Abstract
Autophagy is a primary mechanism for maintaining cellular homeostasis. The synergistic actions of autophagy-related (ATG) proteins strictly regulate the whole autophagic process. Therefore, accurate identification of ATGs is a first and critical step to reveal the molecular mechanism underlying the regulation of autophagy. Current computational methods can predict ATGs from primary protein sequences, but owing to the limitations of algorithms, significant room for improvement still exists. In this research, we propose EnsembleDL-ATG, an ensemble deep learning framework that aggregates multiple deep learning models to predict ATGs from protein sequence and evolutionary information. We first evaluated the performance of individual networks for various feature descriptors to identify the most promising models. Then, we explored all possible combinations of independent models to select the most effective ensemble architecture. The final framework was built and maintained by an organization of four different deep learning models. Experimental results show that our proposed method achieves a prediction accuracy of 94.5 % and MCC of 0.890, which are nearly 4 % and 0.08 higher than ATGPred-FL, respectively. Overall, EnsembleDL-ATG is the first ATG machine learning predictor based on ensemble deep learning. The benchmark data and code utilized in this study can be accessed for free at https://github.com/jingry/autoBioSeqpy/tree/2.0/examples/EnsembleDL-ATG.
Keywords: Autophagy protein, Ensemble deep learning, Position-specific scoring matrix, AutoBioSeqpy
Graphical Abstract
Highlights
-
•
We proposed a novel ensemble deep learning framework called EnsembleDL-ATG for effectively identifying autophagy proteins.
-
•
Experimental results demonstrate that the performance of EnsembleDL-ATG is superior to that of preexisting methods.
-
•
We provided the details of encoding methods and deep learning models with a step-by-step guide on how to execute them.
-
•
We visualized the representations of ATGs learned by the ensemble deep learning framework using layerUMAP.
1. Introduction
In 1963, at the CIBA Foundation Symposium on Lysosomes, Christian de Duve created the term “autophagy” to refer to the cellular process of breaking down its own cytoplasmic components using lysosomes [1]. The 2016 Nobel Prize in Physiology or Medicine was bestowed upon Yoshinori Ohsumi for his groundbreaking discoveries related to the mechanisms involved in autophagy [2]. Maintaining cellular homeostasis and adapting to diverse cellular stresses are critical functions of autophagy, which is a lysosome-dependent intracellular catabolic process that is highly conserved among eukaryotes [1], [3]. Autophagy can be categorized into three primary types based on how the cargo is transported to the lysosome or vacuole [4]: macroautophagy [5], microautophagy [6], and chaperone-mediated autophagy [7]. In addition, it can also be selective or nonselective, depending on the type of cargo that is sequestered. Most cytoplasmic contents are turned over by nonselective autophagy, while substrates and organelles can be degraded via selective autophagy in some cases, such as aggrephagy, ferritinophagy, glycophagy, lipophagy, lysophagy, mitophagy, nucleophagy, pexophagy, reticulophagy, ribophagy, and xenophagy [8], [9], [10]. Increasing evidence suggests that autophagy dysregulation has been implicated in a broad spectrum of human diseases [11], [12], including cancer [13], [14], [15], [16], [17], [18], neurodegenerative diseases [19], [20], [21], [22], infectious disease [23], [24], and autoimmune disorders [25], [26].
Over the past decade, numerous efforts have been made to elucidate the molecular mechanisms underlying autophagy regulation [3]. The initiation of autophagy involves the creation of a double-membrane vesicle, known as an autophagosome, which engulfs portions of the cytoplasm. This process involves the coordinated action of a group of conserved proteins coded by autophagy-related (ATG) genes, and among them, the Atg1/unc-51-like kinase (ULK) kinase complex is the most advanced part that plays a vital role in initiating autophagosome formation [27], [28], [29]. During autophagosome formation, the LC3/Atg8-phosphatidylethanolamine (PE) and the ATG12-ATG15 ubiquitin-like protein conjugation systems, as well as the ATG9/Atg9 cycling system, are crucial components [30], [31]. Furthermore, in addition to autophagosome formation, ATGs are also involved in other sequential steps of the autophagy process by forming different protein complexes, including cargo recognition and engulfment, autophagosome-lysosome fusion, and autophagosome elongation, maturation, and degradation [32], [33]. In general, autophagy is highly dependent on the availability of ATGs, which are the major executor and organizer of the autophagy machinery [34]. Therefore, detailed knowledge of the molecular mechanisms mediated by these ATGs would provide valuable insight into the intact autophagy process.
There is a growing interest in exploring the role of ATGs in autophagy regulation mechanisms, particularly in their involvement in constructing and predicting related biological networks. For example, Türei et al. developed the Autophagy Regulatory Network (ARN) to comprehensively collect interactions between human autophagy components or their protein regulators with transcription factors and miRNAs [35]. Wu et al. developed ncRDeathDB to archive ncRNA-associated cell death interactions, including apoptosis and autophagy [36]. Zhang et al. constructed GAMDB to manually curate and predicted the intricate relationships between microRNA-regulated autophagic mechanisms and gerontology [37]. Jacomin et al. developed the iLIR database to store all putative canonical Atg8-interacting proteins predicted from the proteomes of eight model organisms [38]. Deng et al. developed THANATOS to integrate biological information about proteins and posttranslational modifications in the regulation of autophagy [39]. As research has progressed, numerous ATGs have been identified in various organisms, especially mammals. However, it is likely that there are other proteins, perhaps some yet undiscovered ATGs, involved in the molecular control of autophagy. Wet-lab identification experiments can characterize the biological activities of ATGs, but they are expensive, time-consuming, or both. Since ATGs are highly conserved genes, their paralogs as well as orthologs can be readily identified in different species. Taking advantage of this fact, several machine learning-based computational models have been presented to extract discriminative features of ATGs and non-ATGs, and make large-scale predictions for multiple species [40]. One representative example is ATGPred-FL, which was constructed based on optimal 14-dimensional sequence-derived features and a support vector machine algorithm [41]. ATGPred-FL performed favorably on the benchmark datasets with high classification accuracy. Nevertheless, there is still much room for improvement on autophagy-related protein prediction due to the lack of important evolutionary information on ATGs.
To address the above limitations and further improve the prediction performance, we have proposed a new method, EnsembleDL-ATG (https://github.com/jingry/autoBioSeqpy/tree/2.0/examples/EnsembleDL-ATG), which uses a novel ensemble framework to integrate protein sequence and evolutionary information from complementary deep learning models to identify ATGs. For sequence information, we designed five sequence-level deep learning (DL) models, including convolutional neural networks (CNNs), recurrent neural networks (RNNs) with bidirectional long short-term memory (BiLSTM) or bidirectional gated recurrent units (BiGRU), and the combination of the two networks (CNN-BiLSTM and CNN-BiGRU), to progressively extract higher-level abstract features from ATG primary sequences. On the other hand, to fully characterize the evolutionary history of a given ATG, we generated nine types of feature descriptors based on evolutionary information embedding in a position-specific scoring matrix (PSSM), including AAC-PSSM, DP-PSSM, DPC-PSSM, Pse-PSSM, PSSM-AC, PSSM400, SVD_PSSM, Single_Average, and DFMCA_PSSM. The PSSM contains the probability of occurrence of each type of amino acid residues at each position and hence can be considered as a measure of residue conservation at a given position [42]. We used several densely connected neural networks (DNNs) to further extract more information from each PSSM-based feature vector. We first investigated various single deep learning models and performed a systematic grid search in the hyperparameter space of layer depth and neuron width to generate optimized, more informative prediction models. After that, all single models with the best performance were combined to establish the ensemble architecture framework. Here, we enumerated and tested all possible combinations of single deep learning models, which would involve 511 combinations. Instead of averaging results from individual models, as is traditionally done [43], we weighed the outcomes of each model to maximize the model’s respective strengths. We showed that this ensemble deep learning approach can accurately identify autophagy proteins and outperform the existing method on the same task. Finally, our ensemble deep learning framework was developed, trained, and evaluated in the autoBioSeqpy tool. We also provided detailed descriptions of various deep learning models, as well as a step-by-step guide on how to execute them.
2. Materials and methods
2.1. Datasets
To maintain consistency with the previous ATGPred-FL approach, we utilized the datasets constructed by Jiao et al. to develop, train, and assess deep learning models that predict ATGs [41]. Initially, the authors gathered the ATG sequences, which possessed experimentally verified functional annotation data, from the most recent version of the Universal Protein KnowledgeBase (UniProtKB) [44]. Second, non-ATG sequences were collected and filtered from the protein family database (PFAM) [45] based on two principles: (i) protein families associated with ATGs were removed; (ii) each protein was the longest sequence from the remaining protein families. Third, the CD-HIT program was applied to avoid homology bias [46]. Finally, a benchmark dataset containing 986 sequences was obtained, including 493 ATGs and 493 non-ATGs. According to Jiao et al.’s work, four-fifths of the sequences were used for training the model, and the remaining data were used as independent samples to test the model (see Table S1).
2.2. Feature representation algorithms
In this study, the dictionary-embedding encoding method and nine PSSM-based features were used to encode the protein sequences in the datasets.
2.2.1. Dictionary-embedding encoding
Each protein primary sequence is considered a sentence, and the amino acid symbols are used as words that make up the sentence. We created a dictionary of amino acid symbols to map each residue by integer encoding, where naturally occurring residues were transformed into index-based integers in the range of 1–20 (e.g., A is given a value of 1) [47]. In addition, pseudo-amino acids or unknown “X” symbols were all assigned to 0. Such encoded sequences were then passed to the embedding layer, and a lookup table was used to map these inputs into low-level features. Specifically, each protein sequence was converted into a 2D vector (128, L), in which L represents the length of the input sequence and 128 is the output dimension. The embedding weight matrix was initialized with random weights, and these weights were learned during training.
2.2.2. Evolutionary-based descriptors
As the most commonly used descriptor for characterizing the evolutionary information of protein sequences, position-specific scoring matrix (PSSM) is a special substitution matrix that contains the alignment position information. Several previous studies have reported that PSSM-based descriptors could enhance the performance of protein property predictors [48], [49], [50]. First, a search for homologous protein sequences was carried out with the PSI-BLAST program [51] against nonredundant sequence databases such as NCBI or Swiss-Prot. Having multiple sequence alignments performed, an original PSSM matrix was subsequently created, where the rows indicate the positions of amino acid residues in an alignment, the columns stand for the names of residues, and the values inside are binary logarithms of residues derived from multiple alignment scores. In such a matrix, positive values denote identical or similar sequences that have been aligned, while negative values mean nonconserved alignments. Finally, different types of derived features can be extracted directly from the original PSSM matrix by three forms of matrix transformations, including row transformations, column transformations, and their combinations. Here, we selected 9 PSSM-based features to describe the evolutionary information of ATGs, which are AAC-PSSM, DP-PSSM, DPC-PSSM, PSe-PSSM, PSSM-AC, PSSM400, SVD_PSSM, Single_Average, and DFMCA_PSSM. All these features were implemented by the PSSMCOOL package [52], and a detailed description of them is given below.
2.2.2.1. AAC-PSSM
This descriptor is the column average of a PSSM matrix, and each protein sequence is transformed into a 20D vector. It is calculated by Eq. (1).
| (1) |
2.2.2.2. DPC-PSSM
As a variant of the dipeptide composition (DPC), this descriptor is also defined as a 400-dimensional vector. When computing it, the elements of two consecutive rows and two different columns are multiplied together. It is worth noting that the action is run on different rows and columns. Later, a summation operation is implemented for the calculated values. For each two consecutive rows, the summation value is divided by L-1. The formula for generating this descriptor is provided in Eq. (2).
| (2) |
2.2.2.3. PSSM-AC
This descriptor represents autocovariance transformation, and computes the correlations of the same attribute between two amino acid residues spaced along the protein sequence as L-g. The length of it is determined by the parameter L-g. When L-g is set to 10, all protein sequences are translated into 200D vectors. The formula for creating this feature is given in Eq. (3).
| (3) |
2.2.2.4. PSe-PSSM
This feature vector is a combination of the AAC-PSSM feature vector and a vector of correlation factors corresponding to 20 columns in the PSSM matrix. The values of correlation factor vectors are calculated for each column, and the parameter lag could be set to any integer between 1 and 15. Thus, for all protein sequences, (20 + 20 × lag)-dimensional feature vectors can be obtained by this way. The formula for generating this descriptor is afforded in Eq. (4).
| (4) |
2.2.2.5. PSSM400
For each standard amino acid, all corresponding rows in a PSSM matrix are first collected to generate a submatrix. Computing the average of the columns in this submatrix, a 20-dimensional feature vector is achieved for each amino acid. By combining the feature vectors of all 20 amino acids, a 400-dimensional vector could be gained finally.
2.2.2.6. DP-PSSM
This feature first calculates the average of the positive and negative values in each column of the PSSM matrix and concatenates them together. Next, the differences between the values in the rows with distance k are computed. Finally, the square averages for these differences with positive and negative scores are computed respectively.
2.2.2.7. DFMCA_PSSM
The PSSM matrix columns could be viewed as 20-time series. Based on this assumption, this feature uses the detrended moving-average cross-correlation analysis (DMCA) algorithm for each column and each pair of columns of the PSSM matrix to measure the level of cross-correlation between two detrended nonstationary time series.
2.2.2.8. SVD_PSSM
As a popular matrix decomposition method for effectively reducing a matrix, the singular value decomposition (SVD) has been successfully applied in many fields. In computational biology, it is frequently used to reduce the dimensionality of a protein matrix to make some subsequent matrix calculations easier. Through this method, each protein sequence is finally transformed into a 20D vector.
2.2.2.9. Single_Average
This feature is developed to combine the rows associated with the same amino acid in a PSSM matrix, and the domains with analogous conservation rates are taken into account for each protein sequence. The formula for generating this descriptor is offered in Eq. (5).
| (5) |
2.3. Single model architectures
We first evaluated four types of neural network architectures and trained a total of fourteen single models to predict ATGs. For the CNN, RNN, and CNN-RNN architectures, each primary protein sequence is coded by the dictionary encoding method and a score between 0 and 1 is exported, which represents the probability of the query protein being an ATG or a non-ATG. The fourth DNN architecture takes PSSM-based features as inputs and gives a probability score. To obtain the best possible results from each model, a grid search approach was used to determine the combination of hyperparameters that maximizes the performance (Fig. S1-S3). Details about the model architectures are provided below (Table S2).
2.3.1. Convolutional neural network architecture (CNN)
-
(1)
Embedding layer (input_dim: 26; output_dim: 128; input_length: 2000)
-
(2)
Dropout layer (20 % dropout)
-
(3)
Convolution layer (250 filters; kernel size: 13; ReLU activation; 0 % dropout; step size, 1)
-
(4)
Pooling layer (global maximum value)
-
(5)
Fully connected layer (125 units)
-
(6)
Dropout layer (20 % dropout)
-
(7)
Activation layer (ReLU activation)
-
(8)
Output layer (1 units, sigmoid activation)
2.3.2. Bidirectional long short-term memory architecture (BiLSTM)
-
(1)
Embedding layer (input_dim: 26; output_dim: 128; input_length: 2000)
-
(2)
Bidirectional LSTM layer (64 units, tanh activation; sigmoid recurrent activation; 0% dropout)
-
(3)
Bidirectional LSTM layer (64 units, tanh activation; sigmoid recurrent activation; 0% dropout)
-
(4)
Pooling layer (global maximum value)
-
(5)
Fully connected layer (125 units)
-
(6)
Dropout layer (20 % dropout)
-
(7)
Activation layer (ReLU activation)
-
(8)
Output layer (1 units, sigmoid activation)
2.3.3. Bidirectional gated recurrent unit architecture (BiGRU)
-
(1)
Embedding layer (input_dim: 26; output_dim: 128; input_length: 2000)
-
(2)
Bidirectional GRU layer (64 units, tanh activation; sigmoid recurrent activation; 0% dropout)
-
(3)
Pooling layer (global maximum value)
-
(4)
Fully connected layer (125 units)
-
(5)
Dropout layer (20 % dropout)
-
(6)
Activation layer (ReLU activation)
-
(7)
Output layer (1 units, sigmoid activation)
2.3.4. Convolutional-bidirectional long short-term memory architecture (CNN-BiLSTM)
-
(1)
Embedding layer (input_dim: 26; output_dim: 128; input_length: 2000)
-
(2)
Convolution layer (250 filters; kernel size: 11; ReLU activation; 0% dropout; step size, 1)
-
(3)
Bidirectional LSTM layer (64 units, tanh activation; sigmoid recurrent activation; 0% dropout)
-
(4)
Pooling layer (global maximum value)
-
(5)
Fully connected layer (125 units)
-
(6)
Dropout layer (20 % dropout)
-
(7)
Activation layer (ReLU activation)
-
(8)
Output layer (1 units, sigmoid activation)
2.3.5. Convolutional-bidirectional gated recurrent unit architecture (CNN-BiGRU)
-
(1)
Embedding layer (input_dim: 26; output_dim: 128; input_length: 2000)
-
(2)
Convolution layer (250 filters; kernel size: 15; ReLU activation; 0% dropout; step size, 1)
-
(3)
Bidirectional GRU layer (64 units, tanh activation; sigmoid recurrent activation; 0% dropout)
-
(4)
Fully connected layer (125 units)
-
(5)
Dropout layer (20 % dropout)
-
(6)
Activation layer (ReLU activation)
-
(7)
Output layer (1 units, sigmoid activation)
2.3.6. Densely connected neural networks with AAC-PSSM (DNNs- AAC-PSSM)
-
(1)
Fully connected layer (15 units, ReLU activation)
-
(2)
Dropout layer (20 % dropout)
-
(3)
Fully connected layer (10 units, ReLU activation)
-
(4)
Dropout layer (20 % dropout)
-
(5)
Activation layer (ReLU activation)
-
(6)
Output layer (1 units, sigmoid activation)
2.3.7. Densely connected neural networks with DPC-PSSM (DNNs- DPC-PSSM)
-
(1)
Fully connected layer (350 units, ReLU activation)
-
(2)
Dropout layer (20 % dropout)
-
(3)
Fully connected layer (300 units, ReLU activation)
-
(4)
Dropout layer (20 % dropout)
-
(5)
Fully connected layer (250 units, ReLU activation)
-
(6)
Dropout layer (20 % dropout)
-
(7)
Activation layer (ReLU activation)
-
(8)
Output layer (1 units, sigmoid activation)
2.3.8. Densely connected neural networks with PSSM-AC (DNNs- PSSM-AC)
-
(1)
Fully connected layer (150 units, ReLU activation)
-
(2)
Dropout layer (20 % dropout)
-
(3)
Fully connected layer (100 units, ReLU activation)
-
(4)
Dropout layer (20 % dropout)
-
(5)
Fully connected layer (50 units, ReLU activation)
-
(6)
Dropout layer (20 % dropout)
-
(7)
Activation layer (ReLU activation)
-
(8)
Output layer (1 units, sigmoid activation)
2.3.9. Densely connected neural networks with PSe-PSSM (DNNs- PSe-PSSM)
-
(1)
Fully connected layer (35 units, ReLU activation)
-
(2)
Dropout layer (20 % dropout)
-
(3)
Fully connected layer (30 units, ReLU activation)
-
(4)
Dropout layer (20 % dropout)
-
(5)
Activation layer (ReLU activation)
-
(6)
Output layer (1 units, sigmoid activation)
2.3.10. Densely connected neural networks with PSSM400 (DNNs- PSSM400)
-
(1)
Fully connected layer (350 units, ReLU activation)
-
(2)
Dropout layer (20 % dropout)
-
(3)
Activation layer (ReLU activation)
-
(4)
Output layer (1 units, sigmoid activation)
2.3.11. Densely connected neural networks with DP-PSSM (DNNs- DP-PSSM)
-
(1)
Fully connected layer (200 units, ReLU activation)
-
(2)
Dropout layer (20 % dropout)
-
(3)
Fully connected layer (150 units, ReLU activation)
-
(4)
Dropout layer (20 % dropout)
-
(5)
Fully connected layer (100 units, ReLU activation)
-
(6)
Dropout layer (20 % dropout)
-
(7)
Activation layer (ReLU activation)
-
(8)
Output layer (1 units, sigmoid activation)
2.3.12. Densely connected neural networks with DFMCA_PSSM (DNNs- DFMCA_PSSM)
-
(1)
Fully connected layer (200 units, ReLU activation)
-
(2)
Dropout layer (20 % dropout)
-
(3)
Fully connected layer (150 units, ReLU activation)
-
(4)
Dropout layer (20 % dropout)
-
(5)
Activation layer (ReLU activation)
-
(6)
Output layer (1 units, sigmoid activation)
2.3.13. Densely connected neural networks with SVD_PSSM (DNNs- SVD_PSSM)
-
(1)
Fully connected layer (15 units, ReLU activation)
-
(2)
Dropout layer (20 % dropout)
-
(3)
Fully connected layer (10 units, ReLU activation)
-
(4)
Dropout layer (20 % dropout)
-
(5)
Fully connected layer (5 units, ReLU activation)
-
(6)
Dropout layer (20 % dropout)
-
(7)
Activation layer (ReLU activation)
-
(8)
Output layer (1 units, sigmoid activation)
2.3.14. Densely connected neural networks with Single_Average (DNNs- Single_Average)
-
(1)
Fully connected layer (350 units, ReLU activation)
-
(2)
Dropout layer (20 % dropout)
-
(3)
Fully connected layer (300 units, ReLU activation)
-
(4)
Dropout layer (20 % dropout)
-
(5)
Fully connected layer (250 units, ReLU activation)
-
(6)
Dropout layer (20 % dropout)
-
(7)
Fully connected layer (200 units, ReLU activation)
-
(8)
Dropout layer (20 % dropout)
-
(9)
Fully connected layer (150 units, ReLU activation)
-
(10)
Dropout layer (20 % dropout)
-
(11)
Activation layer (ReLU activation)
-
(12)
Output layer (1 units, sigmoid activation)
2.4. Ensemble deep learning framework design
An ensemble deep learning framework was developed to effectively improve the performance of the base model and has been successfully applied to the classification of various biological sequences in recent years [53], [54], [55], [56], [57]. In present study, the ensemble DL model was established depending on the kind of input dataset. The ATG primary sequences were coded using the dictionary-embedding representation. The coding array was employed in the CNN and CNN-RNN layers to automatically gain sequence information, which not only includes the local information from the convolution layers, but also the global information from RNN layers. Subsequently, the DNN layers were utilized as the projection layers to deal with the 9 discrete features. In this process, all 9 PSSM-based descriptors were used for prediction independently. Ultimately, by performing multiple concatenation operations in the descriptor dimension, we tested all possible combinations of the sequential and discrete descriptors for prediction. While different feature combinations of DNN were chosen as additional information, connecting to the last fully connected layer of the core architecture. Considering all possible combinations of the 9 PSSM-based descriptors (AAC-PSSM, DP-PSSM, DPC-PSSM, PSe-PSSM, PSSM-AC, PSSM400, SVD_PSSM, Single_Average, and DFMCA_PSSM), 511 (i.e., 29-1) DNN models were built to link to the core architecture. To find the best performing core architecture, we used the same training dataset to test the performance of five sequence-level DL models (CNN, BiLSTM, BiGRU, CNN-BiLSTM, and CNN-BiGRU). Later, the core architecture with the best performance was selected to connect the DNN models. For all models, regardless of whether they are single or multi-model, during the training phase, the data were initially re-shuffled and then randomly divided into three segments: training (70 %), validation (10 %), and testing (20 %). The validation set served the purpose of assessing the binary cross-entropy loss after each epoch, while the test set was employed for model evaluation. To mitigate the influence of random variables in data sampling and model fitting, each training process was replicated five times. The final prediction was derived by averaging the results from these five separately trained models. As a result, a total of 2625 model training sessions were conducted, with 70 performed for an individual model and 2555 for the ensemble deep learning framework.
2.5. Implementation
We employed the autoBioSeqpy tool [58] with the Keras backend [59] to design, train, and evaluate both the individual deep learning models and the ensemble deep learning framework mentioned above. The experimentation was conducted on a workstation running Windows 10, equipped with an NVIDIA GeForce RTX GPU and CUDA 10.2.95.
2.6. Performance measurement
Several standard metrics and plots are employed, including accuracy (ACC), precision (PRE), F value, recall, specificity (SPE), Matthew’s correlation coefficient (MCC), receiver operating characteristic (ROC), precision-recall (PR) accuracy and loss (acc-loss) curves. All metrics are stated as below.
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
where TP, TN, FP, and FN are the numbers of true positives, true negatives, false positives, and false negatives, respectively, of a binary classification. ROC is utilized to make a visual performance comparison, and PR plots precision against the recall at all possible thresholds. The areas under the two curves fall in the interval of [0,1] (0.5 = random choice, 1 = perfect classification), which can reflect the overall performance of the model.
3. Results
3.1. Performance comparison of different single DL models
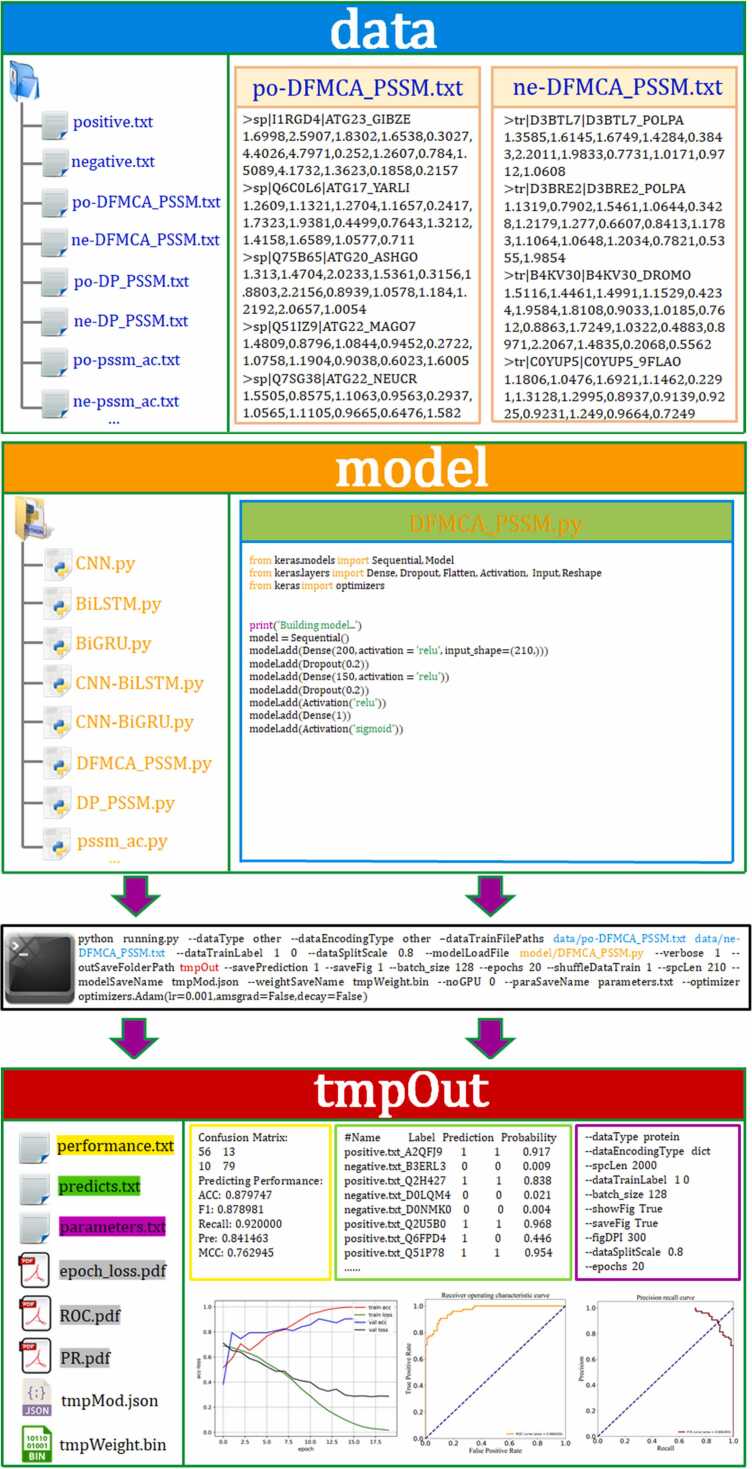
The performance of 14 single DL models was first estimated, including 1 CNN model, 2 RNN models, 2 CNN-RNN models and 9 DNN models. In order to achieve optimal performance on the training dataset, we fine-tuned the hyperparameters and optimized the architectures of each individual DL model. Next, we will provide a detailed explanation of how to implement and train these individual models in autoBioSeqpy by presenting two representative examples. Fig. 1 first shows an example view of the CNN in use. The protein primary sequence was designated as the input data type (--dataType protein). Using the parameter “--dataEncodingType dict”, each sequence was transformed as a 2D vector (1, L), in which L stands for the length of a protein sequence. To match the longest ATG and non-ATG in the training dataset, we set the protein sequence length to 2000 (--spcLen 2000). The autoBioSeqpy splits the input datasets into training-validation and test sets, with a split scale of 0.8 and stratification by category, using the “--dataSplitScale” parameter. Specifically, label 1 represents positive samples (ATGs), and label 0 represents negative samples (non-ATGs), which are stratified using the “--dataTrainLabel 1 0″ parameter. To avoid overfitting and ensure that the model is exposed to different examples at each run, the order of the training data, including positive and negative samples, is shuffled using the “--shuffleDataTrain 1″ parameter. During training, the model was trained for 20 epochs with a batch size of 128 using the “--epochs 20″ and “--batch_size 128″ parameters, respectively. The training process was performed on an NVIDIA GeForce RTX 3070 GPU, as specified by the “--noGPU 0″ parameter. The Adam optimizer was used to minimize the categorical cross-entropy loss between the target and predicted outputs, and the learning rate of the optimizer was set to 0.001 using the “--optimizer optimizers.Adam(lr=0.001,amsgrad=False, decay=False)” parameter. Once the training is completed, autoBioSeqpy saves all result files, including prediction performance, prediction probabilities, command-line arguments, epoch-loss curve, ROC curve, PR curve, and model weights, in the “tmpOut” folder (--outSaveFolderPath tmpOut). It’s worth noting that the above example of the CNN model can also be applied to other sequence-level deep learning models, such as BiLSTM, BiGRU, CNN-BiGRU, and CNN-BiLSTM. This means that users don’t need to modify most of the command-line arguments, they only need to change the model file name, for instance, from ‘CNN.py’ to ‘BiLSTM.py’. Experimental results of all five sequence-level DL models are depicted in Table 1. Obviously, the CNN model with dictionary encoding gained the best overall performance among the five models, providing the highest average scores of ACC (87.0 %), F value (87.1 %), Recall (88.9 %), SEP (85.0 %), PRE (85.4 %), and MCC (0.740). This architecture is composed of seven layers such as the input, embedding, dropout, convolutional, global max pooling, fully connected, and output layers in order. The input sequence is converted into a 128-dimensional vector by the dictionary encoding method in the embedding layer. To avoid over-fitting, a dropout of 0.2 is set to the embedding layer. A total of 250 filters exist in the convolutional layer, and the kernel size is assigned as 13. After that, the global max pooling is carried out to acquire the maximum value for each filter. After the pooling operation, the resulting features are passed to a fully connected layer with 125 hidden neurons. The output of this layer is then fed into a sigmoid activation function to obtain the final prediction probability.
Fig. 1.
The use of autoBioSeqpy is illustrated in a schematic diagram. The “data” fold requires the user to provide the data that will be used for modeling in fasta format, while the “model” fold necessitates the provision of the neural network model as Python code. Upon completing these prerequisites, the user can perform a series of operations by using special commands from the command line. These operations include data reading, sequence encoding, model loading, initialization, training, prediction, evaluation, and visualization. Once completed, all the results are stored in the tmpOut folder, which includes prediction results, prediction probabilities, training parameters, and various graphical representations.
Table 1.
Performance comparison of different deep learning models on the training dataset.
| Encoding | Architecture | ACC (%) | F-value (%) | Recall (%) | SEP (%) | PRE (%) | MCC |
|---|---|---|---|---|---|---|---|
| Dictionary | CNN | 87.0 ± 1.6 | 87.1 ± 2.0 | 88.9 ± 2.5 | 85.0 ± 1.8 | 85.4 ± 4.3 | 0.740 ± 0.032 |
| Dictionary | RNN (BiLSTM) | 76.5 ± 3.6 | 77.5 ± 4.0 | 85.4 ± 8.2 | 63.1 ± 9.4 | 71.6 ± 5.5 | 0.546 ± 0.057 |
| Dictionary | RNN (BiGRU) | 75.1 ± 1.5 | 76.0 ± 5.8 | 80.9 ± 16.4 | 69.6 ± 8.4 | 74.1 ± 6.7 | 0.519 ± 0.035 |
| Dictionary | CNN-RNN (BiLSTM) | 79.5 ± 0.6 | 79.6 ± 2.6 | 83.8 ± 7.3 | 74.7 ± 5.2 | 76.2 ± 2.5 | 0.595 ± 0.016 |
| Dictionary | CNN-RNN (BiGRU) | 81.6 ± 3.3 | 83.1 ± 2.8 | 86.9 ± 5.0 | 76.5 ± 4.3 | 79.6 ± 2.2 | 0.635 ± 0.070 |
| AAC-PSSM | DNN | 60.2 ± 7.0 | 65.1 ± 7.6 | 77.9 ± 19.7 | 47.4 ± 14.2 | 58.4 ± 6.2 | 0.250 ± 0.131 |
| DPC-PSSM | DNN | 90.2 ± 3.7 | 90.7 ± 3.3 | 94.9 ± 2.7 | 86.8 ± 2.8 | 87.0 ± 5.0 | 0.809 ± 0.070 |
| PSSM-AC | DNN | 92.0 ± 2.0 | 92.0 ± 2.2 | 91.6 ± 4.3 | 94.3 ± 3.7 | 92.4 ± 2.2 | 0.841 ± 0.039 |
| PSe-PSSM | DNN | 69.9 ± 6.4 | 72.6 ± 6.6 | 80.4 ± 13.4 | 61.8 ± 9.1 | 66.8 ± 3.2 | 0.416 ± 0.146 |
| PSSM400 | DNN | 81.8 ± 3.3 | 80.5 ± 5.1 | 76.5 ± 10.7 | 86.1 ± 7.5 | 86.1 ± 5.9 | 0.645 ± 0.069 |
| DP-PSSM | DNN | 83.8 ± 3.9 | 84.0 ± 4.6 | 88.4 ± 11.5 | 75.9 ± 9.2 | 81.6 ± 8.6 | 0.694 ± 0.059 |
| DFMCA_PSSM | DNN | 82.9 ± 8.8 | 84.3 ± 5.6 | 91.3 ± 4.9 | 71.4 ± 4.6 | 79.0 ± 9.6 | 0.665 ± 0.171 |
| SVD_PSSM | DNN | 63.4 ± 10.1 | 69.2 ± 6.5 | 76.0 ± 16.0 | 56.9 ± 13.3 | 65.5 ± 9.1 | 0.329 ± 0.193 |
| Single_Average | DNN | 82.5 ± 1.9 | 78.4 ± 2.5 | 69.1 ± 9.4 | 89.4 ± 5.9 | 93.1 ± 10.8 | 0.674 ± 0.038 |
Fig. 2 shows another use case and example application of autoBioSeqpy. The DNN model used feature vectors as input data format (--dataType other). To enable this, the “--dataEncodingType” parameter was set to “other” in autoBioSeqpy. This turns off the built-in encoding method (dictionary) and allows the program to read in the externally calculated features directly. The 210-dimensional DFMCA_PSSM features were calculated and used as input to the DNN model. The example of the DNN model is also applicable to other PSSM-based features, and the results of all single DNN models with different PSSM-based descriptors are listed in Table 1. Clearly, PSSM-AC got the best overall predictive performance and offered the highest average ACC (92.0 %), F value (92.0 %), SEP (94.3 %) and MCC (0.841). This architecture is established from three fully connected layers with 150, 100, and 50 hidden units each. Two dropout interfaces exist between the three layers, the dropout probability of which are set to 0.2. Furthermore, the DPC-PSSM model, which also comprises three fully connected layers with 350, 300, and 250 hidden units, achieved the second-best prediction performance, with an average ACC of 90.2 %, F value (90.7 %), and MCC of 0.809 (Table 1).
Fig. 2.
The autoBioSeqpy supports the input of external features, in addition to the fasta format, as depicted in a manner similar to Fig. 1.
3.2. The ensemble deep learning framework further improve predictive performance
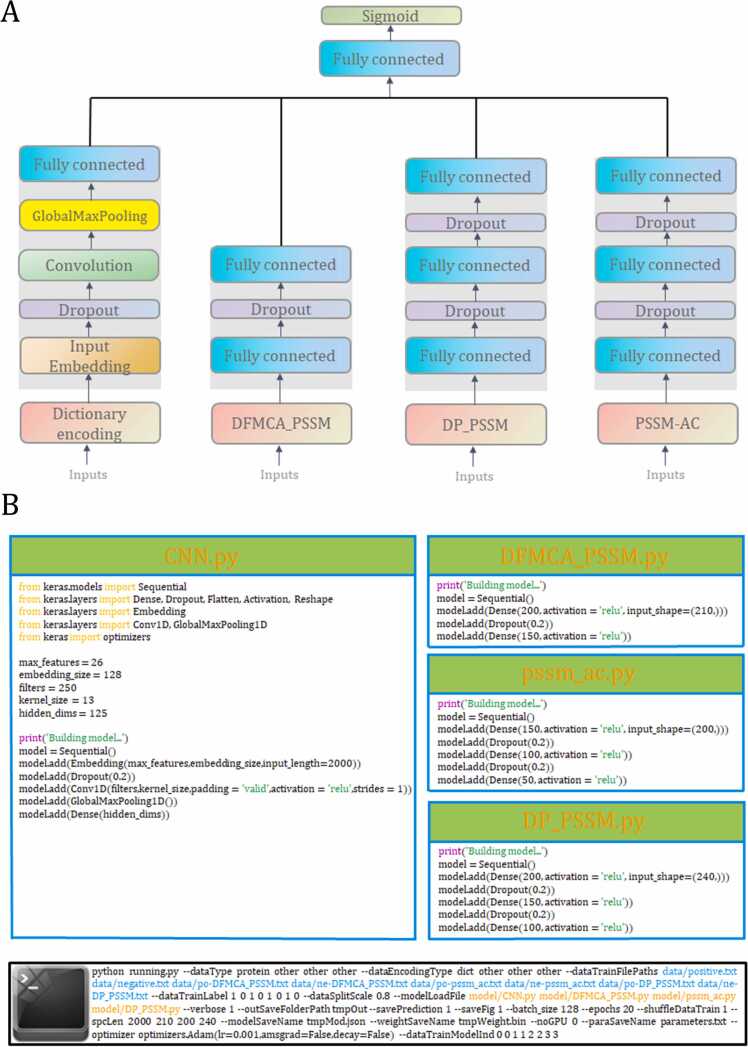
Following a thorough analysis of the prediction results obtained from the 14 individual deep learning models, we propose an ensemble deep learning framework that effectively combines the strengths of various DL algorithms. All possible combinations of the 9 PSSM-based descriptors (AAC-PSSM, DP-PSSM, DPC-PSSM, PSe-PSSM, PSSM-AC, PSSM400, SVD_PSSM, Single_Average, and DFMCA_PSSM) were generated using the concatenated DNN architectures and the best performing core architecture (CNN with dictionary encoding). By doing this, a total of 511 combinations were obtained. For each combination, the training-testing procedure was also repeated five times, and the average of outputs was considered as the final result for comparison. The performance of the ensemble deep learning framework with different combinations of descriptors is outlined in Table S3. It is clear that different performances could be achieved from different combinations of descriptors, and the ensemble deep learning framework outperforms any of single DL models. The ensemble deep learning framework with the ‘DFMCA_PSSM+DP-PSSM+PSSM-AC’ descriptor group gained the best results, providing an average ACC (96.2 %), F value (96.0 %), Recall (95.8 %), SEP (96.6%), PRE (96.4 %), and MCC (0.924) (Table 2). Fig. 3A depicts the optimal ensemble deep learning framework. In practical applications, the new version of autoBioSeqpy supports the use of multiple models for different types of data. The output layers of each model are merged into a new output layer through a dense layer. For example, if users want to integrate the above single models to construct the ensemble framework, they can use the following command-line commands (Fig. 3B). The parameter ‘--dataTrainModelInd’ is designed to specify which model the dataset belongs to, so the value should not be greater than the model’s index. Specifically, the parameter ‘0 0 1 1 2 2 3 3′ after ‘--dataTrainModelInd’ means that the first two dataset files (positive.txt and negative.txt) will be fed into the first model file (CNN.py), and the remaining six dataset files (po-DFMCA_PSSM.txt, ne-DFMCA_PSSM.txt, po-pssm_ac.txt, ne-pssm_ac.txt, po-DP_PSSM.txt, and ne-DP_PSSM.txt) will be fed into DFMCA_PSSM.py, pssm_ac.py, and DP_PSSM.py, respectively.
Table 2.
Performance of the best ensemble deep learning framework on the training dataset.
| Round | ACC (%) | F-value (%) | Recall (%) | SEP (%) | PRE (%) | MCC |
|---|---|---|---|---|---|---|
| 1 | 95.6 | 95.6 | 96.2 | 95.1 | 95.1 | 0.911 |
| 2 | 96.2 | 96.0 | 96.0 | 96.2 | 96.0 | 0.924 |
| 3 | 96.2 | 95.6 | 93.0 | 98.9 | 98.5 | 0.924 |
| 4 | 96.2 | 96.1 | 97.4 | 95.0 | 94.9 | 0.924 |
| 5 | 96.8 | 96.9 | 96.3 | 97.6 | 97.5 | 0.937 |
| Average | 96.2 | 96.0 | 95.8 | 96.6 | 96.4 | 0.924 |
Fig. 3.
The computational framework of the optimal ensemble deep learning architecture. (A) The optimal ensemble framework leverages the power of multiple neural network models to achieve higher prediction accuracy and robustness. The framework consists of several neural network architectures, which are arranged in a specific order from left to right: Convolutional Neural Network (CNN), Deep Neural Networks (DNNs) using DFMCA_PSSM, DNNs using PSSM-AC, and DNNs using DP-PSSM, respectively. (B) An example that implements the architectures described in (A) is available using Keras libraries and autoBioSeqpy. See ‘Materials and methods’ section for more details.
3.3. Performance evaluation of the ensemble deep learning framework with the independent test dataset
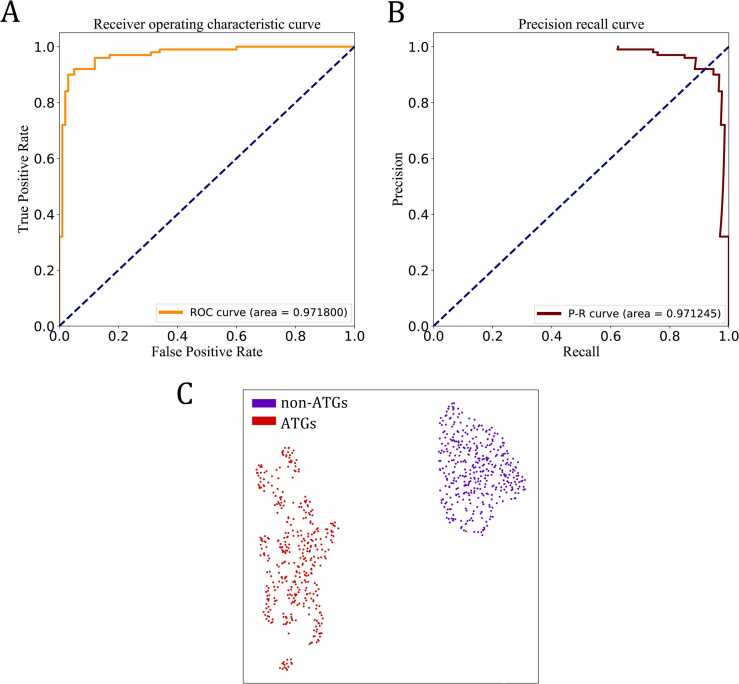
To assess the ability of the proposed ensemble deep learning framework to generalize, we tested it on an independent dataset consisting of 100 ATGs and 100 non-ATGs. Our ensemble approach exhibits excellent performance, providing an overall ACC of 94.5 %, F value of 94.4 %, Recall of 93.0 %, SEP of 96.0 %, PRE of 95.9 %, and MCC of 0.890. Moreover, the ROC and PR curves are selected to further estimate its performance (Fig. 4A and B), and the concrete values of the area under the two curves are also given here (0.972 for ROC and 0.971 for PR). Finally, we used layerUMAP [60] to dissect the ensemble deep learning framework. Fig. 4C displays the 2D UMAP maps that represent the distribution of ATGs (labeled as 1 and shown in red) and non-ATGs (labeled as 0 and shown in purple) in the latent space of the last hidden layer. The UMAP parameters n_neighbors, min_dist, and metric were set to 28, 0.8, and cosine, respectively, spread out the distribution of data points in the projection (Fig. S4). We observed that the features extracted by the ensemble deep learning framework make a clear separation between ATGs and non-ATGs. Taken together, these results validate the effectiveness and stability of the ensemble deep learning approach, which is called EnsembleDL-ATG in this work.
Fig. 4.
Visualizations generated by using autoBioSeqpy to train and apply the best ensemble deep learning model (The architecture are shown in Fig. 3) to the independent test dataset. (A and B) Performance of the best ensemble deep learning framework when differentiating 100 ATGs from 100 non-ATGs, obtaining AUC values of 0.9718 and 0.9712 for the ROC and PR curves, respectively. (C) UMAP visualization of the last hidden layer representations in the ensemble framework for two protein classes.
3.4. Performance comparison of EnsembleDL-ATG with the state-of-the-art method
Due to training and testing on the same datasets, we compared the performance of our proposed EnsembleDL-ATG with the exiting tool ATGPred-FL. Table 3 shows the prediction results of these two methods, the latter of which are acquired from Jiao et al.’ work [41]. Clearly, EnsembleDL-ATG yields higher ACC (96.2 % and 94.5 %), Recall (95.8 % and 93.0 %), SEP (96.6 %, 96.0 %) and MCC (0.924 and 0.890) scores for the training and independent test datasets, respectively. In addition to the comparison of predictive performance, we further analyzed the differences in the prediction results between the two methods (Table S4). We identified five ATG proteins that were missed by ATGPred-FL but correctly predicted by EnsembleDL-ATG. These proteins are ULK1, ATG7, ATG9A, BECN2, and ATG9. Further analysis of the functions of these proteins revealed that they are all classic autophagy-related proteins (Table S5). The comparison results again demonstrate the strong predictive power of our ensemble deep learning framework for ATGs.
Table 3.
Performance comparison of EnsembleDL-ATG and ATGPred-FL on the same training and independent test datasets.
| Tool | ACC (%) | F-value (%) | Recall (%) | SEP (%) | PRE (%) | MCC |
|---|---|---|---|---|---|---|
| Training dataset | ||||||
| ATGPred-FL | 94.4 | - | 94.2 | 94.7 | - | 0.888 |
| EnsembleDL-ATG | 96.2 | 96.0 | 95.8 | 96.6 | 96.4 | 0.924 |
| Independent test dataset | ||||||
| ATGPred-FL | 90.5 | 91.8 | 89.0 | 92.0 | 90.4 | 0.810 |
| EnsembleDL-ATG | 94.5 | 94.4 | 93.0 | 96.0 | 95.9 | 0.890 |
4. Discussion and conclusion
As a self-degradative and evolutionarily conserved process, autophagy plays essential roles in multiple cellular physiological activities, and has been associated with various human diseases. This process is regulated by autophagy proteins (ATGs), which are involved in a variety of diseases and have received increasing attention in recent years [61], [62], [63], [64]. Several useful databases have been developed to advance the study of ATGs and their functions, such as the aforementioned ARN [35], ncRDeathDB [36], GAMDB [37], iLIR [38], and THANATOS [39]. Meanwhile, network biology approaches and multiomics techniques are commonly used for the systematic study of ATGs, but only one machine learning tool (ATGPred-FL) has been proposed to identify ATGs from protein sequences thus far [41].
In the present study, our aim is to propose a new computational tool for identifying ATGs using deep learning algorithms. Deep learning is widely recognized for its remarkable ability to approximate nearly any function and has shown impressive predictive accuracy that often exceeds that of human experts [65]. Despite the remarkable accuracy of deep learning models, they have certain limitations. These models are prone to overfitting, which may lead to high variance, and they can easily get stuck in local optima during training. To overcome these challenges, ensemble methods that combine the predictions of multiple deep learning models have been shown to achieve superior generalizability compared to a single model [66], [67], [68]. Therefore, we try to adopt the idea of ensemble deep learning, where multiple and often independent deep learning models are combined to enable multifaceted abstraction of data. To establish an effective ensemble deep learning framework, six different types of neural network architectures and nine PSSM-derived features were first employed to develop the single-model classifier for ATG prediction. Subsequently, an enumeration strategy was adopted to optimize the ensemble framework, and the final prediction model EnsembleDL-ATG was trained on the aggregation of a CNN model and three DNN models (DFMCA_PSSM, DP-PSSM, and PSSM-AC). The results suggest that the ensemble framework combining heterogeneous models leads to better learning outcomes. A comparative study of the proposed EnsembleDL-ATG with the previous method was conducted to validate the efficiency of the ensemble deep learning strategy.
Although EnsembleDL-ATG has achieved highly competitive performance for the identification of ATGs, we believe that there is still room for further improvement, and more work should be focused on the study of ATGs and their functions. To ascertain the real-world effectiveness of EnsembleDL-ATG, we undertook a rigorous reprocessing of the training data, resulting in a less redundant dataset with an inherent imbalance between ATGs and non-ATGs. Subsequently, we retrained EnsembleDL-ATG on this updated dataset. Notably, we observed that EnsembleDL-ATG’s performance remained unaltered throughout this process, reaffirming the robustness and reliability of our methodology (Table S6 and S7). ATGs can regulate the autophagic process by forming various complexes through interactions with other proteins, but there are still many questions regarding the specific interactions and coordination processes. These questions are the focus of our next research.
To summarize, our proposed ensemble deep learning method effectively predicts ATGs by incorporating the strengths of multiple DL models and achieving better generalizability. By leveraging primary sequences and evolutionary information, our method has the potential to improve our understanding of autophagy, diagnose and treat related diseases, and facilitate the design of novel drugs.
CRediT authorship contribution statement
Lezheng Yu: Conceptualization, Methodology, Data curation, Formal analysis, Writing - original draft, Writing - review & editing, Funding acquisition. Yonglin Zhang: Software, Formal analysis, Validation, Visualization, Investigation. Li Xue: Formal analysis, Validation, Supervision. Fengjuan Liu: Formal analysis, Supervision. Runyu Jing: Methodology, Software, Formal analysis, Validation, Visualization, Writing - review & editing. Jiesi Luo: Conceptualization, Methodology, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 22203057), Fund of Science and Technology Department of Guizhou Province ([2017]5790–07), Natural Science Foundation of Department of Education of Guizhou Province ([2021]021), Joint project of Luzhou Municipal People’s Government and Southwest Medical University (2020LZXNYDJ39), Research and Development Fund Project of Sichuan North Medical College (CBY22-QNA38).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.09.036.
Contributor Information
Runyu Jing, Email: jingry@scu.edu.cn.
Jiesi Luo, Email: ljs@swmu.edu.cn.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Data availability
The source code and benchmark data of our method are available at https://github.com/jingry/autoBioSeqpy/tree/2.0/examples/EnsembleDL-ATG.
References
- 1.Jiang P., Mizushima N. Autophagy and human diseases. Cell Res. 2014;24:69–79. doi: 10.1038/cr.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B., Klionsky D.J. Autophagy wins the 2016 Nobel Prize in physiology or medicine: breakthroughs in baker's yeast fuel advances in biomedical research. Proc Natl Acad Sci USA. 2017;114:201–205. doi: 10.1073/pnas.1619876114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dash S., Aydin Y., Moroz K. Chaperone-mediated autophagy in the liver: good or bad? Cells. 2019;8:1308. doi: 10.3390/cells8111308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L., Klionsky D.J., Shen H.M. The emerging mechanisms and functions of microautophagy. Nat Rev Mol Cell Biol. 2023;24:186–203. doi: 10.1038/s41580-022-00529-z. [DOI] [PubMed] [Google Scholar]
- 7.Kaushik S., Cuervo A.M. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19:365–381. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condello M., Pellegrini E., Caraglia M., Meschini S. Targeting autophagy to overcome human diseases. Int J Mol Sci. 2019;20:725. doi: 10.3390/ijms20030725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdrakhmanov A., Gogvadze V., Zhivotovsky B. To eat or to die: deciphering selective forms of autophagy. Trends Biochem Sci. 2020;45:347–364. doi: 10.1016/j.tibs.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Guan X., Iyaswamy A., Sreenivasmurthy S.G., Su C., Zhu Z., Liu J., et al. Mechanistic insights into selective autophagy subtypes in Alzheimer's disease. Int J Mol Sci. 2022;23:3609. doi: 10.3390/ijms23073609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider J.L., Cuervo A.M. Autophagy and human disease: emerging themes. Curr Opin Genet Dev. 2014;26:16–23. doi: 10.1016/j.gde.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klionsky D.J., Petroni G., Amaravadi R.K., Baehrecke E.H., Ballabio A., Boya P., et al. Autophagy in major human diseases. EMBO J. 2021;40 doi: 10.15252/embj.2021108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang F., Wang B.R., Wang Y.G. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol. 2018;24:4643–4651. doi: 10.3748/wjg.v24.i41.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onorati A.V., Dyczynski M., Ojha R., Amaravadi R.K. Targeting autophagy in cancer. Cancer. 2018;124:3307–3318. doi: 10.1002/cncr.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun C.W., Jeon J., Go G., Lee J.H., Lee S.H. The dual role of autophagy in cancer development and a therapeutic strategy for cancer by targeting autophagy. Int J Mol Sci. 2020;22:179. doi: 10.3390/ijms22010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia H., Green D.R., Zou W. Autophagy in tumour immunity and therapy. Nat Rev Cancer. 2021;21:281–297. doi: 10.1038/s41568-021-00344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei Y., Zhang E., Bai L., Li Y. Autophagy in cancer immunotherapy. Cells. 2022;11:2996. doi: 10.3390/cells11192996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng R., Weng S., Xu J., Li Z., Wang Y., Aizimuaji Z., et al. Autophagy and biotransformation affect sorafenib resistance in hepatocellular carcinoma. Comput Struct Biotechnol J. 2023;21:3564–3574. doi: 10.1016/j.csbj.2023.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan C.C., Yu J.T., Tan M.S., Jiang T., Zhu X.C., Tan L. Autophagy in aging and neurodegenerative diseases: implications for pathogenesis and therapy. Neurobiol Aging. 2014;35:941–957. doi: 10.1016/j.neurobiolaging.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Li S., Le W. An insight review of autophagy biology and neurodegenerative diseases: machinery, mechanisms and regulation. Sci China Life Sci. 2017;60:1457–1459. doi: 10.1007/s11427-017-9214-7. [DOI] [PubMed] [Google Scholar]
- 21.Milner M.T., Maddugoda M., Götz J., Burgener S.S., Schroder K. The NLRP3 inflammasome triggers sterile neuroinflammation and Alzheimer's disease. Curr Opin Immunol. 2021;68:116–124. doi: 10.1016/j.coi.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Lu R., Zhang L., Yang X. Interaction between autophagy and the NLRP3 inflammasome in Alzheimer's and Parkinson's disease. Front Aging Neurosci. 2022;14 doi: 10.3389/fnagi.2022.1018848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabli I.K., Sancho-Shimizu V. Inborn errors of autophagy and infectious diseases. Curr Opin Immunol. 2021;72:272–276. doi: 10.1016/j.coi.2021.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Niu H., Deng M. Editorial: the role of autophagy in infectious diseases. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.1039282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y., Shen J., Ran Z. Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy. 2020;16:3–17. doi: 10.1080/15548627.2019.1603547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin M., Zhang Y. Autophagy and autoimmune diseases. Adv Exp Med Biol. 2020;1207:405–408. doi: 10.1007/978-981-15-4272-5_28. [DOI] [PubMed] [Google Scholar]
- 27.Lamb C.A., Yoshimori T., Tooze S.A. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 28.Zhuang X., Chung K.P., Jiang L. Origin of the autophagosomal membrane in plants. Front Plant Sci. 2016;7:1655. doi: 10.3389/fpls.2016.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esser L.M., Schmitz K., Hillebrand F., Erkelenz S., Schaal H., Stork B., et al. Phosphorylation of pICln by the autophagy activating kinase ULK1 regulates snRNP biogenesis and splice activity of the cell. Comput Struct Biotechnol J. 2023;21:2100–2109. doi: 10.1016/j.csbj.2023.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 31.Yu L., Chen Y., Tooze S.A. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14:207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis S., Wang J., Ferro-Novick S. Crosstalk between the secretory and autophagy pathways regulates autophagosome formation. Dev Cell. 2017;41:23–32. doi: 10.1016/j.devcel.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu G., Rios L., Yan Z., Jasper A.M., Luera D., Luo S., et al. Autophagy regulator Atg9 is degraded by the proteasome. Biochem Biophys Res Commun. 2020;522:254–258. doi: 10.1016/j.bbrc.2019.11.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rožman S., Yousefi S., Oberson K., Kaufmann T., Benarafa C., Simon H.U. The generation of neutrophils in the bone marrow is controlled by autophagy. Cell Death Differ. 2015;22:445–456. doi: 10.1038/cdd.2014.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Türei D., Földvári-Nagy L., Fazekas D., Módos D., Kubisch J., Kadlecsik T., et al. Autophagy regulatory network - a systems-level bioinformatics resource for studying the mechanism and regulation of autophagy. Autophagy. 2015;11:155–165. doi: 10.4161/15548627.2014.994346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu D., Huang Y., Kang J., Li K., Bi X., Zhang T., et al. ncRDeathDB: a comprehensive bioinformatics resource for deciphering network organization of the ncRNA-mediated cell death system. Autophagy. 2015;11:1917–1926. doi: 10.1080/15548627.2015.1089375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., Xie T., Tian M., Li J., Song S., Ouyang L., et al. GAMDB: a web resource to connect microRNAs with autophagy in gerontology. Cell Prolif. 2016;49:246–251. doi: 10.1111/cpr.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacomin A.C., Samavedam S., Promponas V., Nezis I.P. iLIR database: a web resource for LIR motif-containing proteins in eukaryotes. Autophagy. 2016;12:1945–1953. doi: 10.1080/15548627.2016.1207016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng W., Ma L., Zhang Y., Zhou J., Wang Y., Liu Z., et al. THANATOS: an integrative data resource of proteins and post-translational modifications in the regulation of autophagy. Autophagy. 2018;14:296–310. doi: 10.1080/15548627.2017.1402990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng L., Zeng Y., Hu S., Zhang N., Cheung K.C.P., Li B., et al. Systematic prediction of autophagy-related proteins using Arabidopsis thaliana interactome data. Plant J. 2021;105:708–720. doi: 10.1111/tpj.15065. [DOI] [PubMed] [Google Scholar]
- 41.Jiao S., Chen Z., Zhang L., Zhou X., Shi L. ATGPred-FL: sequence-based prediction of autophagy proteins with feature representation learning. Amino Acids. 2022;54:799–809. doi: 10.1007/s00726-022-03145-5. [DOI] [PubMed] [Google Scholar]
- 42.Kumari B., Kumar R., Kumar M. PalmPred: an SVM based palmitoylation prediction method using sequence profile information. PLOS One. 2014;9 doi: 10.1371/journal.pone.0089246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jing R., Wen T., Liao C., Xue Li, Liu F., Yu L., et al. DeepT3 2.0: improving type III secreted effector predictions by an integrative deep learning framework. NAR Genom Bioinform. 2021;3 doi: 10.1093/nargab/lqab086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:480–489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49:412–419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veltri D., Kamath U., Shehu A. Deep learning improves antimicrobial peptide recognition. Bioinformatics. 2018;34:2740–2747. doi: 10.1093/bioinformatics/bty179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo J., Yu L., Guo Y., Li M. Functional classification of secreted proteins by position specific scoring matrix and auto covariance. Chemom Intell Lab Syst. 2012;110:163–167. doi: 10.1016/j.chemolab.2011.11.008. [DOI] [Google Scholar]
- 49.Yu L., Liu F., Li Y., Luo J., Jing R. DeepT3_4: a hybrid deep neural network model for the distinction between bacterial type III and IV secreted effectors. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.605782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu L., Xue L., Liu F., Li Y., Jing R., Luo J. The applications of deep learning algorithms on in silico druggable proteins identification. J Adv Res. 2022;41:219–231. doi: 10.1016/j.jare.2022.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul S.F., Koonin E.V. Iterated profile searches with PSI-BLAST -- a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/S0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 52.Mohammadi A., Zahiri J., Mohammadi S., Khodarahmi M., Arab S.S. PSSMCOOL: a comprehensive R package for generating evolutionary-based descriptors of protein sequences from PSSM profiles. Biol Methods Protoc. 2022;7 doi: 10.1093/biomethods/bpac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J., Zhao Y., Gong W., Liu Y., Wang M., Huang X., et al. EDLMFC: an ensemble deep learning framework with multi-scale features combination for ncRNA-protein interaction prediction. BMC Bioinform. 2021;22 doi: 10.1186/s12859-021-04069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L., Li G., Li X., Wang H., Chen S., Liu H. EDLm6APred: ensemble deep learning approach for mRNA m6A site prediction. BMC Bioinform. 2021;22 doi: 10.1186/s12859-021-04206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H., Liu H., Huang T., Li G., Zhang L., Sun Y. EMDLP: Ensemble multiscale deep learning model for RNA methylation site prediction. BMC Bioinform. 2022;23 doi: 10.1186/s12859-022-04756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akpokiro V., Martin T., Oluwadare O. EnsembleSplice: ensemble deep learning model for splice site prediction. BMC Bioinform. 2022;23 doi: 10.1186/s12859-022-04971-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aybey E., Gümüş Ö. SENSDeep: an ensemble deep learning method for protein-protein interaction sites prediction. Interdiscip Sci. 2022 doi: 10.1007/s12539-022-00543-x. [DOI] [PubMed] [Google Scholar]
- 58.Jing R., Li Y., Xue L., Liu F., Li M., Luo J. autoBioSeqpy: a deep learning tool for the classification of biological sequences. J Chem Inf Model. 2020;60:3755–3764. doi: 10.1021/acs.jcim.0c00409. [DOI] [PubMed] [Google Scholar]
- 59.Chollet F. Keras: Deep learning library for theano and tensorflow. 2015. https://keras.io/2015.
- 60.Jing R., Xue L., Li M., Yu L., Luo J. layerUMAP: a tool for visualizing and understanding deep learning models in biological sequence classification using UMAP. iScience. 2022;25 doi: 10.1016/j.isci.2022.105530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hain A.U.P., Bosch J. Autophagy in plasmodium, a multifunctional pathway? Comput Struct Biotechnol J. 2013;8 doi: 10.5936/csbj.201308002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heckmann B.L., Teubner B.J.W., Tummers B., Boada-Romero E., Harris L., Yang M., et al. LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer's disease. Cell. 2019;178:536–551. doi: 10.1016/j.cell.2019.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heckmann B.L., Teubner B.J.W., Boada-Romero E., Tummers B., Guy C., Fitzgerald P., et al. Noncanonical function of an autophagy protein prevents spontaneous Alzheimer's disease. Sci Adv. 2020;6 doi: 10.1126/sciadv.abb9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rickman A.D., Hilyard A., Heckmann B.L. Dying by fire: noncanonical functions of autophagy proteins in neuroinflammation and neurodegeneration. Neural Regen Res. 2022;17:246–250. doi: 10.4103/1673-5374.317958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 66.Lv L., Chen T., Dou J., Plaza A. A hybrid ensemble-based deep-learning framework for landslide susceptibility mapping. Int J Appl Earth Obs. 2022;108 doi: 10.1016/j.jag.2022.102713. [DOI] [Google Scholar]
- 67.Peng L., Wang C., Tian G., Liu G., Li G., Lu Y., et al. Analysis of CT scan images for COVID-19 pneumonia based on a deep ensemble framework with DenseNet, Swin transformer, and RegNet. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.995323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tian G., Wang Z., Wang C., Chen J., Liu G., Xu H., et al. A deep ensemble learning-based automated detection of COVID-19 using lung CT images and vision transformer and ConvNeXt. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1024104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Data Availability Statement
The source code and benchmark data of our method are available at https://github.com/jingry/autoBioSeqpy/tree/2.0/examples/EnsembleDL-ATG.