Abstract
The recent emergence of SARS-CoV-2 Omicron (B.1.1.529 lineage) variants possessing numerous mutations has raised concerns of decreased effectiveness of current vaccines, therapeutic monoclonal antibodies and antiviral drugs for COVID-19 against these variants1,2. The original Omicron lineage, BA.1, prevailed in many countries, but more recently, BA.2 has become dominant in at least 68 countries3. Here we evaluated the replicative ability and pathogenicity of authentic infectious BA.2 isolates in immunocompetent and human ACE2-expressing mice and hamsters. In contrast to recent data with chimeric, recombinant SARS-CoV-2 strains expressing the spike proteins of BA.1 and BA.2 on an ancestral WK-521 backbone4, we observed similar infectivity and pathogenicity in mice and hamsters for BA.2 and BA.1, and less pathogenicity compared with early SARS-CoV-2 strains. We also observed a marked and significant reduction in the neutralizing activity of plasma from individuals who had recovered from COVID-19 and vaccine recipients against BA.2 compared to ancestral and Delta variant strains. In addition, we found that some therapeutic monoclonal antibodies (REGN10987 plus REGN10933, COV2-2196 plus COV2-2130, and S309) and antiviral drugs (molnupiravir, nirmatrelvir and S-217622) can restrict viral infection in the respiratory organs of BA.2-infected hamsters. These findings suggest that the replication and pathogenicity of BA.2 is similar to that of BA.1 in rodents and that several therapeutic monoclonal antibodies and antiviral compounds are effective against Omicron BA.2 variants.
The Omicron variant of SARS-CoV-2, the virus responsible for COVID-19, was first detected in late November 2021 and has spread rapidly around the world. Omicron variants have been classified into four different sublineages: BA.1, BA.1.1, BA.2 and BA.3. The original Omicron lineage, BA.1, rapidly became the prevailing variant circulating in many countries; however, BA.2 variants have become dominant in at least 68 countries3. Moreover, the prevalence of BA.2 is increasing rapidly in several other countries including South Africa, Sweden, Austria, Singapore, Georgia and Sri Lanka (https://covariants.org/per-variant). Preliminary data indicate that the BA.2 variant may be more transmissible than the BA.1 variant5,6.
Recently, we and others have shown that BA.1 variants are less pathogenic in animal models than previously circulating variants of concern7–9 (VOC), consistent with preliminary clinical data in humans10. Moreover, other studies have reported that BA.1 variants show reduced sensitivity to vaccine- or infection-induced antibodies, as well as some therapeutic monoclonal antibodies11–15. The spike (S) protein of SARS-CoV-2 mediates viral receptor binding and membrane fusion, both of which are essential for viral infection of host cells. The S protein is also the principal antigen targeted by the host neutralizing antibody response16. Notably, mutations in the S protein, such as E484K, N501Y, D614G and P681H/R, have been shown to affect the infectivity, pathogenicity, transmissibility, species tropism and/or antigenicity of SARS-CoV-217–21. Compared with the reference strain Wuhan/Hu-1/2019, the BA.1 and BA.2 variants have 36 and 31 amino acid substitutions in the S protein, respectively. Although the BA.1 and BA.2 variants share 20 of these substitutions, BA.2 possesses 11 amino acid changes that are not found in BA.1. These findings suggest that the replicative capacity, pathogenicity, transmissibility and antigenicity of BA.2 variants may differ from those of BA.1 variants. Here we characterized the functional activity of BA.2 variants in vivo. In addition, we evaluated the efficacy of therapeutic monoclonal antibodies and antiviral drugs for COVID-19 against BA.2 variants in vivo.
BA.2 infection in mice
We isolated the following BA.2 variants in VeroE6/TMPRSS2 cells: hCoV-19/Japan/UT-NCD1288-2N/2022 (NCD1288), hCoV-19/Japan/UT-HP353-1N/2022 (HP353), hCoV-19/Japan/UT-HP354-1N/2022 (HP354), and hCoV-19/Japan/TY40-385/2022 (TY40-385). NCD1288 and TY40-385 were isolated from travellers arriving in Japan from India. HP353 and HP354 were isolated from residents in Japan. These isolates contain 31 amino acid changes in their S proteins compared to the reference strain Wuhan/Hu-1/2019. These differences include 7 changes in the N-terminal domain (NTD), including substitutions and deletions (T19L, Δ24-27S, G142D and V213G), 16 substitutions in the receptor-binding domain (RBD) (G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y and Y505H), the D614G mutation, three substitutions close to the furin cleavage site (H655Y, N679K and P681H), and four substitutions in the S2 region (N764K, D796Y, Q954H and N969K).
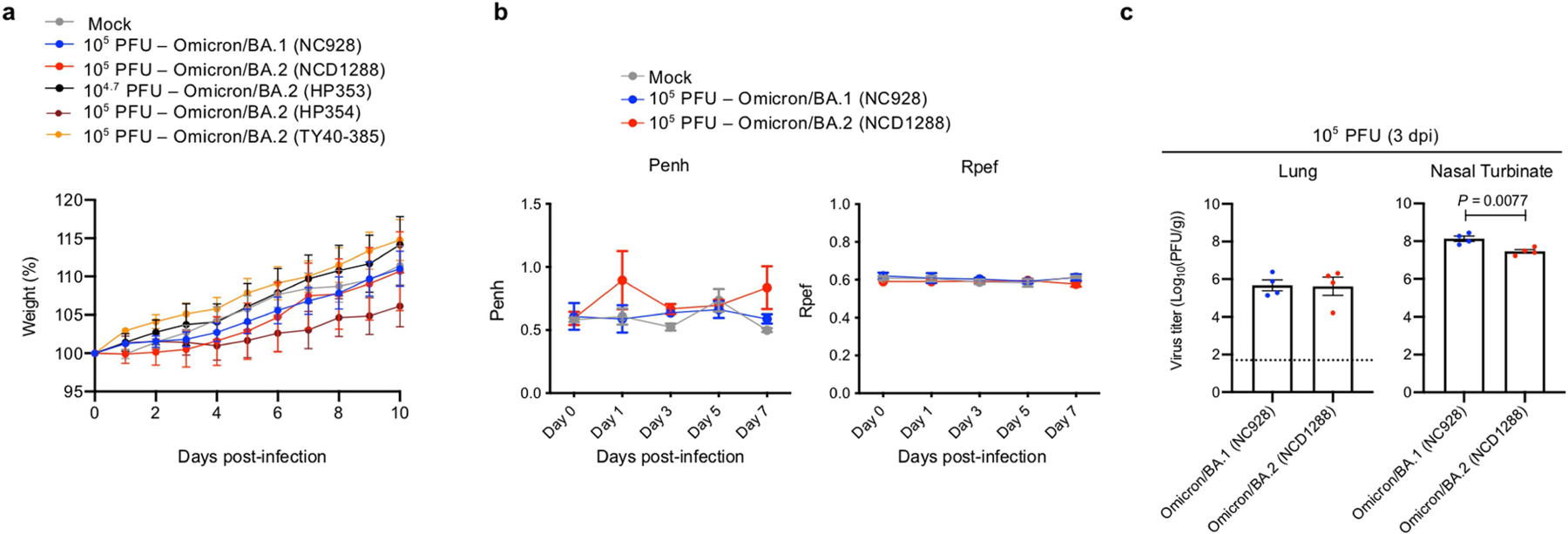
Given that the BA.2 variant possesses substitutions including K417N, E484K, and N501Y in its S protein, and that these amino acids substitutions are key for mouse adaptation22–24, we predicted that this variant would infect immunocompetent mice and replicate in their respiratory organs as is seen with BA.1 variants. We inoculated female BALB/c mice with 105 plaque-forming units (PFU) of BA.1 (NC928), BA.2 (NCD1288) or PBS (mock), and assessed their body weights for 10 days. Intranasal inoculation of BALB/c mice with BA.1 or BA.2 did not cause body weight reduction (Fig. 1a). We also measured pulmonary function in the infected mice by measuring Penh and Rpef, which are surrogate markers for bronchoconstriction and airway obstruction, respectively, using a whole-body plethysmography system. No changes were observed in the Penh or Rpef of the BA.1- or BA.2-infected groups compared with the mock-infected group at any timepoint after infection (Fig. 1b). By contrast, our previous data showed that B.1.351 (Beta variant), which replicated to a high titre in the lungs of wild-type BALB/c mice, caused a significant increase in Penh and a decrease in Rpef at 2 days post-infection7 (dpi). At 2 dpi, BALB/c mice infected with BA.1 or BA.2 exhibited similar viral titres in nasal turbinates; however, the mean virus titre of BA.2 in the lungs (mean titre 6.9 log10 PFU g−1) was slightly but significantly higher than that of BA.1 (mean titre 6.4 log10 PFU g−1) (Fig. 1c, left). At 5 dpi, the lung titres in the BA.2-infected group were lower (33-fold, P < 0.001) than those in the BA.1-infected group, although no differences in viral titres in the nasal turbinates were observed between the two groups at this timepoint (Fig. 1c, right).
Fig. 1 |. BA.2 and BA.1 show similar infectivity and pathogenicity in BALB/c mice.
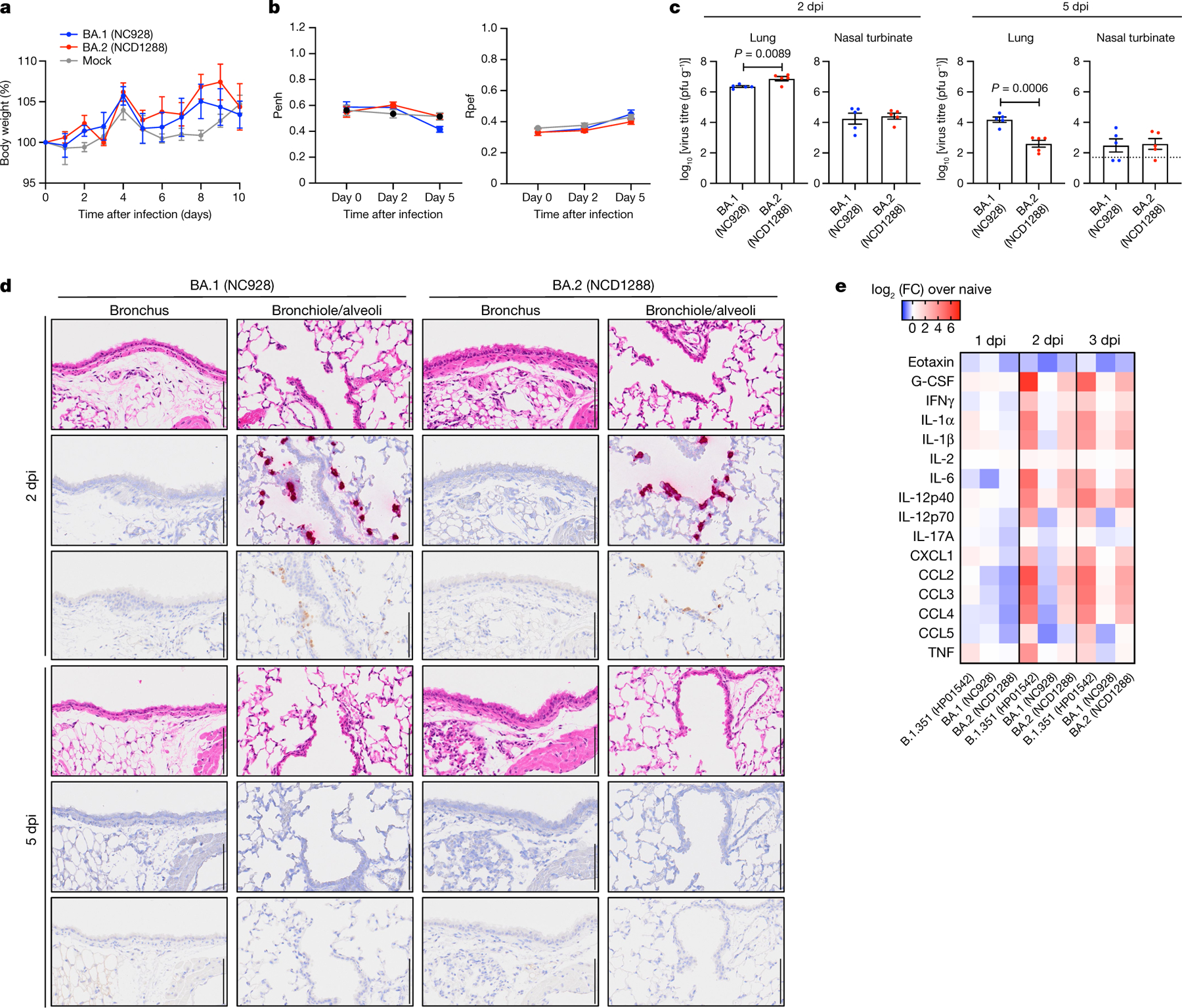
a–c, Mice were inoculated intranasally with 105 PFU BA.1 (NC928), BA.2 (NCD1288) or PBS (mock). a, Body weights of virus-infected (n = 5) and mock-infected (n = 5) mice were monitored daily for 10 days after viral infection. Data are mean percentage ± s.e.m. of the starting weight. b, Pulmonary function analyses in virus-infected (n = 5) and mock-infected (n = 5) mice. Penh and Rpef were measured by whole-body plethysmography. Data are mean ± s.e.m. c, Virus replication in infected mice. Mice (n = 5) were euthanized at 2 and 5 dpi for virus titration. Virus titres in the nasal turbinates and lungs were determined by plaque assay. Data are mean ± s.e.m.; points represent data from individual mice. The lower limit of detection is indicated by the horizontal dashed line. Data were analysed with the Mann–Whitney test. d, Histopathological examination of the lungs of infected mice. Three mice per group were infected with 105 PFU BA.1 (NC928) or BA.2 (NCD1288) and euthanized at 2 or 5 dpi for histopathological examination. Representative images of the bronchi, and bronchioles and alveoli of mice infected with BA.1 or BA.2 are shown. Top row, haematoxylin and eosin (H&E) staining. Middle row, in situ hybridization targeting the nucleocapsid gene of SARS-CoV-2. Bottom row, immunohistochemistry using a rabbit polyclonal antibody that detects SARS-CoV-2 nucleocapsid protein. Scale bars, 100 μm. e, Heat map of cytokine and chemokine concentrations in the lungs of mice (n = 4) infected with 105 PFU BA.1 (NC928), BA.2 (NCD1288) or Beta B.1.351 (HP01542) at 1, 2 and 3 dpi (see Extended Data Fig. 1). Data are from one experiment. FC, fold change.
We performed a histopathological analysis of the lungs of BALB/c mice infected with BA.1 (NC928) or BA.2 (NC1288). In both BA.1- and BA.2-infected mice, inflammatory cell infiltration around the bronchi and bronchioles, and in the alveolar spaces was minimal at 2 and 5 dpi (Fig. 1d). In situ hybridization revealed that viral RNA was present in the bronchiolar and alveolar epithelium of both BA.1- and BA.2-infected mice, with no differences between the infecting viruses at 2 dpi (Fig. 1d). The distribution of viral antigen, as determined by immunohistochemistry, was similar to that of viral RNA in both BA.1- and BA.2-infected mice (Fig. 1d). In both BA.1- and BA.2-infected mice, the amount of detectable viral RNA and antigen decreased over time (Fig. 1d). These results suggest that although both BA.1 and BA.2 infect bronchiolar and alveolar epithelium in the lungs of BALB/c mice, they display substantially less infectivity in the lung of these mice than the B.1.351 variant7.
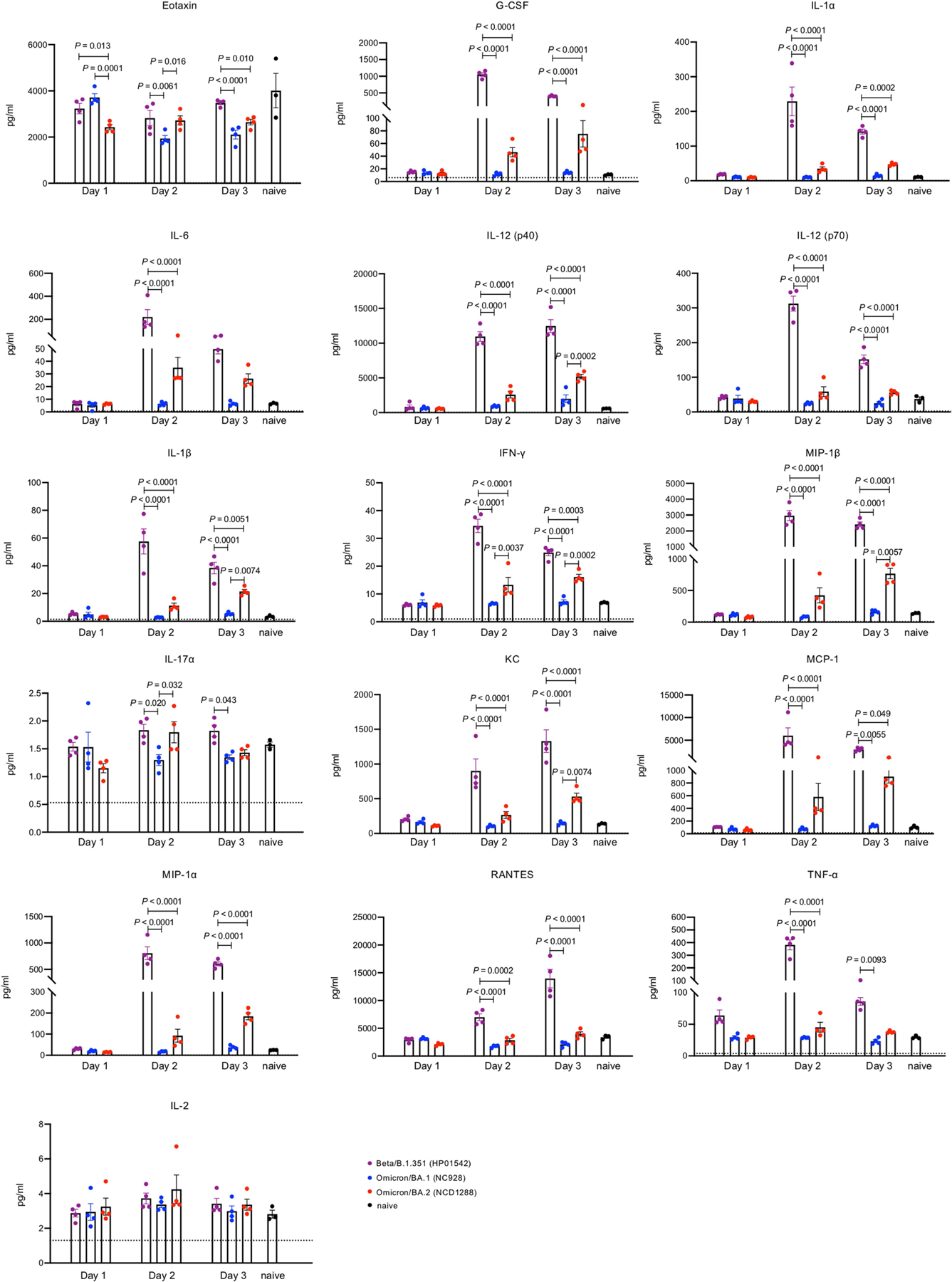
We also assessed the inflammatory responses in the lungs of BALB/c mice. Consistent with our previous study7, BA.1 (NC928)-infected BALB/c mice showed similar cytokine and chemokine levels at 1, 2, or 3 dpi as naive mice. Mice infected with BA.2 (NC1288) had significantly higher levels of several pro-inflammatory cytokines and chemokines, such as IL-1β, IFNγ, and MIP-1β, compared with mice inoculated with BA.1 at 2 or 3 dpi. However, cytokine and chemokine levels were much lower in the lungs of mice infectedwith BA.2 thanthose infectedwith B.1.351 (HP01542) (Fig. 1e and Extended Data Fig. 1). These results demonstrate that infection with BA.2 induces a more limited inflammatory response in the lungs of mice than B.1.351, probably owing to its lower replicative ability.
Next, we evaluated the replication of BA.2 in transgenic mice expressing human ACE2 (hACE2) under the control of an epithelial cytokeratin promoter (K18-hACE2 mice), a more susceptible mouse model25,26. At 3 dpi, the infectious virus titres in the lungs and nasal turbinates were substantially (270- to 830-fold) lower in the respiratory tract of mice infected with BA.2 or BA.1 compared with mice infected with WA1/2020 D614G; viral RNA levels in the lungs, nasal turbinates and nasal washes were similarly lower (Fig. 2a,b). We also assessed the inflammatory responses in the lungs of K18-hACE2 mice at 3 dpi. Although K18-hACE2 mice infected with BA.2 (NC1288) had significantly higher levels of some pro-inflammatory cytokines and chemokines, such as IL-1α, TNF and MIP-1α, compared with mice inoculated with BA.1 at 3 dpi, levels of many inflammatory cytokines and chemokines including IL-1β, IL-6, MCP-1, IP-10 and MIP-1β, were lower in the lungs of both BA.1- and BA.2- infected mice than in those of WA1/2020 D614G-infected mice (Fig. 2c and Extended Data Fig. 2). Together, these findings indicate that BA.2 is less pathogenic in both wild-type and hACE2 transgenic mice, similar to findings for BA.17–9.
Fig. 2 |. BA.2 and BA.1 show similar infectivity and pathogenicity in K18-hACE2 mice.
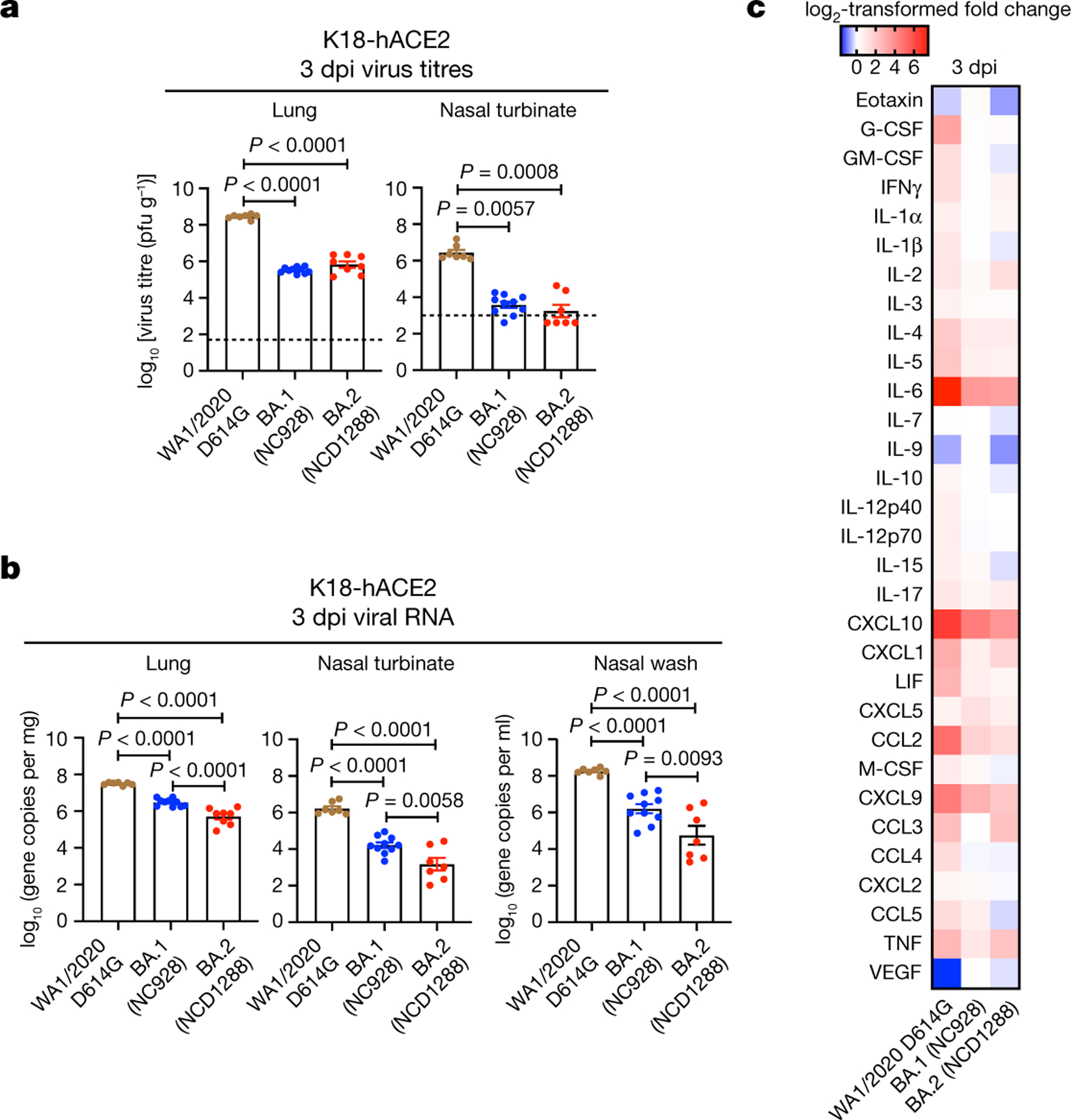
a–c, K18-hACE2 mice were inoculated intranasally with 103 PFU WA1/2020 D614G, BA.1 (NC928) or BA.2 (NCD1288) (D614G: n = 7, BA.1 (NC928): n = 10, BA.2 (NCD1288): n = 8). Viral titres (a) and RNA levels (b) were measured at 3 dpi. Viral titres in the nasal turbinates and lungs were determined by performing plaque assays. Data are mean ± s.e.m. Points represent data from individual mice; the lower limit of detection is indicated by the horizontal dashed line. Data were analysed with the Kruskal–Wallis test with Dunn’s multiple comparisons (titres in the nasal turbinate of infected hamsters) or a one-way ANOVA with Tukey’s multiple comparisons test (titres in the lungs of mice, RNA levels in the lungs, nasal turbinates and nasal washes of infected mice). c, Heat map of cytokine and chemokine concentrations (see Extended Data Fig. 2) in the lungs of infected K18-hACE2 mice at 3 dpi. Data are from one (c) or two (a,b) experiments.
BA.2 infection in hamsters
We next evaluated the replication and pathogenicity of BA.2 variants in Syrian hamsters, a well-established small animal model for the study of COVID-1927–29. Syrian hamsters were inoculated with 103 PFU of BA.1 (NC928), BA.2 (NCD1288) or BA.2 (HP353). No differences in weight change were observed among the three groups, with all animals gaining weight (Fig. 3a). We also evaluated pulmonary function in the infected hamsters using the whole-body plethysmography system. Infection with the BA.2 (HP353), BA.2 (NCD1288) or BA.1 (NC928) variants did not cause substantial changes in either Penh or Rpef (Fig. 3b). Inoculation with a higher dose of BA.1 (105 PFU) or BA.2 (105 PFU of NCD1288, HP354 or TY40-385, or 104.7 PFU of HP353) did not cause any difference in weight change among the five groups (Extended Data Fig. 3a). In addition, inoculation of hamsters with 105 PFU of BA.1 (NC928) or BA.2 (NCD1288) did not cause substantial changes in Penh and Rpef in either hamster group (Extended Data Fig. 3b).
Fig. 3 |. BA.2 and BA.1 show similar infectivity and pathogenicity in wild-type hamsters.
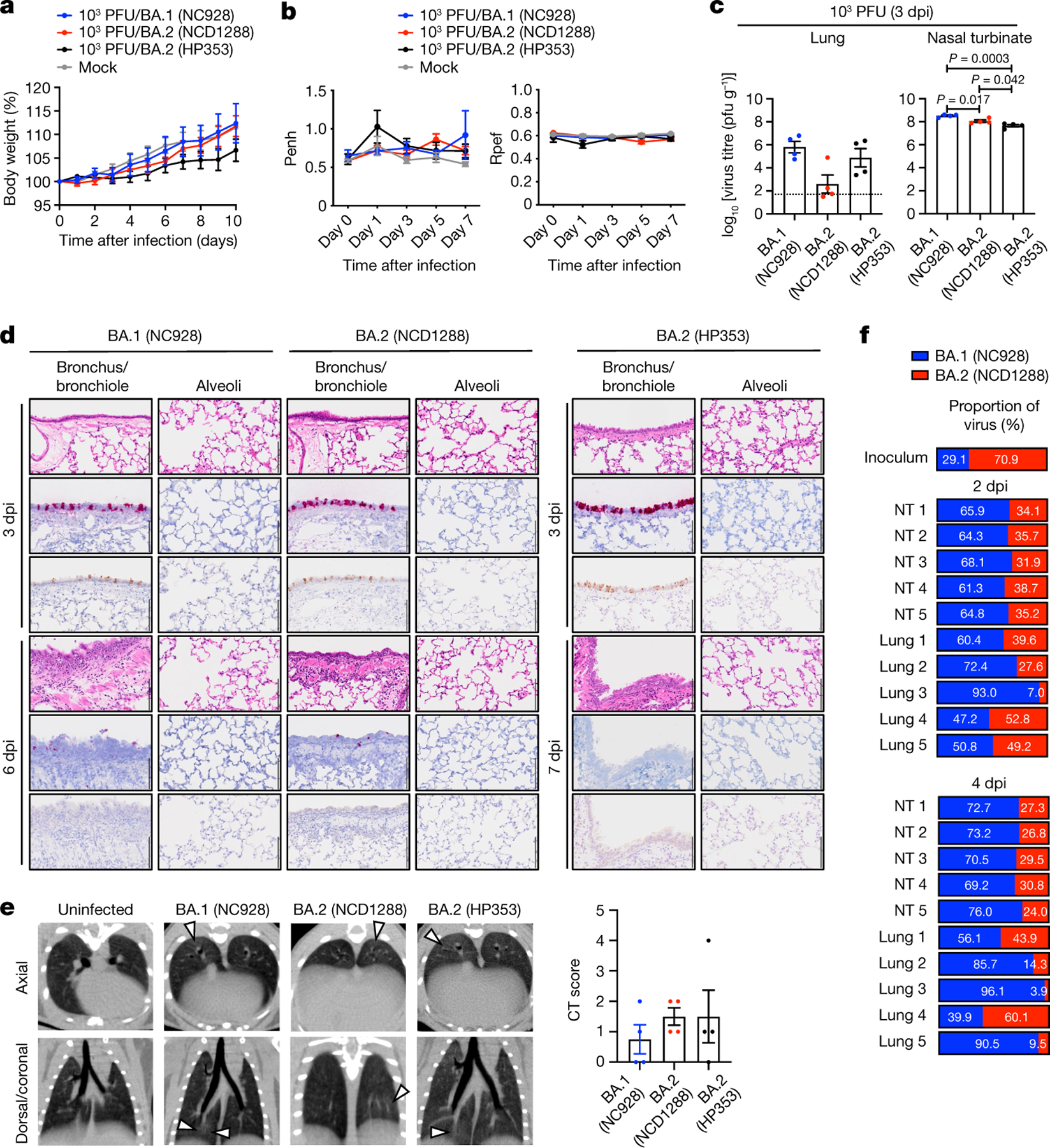
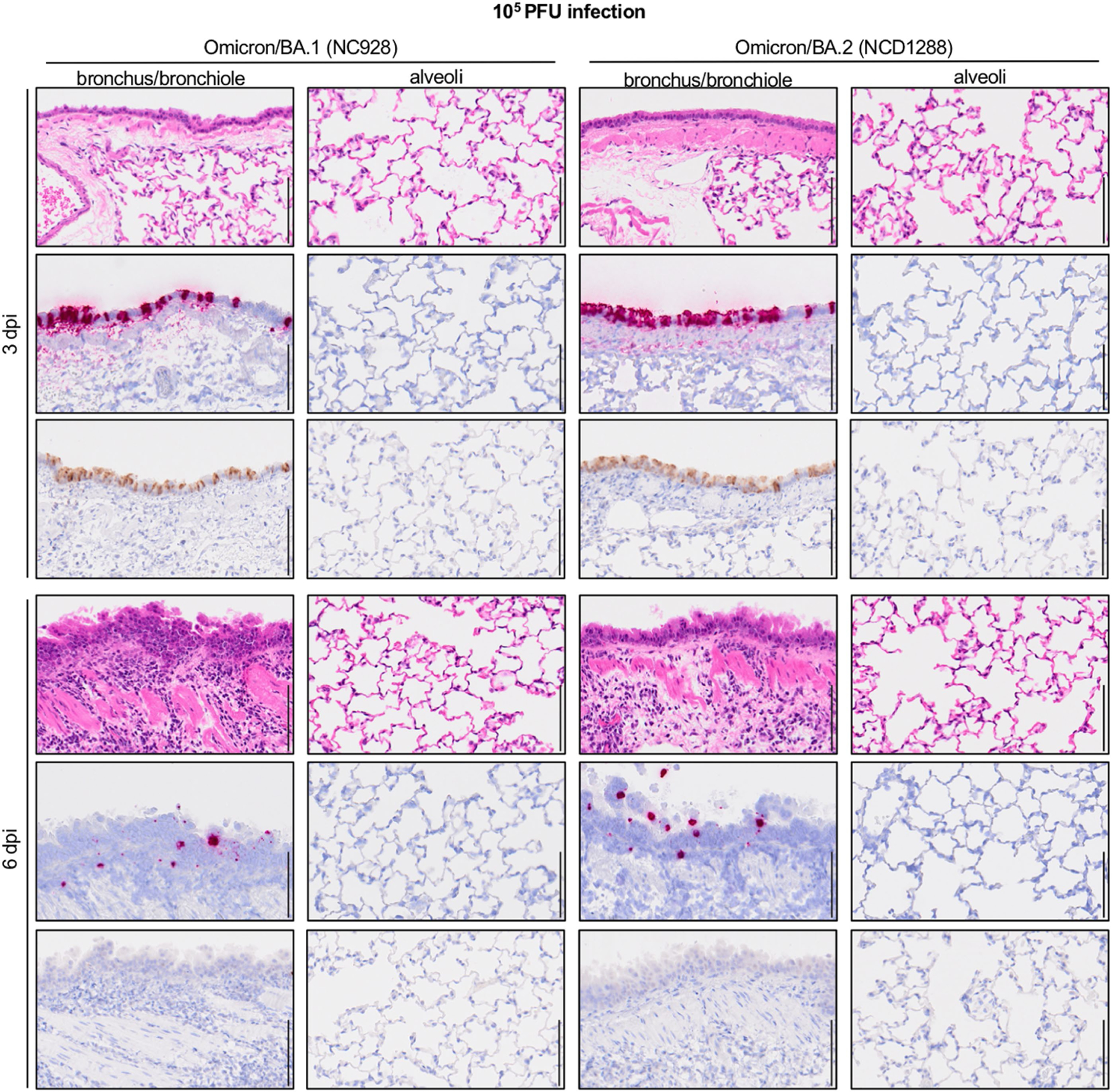
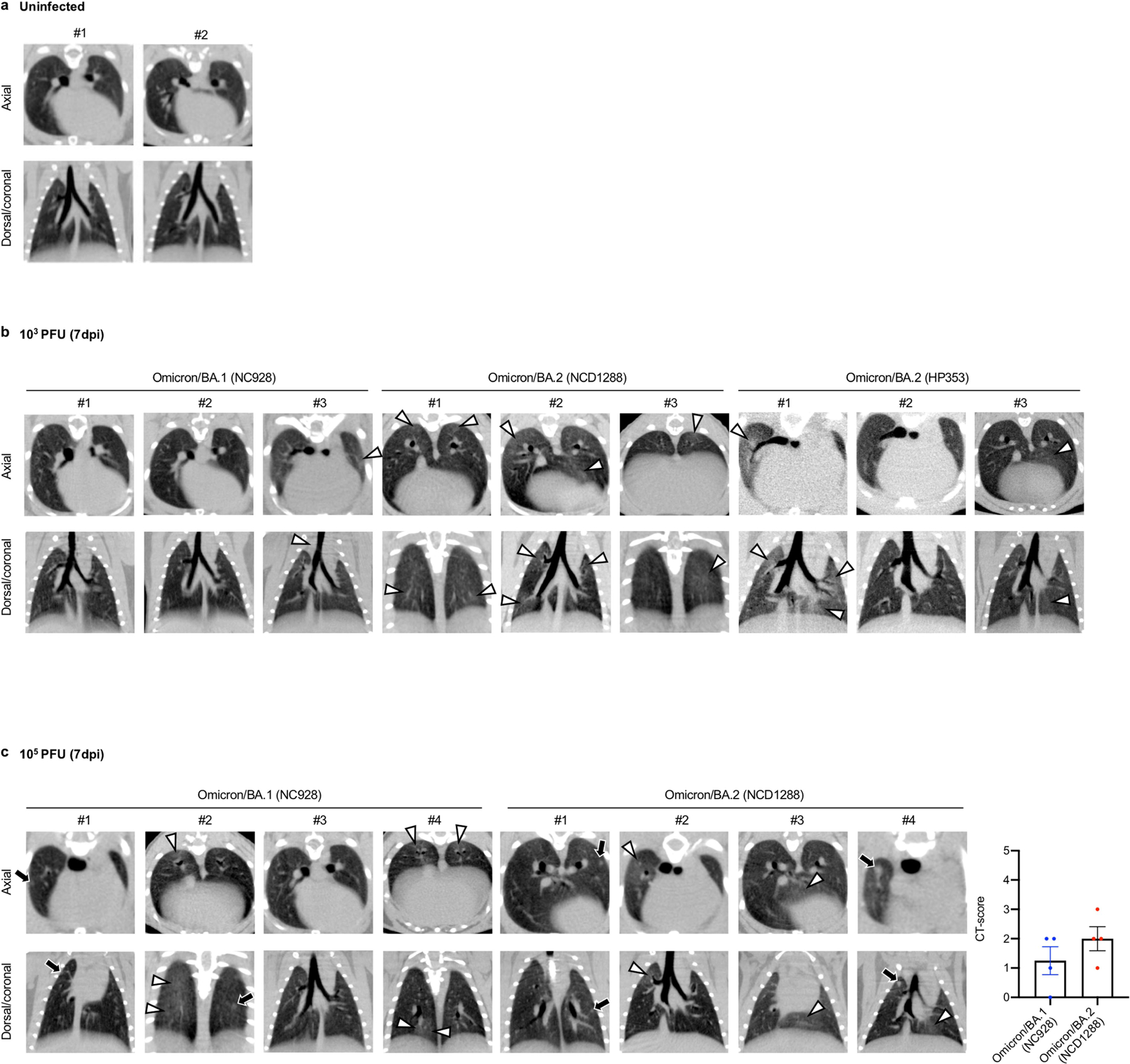
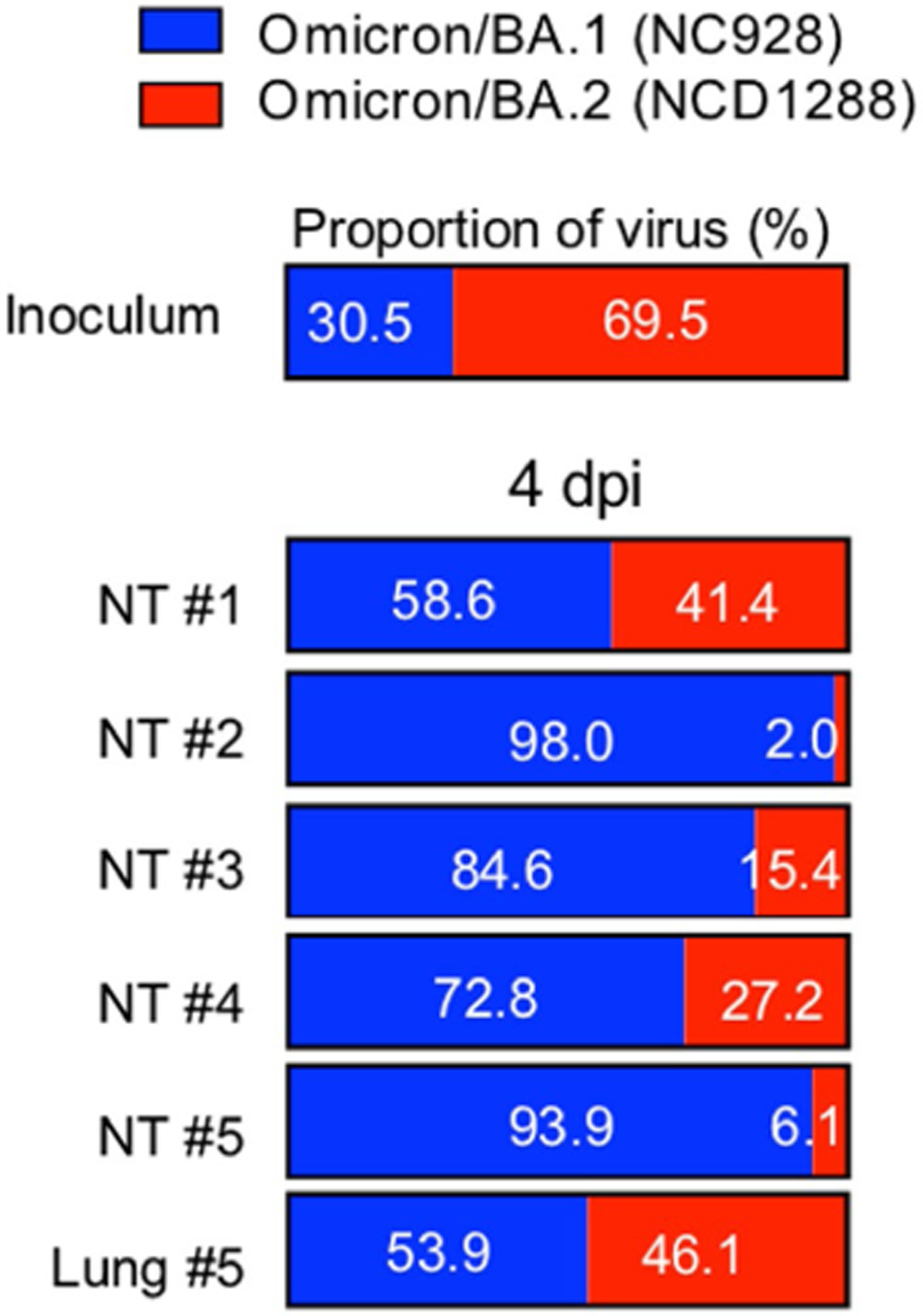
a–e, Syrian hamsters were inoculated intranasally with 103 PFU BA.1 (NC928), BA.2 (NCD1288), BA.2 (HP353) or PBS (mock). a, Changes in body weight of virus-infected (n = 4) or mock-infected (n = 8) hamsters. b, Pulmonary function analyses by whole-body plethysmography in virus-infected (n = 4) or mock-infected (n = 3) hamsters. c, Infectious virus titres in infected hamsters at 3 dpi (n = 4). Data were analysed with the Kruskal–Wallis test with Dunn’s multiple comparisons (lungs) or one-way ANOVA with Tukey’s multiple comparisons (nasal turbinates). d, Histopathological examination of the lungs of infected hamsters at 3, 6 or 7 dpi. Hamsters were infected with 103 PFU BA.1 (NC928) (n = 4), BA.2 (NCD1288) (n = 4) or BA.2 (HP353) (n = 3). Representative images of the bronchi and bronchioles, and alveoli of infected hamsters are shown. Top row, H&E staining. Middle row, in situ hybridization for SARS-CoV-2 viral RNA. Bottom row, immunohistochemistry for SARS-CoV-2 nucleocapsid protein. Scale bars, 100 μm. e, Micro-CT images of the lungs of mock-infected (n = 3) or virus-infected (n = 4) hamsters at 7 dpi. Lung abnormalities included minimal, patchy, ill-defined, peri-bronchial ground glass opacity (white arrowheads). CT severity scores for infected hamsters (n = 4). f, Co-infection with BA.1 and BA.2. BA.1 (NC928) and BA.2 (NCD1288) were mixed at an equal ratio on the basis of their infectious titres, and the virus mixture (total 2 × 105 PFU) was inoculated into hamsters (n = 5). Shown are the relative proportions of BA.1 and BA.2 in the lung and nasal turbinate (NT) of infected hamsters 1–5 at 2 and 4 dpi. Data are from one experiment. Data in a–c,e are mean ± s.e.m.
We also evaluated levels of infection in the respiratory tract of the hamsters. At 3 dpi, in contrast to our observations in mice, we found that virus titres in the nasal turbinates of hamsters infected with 103 PFU of BA.1 (NC928) were slightly but significantly higher than in those infected with 103 PFU of BA.2 (NCD1288) or BA.2 (HP353) (mean titres 8.5, 8.1 or 7.7 log10 PFU g−1, respectively) (Fig. 3c). The lung titres in the BA.1 (NC928)-infected group (mean titres 5.8 log10 PFU g−1) were not significantly different from those in the BA.2 (NCD1288)- or BA.2 (HP353)-infected groups (mean titres 2.6 or 4.9 log10 PFU g−1, respectively). For hamsters infected with 105 PFU of virus, similar titres were detected in the lungs of animals inoculated with BA.1 (NC928) or BA.2 (NCD1288) at 3 dpi; however, BA.1 replicated more efficiently than BA.2 in the nasal turbinates of infected animals, similar to our findings in animals infected with the low 103 PFU dose (Extended Data Fig. 3c).
We performed histopathological analysis of the lungs of the BA.1- and BA.2-infected hamsters. This examination revealed that neutrophils and mononuclear cells infiltrated the bronchial and bronchiolar epithelium and subepithelial connective tissues in the bronchi and bronchioles of all hamsters infected with BA.1 (NC928), BA.2 (NCD1288) or BA.2 (HP353) at 6 dpi or 7 dpi, despite minimal inflammation at 3 dpi (Fig. 3d and Extended Data Fig. 4). By contrast, there was negligible infiltration of inflammatory cells into the alveolar space of the BA.1- and BA.2-infected hamsters at any timepoint examined (Fig. 3d and Extended Data Fig. 4). Furthermore, there were no obvious pathological differences between the different viral doses. Viral RNA-positive or antigen-positive cells were detected in the bronchial epithelium at 3 dpi in all of the BA.1 (NC928)-, BA.2 (NCD1288)- and BA.2 (HP353)-infected hamsters, and more positive cells were seen in the hamsters injected with 105 PFU than in those infected with 103 PFU (Fig. 3d and Extended Data Fig. 4). The number of viral RNA-positive or antigen-positive cells decreased over time in all of the BA.1 (NC928)-, BA.2 (NCD1288)- and BA.2 (HP353)-infected animals. Overall, there was no obvious difference in the magnitude of inflammation in the bronchi and bronchioles or the distribution of viral RNA and antigen among the BA.1- and BA.2-infected animals. These observations suggest that BA.1 and BA.2 mainly affect the bronchi, resulting in bronchitis and bronchiolitis, and that the pathogenicity of BA.1 and BA.2 is reduced compared with earlier viruses in the hamster model.
We performed microcomputed tomography (micro-CT) to assess for lung abnormalities in hamsters at 7 dpi. We used a previously defined CT severity score (Methods) to evaluate animals for ground glass opacities, nodules and regions of lung consolidation30. Micro-CT analysis revealed minimal lung abnormalities in all of the BA.2 (NCD1288)- or BA.2 (HP353)-infected hamsters, and in 62.5% (5 out of 8) of the BA.1 (NC928)-infected hamsters, including minimal, patchy, ill-defined, peri-bronchial ground glass opacity and a few, small, focal rounded or nodular regions, consistent with minimal pneumonia (Fig. 3e and Extended Data Fig. 5). These imaging features can be seen in COVID-19 pneumonia, but can also occur with a variety of infectious and non-infectious processes30. BA.2-infected hamsters had slightly higher CT severity scores (mean of 1.5 for hamsters inoculated with 103 PFU BA.2 (NCD1288), 1.5 for hamsters inoculated with 103 PFU BA.2 (HP353), and 2 for hamsters inoculated with 105 PFU BA.2 (NCD1288)) than BA.1 (NC928)-infected hamsters (mean of 0.75 for hamsters inoculated with 103 PFU and 1.25 for hamsters inoculated with 105 PFU), owing to lung abnormalities being present in a slightly higher number of lobes in BA.2-infected hamsters. Thus, the differences in lung abnormalities between BA.2- and BA.1-infected hamsters were subtle, based on micro-CT analysis. In contrast to commonly reported imaging features of COVID-19 pneumonia, Syrian hamsters infected with BA.1 or BA.2 showed minimal lung abnormalities and no consolidation, in contrast with previous studies with ancestral or other VOC20,27–29,31, consistent with our previous analysis of BA.1 infection7.
To compare the relative fitness and infectivity of BA.1 and BA.2, we inoculated 5 hamsters with 2 × 105 PFU of a mixture (1:1) of BA.1 (NC928) and BA.2 (NCD1288). At 2 and 4 dpi, the nasal turbinates and lungs of the infected hamsters were collected and assessed by Next Generation Sequencing (NGS) to determine the ratio of BA.1 to BA.2. The ratio was calculated on the basis ofthe differences between these twoviruses across nine regions in the S protein. NGS analysis revealed that BA.1 was dominant in the nasal turbinates of all 5 infected animals at both 2 and 4 dpi (Fig. 3f). The lung samples showed a greater prevalence of BA.1, except for the samples from the fourth animal at 2 dpi and 4 dpi; even in this animal, the prevalence of BA.1 (39.9%) at 4 dpi was higher than that in the inoculum (29.1%). Asimilar trendwasseen when hamsters were inoculated with a lower dose (2 × 103 PFU) of the 1:1 mixture of BA.1 (NC928) and BA.2 (NCD1288) (Extended Data Fig. 6). These results show that BA.1 outcompetes BA.2 during upper and lower airway tract infection in hamsters.
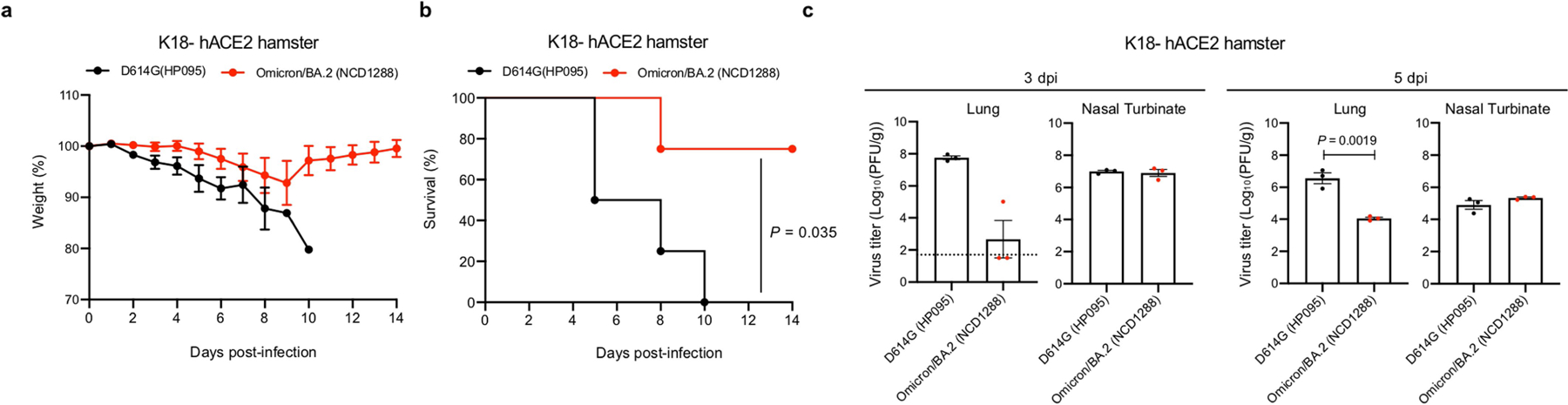
We also investigated the infection and pathogenicity of BA.2 using a more susceptible hamster model: transgenic hamsters expressing hACE2 (line M41) under the control of an epithelial cytokeratin-18 promoter32. Intranasal inoculation of hamsters with 103 PFU of D614G (HP095) virus caused remarkable weight loss (> 10%) within the first week (Fig. 4a) and resulted in 100% mortality at 5 dpi (Fig. 4b). By contrast, most animals infected with BA.1 (WI221686) or BA.2 (NCD1288) survived (75% or 100%, respectively). The lung titres in the BA.2-infected group were more than 100-fold lower than those in the BA.1-infected group and more than 10,000-fold lower than those in the D614G (HP095)-infected group at 3 and 5 dpi (Fig. 4c), although all viruses replicated to similar levels in the nasal turbinates. Similar results were observed in a different line (line M51) of hACE2 transgenic hamsters that is less susceptible than line M41 (Extended Data Fig. 7). These results demonstrate that the pathogenicity of BA.2 is similar to that of BA.1 in both wild-type and hACE2 transgenic hamsters and that BA.2 is attenuated in its replication in their lower respiratory tracts.
Fig. 4 |. BA.2 and BA.1 show similar infectivity and pathogenicity in hACE2-expressing hamsters.
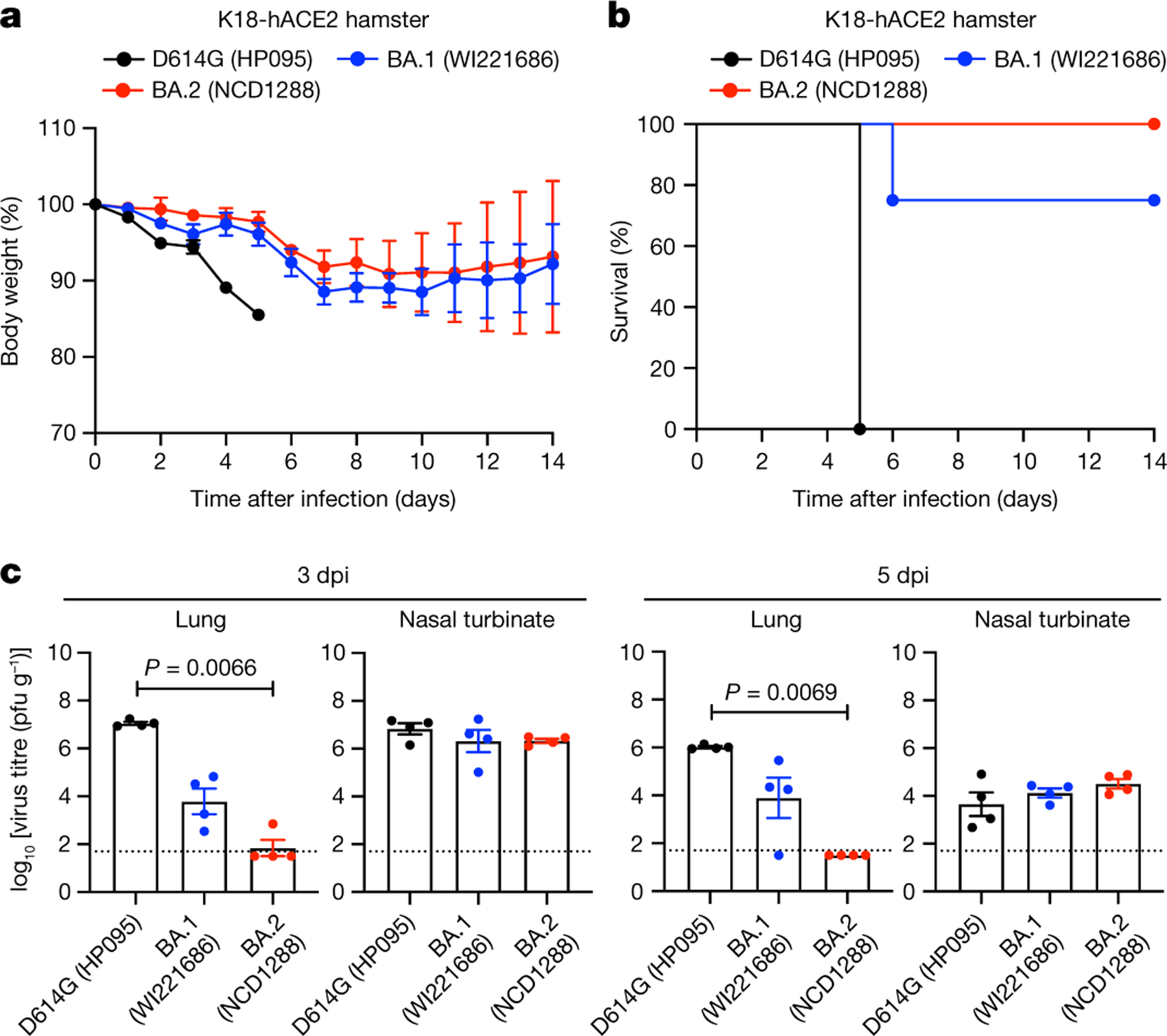
a–c, hACE2-expressing Syrian hamsters were inoculated intranasally with 103 PFU of D614G (HP095), BA.1 (WI221686) or BA.2 (NCD1288). a,b, Body weight (a) and survival (b) were monitored daily for 14 days. Data are mean percentage of the starting weight ± s.e.m. Survival data were analysed with the log-rank (Mantel–Cox) test. c, Four hamsters per group were euthanized at 3 or 5 dpi for virus titration. Virus titres in the nasal turbinates and lungs were determined using plaque assay. Data are mean ± s.e.m.; points represent data from individual hamsters; the lower limit of detection is indicated by the horizontal dashed line. Data were analysed with the Kruskal–Wallis test with Dunn’s multiple comparisons (titres in the lungs of infected hamsters) or one-way ANOVA with Tukey’s multiple comparisons (titres in the nasal turbinate). Data are from one experiment.
Antibody responses to BA.2 variants
Previous studies have established that both BA.1 and BA.1.1 variants show reduced sensitivity to antibodies in sera and/or plasma from individuals who have recovered from COVID-19 (convalescent individuals) and vaccinated individuals compared with the ancestral strains and other VOC12,33–37. We evaluated whether antibodies in plasma from convalescent individuals and vaccine recipients retain neutralizing activity against BA.2. We obtained plasma from four different groups: individuals (at least 14 days after their third dose of vaccine; n = 39) who received three doses of the mRNA vaccine BNT162b2 (Pfizer-BioNTech); individuals (1 (n = 13), 3 (n = 11) or 6 (n = 12) months after a second dose) who received two doses of BNT162b2 after previous infection during the first wave; individuals (n = 20) who received two doses of BNT162b2 before a Delta breakthrough infection; and individuals (n = 10) who received two doses of BNT162b2 or the mRNA-1273 (Moderna) vaccine before Omicron breakthrough infection. Neutralization titres against BA.2 were determined using authentic BA.2 (NCD1288) and a focus reduction neutralization test (FRNT), and compared it with plasma raised against an ancestral SARS-CoV-2 strain (SARS-CoV-2/UT-NC002-1T/Human/2020/Tokyo; NC002) from February 2020, Delta (UW-5250), BA.1 (NC928) and BA.1.1 (NC929) (Fig. 5a–d).
Fig. 5 |. Antibody responses to BA.2 variants.
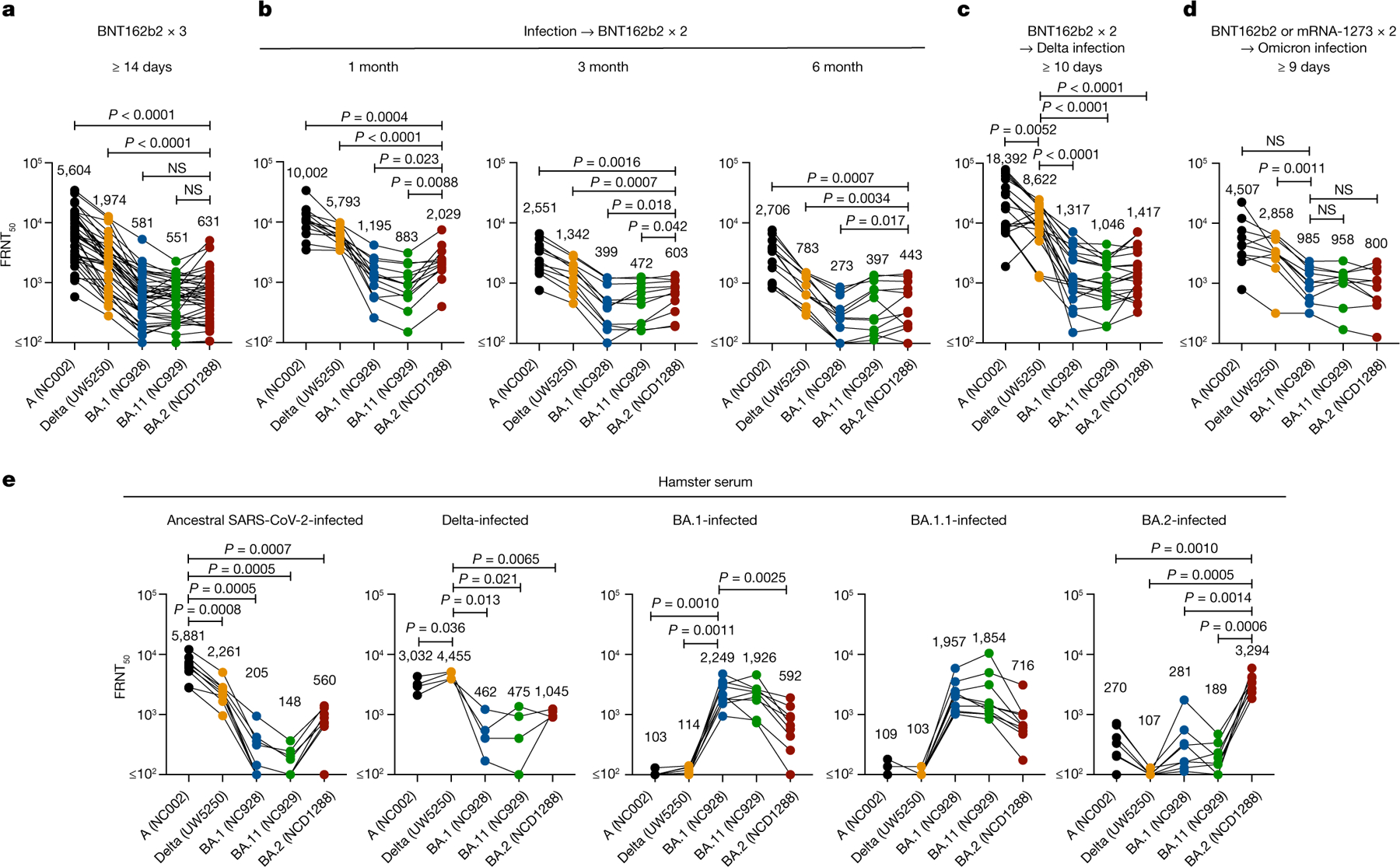
a, Neutralizing antibody titres of human plasma or serum obtained from individuals immunized with three doses of the BNT162b2 vaccine (n = 39). Samples were collected at least 14 days after the third dose. b, Neutralizing antibody titres of human plasma obtained from individuals immunized with two doses of the BNT162b2 vaccine after previous infection. Samples were collected 1, 3 or 6 months after the second immunization (n = 13, 11 or 12, respectively). c, Neutralizing antibody titres of human plasma or serum obtained from individuals who were infected with the Delta variant after two doses of the BNT162b2 vaccine (n = 20). Samples were collected at least ten days after symptom onset. d, Neutralizing antibody titres of human plasma obtained from individuals who were infected with the Omicron variant after two doses of the BNT162b2 or mRNA-1273 vaccine (n = 10). Samples were collected 9–16 days after symptom onset. Individual titres and detailed information about the participants are shown in the source data. e, Neutralizing antibody titres of serum obtained from hamsters infected with ancestral SARS-CoV-2 (NC002) (n = 9), Delta B.1.627.2 (UW-5250) (n = 4), BA.1 (NC928) (n = 9), BA1.1 (NC929) (n = 10) or BA.2 (NCD1288) (n = 8). Samples were collected three weeks after infection. P-values were calculated by one-way ANOVA with Dunnett’s multiple comparisons test. Each dot represents data from one individual. Geometric mean titres are shown.
The geometric mean 50% FRNT (FRNT50) titres of plasma from individuals immunized with a third dose of the BNT162b2 vaccine against BA.1, BA1.1 and BA.2 were significantly reduced (by 3.1- to 10.2-fold) compared with those against the ancestral and Delta strains (Fig. 5a). FRNT50 geometric mean titres against BA.1, BA1.1 and BA.2 after administration of a second dose of the BNT162b2 vaccine in previously infected individuals were 2.9- to 11.3-fold lower than those against the ancestral and Delta strains at 1 month after vaccination; however, this reduction in neutralizing titres was larger for BA.1 and BA.1.1 than for BA.2 at 1, 3 and 6 months after vaccination (Fig. 5b). Most of the plasma samples from the COVID-19 vaccine recipients who experienced Delta breakthrough infections showed high FRNT50 values against the Delta variant (Fig. 5c). However, this breakthrough infection induced low levels of neutralizing antibodies against BA.1, BA.1.1 and BA.2 compared with the Delta variant. For the samples from the COVID-19 vaccine recipients who experienced Omicron breakthrough infection, the FRNT50 geometric mean titres against the three Omicron strains were 2.9- to 3.6-fold lower than against the Delta variant (Fig. 5d), even though these individuals were infected with Omicron variants. Together, these results suggest that antibodies elicited by the COVID-19 mRNA vaccine or SARS-CoV-2 infection have reduced neutralizing activity against BA.2 compared with the ancestral and Delta strains, and that BA.2 may be antigenically different from other Omicron strains38.
To analyse the antigenic properties of Omicron variants, we performed an FRNT using post-infection hamster antisera raised against the ancestral (NC002), Delta (UW-5250), BA.1 (NC928), BA.1.1 (NC929) or BA.2 (NCD1288) strains. Sera raised against the ancestral or Delta strains exhibited significantly lower levels of neutralizing activities against the three Omicron strains, compared to each homologous strain (Fig. 5e). Antisera raised against BA.1, BA.1.1 or BA.2 showed markedly low FRNT50 geometric mean titres against the ancestral and Delta strains. For the antisera raised against BA.1, the FRNT50 geometric mean titres against BA.2 were significantly (3.8- and 3.3-fold) lower than against BA.1 and BA.1.1. The antisera raised against BA.2 had less (11.7- and 17.4-fold) neutralizing activity against BA.1 and BA.1.1 than against the BA.2 virus (Fig. 5e). Collectively, these results suggest that BA.2 is antigenically different from BA.1 and BA.1.1 as well as the ancestral and Delta strains.
Antibody efficacy against BA.2 variants
We recently evaluated the therapeutic effect of FDA-approved monoclonal antibodies against a BA.1 variant in hamsters39. Our data suggest that certain monoclonal antibodies may lose efficacy against this variant. We therefore tested whether therapeutic monoclonal antibodies can inhibit infection of BA.2 variants in hamsters. As of April 2022, the US Food and Drug Administration (FDA) has authorized the emergency use of four monoclonal antibodies for the treatment and/or prevention of COVID-19: REGEN-COV, a combination of imdevimab (REGN10987) and casirivimab (REGN10933); Xevudy, which is sotrovimab (VIR-7831); Evusheld (AZD7442), a combination of tixagevimab (COV2-2196 (also known as AZD8955)) and cilgavimab (COV2-2130 (also known as AZD1061)); and bebtelovimab (LY-CoV1404). We tested REGN10987 + REGN10933, S309 (the precursor of sotrovimab) and COV2-2196 + COV2-2130 for their therapeutic efficacy against BA.2. We synthesized these monoclonal antibodies according to publicly available sequences without modifications in their Fc regions, as described previously15,39. We inoculated Syrian hamsters with 103 PFU of CoV-2/UT-HP095-1N/Human/2020/Tokyo (D614G; HP095) or BA.2 (NCD1288), and one day later injected them intraperitoneally with 5 mg kg−1 of S309, 2.5 mg kg−1 each of REGN10987 and REGN10933, or 2.5 mg kg−1 each of COV2-2196 and COV2-2130 (Fig. 6a). A monoclonal antibody specific to influenza B virus haemagglutinin was used as a control. Sera were also collected at this timepoint and tested in an ELISA for RBD-specific IgG antibodies, which confirmed successful antibody transfer. Similar to our previous experiment39, in the D614G (HP095)-infected groups, treatment with REGN10987 + REGN10933 or COV2-2196 + COV2-2130 significantly reduced virus titres in the lungs compared with infected hamsters treated with the control monoclonal antibody (Fig. 6b). In addition, treatment with S309 resulted in a 62-fold reduction in D614G virus titres in the lungs, although this did not attain statistical significance. For the BA.2 (NCD1288)-infected groups, COV2-2196 + COV2-2130 treatment suppressed virus infection in the lungs (mean reduction in viral titre of 2.8 log10 PFU g−1) (Fig. 6c), similar to our previous findings with BA.139 (NC928). In addition, S309 and REGN10987 + REGN10933, which wepreviously showed were not effective against BA.1, reduced lung virus titres (mean reduction in viral titre of 2.7 and 2.2 log10 PFU g−1, respectively), although the difference was not statistically significant between the REGN10987 + REGN10933-treated and control monoclonal antibody-treated groups. None of the monoclonal antibodies that we tested affected the virus titres in the nasal turbinates of the hamsters. These results suggest that REGN10987 + REGN10933, COV2-2196 + COV2-2130 and S309 can restrict viral replication in the lungs of hamsters infected with BA.2 when these monoclonal antibodies are administrated therapeutically.
Fig. 6 |. Therapeutic effects of monoclonal antibodies and antiviral compounds against BA.2 variants.
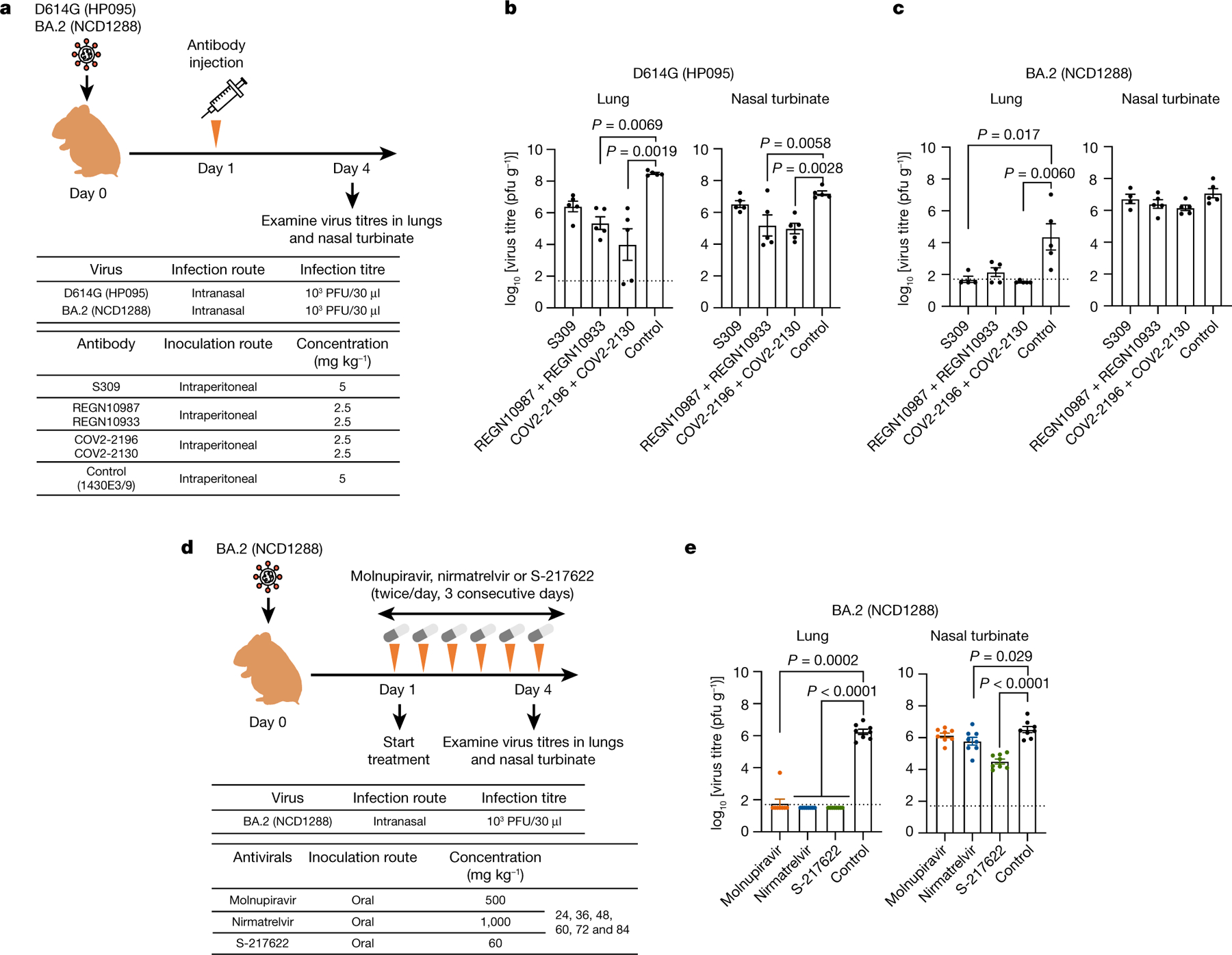
a, Schematic of the experimental workflow for assessing the therapeutic effects of monoclonal antibodies. b, Syrian hamsters were inoculated intranasally with 103 PFU of BA.2 (NCD1288) or D614G (HP095). One day after infection, hamsters were injected intraperitoneally with a single dose of REGN10987 + REGN10933 or COV2-2196 + COV2-2130 2.5 mg kg−1 each) or S309 as monotherapy (5 mg kg−1). A human monoclonal antibody (1430E3/9) against influenza B virus haemagglutinin was injected as a control. Four to five hamsters per group were euthanized at 4 dpi for virus titration. c, Schematic of the experimental workflow for assessing the therapeutic effects of antiviral compounds. d, Syrian hamsters were intranasally inoculated with 103 PFU of BA.2 (NCD1288). One day after infection, hamsters were treated with: 500 mg kg−1 molnupiravir, 1,000 mg kg−1 nirmatrelvir or 60 mg kg−1 S-217622 orally twice daily for 3 days. Methylcellulose served as a control for oral treatment. Eight hamsters per group were euthanized at 4 dpi for virus titration. Viral titres in the nasal turbinates and lungs were determined by plaque assay. Data are mean ± s.e.m.; points indicate data from individual hamsters; the lower limit of detection is indicated by the horizontal dashed line. For comparison of the lung and nasal turbinate titres of BA.2 (NCD1288)- and D614G (HP095)-infected hamster groups, we used a Kruskal–Wallis test with Dunn’s multiple comparisons and one-way ANOVA with Dunnett’s multiple comparisons, respectively. Data are from one experiment.
Antiviral efficacy against BA.2 variants
Molnupiravir, an inhibitor of the RNA-dependent RNA polymerase of SARS-CoV-2, and nirmatrelvir40, an inhibitor of the main protease (also called 3CLpro) of SARS-CoV-2, have been authorized for emergency use to treat COVID-19 by the FDA. In addition, S-217622, another inhibitor of 3CLpro, is currently in clinical trials39,41. We assessed the therapeutic efficacy of these compounds in hamsters infected with BA.2 (NCD1288). Hamsters inoculated intranasally with 103 PFU of BA.2 (NCD1288) were treated via oral gavage twice daily (at 12-h intervals) for 1, 2 or 3 days with 500 mg kg−1 per 12 h (1,000 mg kg−1 day−1) of molnupiravir, 60 mg kg−1 per 12 h (120 mg kg−1 day−1) of S-217622, or 1,000 mg kg−1 per 12 h (2,000 mg kg−1 day−1) of nirmatrelvir, beginning 24 h afterinfection (Fig. 6d). These dosages were selectedon the basis of previous studies in mice or hamsters39,42,43. Although no differences in viral titres in the nasal turbinates were detected between the animals treated with molnupiravir or nirmatrelvir and those treated with methylcellulose (control) at 4 dpi, treatment with S-217622 significantlyreducedthe virus titres in the nasal turbinates at 4 dpi. Moreover, all of the compounds tested considerably reduced the lung virus titres; no infectious virus was recovered at 4 dpi from the lungs of animals treated with nirmatrelvir or S-217622 (Fig. 6e). Our data suggest that the three antiviral compounds tested effectively inhibit BA.2 replication in the lower respiratory tract of hamsters. In addition, hamsters treated with S-217622 showed reduced infection in the upper respiratory tract, consistent with a previous study39.
Discussion
Here we show that the Omicron sublineage BA.2 variants of SARS-CoV-2 are less pathogenic in rodent models than previous SARS-CoV-2 variants, as has been reported for Omicron sublineage BA.1 variants7–9. We observed that BA.2 variants were limited in their infectivity of the lungs of mice and hamsters compared with SARS-CoV-2 strains from early in the pandemic. Moreover, infection with BA.2 induced a limited pro-inflammatory cytokine and chemokine response in mouse lungs, probably owing to the diminished replicative ability of BA.2 in the lungs compared with that of previous SARS-CoV-2 variants, such as the ancestral strain or B.1.351 variant. Notably, histopathological examination revealed that viral RNA and antigen were rarely detected in the alveoli of mice and hamsters after infection with BA.2. Thus, the limited infectivity of BA.2 variants in the lung might contribute to lower disease severity in animal models. Further investigation is required to determine whether BA.2 variants can replicate efficiently and equivalently in human alveolar epithelial cells.
Our study suggests that the pathogenicity and replicative ability of BA.2 variants are similar to those of BA.1 variants in rodent models. By contrast, a recent study showed that a recombinant, chimeric SARS-CoV-2 possessing the BA.2 S gene in the background of an ancestral SARS-CoV-2 strain was more pathogenic than a recombinant virus bearing the BA.1 S gene in the same background4, suggesting that the differences in the BA.1 and BA.2 S genes are responsible for the difference in pathogenicity between these chimeric viruses; on the basis of these data, the authors concluded that BA.2 is more pathogenic than BA.1. However, in natural isolates, the genomes of BA.1 and BA2 also differ at locations other than the S gene. Given that the amino acid sequence of the S protein of the recombinant BA.2 virus that these authors tested is the same as that of the BA.2 isolate that we tested, differences at locations other than the S gene could have epistatic or independent effects that offset differences in the pathogenic potential of the S gene44.
Omicron variants are less likely than Delta variants to be associated with pneumonia in human patients with COVID-1945. We observed that BA.2 infection in the lung was less pathogenic in mice and hamsters than SARS-CoV-2 strains from early in the pandemic (Figs. 1–4), similar toreports on BA.17–9. In human nasal epithelial cultures, Omicron and Delta variants have been reported toreplicate with similar efficiency; however, in lower airway organoids and Calu-3 lung cells, Omicron variants replicate tolower titres than Delta variants46. The human protein transmembrane protease serine 2 (TMPRSS2) activates the S protein of SARS-CoV-2, which mediates viral entry into host cells. This protease is expressed at high levels in human and hamster lung47,48. The S protein of Omicron variants has been shown to use TMPRSS2 less efficiently46; therefore, the reduced TMPRSS2 usage of the Omicron S protein may affect the replication of BA.1 and BA.2 variants in the lungs of humans and hamsters.
The RBD of the S protein is the primary target for therapeutic antibodies and antibodies elicited by vaccination or infection. BA.2 has 16 amino acid substitutions in its RBD compared with the reference strain Wuhan/Hu-1/2019. BA.2 and BA.1 variants share 12 of these 16 substitutions; BA.2 possesses four RBD substitutions (S371F, T376A, D405N and R408S) that differ from those in BA.1, which could affect the efficacy of therapeutic monoclonal antibodies and mRNA vaccines against the different Omicron sublineages. We found that S309, REGN10987 + REGN10933 and COV2-2196 + COV2-2130 inhibited BA.2 replication in the lungs of hamsters. By contrast, ourpreviousstudyshowedthatthetherapeuticadministration of S309 or REGN10987 + REGN10933 had no effect on the virus titres in the lungs of hamsters infected with BA.139. The disparity between the in vivo findings with S309 has also been observed by others49 and may be owing, in part, to the attenuated infectivity (approximately 100-fold lower levels) of BA.2 compared to BA.1 in hamster lungs (Fig. 3c). These findings highlight that, when treating COVID-19 patients with monoclonal antibody-based therapy, the prevailing Omicron sublineage should be considered.
A marked and significant reduction in neutralizing activity was observed against BA.2 compared with the ancestral and Delta strains for convalescent individuals and vaccinees. Of note, plasma from individuals infected with SARS-CoV-2 strains from early in the pandemic and then vaccinated with the BNT162b2 vaccine showed higher neutralizing activity against BA.2 than against BA.1 and BA.1.1 (Fig. 5b). In addition, we found that BA.2 was less well recognized than BA.1 by sera from hamsters infected with BA.1 (Fig. 5e), which is consistent with the data obtained with sera from hamsters previously infected with BA.14. We also observed that BA.1 and BA.1.1 were poorly recognized by hamster antisera raised against BA.2 (Fig. 5e). A recent study using in vitro neutralization assays reported that there were differences in the reactivity of several monoclonal antibodies against these different Omicron sublineages38. These observations suggest that BA.2 are antigenically distinct from BA.1 and BA.1.1.
We note several limitations in this study. (1) In wild-type mice, we characterized a BA.2 isolate (NCD1288) with a single experiment. Therefore, further studies are required to determine whether the results obtained are reproducible and whether the other BA.2 strains exhibit infectivity and pathogenicity similar to the isolate tested. (2) The relationships between neutralizing titres and protection against COVID-19 in humans remains unclear. Future clinical studies should assess how much neutralizing activity is required to prevent Omicron variant infections and predict vaccine efficacy. (3) Although hamsters are among the most susceptible animals to SARS-CoV-2 of those tested, including mice and non-human primates, the BA.2 variant is attenuated in the lungs of hamsters. It is unclear whether the BA.2 variant in humans is attenuated to the degree that it is in hamsters. Differences in replication of Omicron variants in humans and hamsters could affect the effectiveness of the monoclonal antibodies and antiviral compounds. Clinical data on the efficacy of the monoclonal antibodies and antiviral drugs for the treatment of patients infected with BA.2 are needed to corroborate the findings from the hamster model.
In summary, our data suggest that the Omicron BA.2 variant of SARS-CoV-2 has similar infectivity and pathogenicity as the Omicron BA.1 variant in rodent models and is susceptible to restriction by several therapeutic monoclonal antibodies and antiviral compounds in clinical use or development.
Methods
Cells
VeroE6/TMPRSS2 (JCRB 1819) cells31,50 were propagated in the presence of 1 mg ml−1 geneticin(G418; Invivogen) and 5 μg ml−1 plasmocinprophylactic (Invivogen) in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS). Vero-hACE2-TMPRSS2 cells were cultured in DMEM supplemented with 10% FCS, 10 mM HEPES pH 7.3, and 100 U ml−1 of penicillin–streptomycin, 10 μg ml−1 of puromycin. VeroE6/TMPRSS2 and Vero-hACE2-TMPRSS2 cells were maintained at 37 °C with 5% CO2.
Chinese hamster ovary (CHO) cells were maintained in DMEM containing 10% FCS and antibiotics at 37 °C with 5% CO2. Expi293F cells (Thermo Fisher Scientific) were maintained in Expi293 expression medium (Thermo Fisher Scientific) at 37 °C under 8% CO2. The cells were regularly tested for mycoplasma contamination using PCR, and confirmed to be mycoplasma-free.
Clinical specimens
After informed consent was obtained, respiratory specimens were collected from SARS-CoV-2-infected individuals and plasma specimens were collected from COVID-19 convalescent individuals and vaccinees. The research protocol was approved by the Research Ethics Review Committee of the Institute of Medical Science of the University of Tokyo (approval number: 2019–71–0201 and 2020-74-0226).
After informed consent was obtained, specimens were collected from individuals who received the mRNA vaccine BNT162b2 (Pfizer-BioNTech) in the Immunity-Associated with SARS-CoV-2 (IASO) cohort study. The research protocol was approved by the institutional review board at the University of Michigan Medical School (protocol number HUM00184533).
Viruses
hCoV-19/Japan/UT-NCD1288-2N/2022 (Omicron BA.2; NCD1288), hCoV-19/Japan/UT-HP353-1N/2022 (Omicron BA.2; HP353), hCoV-19/Japan/UT-HP354-1N/2022 (Omicron BA.2; HP354), hCoV-19/Japan/TY40-385/2022 (Omicron BA.2; TY40-385), hCoV-19/Japan/NC928-2N/2021 (Omicron BA.1; NC928)15, hCoV-19/USA/WI-WSLH-221686/2021 (Omicron BA.1; WI221686)15, hCoV-19/Japan/NC929-1N/2021 (Omicron BA.1.1; NC929)39, SARS-CoV-2/UT-HP095-1N/Human/2020/Tokyo (D614G; HP095)31, hCoV-19/USA/MD-HP01542/2021 (Beta; HP01542), hCoV-19/USA/WI-UW-5250/2021 (Delta; UW-5250) and SARS-CoV-2/UT-NC002-1T/Human/2020/Tokyo (NC002) were propagated in VeroE6/TMPRSS2 cells in VP-SFM (Thermo Fisher Scientific). The SARS-CoV-2/USA-WA1/2020 (WA1/2020) recombinant strain with substitution (D614G) was described previously51. BA.2 (NCD1288), BA.2 (HP353), BA.2 (HP354) and BA.2 (TY40-385) were subjected to NGS as described15,52.
All experiments with SARS-CoV-2 were performed in enhanced biosafety level 3 (BSL3) containment laboratories at the University of Tokyo and the National Institute of Infectious Diseases, Japan, which are approved for such use by the Ministry of Agriculture, Forestry, and Fisheries, Japan. Animal studies at Washington University were performed in approved A-BSL3 facilities using appropriate personal protection and positive pressure respirators.
Antibodies
Amino acid sequences for the variable region ofthe heavy and light chains of the following human monoclonal antibodies againstthe S proteinwere used for gene synthesis: clones tixagevimab (COV2-2196/AZD8895; GenBank accession numbers QLI33947 and QLI33948), casirivimab (REGN10933; PDB accession numbers 6XDG_B and 6XDG_D), cilgavimab (COV2-2130/AZD1061; GenBank accession numbers QKY76296 and QKY75909), imdevimab (REGN10987; PDB accession numbers 6XDG_A and 6XDG_A), and S309 (PDB accession numbers 6WS6_A and 6WS6_F). An artificial signal sequence and the constant gamma heavy (IgG1, UniProtKB/Swiss-Prot accession number P01857) and kappa (UniProtKB/Swiss-Prot accession number P01834) or lambda (UniProtKB/Swiss-Prot accession number P0DOY2) light chain coding sequences were added before and after each variable region. Codon usage was optimized for expression in CHO cells. The synthesized genes were cloned into a plasmid for protein expression and transfected into CHO cells. Cell culture media were collected after incubation for 10–14 days at 37 °C. A human monoclonal antibody (1430E3/9) against the haemagglutinin of influenza B virus was previously cloned into the expression vector Mammalian Power Express System (TOYOBO)53 and was transiently expressed by Expi293F cells. Monoclonal antibodies were purified by using MabSelect SuRe LX (Cytiva) or a protein Acolumn. Purity was confirmed by SDS–PAGE and/or HPLC before use. The reactivities of these antibodies against SARS-CoV-2, including the Alpha, Beta, Delta, Gamma and Omicron variants, have been tested previously.
Antiviral compounds
Molnupiravir (EIDD-2801) and nirmatrelvir (PF-07321332) were purchased from MedChemExpress. S-217622 was kindly provided by Shionogi. All compounds were dissolved in 0.5% methylcellulose prior to use in in vivo experiments.
Animal experiments and approvals
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01), University of Wisconsin, Madison (V006426) and the Animal Experiment Committee of the Institute of Medical Science, the University of Tokyo (approval numbers PA19–72 and PA19–75). All animals were housed under specific pathogen-free conditions in a temperature control environment with a 12 h: 12h light: dark cycle, with 50% humidity and ad libitum access to water and standard laboratory chow. Virus inoculations were performed under anaesthesia, and all efforts were made to minimize animal suffering. In vivo studies were not blinded, and animals were randomly assigned to infection groups. No sample-size calculations were performed to power each study. Instead, sample sizes were determined based on prior in vivo virus challenge experiments.
Experimental infection of Syrian hamsters
Five- to six-week-old male wild-type Syrian hamsters ( Japan SLC) were used in this study. Baseline body weights were measured before infection. Under isoflurane anaesthesia, four hamsters per group were intranasally inoculated with 103 PFU (in 30 μl), 104.7 PFU (in 30 μl), or 105 PFU (in 30 μl) of BA.2 (NCD1288), BA.2 (HP353), BA.2 (HP354), BA.2 (TY40-385) or BA.1 (NC928). Body weight was monitored daily for 10 days. For virological and pathological examinations, 4 hamsters per group were intranasally infected with 103 PFU or 105 PFU of virus; 3 and 6 dpi, the animals were euthanized and nasal turbinates and lungs were collected. The virus titres in the nasal turbinates and lungs were determined by use of plaque assays on VeroE6/TMPRSS2 cells.
For co-infection studies, BA.1 (NC928) and BA.2 (NCD1288) were mixed at an equal ratio on the basis of their titres, and the virus mixture (total 2 × 105 PFU in 60 μl or 2 × 103 PFU in 30 μl) was inoculated into hamsters. The animals were euthanized and nasal turbinates and lungs were collected at 4 dpi.
The K18-hACE2 transgenic hamster lines (line M51; the same line as our previous study7 and line M41) were developed with a piggyBac-mediated transgenic approach. The K18-hACE2 cassette from the pK18-hACE2 plasmid was transferred into a piggyBac vector, pmhyGENIE-3, for pronuclear injection. hACE2 transgenic hamsters will be described in detail elsewhere32. Eight- to nine-week-old female (M41) or 16-month-old male (M51) K18-hACE2 transgenic hamsters, whose hACE2 expression was confirmed, were intranasally inoculated with 103 PFU of D614G (HP095), BA.1 (WI221686) or BA.2 (NCD1288) in 30 μl. Body weight and survival were monitored daily, and nasal turbinates and lungs were collected at 3 and 5 dpi for virological analysis.
Experimental infection of mice
Heterozygous K18-hACE2 C57BL/6J mice (strain 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from the Jackson Laboratory. BALB/c mice were purchased from Japan SLC.
Twelve-week-old female K18-hACE2 mice were inoculated via the intranasal route with 103 PFU (in 50 μl) of BA.1 (NC928), BA.2 (NCD1288) or WA1/2020 D614G; At 3 dpi, the mice were euthanized and lungs were collected. The virus titres in the nasal turbinates and lungs were determined by plaque assays on VeroE6/TMPRSS2-hACE2 cells. To measure viral RNA levels, nasal washes were collected in 0.5 ml of PBS, and tissues were weighed and homogenized with zirconia beads in a MagNA Lyser instrument (Roche Life Science) in 1 ml of DMEM supplemented with 2% FBS. Tissue homogenates were clarified by centrifugation at 10,000 rpm for 5 min and stored at −80 °C. Viral RNA from homogenized tissues or nasal washes was isolated by using the MagMAX Viral RNA Isolation Kit (Thermo Fisher) and measured by TaqMan one-step quantitative PCR with reverse transcription on an ABI 7500 Fast Instrument. Copies of SARS-CoV-2 N gene RNA in samples were determined by using a previously published assay54. In brief, a TaqMan assay was designed to target a highly conserved region of the N gene (forward primer: ATGCTGCAATCGTGCTACAA; reverse primer: GACTGCCGCCTCTGCTC; probe: 56-FAM–TCAAGGAAC–ZEN–AACATTGCCAA–3IABkFQ). This region was included in an RNA standard to allow for copy number determination down to 10 copies per reaction. The reaction mixture contained final concentrations of primers and probe of 500 and 100 nM, respectively.
Five-week-old female BALB/c mice were intranasally inoculated with 105 PFU (in 50 μl) of BA.1 (NC928) or BA.2 (NCD1288) under isoflurane anaesthesia. Body weights were measured before infection and then daily. At 2 and 5 dpi, the mice were euthanized and nasal turbinates and lungs were collected. The virus titres in the nasal turbinates and lungs were determined by plaque assays on VeroE6/TMPRSS2 cells.
Lung function
Respiratory parameters were measured by using a whole-body plethysmography system (PrimeBioscience) according to the manufacturer’s instructions. In brief, hamsters were placed in the unrestrained plethysmography chambers and allowed to acclimatize for 1 min before data were acquired over a 3-min period by using FinePointe software. Mice were placed in the unrestrained plethysmography chambers and allowed to acclimatize for 5 min before data were acquired over a 5-min period by using FinePointe software.
Micro-CT imaging
Hamsters were inoculated intranasally with 103 PFU (in 30 μl) or 105 PFU (in 30 μl) of BA.1 (NC928), BA.2 (NCD1288), BA.2 (HP353) or PBS. Lungs of infected animals were imaged by using an in vivo micro-CT scanner (CosmoScan FX; Rigaku). Under ketamine-xylazine anaesthesia, the animals were placed in the image chamber and scanned for 2 min at 90 kV, 88 μA, FOV 45 mm, and pixel size 90.0 μm. After scanning, the lung images were reconstructed by using the CosmoScan Database software of the micro-CT (Rigaku Corporation) and analysed using the manufacturer-supplied software.
A CT severity score, adapted from a human scoring system, was used to grade the severity of the lung abnormalities55. Each lung lobe was analysed for degree of involvement and scored from 0–4 depending on the severity: 0 (none, 0%), 1 (minimal, 1%–25%), 2 (mild, 26%–50%), 3 (moderate, 51%–75%) or 4 (severe, 76%–100%). Scores for the 5 lung lobes were summed to obtain a total severity score of 0–20, reflecting the severity of abnormalities across the 5 groups. Images were anonymized and randomized; the scorer was blinded to the group allocation.
Pathology
Excised animal tissues were fixed in 4% paraformaldehyde in PBS, and processed forparaffin embedding. The paraffin blocks were cut into3-μm-thick sections and mounted on silane-coated glass slides for histopathological examination. SARS-CoV-2 RNA was detected by in situ hybridization using an RNAscope 2.5 HD Red Detection kit (Advanced Cell Diagnostics) with an antisense probe targeting the nucleocapsid gene of SARS-CoV-2 (Advanced Cell Diagnostics; RNAscope Probe V-nCoV-N, 846081) and following the manufacturer’s instructions as previously described7. Tissue sections were alsoprocessedforimmunohistochemicalstainingwitharabbitpolyclonal antibody for SARS-CoV nucleocapsid protein (ProSpec; ANT-180, 1:500 dilution, Rehovot), which cross-reacts with SARS-CoV-2 nucleocapsid protein. Specific antigen-antibody reactions were visualized by means of 3,3’-diaminobenzidine tetrahydrochloride staining using the Dako Envision system (Dako Cytomation; K4001, 1:1 dilution).
Analysis of the ratio of BA.1 and BA.2 viruses after co-infection
Viral RNA was extracted by using a QIAamp Viral RNA Mini Kit (QIAGEN). Thewholegenomeof SARS-CoV-2 wasamplifiedbyusingamodified ARTIC network protocol in which some primers were replaced or added56,57. In brief, viral cDNA was synthesized from the extracted RNA by using a Lunar-Script RT SuperMix Kit (New England BioLabs). The DNA was amplified by performing a multiplexed PCR in two pools using the ARTIC-N1 primers v558 and the Q5 Hot Start DNA polymerase (New England BioLabs). The DNA libraries for Illumina NGS were prepared from pooled amplicons by using a QIAseq FX DNA Library Kit (QIAGEN) and were then analysed using the MiSeq or iSeq 100 System (Illumina). The reads were assembled by the CLC Genomics Workbench (version 21, Qiagen) with the Wuhan/Hu-1/2019 sequence (GenBank accession no. MN908947) as a reference. The ratio of BA.1 and BA.2 was calculated from the differences between these two viruses across 9 regions. The nucleotide numbers in these 9 regions are: 21762, 21765–21770, 21846, 21987–21995, 22775, 22786, 23202, 24130 and 24503 in the SARS-CoV-2 genome, which correspond to amino acids 67, 69–70, 95, 143–145, 405, 408, 547, 856 and 981 in the spike protein, respectively. Samples with read depths of more than 100 were analysed.
Evaluation of therapeutic efficacy of monoclonal antibodies and antiviral compounds in Syrian hamsters
Five- to six-week-old male Syrian hamsters ( Japan SLC) were used in this study. For the evaluation of monoclonal antibody efficacy in hamsters, under isoflurane anaesthesia, 4 or 5 hamsters per group were inoculated intranasally with 103 PFU (in 30 μl) of BA.2 (NCD1288) or D614G (HP095). Twenty-four hours after infection, the hamsters were injected intraperitoneally with 1 ml of a monoclonal antibody preparation (5 mg kg−1). The hamsters were euthanized at 4 dpi, and the virus titres in the nasal turbinates and lungs were determined by plaque assays on VeroE6/TMPRSS2 cells.
For the evaluation of antiviral compound efficacy in hamsters, under isoflurane anaesthesia, 8 hamsters per group were inoculated intranasally with 103 PFU (in 30 μl) of BA.2 (NCD1288). At 24 h after inoculation, hamsters were treated with the following antiviral compounds: (1) molnupiravir, 500 mg kg−1 (in 1 ml) administered orally twice daily; (2) nirmatrelvir, 1,000 mg kg−1 (in 1 ml) administered orally twice daily; (3) S-217622, 60 mg kg−1 (in 1 ml) administered orally twice daily; or (4) methylcellulose (1 ml) as a control for oral treatment. The animals were euthanized at 4 dpi, and the virus titres in the nasal turbinates and lungs were determined by plaque assays on VeroE6/TMPRSS2 cells.
Cytokine and chemokine measurement
Under isoflurane anaesthesia, five-week-old female BALB/c mice were intranasally inoculated with 105 PFU (in 50 μl) of Beta (HP01542), BA.1 (NC928) or BA.2 (NCD1288); 1, 2 and 3 dpi, the mice were euthanized, and lungs were collected. For cytokine and chemokine measurement, homogenates of mouse lungs were processed with the Bio-Plex Mouse Cytokine 23-Plex (Bio-Rad Laboratories).
For studies with K18-hACE2 mice, lung homogenates were incubated with Triton X-100 (1% final concentration) for 1 h at room temperature to inactivate SARS-CoV-2. Homogenates were analysed for cytokines and chemokines by Eve Technologies Corporation using their Mouse Cytokine Array/Chemokine Array 31-Plex (MD31) platform.
Enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assays (ELISAs) were performed as previously reported59. In brief, 96-well Maxisorp microplates (Nunc) were incubated with the recombinant RBD of the S protein or HexaPro prefusion-stabilized versions of the S ectodomain (S6pro) (prototype virus or omicron variant) (50 μl per well at 2 μg ml−1), or with PBS at 4 °C overnight and were then incubated with 5% skim milk in PBS containing 0.05% Tween-20 (PBS-T) for 1 h at room temperature. The microplates were reacted for 1 h at room temperature with hamster serum samples that has been diluted 40-fold or with 1 μg ml−1 monoclonal antibody in PBS-T containing 5% skim milk and subsequently serially diluted twofold, followed by peroxidase-conjugated goat anti-human IgG, Fcγ Fragment specific antibody ( Jackson Immuno-Research) (1:5,000 dilution) for 1 h at room temperature. Then, 1-Step Ultra TMB-Blotting Solution (Thermo Fisher scientific) was added to each well and incubated for 3 min at room temperature. The reaction was stopped by the addition of 2 MH2SO4 and the optical density at 450 nm (OD450) was immediately measured. The average OD450 values oftwo PBS-wells were subtracted from the average OD450 values of the two RBD or S6pro wells for background correction. Asubtracted OD450 value of 0.1 or more was regarded as positive; the minimum dilution to give a positive result was used as the ELISA titre (hamster serum) or minimum concentration to bind the S protein (monoclonal antibodies).
Focus reduction neutralization test
Neutralization activities of SARS-CoV-2 were determined by using an FRNT as previously described15. Serial dilutions of plasma were mixed with 1,000 focus-forming units of virus per well and incubated for 1 h at 37 °C. The antibody-virus mixture was inoculated on VeroE6/TMPRSS2 cells in 96-well plates in duplicate and incubated for 1 h at 37 °C. An equal volume of 1.2% Avicel RC-581 (DuPont Nutrition) in culture medium was added to each well. The cells were incubated for 24 h at 37 °C and then fixed with formalin. After the formalin was removed, the cells were immunostained with a mouse monoclonal antibody against SARS-CoV-1/2 nucleoprotein (clone 1C7C7 (Sigma-Aldrich)) (1:2,000 dilution), followed bya horseradish peroxidase-labelled goat anti-mouse immunoglobulin (SeraCare Life Sciences) (1:2,000 dilution). The infected cells were stained with TrueBlue Substrate (SeraCare Life Sciences) and then washed with distilled water. After cell drying, the focus numbers were quantified by using an ImmunoSpot S6 Analyzer, ImmunoCapture software, and BioSpot software (Cellular Technology). The results are expressed as the 50% focus reduction neutralization titre (FRNT50). The FRNT50 values were calculated by using GraphPad Prism (GraphPad Software).
Reagent availability
All materials are available from the authors or from commercially available sources, except for the clinical specimens. Owing to the extremely limited amount of these clinical specimens, we are unable to make them available.
Statistical analysis
GraphPad Prism was used to analyse all of the data. Statistical analysis included unpaired t-tests, Mann–Whitney tests, log-rank (Mantel–Cox) tests and ANOVA with multiple corrections post-test. Differences among groups were considered significant for P < 0.05.
Extended Data
Extended Data Fig. 1 |. Host responses in the lungs of Balb/c mice infected with SARS-CoV-2 Omicron/BA.2, related to Fig. 1e.
Balb/c mice were intranasally infected with Beta/B.1.351 (HP01542), Omicron/BA.1 (NC928), or Omicron/BA.2 (NCD1288). Pro-inflammatory cytokine/chemokine responses in the lungs of the infected mice were assessed at 1, 2, or 3 dpi (infected mice, n = 4; naïve mice, n = 3). Vertical bars show the mean ± s.e.m. Points indicate data from individual mice. Data were analyzed by two-way ANOVA with Tukey’s multiple comparisons test. Data are from one experiment.
Extended Data Fig. 2 |. Host responses in the lungs of K18-hACE2 mice infected with SARS-CoV-2 Omicron/BA.2, related to Fig. 2c.
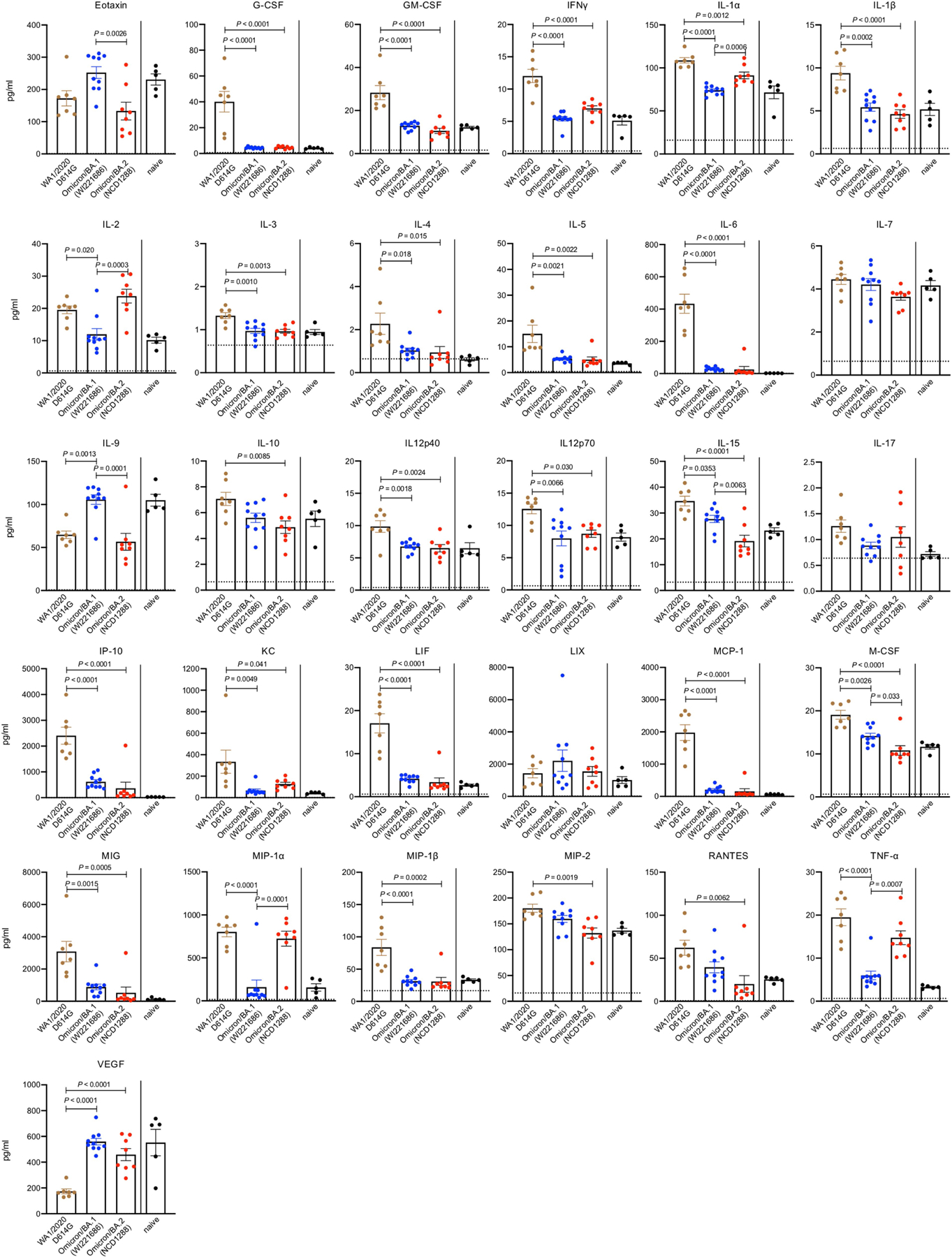
K18-hACE2 mice were intranasally inoculated with 103 PFU in 50 μL of WA1/2020 D614G, Omicron/BA.1 (NC928), or BA.2 (NCD1288). Pro-inflammatory cytokine/chemokine responses in the lungs of the infected mice were assessed at 3 dpi [WA1/2020 D614G-infected mice, n = 7; Omicron/BA.1 (NC928)-infected mice, n = 10; Omicron/BA.2 (NCD1288)-infected mice, n = 8; naïve mice, n = 5]. Vertical bars show the mean ± s.e.m. Points indicate data from individual mice. Data were analyzed by using a two-way ANOVA with Tukey’s multiple comparisons test. Cytokine and chemokine data were run in a single experiment but were obtained mice harvested in two independent experiments.
Extended Data Fig. 3 |. Pathogenicity in hamsters infected with SARS-CoV-2 Omicron/BA.2, related to Fig. 3a–c.
a, Syrian hamsters were intranasally inoculated with 105 PFU in 30 μL of Omicron/BA.1 (NC928), Omicron/BA.2 (NCD1288), Omicron/BA.2 (HP354), Omicron/BA.2 (TY40–385), 104.7 PFU of Omicron/BA.2 (HP353), 103 PFU of Delta/B.1.627.2 (UW5250), or PBS (mock). Body weights of virus-infected (n = 4–9) and mock-infected hamsters (n = 8, same data used in Fig. 3a) were monitored daily for 10 days. Data are presented as the mean percentages of the starting weight (± s.e.m.). b, Pulmonary function analyses in infected hamsters. Penh and Rpef were measured by using whole-body plethysmography. Mean ± s.e.m. (Omicron/BA.1 (NC928)- or Omicron/BA.2 (NCD1288)-infected hamsters, n = 4; mock-infected hamsters, n = 3). c, Virus replication in Omicron/BA.1 (NC928)- or Omicron/BA.2 (NCD1288)-infected Syrian hamsters. Hamsters (n = 4) were euthanized at 3 dpi for virus titration. Virus titers in the nasal turbinate and lungs were determined by use of plaque assays. Vertical bars show the mean ± s.e.m. Points indicate data from individual hamsters. The lower limit of detection is indicated by the horizontal dashed line. Data were analyzed by use of a one-way ANOVA with Tukey’s multiple comparisons test. Data are from one experiment.
Extended Data Fig. 4 |. Pathological findings in the lungs of SARS-CoV-2-infected animals, related to Fig. 3d.
Four hamsters per group were inoculated with 105 PFU of Omicron/BA.1 (NC928) or Omicron/BA.2 (NCD1288) and sacrificed at 3 or 6 dpi for histopathological examination. Representative images of the bronchi/bronchioles and alveoli of hamsters infected with BA.1 or BA.2 are shown. Upper panels, hematoxylin and eosin (H&E) staining. Middle panels, in situ hybridization targeting the nucleocapsid gene of SARS-CoV-2. Lower panels, immunohistochemistry with a rabbit polyclonal antibody that detects SARS-CoV-2 nucleocapsid protein. Scale bars, 100 μm.
Extended Data Fig. 5 |. micro-CT images of the lungs of SARS-CoV-2-infected Syrian hamsters, related to Figure 3e.
a-b, Syrian hamsters were intranasally inoculated with PBS (mock) (a), or with 103 PFU of Omicron/BA.1 (NC928), Omicron/BA.2 (NCD1288) or Omicron/BA.2 (HP353) (b). In addition to the images shown in Fig. 3e, the remaining representative Micro-CT images of the lungs of mock-infected hamsters (n = 2) and virus-infected hamsters (n = 3) at 7 dpi are shown. c, Representative micro-CT axial and coronal images of the lungs of four hamsters per group inoculated with 105 PFU of Omicron/BA.1 (NC928) or 105 PFU of Omicron/BA.2 (NCD1288) at 7 dpi. Lung abnormalities included minimal, patchy, ill-defined, peri-bronchial ground glass opacity (white arrowheads), and few, small, focal rounded/nodular regions (black arrows), consistent with minimal pneumonia. Coronal CT images were reformatted to optimize lesion visualization. CT severity scores for hamsters inoculated with 105 PFU of Omicron/BA.1 (n = 4) or Omicron/BA.2 (n = 4) were analyzed by using the unpaired student’s t-test. Vertical bars show the mean ± s.e.m. Points indicate data from individual hamsters. Data are from one experiment.
Extended Data Fig. 6 |. Comparison the relative fitness and infectivity of SARS-CoV-2 Omicron/BA.1 and Omicron/BA.2, related to Fig. 3f.
Omicron/BA.1 and Omicron/BA.2 co-infection. BA.1 (NC928) and BA.2 (NCD1288) were mixed at an equal ratio on the basis of their infectious titers, and the virus mixture (total 2 × 103 PFU) was inoculated into five hamsters. Nasal turbinates and lungs were collected from the infected animals at 4 dpi and analyzed by using next generation sequencing (NGS). Shown are the relative proportions of BA.1 and BA.2 in the infected animals. Data are from one experiment.
Extended Data Fig. 7 |. Pathogenicity and replication in hACE2-expressing hamsters infected with SARS-CoV-2 Omicron/BA.2, related to Fig. 4.
a–c, hACE2-expressing Syrian hamsters (line M51) were intranasally inoculated with 103 PFU in 30 μL of D614G (HP095) or Omicron/BA.2 (NCD1288) a, b, Body weights (a) and survival (b) were monitored daily for 14 days. The values for body weights are presented as the mean percentages of the starting weight ± s.e.m. Survival data were analyzed by using the Log-rank (Mantel-Cox) test. c, Three hamsters per group were euthanized at 3 or 5 dpi for virus titration. Virus titers in the nasal turbinates and lungs were determined by use of plaque assays. Vertical bars show the mean ± s.e.m. Points indicate data from individual hamsters. The lower limit of detection is indicated by the horizontal dashed line. Data were analyzed by using the Mann-Whitney test (titers in the lungs at 3 dpi) or unpaired student’s t-test (titers in the lungs at 3 dpi, and those in the nasal turbinates at 3 and 5 dpi). Data are from one experiment.
Supplementary Material
Acknowledgements
We thank Shionogi for providing S-217622; S. Watson for scientific editing; S. Matsubara, K. Yokota, N. Mizutani, H. Morita, H. Miura, S. Nagata and Y. Sato for technical assistance along with the IASO study team for their efforts with the organization and collection of blood samples from individuals after immunization. This work was supported by a Research Program on Emerging and Re-emerging Infectious Diseases (JP20fk0108412, JP20fk0108502, JP21fk0108615, JP20fk0108472 and JP21fk0108104), a Project Promoting Support for Drug Discovery (JP20nk0101632), the Japan Program for Infectious Diseases Research and Infrastructure (JP22wm0125002) from the Japan Agency for Medical Research and Development (AMED), the National Institutes of Allergy and Infectious Diseases Center for Research on Influenza Pathogenesis (HHSN272201400008C), R01 AI157155 (NIAID), the Center for Research on Influenza Pathogenesis and Transmission (CRIPT) (75N93021C00014), the Collaborative Influenza Vaccine Innovation Center (75N93019C00051), and a Grant-in-Aid for Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labour and Welfare, Japan (JPMH20HA1006, JPMH 20HA2007). This work was also supported in part by the Intramural Research Program of National Center for Global Health and Medicine, and in part by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
IASO study team
Pamela Bennett-Baker25, Zijin Chu25, Dawson Davis25, Theresa Kowalski-Dobson25, Ashley Eckard25, Carmen Gherasim25, Wolf Gremel25, Kathleen Lindsey25, David Manthei25, Alyssa Meyers25, Julio Zuniga Moya25, Aaron Rico25, Emily Stoneman25, Victoria Blanc25, Savanna Sneeringer25, Lauren Warsinske25
25 University of Michigan, Ann Arbor, MI, USA.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-022-04856-1.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Competing interests Y.K. has received unrelated funding support from Daiichi Sankyo Pharmaceutical, Toyama Chemical, Tauns Laboratories, Shionogi, Otsuka Pharmaceutical, KM Biologics, Kyoritsu Seiyaku, Shinya Corporation and Fuji Rebio. M.S.D. is a consultant for Inbios, Vir Biotechnology, Senda Biosciences and Carnival Corporation, and is on the scientific advisory boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, Immunome and Emergent BioSolutions. The other authors declare no competing interests.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-022-04856-1.
Peer review information Nature thanks Charaf Benarafa, Timothy Sheahan and the other, anonymous, reviewers for their contribution to the peer review of this work.
Data availability
All data supporting the findings of this study are available in the paper. There are no restrictions in obtaining access to primary data. Source data are provided with this paper. The sequences of the viruses (hCoV-19/Japan/UT-HP353-1N/2022 (Omicron BA.2; HP353), hCoV-19/Japan/UT-HP354-1N/2022 (Omicron BA.2; HP354) and hCoV-19/Japan/TY40-385/2022 (Omicron BA.2; TY40-385)) were deposited in the Global Initiative on Sharing All Influenza Data (GISAID) database (https://www.gisaid.org/) with accession ID EPI_ISL_12438431 (HP353), EPI_ISL_12438433 (HP354), and EPI_ISL_12438432 (TY40-385), respectively. Source data are provided with this paper.
References
- 1.Callaway E & Ledford H How bad is Omicron? What scientists know so far. Nature 600, 197–199 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Flemming A Omicron, the great escape artist. Nat. Rev. Immunol 22, 75 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Weekly epidemiological update on COVID-19, 5 April 2022, https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19-5-april-2022 (2022).
- 4.Yamasoba D et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell 185,2103–2115.e19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyngse F et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. Preprint at medRxiv 10.1101/2022.01.28.22270044 (2022). [DOI] [Google Scholar]
- 6.Elliott P et al. Post-peak dynamics of a national Omicron SARS-CoV-2 epidemic during January 2022. Preprint at medRxiv 10.1101/2022.02.03.22270365 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halfmann PJ et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603, 687–692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shuai H et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603, 693–699 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Suzuki R et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 603, 700–705 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.SARS-CoV-2 Variants of Concern and Variants Under Investigation in England, Technical Briefing 36 (UK Health Security Agency, 2022). [Google Scholar]
- 11.Planas D et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Cameroni E et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 602, 664–670 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Y et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602, 676–681 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Takashita E et al. Efficacy of antibodies and antiviral drugs against Covid-19 Omicron variant. N. Engl. J. Med 386, 995–998 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walls AC et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou YJ et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie X et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med 27, 620–621 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Lubinski B et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 25, 103589 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito A et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 602, 300–306 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 602, 294–299 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leist SR et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085.e1012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinnon KH 3rd et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586, 560–566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muruato A et al. Mouse-adapted SARS-CoV-2 protects animals from lethal SARS-CoV challenge. PLoS Biol 19, e3001284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winkler ES et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol 21, 1327–1335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCray PB Jr et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol 81, 813–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai M et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl Acad. Sci. USA 117, 16587–16595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sia SF et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583, 834–838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan JF et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis 71, 2428–2446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson S et al. Radiological Society of North America Expert Consensus Statement on Reporting Chest CT Findings Related to COVID-19. Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA—Secondary Publication. J. Thorac. Imaging 35, 219–227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imai M et al. Characterization of a new SARS-CoV-2 variant that emerged in Brazil. Proc. Natl Acad. Sci. USA 118, e2106535118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilliland T et al. Protection of human ACE2 transgenic Syrian hamsters from SARS CoV-2 variants by human polyclonal IgG from hyper-immunized transchromosomic bovines. Preprint at bioRxiv, 10.1101/2021.07.26.453840 (2021). [DOI] [Google Scholar]
- 33.Gruell H et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med 28, 477–480 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng SMS et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat. Med 28, 486–489 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou J et al. Neutralization against Omicron SARS-CoV-2 from previous non-Omicron infection. Nat. Commun 13, 852 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossler A, Riepler L, Bante D, von Laer D & Kimpel J SARS-CoV-2 Omicron variant neutralization in serum from vaccinated and convalescent persons. N. Engl. J. Med 386, 698–700 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carreno JM et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 602, 682–688 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Iketani S et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 604, 553–556 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uraki R et al. Therapeutic efficacy of antibodies and antivirals against a SARS-CoV-2 Omicron variant. Research Square 10.21203/rs.3.rs-1240227/v1 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Hammond J et al. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N. Engl. J. Med 386, 1397–1408 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unoh Y et al. Discovery of S-217622, a non-covalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J. Med. Chem 65, 6499–6512 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahl A et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature 591, 451–457 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owen DR et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 374, 1586–1593 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Majumdar S & Sarkar R Mutational and phylogenetic analyses of the two lineages of the Omicron variant. J. Med. Virol 10.1002/jmv.27558 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vieillard-Baron A et al. Epidemiological characteristics and severity of Omicron variant cases in the APHP critical care units. Preprint at medRxiv 10.1101/2022.01.25.22269839 (2022). [DOI] [Google Scholar]
- 46.Meng B et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603, 706–714 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferren M et al. Hamster organotypic modeling of SARS-CoV-2 lung and brainstem infection. Nat. Commun 12, 5809 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomris I et al. Distinct spatial arrangements of ACE2 and TMPRSS2 expression in Syrian hamster lung lobes dictates SARS-CoV-2 infection patterns. PLoS Pathog 18, e1010340 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Case J et al. Resilience of S309 and AZD7442 monoclonal antibody treatments against infection by SARS-CoV-2 Omicron lineage strains. Preprint at bioRxiv 10.1101/2022.03.17.484787 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuyama S et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl Acad. Sci. USA 117, 7001–7003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen RE et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med 27, 717–726 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corbett KS et al. mRNA-1273 protects against SARS-CoV-2 Beta infection in nonhuman primates. Nat. Immunol 22, 1306–1315 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yasuhara A et al. Antigenic drift originating from changes to the lateral surface of the neuraminidase head of influenza A virus. Nat. Microbiol 4, 1024–1034 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Case JB, Bailey AL, Kim AS, Chen RE & Diamond MS Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology 548, 39–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chung M et al. CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 295, 202–207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itokawa K, Sekizuka T, Hashino M, Tanaka R & Kuroda M Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS ONE 15, e0239403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quick J nCoV-2019 sequencing protocol protocols.io https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye (2020).
- 58.Itokawa K et al. nCoV-2019 sequencing protocol for Illumina V.5 protocols.io https://www.protocols.io/view/ncov-2019-sequencing-protocol-for-illumina-b2msqc6e?version_warning=no (2021).
- 59.Yamayoshi S et al. Antibody titers against SARS-CoV-2 decline, but do not disappear for several months. eClinicalMedicine 32, 100734 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available in the paper. There are no restrictions in obtaining access to primary data. Source data are provided with this paper. The sequences of the viruses (hCoV-19/Japan/UT-HP353-1N/2022 (Omicron BA.2; HP353), hCoV-19/Japan/UT-HP354-1N/2022 (Omicron BA.2; HP354) and hCoV-19/Japan/TY40-385/2022 (Omicron BA.2; TY40-385)) were deposited in the Global Initiative on Sharing All Influenza Data (GISAID) database (https://www.gisaid.org/) with accession ID EPI_ISL_12438431 (HP353), EPI_ISL_12438433 (HP354), and EPI_ISL_12438432 (TY40-385), respectively. Source data are provided with this paper.


