Abstract
The ProTide prodrug design is a powerful tool to improve cell permeability and enhance the intracellular activation of nucleotide antiviral analogues. Previous in vitro studies showed that the activation of ProTide prodrugs varied in different cell lines. In the present study, we investigated the activation profiles of two antiviral prodrugs tenofovir alafenamide (TAF) and sofosbuvir (SOF) in five cell lines commonly used in antiviral research, namely, Vero E6, Huh-7, Calu-3, A549, and Caco-2. We found that TAF and SOF were activated in a cell-dependent manner with Vero E6 being the least efficient and Huh-7 being the most efficient cell line for activating the prodrugs. We also demonstrated that TAF was activated at a significantly higher rate than SOF. We further analyzed the protein expressions of the activating enzymes carboxylesterase 1, cathepsin A, histidine triad nucleotide-binding protein 1, and the relevant drug transporters P-glycoprotein and organic anion-transporting polypeptides 1B1 and 1B3 in the cell lines using the proteomics data extracted from the literature and proteome database. The results revealed significant differences in the expression patterns of the enzymes and transporters among the cell lines, which might partially contribute to the observed cell-dependent activation of TAF and SOF. These findings highlight the variability of the abundance of activating enzymes and transporters between cell lines and emphasize the importance of selecting appropriate cell lines for assessing the antiviral efficacy of nucleoside/nucleotide prodrugs.
Keywords: ProTide, prodrug, tenofovir alafenamide, sofosbuvir
Nucleosides and nucleotide analogues are an essential class of therapeutics for treating viral infections. Nucleoside/nucleotide analogues need to be converted to their active metabolites, triphosphate nucleosides (TP-Nuc), to exert their intended antiviral activity.1 Thus, the antiviral activity depends on the intracellular formation of TP-Nuc.2−4 However, due to the limited cell permeability and slow intracellular activation rate, nucleoside/nucleotide analogues often cannot generate sufficient TP-Nuc in the intracellular space, limiting their antiviral efficacy. As a result, nucleoside/nucleotide analogues are frequently structurally modified to ester prodrugs to enhance intracellular accumulation and activation. Among the ester prodrug designs, the ProTide technology, also termed as McGuigan prodrugs, relies on the masking of the phosphate and phosphonate groups with an amino acid ester or an aryl group and has led to the development of several FDA-approved antiviral prodrugs.5,6
Various cell lines have been used as in vitro models to evaluate the antiviral activity of nucleoside/nucleotide prodrugs. Interestingly, the antiviral efficacy of a nucleoside/nucleotide prodrug could vary significantly among different cell lines.2,4 We hypothesize that the varied antiviral activity is partially due to the fact that those cell lines may exhibit different capabilities in activating nucleoside/nucleotide prodrugs. The present study aims to explore the prodrug activation patterns in various cell lines using two typical ProTide prodrugs, tenofovir alafenamide (TAF) and sofosbuvir (SOF) (Figure 1). TAF is a commonly used antihuman immunodeficiency virus (HIV) and antihepatitis B virus (HBV) agent,7,8 and sofosbuvir (SOF) is mainly used for treating hepatitis C virus (HCV) infections.9 Both TAF and SOF are broad-spectrum antivirals with the potential to inhibit other viruses.
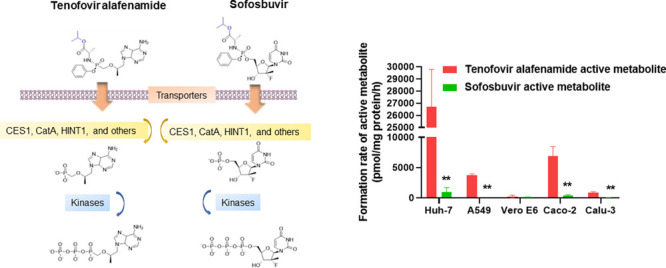
Figure 1.
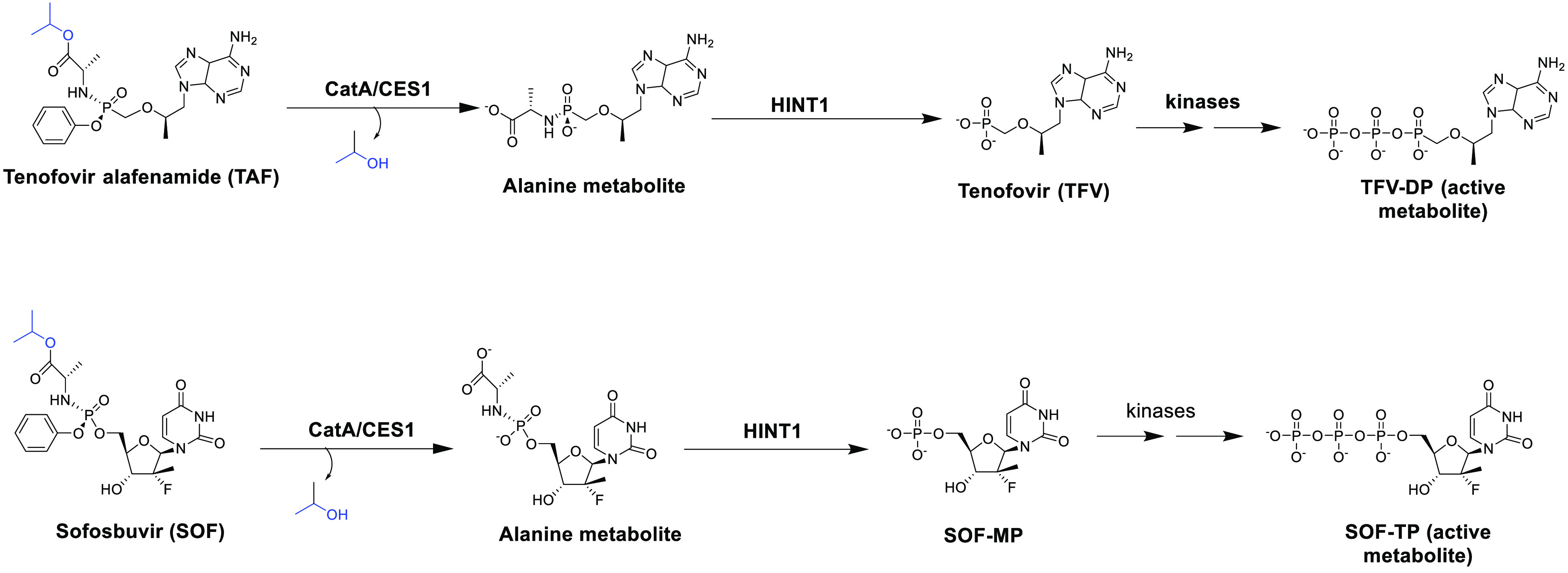
Activation pathways of TAF and SOF. After entering the cell, TAF and SOF are first hydrolyzed to their alanine metabolites by CES1 and CatA. The alanine metabolites are then metabolized by HINT1 to form the monophosphate products (MP-Nuc). MP-Nuc metabolites are subsequently phosphorylated by native kinases to generate the active metabolites TP-Nuc, namely TFV-DP for TAF and SOF-TP for SOF.
The activation process of TAF and SOF is initiated by the cleavage of the prodrug anionic phosphate moiety to form monophosphate nucleoside (MP-Nuc), which will be further phosphorated by host kinases to form the active metabolite TP-Nuc (Figure 1).10 The responsible esterases for hydrolyzing the prodrugs are mainly carboxylesterase 1 (CES1) and cathepsin A (CatA).11−15 Moreover, histidine triad nucleotide-binding protein 1 (HINT1) cleaves the P–N bond, leading to the formation of the monophosphate nucleoside metabolite.11−13
TAF and the SOF can enter cells via passive diffusion. In addition, drug transporters may play a role in the intracellular accumulation of the two prodrugs. TAF is the substrate of the influx transporters organic anion-transporting polypeptides 1B1 and 1B3 (OATP1B1 and OATP1B3) and the efflux transporter P-glycoprotein (P-gp),14,16 and SOF is a P-gp substrate.17 Therefore, in addition to the activating enzymes, drug transporters could also influence the intracellular levels of the active metabolites of TAF and SOF.
In this study, we performed in vitro studies to investigate the activation of TAF and SOF in five cell lines commonly used in virology research, namely Vero E6, Huh-7, Calu-3, A549, and Caco-2.18 Additionally, we analyzed the protein expression profiles of relevant activating enzymes and transporters to shed light on the mechanisms underlying the cell-line-dependent activation of nucleosides/nucleotide antiviral prodrugs. Our findings emphasize the importance of selecting appropriate cell lines for the study of nucleoside and nucleotide antiviral prodrugs.
Methods
Materials
The African monkey kidney cell line Vero E6 cells were provided by Dr. Christiane E. Wobus at the University of Michigan and grown in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% fetal bovine serum (FBS) (GIBCO, ThermoFisher Scientific). Human hepatocellular carcinoma cell line Huh7 cells were provided by Dr. Lei Yin at the University of Michigan and grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS. Human lung adenocarcinoma epithelial cells Calu-3 were provided by Dr. Daniel Goldstein at the University of Michigan and grown in EMEM supplemented with 10% FBS. Human pneumocyte type II carcinoma cells A549 (ATCC CCL-185) were purchased from ATCC and grown in F-12K supplemented with 10% FBS. Human epithelial colorectal adenocarcinoma cell Caco-2 (ATCC HTB-37) were purchased from ATCC (Manassas, VA) and grown in EMEM supplemented with 20% FBS.
TAF (≥99%), TFV (≥99%), and SOF (≥99%) were purchased from MedChem Express (Monmouth Junction, NJ). TFV-DP (≥95%) was obtained from Cayman Chemical (Ann Arbor, MI). Sofosbuvir-5′-triphosphate (SOF-TP) (≥95%) and sofosbuvir-5′-monophosphate (SOF-MP) (≥95%) were purchased from Sierra Bioresearch (Tucson, AZ). PhosSTOP EASYpack (PhosSTOP) was purchased from Roche (Basel, Switzerland). Dimethyl sulfoxide (DMSO), 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB, ≥ 98%), 2-chloroadenosine, and adenosine-15N5 5′-triphosphate disodium salt solution were purchased from Sigma-Aldrich (St. Louis, MO, USA). Qubit protein assay kits and phosphate-buffered saline (PBS) were obtained from Thermo Fisher Scientific (Waltham, MA). The MTS Assay Kit (ab197010) was purchased from Abcam (Cambridge, MA, USA). All other chemicals and reagents were analytical-grade and commercially available. Deionized water was generated from a Milli-Q system (Millipore Corporation, Billerica, MA, USA).
In Vitro Metabolism of TAF and SOF in Various Cell Lines
Vero E6, Huh7, Calu-3, A549, and Caco-2 cells were seeded in a six-well plate at a density of 0.5 × 106 cells/well. When the cells reached 80–90% confluence, the cell culture medium was replaced with the medium containing 10 μM TAF or 10 μM SOF and incubated at 37 °C. We conducted a cytotoxicity study and revealed that both TAF and SOF did not exhibit cytotoxicity at a concentration of 10 μM (Supporting Information). At 6, 12, and 24 h following the incubation, the medium was discarded, and the cells were washed twice with 2 mL of ice-cold PBS. 0.3 mL of ice-cold lysis buffer was then added to each well, and the cells were scraped and transferred to a 2 mL tube. The lysis buffer contained 70% methanol (v/v, methanol:water = 7:3), 0.5 μM 2-chloroadenosine, 5 μM adenosine-15N5 5′-triphosphate (internal standards), 2% formic acid, 10% PhosStop, and 50 μM DTNB as the protectors for the MP-Nuc and TP-Nuc metabolites.21 Then, each well was washed with 0.3 mL of water, and the solution was combined with the lysate from the well. The protein concentration was determined using the Qubit protein assay kit according to the manufacturer’s instructions. The samples were mixed on a shaker (Benchmark, Multi-Therm) at 1500 rpm for 5 min and stored at −80 °C until use. For LC-MS/MS analysis, samples were thawed on ice and centrifuged twice at 21,130g for 10 min at 4 °C. The supernatants were transferred to autosampler vials for LC-MS/MS analysis.
LC-MS/MS Analysis for TAF, SOF, and Their Monophosphate and Triphosphate Metabolites
We performed the LC-MS/MS analysis of TAF, SOF, and their monophosphate and triphosphate metabolites using methods similar to those previously published with some minor modifications.18−20 TAF and SOF were analyzed by using a reverse phase LC system coupled with a Sciex 3000 Triple Quadrupole mass spectrometer. We used a RESTEK Ultra II C18 column (5 μm, 50 cm × 2.1 mm, Bellefonte, PA, USA) with a RESTEK UltraShield UHPLC precolumn filter (0.2 μm frit, Bellefonte, PA, USA) for chromatographic separation at 45 °C. The mobile phases comprised water containing 0.1% formic acid (mobile phase A) and acetonitrile containing 0.1% formic acid (mobile phase B) with a flow rate of 0.5 mL/min and the following gradient for the mobile phase B: 0–0.1 min 5%, 0.1–2 min 5% to 90%, 2–3 min 90%, 3–3.1 min 90% to 5%, 3.1–5 min 5%. The total run time was 5 min.
The monophosphate and triphosphate metabolites (TFV, TFV-DP, SOF-MP, and SOF-TP) were analyzed by using an ion-exchange LC system coupled with a Sciex 3000 Triple Quadrupole mass spectrometer. We used a BioBasic AX column (2.1 × 50 mm, 4.6 μm; ThermoFisher) with a RESTEK UltraShield UHPLC precolumn filter (0.2 μm frit, Bellefonte, PA, USA) for chromatographic separation at 50 °C. The mobile phases consisted of 30% acetonitrile (pH 6.0, mobile phase A) and 30% acetonitrile (pH 10.0, mobile phase B) with a flow rate of 0.6 mL/min and the following gradient for the mobile phase B: 0–0.1 min 2%, 0.1–0.4 min 2% to 50%, 0.4–0.8 min 50%, 0.8–1 min 50% to 95%, 1–3.5 min 95%, 3.5–3.7 min 95% to 2%, 3.7–5 min 2%. The total run time was 5 min. 2-Chloroadenosine and adenosine-15N5 5′-triphosphate were used as the internal standards for the prodrugs and metabolites, respectively. The MS parameters can be found in the Supporting Information.
Extraction of Proteomics Data of Key Enzymes and Transporters Involved in TAF and SOF Activation and Disposition
We analyzed the expressions of key enzymes and transporters involved in TAF and SOF activation and disposition in the cell lines by extracting proteomics data from the literature and the Human Protein Atlas (HPA) database.21,22 The target activating enzymes include CES1, CatA, and HINT1,11−14 and the target transporters are the influx transporters OATP1B1 and OATP1B3 and the efflux transporter P-gp, given that OATP1B1 and OATP1B3 facilitate the cellular uptake of TAF, and both TAF and SOF are the substrates of P-gp.14,16,17
Data Analysis
The student t-test was utilized to compare the metabolism profiles of TAF and SOF. A P-value of less than 0.05 was considered statistically significant. GraphPad Prism v8.3.0 (GraphPad Software) was used for generating graphs. For the LC-MS/MS analysis, the analyte-to-internal standard ratios were calculated for quantification.
Results
Cell-Dependent Activation of TAF and SOF
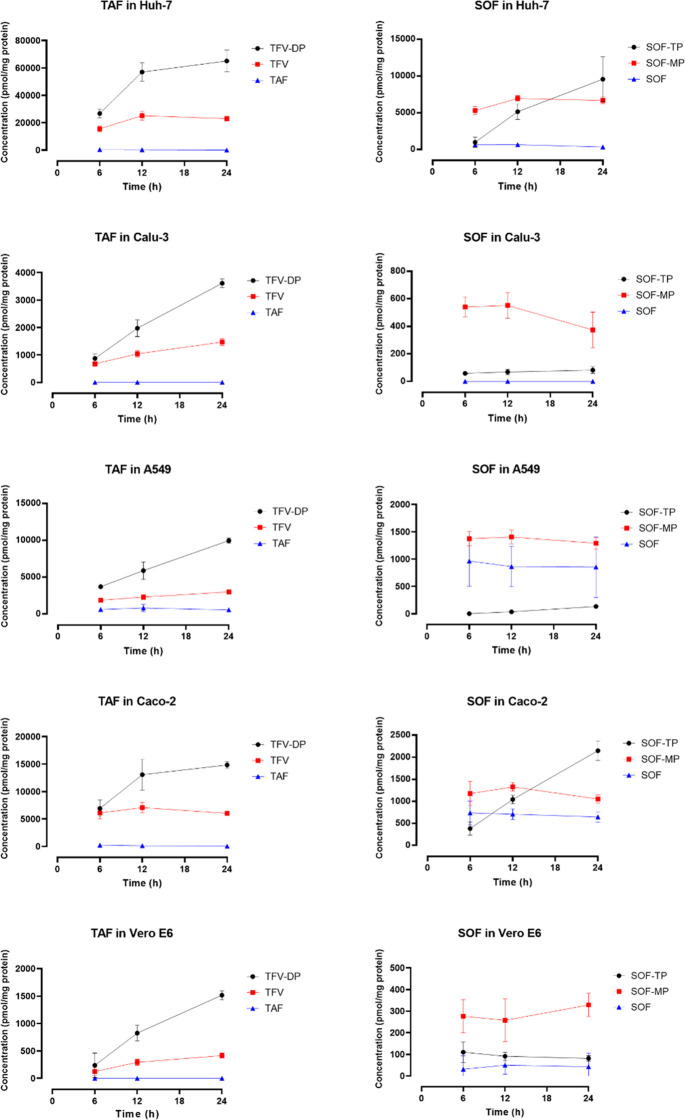
The time–concentration profiles of the prodrugs TAF and SOF and their MP-Nuc and TP-Nuc metabolites in the five tested cell lines are illustrated in Figure 2. Both TAF and SOF showed a marked cell-dependent activation pattern, with Huh-7 being the most efficient cell line and Vero E6 being the least efficient cell line activating the prodrugs. TAF was able to generate a noticeably higher amount of the active metabolite TP-Nuc than SOF in all the cell lines.
Figure 2.
Time–concentration profiles of TAF, SOF, and their metabolites (TFV, TFV-DP, SOF-MP, and SOF-TP) after incubation with Huh-7, Calu-3, A549, Caco-2, and Vero-E6 cells. The intracellular concentrations were measured at 6, 12, and 24 h following incubation with 10 μM of the prodrugs. Data are presented as the mean ± SD of four independent experiments.
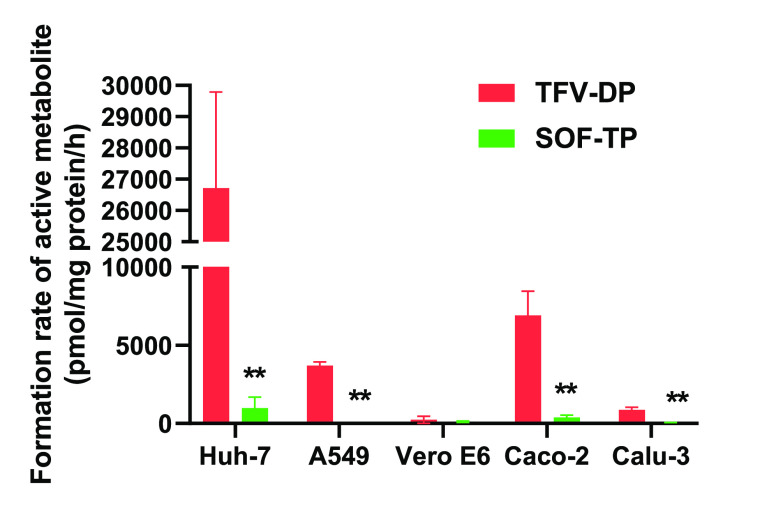
At six hours following incubation, the formation rates of the active TP-Nuc metabolite (TFV-DP) from TAF were significantly higher compared to SOF in Huh-7, A549, Caco-2, and Calu-3 cells (Figure 3).
Figure 3.
Activation rates of TAF and SOF after incubation with Huh-7, A549, Vero E6, Caco-2, and Calu-3 cells for 6 h. The activation rates were determined by measuring the intracellular formation of the TP metabolites. The values were normalized by cellular protein concentrations. ** P < 0.01, n = 4.
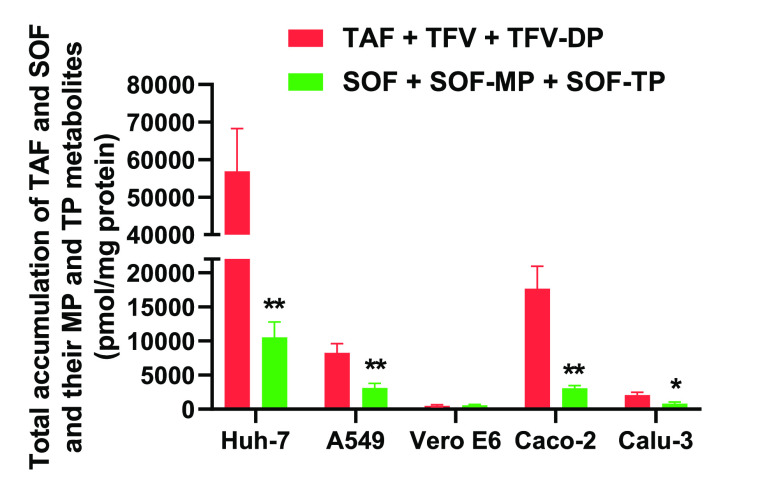
We further calculated the total intracellular accumulation of each prodrug and its MP-Nuc and TP-Nuc metabolites in the tested cell lines after incubation for 6 h. Similar to the activation rates, the total accumulation of TAF and its metabolites was also significantly higher than the total accumulation of SOF and its metabolites in all cell lines except for Vero E6 (Figure 4).
Figure 4.
Total intracellular accumulations of TAF and SOF and their MP-Nuc and TP-Nuc metabolites in Huh-7, A549, Vero E6, Caco-2, and Calu-3 cells after incubation for 6 h. The total accumulations are the sum of the prodrug and its MP and TP metabolites and are normalized by cellular protein concentrations. ** P < 0.01, n = 4.
The metabolism of TAF and SOF to their intermediate metabolite MP-Nuc is primarily catalyzed by CES1, CatA, and HINT1, whereas the conversion of MP-Nuc to the active metabolite TP-Nuc is mediated by kinases (Figure 1). We calculated the ratios of the intracellular concentration of MP-Nuc to prodrug and the ratios of TP-Nuc to MP-Nuc (Table 1) for both TAF and SOF to evaluate the metabolizing efficiencies of the prodrugs at each metabolic step in the cell lines. Interestingly, the mean MP-Nuc to TAF ratio was about 10 times the mean MP-Nuc to SOF ratio (36 vs 3.5), and the mean TP-Nuc to MP-Nuc ratio for TAF ratio was approximately 5 times the ratio for SOF (1.6 vs 0.27), indicating that the catalytic efficiencies of the metabolizing enzymes are higher for TAF compared to SOF at both metabolic steps.
Table 1. Monophosphate Metabolite to Prodrug Concentration ratios (TFV:TAF and SOF-MP:SOF) and Triphosphate Metabolite to Monophosphate Metabolite Concentration Ratios (TFV-DP:TFV and SOF-TP:SOF-MP) in Different Cell Lines after a 6-Hour Incubationa.
| Huh-7 | A549 | Vero E6 | Caco-2 | Calu-3 | Mean ± SD | |
|---|---|---|---|---|---|---|
| TFV:TAF | 45 ± 16 | 3.4 ± 1.4 | NAb | 58 ± 50 | NAb | 36 ± 29 |
| SOF-MP:SOF | 6.6 ± 4.0* | 1.9 ± 1.2 | NAb | 1.9 ± 1.2 | NAb | 3.5 ± 2.7 |
| TFV-DP:TFV | 1.8 ± 0.44 | 2.0 ± 0.2 | 1.5 ± 1.1 | 1.1 ± 0.1 | 1.3 ± 0.3 | 1.6 ± 0.3 |
| SOF-TP:SOF-MP | 0.18 ± 0.11** | NAb | 0.42 ± 0.19 | 0.36 ± 0.21** | 0.11 ± 0.03** | 0.27 ± 0.15** |
* P < 0.05, ** P < 0.01, n = 4 (TFV:TAF vs. SOF-MP:SOF, TFV-DP:TFV vs. SOF-TP:SOF-MP).
Monophosphate metabolite to prodrug concentration ratios were not determined because the analyte concentrations were below the low limits of detection after 6 h of incubation.
Expression of TAF and SOF Activating Enzymes and Transporters
Relative protein expression levels were normalized to those in the Calu-3 cells (Table 2). The expression levels of the proteins of interest differ significantly among the five cell lines. For CES1, its expression levels in A549 and Caco-2 are much higher than those in Huh-7 and Calu-3, and CES1 is absent in Vero E6. For CatA and HINT1, the differences in their expression levels across the cell lines are less pronounced than CES1. The efflux transporter P-gp is expressed at the highest level in the Caco-2, Huh-7, and Vero E6 cells. The influx transporters OATP1B1 and OATP1B3 are either absent or expressed at a very low level in the Caco-2 and Vero E6 cells.
Table 2. Relative Protein Expression Levels of TAF and SOF Activating Enzymes and Transporters in Different Cell Linesa.
| Calu-3 | A549 | Caco-2 | Huh-7 | Vero E6 | |
|---|---|---|---|---|---|
| CES1 | 1.00 | 269 | 106 | 6.75 | ULD |
| CatA | 1.00 | 0.67 | 2.03 | 1.00 | 0.84 |
| HINT1 | 1.00 | 1.33 | 1.05 | 0.92 | 1.50 |
| P-gp | 1.00 | ULD | 48.5 | 17.3 | 26.7 |
| OATP1B1 | 1.00 | 2.00 | ULD | 0.60 | ULD |
| OATP1B3 | 1.00 | 1.02 | 0.01 | 0.18 | ULD |
Note: The protein expression levels are normalized to that in the Calu-3 cells. ULD: Under the Limit of Detection.
Discussion
Vero E6, A549, Calu-3, Caco-2, and Huh-7 are among the most commonly used cell lines for evaluating the antiviral activity of antiviral drugs in virology studies.23 Among these, Vero E6 cells have been widely used as an in vitro cell model for anti-SARS-CoV-2 research due to its outstanding efficiency in propagating SARS-CoV-2.24,25 The lung cell lines A459 and Calu-3 are commonly used as in vitro pneumonocyte models for the research of pulmonary infections such as COVID-19.26−28 The human colon cell line Caco-2 and human liver cell line Huh-7 have been adopted as in vitro models for the study of colon and hepatitis virus infections.2,23 The antiviral activity of a prodrug depends on the intracellular accumulation and activation of the prodrug. Previous in vitro studies have shown that the activation and antiviral efficacy of nucleoside/nucleotide prodrugs, such as remdesivir, were cell-line-dependent.2,4 Thus, the in vitro antiviral activity results could be misleading when the prodrug is not adequately activated in the selected cells.
In this study, we determined the activation patterns of two widely used antiviral prodrugs, TAF and SOF, in the five cell lines to illustrate the cell-dependent activation of nucleoside/nucleotide prodrugs. Moreover, we analyzed the protein expressions of the enzymes and transporters involved in the activation and disposition of the two prodrugs, including CES1, CatA, HINT1, P-gp, OATP1B1, and OATP1B3. CES1 and CatA are the major enzymes catalyzing the hydrolysis of TAF and SOF, the first metabolic activation step of the prodrugs. We previously reported that TAF was a much more efficient substrate of CES1 and CatA than SOF (CES1: 772 vs 9.79 pmol/min/μg protein and CatA: 3941 vs 8.35 pmol/min/μg protein).14 Consistent with the findings, the present study demonstrated that TAF was activated at a higher rate than that of SOF in the cell lines (Figures 2 and 3). Moreover, the mean ratio of the MP-metabolite (TFV) to TAF was approximately 10-fold higher than the ratio of SOF-MP to SOF, further supporting that TAF was hydrolyzed at a significantly higher rate in the cells compared to SOF. We also observed that the triphosphate metabolite to monophosphate metabolite intracellular concentration ratio of TAF (i.e., TFV-DP/TFV) was significantly greater than that of SOF, indicating that the conversion of TAF monophosphate metabolite to its active metabolite is more efficient compared to SOF-MP.
The study demonstrated a marked cell-dependent activation pattern. The rank of the activation rates after a 6 h incubation period for both TAF and SOF is as follows: Huh-7 > Caco-2 > A549 > Calu-3 > Vero E6. Interestingly, the proteomics analysis revealed that CES1 protein expression levels in the Huh-7, Caco-2, and A549 cells were much higher than those in Calu-3, while CES1 is absent in Vero E6 (Table 2). In addition, CatA and HINT1 expressions were comparable among the five cell lines. The results suggest that the higher activation rates of TAF and SOF in Huh-7, Caco-2, and A549 might be due to the higher expression levels of CES1 in these cell lines.
The primary rationale for nucleoside/nucleotide prodrug design is to increase cell membrane permeability and enhance intracellular drug accumulation.29 We estimated the transmembrane efficiency of TAF and SOF in the five cell lines by calculating the total intracellular accumulation of the prodrugs and their MP-Nuc and TP-Nuc metabolites after incubation for 6 h. Similar to the rates of activation, TAF was accumulated at significantly higher rates than SOF, and the accumulations were higher in Huh-7, Caco-2, and A549 than in Calu-3 and Vero E6 for both TAF and SOF. It has been well established that, besides the physicochemical property of a drug, drug transporters also play an important role in intracellular drug accumulation.30 Thus, we analyzed the protein expressions of P-gp, OATP1B1, and OATP1B3 in the cell lines (Table 2) since both TAF and SOF are substrates of P-gp, and TAF is also a substrate of OATP1B1 and OATP1B3.14,16,17 Interestingly, Vero E6, which exhibited the lowest levels of intracellular drug accumulation, showed the second highest expression of the efflux transporter P-gp and no expression of the influx transporters OATP1B1 and OATP1B3. The findings indicate that the differences in drug transporter expression among the cell lines could also play a role in the observed cell-dependent prodrug accumulation and activation.
In summary, our study demonstrated a significant cell-dependent pattern of intracellular accumulation and activation of nucleoside and nucleotide prodrugs in five cell lines commonly used in antiviral research and revealed differences in the expressions of activating enzymes and transporters among the cell lines. The findings suggest that the careful selection of in vitro cell models is essential for studying the antiviral activity of nucleoside/nucleotide prodrugs. In particular, Vero E6, one of the most commonly used cell lines for antiviral research, appears to be inappropriate for evaluating the antiviral efficacy of a nucleoside/nucleotide prodrug because of its inability to activate ester-containing prodrugs. The results also indicate that the activation levels of a prodrug could vary significantly in different organs due to the expressions of relevant transporters and activating enzymes differing across these organs, which could lead to unintended therapeutic outcomes.31
Glossary
Abbreviations
- CatA
Cathepsin A
- CE
collision energy
- CES1
carboxylesterase 1
- COVID-19
coronavirus disease 2019
- CXP
collision cell exit potential
- DMEM
Dulbecco’s Modified Eagle Medium
- DMSO
dimethyl sulfoxide
- DP
delustering potential
- DTNB
5,5′-dithiobis (2-nitrobenzoic acid)
- EMEM
Eagle’s Minimum Essential Medium
- EP
entrance potential
- FBS
fetal bovine serum
- FP
focusing potential
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HINT1
histidine triad nucleotide-binding protein 1
- HIV
human immunodeficiency virus
- IV
intravenous
- MP-Nuc
monophosphate nucleoside
- PBS
phosphate-buffered saline
- P-gp
P-glycoprotein 1
- RdRp
RNA-dependent RNA polymerase
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SOF
sofosbuvir
- SOF-TP
sofosbuvir triphosphate
- TAF
tenofovir alafenamide
- TFV-DP
tenofovir diphosphate
- TP-Nuc
triphosphate nucleoside
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.3c00050.
Cytotoxicity assay and the mass spectrometer parameters for the LC-MS/MS assay (PDF)
Author Contributions
Y.L., J.L., and H.J.Z. wrote the manuscript. Y.L., S.S., and J. L. searched for, extracted, and analyzed data, and generated figures. Y.L., S.S., and J.L. conducted cell culture, prepared the samples, and performed the LC-MS/MS analysis. W.W. facilitated the Qubit protein assay. H.J.Z. obtained grant funding that supported this work.
Author Contributions
# The authors equally contributed to the work.
This work was partly supported by the National Institute of Allergy and Infectious Diseases (R21 AI163425).
The authors declare no competing financial interest.
Supplementary Material
References
- De Schutter C.; Ehteshami M.; Hammond E. T.; Amblard F.; Schinazi R. F. J. C. p. d. Metabolism of Nucleosides and Nucleotides Prodrugs. Curr Pharm Des 2018, 23, 6984–7002. 10.2174/1381612823666171011104158. [DOI] [PubMed] [Google Scholar]
- Mumtaz N.; Jimmerson L. C.; Bushman L. R.; Kiser J. J.; Aron G.; Reusken C. B. E. M.; Koopmans M. P. G.; van Kampen J. J. A. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Antiviral Res. 2017, 146, 161–163. 10.1016/j.antiviral.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Mackman R. L. Phosphoramidate Prodrugs Continue to Deliver, The Journey of Remdesivir (GS-5734) from RSV to SARS-CoV-2. ACS Med Chem Lett 2022, 13, 338–347. 10.1021/acsmedchemlett.1c00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijssers A. J.; George A. S.; Schäfer A.; Leist S. R.; Gralinksi L. E.; Dinnon K. H.; Yount B. L.; Agostini M. L.; Stevens L. J.; Chappell J. D.; Lu X.; Hughes T. M.; Gully K.; Martinez D. R.; Brown A. J.; Graham R. L.; Perry J. K.; Du Pont V.; Pitts J.; Ma B.; Babusis D.; Murakami E.; Feng J. Y.; Bilello J. P.; Porter D. P.; Cihlar T.; Baric R. S.; Denison M. R.; Sheahan T. P. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Reports 2020, 32, 107940. 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanazi A. S.; James E.; Mehellou Y. The ProTide Prodrug Technology: Where Next?. ACS Medicinal Chemistry Letters 2019, 10, 2–5. 10.1021/acsmedchemlett.8b00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehellou Y.; Rattan H. S.; Balzarini J. The ProTide Prodrug Technology: From the Concept to the Clinic. J. Med. Chem. 2018, 61, 2211–2226. 10.1021/acs.jmedchem.7b00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L. J.; Chan H. L. Y. Tenofovir Alafenamide: A Review in Chronic Hepatitis B. Drugs 2017, 77, 1017–1028. 10.1007/s40265-017-0754-9. [DOI] [PubMed] [Google Scholar]
- Gibson A. K.; Shah B. M.; Nambiar P. H.; Schafer J. J. Tenofovir Alafenamide. Ann Pharmacother 2016, 50, 942–952. 10.1177/1060028016660812. [DOI] [PubMed] [Google Scholar]
- Keating G. M. Sofosbuvir: a review of its use in patients with chronic hepatitis C. Drugs 2014, 74, 1127–1146. 10.1007/s40265-014-0247-z. [DOI] [PubMed] [Google Scholar]
- Eastman R. T.; Roth J. S.; Brimacombe K. R.; Simeonov A.; Shen M.; Patnaik S.; Hall M. D. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent Sci 2020, 6, 672–683. 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkus G.; Wang R.; Liu X.; Kutty N.; MacArthur H.; Cihlar T.; Gibbs C.; Swaminathan S.; Lee W.; McDermott M. J. A. a. chemotherapy. Cathepsin A is the major hydrolase catalyzing the intracellular hydrolysis of the antiretroviral nucleotide phosphonoamidate prodrugs GS-7340 and GS-9131. Antimicrob. Agents Chemother. 2007, 51, 543–550. 10.1128/AAC.00968-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkus G.; Kutty N.; He G.-X.; Mulato A.; Lee W.; McDermott M.; Cihlar T. J. M. p. Activation of 9-[(R)-2-[[(S)-[[(S)-1-(Isopropoxycarbonyl) ethyl] amino] phenoxyphosphinyl]-methoxy] propyl] adenine (GS-7340) and other tenofovir phosphonoamidate prodrugs by human proteases. Mol. Pharmacol. 2008, 74, 92–100. 10.1124/mol.108.045526. [DOI] [PubMed] [Google Scholar]
- Murakami E.; Tolstykh T.; Bao H.; Niu C.; Steuer H. M.; Bao D.; Chang W.; Espiritu C.; Bansal S.; Lam A. M.; Otto M. J.; Sofia M. J.; Furman P. A. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J. Biol. Chem. 2010, 285, 34337–34347. 10.1074/jbc.M110.161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Liu S.; Shi J.; Zhu H. J. Activation of Tenofovir Alafenamide and Sofosbuvir in the Human Lung and Its Implications in the Development of Nucleoside/Nucleotide Prodrugs for Treating SARS-CoV-2 Pulmonary Infection. Pharmaceutics 2021, 13, 1656. 10.3390/pharmaceutics13101656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanazi A. S.; Miccoli A.; Mehellou Y. Aryloxy Pivaloyloxymethyl Prodrugs as Nucleoside Monophosphate Prodrugs. J. Med. Chem. 2021, 64, 16703–16710. 10.1021/acs.jmedchem.1c01490. [DOI] [PubMed] [Google Scholar]
- Begley R.; Das M.; Zhong L. J.; Ling J.; Kearney B. P.; Custodio J. M. Pharmacokinetics of Tenofovir Alafenamide When Coadministered With Other HIV Antiretrovirals. Jaids-J Acq Imm Def 2018, 78, 465–472. 10.1097/QAI.0000000000001699. [DOI] [PubMed] [Google Scholar]
- Kirby B. J.; Symonds W. T.; Kearney B. P.; Mathias A. A. Pharmacokinetic, Pharmacodynamic, and Drug-Interaction Profile of the Hepatitis C Virus NS5B Polymerase Inhibitor Sofosbuvir. Clin Pharmacokinet 2015, 54, 677–690. 10.1007/s40262-015-0261-7. [DOI] [PubMed] [Google Scholar]
- Rower J. E.; Jimmerson L. C.; Chen X.; Zheng J. H.; Hodara A.; Bushman L. R.; Anderson P. L.; Kiser J. J. Validation and Application of a Liquid Chromatography-Tandem Mass Spectrometry Method To Determine the Concentrations of Sofosbuvir Anabolites in Cells. Antimicrob. Agents Chemother. 2015, 59, 7671–7679. 10.1128/AAC.01693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.; Chang L.; Ke C.; Xie Y.; Shen J.; Tan B.; Liu J. Challenges and stepwise fit-for-purpose optimization for bioanalyses of remdesivir metabolites nucleotide monophosphate and triphosphate in mouse tissues using LC-MS/MS. J Pharm Biomed Anal 2021, 194, 113806. 10.1016/j.jpba.2020.113806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. J.; Chang L.; Yang Y.; Wang X.; Xie Y. C.; Shen J. S.; Tan B.; Liu J. Pharmacokinetics and tissue distribution of remdesivir and its metabolites nucleotide monophosphate, nucleotide triphosphate, and nucleoside in mice. Acta Pharmacol Sin 2021, 42, 1195–1200. 10.1038/s41401-020-00537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; He M.; Xie Q.; Su A.; Yang K.; Liu L.; Liang J.; Li Z.; Huang X.; Hu J.; Liu Q.; Song B.; Hu C.; Chen L.; Wang Y. Predicting In Vitro and In Vivo Anti-SARS-CoV-2 Activities of Antivirals by Intracellular Bioavailability and Biochemical Activity. ACS Omega 2022, 7, 45023–45035. 10.1021/acsomega.2c05376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Protein Atlas, https://www.proteinatlas.org/.
- Zecha J.; Lee C.-Y.; Bayer F. P.; Meng C.; Grass V.; Zerweck J.; Schnatbaum K.; Michler T.; Pichlmair A.; Ludwig C.; Kuster B. Data, Reagents, Assays and Merits of Proteomics for SARS-CoV-2 Research and Testing. Molecular & Cellular Proteomics 2020, 19, 1503–1522. 10.1074/mcp.RA120.002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essaidi-Laziosi M.; Perez Rodriguez F. J.; Hulo N.; Jacquerioz F.; Kaiser L.; Eckerle I. Estimating clinical SARS-CoV-2 infectiousness in Vero E6 and primary airway epithelial cells. Lancet Microbe 2021, 2, e571 10.1016/S2666-5247(21)00216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. D.; Shang B.; Yang R. F.; Yu H.; Ma Z. H.; Shen X.; Ji Y. Y.; Lin Y.; Wu Y. D.; Lin G. M.; Tian L.; Gan X. Q.; Yang S.; Jiang W. H.; Dai E. H.; Wang X. Y.; Jiang H. L.; Xie Y. H.; Zhu X. L.; Pei G.; Li L.; Wu J. R.; Sun B. The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells. Cell Res 2004, 14, 400–406. 10.1038/sj.cr.7290240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodi G.; Gentili V.; Casciano F.; Romani A.; Zauli G.; Secchiero P.; Zauli E.; Simioni C.; Beltrami S.; Fernandez M.; Rizzo R.; Voltan R. Cell cycle block by p53 activation reduces SARS-CoV-2 release in infected alveolar basal epithelial A549-hACE2 cells. Front Pharmacol 2022, 13, 1018761. 10.3389/fphar.2022.1018761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiewsakun P.; Phumiphanjarphak W.; Ludowyke N.; Purwono P. B.; Manopwisedjaroen S.; Srisaowakarn C.; Ekronarongchai S.; Suksatu A.; Yuvaniyama J.; Thitithanyanont A. Systematic exploration of SARS-CoV-2 adaptation to Vero E6, Vero E6/TMPRSS2, and Calu-3 cells. Genome Biol Evol 2023, 10.1093/gbe/evad035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossegesse M.; Bourquain D.; Neumann M.; Schaade L.; Schulze J.; Mache C.; Wolff T.; Nitsche A.; Doellinger J. Deep Time Course Proteomics of SARS-CoV- and SARS-CoV-2-Infected Human Lung Epithelial Cells (Calu-3) Reveals Strong Induction of Interferon-Stimulated Gene Expression by SARS-CoV-2 in Contrast to SARS-CoV. J Proteome Res 2022, 21, 459–469. 10.1021/acs.jproteome.1c00783. [DOI] [PubMed] [Google Scholar]
- Sinokrot H.; Smerat T.; Najjar A.; Karaman R. Advanced Prodrug Strategies in Nucleoside and Non-Nucleoside Antiviral Agents: A Review of the Recent Five Years. Molecules 2017, 22, 1736. 10.3390/molecules22101736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X.; Korzekwa K.; Elsby R.; Fenner K.; Galetin A.; Lai Y.; Matsson P.; Moss A.; Nagar S.; Rosania G. R.; Bai J. P.; Polli J. W.; Sugiyama Y.; Brouwer K. L. Intracellular drug concentrations and transporters: measurement, modeling, and implications for the liver. Clinical pharmacology and therapeutics 2013, 94, 126–141. 10.1038/clpt.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan V. C.; Muller F. L. Advantages of the Parent Nucleoside GS-441524 over Remdesivir for Covid-19 Treatment. ACS Med Chem Lett 2020, 11, 1361–1366. 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.