucleated AT2 cells followed by replication of mononucleated AT2 cells. M phase entry of binucleated and S phase entry of mononucleated cells were both triggered by AT1-produced hbEGF signaling via EGFR to Wnt-active AT2 cells. Repeated AT1 cell killing elicited exuberant AT2 proliferation, generating aberrant daughter cells that ceased surfactant function yet failed to achieve AT1 differentiation. This hyperplasia eventually resolved, yielding normal-appearing alveoli. Overall, this specialized regenerative program confers a delicate simple epithelium with functional resiliency on par with the physical durability of thicker, pseudostratified, or stratified epithelia.
Introduction
A primary function of epithelia is to provide a protective barrier against the outside world. The distal lung epithelium must additionally and continuously prevent fluid leakage from underlying capillaries into gas exchange air sacs (alveoli), a hallmark of acute respiratory distress syndrome (ARDS; Matthay et al., 2019; Szidon et al., 1972). Yet, unlike other epithelia that maximize barrier resilience physically by maintaining a stratified or pseudostratified organization that can withstand acute cell loss, the alveolar epithelium must remain ultrathin to continuously co-execute its essential gas exchange function. Alveoli must also continuously generate surfactant detergent to minimize surface tension and thereby prevent respiratory failure from air sac collapse. Considering these constraints, the alveolar epithelium has evolved as a delicate simple monolayer composed of cuboidal, surfactant-producing alveolar type II (AT2) cells and ultrathin alveolar type I (AT1) cells. AT1 cells are less abundant but comprise ∼97% of the surface area due to their expansive morphology (Weibel, 2015).
Despite this physical fragility, the alveolar epithelium demonstrates a remarkable capacity for scar-free repair (Logan and Desai, 2015). Classical studies suggest a model whereby alveolar repair is initiated by AT2 cell replication that serves to maintain surfactant-producing cells while simultaneously generating progeny that differentiate into AT1 cells (Evans et al., 1973; Adamson and Bowden, 1974). AT2 cells have since been shown by lineage tracing to act as stem cells that self-renew to generate new AT2 and AT1 cells (Barkauskas et al., 2013; Desai et al., 2014). AT2 cell fate-mapping at specific intervals after injury suggests that AT1 cell regeneration does not begin until multiple days after the onset of proliferation, further supporting stem cell division prior to AT1 regeneration (Jansing et al., 2017; LaCanna et al., 2019). However, this model in which AT1 cell regeneration begins five or more days after alveolar injury is difficult to reconcile with the notable paucity of epithelial denudation over this interval (Jansing et al., 2017; Crapo et al., 1980). As shown below, we found that a fluorescent lineage tag cannot be detected for several days during active AT1 differentiation. This transient lineage mark infidelity called into doubt the dynamics of AT1 regeneration inferred from AT2 cell lineage tracing in acute injury, and challenged the current paradigm that AT2 stem cell division must precede AT1 cell regeneration.
To map the kinetics of alveolar repair with temporal precision, we exploited genetic and chemical approaches to selectively and synchronously ablate AT1 cells without injuring either AT2 stem cells or their fibroblast Wnt niches (Nabhan et al., 2018; Zacharias et al., 2018; Juul et al., 2020). Electron microscopic and immunohistological analysis of the immediate response to widespread AT1 cell ablation revealed near-continuous barrier maintenance by ultrarapid flattening of AT2 cells. Measurements of blood oxygenation and capillary leak into alveolar airspaces corroborated global preservation of gas exchange and barrier function, respectively. Hence, we found the initial step of repair involves rapid flattening and AT1 differentiation of adjacent AT2 cells.
Following depletion by AT1 differentiation, AT2 cells were restored by two phases of proliferation—rapid ploidy reduction of binucleated AT2 cells followed by DNA replication and division of mononucleated AT2 cells. EGFR regulated both M phase entry of binucleated AT2 cells and S phase entry of mononucleated AT2 cells in a Wnt-dependent manner. The operative epidermal growth factor receptor (EGFR) ligand was heparin-binding EGF-like growth factor (hbEGF), presumably available for rapid release by enzymatic cleavage upon injury (Greenlee et al., 2007; Kheradmand et al., 2002). Repeated AT1 cell killing did not result in significant denudation or cellular insufficiency, but rather elicited substantial hyperplasia. The cells were of AT2 cell origin and often clonal, generating densely packed clusters of proliferative daughter cells that lacked surfactant function. The hyperplasia eventually resolved after cessation of injury, leaving normal-appearing alveoli.
Altogether, our findings illuminate a specialized program by which a delicate, simple epithelium continuously co-executing essential gas exchange and capillary barrier functions is endowed with resiliency to severe acute injury.
Results
Near-continuous barrier maintenance and rapid AT1 cell regeneration by differentiation of AT2 cells
Classical electron microscopy (EM) studies suggest that AT1 cells are highly vulnerable to alveolar injuries, presumably due to their delicate morphology and exposure encompassing most of the surface area. Whether quiescent AT2 cells can undergo AT1 differentiation in vivo without first proliferating is not known, but as mentioned above, dynamic AT2 cell lineage tracing in acute lung injury suggests they do not (Aspal and Zemans, 2020). However, since cell cycle transit of AT2 cells has been estimated to take hours, a postproliferative AT1 differentiation model is difficult to reconcile with the paucity of denudation observed at earlier time points after acute injury (Uhal, 1997).
To quantitatively analyze AT1 cell death and regeneration, we exploited a genetic approach to broadly mark AT1 cells with a membrane green fluorescent protein (mGFP) tag (R26mTmG) using the Hopx-CreER allele. We then employed two complementary methods of AT1 cell ablation. Co-expression of the diphtheria toxin (DT) receptor (R26iDTR) enabled precisely timed and synchronous ablation of marked AT1 cells while butylated hydroxytoluene (BHT) injury preserved the CreER system for AT2 cell lineage tracing and genetic manipulations (Adamson et al., 1977; Hirai et al., 1977). We found that 150 ng of DT or 450 mg/kg of BHT eliminated the majority of AT1 cells, yet immunostaining as early as 2 h after toxin administration revealed an intact epithelium comprised of regenerated (mGFP-negative) AT1 cells integrated in a monolayer with unablated (mGFP-positive) AT1 cells (Fig. 1, A–E; and Fig. S1, A–E).
Figure 1.
Rapid AT1 cell regeneration after widespread targeted ablation. (A) Strategy for acute genetic or chemical ablation of pulse-marked (mGFP) AT1 cells. (B and C) Endogenous fluorescence (B) and immunostaining (C) shows initial broad labeling of AT1 population (mGFP+Ager+, control) with widespread AT1 cell loss and reconstitution (mGFP−Ager+) at 2 h after DT and 2 d after BHT administration. (D and E) Immunostaining (D) and quantitation (E) of AT1 cells (Lamp3−Nkx2.1+) before and after AT1 cell ablation with DT or BHT. Note representative marked AT1 cell (mGFP+, arrow) in control and multiple unmarked AT1 cells (mGFP−, arrowhead) after acute ablation (control: 292 AT1 cells scored; DT: 290 AT1 cells scored; BHT: 308 AT1 cells scored; n = 3 mice per condition). Scale bars: 100 µm (B), 25 µm (C), 10 µm (D). DTR, diphtheria toxin receptor; TMX, tamoxifen. Data are mean ± SEM. P values calculated by two-sided Student’s t test.
Figure S1.
Nkx2.1 is a pan-alveolar epithelial marker that enables quantitation of AT2 and AT1 cells. (A–C) Representative image (A) and quantitation (B and C) of co-staining for a pan-epithelial nuclear lineage tag driven by Shh-Cre (H2B-GFP, green) and Nkx2.1 (yellow) shows 100% concordance, with Lamp3 (magenta) distinguishing AT2 (Lamp3+) and AT1 (Lamp3−) cells (2,234 Nkx2.1+ cells scored in n = 3 mice). (D and E) Representative image (D) and quantitation (E) of efficiency and specificity of AT1 cell targeting by Hopx-CreER after three doses of 4 mg of tamoxifen (532 Nkx2.1+Lamp3+ AT2 cells scored and 292 Nkx2.1+Lamp3− AT1 cells scored, n = 3 mice). Scale bars, 20 µm (A), 15 µm (D). Data are mean ± SEM.
We next co-stained for protein markers of AT2 and AT1 cells along with the injury marker Keratin 8 (Krt8) at specific time points after DT administration. Intense Krt8 protein staining characterizes AT2-to-AT1 intermediates identified in single-cell transcriptomic studies of bleomycin lung injury (Strunz et al., 2020; Choi et al., 2020). The results at 0.5 h showed emergence of strong Krt8 labeling in surfactant-producing cells with long, flattened extensions. At 2 h after DT, Krt8+ cells lacked AT2 markers but were positive for AT1 marker proteins (Fig. 2 A). Epithelial cell quantitation at 2 h after toxin showed a substantial reduction in the number of AT2 cells but no difference in the number of AT1 cells compared with uninjured lungs (Fig. 2, B and C). Given the short time scale, these results collectively suggested that ablated AT1 cells were rapidly regenerated by differentiation of nearby AT2 cells (Fig. 2 D).
Figure 2.
AT2 cell differentiation regenerates AT1 cells without stem cell proliferation. (A) Krt8 immunostaining 0.5 and 2 h after DT shows emergence of early (Krt8+SpC+Ppdn−, arrows) and late (Krt8+SpC−Ppdn+, arrowheads) intermediates in AT2-to-AT1 cell differentiation. (B–D) Immunostaining (B) for AT2 (Lamp3+Nkx2.1+) and AT1 (Lamp3−Nkx2.1+) cells with quantitation (C) and model (D) shows no reduction in AT1 (Lamp3−Nkx2.1+) but diminished AT2 (Lamp3+Nkx2.1+) cells at 2 h after AT1 ablation by DT (control: 2,234 Nkx2.1+ cells scored; DT: 2,039 Nkx2.1+ cells scored; n = 3 mice per condition). (E) Strategy for AT2 lineage trace with cell cycle mapping after AT1 cell ablation. (F) Representative images of AT2-to-AT1 differentiation without DNA replication (left, H2B-mCherry+Lamp3−Ager+EdU−) and AT2 cell cycle entry (right, H2B-mCherry+Lamp3+Ager−EdU+). Note, Ager is a basal surface marker of AT1 cells. (G and H) (G) Quantitation of lineage-marked AT2 cells that generated AT1 cells (H2B-mCherry+Lamp3−) and (H) the proportions with and without preceding DNA replication (H2B-mCherry+Lamp3−EdU+ or −) (control: 539 cells scored; BHT: 681 cells scored; n = 3 mice per condition). (I–K) Representative images (I) of mGFP+ AT2 cell clone with AT2 phenotype (left, uninjured lung) and AT1 cell phenotype (right, post-injury). Quantitation (J) of clones as isolated AT1 or AT1 with accompanying parent AT2 cell and model (K) for AT2-to-AT1 differentiation without preceding proliferation as primary mechanism for AT1 cell regeneration (304 mGFP+ AT1 cells scored, n = 3 mice). Scale bars, 15 µm (A), 50 µm (B), 10 µm (F), 25 µm (I). Dox, doxycycline. Data are mean ± SEM. P values calculated by two-sided Student’s t test.
To visualize dynamic flattening of AT2 cells by lineage tracing, we combined the SftpC-CreER-rtTA knock-in and R26mTmG alleles to broadly mark AT2 cells with mGFP prior to BHT administration. However, at 60 h after BHT—12 h after the peak of both AT1 cell death and AT2 cell proliferation as previously characterized by thymidine labeling and ultrastructural analyses—we did not detect the AT2-lineage mark in AT1 cells. This result suggested lineage tracer dysfunction, and we hypothesized the fluorescent protein tag may be lost during AT1 cell differentiation. To test this, we combined the Shh-Cre-GFP knock-in, R26iDTR, and R26mTmG alleles to ubiquitously mark all AT1, AT2, and airway cells prior to genetic ablation. 24 h after toxin administration, the lineage reporter was not detectable in regenerated AT1 cells by either visualization of endogenous fluorescence or antibody staining against GFP (Fig. S2, A–D). To ensure that loss of the lineage mark was not due to the toxin or an injury effect, we similarly analyzed its performance at embryonic day 18, when bipotent progenitors mature into AT1 cells during a process called sacculation (Desai et al., 2014). These results reproduced loss of the lineage mark during flattening of nascent AT1 cells (Fig. S2, E–G). The R26mTmG lineage marker becomes detectable after several days in the newly generated AT1 cells in both contexts, as previously demonstrated (Desai et al., 2014; Nabhan et al., 2018). Hence, AT2 cell lineage tracing during acute injury repair is not an accurate way to determine the timing of AT1 cell regeneration. Because the dysfunction only lasts several days, however, later emergence of the lineage mark in AT1 cells does confirm that they arose from AT2 cells (see below).
Figure S2.
Rapid loss of mGFP fluorescent lineage marker in flattening epithelial cells during AT1 differentiation in injury and embryonic development. (A) Strategy for pan-alveolar epithelial lineage marking with mGFP prior to pulsed epithelial ablation. (B and C) Direct fluorescence (top row) and co-staining for pan-epithelial lineage marker (mGFP) and AT1 cells (Pdpn; bottom row) in control adult lung (B) and 24 h after DT administration (C) shows the absence of the pan-epithelial mGFP in the majority of AT1 cells by direct fluorescence and antibody staining. (D) Low- (left column) and high- (right column) magnification micrographs of co-staining for pan-epithelial lineage marker (mGFP), AT1 cells (Pdpn), and AT2 cell injury marker (Krt8) show loss of mGFP protein in AT1 cells, including ones co-expressing the AT2 cell injury marker (arrows; n = 5 control and 5 DT-treated mice). (E–G) Co-staining for pan-epithelial lineage marker (mGFP), AT1 cells (Pdpn), and AT2 cells (pro-SpC) in sacculating embryonic day 18 lung (E) with separated channels of boxed regions (F and G) shows mGFP in nascent AT2 cells (asterisks) but patchy absence in newly forming AT1 cells at varying stage of flattening (arrows; n = 5 mice). Scale bars, 100 µm (B top, C top, D left), 50 µm (E), 25 µm (B bottom, C bottom, D right), 10 µm (F and G).
To directly test if AT2 cells differentiate into AT1 cells without cellular replication, we combined 5-ethynyl-2′-deoxyuridine (EdU) DNA incorporation with AT2 lineage tracing after BHT injury. The results showed that newly generated AT1 cells carrying the AT2 lineage tag after injury were EdU− (Fig. 2, E–H). We also performed clonal analysis of random AT2 cells by exploiting the sparse ligand-independent recombination of the SftpC-CreER-rtTA knock-in allele. Analysis by whole-mount confocal microscopy in thick tissue slices confirmed that the vast majority of AT1 cells carrying the AT2 cell lineage mark were spatially isolated without a nearby marked “parent” AT2 cell (Fig. 2, I–K). Together, these results support that ablated AT1 cells were regenerated by differentiation of AT2 cells without antecedent proliferation.
We next performed EM analysis of lungs collected at 0.5 h after toxin-mediated genetic ablation to visualize alveolar structure and assess epithelial integrity at high resolution. We observed necrotic AT1 cells as expected, many of which remained attached to the basal lamina. However, most of the alveolar surface was covered by a thick squamous epithelium with electron-lucent cytoplasm containing lamellar bodies, the hallmark of surfactant-producing AT2 cells. Also apparent were electron-dense, mature-appearing intercellular tight junctions between the flattened AT2 cell extensions and adjacent healthy AT1 and AT2 cells. There was a notable absence of open epithelial cell junctions or exposed basal lamina (Fig. 3 A). These ultrastructural features suggested that epithelial barrier and gas diffusion functions were preserved, at least in the local regions we surveyed by EM. To globally measure alveolar function, we next quantified protein and leak of fluorescently tagged albumin into the airspaces and the arterial oxygen saturation after DT and BHT. The results showed no impairment in global alveolar barrier and gas exchange functions with widespread synchronous AT1 cell ablation using DT. BHT however, which kills both AT1 and capillary endothelial cells, resulted in a slight but statistically significant increase in capillary leak without affecting oxygenation (Fig. 3, B–E).
Figure 3.
Ultrarapid AT2 cell flattening sustains barrier and gas exchange functions after AT1 cell killing. (A) Transmission EM images of healthy (black arrowheads) and dying (black arrow) AT1 cells and of flattening AT2 cells with electron-lucent cytoplasm (red arrowheads) and occasional residual lamellar bodies (red arrows) at 30 min after ablation. Note the presence of mature-appearing electron-dense tight junctions (purple arrows) between flattening AT2 and neighboring epithelial cells. (B) Strategy for quantitating alveolar function after acute AT1 cell ablation (genetic, chemical) or generalized (infectious) injury. (C–E) Epithelial permeability (C and D) and oxygenation (E) are not disrupted with widespread AT1 cell killing, whereas generalized injury induces capillary leak and impairs gas exchange (control, n = 4 mice; DT, n = 4 mice; BHT; n = 4 mice; infection, n = 3 [C] or 5 [D and E] mice). Scale bars, 1 µm (A top, A center), 3 µm (A middle left, A middle right), 3 µm (A bottom). P values calculated by two-sided Student’s t test.
Collectively, the results above illuminate the remarkably rapid and comprehensive capacity of the alveolar epithelium to maintain both barrier and gas exchange functions in the setting of widespread AT1 cell loss. Overall, our results suggest that this resilience is enabled by ultrarapid flattening of AT2 cells that simultaneously restores epithelial integrity and generates a thin diffusion barrier that culminates in AT1 cell differentiation.
Mitosis of binucleated AT2 cells initiates AT2 cell repletion
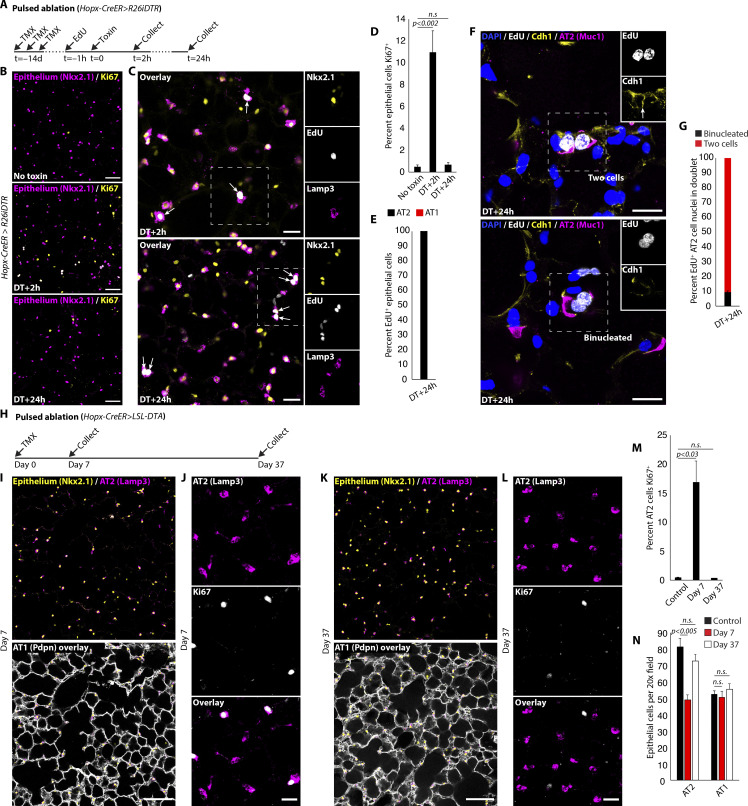
We next explored the kinetics of AT2 cell repopulation by staining for proliferation markers at sequential time points after synchronous AT1 cell ablation. The percentage of alveolar epithelial cells active in the cell cycle (Nkx2.1+Ki67+) increased from 0.5 ± 0.2% (n = 5,373 Nkx2.1+ cells scored in four mice) in the uninjured state to 6.8 ± 1.1% at 0.5 h (n = 2,301 Nkx2.1+ cells scored in two mice) and 11.0 ± 2.0% at 2 h (4,292 Nkx2.1+ cells scored in three mice) after DT, then declined to baseline (0.7 ± 0.2%; 2,392 Nkx2.1+ cells scored in three mice) by 24 h. Curiously, 62.7 ± 4.7% of Ki67+ epithelial cell nuclei (900 Nkx2.1+Ki67+ nuclei scored in three mice) were observed in “doublets” at 2 h after DT, a time point seemingly too early for passage through the entire cell cycle (Uhal, 1997). This finding suggested activation of a latent polyploid population of AT2 cells. Immunostaining analysis and FACS quantitation of uninjured lungs revealed that ∼2.4 ± 0.3% of the healthy AT2 cell population has 4n nuclear content, consisting primarily of binucleated cells (Fig. 4, A and B). To determine if binucleation is enriched in the Wnt-active subset of AT2 cells previously identified to harbor stem-cell function, we quantified their proportion in general (by Sftpc-CreER) and Axin2+ (by Axin2-CreER) AT2 cells in whole mount slices of intact, uninjured lung. This experiment revealed significant enrichment of binucleation in Wnt-active (Axin2+) AT2 cells, corroborated by FACS analysis, employing Axin2-rtTA-nls-GFP to identify the Wnt-active AT2 subset (Fig. 4, C–E; and Fig. S3). Binucleated AT2 cells were quiescent (Ki67−) in health and then rapidly entered the cell cycle (Ki67+) after AT1 cell killing (Fig. 4, F and G). Nuclear localization of β-catenin protein confirmed active canonical Wnt signaling (Fig. 4, F and H).
Figure 4.
Quiescent Axin2+ binucleated AT2 cells are activated by AT1 cell death. (A and B) Binucleated AT2 cells in uninjured mouse lung by immunostaining (A) and quantitation of ploidy by FACS (B) with micrograph of sorted 4n AT2 cell (2.4 ± 0.3% 4n, n = 4 mice). (C–E) Lineage marking of general (Sftpc-CreER) (C) or Axin2+ (Axin2-CreER) (D) AT2 cells with quantitation (E) reveals strong enrichment of binucleation (dual arrows) in Axin2+ AT2 population (Sftpc-CreER: 974 Tomato+ cells scored in n = 4 mice; Axin2-CreER: 176 Tomato+Lamp3+ cells scored in n = 3 mice). (F–H) Immunostaining (F) shows cell cycle entry (Ki67+) of Wnt-active (nuclear-localized β-catenin) binucleated AT2 cell at 0.5 h after DT, with quantitation of proliferation (G) (control: 5,273 Nkx2.1+ cells scored, n = 4 mice; 0.5 h: 2,301 Nkx2.1+ cells scored, n = 2 mice; 2 h: 4,292 Nkx2.1+ nuclei scored, n = 3 mice) and Wnt activity (H) (n = 307 Nkx2.1+Ki67+ cells scored, n = 3 mice). Scale bars, 5 µm (B), 15 µm (C, D, and F), 40 µm (A). PI, propidium iodide. Data are mean ± SEM. P values calculated by two-sided Student’s t test.
Figure S3.
Axin2+ AT2 cell population is enriched for 4n nuclear content. FACS gating strategy (top) and quantitation (bottom) of DNA content in GFP+ and GFP− AT2 cells from mice expressing GFP under the control of the Axin2 promoter (GFP− AT2 cells: 1.89 ± 0.66% 4n; GFP+ AT2 cells: 6.21 ± 0.10% 4n; n = 3 mice; P < 0.003). Note enrichment of 4n cells in Axin2+ AT2 cells by FACS (6.12% versus 3.11% in Axin2− AT2 cells), supporting results by whole-mount immunofluorescence in Fig. 4, C–E.
Adult progenitor cells are widely believed to arrest at G0 of the cell cycle, prior to S phase entry and DNA replication upon reactivation (Cheung et al., 2013). However, recent reports in less-derived organisms suggest alternative points of cell cycle entry (Otsuki and Brand, 2018; Nguyen et al., 2017). To map cell cycle transit of binucleated AT2 cells, we co-stained for DNA replication (EdU), cell cycle activity (Ki67), and mitosis (phosphorylated histone H3, pH3) at serial time points after AT1 cell ablation (Fig. 5 A). The results indicated that S phase entry and DNA replication are restricted to mononucleated AT2 cells (Ki67+ nuclear “singlets”) and that activated binucleated AT2 cells (Ki67+ nuclear “doublets”) do not enter S phase (Fig. 5, B and C). We next asked if EdU−/Ki67+ binucleated AT2 cells served as functional progenitors or were merely remnants of failed mitosis. We did so by co-staining for Ki67 and the adherens junction marker E-cadherin (Cdh1) to determine successful cell division. While at 0.5 h after DT nearly all Ki67+ doublets were binucleated cells, by 2 h many had completed division, evidenced by E-cadherin demarcating two daughter AT2 cells (Fig. 5, D and E). EdU− cells were in M phase (pH3+ with formation of sister chromatids) prior to division (Fig. 5, F and G), suggesting binucleated AT2 cell division by mitotic entry and not amitosis. Together, these data demonstrate that direct M phase entry from quiescence of binucleated AT2 cells initiates AT2 cell restoration prior to conventional stem cell division (Fig. 5 H).
Figure 5.
Binucleated AT2 cells enter the M phase from quiescence and complete mitosis. (A) Strategy for cell cycle mapping after AT1 cell ablation. (B and C) Immunostaining (B) with quantitation (C) shows that cell cycle–active AT2 cell doublets (arrows) have not entered S phase (Nkx2.1+Ki67+EdU−) and that the cell cycle–active cells in S phase (Nkx2.1+Ki67+EdU+) are all singlets (arrowheads) at 2 h after DT (left bar: 456 Nkx2.1+Ki67+ cells scored, n = 3 mice; right bar: EdU scored in 404 Nkx2.1+Ki67+ cells, n = 3 mice). (D and E) E-cadherin (Cdh1) immunostaining (D) with quantitation (E) reveals a reduction in binucleated and corresponding increase in post-mitotic AT2 cells between 0.5 and 2 h after DT (0.5 h: 112 Ki67+ nuclei scored; 2 h: 116 Ki67+ nuclei scored; n = 3 mice per time point). (F and G) Immunostaining (F) with quantitation (G) reveals entry of AT2 cell into mitosis without antecedent DNA replication at 2 h after DT (Lamp3+pH3+EdU−). Note adjacent AT2 cell in S phase that has not yet entered M phase (87 cells scored, n = 3 mice). (H) Schematic of binucleated AT2 cell cycle transit after activation from quiescence. Scale bars, 50 µm (B), 20 µm (D), 5 µm (F). TMX, tamoxifen. Data are mean ± SEM. P values calculated by two-sided Student’s t test.
AT1 cell–derived hbEGF drives proliferation of mononucleated and binucleated Wnt-active AT2 cells via EGFR activation
Axin2 expression and nuclear localization of β-catenin protein indicate active Wnt signaling in quiescent binucleated AT2 cells, consistent with prior observations that Wnt is necessary but not sufficient for AT2 cell proliferation (Nabhan et al., 2018). EGFR activation has long been believed to drive proliferation of AT2 cells based on in vitro studies and demonstration that oncogenic KrasG12D rapidly transforms AT2 cells (Desai et al., 2014; Lin et al., 2012; Xu et al., 2012). However, a physiologic role for EGFR in AT2 proliferation has not been demonstrated in vivo and the putative ligand(s) and cellular source(s) are unknown. It is also unknown if other (non-EGFR) signaling pathways contribute to AT2 cell proliferation in vivo, including ones that may not require concomitant Wnt activity. To examine the mechanism(s) of AT2 cell proliferation in vivo, we systemically inhibited either Wnt or EGFR prior to AT1 cell ablation and then asked whether each pathway was necessary for activation of binucleated and mononucleated AT2 cells (Fig. 6 A). Receptor tyrosine kinase activity (phosphorylated Erk1/2 [pErk1/2]) was detected in both binucleated and mononucleated AT2 cells after injury, consistent with EGFR activation (Fig. 6 B). Inhibition of either Wnt or EGFR abrogated activation of mononucleated and binucleated AT2 cells, evidenced by a significant reduction in both Ki67+ singlets and doublets shortly after DT administration (Fig. 6, C–E). These data suggest that both EGFR and Wnt are necessary for cell cycle activation and that EGFR can trigger AT2 cell entry into either M or S phase, as appropriate.
Figure 6.
EGFR and Wnt are necessary for activation of mononucleated and binucleated AT2 cells. (A) Strategy for systemic Wnt (C59) or EGFR (Erlotinib) inhibition prior to AT1 cell ablation by DT. (B) pErk1/2 staining shows MAPK signaling in activated mononucleated (top) and binucleated (bottom) AT2 cells at 0.5 h after DT. (C–E) Representative images (C) and quantitation of mononucleated (Ki67+/Nkx2.1+ singlets) (D) and binucleated (Ki67+/Nkx2.1+ doublets) (E) AT2 cell activation at 2 h after DT in controls and with inhibition of Wnt (C59) or EGFR (Erlotinib) (no toxin: 5,273 Nkx2.1+ nuclei scored in n = 4 mice; control: 4,292 Nkx2.1+ nuclei scored in n = 3 mice; C59: 4,258 Nkx2.1+ nuclei scored in n = 2 mice; Erlotinib: 2,171 Nkx2.1+ nuclei scored in n = 3 mice). Scale bars, 5 µm (B), 50 µm (C). TMX, tamoxifen. Data are mean ± SEM. P values calculated by two-sided Student’s t test.
To identify candidate EGFR ligand(s) and their cellular source(s), we examined single-cell RNA sequencing (scRNA-seq) profiles of alveolar cell types. This analysis revealed only hbEGF expressed by AT1 cells and pericytes, and we confirmed AT1 cell expression by RNA in situ hybridization (Fig. 7, A and B; Nagendran et al., 2018; Tabula Muris Consortium, 2020). Hence, hbEGF protein is presumably tethered to the AT1 cell surface and/or extracellular matrix until it is rapidly released by activity of matrix metalloproteinases reported to be induced with lung injury, whereupon it can signal to nearby EGFR-expressing AT2 cells (Greenlee et al., 2007; Kheradmand et al., 2002). We used an EGFR-Emerald fusion protein knock-in allele to visualize EGFR protein localization and directly confirm signaling in AT2 cells after injury by visualizing receptor clustering and endocytosis (Yang et al., 2017). These experiments identified AT2 cells with diffuse apical localization of EGFR at baseline and punctate cytoplasmic co-localization with the adaptor protein growth factor receptor bound protein 2 (GRB2) after AT1 cell ablation (Fig. 7, C–F; Wang and Moran, 1996). MAPK signaling activity in AT2 cells was also demonstrated after BHT injury by immunostaining for pErk1/2 (Fig. 7 G). To definitively test for a putative AT1-to-AT2 hbEGF-EGFR signaling axis, we deleted hbEGF in AT1 cells or EGFR in AT2 cells and then quantified proliferation following AT1 cell killing (Fig. 7 H). For these experiments, we employed BHT to avoid ectopic expression of simian hbEGF (R26iDTR) in AT1 cells (Naglich et al., 1992). Both approaches demonstrated significantly reduced AT2 cell proliferation, confirming that AT1 cell–produced hbEGF is a key ligand that drives AT2 cell activation via EGFR in acute injury in vivo (Fig. 7, I–K). Together, these data implicate AT1 cells as the AT2 proliferative niche that is poised to rapidly provide hbEGF upon alveolar injury.
Figure 7.
AT1 cells constitute an hbEGF-EGFR signaling niche for AT2 cell proliferation. (A and B) scRNA-seq (A) and in situ hybridization (B) shows hbEGF expression by AT1 cells. (C and D) EGFR-Emerald allele shows EGFR protein (Emerald-GFP) restricted to AT2 cell apical membrane (Muc1) in control (C) and internalized on endosomes (Grb2 protein co-localization) after BHT (D). (E and F) Representative immunostaining (E) of inactive (no toxin, apical) and activated (BHT, endocytosed) EGFR with quantitation (F) of EGFR-active AT2 cells in controls and after BHT (n = 300 Lamp3+ cells scored in uninjured and n = 358 Lamp3+ cells scored 40 h after BHT; n = 3 mice per condition). (G) pErk1/2+ AT2 cell (Lamp3+) after BHT. (H) Strategy for deletion of hbEGF in AT1 or EGFR in AT2 cells prior to AT1 killing, with EdU administration 1 h prior to evaluation. (I–K) Representative images (I and J) and quantitation (K) of AT2 cell proliferation (Lamp3+EdU+) in control lungs and 2 d after BHT (hbEGF+/del: 2,181 Lamp3+ cells scored in n = 4 mice; hbEGFlox/del: 3,410 Lamp3+ cells scored in n = 5 mice; EGFR+/del: 3,432 Lamp3+ cells scored in n = 3 mice; EGFRlox/del: 2,962 Lamp3+ cells scored in n = 3 mice). Scale bars, 5 µm (B–D and G), 15 µm (E), 25 µm (I and J). Em, Emerald GFP; TMX, tamoxifen. Data are mean ± SEM. P values calculated by two-sided Student’s t test.
Repeated cycles of AT1 cell ablation drive AT2-derived hyperplasia that provides barrier but compromises surfactant and gas exchange functions
Synchronous AT1 cell ablation by one-time DT administration resulted in rapid and complete AT1 cell reconstitution, even after widespread AT1 cell elimination. We next used EdU to track the fate of the daughter cells generated by AT2 cell division (Fig. 8 A). With a single pulse of AT1 cell ablation by DT administration, AT2 cell proliferation resolved to baseline within 24 h (Fig. 8, B and D). Detection of replicated DNA demonstrated that 92.1 ± 0.5% (n = 110 Nkx2.1+/EdU+ nuclei scored, n = 2 mice) of AT2 cells that entered S phase completed karyokinesis by 24 h (Nkx2.1+EdU+ doublets). As expected, since AT1 cells had already been regenerated, daughter cells of AT2 proliferation exclusively retained an AT2 cell (Lamp3+) phenotype, a minor subset of which became binucleated via endomitosis (Fig. 8, E–G).
Figure 8.
Kinetics of AT2 cell repletion following depletion by differentiation. (A) Strategy for pulsed AT1 cell ablation by DT. (B and D) Representative images (B) and quantitation (D) show that proliferation observed at 2 h after DT subsides within 24 h (no toxin: 5,273 Nkx2.1+ nuclei scored in n = 4 mice; 2 h: 4,292 Nkx2.1+ nuclei scored in n = 3 mice; 24 h: 2,392 Nkx2.1+ nuclei scored in n = 3 mice). (C and E) Immunostaining (C) and quantitation (E) show AT2 cells that enter S phase at 2 h (top, Nkx2.1+EdU+ singlets) and undergo karyokinesis by 24 h (bottom, Nkx2.1+EdU+ doublets) retain an AT2 cell (Lamp3+) phenotype (n = 221 Nkx2.1+/EdU+ nuclei scored in n = 3 mice). (F and G) (F) E-cadherin (Cdh1) immunostaining with (G) quantitation reveals that a minor subset of AT2 cells that entered S phase (EdU+) undergo karyokinesis without completion of cytokinesis at 24 h (n = 68 EdU+ nuclei in doublet scored, n = 2 mice). (H) Strategy for targeted AT1 cell ablation by inducible intracellular toxin expression (R26DTA). (I–L) Immunostaining shows AT1 (Nkx2.1+Lamp3−-Pdpn+) and AT2 cells (Nkx2.1+Lamp3+), including a proliferating AT2 cell subset (Lamp3+Ki67+), at 7 d (I and J) and 37 d (K and L) after AT1 cell ablation. (M) Quantitation of proportion of AT2 cells proliferating in controls and at both time points (control: 1,709 Nkx2.1+Lamp3+ cells scored; day 7: 1,323 Nkx2.1+Lamp3+ cells scored; day 37: 1,139 Nkx2.1+Lamp3+ cells scored, n = 3 mice per condition). (N) Quantitation shows significant reduction in AT2 (Lamp3+Nkx2.1+) but not AT1 (Lamp3−Nkx2.1+) cells at 7 d that is resolved by 37 d after AT1 cell ablation (control: 2,234 Nkx2.1+ cells scored; day 7: 1,794 Nkx2.1+ cells scored; day 37: 3,748 Nkx2.1+ cells scored, n = 3 mice per condition). Scale bars, 100 µm (I and K), 50 µm (B), 25 µm (C, J, and L), 15 µm (F). TMX, tamoxifen. Data are mean ± SEM. P values calculated by two-sided Student’s t test.
We next tested whether rapidly iterated cycles of widespread AT1 ablation would exhaust or deplete AT2 cell proliferative capacity. To do so, we modified our genetic approach to enable repeated rounds of AT1 cell killing. In Hopx-CreER mice carrying a conditional DT A subunit allele (R26DTA), tamoxifen administration results in AT1 cell ablation. Using this strain, newly regenerated AT1 cells can be eliminated by administering a second dose of tamoxifen since they induce Hopx expression during AT1 differentiation. We performed one or three cycles of AT1 cell ablation prior to analyzing the epithelium (Fig. 8 H and Fig. 9 A). One-time administration of tamoxifen transiently depleted AT2 cells, without an observed change in the number of AT1 cells (Fig. 8, I–N), recapitulating the results of our DTR experiments. To our surprise, repeated cycles of AT1 ablation resulted in dramatic hyperplasia comprised of highly proliferative and frequently multinucleated epithelial cells that lacked surfactant function (Fig. 9, B–E). The hyperplastic cells ranged from cuboidal to squamous, co-expressed injury (Krt8), AT2 (Muc1) and AT1 (Pdpn, lycopersicon esculentum lectin [LEL]) markers, and were frequently polyploid, but did not contain lamellar bodies or stain positive for surfactant protein C (pro-SpC; Fig. 9, F–H). The end-result was a thick and cellularly dense alveolar epithelium that presumably maximized barrier strength at the expense of surfactant production and efficient gas diffusion.
Figure 9.
Repeated AT1 cell ablation elicits epithelial hyperplasia with dysregulated molecular phenotypes and ploidy. (A) Strategy for pulsed or repeated genetic ablation by inducible, intracellular toxin expression (R26DTA). (B–D) Immunostaining (B and C) shows hyperplasia and absence of surfactant function in proliferating epithelial cells (Ki67+/Nkx2.1+/Lamp3−) after repeated but not pulsed ablation with quantitation (D) (pulsed: 195 Nkx2.1+Ki67+ cells scored; repeated: 277 Nkx2.1+Ki67+ cells scored; n = 3 mice per condition). (E) Quantitation of AT2 (Lamp3+) and total epithelial (Nkx2.1+) cells (control: 2,234 Nkx2.1+ cells scored; pulsed: 1,794 Nkx2.1+ cells scored; repeated: 2,904 Nkx2.1+ cells scored; n = 3 mice per condition). (F) Electron micrographs show AT2 cell with cytoplasmic lamellar bodies after pulsed ablation (left) and hyperplastic epithelium that lacks surfactant organelles after repeated ablation (right). (G) Hyperplastic cells are morphologically diverse and lack surfactant protein (pro-SpC−) while expressing an AT2 cell injury marker (Krt8) and variable levels of AT2 (Muc1) and AT1 (Pdpn, LEL) cell surface markers. (H) Multinucleation by transmission EM (numbers indicate nuclei); 9.4 ± 0.8% of hyperplastic epithelial cells are binucleated, while 0.53 ± 0.3% contain greater than two nuclei (776 Krt8hi epithelial cells scored, n = 3 mice). Scale bars, 50 µm (B), 10 µm (C and G), 3 µm (F), 5 µm (H). TMX, tamoxifen. P values calculated by two-sided Student’s t tests (D and E).
BHT similarly induced AT1 cell killing over multiple days, allowing us to track the contribution of AT2 cells to epithelial hyperplasia by lineage tracing. AT2 cell clonal analysis using a multicolor reporter allele (R26iMb-Mosaic) revealed monoclonal expansion capable of generating more than 30 new cells from a single AT2 cell over 5 d (Fig. 10, A and B). Despite the magnitude of these histological abnormalities, AT2 cell–derived hyperplasia eventually regressed without obvious consequences to alveolar architecture or epithelial composition by 1 mo later (Fig. 10, C and D). Broad AT2 cell lineage marking using the mGFP tag (R26mTmG) confirmed that the regenerated AT1 cells had derived from AT2 cells (Fig. 10, E and F). Together, these findings suggest that repeated cycles of AT1 ablation indirectly drive multiple rounds of AT2 cell division, perhaps including newly born daughter cells that would otherwise exit the cell cycle (Fig. 10 G). However, many daughter cells were molecularly dysregulated, having lost AT2 surfactant function without achieving AT1 cell identity. Wnt inhibition during repeated AT1 cell ablation abrogated the hyperplastic response and significantly depleted AT2 cells, yet AT1 cell regeneration by direct AT2-to-AT1 cell differentiation proceeded, as expected (Fig. S4, A–E). Collectively, these findings suggest that the alveolar epithelium has a tremendous proliferative reserve and prioritizes barrier preservation over gas exchange and surfactant production.
Figure 10.
AT2 cell–derived hyperplasia spontaneously resolves with injury resolution. (A) Tomato+ AT2 cell clones from multicolor reporter without injury (left top) and after BHT (left bottom) shows local expansion with proliferative progeny that induced an injury marker (Krt8+) and lost surfactant protein C (proSpC−) expression (small panels). (B) Quantitation of clone size distribution after sustained chemical injury (89 Tomato+ AT2 cell clones scored). (C and D) Epithelial hyperplasia at 5 d after BHT resolves by 32 d shown by staining (C) and with quantitation (D) (control: 2,234 Nkx2.1+ cells scored, n = 3 mice; day 5: 4,254 Nkx2.1+ cells scored, n = 3 mice; 1 mo: 2,457 Nkx2.1+ cells scored, n = 4 mice). (E and F) AT2 cell lineage tag marks only cuboidal AT2 cells in uninjured lung (E, top left and middle column) whereas 38 d after BHT nearly all AT1 cells carry the AT2 lineage mark (mGFP+Ager+) (E, bottom left and right column) with quantitation of AT2-derived AT1 cells (F) (control: 404 AT1 cells scored; BHT: 268 AT1 cells scored, n = 3 [control] and 2 [BHT] mice). (G) Cartoon of outcomes of pulsed versus repeated AT1 cell ablation. Scale bars, 100 µm (A left, C, and E left), 10 µm (A middle/right), 25 µm (E middle/right). Data are mean ± SEM. P values calculated by two-sided Student’s t test.
Figure S4.
Wnt signaling is necessary for AT2 cell repletion but not for AT2-to-AT1 cell differentiation. (A) Strategy for AT2 cell lineage tracing with pan-Wnt inhibition (C59) during sustained AT1 cell killing. (B and C) AT1 (Pdpn) and AT2 (Lamp3) marker staining shows that pan-Wnt inhibition for days 2–5 after BHT administration results in a severe reduction of AT2 cells but broad AT1 cell coverage (B). AT2 cell lineage tracing shows differentiation into AT1 cells (mGFP+, flat morphology) with maintenance of epithelial (HPA+) coverage over the capillaries (Emcn+) (C). (D and E) Quantitation of epithelial (Nkx2.1+) cells and AT2 (Nkx2.1+Lamp3+) cells after BHT with and without pan-Wnt inhibition (control: 2,234 Nkx2.1+ cells; BHT+5 d: 4,254 Nkx2.1+ cells scored; BHT+5 d [+C59]: 1,217 Nkx2.1+ cells scored; n = 3 mice per condition). Scale bars, 100 µm (B), 25 µm (C). Emcn, endomucin; HPA, helix pomatia agglutinin lectin; TMX, tamoxifen. Data are mean ± SEM. P values calculated by two-sided Student’s t test.
Discussion
Our study illuminates a temporally coordinated program for regeneration of the lung alveolar epithelium involving discrete mechanisms. Ultrarapid AT2 cell flattening preserves alveolar barrier and gas exchange functions while rapidly regenerating lost AT1 cells. While we could not successfully visualize dynamic AT2 cell flattening by short-term lineage tracing due to transient loss of the reporter fluorophore, longer chase periods confirm the AT2 cell origin of regenerated AT1 cells. Surfactant-producing AT2 cells are then replenished by proliferation of Wnt-active binucleated and mononucleated AT2 cells, both regulated by EGFR activation. AT2 cell activation is primed with constitutive hbEGF transcription by AT1 cells presumably ensuring protein is poised for release by injury-induced protease activity. This regenerative program endows a delicate simple epithelium with a functional resiliency that compensates for the lack of physical durability intrinsic to stratified and pseudostratified epithelial barriers elsewhere in the body.
In stratified esophagus and skin epithelia, wound closure is accomplished by modulation of proliferation, differentiation, and cell migration (Doupé et al., 2012; Park et al., 2017). However, rapid flattening of nearby cells for barrier reconstitution, as we observed in alveoli, has been reported with severe acute injury in non-simple epithelia, for instance in the stratified zebrafish epidermis and pseudostratified mouse airways (Erjefält et al., 1995; Rawlins et al., 2007; Chen et al., 2016). Real-time imaging of epithelial repair after targeted single-cell ablation in live zebrafish directly visualizes remarkably rapid cell elimination and nearly “gap-free” breach repair within minutes (Shkarina et al., 2022). In our AT1 ablation experiments, epithelial tight junctions between flattening AT2 cells and their neighbors were visible by electron microscopy within 30 min of injury. Given the primal importance of maintaining a continuous leak-proof barrier intrinsic to pulmonary alveoli, evolution of a mechanism for ultrarapid barrier regeneration is perhaps not surprising.
In the epidermis and airway injury models mentioned above, the first responder cells execute only a barrier restoration role—they are not stem cells and do not undergo differentiation. Thus, their acute depletion does not reduce the pool of local stem cells. In alveolar injury, by contrast, AT2 cells are depleted through differentiation, which reduces the number available for proliferation. Are constitutive (Axin2+) AT2 stem cells preferentially safeguarded against depletion? It appears possible since experimentally sustained Wnt activity in vivo during aging and Wnt supplementation in vitro have been found to inhibit AT2 to AT1 differentiation (Nabhan et al., 2018). Thus, constitutive AT2 stem cells may be simultaneously poised to proliferate and safeguarded from depletion by virtue of their Wnt-active state. Other tissues have similarly evolved customized cellular programs for epithelial repair that precede and complement the relatively slow process of stem cell proliferation (Heller et al., 2014; Aragona et al., 2017; Tai et al., 2019).
In some contexts, binucleation can impair cell cycle progression and binucleated AT2 cells lacking proliferative competence were recently observed as a pathologic consequence of genotoxic lung injury (Weng et al., 2022; Ganem and Pellman, 2007). By contrast, the binucleated AT2 cells we identified that reside in healthy lungs are adaptive. Because they are poised to directly enter mitosis from quiescence, their rapid division may mitigate AT2 cell depletion and the consequent surfactant deficiency that can worsen hypoxemia from alveolar collapse. Determining their specific role(s) will require new genetic tools to mark and manipulate them in vivo. Functional ploidy reduction as we describe here in mammalian lungs has been identified as a regenerative mechanism in other tissues and in different species (Otsuki and Brand, 2018; Matsumoto et al., 2020; Chan et al., 2022; Nguyen et al., 2017; Bailey et al., 2021). During injury resolution, new binucleated AT2 cells are generated by endoreplication, potentially restoring this population for future injuries. Interestingly, binucleated AT2 cell activation and mitosis are both Wnt-dependent and EGFR-driven by AT1 cell–produced hbEGF, the same program that drives proliferation of mononucleated AT2 cells. It will be interesting to learn if ploidy reduction and cellular replication in other tissues are also co-regulated, and whether resident polyploid cells are differentially susceptible to oncogenesis compared with conventional diploid stem cells.
We were surprised to find relatively few denuded AT1 cells and macrophages in the alveolar lumen after targeted AT1 cell killing, raising the possibility that AT2 cells or their progeny could be phagocytosing dying AT1 cells and thereby minimizing inflammation. Another surprise was the finding that several cycles of AT1 cell ablation resulted in epithelial hyperplasia. Since AT2 cells were not themselves being injured, their excessive proliferation is presumably driven by external signals. Many of the hyperplastic cells had ceased AT2 cell surfactant activity yet failed to acquire a flattened AT1 cell morphology for efficient gas exchange. This concept of cells “stuck” in a normally transient state between AT2 and AT1 identities is proposed to contribute to defective alveolar maintenance in ARDS and pulmonary fibrosis (Strunz et al., 2020; Jiang et al., 2020; Kobayashi et al., 2020; Chan et al., 2022). It is puzzling that despite not being injured themselves, some AT2 cells underwent massive endoreplication. Perhaps continuous barrier integrity for preventing pulmonary edema and/or scar initiation is of such importance that suspending quality control of replication during repetitive passage through the cell cycle is adaptive at the organismal level. Classical studies have proposed that disruption of the alveolar epithelial barrier drives fibrotic remodeling, so overproliferation of AT2 cells could mitigate this risk (Haschek and Witschi, 1979). Interestingly, alveolar epithelial hyperplasia has long been considered a nonspecific injury response in human lung and is also a histologic feature of ARDS (Bachofen and Weibel, 1974; Tomashefski, 2000). If the pathophysiology in humans is similar to that in mouse, it may be worth considering the possibility that enhancing cellular differentiation may be more impactful than promoting stem cell proliferation for prevention or treatment of ARDS.
Materials and methods
Mouse strains
Wild type animals were C57BL/6 (Jackson Laboratories). Expression of estrogen-inducible Cre recombinase (Cre-ERT2) for conditional, cell type–specific targeting in vivo in mixed strain backgrounds employed the following gene-targeted Cre alleles: Hopx-CreER (Takeda et al., 2011; #017606; JAX Strain) for AT1 cells; Sftpc-CreER (Van Keymeulen et al., 2011; #028054; JAX Strain) and Sftpc-CreER-rtTA (Chapman et al., 2011) for AT2 cells; and Axin2-CreER (van Amerongen et al., 2012; #018867; JAX Strain) for Wnt/β-catenin-responsive cells. Shh-Cre (Harfe et al., 2004; #005622; JAX Strain) was used to irreversibly mark the entire endoderm-derived lung epithelium at the onset of development. Cre-dependent deletion of hbEGF (Iwamoto et al., 2003) and EGFR (Choi et al., 2020) was performed using conditional “floxed” alleles crossed with germline deletion alleles (hbEGFf/d and EGFRf/d). Heterozygous wild type and deletion allele combinations served as littermate controls (hbEGF+/d and EGFR+/d). Conditional fluorescent Cre reporter alleles included: R26H2B-EGFP (Abe et al., 2011; CBD0238K; RIKEN); R26Tomato (Madisen et al., 2010; #007914; JAX Strain); R26mTmG (Muzumdar et al., 2007; #007676; JAX Strain), in which recombination switches expression from membrane-targeted Tomato to mGFP; and the R26iMb-Mosaic (Pontes-Quero et al., 2017; #031298; JAX Strain) allele, in which recombination induces stochastic expression of either membrane-targeted YFP, Tomato, or Kate2. Doxycycline-induced (1 mg/ml doxycycline in 1% sucrose drinking water; Abcam) fluorescent cell nuclear labeling was performed using Sftpc-CreER-rtTA with Col1a1TetoH2BCherry (Egli et al., 2007; #014592; JAX Strain) for AT2 cells and Axin2-rtTA-nls-GFP (van de Moosdijk et al., 2020; #036013; JAX Strain) with tetO-H2B-GFP (Tumbar et al., 2004; #005104; JAX Strain) for Wnt/β-catenin–responsive cells. Inducible, gene-targeted cell ablation employed the R26iDTR (Buch et al., 2005; #007900; JAX Strain) allele, which sensitizes cells to DT by conditional expression of the toxin receptor, and the R26DTA (Voehringer et al., 2008; #009669; JAX Strain) allele, which kills cells by conditionally inducing synthesis of the cytotoxic protein fragment. A chimeric EGFR-GFP (EGFR-Emerald) knock-in allele (Yang et al., 2017) was used to localize EGFR protein. Induction of Cre-ERT2 alleles was accomplished by intraperitoneal injection of 4 mg tamoxifen (Sigma-Aldrich), dissolved in corn oil by sonication and stored in aliquots at −80°C, once per day for three consecutive days. Genotyping was performed by PCR of DNA extracted from ear or toe clips using published primer sets. All experiments were performed using adult male and female animals between 2 and 8 mo of age. Mice were housed in filtered cages, and all experiments were approved by the Institutional Animal Care and Use Committee at Stanford University.
For animal strains not distributed publicly by the Jackson Laboratories or the RIKEN Center, PCR primers used for genotyping were as follows:
Sftpc-CreER-rtTA
Mutant forward: 5′-TCGCCTTCTATCGCCTTCTTG-3′
Communal reverse: 5′-CCTTTTGCTCTGTTCCCCATTA-3′
Wild type forward: 5′-TGGTTCCGAGTCCGATTCTTC-3′
hbEGF f’lox or del
Wild type forward: 5′-CATGATGCTCCAGTGAGTAGGCTCTGATTAC-3′
Wild type reverse: 5′-AGGGCAAGATCATGTGTCCTGCCTCAAGCC-3′
Mutant forward: 5′-CGGACAGTGCCTTAGTGGAACCTC-3′
Mutant reverse: 5′-GCTTCTTCTTAGGAGGGATCTTGGC-3′
EGFR f’lox or del
Wild type forward: 5′-CTTTGGAGAACCTGCAGATC-3′
Wild type reverse: 5′-CTGCTACTGGCTCAAGTTTC-3′
Mutant forward: 5′-CTCAGCCAGATGATGTTGAC-3′
Mutant reverse: 5′-CCTCGTCTGTGGAAGAACTA-3′
EGFR-Emerald
Mutant forward: 5′-TGGTGCAGATGAACTTCAGG-3′
Communal reverse: 5′-CCTCACCATGAGGCAAACTT-3′
Wild type forward: 5′-CCACAGCTGAAAATGCAGAG-3′
Toxin-mediated ablation and generalized infectious alveolar injury
Gene-targeted and synchronous cell ablation by R26iDTR was performed by intraperitoneal injection of 4 mg of tamoxifen once per day for three consecutive days, followed by a single intraperitoneal injection of 150 ng DT (Sigma-Aldrich, diluted in PBS and stored in 200-μl aliquots at −80°C) ∼2 wk later. Littermates lacking only the R26iDTR allele also received tamoxifen and DT and served as controls. To quantitate AT1 cell ablation kinetics and efficiency, the Hopx-CreER allele was used to recombine R26mTmG simultaneously with R26iDTR, which demonstrated depletion of the AT1 cell epithelial surface by DT, relative to controls and consistent with epithelial cell quantification. For pan-epithelial cell ablation, the Shh-Cre allele was combined with R26iDTR and R26mTmG alleles, and mice were treated with a single intraperitoneal injection of 150 ng DT at 2 mo of age. To inhibit Wnt secretion and signaling, the Porcupine inhibitor C59 (Tocris) was administered by oral gavage at a dose of 50 mg/kg body weight 7 h before DT injection. To inhibit EGFR signaling, Erlotinib (Selleck) was administered by oral gavage at a dose of 100 mg/kg body weight 2 h before DT injection. C59 and Erlotinib were prepared in a solution of 0.5% carboxy methyl cellulose/0.1% Tween 80 that was sonicated for 20 min immediately prior to administration in 100 μl volumes. Gene-targeted cell ablation by R26DTA was performed by a single intraperitoneal injection of 4 mg tamoxifen or by 4 mg tamoxifen injections once per day for three consecutive days. Animals were sacrificed 7 d after the final dose of tamoxifen. Chemical ablation of AT1 cells was performed by a single intraperitoneal injection of BHT (Thermo Fisher Scientific) dissolved in corn oil with gentle warming and stored in 500 μl aliquots at −80°C) at a dose of 450 mg/kg (Figs. 1, 2, and 7) or 100 mg/kg body weight (Figs. 10 and S4). Although surviving animals fully recovered, 100 mg/kg BHT resulted in significant respiratory mortality within 1 wk of administration (33.3%; n = 12 animals). To inhibit Wnt signaling during BHT injury, C59 (Tocris) was administered by oral gavage at a dose of 50 mg/kg body weight once every 12 h beginning 24 h after BHT injection. Generalized infectious alveolar injury was performed as previously described (Gotts et al., 2019). Streptococcus pneumoniae serotype 19F (49619; American Type Culture Collection) was grown in brain–heart broth (Becton Dickinson) and harvested at the mid-log phase (optical density 0.50 at 600 nm). The bacterial culture was then pelleted and resuspended in PBS at three different serial dilutions to enable aliquot of 1.5 colony-forming units of bacteria. Mice were inoculated intranasally with 1.5 colony-forming units of bacteria suspended in 50 μl of solution.
Bronchoalveolar lavage (BAL) and pulse oximetry
For experimental assessment of vascular permeability in alveoli, mice underwent anesthetization by isoflurane and were injected retro-orbitally 4 h prior to evaluation with 2 mg of FITC-conjugated albumin (Thermo Fisher Scientific) suspended in 200 μl of PBS. Mice were euthanized by carbon dioxide inhalation, and after exsanguination, the trachea was cannulated and the lungs were lavaged twice with 250 μl PBS. BAL fluid was centrifuged at 500 g for 10 min and supernatant was separated from pellet to isolate protein from cells. BAL protein was measured with the BCA Protein Assay (Thermo Fisher Scientific), and FITC fluorescence intensity was measured using a fluorescence plate reader (excitation 490 nm and emission 525 nm) and serial dilution techniques. To evaluate gas exchange, the MouseOx+ cervical collar system (Starr Life Sciences) was used to measure arterial oxygen saturation continuously for 5 min prior to euthanasia and a mean oxygen saturation was calculated and reported.
Lung isolation, fixation, staining, and imaging
Mice were euthanized by carbon dioxide inhalation, the abdominal aorta was severed, and sternotomy was performed. Phosphate-buffered saline (PBS; Ca2+- and Mg2+-free, pH 7.4) was perfused at a flow rate of 7.5 ml/min into the right ventricle using a 21-gauge needle attached to a digital pump (Ismatec) to clear the pulmonary vasculature. For developmental analysis, embryos were staged by vaginal plugging of the mother, with noon on the day of appearance of the plug taken as embryonic day 0.5. Individual embryos were also staged by fetal crown-rump length at the time of euthanasia. Fetal lungs were removed en bloc and a scalpel was used to excise the tip of the accessory lobe, after which lungs and the tip were fixed in methanol (Me):dimethylsulphoxide (DMSO; 4:1) with rocking overnight at 4°C, bleached in Me:DMSO:30% H202 (4:1:1) for 5 h at room temperature, then dehydrated, and stored in Me at 20°C until staining.
For analysis of postnatal lung, the trachea was cannulated and the lungs were gently inflated with molten 2% ultralow gelling temperature agarose (Sigma-Aldrich) until visualization of complete distention of the accessory lobe, then removed en bloc and submerged in ice-cold PBS. Individual lobes were dissected and fixed in three different conditions. For whole-mount imaging, the left and right upper lobes were separated and trimmed to create a flat base, glued to a tissue disk with cyanoacrylate, and then submerged in ice-cold PBS and sliced at a thickness of 600 µm using a vibrating microtome (Leica). For direct imaging without immunostaining, slices were fixed in paraformaldehyde (PFA, 4% in PBS) overnight at 4°C then stored in 80% glycerol at 4°C. For whole-mount immunofluorescence staining, slices were fixed in Me:DMSO (4:1) with rocking overnight at 4°C, then dehydrated in Me and stored at −20°C until staining. For immunohistochemistry and RNA in situ hybridization, the right middle, right lower, and accessory lobes were immediately fixed in PFA, 4% in PBS overnight at 4°C, transferred to 30% sucrose in PBS at 4°C overnight, then submerged in optimal cutting temperature compound (Thermo Fisher Scientific) in an embedding mold, frozen on dry ice, and stored at −80°C. 14-µm sections were then obtained using a cryostat (CM3050; Leica) and collected on glass slides.
Whole-mount immunofluorescence was performed as previously described (Desai et al., 2014). Vibratome sections fixed in Me were transferred into solutions containing 0.1% Tween-20 mixed with PBS (PBST) and sequentially shaken for 10 min in 75% Me-PBST, 50% Me-PBST, 25% Me-PBST, and 0% Me-PBST solutions. Samples were subsequently transferred to blocking solution composed of 10% goat serum, 3% BSA, and 87% 0.1% Triton X-100 mixed in PBS, and shaken twice for 1 h at room temperature. Primary antibodies were then diluted in blocking solution and transferred into sample-containing Eppendorf tubes prior to incubation overnight at 4°C. The following day, samples were shaken at 4°C in blocking solution for 1 h, for a total of five washes. Samples were then exposed to secondary antibodies or fluorescent conjugates diluted in blocking solution and then were incubated overnight by shaking at 4°C. On the third day, samples were washed for 30 min for a total of five washes in PBS at room temperature and then placed in 80% glycerol solution mixed in PBS. For immunofluorescent staining of cryosections on slides, identical solutions were used except blocking solution contained 5% goat serum. Primary antibody incubation occurred at room temperature for 2 h, followed by five 10-min washes in blocking solution. Secondary antibody incubation occurred at 4°C overnight, again followed by five 10-min washes in PBS. In both whole mount and cryosection immunofluorescence, DAPI was incubated with samples during the final wash step.
Secondary antibodies conjugated to an Alexa Fluor dye (A488, A555, or A633; Invitrogen) were used. DAPI (5 ng/ml; Sigma-Aldrich) was included during incubation with secondary antibodies. For detection of EGFR-Emerald and pERK1/2, a horseradish peroxidase–conjugated secondary antibody (Invitrogen) was used against an anti-GFP or anti-pERK1/2 antibody and detected by tyramide signal amplification (Biotium). Lycopersicon esculentum lectin (1:500, DL11781; Vector) and Helix pomatia agglutinin lectin (1:500, L32454; Thermo Fisher Scientific) were preconjugated to DyLight-649 and A647, respectively. Primary antibodies against the following antigens were: Ager (rat, 1:500, MAB1179; R&D), GFP (chicken, 1:500, 13970; Abcam), RFP (rabbit, 1:500, 42872; Rockland), Pdpn (hamster, 2 µg/ml, 8.1.1; Developmental Studies Hybridoma Bank [DSHB]), pro-SpC (rabbit, 1:500, AB3786; Millipore), Keratin 8 (rat, 0.2 µg/ml, TROMA-I; DSHB), Lamp3 (guinea pig, 1:500, #391005; Synaptic Systems), Mucin 1 (rabbit, 1:500, NBP160046; Novus), Nkx2.1 (rabbit, 1:200, EPR5955; Abcam), pH3 (rabbit, 1:500, 06-570; Millipore), Cdh1 (rat, 1:200, ECCD-2; Invitrogen), Ki67 (rat, 1:100, SolA15; eBioscience), Ki67 (rabbit, 1:200, ab16667; Abcam), Grb2 (rabbit, 1:200, ab32037; Abcam), pErk1/2 (rabbit, 1:200, 9101; Cell Signaling Technology), ß-catenin-FITC (mouse, 1:250, 14; BD Transduction Laboratories), and Endomucin (rat, 1:250, V.5C7; Millipore). EdU (Carbosynth), injected as a single intraperitoneal injection (0.1 mg/g) or provided in drinking water (1 mg/ml in 1% sucrose water) as indicated, was detected prior to application of primary antibodies using the Click-iT EdU Cell Proliferation Kit (Thermo Fisher Scientific).
Multiplexed RNA in situ hybridization was performed by Proximity Ligation In Situ Hybridization as previously described (Nagendran et al., 2018). Comprehensive methods and information on optimization of reagents, incubation time, and solution are available open-access on Bio-protocols (Nagendran et al., 2020). The following sets of probe pairs were used to detect transcripts of the indicated genes:
hbEGF
5′-AGGTCAGGAATACCAGGTTGTAATGGAGGACCCAGCGTGGCATTGA-3′
5′-CGATGGGTTACAGAGCAAATTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGAACGAACCACTGTCTCAGAA-3′
5′-CTCCTCTATGTAGAACGTGTTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGATAGGGGATTCTCCACTGGT-3′
5′-TGTAGTGTGGTCATATGTGTTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGAGCTGACACTCTAATCCAGA-3′
5′-TCCTGGAGCATATGGAAACTTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGTGTCCCTTCCAAGTCCTGGA-3′
5′-ACTTTGAAAAGGTTCAGATCTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGAGGTATCAATGACCAAGAAT-3′
5′-CCTTCCCTATAGTAATTGATTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGTTTGGATGCGGCGGCCACTG-3′
5′-GACGGGGAGGCAGCGATCACTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGTTGCGGCTACTTGAACACAT-3′
5′-GGGTCCAAACAGCAGATCCCTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGTTGCGGTATTTCACAGTTAG-3′
5′-AAACTCAAGAGTTCTCGAGCTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGTTGCGGCCGCGGTTCGTCTG-3′
5′-TGAGATCCTGTCTGGGAACCTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGTCACCAACGCGGACAACACT-3′
5′-CCGCTCCAGACTCTCACCGGTTAGTATGATGACAATATAGCCAGGTT-3′
5′-AGGTCAGGAATACCAGGTTGTAATGGTAGACAAAAATCACCTAGGC-3′
5′-GGCTTTCGGAACACGAACGGTTAGTATGATGACAATATAGCCAGGTT-3′
Ager
5′-AGGTCAGGAATACTTAGCTATTGATGGTCATCGACAATTCCAGTGGC-3′
5′-CCCGACACCGGAAAGTCCCCTTCTGTGTAGACGACTATAGCCAGGTT-3′
5′-AGGTCAGGAATACTTAGCTATTGATGGTTTGCCATCGGGAATCAGAA-3′
5′-TCTTCCTTCACGAGTGTTTCTTCTGTGTAGACGACTATAGCCAGGTT-3′
After staining, vibratome slices were immersed in Vectashield (Vector) and placed onto Chambered Coverglass (#1.5; Lab-Tek). Cryotome sections were sealed by #1.5 Coverglass (Thermo Fisher Scientific) with nail hardener (Sally Hansen). Images were acquired using a Leica Sp5 inverted laser scanning confocal microscope with LAS X Software. Leica glycerol objectives were used for 20× and 63× magnification at room temperature. Images were processed (pseudocolored, contrast-adjusted, and overlaid) using ImageJ (National Institutes of Health). Quantification of nuclear stains was performed in QuPath (University of Edinburgh), while cytoplasmic stains were quantified manually using the cell counter function in ImageJ.
Transmission EM and ultrastructural analysis
To visualize alveolar wall ultrastructure following gene-targeted ablation of AT1 cells, lungs from mice carrying Hopx-CreER and R26iDTR or R26DTA alleles were isolated as described above. Vibratome slices (thickness of 600 µm) were fixed (2% glutaraldehyde [EMS] and 4% PFA [EMS] in 0.1 M sodium cacodylate [EMS], pH 7.4) overnight, post-fixed in 1% OsO4 for 1 h at 4°C, washed, stained in 1% uranyl acetate overnight at 4°C, dehydrated to 100% ethanol, infiltrated with Embed 812 resin (EMS), then oriented in casts and polymerized at 65°C for 24 h. Blocks were sectioned to a thickness of 80 nm using an ultramicrotome (UC7; Leica), stained for 40 s in 3.5% uranyl acetate in 50% acetone followed by staining in Sato’s Lead Citrate for 2 min, and visualized with a JEM-1400 transmission electron microscope (JOEL) with a 120-kV beam and imaged with an Orius SC1000 (Gatan) digital camera. Epithelial cells were scored for the presence of lamellar bodies, cuboidal versus squamous morphology, and cytoplasmic electron density. Septal walls were traced throughout each individual section to determine the presence or absence of exposed (denuded) basal lamina.
Clonal analysis
The Sftpc-ER-rtTA allele induced sparse (∼1%) ligand-independent recombination of the R26mTmG allele in AT2 cells, which resulted in isolated single-cell AT2 cell clones marked by the membrane GFP. 2-mo-old Sftpc-ER-rtTA > R26mTmG animals were treated with 450 mg/kg bodyweight BHT and euthanized 72 h later. AT1 cells expressing the AT2 cell lineage tag were subsequently identified in 600-µm-thick vibratome sections by imaging endogenous GFP fluorescence using confocal microscopy (20× objective) and were scored for the presence of a neighboring lineage-positive AT2 cell within three contiguous airspaces (∼100–150 µm) in the x, y, and z coordinate planes. To quantify clonal expansion of AT2 cells following repeated AT1 cell ablation, Sftpc-ER-rtTA > R26iMb-Mosaic animals were treated with intraperitoneal injections of 4 mg tamoxifen once per day for three consecutive days, followed by a single intraperitoneal injection of 100 mg/kg bodyweight BHT at least 2 wk after the final dose of tamoxifen. 5 d later, proliferating AT2 cell foci were scored by whole-mount immunofluorescence for the number of contiguous cells within each clone by analyzing mTomato, amplified by an anti-RFP antibody, and imaged by confocal microscopy (63× objective) to delineate single cells.
Flow cytometry and ploidy analysis
Lung tissue was isolated from Sftpc-CreER > R26mTmG animals treated with intraperitoneal injections of 4 mg tamoxifen once per day for three consecutive days to fluorescently mark AT2 cells or Axin2-rtTA-nls-GFP > TetO-H2b-GFP animals treated with doxycycline in drinking water for 10 consecutive days prior to analysis. Lungs were exposed and perfused 2 wk after the final tamoxifen injection as described above and inflated with digestion buffer (5 U/ml dispase, 200 U/ml collagenase I, 4 U/ml elastase, and 4 U/ml DNAse in HBSS). Following ligation of the trachea, the heart–lung block was carefully dissected and placed into a sterile Petri dish at 20°C for 45 min. The heart and large airways were discarded, and the remaining lung tissue was manually homogenized using a single-edge blade, suspended in digestion buffer, dissociated by GentleMACS (Miltenyi Biotec), and sequentially washed through 100 and 40 µm filters after red blood cell (RBC) lysis (ACK lysis buffer; Gibco). The peripheral lung single-cell suspension was then incubated with a cell viability stain (Sytox Orange; Invitrogen) and antibodies to fluorescently label RBCs (Ter119; 7436; BD), immune cells (anti-CD45, 30-F11; Biolegend), endothelial cells (anti-PECAM, 390; Biolegend), and epithelial cells (anti-EPCAM, G8.8; Biolegend). After exclusion of RBCs and dead cells, AT2 cells were enriched by negative selection of CD45+ and PECAM+ cells and positive selection of EPCAM+ cells using a FACSAria Fusion flow cytometer (BD). Axin2+ (GFP+) AT2 cells were further enriched by MHC-II staining (107632; Biolegend; Hasegawa et al., 2017). Sorted EPCAM+mGFP+ or MHCII+EPCAM+GFP+ cells were collected, fixed in 70% ethanol at −20°C, rehydrated in PBS, incubated in FxCycle PI/RNase staining solution (Invitrogen) for at least 30 min, and returned to the flow cytometer for single-cell quantitation of propidium iodide fluorescence intensity. FACS data analyses and plotting were done in FlowJo (BD).
Analysis of scRNA-seq data
scRNA-seq read counts for mouse lung were obtained from Tabula Muris Consortium (2020). Mean log-transformed read counts were calculated and a heatmap of these data for selected genes and lung alveolar cell types was generated using Seaborn v.0.11.2.
Statistical analysis
Replicate experiments were all biological replicates with different animals and quantitative values are presented as mean ± SEM unless indicated otherwise. Student’s t tests were two-sided and used to determine statistical significance where indicated. No statistical method was used to predetermine sample size, and data distribution was tested for normality prior to statistical analysis and plotting. Both male and female animals were used in experiments and subjects were age- and gender-matched in biological replicates and in comparison of different groups.
Online supplemental material
Fig. S1 shows Nkx2.1 is a pan-alveolar epithelial marker that enables quantitation of AT2 and AT1 cells. Fig. S2 shows rapid loss of mGFP fluorescent lineage marker in flattening epithelial cells during AT1 differentiation in injury and embryonic development. Fig. S3 shows Axin2+ AT2 cell population is enriched for 4n nuclear content. Fig. S4 shows Wnt signaling is necessary for AT2 cell repletion but not for AT2-to-AT1 cell differentiation.
Acknowledgments
We thank members of the Desai lab, R. Zemans, and R. Nusse for discussions and comments on the manuscript; A. Straight, E. Gronroos, and C. Logan for discussions on polyploidy; D. Peterson for assistance with analysis of transcriptomic data; and the Stanford University Stem Cell Institute FACS Core and Cell Sciences Imaging Core facilities.
This work is supported by the National Heart Lung and Blood Institute, the National Institutes of Health, the National Cancer Institute, and the Ludwig Cancer Institute (grants 5R01HL14254902, 5UG3HL14562302, and R35197750). T.J. Desai is the Woods Family Endowed Faculty Scholar in Pediatric Translational Medicine of the Stanford Child Health Research Institute and the Robert A. and Gertrude T. Hudson Endowed Professor. J. Guild was supported by the National Heart Lung and Blood Institute and the Stanford University Medical Scientist Training Program (grant 1F30HL16015601).
Author contributions: conceptualization: J. Guild, T.J. Desai; methodology: J. Guild, T.J. Desai, M.A. Matthay; investigation: J. Guild, N.H. Juul, A. Andalon, H. Taenaka; visualization: J. Guild, N.H. Juul, A. Andalon, H. Taenaka; funding acquisition: J. Guild, T.J. Desai, R.J. Coffey; project administration: T.J. Desai; supervision: T.J. Desai; writing—original draft: J. Guild, T.J. Desai; and writing—review & editing: J. Guild, T.J. Desai.
References
- Abe, T., Kiyonari H., Shioi G., Inoue K.I., Nakao K., Aizawa S., and Fujimori T.. 2011. Establishment of conditional reporter mouse lines at ROSA26 locus for live cell imaging. Genesis. 49:579–590. 10.1002/dvg.20753 [DOI] [PubMed] [Google Scholar]
- Adamson, I.Y., and Bowden D.H.. 1974. The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab. Invest. 30:35–42. [PubMed] [Google Scholar]
- Adamson, I.Y., Bowden D.H., Cote M.G., and Witschi H.. 1977. Lung injury induced by butylated hydroxytoluene: Cytodynamic and biochemical studies in mice. Lab. Invest. 36:26–32. [PubMed] [Google Scholar]
- Aragona, M., Dekoninck S., Rulands S., Lenglez S., Mascré G., Simons B.D., and Blanpain C.. 2017. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat. Commun. 8:14684. 10.1038/ncomms14684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspal, M., and Zemans R.L.. 2020. Mechanisms of ATII-to-ATI cell differentiation during lung regeneration. Int. J. Mol. Sci. 21:3188. 10.3390/ijms21093188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachofen, M., and Weibel E.R.. 1974. Basic pattern of tissue repair in human lungs following unspecific injury. Chest. 65:14S–19S. 10.1378/chest.65.4_Supplement.14S [DOI] [PubMed] [Google Scholar]
- Bailey, E.C., Kobielski S., Park J., and Losick V.P.. 2021. Polyploidy in tissue repair and regeneration. Cold Spring Harb. Perspect. Biol. 13:a040881. 10.1101/cshperspect.a040881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas, C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., and Hogan B.L.M.. 2013. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123:3025–3036. 10.1172/JCI68782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch, T., Heppner F.L., Tertilt C., Heinen T.J.A.J., Kremer M., Wunderlich F.T., Jung S., and Waisman A.. 2005. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods. 2:419–426. 10.1038/nmeth762 [DOI] [PubMed] [Google Scholar]
- Chan, K.Y., Yan C.C.S., Roan H.Y., Hsu S.C., Tseng T.L., Hsiao C.D., Hsu C.P., and Chen C.H.. 2022. Skin cells undergo asynthetic fission to expand body surfaces in zebrafish. Nature. 605:119–125. 10.1038/s41586-022-04641-0 [DOI] [PubMed] [Google Scholar]
- Chapman, H.A., Li X., Alexander J.P., Brumwell A., Lorizio W., Tan K., Sonnenberg A., Wei Y., and Vu T.H.. 2011. Integrin α6β4 identifies an adult distal lung epithelial population with regenerative potential in mice. J. Clin. Invest. 121:2855–2862. 10.1172/JCI57673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.H., Puliafito A., Cox B.D., Primo L., Fang Y., Di Talia S., and Poss K.D.. 2016. Multicolor cell barcoding technology for long-term surveillance of epithelial regeneration in zebrafish. Dev. Cell. 36:668–680. 10.1016/j.devcel.2016.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, W.K.C., Zhao M., Liu Z., Stevens L.E., Cao P.D., Fang J.E., Westbrook T.F., and Nguyen D.X.. 2013. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 23:725–738. 10.1016/j.ccr.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J., Park J.E., Tsagkogeorga G., Yanagita M., Koo B.K., Han N., and Lee J.H.. 2020. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell. 27:366–382.e7. 10.1016/j.stem.2020.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crapo, J.D., Barry B.E., Foscue H.A., and Shelburne J.. 1980. Structural and biochemical changes in rat lungs occurring during exposures to lethal and adaptive doses of oxygen. Am. Rev. Respir. Dis. 122:123–143. 10.1164/arrd.1980.122.1.123 [DOI] [PubMed] [Google Scholar]
- Desai, T.J., Brownfield D.G., and Krasnow M.A.. 2014. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 507:190–194. 10.1038/nature12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupé, D.P., Alcolea M.P., Roshan A., Zhang G., Klein A.M., Simons B.D., and Jones P.H.. 2012. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science. 337:1091–1093. 10.1126/science.1218835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli, D., Rosains J., Birkhoff G., and Eggan K.. 2007. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 447:679–685. 10.1038/nature05879 [DOI] [PubMed] [Google Scholar]
- Erjefält, J.S., Erjefält I., Sundler F., and Persson C.G.. 1995. In vivo restitution of airway epithelium. Cell Tissue Res. 281:305–316. 10.1007/BF00583399 [DOI] [PubMed] [Google Scholar]
- Evans, M.J., Cabral L.J., Stephens R.J., and Freeman G.. 1973. Renewal of alveolar epithelium in the rat following exposure to NO2. Am. J. Pathol. 70:175–198. [PMC free article] [PubMed] [Google Scholar]
- Ganem, N.J., and Pellman D.. 2007. Limiting the proliferation of polyploid cells. Cell. 131:437–440. 10.1016/j.cell.2007.10.024 [DOI] [PubMed] [Google Scholar]
- Gotts, J.E., Bernard O., Chun L., Croze R.H., Ross J.T., Nesseler N., Wu X., Abbott J., Fang X., Calfee C.S., and Matthay M.A.. 2019. Clinically relevant model of pneumococcal pneumonia, ARDS, and nonpulmonary organ dysfunction in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 317:L717–L736. 10.1152/ajplung.00132.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee, K.J., Werb Z., and Kheradmand F.. 2007. Matrix metalloproteinases in lung: Multiple, multifarious, and multifaceted. Physiol. Rev. 87:69–98. 10.1152/physrev.00022.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe, B.D., Scherz P.J., Nissim S., Tian H., McMahon A.P., and Tabin C.J.. 2004. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 118:517–528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Haschek, W.M., and Witschi H.. 1979. Pulmonary fibrosis--a possible mechanism. Toxicol. Appl. Pharmacol. 51:475–487. 10.1016/0041-008X(79)90372-7 [DOI] [PubMed] [Google Scholar]
- Hasegawa, K., Sato A., Tanimura K., Uemasu K., Hamakawa Y., Fuseya Y., Sato S., Muro S., and Hirai T.. 2017. Fraction of MHCII and EpCAM expression characterizes distal lung epithelial cells for alveolar type 2 cell isolation. Respir. Res. 18:150. 10.1186/s12931-017-0635-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller, E., Kumar K.V., Grill S.W., and Fuchs E.. 2014. Forces generated by cell intercalation tow epidermal sheets in mammalian tissue morphogenesis. Dev. Cell. 28:617–632. 10.1016/j.devcel.2014.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai, K.I., Witschi H., and Côté M.G.. 1977. Electron microscopy of butylated hydroxytoluene-induced lung damage in mice. Exp. Mol. Pathol. 27:295–308. 10.1016/0014-4800(77)90002-8 [DOI] [PubMed] [Google Scholar]
- Iwamoto, R., Yamazaki S., Asakura M., Takashima S., Hasuwa H., Miyado K., Adachi S., Kitakaze M., Hashimoto K., Raab G., et al. 2003. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc. Natl. Acad. Sci. USA. 100:3221–3226. 10.1073/pnas.0537588100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansing, N.L., McClendon J., Henson P.M., Tuder R.M., Hyde D.M., and Zemans R.L.. 2017. Unbiased quantitation of alveolar type II to alveolar type I cell transdifferentiation during repair after lung injury in mice. Am. J. Respir. Cell Mol. Biol. 57:519–526. 10.1165/rcmb.2017-0037MA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, P., Gil De Rubio R., Hrycaj S.M., Gurczynski S.J., Riemondy K.A., Moore B.B., Omary M.B., Ridge K.M., and Zemans R.L.. 2020. Ineffectual type 2-to-Type 1 alveolar epithelial cell differentiation in idiopathic pulmonary fibrosis: Persistence of the KRT8hi transitional state. Am. J. Respir. Crit. Care Med. 201:1443–1447. 10.1164/rccm.201909-1726LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juul, N.H., Stockman C.A., and Desai T.J.. 2020. Corrigendum: Niche cells and signals that regulate lung alveolar stem cells in vivo. Cold Spring Harb. Perspect. Biol. 12:a040303. 10.1101/cshperspect.a040303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradmand, F., Rishi K., and Werb Z.. 2002. Signaling through the EGF receptor controls lung morphogenesis in part by regulating MT1-MMP-mediated activation of gelatinase A/MMP2. J. Cell Sci. 115:839–848. 10.1242/jcs.115.4.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Tata A., Konkimalla A., Katsura H., Lee R.F., Ou J., Banovich N.E., Kropski J.A., and Tata P.R.. 2020. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22:934–946. 10.1038/s41556-020-0542-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCanna, R., Liccardo D., Zhang P., Tragesser L., Wang Y., Cao T., Chapman H.A., Morrisey E.E., Shen H., Koch W.J., et al. 2019. Yap/Taz regulate alveolar regeneration and resolution of lung inflammation. J. Clin. Invest. 129:2107–2122. 10.1172/JCI125014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., Song H., Huang C., Yao E., Gacayan R., Xu S.M., and Chuang P.T.. 2012. Alveolar type II cells possess the capability of initiating lung tumor development. PLoS One. 7:e53817. 10.1371/journal.pone.0053817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan, C.Y., and Desai T.J.. 2015. Keeping it together: Pulmonary alveoli are maintained by a hierarchy of cellular programs. BioEssays. 37:1028–1037. 10.1002/bies.201500031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen, L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, T., Wakefield L., Tarlow B.D., and Grompe M.. 2020. In Vivo lineage tracing of polyploid hepatocytes reveals extensive proliferation during liver regeneration. Cell Stem Cell. 26:34–47.e3. 10.1016/j.stem.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay, M.A., Zemans R.L., Zimmerman G.A., Arabi Y.M., Beitler J.R., Mercat A., Herridge M., Randolph A.G., and Calfee C.S.. 2019. Acute respiratory distress syndrome. Nat. Rev. Dis. Primers. 5:18. 10.1038/s41572-019-0069-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar, M.D., Tasic B., Miyamichi K., Li L., and Luo L.. 2007. A global double-fluorescent Cre reporter mouse. Genesis. 45:593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nabhan, A.N., Brownfield D.G., Harbury P.B., Krasnow M.A., and Desai T.J.. 2018. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 359:1118–1123. 10.1126/science.aam6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagendran, M., Andruska A.M., Harbury P.B., and Desai T.J.. 2020. Advances in proximity ligation in situ hybridization (PLISH). Bio Protoc. 10:e3808. 10.21769/BioProtoc.3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagendran, M., Riordan D.P., Harbury P.B., and Desai T.J.. 2018. Automated cell-type classification in intact tissues by single-cell molecular profiling. Elife. 7:e30510. 10.7554/eLife.30510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglich, J.G., Metherall J.E., Russell D.W., and Eidels L.. 1992. Expression cloning of a diphtheria toxin receptor: Identity with a heparin-binding EGF-like growth factor precursor. Cell. 69:1051–1061. 10.1016/0092-8674(92)90623-K [DOI] [PubMed] [Google Scholar]
- Nguyen, P.D., Gurevich D.B., Sonntag C., Hersey L., Alaei S., Nim H.T., Siegel A., Hall T.E., Rossello F.J., Boyd S.E., et al. 2017. Muscle stem cells undergo extensive clonal drift during tissue growth via Meox1-mediated induction of G2 cell-cycle arrest. Cell Stem Cell. 21:107–119.e6. 10.1016/j.stem.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Otsuki, L., and Brand A.H.. 2018. Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science. 360:99–102. 10.1126/science.aan8795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S., Gonzalez D.G., Guirao B., Boucher J.D., Cockburn K., Marsh E.D., Mesa K.R., Brown S., Rompolas P., Haberman A.M., et al. 2017. Tissue-scale coordination of cellular behaviour promotes epidermal wound repair in live mice. Nat. Cell Biol. 19:155–163. 10.1038/ncb3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes-Quero, S., Heredia L., Casquero-García V., Fernández-Chacón M., Luo W., Hermoso A., Bansal M., Garcia-Gonzalez I., Sanchez-Muñoz M.S., Perea J.R., et al. 2017. Dual ifgMosaic: A versatile method for multispectral and combinatorial mosaic gene-function analysis. Cell. 170:800–814.e18. 10.1016/j.cell.2017.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins, E.L., Ostrowski L.E., Randell S.H., and Hogan B.L.M.. 2007. Lung development and repair: Contribution of the ciliated lineage. Proc. Natl. Acad. Sci. USA. 104:410–417. 10.1073/pnas.0610770104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkarina, K., Hasel de Carvalho E., Santos J.C., Ramos S., Leptin M., and Broz P.. 2022. Optogenetic activators of apoptosis, necroptosis, and pyroptosis. J. Cell Biol. 221:e202109038. 10.1083/jcb.202109038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunz, M., Simon L.M., Ansari M., Kathiriya J.J., Angelidis I., Mayr C.H., Tsidiridis G., Lange M., Mattner L.F., Yee M., et al. 2020. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11:3559. 10.1038/s41467-020-17358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szidon, J.P., Pietra G.G., and Fishman A.P.. 1972. The alveolar-capillary membrane and pulmonary edema. N. Engl. J. Med. 286:1200–1204. 10.1056/NEJM197206012862208 [DOI] [PubMed] [Google Scholar]
- Tabula Muris Consortium . 2020. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 583:590–595. 10.1038/s41586-020-2496-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, K., Cockburn K., and Greco V.. 2019. Flexibility sustains epithelial tissue homeostasis. Curr. Opin. Cell Biol. 60:84–91. 10.1016/j.ceb.2019.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., and Epstein J.A.. 2011. Interconversion between intestinal stem cell populations in distinct niches. Science. 334:1420–1424. 10.1126/science.1213214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomashefski, J.F., Jr. 2000. Pulmonary pathology of acute respiratory distress syndrome. Clin. Chest Med. 21:435–466. 10.1016/S0272-5231(05)70158-1 [DOI] [PubMed] [Google Scholar]
- Tumbar, T., Guasch G., Greco V., Blanpain C., Lowry W.E., Rendl M., and Fuchs E.. 2004. Defining the epithelial stem cell niche in skin. Science. 303:359–363. 10.1126/science.1092436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhal, B.D. 1997. Cell cycle kinetics in the alveolar epithelium. Am. J. Physiol. 272:L1031–L1045. 10.1152/ajplung.1997.272.6.L1031 [DOI] [PubMed] [Google Scholar]
- van Amerongen, R., Bowman A.N., and Nusse R.. 2012. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 11:387–400. 10.1016/j.stem.2012.05.023 [DOI] [PubMed] [Google Scholar]
- van de Moosdijk, A.A.A., van de Grift Y.B.C., de Man S.M.A., Zeeman A.L., and van Amerongen R.. 2020. A novel Axin2 knock-in mouse model for visualization and lineage tracing of WNT/CTNNB1 responsive cells. Genesis. 58:e23387. 10.1002/dvg.23387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keymeulen, A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., and Blanpain C.. 2011. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 479:189–193. 10.1038/nature10573 [DOI] [PubMed] [Google Scholar]
- Voehringer, D., Liang H.E., and Locksley R.M.. 2008. Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J. Immunol. 180:4742–4753. 10.4049/jimmunol.180.7.4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., and Moran M.F.. 1996. Requirement for the adapter protein GRB2 in EGF receptor endocytosis. Science. 272:1935–1939. 10.1126/science.272.5270.1935 [DOI] [PubMed] [Google Scholar]
- Weibel, E.R. 2015. On the tricks alveolar epithelial cells play to make a good lung. Am. J. Respir. Crit. Care Med. 191:504–513. 10.1164/rccm.201409-1663OE [DOI] [PubMed] [Google Scholar]
- Weng, A., Maciel-Herrerias M., Watanabe S., Welch L.C., Flozak A.S., Grant R.A., Aillon R.P., Dada L.A., Han S.H., Hinchcliff M., et al. 2022. Lung injury induces alveolar type 2 cell hypertrophy and polyploidy with implications for repair and regeneration. Am. J. Respir. Cell Mol Biol. 66:564–576. 10.1165/rcmb.2021-0356OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Rock J.R., Lu Y., Futtner C., Schwab B., Guinney J., Hogan B.L.M., and Onaitis M.W.. 2012. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proc. Natl. Acad. Sci. USA. 109:4910–4915. 10.1073/pnas.1112499109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.P., Ma H., Starchenko A., Huh W.J., Li W., Hickman F.E., Zhang Q., Franklin J.L., Mortlock D.P., Fuhrmann S., et al. 2017. A chimeric Egfr protein reporter mouse reveals Egfr localization and trafficking in vivo. Cell Rep. 19:1257–1267. 10.1016/j.celrep.2017.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias, W.J., Frank D.B., Zepp J.A., Morley M.P., Alkhaleel F.A., Kong J., Zhou S., Cantu E., and Morrisey E.E.. 2018. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 555:251–255. 10.1038/nature25786 [DOI] [PMC free article] [PubMed] [Google Scholar]