Highlights
The lipidome of cells and organelles is more complex than originally anticipated. It has become clear that biomembranes are active materials bearing a tremendous regulatory potential. Cells control the collective biophysical properties of their organelle membranes in order to maintain the organization and functionality of the membrane proteome.
The concept of homeoviscous adaptation provided an intuitive interpretation for the changes of membrane compositions with temperature. However, the mechanistic relevance of membrane viscosity for many cellular processes including those regulating the lipid fatty acyl chain composition has been recently challenged.
The crucial role of membrane compressibility and thickness in organizing the membrane proteome, however, has gained fresh emphasis. The transverse membrane compressibility of the endoplasmic reticulum (ER) membrane modulates crucial aspects of transmembrane protein biology including their bilayer insertion and extraction, their conformational dynamics and activity, as well as their sorting along the secretory pathway and inheritance from mother to daughter cells.
The unfolded protein response (UPR) can sense aberrant ER membrane stiffening and surfaces as a prime candidate for balancing membrane lipid and protein production at the ER as a mechanism of biophysical membrane homeostasis during cellular stress.
Keywords: membrane fluidity, membrane thickness, membrane compressibility, homeoviscous adaptation, unfolded protein response
Abstract
Biomembranes are complex materials composed of lipids and proteins that compartmentalize biochemistry. They are actively remodeled in response to physical and metabolic cues, as well as during cell differentiation and stress. The concept of homeoviscous adaptation has become a textbook example of membrane responsiveness. Here, we discuss limitations and common misconceptions revolving around it. By highlighting key moments in the life cycle of a transmembrane protein, we illustrate that membrane thickness and a finely regulated membrane compressibility are crucial to facilitate proper membrane protein insertion, function, sorting, and inheritance. We propose that the unfolded protein response (UPR) provides a mechanism for endoplasmic reticulum (ER) membrane homeostasis by sensing aberrant transverse membrane stiffening and triggering adaptive responses that re-establish membrane compressibility.
Biomembranes are complex materials composed of lipids and proteins that compartmentalize biochemistry. They are actively remodeled in response to physical and metabolic cues, as well as during cell differentiation and stress. The concept of homeoviscous adaptation has become a textbook example of membrane responsiveness. Here, we discuss limitations and common misconceptions revolving around it. By highlighting key moments in the life cycle of a transmembrane protein, we illustrate that membrane thickness and a finely regulated membrane compressibility are crucial to facilitate proper membrane protein insertion, function, sorting, and inheritance. We propose that the unfolded protein response (UPR) provides a mechanism for endoplasmic reticulum (ER) membrane homeostasis by sensing aberrant transverse membrane stiffening and triggering adaptive responses that re-establish membrane compressibility.
Biomembranes are complex, composite materials
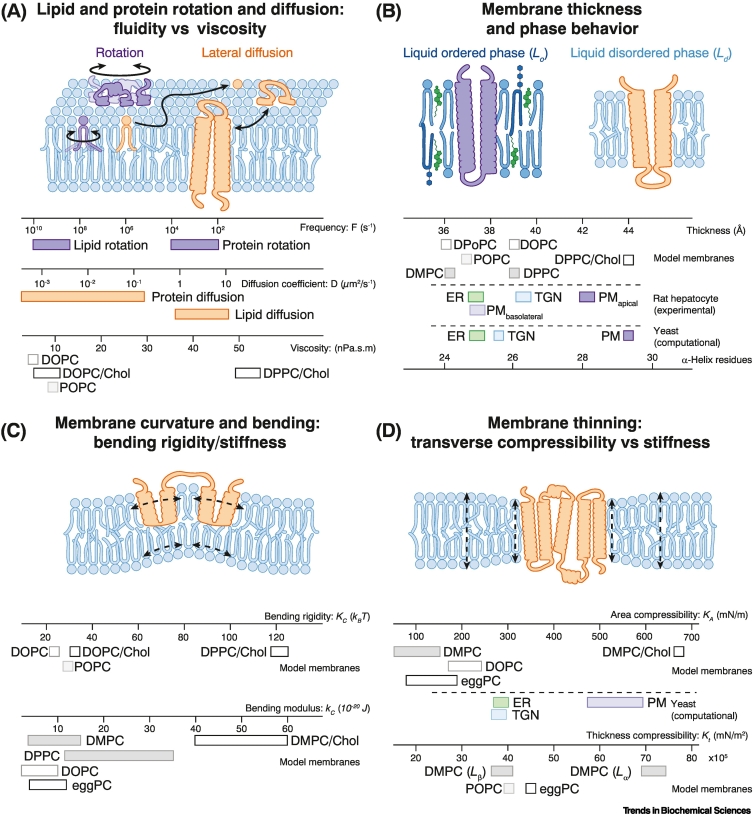
Biomembranes are fascinating materials composed of proteins embedded in a bilayer of lipids. These semipermeable barriers of 3–4.5 nm thickness encapsulate cells and organelles in various shapes to compartmentalize biochemistry. Because most lipids are not covalently linked to proteins, they can form fluid bilayers that self-repair and undergo extreme topological transitions during fusion and fission. Individual lipids can regulate membrane protein function through stereo-specific interactions or contribute to cellular signaling as second messengers. Such highly specific protein–lipid interactions are complemented by bulk membrane properties such as membrane fluidity (see Glossary), membrane bending stiffness/rigidity, membrane thickness, and membrane compressibility (Figure 1A–D), which globally and potently modulate virtually every aspect of membrane protein biology.
Figure 1.
Selected biophysical membrane properties discussed in this review.
A schematic representation of biophysical membrane properties (top) is shown along with experimentally and computationally determined values on model membranes and biomembranes (bottom). (A) Bulk membrane viscosity modulates rotation and lateral diffusion of lipids and proteins in biomembranes. The indicated range of values for membrane properties is based on rotation frequencies [7], diffusion rates [8., 9., 10., 11., 12.], and viscosities [13]. (B) Membrane phase separation gives rise to membrane (nano)domains differing in thickness and lipid order. Different membrane lipids and proteins partition differently between coexisting domains. The indicated ranges for membrane thickness were determined for membranes composed of only one [14] or two lipids [15], for complex rat hepatocyte membranes [16], or, via molecular dynamics simulations, for yeast organelle membranes [17]. (C) Membrane bending rigidity refers to the resistance of a biomembrane against bending and curvature. The respective bending rigidities and bending moduli are plotted below [13]. (D) Transverse membrane stiffness refers to the resistance of a bilayer to a force applied perpendicular to the membrane plane causing either bilayer compression or stretching. Typically, cholesterol can be expected to increase membrane viscosity, bending rigidity, thickness, acyl chain order, and transverse membrane stiffness in most fluid, lamellar biomembranes. Thickness compressibility and area compressibility are two closely related aspects of membrane compressibility [18]. Hence, we provide values for both thickness compressibility and area compressibility of model membranes [13] along with the area compressibility of yeast organelle membranes derived from molecular dynamics simulations [17]. Abbreviations: Chol, cholesterol; DMPC, dimyrisoylphosphatidylcholine (C14:1/C14:1 PC); DOPC, dioleoylphosphatidylcholine (C18:1/C18:1 PC); DPoPC, dipalmitoleoylphosphatidylcholine (C16:1/C16:1 PC); DPPC, dipalmitoylphosphaticylcholine (C16:0/C16:0 PC); eggPC, egg yolk isolated phosphatidylcholine; ER, endoplasmic reticulum; PM, plasma membrane; POPC sn-1-palmitoyl-sn-2-oleoylphosphatidylcholine (C16:0/C18:1 PC); TGN, trans Golgi network.
Even though a fluid lipid bilayer can be generated in vitro using a single lipid species, biomembranes found in vivo consist of a plethora of lipids that are distinct in their structures, properties, and functions. Lipidome complexity ranges from dozens of identified lipid species in some bacteria [1] to several hundred in Saccharomyces cerevisiae [2], and over a thousand in mammalian cells [3]. The lipid composition of biomembranes differs between organisms, cell types, and organelles [4] and is dynamically remodeled in response to both physical and metabolic cues. Hence, biomembranes are complex and adaptive materials that regulate protein function, organize cellular signaling, and establish organelle identity.
While we are far from understanding the full relevance of lipid diversity, the complex composition of biomembranes and the resulting biophysical properties encode a wealth of information with great regulatory potential. Integral membrane protein folding, topology, structure, dynamics, localization, function, and stability are affected by specific protein–lipid interactions and collective biophysical membrane properties [5,6]. Although being only vaguely defined, membrane fluidity is arguably the most famous membrane property and often used to rationalize how cellular phenotypes affected by the membrane lipid composition. Here, we discuss new trends at the intersection of membrane biochemistry, biophysics, and cell biology by critically dissecting the role of membrane fluidity and highlighting the contribution of membrane compressibility to key aspects of cellular physiology. We aim to raise an awareness for the entangled yet distinct impact of biophysical membrane properties on membrane proteins both at the molecular and the cellular level and suggest a crucial role of the unfolded protein response (UPR) in surveilling endoplasmic reticulum (ER) membrane compressibility.
Membrane fluidity and the dogma of homeoviscous adaptation
Biomembranes are liquid-crystalline assemblies allowing lipids and proteins to rotate along their longitudinal axis and to diffuse laterally, thereby exchanging their neighbors and exploring the membrane (Figure 1A). Membrane viscosity is modulated by the membrane composition and is a common measure for fluidity [13]. Membrane fluidity, however, remains vaguely defined, but generally refers to the dynamic behavior of membrane constituents. It must not be confused with a structural property such as the lipid packing density. The translational diffusivity of fluorescent proteins and lipid analogues often correlates with the lipid packing density, but also depends on other factors such as temperature, the ionic strength, the pH, and the exact lipid composition (Box 1). The diffusion coefficient (D) of translational motion can be determined by a variety of techniques such as fluorescence recovery after photobleaching (FRAP), single particle tracking, and scanning stimulated emission depletion fluorescence correlation spectroscopy (STED-FCS) featuring different temporal and spatial resolution [8,9,19]. Fluorescent lipid analogues diffuse much faster in artificial membranes (D ≈ 8.5 μm2/s) than in biomembranes (D ≈ 1.2 μm2/s) that are crowded and contain both obstacles and barriers [10]. The diffusion coefficient of membrane proteins in biomembranes ranges from ~0.0002 to 0.5 μm2/s and are typically 10–100-fold higher in artificial, uncrowded membranes lacking confinements [8,11,12]. Even though lipid bilayers are often regarded as 2D fluids, membrane proteins do not distribute randomly in biomembranes [20,21]. Hydrophobicity, charge density, and lateral stiffness can vary dramatically along the third dimension of the lipid bilayer (the bilayer thickness) thereby affecting membrane protein localization and function in multiple ways.
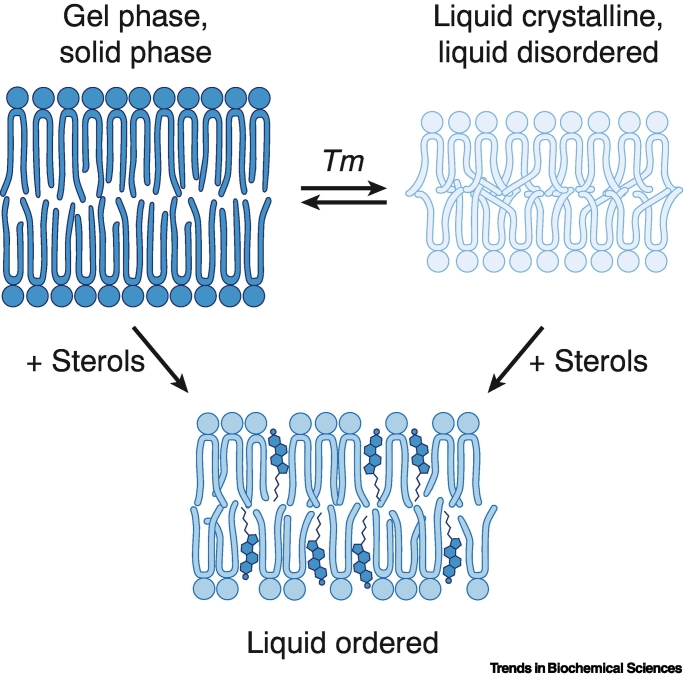
Box 1. Lipid packing and membrane fluidity.
Biomembranes mainly exist as fluid lamellar bilayers. Depending on the lipid composition and environmental factors (e.g., pH or temperature), the packing of lipids and the conformation of the lipid acyl chains can vary (Figure I). Differences in lipid packing leads to the formation of different lamellar phases. Most biological membranes are rich in unsaturated lipids and adopt a fluid and disordered phase at room temperature known as the liquid crystalline phase (Lα) or the liquid disordered phase (LD). Membranes that mainly consist of saturated lipids can adopt at low temperatures an ordered and rigid gel phase (Lβ) also known as the solid ordered phase (SO). Including cholesterol in complex lipid mixtures leads to ordering of the lipid acyl chains and the formation of a liquid ordered phase (LO) that is relatively fluid despite the high lipid order.
In model membranes consisting of a single (or few) lipid species, the transition between Lβ to the Lα phase (gel-to-liquid crystalline) can be observed by slowly heating or cooling the membranes thereby crossing the phase transition temperature known as the lipid melting temperature Tm. Longer, saturated lipid acyl chains result in higher Tm values due to their tight packing and increased van der Waals interactions [14]. Shorter acyl chains, unsaturated acyl chains pack less tightly and lower the Tm value. In addition to the lipid acyl chains, other factors influence the Tm such as the lipid headgroups, the presence of sterols, the ionic strength and the pH. From the Tm of a lipid species, one can predict the phase behavior at a given temperature. As such, it is a good indication for the contribution of this lipid species to lipid ordering. In general, tight packing lipids (saturated phospholipids, sphingolipids) and cholesterol increase membrane lipid order and make them stiffer, whereas loosely packing lipids (unsaturated phospholipids) make a membrane more fluid [14].
Figure I.
The lipid composition and environmental factors determine membrane phase behavior.
Lamellar lipid bilayers come in distinct flavors differing in lipid packing and fluidity. Lipids with long, saturated acyl chains tend to form a non-fluid gel phase (Lβ) also known as solid ordered phase (SO). Most biological membranes at physiological temperatures are in the liquid disordered phase (LD) charaterized by a high degree of membrane fluidity with respect to lateral diffusion. Sterols can order lipid acyl chains and increase lipid packing thereby giving rise to a liquid ordered phase (LO), which can coexist with LD domains in the same biomembrane. Furthermore, sterols can ‘fluidize’ gel domains.
Alt-text: Box 1
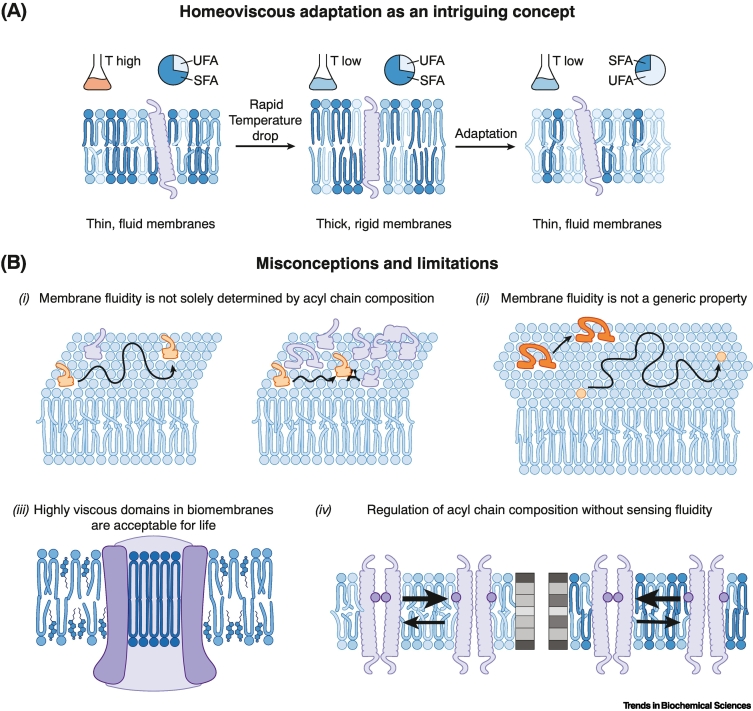
Nevertheless, membrane fluidity/viscosity has gained a ‘celebrity status’ among the bulk membrane properties since it was proposed to be homeostatically regulated in response to environmental temperature [22]. When liposomes formed from Escherichia coli lipid extracts are cooled below their melting temperature, they undergo a fluid-to-solid phase transition and form a gel phase [22] (Figure 2A). However, this does not happen with lipid extracts from cold-adapted E. coli cultivated at low temperatures [22], likely due to the increased abundance of unsaturated acyl chains that reduces lipid packing [23]. Based on these observations, it was proposed that E. coli maintains membrane fluidity by adapting the lipid acyl chain composition in a process referred to as ‘homeoviscous adaptation‘ (Figure 2A). The concept of homeoviscous adaptation inspired hundreds of studies on temperature-dependent changes of the lipidome across the tree of life and has become the textbook example of membrane responsiveness.
Figure 2.
The homeoviscous adaptation concept and common misconceptions.
(A) The homeoviscous adaptation concept as proposed in its original form [22]. Bacteria cultivated at a given temperature established a characteristic lipid composition with a defined ratio of saturated (SFA) to unsaturated (UFA) lipid acyl chains. When exposed to a sudden drop of temperature, these lipids can no longer support membrane fluidity, lipid packing increases, and the membrane solidifies. However, cold-adapted bacteria can maintain membrane fluidity in the cold due to an increased proportion of unsaturated fatty acyl chains in their membranes. (B) Membrane fluidity is only a vaguely defined term, but frequently used to refer to rotational or translational motions of membrane constituents, membrane phase behavior, lipid packing, membrane compositions, or a combination of those. Important misconceptions remain even if membrane fluidity is used to refer to only translational motions. (i) Lipid acyl chains are not the only regulators of membrane viscosity. Membrane crowding, for example, has a significant impact on bulk membrane viscosity. (ii) Membrane fluidity is not a generic property. It is distinct for different molecules (here lipids versus proteins) and affected both by the length and time-scale of observation. (iii) The formation of solid-like gel phases is often considered as being incompatible with life. Yet, a reversible formation of solid-like gel phases has been observed in bacteria and mammalian cells, while highly ordered, sphingolipid-rich, but cholesterol-poor membrane patches were structurally resolved in an hexamer of the proton pump Pma1 from yeast [35], thereby providing also structural evidence for gel domains in the plasma membrane as previously predicted based by fluorescence spectroscopy [36]. (iv) The sensory mechanism regulating the proportion of saturated and unsaturated fatty acyl chains in biomembranes does not sense membrane fluidity. The lipid saturation sensor Mga2, for example, uses a bulky tryptophan residue (purple circle) to sense a small portion of the lateral stiffness profile in the membrane (indicated by different shades of gray) and its degree of activation does not correlate with membrane fluidity [37].
Lipid remodeling in response to environmental temperature has been observed in a broad range of poikilothermic organisms, including bacteria [1], archaea [24], yeast [2,25], plants [26], worms [27], and fish [28]. Remarkably, body temperature and lipid compositions do not only correlate, but they also seem to be interdependent. When fruit flies are exposed to colder temperatures, they adjust their food preference from yeast to plants to obtain the polyunsaturated fatty acids required to maintain membrane fluidity [29]. Ectothermic lizards can adjust their body temperature by rocking back and forth [30], but the body temperature set-point decreases by several degrees when they are on a diet rich in polyunsaturated fatty acids [31]. Even warm-blooded mammals adjust their lipid composition with temperature. This is illustrated by the outer extremities of arctic reindeer, which are exposed to permafrost and feature high concentrations of unsaturated fatty acyl chains [32]. Hence, it is not surprising that lipidome remodeling for biophysical membrane homeostasis is also observed in cultured mammalian cells [33]. Climate change puts a new spotlight on the concept of homeoviscous adaptation as a basis for the global remodeling of biomembranes in poikilotherms with unpredictable consequences on biodiversity [34].
Homeoviscous adaptation – limitations and misconceptions
The concept of homeoviscous adaptation provides an intuitive interpretation for the ubiquitously observed changes of membrane compositions with temperature, but its validity has been challenged [37,38]. Here, we discuss some of its main limitations and common misconceptions revolving around it (Figure 2B).
Membrane fluidity is not solely determined by the acyl chain composition
The unsaturation of the lipid acyl chains is often regarded as the key determinant of membrane fluidity and the main player in homeoviscous adaptation [22]. However, lipid acyl chains are relevant for membrane functions well beyond their impact on membrane fluidity [39]. Bulk membrane fluidity is determined not only by acyl chain unsaturation, but also by the lipid composition at various other levels, including the lipid headgroup composition, acyl chain length, localization of the double bond, and the abundance of sphingolipid and sterols [14,40]. Crowding of membrane proteins significantly reduces their diffusion [41]. In fact, molecular dynamics simulations indicate that cholesterol concentration and protein crowding have a larger impact on membrane viscosity than the acyl chain composition [42] (Figure 2B, i). Thus, both proteins and lipids are essential determinants of membrane fluidity.
Membrane fluidity is not a single, generic property of a biomembrane
Fluorescent probes can elegantly uncover diverse dynamic processes in biomembranes via their rotational or translational mobilities. Each probe, however, reports on distinct aspects of membrane fluidity depending on their size, shape, and preferred position in the lipid bilayer. For example, the anisotropy of different fluorescent probes is employed to report on rotational motions a different depths in the lipid bilayer [43], while FRAP experiments assess translational motions, diffusion coefficients, and physically defined membrane viscosities. Hence, a single biomembrane features different viscosities for each molecule and at different length scales [13] (Figure 2B, ii). Even though the diffusion of objects is only weakly dependent on the size of the hydrophobic membrane insertion [44,45], protein diffusion in biomembranes is typically more constrained than lipid diffusion resulting in 10–100-fold lower diffusion coefficients for proteins [8,11]. Some rhomboid intramembrane proteases have been observed to diffuse unusually fast, presumably by locally distorting the lipid bilayer [12].
Solid-like gel domains are acceptable for life
Solid-like membrane domains (gel domains) are often assumed to be incompatible with life. In fact, the photosynthetic activity in the cyanobacterium Anacystis nidulans was observed to cease at a temperature that coincides with the onset of gel phases in the plasma membrane [46]. While a minimum of membrane fluidity is obviously essential to maintain cellular signaling and functional electron transport chains for energy production, this does not preclude the possibility that gel domains may coexist with fluid membrane domains in complex biomembranes as observable in model membranes. Indeed, bacterial membranes depleted of fluidity-promoting unsaturated lipids can separate into coexisting membrane domains featuring vastly distinct viscosities [47]. Under these conditions, membrane proteins accumulate in the fluid membrane domain to maintain their vital activities, while gel domains are virtually depleted of membrane proteins [47]. A similar exclusion of transmembrane proteins from a membrane domain has also been observed in the plasma membrane of phosphatidylserine-deficient yeast [48] and in the yeast vacuole upon nutrient starvation [49]. Even though the yeast vacuole membrane seems to phase-separate into two fluid membrane domains, a liquid disordered (Ld) and liquid ordered (Lo) one, the exclusion of membrane proteins in the Lo domain raises several questions. By what mechanism are integral membrane proteins excluded? How do the Lo domains contribute to the turnover of lipid droplets by micro-autophagy/lipophagy for energy homeostasis under nutrient starvation [50]? What, if anything, is required for observing gel phases in the vacuole membranes?
It is remarkable that when one analyzes the thermotropic properties of biological membranes (or membrane extracts) from an organism, the phase transition temperature of the membranes often correlates with the environmental temperature [38,49,51]. For example, the main transition temperature Tm of E. coli membranes perfectly follows the temperature of cultivation [22,52]. Hence, phase transition temperatures are kept close and at a constant distance to the cultivation temperature thereby suggesting a tight regulation and a possible physiological relevance.
Solid-like domains of high lipid order, which likely represent gel domains, were observed in the ER of mammalian cells exposed either to saturated fatty acids [53] or to cold [54], further indicating that the formation of gel phases is reversible and does not contradict life. Particularly strong, complementary evidence for the existence of gel (nano)domains has been collected in yeast. Fluorescence spectroscopy and confocal microscopy suggested the presence of gel domains in the yeast plasma membrane, which is composed of a variety of coexisting compartments including the microcompartment containing Pma1 [36,55]. Cryoelectron microscopy of the hexameric proton pump Pma1 revealed a highly ordered membrane patch composed of 57 lipids, which is surrounded by the protein even after its isolation via a nonionic detergent [35] (Figure 2B, iii). Isolating microcompartments containing Pma1 via styrene–maleic acid–lipid-particles revealed by lipidomic analyses an increased abundance of sphingolipids, but not ergosterol [56]. These observations provide strong evidence for highly ordered, sphingolipid-enriched, but sterol-depleted membrane patches in the yeast plasma membrane, which to all likelihood represent a gel nanodomain. Hence, solid-like domains may be more prevalent in biology than often anticipated.
Lipid acyl chain composition is regulated without sensing membrane viscosity
A critical role of the bulk membrane fluidity has been established for the diffusion of electron carriers in the electron transport chains of the bacterial cell membrane and the inner mitochondrial membrane of yeast [57]. Membrane lipid perturbation impairs the diffusion of ubiquinol and thus electron transport, thereby suggesting that membrane fluidity is indeed limiting for energy production when lipid metabolism is perturbed [57].
However, physiological changes of biomembrane compositions can affect cellular processes without a relevant impact on lipid and membrane protein diffusion. This is the case even for machineries that regulate the lipid acyl chain composition, often assumed to be the main determinant of membrane fluidity [37,58]. Arguably the best understood system regulating membrane fluidity is the OLE pathway in S. cerevisiae, which controls the production of unsaturated fatty acids by regulating the expression of OLE1 that encodes the sole fatty acid desaturase Ole1 [59]. The membrane-embedded sensor protein Mga2 responds to changes in the lateral stiffness profile from saturated lipids by populating distinct rotational configurations of its dimeric transmembrane domain (TMD) [60] (Figure 2B, iv). These changes in the conformational dynamics of the TMD trigger the ubiquitylation of the sensor protein, which is then cleaved by the proteasome to release a transcriptionally active fragment that upregulates OLE1 expression [37,59]. Increased fatty acid desaturation and the incorporation of unsaturated fatty acids in membrane lipids should then decrease lipid packing and membrane viscosity [59]. Reconstituting key steps of this pathway in vitro revealed that the ubiquitylation of the Mga2 sensor does not correlate with membrane viscosity, thereby demonstrating that changes in membrane fluidity cannot be the trigger for activating the OLE pathway [37]. Notably, an analogous sensory system regulating lipid saturation in Bacillus subtilis relies on the transmembrane protein DesK, which uses membrane thickness as a cue to upregulate lipid desaturation [58]. If membrane viscosity is not even relevant for the sensors that regulate the production of fluidity-promoting, unsaturated lipids, we should carefully re-evaluate the significance of membrane viscosity also in other cellular processes.
Membrane thickness and compressibility orchestrate the membrane proteome
The TMDs of integral membrane proteins interface with the hydrophobic tails of membrane lipids. Normally, the hydrophobic thickness of a TMD and the lipid bilayer tend to match and each hydrophobic mismatch causes a local bilayer deformation thereby creating a fingerprint in the membrane with a perturbed thickness and curvature [61] (Box 2).
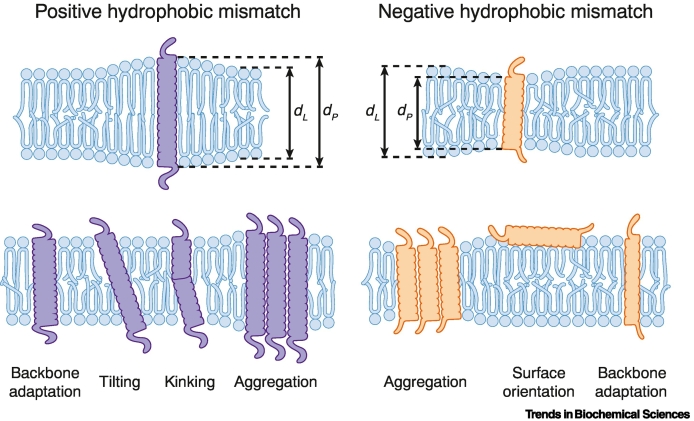
Box 2. Membrane thickness and hydrophobic mismatch.
Hydrophobic mismatch refers to a situation where the hydrophobic length of a protein TMD (dP) does not match the thickness of the lipid bilayer (dL) and the membrane gets locally deformed.
A positive hydrophobic mismatch (a long TMD in a thin bilayer or dP> dL) can be compensated either by tilting a single-pass transmembrane protein or by stretching of the lipid acyl chains (Figure I, top left). A negative hydrophobic mismatch (a short TMD in a thick bilayer) induces a local membrane thinning associated with a distortion of the bilayer and lipid acyl chain disordering (Figure I, top right). The local distortion of the bilayer can affect the conformation, stability, and activity of membrane proteins. Adaptation of proteins to hydrophobic mismatch is best studied for single pass membrane proteins and model peptides [111]. In general TMDs with either positive or negative hydrophobic mismatch tend to aggregate to reduce the energetic penalty from bilayer distortion (Figure I, bottom middle). TMDs under positive hydrophobic mismatch conditions can show backbone conformational changes in order to reduce the effective TMD length. Alternatively, the TMD can tilt or kink to reduce the effective length (Figure I, bottom left). TMDs under negative hydrophobic mismatch conditions can also adjust their confirmation, similar to TMDs under positive hydrophobic mismatch, in order to extend their effective length (Figure I, bottom right). Short polypeptides may not insert into bilayer and orient at the membrane surface.
Figure I.
Hydrophobic mismatch affects the structure, topology, and oligomerization of integral membrane proteins.
A positive hydrophobic mismatch refers to a situation where an integral membrane protein features a higher hydrophobic thickness (dP) than the surrounding lipid bilayer (dL). Negative hydrophobic mismatch refers to the opposite situation. The impact of hydrophobic mismatch on the structure, topology, and oligomerization of integral membrane proteins is illustrated schematically for two transmembrane helices differing in their hydrophobic thickness.
Alt-text: Box 2
Unlike in synthetic, lipid-only membranes, the thickness of biomembranes is dominated by the proteins residing in it rather than by the lipid composition [16]. This is because biomembranes are crowded with membrane proteins, which are less deformable than the surrounding lipid bilayer [62]. But even though membrane proteins dictate the thickness of biomembranes and the lipid acyl chain order in their vicinity [5,61], lipids are by no means irrelevant when it comes to modulating membrane protein localization and function. Virtually every step in the life cycle of a membrane protein is affected by bilayer thickness and, probably more relevant, transverse membrane compressibility. In the following, we describe different fates for an integral transmembrane protein in the ER and how these fates are crucially affected by bilayer thickness and membrane compressibility.
Membrane protein insertion and extraction
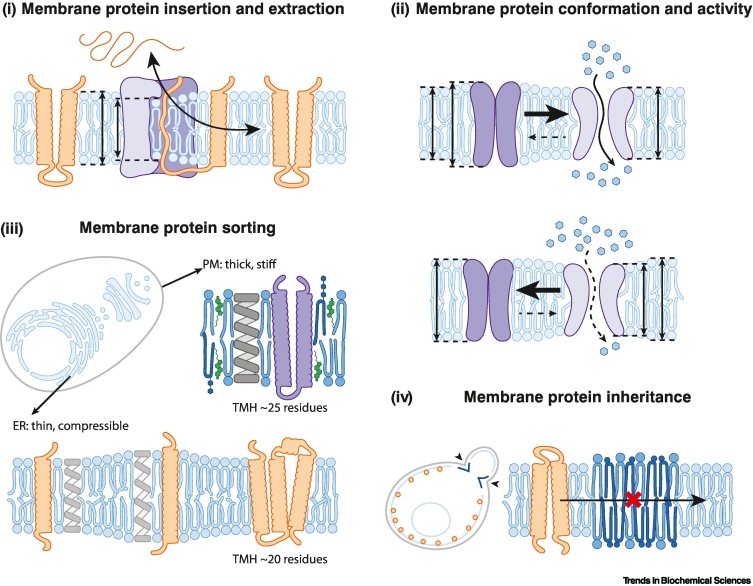
Membrane proteins are thermodynamically most stable with their hydrophobic TMD properly inserted into the lipid bilayer, but only a few of them insert spontaneously into a membrane. This is because the translocation of a polypeptide chain through the membrane requires the energetically unfavorable exposure of hydrophilic amino acid residues to the hydrophobic membrane core. Hence, most membrane proteins of the endomembrane system are inserted in the ER membrane by specialized insertases [63]. Recent studies have established membrane thinning as a common mechanistic denominator of protein translocation and membrane protein insertion systems [64] (Figure 3i). The bilayer distortion and lipid hydrocarbon chain splaying associated with locally induced membrane thinning is thought to lower the energetic barrier for moving hydrophilic portions of a polypeptide chain through the membrane. Membrane thinning is induced both by the ER-localized GET complex inserting tail-anchored proteins with only a single, C-terminal transmembrane helix [65] and the ER membrane complex (EMC) responsible for the co- and post-translational insertion of both tail-anchored proteins and multipass membrane proteins with weakly hydrophobic transmembrane segments [66]. Similar observations have been made for other insertases and protein translocating systems such as the mitochondrial TIM22 complex, the bacterial YidC insertase, and the bacterial twin-arginine translocation (TAT) system [67., 68., 69.].
Figure 3.
The cellular importance of bilayer thickness and of membrane compressibility.
Membrane compressibility affects the energetic penalty associated with a hydrophobic mismatch between a membrane protein and the lipid bilayer. Aberrant thickening and transverse membrane stiffening affects diverse aspects in the life cycle of a membrane protein. (i) The insertion and extraction of membrane proteins requires membrane thinning. (ii) Membrane thickness affects the conformational dynamics of proteins, thereby the population of distinct conformations and the rate of transitioning between them. This dependency can be used to control the activity of mechano-sensitive channels. (iii) Differences in membrane thickness and membrane compressibility (indicated by springs) contribute to the sorting of membrane proteins with transmembrane helices (TMHs) of different lengths along the secretory pathway. Aberrant endoplasmic reticulum (ER) membrane stiffening would lead to missorting of membrane proteins. (iv) Diving cells have to inherit their membrane proteins. Local patches of increased membrane thickness formed by ceramides form a diffusion barrier for some, but not all membrane proteins. By this mechanism the inheritance of aging factors is controlled. Abbreviation: PM, plasma membrane.
Protein removal from the ER and membrane protein extraction also relies on protein complexes that locally distort the lipid bilayer. The Hrd1 retrotranslocation complex, for example, removes misfolded ER-luminal and membrane proteins from the ER, and ubiquitinates them as a signal for their degradation by the proteasome [64]. The E3 ubiquitin ligase Hrd1 associates with Der1, a catalytically inactive rhomboid pseudoprotease to stabilize a locally thinned lipid bilayer, which is crucial for efficient retrotranslocation [70]. Dfm1, another member of the yeast Der1 family, induces local membrane thinning to facilitate membrane protein extraction from the ER [71].
The model whereby membrane thinning supports translocation processes may be even more general and could be extended to other amphiphilic molecules traversing biological membranes. Some lipid scramblases induce membrane distortions to catalyze the exchange of lipids between the two leaflets of a lipid bilayer [72,73]. Reconstitution experiments demonstrate a strong dependence of lipid scrambling activity on the hydrophobic thickness of the bilayer [72].
Given the importance of bilayer thinning for translocating amphiphilic molecules, one could predict that decreasing the thickness membrane compressibility should also inhibit transport. Indeed, protein translocation into ER microsomes stalls when cholesterol is delivered to the membrane, which counteracts the splaying of lipid hydrocarbon chains and thereby increases transverse membrane stiffness [74]. Consistently, the extraction of properly ubiquitylated proteins from the ER membrane proteins is hindered, when ceramides with very long chain, saturated acyl chains accumulate [75]. Even though the lipid dependency of membrane protein translocation processes remains understudied, there is accumulating evidence that the thickness membrane compressibility may be an important factor for the bilayer insertion and extraction of membrane proteins.
Protein function, conformational changes, and oligomerization
A hydrophobic mismatch between a protein and the bilayer causes stress on both bilayer lipids and the protein. While proteins are 100- to 1000-fold less deformable than the lipid bilayer, they can undergo conformational changes and the occupancy of distinct conformational states is affected by forces from the lipid bilayer [62]. Recent advances in electron microscopy make the stunning repertoire of membrane protein conformations increasingly accessible [76] and significantly extend our understanding of how native and synthetic biomembranes affect protein structure and function. The functional relevance of hydrophobic matching is particularly well documented for P-type ATPases such as the sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) or the Na+-K+-ATPase in the plasma membrane. These ion pumps are most active when reconstituted lipid environments matching their hydrophobic thickness [62,77,78] (Figure 3ii). It is likely that different protein conformations during a transport cycle induce different membrane distortions, thereby affecting transport kinetics [77,79]. Similarly, the gating of the bacterial mechano-sensitive channel of large conductance (MscL) is greatly facilitated by lowering bilayer thickness, which also lowers the activation energy between distinct closed channel intermediates [80]. The activity of G-protein-coupled receptors (GPCRs), however, can be modulated by a membrane-dependent oligomerization, which includes hydrophobic mismatch-based mechanisms [81,82]. Hence, lipid composition and bilayer properties affect protein function, by impacting on conformational dynamics and protein oligomerization.
Protein sorting along the secretory pathway
After insertion into the lipid bilayer and attaining their native fold, membrane proteins must be sorted to reach their destination. Proteins destined to the plasma membrane feature significantly longer TMDs than ER or Golgi-localized proteins [83] (Figure 3iii). In fact, the plasma membrane is >10% thicker than the membranes of the early secretory pathway in both yeast and mammals [16,84]. The ER has a low sterol content and a high proportion of monounsaturated fatty acyl chains. Therefore, it is particularly compressible and can host the vast spectrum of TMDs destined for different organelles and thus differing in their hydrophobic thicknesses. An increase of transverse membrane stiffness along the secretory pathway is thought to facilitate a gradual sorting of transmembrane proteins from the ER via the Golgi complex to the plasma membrane based on physicochemical features of the TMD [85,86]. Several cargo recruiting factors show clear preference for molecules differing in their hydrophobic thickness. The coat protein complex II (COP-II) cargo receptor Erv14, for example, preferentially recruits proteins with longer TMDs for forward transport from the ER to the Golgi apparatus [87]. Likewise, glycosylphosphatidylinositol (GPI)-anchored proteins and ceramides differing in the length of their acyl chains are clustered and sorted at specialized ER exit sites prior to their transport to the Golgi apparatus [88,89]. A local enrichment of cholesterol and a transverse membrane stiffening at ER exit sites may provide a mechanism for filtering or concentrating the appropriate cargo for forward transport [90]. Selective retention mechanisms in the Golgi complex [91,92] and the differential partitioning of TMDs in distinct transport carriers at the trans-Golgi network (TGN) provide additional selectivity for proper subcellular trafficking of proteins and lipids [85]. The significantly lower membrane area compressibility in the plasma membrane (KA ≈ 470–570 mN/m) compared with the TGN (KA ≈280 mN/m) suggests that hydrophobic mismatch-based mechanisms might be involved in the late secretory pathway [17]. The striking correlation between plasma membrane localization in vivo and protein affinity for a tightly packed, Lo membrane phase in isolated giant plasma membrane vesicles underscores a central role of hydrophobic matching and membrane compressibility for the sorting of transmembrane proteins along the secretory pathway [20].
Selective inheritance of proteins from mother to daughter cells
Cells that grow and divide must distribute their proteome and organelles between the mother and daughter cell. FRAP experiments have uncovered a diffusion barrier for ER membrane proteins along the future plane of ER membrane cleavage, while luminal proteins can pass freely [93]. This diffusion barrier for membrane proteins is established by tightly packing, saturated ceramides forming a locally thickened ER membrane [94] (Figure 3iv). The separation of the new and old ER allows for an asymmetric inheritance of protein aggregates, chaperones, and nuclear baskets has been observed in dividing yeast and neuronal stem cells [95,96]. Recent work using ER-targeted, fluorescent reporters featuring TMDs of variable hydrophobic lengths allowed for direct detection of locally thickened ER domains in live cells [97]. Regions of increased membrane thickness can also be found at contact sites between the ER and the TGN [97], which represent a main site for ceramide transport from the ER to the Golgi apparatus [98]. Hence, the inherent ability of ceramide to pack tightly with other saturated lipids is functionalized not only to establish diffusion barriers for membrane proteins, but also to generate a platform that recruits lipidic substrates for inter-organelle transport.
The examples of membrane protein insertion/extraction, function, sorting, and inheritance highlight a ubiquitously important role of membrane thickness and compressibility in organizing the membrane proteome.
Controlling ER membrane compressibility via the UPR
Aberrant thickening and a decreased membrane compressibility of the ER membrane interferes with crucial ER functions. Given its crucial role as the gateway to the secretory pathway, ER membrane homeostasis is crucial for handling the variety of transmembrane proteins entering the endomembrane system. Sensitive surveillance systems keep the sterol concentration in the ER low (~5–10 mol%) and the proportion of loosely packing, monounsaturated fatty acyl chains high (>70 mol%) [37,99]. The flux of proteins through the ER and the rate of de novo lipid biosynthesis is greatly modulated by the so-called UPR [100]. Originally identified as an ER-to-nucleus signaling pathway that responds to overloading of the ER with unfolded proteins, it has become clear that the UPR is crucial for biophysical ER membrane homeostasis [101]. Each of the three ER-localized sensors of the UPR in mammalian cells are sensitive to ER membrane aberrancies, generally referred to as lipid bilayer stress [102., 103., 104.]. The most conserved transducer of the UPR, the inositol-requiring enzyme (Ire1) from yeast, induces a local thinning of the ER membrane via its short transmembrane helix and an adjacent amphipathic helix that inserts deep into the hydrophobic core of the lipid bilayer [105]. It was proposed that this unusual TMD architecture of Ire1 increases the membrane footprint, thereby rendering it particularly sensitive to decreased compressibility of the ER membrane [106]. Consequently, Ire1 would be one of the first proteins to oligomerize via a hydrophobic mismatch-based mechanism when the ER membrane stiffens. Oligomerization of Ire1, irrespective of triggering by protein folding issues or lipid bilayer stress, activates the cytosolic effector domains and triggers the UPR [106,107]. The active UPR downregulates translation, and selectively upregulates the production of ER chaperones and redox proteins to support protein folding, protein glycosylation in the ER and the Golgi apparatus, components of the ERAD machinery to remove misfolded proteins, and lipid biosynthesis to expand the ER membrane network [100]. Notably, an active UPR remodels the entire secretory pathway including ER-to-Golgi transport, Golgi-to-ER retrieval, vacuolar targeting, and distal secretion [100].
The ability of the UPR to sense ER membrane thickness compressibility and to balance the production of membrane proteins and lipids make it a prime candidate for controlling ER compressibility in a regime that is acceptable for membrane protein insertion, folding, and sorting.
Concluding remarks
Bulk membrane viscosity and transverse membrane stiffness are often correlated and always entangled with other membrane properties, such as the lipid packing density, membrane permeability, and bending rigidity. It is challenging, but not impossible, to single out the most crucial biophysical membrane property for a given process. Membrane property sensors and curvature-sensing proteins, for instance, exhibit unique structural features rendering most sensitive to distinct biophysical properties [108,109]. In this review, we have shown that key events in the life cycle of transmembrane proteins are regulated by membrane compressibility: membrane protein insertion and extraction, activity and oligomerization, sorting, and inheritance. These examples document the importance of maintaining ER membrane compressibility and thus flexibility to hydrophobic mismatch. We suggest that the UPR, which is equipped with a remarkable sensitivity for transverse membrane stiffening of the ER, serves as a homeostatic regulator of ER membrane compressibility by balancing the ratio of protein and lipid production. Increasing the rate of lipid biosynthesis relative to membrane protein insertion would soften the ER membrane. This basal mode of UPR regulation may contribute to an anticipatory UPR that mounts adaptive responses even before misfolded and/or mislocalized membrane proteins accumulate in the ER [110]. How organelles other than the ER maintain their composition and properties is a matter of active research (see Outstanding questions). A major challenge in studying the impact of biophysical membrane properties in native biomembranes is that they are asymmetric and remarkably complex, which remains challenging to recapitulate in vitro. It will be necessary to characterize membrane proteins in context of their nanoenvironment, which they establish by selectively attracting lipids to minimize energetic penalties from membrane distortion [5,61].
Outstanding questions.
How are membrane physical properties homeostatically maintained in the membranes of organelle other than the ER?
What is the membrane compressibility modulus of the ER and other organellar membranes?
How does membrane compressibility and aberrant ER membrane stiffening affect membrane protein folding and topology?
Does the selective recruitment of lipids to a site of membrane thinning modulate reactions involving a hydrophobic mismatch in complex biomembranes?
Are there other genetically encoded membrane property sensors that can sense in a highly specific manner membrane properties such as membrane tension, permeability, or viscosity?
Is the sensitivity of UPR transducers for the transverse membrane compressibility conserved throughout evolution?
Alt-text: Outstanding questions
Acknowledgments
Acknowledgments
Work by the authors was supported by the EU Horizon 2020 research and innovation program (grant agreement no. 866011), Deutsche Forschungsgemeinschaft in the framework of the CRC1027, and Volkswagen Foundation grant #93089. We thank Kandice Levental, Ilya Levental, Tsing-Young Dora Tang, and Michael Kozlov for critically reading the manuscript, as well as the current and previous lab members for valuable discussions.
Declaration of interests
No interests are declared.
Glossary
- Diffusion coefficient (D)
describes the translational, thermal diffusivity of membrane constituents in the lipid bilayer and relates the mean square displacement <r2> to the time of motion. It is typically expressed in μm2/s. Most molecules in biomembranes undergo non-Brownian motion including anomalous diffusion caused by crowding, and confined motion. Diffusion constants can be directly measured, but the temporal and spatial resolution have an impact on the results. According to the Saffman–Delbrück model, the diffusion coefficient D is barely dependent on the size of the protein, only weakly dependent on the size of the membrane-embedded object, but strongly dependent on membrane viscosity [44].
- Hydrophobic mismatch
refers to the unfavorable situation when the length of a transmembrane domain (dP) does not match with the lipid bilayer thickness (dL) resulting in an energetic penalty that is minimized by protein conformational changes, oligomerization, and/or bilayer deformation.
- Lateral stiffness profile
a measure of the resistance of the membrane to deformation at different depths in the membrane. It is correlated with the average local atom number density of the membrane, where an increase in the atom number density of the membrane generally leads to an increase in its lateral stiffness. It is distinct from the lateral tension/pressure profile.
- Lipid packing density
a structural property of a biomembrane that refers to the degree of compaction of lipid molecules in a membrane.
- Membrane bending stiffness/rigidity
the ability of a membrane to resist bending or curvature under applied force. A low membrane bending stiffness/rigidity can be referred to as deformable.
- Membrane compressibility
a measure of the deformability of a membrane induced by a force applied either perpendicular or parallel to the membrane. Because lipids are practically incompressible, the elastic moduli for area compressibility and thickness compressibility are intimately related and practically identical [18]. Direct measurements of the thickness compressibility are scarce, but it is fair to assume it is very similar to the area compressibility KA. A low thickness membrane compressibility is referred to as transverse membrane stiffness.
- Membrane fluidity
a physical property of biomembranes that allows lipids and proteins to rotate and diffuse laterally. The anisotropy of fluorescent probes such as 1,6-diphenyl-1,3,5-hexatriene is often used as a semiquantitative proxy of membrane fluidity. Membrane viscosity is a clearly defined measure for membrane fluidity.
- Membrane thickness
generally refers to the hydrophobic thickness (nm) of a membrane. Typically, it refers to the thickness of the lipid bilayer acyl chain region (phosphate-to-phosphate distance). The thickness of crowded biomembranes is largely determined by the hydrophobic thickness of the transmembrane domains.
- Membrane viscosity (η)
a key mechanical property of biomembranes that controls time-dependent processes such as diffusion or membrane deformation. It is a measure for the resistance against deformation at a given rate. For artificial, fluid bilayers it can range from ~5 to ~55 nPa∙s∙m and it is modulated by cholesterol content [13].
- Unfolded protein response (UPR)
a signaling pathway from the ER to the nucleus. It is activated when the protein folding capacity of the ER is overwhelmed or upon stiffening of the ER membrane, referred to as lipid bilayer stress.
Contributor Information
Mike F. Renne, Email: mike.renne@uks.eu.
Robert Ernst, Email: robert.ernst@uks.eu.
References
- 1.Chwastek G., et al. Principles of membrane adaptation revealed through environmentally induced bacterial lipidome remodeling. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108165. [DOI] [PubMed] [Google Scholar]
- 2.Ejsing C.S., et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sprenger R.R., et al. Lipid molecular timeline profiling reveals diurnal crosstalk between the liver and circulation. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108710. [DOI] [PubMed] [Google Scholar]
- 4.van Meer G., et al. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levental I., Lyman E. Regulation of membrane protein structure and function by their lipid nano-environment. Nat. Rev. Mol. Cell Biol. 2022;24:107–122. doi: 10.1038/s41580-022-00524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowhan W., et al. Lipid-assisted membrane protein folding and topogenesis. Protein J. 2019;38:274–288. doi: 10.1007/s10930-019-09826-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gennis R.B. In: Biomembranes: Molecular Structure and Function. Gennis R.B., editor. Springer New York; 1989. Membrane dynamics and protein-lipid interactions; pp. 166–198. [Google Scholar]
- 8.Saxton M.J., Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 9.Sezgin E., et al. Measuring nanoscale diffusion dynamics in cellular membranes with super-resolution STED-FCS. Nat. Protoc. 2019;14:1054–1083. doi: 10.1038/s41596-019-0127-9. [DOI] [PubMed] [Google Scholar]
- 10.Schneider F., et al. Diffusion of lipids and GPI-anchored proteins in actin-free plasma membrane vesicles measured by STED-FCS. Mol. Biol. Cell. 2017;28:1507–1518. doi: 10.1091/mbc.E16-07-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobson K., et al. Lateral diffusion of proteins in membranes. Annu. Rev. Physiol. 1987;49:163–175. doi: 10.1146/annurev.ph.49.030187.001115. [DOI] [PubMed] [Google Scholar]
- 12.Kreutzberger A.J.B., et al. Rhomboid distorts lipids to break the viscosity-imposed speed limit of membrane diffusion. Science. 2019;363 doi: 10.1126/science.aao0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faizi H.A., et al. A vesicle microrheometer for high-throughput viscosity measurements of lipid and polymer membranes. Biophys. J. 2022;121:910–918. doi: 10.1016/j.bpj.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh D. CRC Press; 2013. Handbook of Lipid Bilayers. [Google Scholar]
- 15.Wang Y., et al. DPPC-cholesterol phase diagram using coarse-grained molecular dynamics simulations. Biochim. Biophys. Acta. 2016;1858:2846–2857. doi: 10.1016/j.bbamem.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Mitra K., et al. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monje-Galvan V., Klauda J.B. Modeling yeast organelle membranes and how lipid diversity influences bilayer properties. Biochemistry. 2015;54:6852–6861. doi: 10.1021/acs.biochem.5b00718. [DOI] [PubMed] [Google Scholar]
- 18.Terzi M.M., et al. Mechanical properties of lipid bilayers: a note on the Poisson ratio. Soft Matter. 2019;15:9085–9092. doi: 10.1039/c9sm01290g. [DOI] [PubMed] [Google Scholar]
- 19.Axelrod D., et al. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorent J.H., et al. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 2017;8:1219. doi: 10.1038/s41467-017-01328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen J.B., et al. How membrane geometry regulates protein sorting independently of mean curvature. ACS Cent. Sci. 2020;6:1159–1168. doi: 10.1021/acscentsci.0c00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinensky M. Homeoviscous adaptation--a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1974;71:522–525. doi: 10.1073/pnas.71.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marr A.G., Ingraham J.L. Effect of temperature on the composition of fatty acids in Escherichia coli. J. Bacteriol. 1962;84:1260–1267. doi: 10.1128/jb.84.6.1260-1267.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cario A., et al. Membrane homeoviscous adaptation in the piezo-hyperthermophilic archaeon Thermococcus barophilus. Front. Microbiol. 2015;6:1152. doi: 10.3389/fmicb.2015.01152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klose C., et al. Flexibility of a eukaryotic lipidome--insights from yeast lipidomics. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishida I., Murata N. Chilling sensitivity in plants and cyanobacteria: the crucial contribution of membrane lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:541–568. doi: 10.1146/annurev.arplant.47.1.541. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz M., et al. Evolutionarily conserved long-chain Acyl-CoA synthetases regulate membrane composition and fluidity. Elife. 2019;8 doi: 10.7554/eLife.47733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiku P.E., et al. Cold-induced expression of delta 9-desaturase in carp by transcriptional and posttranslational mechanisms. Science. 1996;271:815–818. doi: 10.1126/science.271.5250.815. [DOI] [PubMed] [Google Scholar]
- 29.Brankatschk M., et al. A temperature-dependent switch in feeding preference improves Drosophila development and survival in the cold. Dev. Cell. 2018;46:781–793.e4. doi: 10.1016/j.devcel.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 30.Hammel H.T., et al. Regulation of body temperature in the blue-tongued lizard. Science. 1967;156:1260–1262. doi: 10.1126/science.156.3779.1260. [DOI] [PubMed] [Google Scholar]
- 31.Geiser F., et al. Polyunsaturated dietary lipids lower the selected body temperature of a lizard. J. Comp. Physiol. B. 1992;162:1–4. doi: 10.1007/BF00257929. [DOI] [PubMed] [Google Scholar]
- 32.Irving L. Springer Berlin Heidelberg; 2014. Arctic Life of Birds and Mammals: Including Man. [Google Scholar]
- 33.Levental K.R., et al. Lipidomic and biophysical homeostasis of mammalian membranes counteracts dietary lipid perturbations to maintain cellular fitness. Nat. Commun. 2020;11:1339. doi: 10.1038/s41467-020-15203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm H.C., et al. Global ocean lipidomes show a universal relationship between temperature and lipid unsaturation. Science. 2022;376:1487–1491. doi: 10.1126/science.abn7455. [DOI] [PubMed] [Google Scholar]
- 35.Zhao P., et al. Structure and activation mechanism of the hexameric plasma membrane H+-ATPase. Nat. Commun. 2021;12:6439. doi: 10.1038/s41467-021-26782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos F.C., et al. Sphingolipid-enriched domains in fungi. FEBS Lett. 2020;594:3698–3718. doi: 10.1002/1873-3468.13986. [DOI] [PubMed] [Google Scholar]
- 37.Ballweg S., et al. Regulation of lipid saturation without sensing membrane fluidity. Nat. Commun. 2020;11:756. doi: 10.1038/s41467-020-14528-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hazel J.R. Thermal adaptation in biological membranes: is homeoviscous adaptation the explanation? Annu. Rev. Physiol. 1995;57:19–42. doi: 10.1146/annurev.ph.57.030195.000315. [DOI] [PubMed] [Google Scholar]
- 39.Harayama T., Antonny B. Beyond fluidity: the role of lipid unsaturation in membrane function. Cold Spring Harb. Perspect. Biol. 2023;15 doi: 10.1101/cshperspect.a041409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renne M.F., de Kroon A.I.P.M. The role of phospholipid molecular species in determining the physical properties of yeast membranes. FEBS Lett. 2018;592:1330–1345. doi: 10.1002/1873-3468.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houser J.R., et al. The impact of physiological crowding on the diffusivity of membrane bound proteins. Soft Matter. 2016;12:2127–2134. doi: 10.1039/c5sm02572a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fábián B., et al. Protein crowding and cholesterol increase cell membrane viscosity in a temperature dependent manner. J. Chem. Theory Comput. 2023;19:2630–2643. doi: 10.1021/acs.jctc.3c00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geisler L., et al. Studies on the activity of the sympathetic nervous system and the adrenal gland cortex during acute respiratory acidosis. Klin. Wochenschr. 1971;49:87–91. doi: 10.1007/BF01497305. [DOI] [PubMed] [Google Scholar]
- 44.Saffman P.G., Delbrück M. Brownian motion in biological membranes. Proc. Natl. Acad. Sci. U. S. A. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gambin Y., et al. Lateral mobility of proteins in liquid membranes revisited. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2098–2102. doi: 10.1073/pnas.0511026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murata N. Low-temperature effects on cyanobacterial membranes. J. Bioenerg. Biomembr. 1989;21:61–75. doi: 10.1007/BF00762212. [DOI] [PubMed] [Google Scholar]
- 47.Gohrbandt M., et al. Low membrane fluidity triggers lipid phase separation and protein segregation in living bacteria. EMBO J. 2022;41 doi: 10.15252/embj.2021109800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mioka T., et al. Characterization of micron-scale protein-depleted plasma membrane domains in phosphatidylserine-deficient yeast cells. J. Cell Sci. 2022;135 doi: 10.1242/jcs.256529. [DOI] [PubMed] [Google Scholar]
- 49.Leveille C.L., et al. Yeast cells actively tune their membranes to phase separate at temperatures that scale with growth temperatures. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2116007119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao P.-C., et al. Roles for L o microdomains and ESCRT in ER stress-induced lipid droplet microautophagy in budding yeast. Mol. Biol. Cell. 2021;32:br12. doi: 10.1091/mbc.E21-04-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burns M., et al. Miscibility transition temperature scales with growth temperature in a zebrafish cell line. Biophys. J. 2017;113:1212–1222. doi: 10.1016/j.bpj.2017.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mužić T., et al. Melting transitions in biomembranes. Biochim. Biophys. Acta Biomembr. 2019;1861 doi: 10.1016/j.bbamem.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 53.Shen Y., et al. Metabolic activity induces membrane phase separation in endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 2017;114:13394–13399. doi: 10.1073/pnas.1712555114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King C., et al. ER membranes exhibit phase behavior at sites of organelle contact. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7225–7235. doi: 10.1073/pnas.1910854117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuberth C., Wedlich-Söldner R. Building a patchwork - the yeast plasma membrane as model to study lateral domain formation. Biochim. Biophys. Acta. 2015;1853:767–774. doi: 10.1016/j.bbamcr.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 56.van ’t Klooster, J.S., et al. Periprotein lipidomes of Saccharomyces cerevisiae provide a flexible environment for conformational changes of membrane proteins. Elife. 2020;9 doi: 10.7554/eLife.57003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Budin I., et al. Viscous control of cellular respiration by membrane lipid composition. Science. 2018;362:1186–1189. doi: 10.1126/science.aat7925. [DOI] [PubMed] [Google Scholar]
- 58.Cybulski L.E., et al. Activation of the bacterial thermosensor DesK involves a serine zipper dimerization motif that is modulated by bilayer thickness. Proc. Natl. Acad. Sci. U. S. A. 2015;112:6353–6358. doi: 10.1073/pnas.1422446112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ballweg S., Ernst R. Control of membrane fluidity: the OLE pathway in focus. Biol. Chem. 2017;398:215–228. doi: 10.1515/hsz-2016-0277. [DOI] [PubMed] [Google Scholar]
- 60.Covino R., et al. A eukaryotic sensor for membrane lipid saturation. Mol. Cell. 2016;63:49–59. doi: 10.1016/j.molcel.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 61.Corradi V., et al. Lipid-protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 2018;4:709–717. doi: 10.1021/acscentsci.8b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andersen O.S., Koeppe R.E., 2nd Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 63.Hegde R.S., Keenan R.J. The mechanisms of integral membrane protein biogenesis. Nat. Rev. Mol. Cell Biol. 2022;23:107–124. doi: 10.1038/s41580-021-00413-2. [DOI] [PubMed] [Google Scholar]
- 64.Wu X., Rapoport T.A. Translocation of proteins through a distorted lipid bilayer. Trends Cell Biol. 2021;31:473–484. doi: 10.1016/j.tcb.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McDowell M.A., et al. Structural basis of tail-anchored membrane protein biogenesis by the GET insertase complex. Mol. Cell. 2020;80:72–86.e7. doi: 10.1016/j.molcel.2020.08.012. [DOI] [PubMed] [Google Scholar]
- 66.Pleiner T., et al. Structural basis for membrane insertion by the human ER membrane protein complex. Science. 2020;369:433–436. doi: 10.1126/science.abb5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez F., et al. Structural model for the protein-translocating element of the twin-arginine transport system. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1092–E1101. doi: 10.1073/pnas.1219486110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y., et al. YidC insertase of Escherichia coli: water accessibility and membrane shaping. Structure. 2017;25:1403–1414.e3. doi: 10.1016/j.str.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qi L., et al. Cryo-EM structure of the human mitochondrial translocase TIM22 complex. Cell Res. 2021;31:369–372. doi: 10.1038/s41422-020-00400-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu X., et al. Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science. 2020;368 doi: 10.1126/science.aaz2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nejatfard A., et al. Derlin rhomboid pseudoproteases employ substrate engagement and lipid distortion to enable the retrotranslocation of ERAD membrane substrates. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bushell S.R., et al. The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K. Nat. Commun. 2019;10:3956. doi: 10.1038/s41467-019-11753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Falzone M.E., et al. TMEM16 scramblases thin the membrane to enable lipid scrambling. Nat. Commun. 2022;13:2604. doi: 10.1038/s41467-022-30300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nilsson I., et al. Inhibition of protein translocation across the endoplasmic reticulum membrane by sterols. J. Biol. Chem. 2001;276:41748–41754. doi: 10.1074/jbc.M105823200. [DOI] [PubMed] [Google Scholar]
- 75.Hwang J., et al. The ERAD system is restricted by elevated ceramides. Sci. Adv. 2023;9 doi: 10.1126/sciadv.add8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dyla M., et al. Structure and mechanism of P-type ATPase ion pumps. Annu. Rev. Biochem. 2020;89:583–603. doi: 10.1146/annurev-biochem-010611-112801. [DOI] [PubMed] [Google Scholar]
- 77.Starling A.P., et al. Effects of phosphatidylcholine fatty acyl chain length on calcium binding and other functions of the (Ca(2+)-Mg2+)-ATPase. Biochemistry. 1993;32:1593–1600. doi: 10.1021/bi00057a025. [DOI] [PubMed] [Google Scholar]
- 78.Johannsson A., et al. The effect of bilayer thickness on the activity of (Na+ + K+)-ATPase. Biochim. Biophys. Acta Biomembr. 1981;641:416–421. doi: 10.1016/0005-2736(81)90498-3. [DOI] [PubMed] [Google Scholar]
- 79.Norimatsu Y., et al. Protein-phospholipid interplay revealed with crystals of a calcium pump. Nature. 2017;545:193–198. doi: 10.1038/nature22357. [DOI] [PubMed] [Google Scholar]
- 80.Perozo E., et al. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat. Struct. Biol. 2002;9:696–703. doi: 10.1038/nsb827. [DOI] [PubMed] [Google Scholar]
- 81.Botelho A.V., et al. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys. J. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soubias O., et al. Rhodopsin/lipid hydrophobic matching-rhodopsin oligomerization and function. Biophys. J. 2015;108:1125–1132. doi: 10.1016/j.bpj.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sharpe H.J., et al. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schneiter R., et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simons K., Gerl M.J. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 86.Welch L.G., Munro S. A tale of short tails, through thick and thin: investigating the sorting mechanisms of Golgi enzymes. FEBS Lett. 2019;593:2452–2465. doi: 10.1002/1873-3468.13553. [DOI] [PubMed] [Google Scholar]
- 87.Herzig Y., et al. A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by Erv14. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Castillon G.A., et al. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 2009;10:186–200. doi: 10.1111/j.1600-0854.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- 89.Rodriguez-Gallardo S., et al. Ceramide chain length-dependent protein sorting into selective endoplasmic reticulum exit sites. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weigel A.V., et al. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell. 2021;184:2412–2429.e16. doi: 10.1016/j.cell.2021.03.035. [DOI] [PubMed] [Google Scholar]
- 91.Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 1995;14:4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Quiroga R., et al. Short transmembrane domains with high-volume exoplasmic halves determine retention of Type II membrane proteins in the Golgi complex. J. Cell Sci. 2013;126:5344–5349. doi: 10.1242/jcs.130658. [DOI] [PubMed] [Google Scholar]
- 93.Luedeke C., et al. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 2005;169:897–908. doi: 10.1083/jcb.200412143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Megyeri M., et al. Yeast ceramide synthases, Lag1 and Lac1, have distinct substrate specificity. J. Cell Sci. 2019;132 doi: 10.1242/jcs.228411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shcheprova Z., et al. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 96.Moore D., et al. A mechanism for the segregation of age in mammalian neural stem cells. Science. 2015;349:1334–1338. doi: 10.1126/science.aac9868. [DOI] [PubMed] [Google Scholar]
- 97.Lee Z.Y., et al. Compartmentalization of the endoplasmic reticulum in the early C. elegans embryos. J. Cell Biol. 2016;214:665–676. doi: 10.1083/jcb.201601047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu L.-K., et al. An inducible ER-Golgi tether facilitates ceramide transport to alleviate lipotoxicity. J. Cell Biol. 2017;216:131–147. doi: 10.1083/jcb.201606059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Radhakrishnan A., et al. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 101.Radanović T., Ernst R. The unfolded protein response as a guardian of the secretory pathway. Cells. 2021;10:2965. doi: 10.3390/cells10112965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Volmer R., et al. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tam A.B., et al. The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Dev. Cell. 2018;46:327–343. doi: 10.1016/j.devcel.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Celik C., et al. Endoplasmic reticulum stress and lipids in health and diseases. Prog. Lipid Res. 2023;89 doi: 10.1016/j.plipres.2022.101198. [DOI] [PubMed] [Google Scholar]
- 105.Halbleib K., et al. Activation of the unfolded protein response by lipid bilayer stress. Mol. Cell. 2017;67:673–684.e8. doi: 10.1016/j.molcel.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 106.Väth K., et al. Cysteine cross-linking in native membranes establishes the transmembrane architecture of Ire1. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202011078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Promlek T., et al. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell. 2011;22:3520–3532. doi: 10.1091/mbc.E11-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Antonny B. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 109.Covino R., et al. Integrated functions of membrane property sensors and a hidden side of the unfolded protein response. Mol. Cell. 2018;71:458–467. doi: 10.1016/j.molcel.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 110.Rutkowski D.T., Hegde R.S. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J. Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Killian J.A. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta. 1998;1376:401–415. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]