Abstract
COVID-19 (Coronavirus Disease 2019) is an infectious disease caused by SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) and has globally infected 768 million people and caused over 6 million deaths. COVID-19 primarily affects the respiratory system but increasing reports of neurologic symptoms associated with COVID-19 have been reported in the literature. The exact mechanism behind COVID-19 neurologic pathophysiology remains poorly understood due to difficulty quantifying clinical neurologic symptoms in humans and correlating them to findings in human post-mortem samples and animal models. Thus, robust preclinical experimental models for COVID-19 neurologic manifestations are urgently needed. Here, we review recent advances in in vitro, in vivo, and other models and technologies for studying COVID-19 including primary cell cultures, pluripotent stem cell-derived neurons and organoids, rodents, nonhuman primates, 3D bioprinting, artificial intelligence, and multiomics. We specifically focus our discussion on the contribution, recent advancements, and limitations these preclinical models have on furthering our understanding of COVID-19’s neuropathic physiology. We also discuss these models’ roles in the screening and development of therapeutics, vaccines, antiviral drugs, and herbal medicine, and on future opportunities for COVID-19 neurologic research and clinical management.
Keywords: Animal model, Cell model, Stem cell, SARS-CoV-2, COVID-19, Neurologic disorders
COVID-19 is a viral respiratory illness caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It was first reported in Wuhan, China, in December 2019 and has since become a global pandemic.1 There have been 768,983,095 confirmed cases of COVID-19 and 6,953,743 deaths worldwide as of August 2023 according to the World Health Organization (WHO).2 The virus belongs to the family of coronaviruses and shares 79% genome sequence identity with severe acute respiratory syndrome coronavirus (SARS-CoV) and 50% with Middle East respiratory syndrome virus (MERS-CoV), which are two other viruses in the same virus family.3,4 Similar to SARS-CoV and MERS-CoV, although SARS-CoV-2 is a respiratory virus, a growing number of evidence suggests that COVID-19 can cause long-term neurologic complications, even in patients with mild symptoms.5,6 The most common COVID-19 neurologic symptoms include fatigue, myalgia, taste impairment, smell impairment, and headache.5,7 More severe neurologic symptoms include encephalitis and stroke.8−10 Some patients even experience “long-COVID” neurologic symptoms, having persisting symptoms 4 weeks to three months after confirmation of COVID-19 diagnosis.11,12 The virus’s neural pathophysiology has not been well understood. Given the wide range of neurologic symptoms and complications associated with COVID-19, healthcare providers need to be vigilant in monitoring patients for these clinical presentations. Further research is needed to elucidate the mechanisms by which the virus affects the nervous system and to develop effective treatments for COVID-19 neurologic conditions. Robust preclinical experimental models are urgently needed to strengthen the applicability of COVID-19 neurologic studies.
There have been recent papers published reviewing experimental models for SARS-CoV-2.13−15 For example, Shou et al. provided a succinct overview of using animal models for SARS-CoV-2 studies, specifically studying the pulmonary and systemic pathophysiology.14 Another paper by Bakhtazad et al. reviewed COVID-19 animal models with a more focused discussion on SARS-CoV-2 neuroinvasion mechanisms and how the pathophysiology compares to other viruses in the Coronaviruses family such as SARS-CoV, MERS-CoV, and murine-CoV.15 Our paper also provides a basic overview of available COVID-19 preclinical experimental models; however, the novelty of our review is that we provide a more in-depth discussion comparing the models’ applications specifically for preclinical COVID-19 neurologic studies. In addition, we discuss the significance and challenges of previous studies using these preclinical models, especially in terms of correlation with clinical symptomatology in human patients and implications for therapeutic and vaccine development. Finally, we deliberate on the newest technologies like artificial intelligence, 3D printing, and multiomics and how they can help expand the current landscape of COVID-19 neurologic studies.
1. Mechanism of SARS-CoV-2 Infection
1.1. Viral Infection
SARS-CoV-2 is commonly transmitted through respiratory droplets.16 The virus contains a single-stranded positive-sense RNA, and its envelope contains spike proteins that help the virus bind to angiotensin-converting enzyme 2 (ACE2) receptor.3,4,17,18 ACE2 receptors are expressed in many different cell types, including epithelial cells of the oral and nasal mucosa, pneumocytes of the lung, endothelial cells of arteries and veins, intestinal mucosa cells, kidney cells, immune cells, glial cells, and neurons.19−21 After binding with the receptor, cell proteases, mainly transmembrane protease serine 2 (TMPRSS2), cleave the spike protein and activate the viral membrane.22 The viral membrane then fuses with the host cell, and the virus releases its RNA content, which is then transcribed to produce new virions.19 The new virions then get released and infect other cells.
1.2. Neurologic Disorders of COVID-19 Infection
More than one-third of patients with COVID-19 experience neurologic symptoms during the course of their disease, and patients with more severe clinical manifestations of COVID-19 are more likely to develop neurologic symptoms and conditions.23,24 Symptoms and conditions include dizziness, headache, acute cerebrovascular disease, epilepsy, ataxia, acute disseminated encephalomyelitis (ADEM), viral encephalitis, anosmia (loss of smell), ageusia (loss of taste), myalgia, and fatigue.25 The mechanisms by which these neurologic symptoms and conditions develop are not well understood, and SARS-CoV-2 viral titers are only positive in a minority of patients presenting with neurologic symptoms.26 There have been arguments for direct neurotropism of SARS-CoV-2, and there are other arguments for systemic inflammation, autoimmunity, and metabolic changes as causal factors for neurologic symptoms27 (Figure 1).
Figure 1.
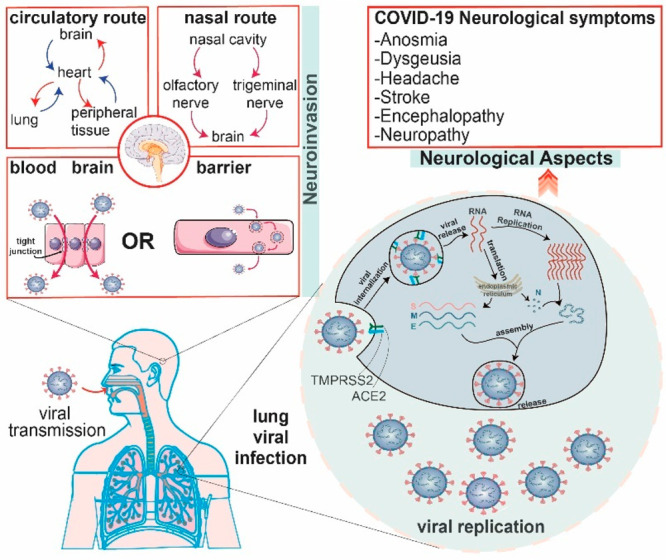
Proposed main mechanisms of SARS-CoV-2 virus cell entry and replication, transmission into the nervous system, and neurologic symptomatology. SARS-CoV-2 is first transmitted nasally, intratracheally, and intraocularly. In the lungs, bronchial epithelial cells have high levels of ACE2 receptors, which the virus uses to enter cells and replicate intracellularly. High levels of respiratory infection can then lead to cytokine storms and other widespread inflammation responses. This response may damage an intact blood–brain barrier, which can help viral entry into the nervous system, and the inflammatory response is associated with pro-thrombotic states, which may lead to occlusion of cerebral vessels and lead to brain injury. In addition, SARS-CoV-2 may also have direct neuroinvasive potential. SARS-CoV-2 viruses may enter nasal epithelium and neuronal cells, such as the olfactory nerve, through ACE2 receptors and then become transmitted into the brain through a neuronal retrograde fashion. SARS-CoV-2 viruses may also be able to enter the nervous system directly through other means, such as direct infection of choroid plexus cells and then dissemination through cerebrospinal fluid. As a result of the systemic inflammatory effects or direct neuroinvasion, patients develop neurologic symptoms such as anosmia and stroke. Some elements were created by Servier Medical Art.
1.2.1. Central Nervous System
Headache and dizziness are two of the most common central nervous system (CNS) symptoms and can present as isolated symptoms after COVID-19 infection.25,28,29 Cerebrovascular accidents (CVA) like stroke are also common, especially in individuals with risk factors.8 The mechanism behind CVA is likely the systemic inflammatory response to SARS-CoV-2 infection, such as toll-like receptor activation, endothelial dysfunction, tissue-factor pathway activation, and von Willebrand factor elevation.30,31 Infected patients commonly develop thrombocytopenia and elevated D-dimer, a reflection of a hypercoagulable state.32
More severe neurologic symptoms from COVID-19 infection may manifest as encephalopathy, with clinical features such as delirium, agitation, or decreased level of consciousness.33 These symptoms tend to be present more in hospitalized patients; for example, impaired consciousness is present in 7.5% of hospitalized patients.34 There can be multiple underlying reasons for a patient with COVID-19 to present with altered sensorium, including viral encephalitis, metabolic perturbation, infectious toxic encephalopathy, seizures with postictal confusion, and stroke. Seizures may follow encephalopathy or may be caused by COVID-19’s systemic effects.35 The systemic release of inflammatory cytokines and colony-stimulating factors, which can trigger neuronal hyperexcitability through glutamate receptor activation, ultimately leads to seizure episodes.36,37 In reported cases of viral meningoencephalitis, cerebral spinal fluid (CSF) analysis showed positive SARS-CoV-2 titers in some but not all patients.9,10
A rare form of encephalopathy, acute necrotizing encephalopathy (ANE), also has been described in the literature and is likely due to the uncontrolled release of cytokines during infection.38 ANE leads to disruption of blood brain barrier (BBB) without direct viral invasion. Finally, there have been limited cases of ataxia and acute disseminated encephalomyelitis (ADEM) reported in COVID-19 patients.39,40
1.2.2. Peripheral Nervous System
Peripheral nervous system (PNS) manifestations after COVID-19 infection include anosmia, ageusia, myositis, and neuropathy. Anosmia and ageusia are the most common presentations, even in patients with mild COVID-19 symptoms. One study showed olfactory and gustatory dysfunctions being present in 85.6% and 88.0% of 417 patients, respectively.41,42 Most patients slowly regain their sense of taste and smell as they recover from SARS-CoV-2 infections.43
Neuropathy and myositis are also complications from COVID-19 infections and can present as cranial nerve deficits, peripheral neuropathy, myalgia, and, in rare cases, Guillain-Barré syndrome.44,45 Myalgia can present in up to one-fourth of patients after COVID-19 infection.45 Neuropathy is a rarer presentation than myalgia, and Guillain-Barré syndrome is a severe form of neuropathy that has been reported in COVID-19 patients.46 CSF analysis is mostly negative for SARS-CoV-2 viruses in COVID-19 patients with Guillain-Barré, which could suggest a more autoimmune pathology rather than a neurotrophic etiology.46
2. In Vitro and In Vivo Models For Investigation of Covid-19 Neurologic Pathophysiology
2.1. Non-neuronal Cell Model
Common animal cell lines used in SARS-CoV-2 studies include Vero E6 (African green monkey kidney epithelial cells, expressing ACE2 receptor), BHK-21 (Syrian hamster kidney cells), and LLC-MK2 (rhesus macaque kidney epithelial cells). Common human cell lines include HEK 293T (human embryo kidney epithelial cells), Huh7 (human hepatocellular carcinoma), Caco2 (human epithelial colorectal adenocarcinoma cells), Calu3 (human lung adenocarcinoma), and HeLa (human cervical cell line) (Figure 2). These non-neuronal in vitro cell lines are used because they are generally easy to grow and become infected with SARS-CoV-2.47−51 Notably, the HEK 293T cells are a variant of the human embryo kidney epithelial cells.52 They express T SV-40 antigens, which enable the amplification of transfected plasmids containing the SV40 origin of replication and thus considerably increase the expression levels of desired gene products like ACE2 receptors.52 Cell models are useful for studying viral infection and propagation mechanisms and for conducting pharmacological research.47−51 However, most of these cell lines are behaviorally different from neural cells and often have tumor-inducing mutations, such as TP53 mutation, that may affect in vitro infection mechanisms and responses.53
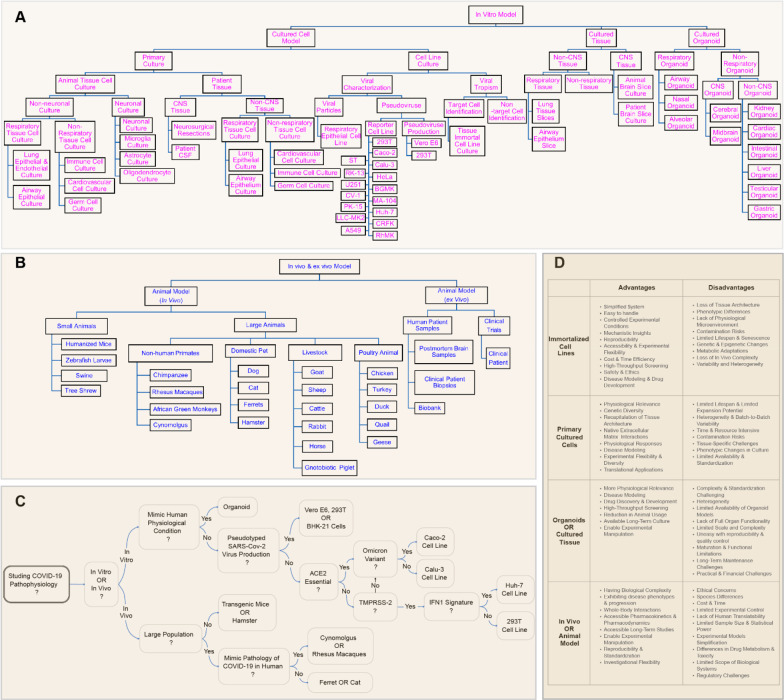
Figure 2.
Common in vitro and in vivo models for studying SARS-CoV-2 viral infection and therapeutics development. (A) In vitro models can be constructed and studied as cells, tissues, or organoids. Common in vitro models are listed. (B) Common in vivo and ex-vivo models include small and large animal models and samples from patients and clinical trials. (C) Flowchart of suggested model based on the purpose and design of SARS-CoV-2 study. (D) Comparison of advantages and disadvantages of in vitro and in vivo models from a cell, tissue, or whole organism level.
2.2. PSC-Derived Neuron and Neuronal Organoid
Pluripotent stem cells are cells that can differentiate into all the derivatives of the three germ cell layers, and they are naturally present in the form of embryonic stem cells in preimplantation embryos.54,55 Through in vitro technology, induced pluripotent stem cells (iPSC) are used to generate various types of neurons and neural supporting cells in a laboratory setting.56−60 Human iPSCs-derived CNS cells have been used to study SARS-CoV-2 replicability in monolayer neuron cultures (Figure 3 and Table 1). The transcriptome of iPSC-derived cells is similar to that of their respective primary counterparts, allowing the cells to respond to immune stimuli.61 One study observed sparse infection through immunostaining in primary astrocyte culture and hiPSC-derived cortical neurons and astrocytes but not in hiPSC-derived microglia.62 The study revealed the ability of SARS-CoV-2 to infect monolayer human cortical neurons and astrocytes, but not microglia, although infections of neurons and astrocytes were sparse.62 Furthermore, the study demonstrated significant productive SARS-CoV-2 viral infection, cell-to-cell viral spread, and apoptosis of primary human choroid plexus epithelial cells.62 Another study confirmed sparse infections in hiPSC cortical neurons and strong infectability in dopaminergic neurons but found sparse infections in microglia.63 It is unclear why the two studies showed different infectability in microglia, but different methodologies for cell preparation and inoculation may have resulted in the slightly elevated infection levels in the second study.63 More studies would be needed to confirm the infectability of microglia cells. Other studies have similarly shown viral replicability in hiPSC neural progenitor cells, cortical neurons, and astrocytes.64,65
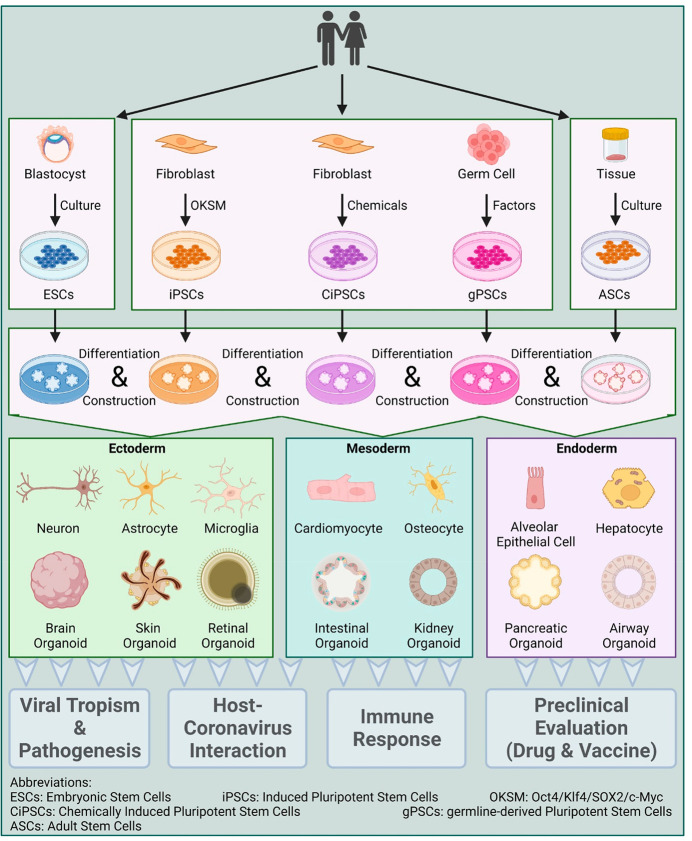
Figure 3.
Organoid generation for viral pathogenesis investigation. Organoid generation can be derived from human or animal tissues or induced pluripotent stem cells (iPSCs). For tissue-derived organoids, tissue samples are first obtained from humans or animals, purified and processed, and then seeded for harvesting. For iPSC-derived organoids, iPSCs are maintained as undifferentiated populations under defined conditions and then induced to form different tissue types (ectoderm, mesoderm, or endoderm). The organoids can then be used for diverse purposes in studies, as they recapitulate in vivo tissue-like structures and functions in vitro. This figure was created with BioRender.
Table 1. SARS-CoV-2 Virus Replicability in Animal and Human Cell Lines.
| animal cell lines | study 1168 | study 2169 | study 3170 | study 4171 |
|---|---|---|---|---|
| Vero E6 (African green monkey kidney) | yes | yes, MOI of 0.001 for 1 h | yes, MOI of 0.1 for 2 h | yes |
| Vero81 (African green monkey kidney) | yes | yes | not tested | yes |
| Vero/Slam (African green monkey kidney) | yes | yes | not tested | not tested |
| MA104 (African green monkey kidney) | yes | not tested | not tested | not tested |
| BGM (African green monkey kidney) | yes | not tested | not tested | not tested |
| MDCK II (dog kidney) | no | yes | no | not tested |
| FRhK4 | not tested | not tested | yes | not tested |
| LLC-MK2 (rhesus macaque kidney epithelium) | no | yes | yes | yes |
| CRFK (cat feline kidney) | not tested | not tested | yes | not tested |
| RK-13 (rabbit kidney) | not tested | not tested | yes | not tested |
| PK-15 (pig porcine kidney) | not tested | not tested | yes | not tested |
| human cell lines | study 1168 | study 2169 | study 3170 | study 4171 |
|---|---|---|---|---|
| Caco-2 (human colorectal adenocarcinoma) | significant level of virus multiplication but no cytopathic effects | yes | yes | not tested |
| Calu3 (human lung adenocarcinoma) | not tested | yes | yes | luciferase activity significant |
| 293T (human embryonic kidney) | no | yes | yes | yes |
| Huh-7 (human hepatocellular carcinoma) | not tested | yes | yes | luciferase activity significant |
| A549 (human lung adenocarcinoma) | not tested | yes | no | luciferase activity significant |
| BEAS-2B (human bronchus normal epithelium) | not tested | yes | not tested | not tested |
| H1299 (human nonsmall cell lung carcinoma) | not tested | yes | not tested | not tested |
| U251 (human neuronal glioblastoma) | not tested | not tested | yes | not tested |
| HeLa (human cervical cancer) | no | not tested | no | yes |
Monolayer primary and hiPSC-derived neural cells share some similar advantages with non-neuronal cell lines, including the relative ease of in vitro growth, infection, and therapeutic testing. There are also opportunities to generate COVID-19 patient-specific iPSCs66 that would closely mimic individuals’ neural conditions and enable personalized therapeutic approaches.67 One limitation of in vitro neural cells, however, is that they do not recapitulate cellular behaviors in a 3D environment. To overcome this disadvantage, neuronal organoids can be used.
Neuronal organoids have been historically used for other viral infections that have neuro-invasive potentials, such as CMV and Zika. For example, iPSC-derived brain organoids were used to model CMV pathology similar to what was found in clinical specimens, such as an impaired generation of cortical layers.68,69 iPSC-derived brain organoids were also used to mimic the abnormal neurogenesis process in Zika-virus infections.70,71 Compared to viruses with well-documented neurotropism and impact on neurogenesis, the pathology behind the neurologic manifestations of SARS-CoV-2 is more complex and controversial. hiPSC-derived organoids have been used to model SARS-CoV-2 infection in many organs, including the lung, intestine, kidney, pancreas, liver, vasculature, and brain.63,72−75 Specifically for the brain (Table 2), one study by Jacob et al. examined SARS-CoV-2 infection on hiPSC-derived cortical, hypothalamic, hippocampal, and midbrain organoids, and only sparse SARS-CoV-2 nucleoproteins were observed in these organoids (around 1%).62 Notably, some of the hippocampal organoids contained regions with choroid plexus epithelial cells, and a greater density of infected cells was observed in those choroid plexus regions.62 To further explore this finding in choroid plexus cells, choroid plexus organoids (CPO) expressing ACE2 and TMPRSS2 were generated.62 After viral inoculation, CPO showed significant SARS-CoV-2 infections and replications (10–20% of the cells), inflammatory cellular responses, and cell deaths.62 Another study used hiPSC-derived cortical organoids and choroid plexus organoids. Similarly, there were productive SARS-CoV-2 infections in choroid plexus organoids and only sparse infections were found in cortical neurons.76 The findings in organoids are consistent with the demonstrated high infectability of monolayer culture of human choroid plexus.62 These findings may suggest that the choroid plexus could serve as a productive infection site and act as a CNS entry site for SARS-CoV-2, leading to viral entry into the CSF. Another finding to support this theory is the high-level expression of SARS-CoV-2 entry factors, such as ACE2 and TMPRSS2, in choroid plexus compared to other brain regions.77 Meanwhile, compared to the two aforementioned organoid studies,62,76 a few other organoid studies64,65,74,78 have shown higher viral proliferation in cortical neurons. More studies would be needed to confirm the infectability of cortical neurons, which would be helpful in better understanding the neurotropism of SARS-CoV-2 and thus viral entry mechanisms in the nervous system.
Table 2. SARS-CoV-2 in Neural Stem Cells and Organoids in Previous Studies.
| ref | model | more details about the model | virus used and dose | virus expression |
|---|---|---|---|---|
| (64) | human iPSC neural progenitor cells (NPC) | iPSCs were differentiated into human neural progenitor cells for a total of 9 days | SARS-CoV-2 HKU-001a isolate from a COVID-19 patient nasopharyngeal specimen, 10 multiplicities of infection (MOI) | SARS-CoV-2 RNA gene number increased significantly at 24- and 48-h post infection (hpi); cell viability significantly decreased at 72 and 120 hpi |
| (62) | hiPSC-derived cortical neurons, astrocytes, and microglia | cortical neurons with MAP2 marker, astrocytes with GFAP, and microglia with PU1 | SARS-CoV-2 USA-WA1/2020 viral isolate | 120 hpi showed sparse infection of cortical neurons and astrocytes; hiPSC-derived microglia showed no infection at 48 and 120 hpi |
| (63) | hiPSC cortical neurons, microglia, and dopaminergic neurons | day 45 cortical neurons with beta III-tubulin, dopaminergic neurons with FOXA2 and MAP2, microglial cells with PU.1 and IBA1 | vesicular stomatitis virus (VSV)-based SARS-CoV-2 pseudoentry virus, inoculation for 24 h | dopaminergic neurons but not microglia or cortical neurons were infected (measured by pseudovirus reporter luciferase activity) |
| (65) | iPSC culture NPCs, neurons, and astrocytes | NPCs with SOX-2, cortical neurons with MAP-2, and astrocytes with GFAP | SARS-CoV-2 USA-WA1/2020 isolate, 2 MOI | at 2 and 48 hpi, all three cell types supported viral replication over time, with the amount of viral RNA being highest in neurons when compared with NPCs and astrocytes |
| (62) | hiPSC-derived organoids | cortical, hippocampal, hypothalamic, and midbrain (CTIP2, PROX1, OTP, and TH as markers, respectively); mixed cell types (e.g., neurons and astrocytes) | SARS-CoV-2 USA-WA1/2020 viral isolate | viral nucleoprotein was detected sparsely in the organoids in a range that averaged between 0.6% and 1.2% of all cells at 24 and 72 hpi; most of the infected cells are neurons |
| (64) | brain organoids | 35-day-old brain organoid, mixed cell types resembling developing cerebral cortex (e.g., neural progenitor cells) | SARS-CoV-2 HKU-001a from a COVID-19 patient nasopharyngeal specimen | extensive SARS-CoV-2 antigen was detected at 72 hpi; infection colocalized with neuronal marker |
| (76) | brain organoids | organoids aged more than 55 days; mixed cell types, (e.g., choroid plexus and cortical tissue) | SARS-CoV-2 spike pseudo virions carrying a 19-amino-acid deletion (c19) from the C terminus | significant increase in viral genome copies in organoid supernatant between 24 and 48 hpi; around 13% of cells in the choroid plexus were infected, and neuronal regions mostly did not get infected |
| (65) | brain organoids | mixed cell types, such as NPCs, neurons, and glial cells characterized by their respective markers SOX-2, MAP-2, and GFAP | SARS-CoV-2 pseudo virus, 2 MOI | infection was more efficient in neurons than in NPCs at 24 hpi |
| (74) | brain organoids | two different age groups of organoids (day 15 and day 60) | SARS-CoV-2 (NRW-42) isolate from a nasopharyngeal and oropharyngeal swab specimen of an infected patient, TCID50/mL of 50 or 17.5 PFU/organoid | at 48 and 96 hpi, day-60 organoid had a significantly higher number of viral-positive cells than day-15 organoid |
| (78) | brain organoids | 9-week-old organoids | SARS-CoV-2 isolate USA-WA1/2020 | at 24 and 96 hpi, there was a significantly increasing number of SARS-CoV-2 positive cells; many infected cells were MAP2-positive mature neurons, but sparse infections of SOX2-positive neural stem cells were also observed; at 96 hpi there was a widespread infection, mostly limited to the regions with high cortical cell density |
| (62) | choroid plexus organoid | 47 and 67 days in vitro | SARS-CoV-2 USA-WA1/2020 viral isolate | many TTR+ choroid plexus cells showed infection |
In addition to modeling in a 3D environment, brain organoids have the advantage of allowing analyses of SARS-CoV-2 infection in real-time, with the ability to introduce therapeutic agents.62 A disadvantage of brain organoids is the lack of interactions with other tissue types and organs in vitro, such as BBB, and this is a drawback monolayer neuronal cell cultures share as well. This disadvantage results in challenges in studying the systemic mechanisms of SARS-CoV-2 infection, such as the effect of hypoxia and systemic inflammation on the CNS. One potential way to remedy this drawback is to incorporate 3D bioprinting of other cell types, such as blood vessels, into the brain organoids, which will be discussed further in a later section.
2.3. Animal Model
2.3.1. Nonprimate Animal Model
There is an urgent need to develop animal models that are useful for studying not only the effects of SARS-CoV-2 on the nervous system but also the systemic and long-term neurologic effects. It is important to note that no single animal model is a perfect representation of human disease, and researchers must carefully consider the strengths and limitations of each model when designing studies (Table 3). Additionally, studies in animal models must be validated by studies in humans to ensure their relevance to human disease (Figure 2).
Table 3. SARS-CoV-2 Infectivity and Pathologic Response of Different Species of Animal Models.
| animal model | ACE2 receptors | neurotropism | pathophysiology of SARS-CoV-2 infections | ref |
|---|---|---|---|---|
| wild type mice | have murine isoform of ACE2 receptor, but it has a low affinity for viral S protein on SAR-CoV-2; modified SARS-CoV-2 strains with higher affinity to murine ACE2 receptor, such as SARS-CoV-2 B.1.351 and SARS-CoV-2 MA, are needed to produce disease states in both young and old mice | not determined | modified SARS-CoV-2 strains produced lung abnormalities; SARS-CoV-2 MA caused more severe disease in older mice | (80, 172−174) |
| K18-hACE2 transgenic mice | transgenic human ACE2 receptor expressed through K18 promoter | increased viral titer after intranasal administration measured by viral nucleocapsid protein antigen | local and systemic elevation of chemokines, including the brain; elevated levels of IFN- λ in the lungs; tissue pathology including vasculitis is shown in the brain; perivascular inflammation is observed | (78, 82, 84, 175−176) |
| HFH4-hACE2 transgenic mice | transgenic human ACE2 receptor expressed through HFH4 | high levels of virus and ACE2 receptors were detected in the brain | severe immune and pathological response like K18-hACE2 mice | (82, 83, 173) |
| Syrian hamster (Mesocricetus auratus) | Syrian hamster ACE2 receptors are homologous to humans | virus or viral RNA in the CNS and neurovascular inflammation have been detected by some studies; retrograde infection from olfactory infection may be possible | mononuclear cell infiltrates, apoptosis, and pro-inflammatory cytokines like IFN-γ are observed in the lungs and airways early after infection (2–3 days) | (100, 102−105, 177, 178) |
| ferret | ferret ACE2 receptors share some amino acid residues with humans | no, but is present in olfactory tissues, such as the nasal turbinate and sustentacular cells of the olfactory epithelium | virus multiplication and antibodies are detectable in the upper respiratory tract a few days after exposure | (110, 179−181) |
| nonhuman primates (gibbon, green monkey, macaque, orangutan, and chimpanzee) | ACE2 receptors have identical sequences in regions important for interaction with SARS-CoV-2 | mixed evidence; some studies demonstrate neuronal damage due to systemic effects, such as strokes due to hypercoagulable states, but no viral level in CSF; other studies show high viral in the brain and olfactory bulb | lung infiltrates and elevated serum cytokine levels (cytokine storm); decreased hematocrit and hemoglobin; rhesus macaques have more severe disease than other macaques; aged primates have more severe disease | (110−113, 117− 120, 179, 182−183) |
2.3.1.1. Mouse
The mouse (Mus musculus) has several advantages as a virological model: low cost, ease of handling, and the possibility of large-scale studies.79 However, wild-type and inbred strains only express the murine isoform of the ACE2 receptor, to which the SARS-CoV-2 virus has low binding affinity.80 To overcome this barrier, SARS-CoV-2 studies have used transgenic mice expressing human ACE2 (hACE2).81
K18 (epithelial cell cytokeratin-18 promoter) is an exogenous promoter that can be used to induce transgenic hACE2 expression in mice.82,83 It was originally developed for studying SARS-CoV and subsequently evaluated for SARS-CoV-2.84 Song et al. used K18-hACE2 transgenic mice and observed increasing viral titers in mice brains after intranasal administration of SARS-CoV-2.78 The virus was widely present in neural cells in the forebrain but was not detected in the cerebellum, except in the pial meninges and dorsal cochlear nucleus. Other regions also contained a relatively low density of infected cells, the dentate gyrus, the globus pallidus, and cortical layer 4.78 Notably, the virus was not detected in the vascular endothelium.78 Compared to SARS-CoV-2 studies, SARS-CoV studies using K18 transgenic mice were more lethal and more symptomatic.85,86
Hepatocyte nuclear factor-3/forkhead homologue 4 (HFH-4/FOXJ) is another exogenous promoter, expressed in developing mice cells’ lung motile cilia.81 Transgenic mice expressing HFH4-hACE2 have been shown to be susceptible to SARS-CoV-2 infection.81 HFH4-hACE2 transgenic mice demonstrated brain infection after intranasal administration of the SARS-CoV-2 virus.87,88 Only ∼60% of mice survived after infection, and interestingly, significant brain viral levels were only detected in the deceased mice.87,88 The specific brain regions that were infected were not discussed.87,88
Adenovirus and adeno-associated virus (AAV) are vector options to deliver exogenous promoters in mice. Compared with adenoviral vectors, the use of AAV tends to elicit less host immune response.89 However, one common drawback for both the use of adenovirus and AAV is the possibility of nonuniform viral transduction and thus expression in the neural tissues. Using AAV virus, Song et al. compared respiratory versus neural routes of administration on SARS-CoV-2 neuroinvasion. Intratracheal administration of AAV virus to express hACE2 receptors followed by intranasal administration of SARS-CoV-2 (respiratory route) was compared with intracisternal AAV and intraventricular SARS-CoV-2 administration (neural route). Intranasally infected mice showed signs of lung pathology but no weight loss or death. However, intraventricularly infected mice showed weight loss and death, even at the challenge virus dose 100-fold lower than that used for intranasal infection.78 This result highlights the potential of SARS-CoV-2 for neuroinvasion through the hACE2 receptors and causing lethality in AAV-hACE2 mice. In addition, Song et al. demonstrated that using AAV induction, hACE2 can be induced in an organ-specific manner.
Some newer transgenic hACE2 mouse models have also been generated by CRISPR-Cas9 and knock-in approaches, thus endogenously expressing human ACE2 receptors. These mouse models with endogenous hACE2 expression may be more clinically similar to humans than the mouse models expressing exogenous genetic materials.90 Intranasal administration of SARS-CoV-2 in CRISPR-generated hACE2 mice has demonstrated significant viral load in the lung and brain.91
One common drawback of all the discussed transgenic mouse models is that unlike in what is naturally seen in human neural tissues, hACE2 is highly expressed in these mice’s nervous system, which may misconstrue the infection mechanisms and presentations.83,84,92,93 Another drawback is the length of time needed to develop these mouse strains.
In short, the need to engineer human ACE2 expression is a limitation of the mouse model because hACE2 receptors in mice may not be expressed in the same cell types and at the same levels as they are in humans. This may lead to higher levels of SARS-CoV-2 neuro-invasion in hACE2 transgenic mice than in the native human nervous system. On the other hand, the silver lining is that transgenic mouse models can show which cell types allow SARS-CoV-2 neuro-invasion when expressing hACE2 receptors, allowing researchers to test mechanisms of neuro-invasion as well as antiviral strategies.94,95 Another advantage of using hACE2 mice is the ability to study long-COVID and its correlation with different acute-phase infection severities. By adjusting hACE2 expressions in mice, researchers can create mouse models of mild infections and see if they develop long-COVID symptoms. This may be important in studying long COVID, as young patients who have only had mild symptoms during the acute infection phase can still develop long-COVID symptoms that impact their quality of life.96 For a more in-depth review of the implications of animal studies on studying the neural impact of long-COVID, Jansen et al. provide a thorough overview and discussion.97
2.3.1.2. Hamster
Golden Syrian hamsters (Mesocricetus auratus) have been used extensively for virological studies, such as for SARS-CoV and influenza virus.98 In a systematic review of hamsters, seven studies investigated whether the virus was present in the brain following SARS-CoV-2 nasal inoculation.99 Three studies found either viral RNA or virus (by plaque formation) in the olfactory bulb or brain at 1 to 14 days postinfection,100−102 with 2 logs lower than in nasal turbinate and with similar levels in the brainstem, cerebral cortex, and cerebellum.102 The other four studies found no evidence of brain infection with antibodies. One advantage of using hamsters for SARS-CoV-2 studies is that the hamster ACE2 gene is highly analogous to human ACE2 and that hamsters can be infected by and transmit SARS-CoV-2.98,104,105 Hamsters exhibit viral pathogenesis and immune responses consistent with those seen in humans.98 While hamsters are easy to handle, the availability of hamster-compatible reagents, such as reagents to examine immune responses and protein expression, is limited.100
2.3.1.3. Ferret
Ferrets (Mustula putorius furo) are another commonly used animal model because of their ability to transmit human respiratory viruses.106,107 In addition to the presence of appropriate viral receptors, histo-anatomical features of the ferret respiratory tract are analogous to that of the human respiratory tract.108,109 Another advantage of ferrets is their larger size than most other rodents. This allows for more volume of blood tests for immunological analyses and monitoring.
In terms of neural studies, SARS-CoV-2 viral RNA has not been found in the brain of ferrets after intranasal administration but is present in olfactory tissues, such as the nasal turbinate and sustentacular cells of the olfactory epithelium.110 These findings are consistent with those of hamsters.100−102
2.3.1.4. Others
Domestic animals such as pigs, dogs, chickens, and ducks may not be good models for studying COVID-19 neurologic effects because they have low susceptibility to SARS-CoV-2 infections. Serum and organ samples of pigs, dogs, chickens, and ducks show low levels of SARS-CoV-2 replication after intranasal viral inoculation.111
2.3.2. Nonhuman Primate Animal Model
Nonhuman primates (NHPs) are phylogenetically close to humans, thus are generally beneficial for studying infections. NHPs including rhesus macaques (Macaca mulatta),112−115 cynomolgus macaques (Macaca fascicularis),114,116 African baboons (Papio hamadryas),115 and African green monkeys (AGMs) (Chlorocebus aethiops).117 NHPs have been demonstrated to be permissive to SARSCoV-2 infection and present symptoms similar to humans such as fever, diarrhea, and pneumonitis.112−116
Studies using NHPs have generated mixed results about whether there are detectable viral loads in different CNS regions. For example, one study by Munster et al. showed that in infected rhesus macaques presenting with respiratory symptoms, no significant SARS-CoV-2 viral loads were detected in frontal lobes, brainstems, or cerebellum.118 Another study done by Rutkai et al. confirms the lack of direct cortical infection and provides an explanation for the route of neurologic manifestations in NHPs.119 Rutkai et al. used Rhesus macaques and African green monkeys to demonstrate neuronal injury, microhemorrhage and strokes, and brain hypoxia after mucosal SARS-CoV-2 infection. Suspected endothelial cell infection was confirmed by colocalization immunofluorescence, but SARS-CoV-2 virus was not identified in the CSF, consistent with most findings among human subjects except in rare cases of encephalitis.119 There were elevated CO2 levels in the animals, suggesting mild hypoxemia and impaired gas exchange in the lungs.119 On the basis of this, Rutkai et al. concluded that the underlying cause of neuronal injury was a lack of oxygen.119 This does not appear to be a direct consequence of virus infection, as only limited virus was seen in brain vasculature and did not appear to involve parenchymal cells.119 Instead, neuronal injury and death most likely occur because of energy failure, which is an early consequence of hypoxic-ischemic events.119
On the contrary, Jiao et al. found evidence of virus RNA and nuclear capsid antigen in the brain of NHPs, including the olfactory bulb, suggesting direct neuro-invasion of SARS-CoV-2.120 However, this study applied an extremely high dose of the virus, about 20 times higher than the other monkey studies.120 Thus, more studies and caution are needed to confirm SARS-CoV-2 neuro-infectability of NHPs.
3. Vaccine and Therapeutics Development Using Pre-Clinical Models
When considering preclinical models for vaccine and therapeutic development, the advantages and disadvantages discussed in earlier sections should be taken into consideration. For example, mice can be used for rapid screening of vaccines, antivirals, and other therapeutics in a pipeline high output approach. However, mice’s structural difference in their ACE2 receptors from human ACE2 receptors results in reduced susceptibility to SARS-CoV-2 infection and thus more difficulty in testing the effectiveness of vaccines and therapeutics. Using hACE2-transgenic mice can be a remedy to the shortcoming as discussed previously. The National Institutes of Health (NIH) Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) Preclinical Working Group has developed animal model summaries and descriptions for studying vaccines and therapeutics that may be helpful for reference.121 A reoccurring theme is that SARS-CoV-2 can infect certain species like nonhuman primates, hamsters, ferrets, and hACE2 transgenic mice, but clinical disease and symptomatology in these models have generally been mild (Table 4). It is important to consider the difference in nature and severity of symptoms between what the animal models capture and what is present in human patients when selecting models for studies.
Table 4. SARS-CoV-2 Symptoms Comparison between Human and Animal Models.
| neuronal and non-neuronal symptoms | human | animals | ref |
|---|---|---|---|
| increased body temperature | yes | rhesus macaque, cynomolgus macaques, and ferret | (184 and 185) |
| chest radiograph abnormality | yes | rhesus macaque and cynomolgus macaques | (185) |
| coughing | yes | rhesus macaque | (118) |
| reduced appetite | maybe | rhesus macaque | (118) |
| weight loss | maybe | rhesus macaque, Syrian hamster | (102 and 118) |
| dysgeusia | yes | Syrian hamster | (102) |
| hyposmia/anosmia | yes | Syrian hamster, hACE2 mice | (95, 102, and 186) |
| fatigue | yes | Syrian hamster | (104 and 187) |
| other neurologic symptoms | yes | unclear |
3.1. Vaccine
There are currently three major vaccines approved by the US Food and Drug Administration (FDA) for emergency use in the US, and each of them has gone through extensive and vigorous preclinical testing using cellular and animal models. BioNTech/Pfizer and Moderna vaccines utilize a novel mRNA (mRNA) approach, in which mRNA encoding a subunit of the SARS-CoV-2 spike protein is delivered with a lipid nanoparticle encapsulation to increase stability and uptake by antigen presenting cells.122 The mRNA is then translated into spike protein, inducing cell and humoral immunity.122 The Johnson & Johnson vaccine, in contrast, uses a replication-deficient adenovirus vector to deliver spike-protein DNA into intracellular space, which then is made into antigenic protein.123
The developers of the BioNTech/Pfizer vaccine (BNT162b2 mRNA) used cells, mice, and nonhuman primates for their preclinical data.122 HEK293T/17 and Expi293F cells were used to test expression levels and structural characteristics of the SARS-CoV-2 spike protein from mRNA incubation.122 Following BNT162b2 transfection, BALB/c mice were used to detect and characterize the immunogenicity of the vaccine. The authors found that BNT162b2 elicits strong virus-neutralizing titers and systemic T cell responses.122 Moderna also similarly used mice, specifically BALB/cJ, C57BL/6J, and B6C3F1/J mice, and demonstrated effective immunogenicity of their mRNA-1273 vaccine.123 Finally, BioNTech/Pfizer used 2- to 4-year-old rhesus macaques to detect antibody development and protection from SARS-CoV-2 infection challenge tests.122 The authors found that vaccination significantly reduced virus levels found in nasal and oropharyngeal swabs and bronchoalveolar lavage.122 Notably, the authors commented that they found no clinical signs of significant disease in virus-challenged nonhuman primates (rhesus macaques).122 Vaccine data from nonhuman animal models must be interpreted with caution, especially from a symptomatology perspective. What is reassuring, however, is the recent data showing that mild clinical manifestations of COVID-19 in macaques, measured by CT scan and histopathological samples, are mostly similar to that seen in humans.124
The Johnson & Johnson Ad26 vaccine was tested on hamsters and rhesus macaques.125 In hamsters, the authors demonstrated that a single immunization with an Ad26-vector-containing spike-protein DNA induced immunogenicity and antibody formation, which protected against severe clinical diseases after the high-dose SARS-CoV-2 challenge.125 The authors chose hamsters as a preclinical model because hamsters could be used to generate severe pneumonia models mimicking clinically severe infections in humans.126 The authors also studied the vaccine on rhesus macaques and showed that a single immunization of their vaccine induced a strong neutralizing antibody response and provided protection against the SARS-CoV-2 challenge, similar to the results in hamsters.125
Vaccines have been shown to be effective in preventing severe COVID-19 infection in human clinical data. For example, the BioNTech/Pfizer vaccine had 95% efficacy in preventing infection and symptom development.122 Neurologic symptoms assessed included myalgia, anosmia, and ageusia.127 Furthermore, vaccination has been shown to reduce stroke incidence after COVID-19 infection.127 This is logical because if vaccines can prevent severe systemic inflammation, vaccines would likely have protective effects against COVID-19 neurologic symptoms. However, there have been controversies about rare cases of serious neurologic side effects from COVID-19 vaccination, possibly due to an autoimmunity etiology. For example, in Fall 2020, two patients developed transverse myelitis after receiving the Oxford/AstraZeneca vaccine.128,129 In another study of the mRNA vaccines by BioNTech/Pfizer and Moderna, 7 cases out of 37,000 vaccine recipients developed Bell’s palsy, though the FDA commented that the indicated incidence of Bell’s palsy was not higher than expected in the general population.130
3.2. Therapeutics
Commonly used drugs to treat COVID-19 include antiviral (remdesivir, Molnupiravir), anti-inflammation (dexamethasone, Baricitinib), anticoagulability, and immunoglobulin drugs (convalescent plasma). Most of these drugs have been shown in clinical trials to reduce the severity of COVID-19 infection and lower the rate of hospitalizations.131−133 Currently, the FDA (Food Drug Administration) has approved tocilizumab, remdesivir, baricitinib, nirmatrelvir, and ritonavir.134
By inference, if these drugs can reduce the severity of COVID-19 infection, they likely reduce COVID-19 neurologic symptoms, as COVID-hospitalization has been shown to be a risk factor for COVID-19 neurologic symptoms.135 Indeed, Grundmann et al. have shown that treatment with dexamethasone and remdesivir is associated with reduced mortality and lower frequency of neurologic complications, notably stroke, seizure, and meningitis.136 Grundmann et al. also showed that combined dexamethasone and remdesivir resulted in a further reduction of neurologic complications than single-drug therapy.136 More studies, both clinical and preclinical, are needed to investigate the mechanisms behind neurologic COVID-19 drug complications and to discover new or off-brand use of drugs. For example, Baricitinib was originally indicated for alopecia and rheumatoid arthritis but has recently been approved for COVID-19 use.137 Preclinical neurologic studies of COVID-19 drugs are especially needed; currently limited studies include that of Wang et al. (2021), which found that the antiviral drug remdesivir effectively inhibited SARS-CoV-2 infection in hiPSC-derived neurons and resulted in improvement of neurite lengths of infected neurons.138
Another area of opportunity for COVID-19 therapeutics is herbal medicine. For instance, prior to the COVID-19 pandemic, the traditional Chinese medicine Lianhuaqingwen was shown to be effective in treating influenza.139,140 Then during the pandemic, there have been clinical trials and case-control studies on Lianhuaqingwen for SARS-CoV-2 infection.141−145 A meta-analysis of these studies showed that Lianhuaqingwen can significantly improve clinical COVID-19 symptoms and slow progression to severe conditions from infection.146 Another systematic review showed that Lianhuaqingwen combined with conventional treatment (e.g., oxygen therapy and antiviral medications) is more effective in treating mild or moderate COVID-19 clinical symptoms than conventional treatment alone.147 However, both the meta-analysis and the systematic-review studies caution about the quality of the included studies, thus more studies are needed to confirm the efficacy of Lianhuaqingwen.146,147 Other herbal medicines have also shown clinical potential, and Alam et al. have provided a comprehensive review and discussion on currently available herbal medicines for COVID-19 and their supporting studies.148
4. Perspective
4.1. Future Directions for Model Generation and Improvement
FDA approves only around 10% of neurology drugs from phase I clinical studies.149 A major reason hindering the success of drug development is the preclinical models’ lack of recapitulation of human pathophysiology. Therefore, there is a strong need for robust modeling of human neurologic diseases.
One direction for future COVID-19 model generation is developing more sophisticated brain organoid models (Figure 3). For example, Gleeson et al. integrated perivascular cells into cortical organoids and found that these organoids had more extensive SARS-CoV-2 infection than the traditional organoids.150 More studies using such “assembloid” models with crosstalk between different tissue types are underway. Future studies can expand the single-organoid model to a multiorgan system by connecting the brain organoid to other organs such as lung organoid63 and cardiac organoid151 through blood circulation. Doing so would allow better modeling of systemic responses to COVID-19 infection, such as inflammation and hypoxia, at the whole organism level and the impact on the nervous system. Another avenue of opportunities is using organoids derived from cells of patients at different stages of COVID-19 infection to draw correlations between pathophysiology and clinical findings.
To facilitate the development of more sophisticated organoids, 3D bioprinting is a technology with great potential. 3D bioprinting offers a platform to address the challenges of integrating multiple cell types in brain organoids. 3D bioprinting allows precise placement of cells and growth factors through a bioprinter on a building block like an organoid.152 A recent study by Ahn et al. developed a microengineered physiological system to model the BBB.153 Their platform contained two compartmental microfluidic channel layers.153 The upper layer is a porous membrane with monolayer endothelial cells above and pericytes below the membrane.153 The lower layer contained astrocytes in a 3D Matrigel matrix.153 The authors reported features of this model resembling native BBB, including low permeability and similar architectural features.153
Another challenge is mimicking the human systemic immune response to COVID-19, such as cytokine storms. One solution is to design immune-competent humanized models. For instance, humanized mice have been made by implanting xenografts of human lung tissue into immunodeficient mice, producing bone marrow/liver/thymus-lung (BLT-L) mice.154 These mice then demonstrated infectability with RSV virus, Zika virus, MERS-CoV, and cytomegalovirus and produced an immune response mimicking humans.154 Challenges of making humanized models include the technical expertise required, long preparation time, and immunocompromised status of the animal models.
Finally, a possibility for the future is xenotransplantation of human donor organs into pigs. Pigs have many similarities with humans in organ systems and size.155 Though there have not been actual cases of transplantation into pigs, the idea comes from recent clinical cases of pig heart and kidney transplantation into humans.156,157 If pig organs can be transplanted into humans, doing so vice versa may also be a possibility. These pig models with humanized organs would more accurately capture neurologic COVID-19 pathophysiology and symptomatology. These models can offer new perspectives that include demonstrating the relationship between infection severity and symptoms and comorbidities such as type 1 diabetes, hypertension, and obesity. Nonetheless, caution certainly must be taken with pursuing this idea, as there would be ethical concerns and public health concerns about zoonotic diseases.
4.2. Artificial Intelligence-Based Drug Discovery
New CNS drugs on average take 15–19 years to advance from discovery to regulatory approval.158 This would be too slow for a new emerging disease causing a global health crisis like COVID-19. Therefore, models and technologies to facilitate a swift generation of effective therapeutic targets and agents would be essential. With the tremendous recent advancements in machine learning (ML), artificial intelligence (AI) shows great potential for facilitating drug discovery and application (Figure 4). Furthermore, taking advantage of AI techniques for modeling COVID-19 pathology can replace, reduce, and refine the use of animal models in studies,159 achieving more humane approaches while ensuring high-quality outcomes.
Figure 4.
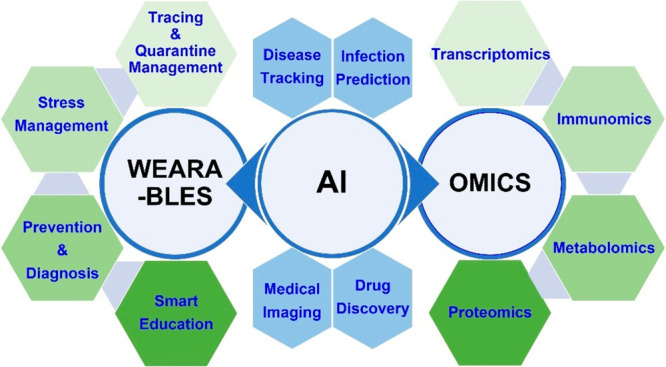
Artificial intelligence application in the COVID-19 pandemic. AI provides a unique opportunity to simultaneously advance infection prevention and management strategies for individual patients and link them to understanding the virus from a population level. For example, studies on “omics” like proteomics and metabolomics can help identify abnormal biomarker levels in infected individual patients and help guide management strategies. Wearable technologies on individuals can also help identify signs (such as elevated body temperature) of infection and offer biophysical and psychological feedback on coping with infections or concerns about infections. Compiling and analyzing these data from individual patients (with consent from users) can then help research agencies identify patterns on prevention/quarantine/virus spread, drug development, and management strategies on a population level using AI algorithms.
In terms of drug discovery, AI/ML has been used at three different early stages of the drug discovery process–target identification, lead generation and optimization, and preclinical development.160,161 In target identification, AI-based approaches have been used to help elucidate molecular mechanisms and pathways underlying diseases and drug activities.160 For lead generation and optimization, algorithms improve the functions and quantitative structure–activity relationship (QSAR) models in screening pipelines and help automate and optimize drug design de novo.160 In preclinical development, AI/ML approaches are utilized to predict physical-chemical properties by processing large amounts of data and to optimize pharmacokinetics and pharmacodynamics.160
An example of using AI to discover off-brand uses of approved medicine is halicin in early 2020.162 Researchers at MIT trained a deep graph-convoluted network (GCN) model based on drug candidates’ molecular features from the Drug-Repurposing Hub, an open-access repository of more than 6,000 compounds many of which the FDA has approved.162 Using the GCN, the team identified halicin, originally labeled as a diabetes drug, as a new potential antibiotic.162 Then, halicin showed a broad-spectrum activity against drug-resistant strains in mice.162 This was the first time an AI/ML-assisted tool was used to identify a new type of antibiotic from scratch. This goes to show that the AI/ML technology holds immense potential for new emerging diseases like COVID-19 and could reduce the burden of animal model usage. Furthermore, with enough data volume, such as a large registry of neurologic symptoms and treatment outcomes from hospitals and clinics, robust AI/ML algorithms can be developed and used to generate management strategies based on each patient’s individualized clinical picture.
4.3. Multiomics in Unraveling the Landscape of SARS-CoV-2 Infection
Given recent advancements in sequencing technologies, access to large-scale omics data sets (e.g., genomics, transcriptomics, proteomics, metabolomics, metagenomics, phenomics) could help revolutionize our understanding of SARS-CoV-2 and its effects on the nervous system.163 With the help of machine learning, multiomics studies can be done to identify neurologic biomarkers that can be used to diagnose and monitor disease progression. For example, one study collected patient blood serum and used proteomics and metabolomics to identify molecular differences from the non-COVID-19 population–dysregulation of macrophage, platelet degranulation, complement system pathways, and metabolic suppression.164 Omics can also help classify the morphological observations underlying COVID-19 neurologic pathology. For example, one study used proteomics to characterize over 10,000 proteins in organ autopsy samples and identified multiple dysregulated proteins involved in fibrosis, coagulation, hypoxia, angiogenesis, and glucose and fatty acid metabolism.165 As more studies in proteomics and metabolomics are done, there may be recurring findings of certain protein and metabolite dysregulation involved in the pathology of patients with neurologic symptoms. Using AI and sampling a large population of patients across different spectra of symptoms and severity, the morphological and metabolic targets can be used to develop and monitor COVID-19 neurologic disease prognostics, progression, therapeutics, and prevention strategies166,167 (Figure 4).
5. Conclusions
Neurologic complications of SARS-CoV-2 infection are common, and a better understanding of neurologic pathophysiology is urgently needed for more effective management and treatment strategies. In this paper, we discussed available preclinical models and their pros and cons for research on disease etiology, vaccines, and therapeutics. We also presented new models and opportunities in the field of COVID-19 neurologic research. This paper has focused on addressing the acute phase of neurologic COVID-19; an area of future discussion is “long-COVID” after the acute phase of infection, which is also highly prevalent.
The authors declare no competing financial interest.
References
- Feng X.; Wang H. Emerging Landscape of Nanobodies and Their Neutralizing Applications against SARS-CoV-2 Virus. ACS Pharmacol Transl Sci. 2023, 6 (7), 925–942. 10.1021/acsptsci.3c00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/ (accessed August 6, 2023).
- Hu B.; Guo H.; Zhou P.; Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol 2021, 19 (3), 141–154. 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R.; Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses 2015, 1282, 1–23. 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsucci D.; Caldarazzo Ienco E.; Nocita G.; Napolitano A.; Vista M. Neurological features of COVID-19 and their treatment: a review. Drugs Context 2020, 9, 1. 10.7573/dic.2020-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshebri M. S.; Alshouimi R. A.; Alhumidi H. A.; Alshaya A. I. Neurological Complications of SARS-CoV, MERS-CoV, and COVID-19. SN Compr Clin Med. 2020, 2 (11), 2037–2047. 10.1007/s42399-020-00589-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S.; Kolappa K.; Prasad M.; Radhakrishnan D.; Thakur K. T.; Solomon T.; Michael B. D.; Winkler A. S.; Beghi E.; Guekht A.; Pardo C. A.; Wood G. K.; Hsiang-Yi Chou S.; Fink E. L.; Schmutzhard E.; Kheradmand A.; Hoo F. K.; Kumar A.; Das A.; Srivastava A. K.; Agarwal A.; Dua T.; Prasad K. Frequency of Neurologic Manifestations in COVID-19: A Systematic Review and Meta-analysis. Neurology 2021, 97 (23), e2269 10.1212/WNL.0000000000012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; He W.; Yu X.; Hu D.; Bao M.; Liu H.; Zhou J.; Jiang H. Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. J. Infect 2020, 80 (6), 639–645. 10.1016/j.jinf.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi T.; Harii N.; Goto J.; Harada D.; Sugawara H.; Takamino J.; Ueno M.; Sakata H.; Kondo K.; Myose N.; Nakao A.; Takeda M.; Haro H.; Inoue O.; Suzuki-Inoue K.; Kubokawa K.; Ogihara S.; Sasaki T.; Kinouchi H.; Kojin H.; Ito M.; Onishi H.; Shimizu T.; Sasaki Y.; Enomoto N.; Ishihara H.; Furuya S.; Yamamoto T.; Shimada S. A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int. J. Infect Dis 2020, 94, 55–58. 10.1016/j.ijid.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. H.; Jiang D.; Huang J. T. SARS-CoV-2 Detected in Cerebrospinal Fluid by PCR in a Case of COVID-19 Encephalitis. Brain Behav Immun 2020, 87, 149. 10.1016/j.bbi.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano J. B.; Murthy S.; Marshall J. C.; Relan P.; Diaz J. V. A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect Dis 2022, 22 (4), e102 10.1016/S1473-3099(21)00703-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah W.; Hillman T.; Playford E. D.; Hishmeh L. Managing the long term effects of covid-19: summary of NICE, SIGN, and RCGP rapid guideline. BMJ. 2021, 372, n136. 10.1136/bmj.n136. [DOI] [PubMed] [Google Scholar]
- Sellner J. Of mice and men: COVID-19 challenges translational neuroscience. Eur. J. Neurol 2020, 27 (9), 1762–1763. 10.1111/ene.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou S.; Liu M.; Yang Y.; Kang N.; Song Y.; Tan D.; Liu N.; Wang F.; Liu J.; Xie Y. Animal Models for COVID-19: Hamsters, Mouse, Ferret, Mink, Tree Shrew, and Non-human Primates. Front Microbiol 2021, 12, 1. 10.3389/fmicb.2021.626553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhtazad A.; Garmabi B.; Joghataei M. T. Neurological manifestations of coronavirus infections, before and after COVID-19: a review of animal studies. J. Neurovirol 2021, 27 (6), 864–884. 10.1007/s13365-021-01014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesudhas D.; Srivastava A.; Gromiha M. M. COVID-19 outbreak: history, mechanism, transmission, structural studies and therapeutics. Infection 2021, 49 (2), 199–213. 10.1007/s15010-020-01516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Moore M. J.; Vasilieva N.; Sui J.; Wong S. K.; Berne M. A.; Somasundaran M.; Sullivan J. L.; Luzuriaga K.; Greenough T. C.; Choe H.; Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426 (6965), 450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K.; Imai Y.; Rao S.; Gao H.; Guo F.; Guan B.; Huan Y.; Yang P.; Zhang Y.; Deng W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005, 11 (8), 875–879. 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z.; Xu Y.; Bao L.; Zhang L.; Yu P.; Qu Y.; Zhu H.; Zhao W.; Han Y.; Qin C. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses 2019, 11 (1), 59. 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig A. M.; Khaleeq A.; Ali U.; Syeda H. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 2020, 11 (7), 995–998. 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- Lukassen S.; Chua R. L.; Trefzer T.; Kahn N. C.; Schneider M. A.; Muley T.; Winter H.; Meister M.; Veith C.; Boots A. W.; Hennig B. P.; Kreuter M.; Conrad C.; Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO Journal 2020, 39 (10), e105114 10.15252/embj.2020105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N. H.; Nitsche A.; Müller M. A.; Drosten C.; Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181 (2), 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L.; Jin H.; Wang M.; Hu Y.; Chen S.; He Q.; Chang J.; Hong C.; Zhou Y.; Wang D.; Miao X.; Li Y.; Hu B. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol 2020, 77 (6), 683–690. 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F.; Deng L.; Zhang L.; Cai Y.; Cheung C. W.; Xia Z. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19). J. Gen Intern Med. 2020, 35 (5), 1545–1549. 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Shen Y.; Li M.; Chuang H.; Ye Y.; Zhao H.; Wang H. Clinical manifestations and evidence of neurological involvement in 2019 novel coronavirus SARS-CoV-2: a systematic review and meta-analysis. J. Neurol 2020, 267 (10), 2777–2789. 10.1007/s00415-020-09974-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B.; Schmidbauer M. L.; Dimitriadis K.; Otto S.; Knier B.; Niesen W.-D.; Hosp J. A.; Gunther A.; Lindemann S.; Nagy G.; Steinberg T.; Linker R. A.; Hemmer B.; Bosel J. Cerebrospinal fluid findings in COVID-19 patients with neurological symptoms. J. Neurol Sci. 2020, 418, 117090. 10.1016/j.jns.2020.117090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghagoli G.; Gallo Marin B.; Katchur N. J.; Chaves-Sell F.; Asaad W. F.; Murphy S. A. Neurological Involvement in COVID-19 and Potential Mechanisms: A Review. Neurocrit Care 2021, 34 (3), 1062–1071. 10.1007/s12028-020-01049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Morales A. J.; Cardona-Ospina J. A.; Gutiérrez-Ocampo E.; Villamizar-Peña R.; Holguin-Rivera Y.; Escalera-Antezana J. P.; Alvarado-Arnez L. E.; Bonilla-Aldana D. K.; Franco-Paredes C.; Henao-Martinez A. F.; Paniz-Mondolfi A.; Lagos-Grisales G. J.; Ramírez-Vallejo E.; Suárez J. A.; Zambrano L. I.; Villamil-Gómez W. E.; Balbin-Ramon G. J.; Rabaan A. A.; Harapan H.; Dhama K.; Nishiura H.; Kataoka H.; Ahmad T.; Sah R. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med. Infect Dis 2020, 34, 101623. 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toptan T.; Aktan C.; Başarı A.; Bolay H. Case series of headache characteristics in COVID-19: headache can be an isolated symptom. Headache 2020, 60 (8), 1788–1792. 10.1111/head.13940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B. M.; Vikse J.; Benoit S.; Favaloro E. J.; Lippi G. Hyperinflammation and derangement of renin-angiotensin-aldosterone system in COVID-19: A novel hypothesis for clinically suspected hypercoagulopathy and microvascular immunothrombosis. Clin. Chim. Acta 2020, 507, 167–173. 10.1016/j.cca.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acherjee T.; Behara A.; Saad M.; Vittorio T. J. Mechanisms and management of prothrombotic state in COVID-19 disease. Ther Adv. Cardiovasc Dis 2021, 15, 1. 10.1177/17539447211053470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-j.; Ni Z.-y.; Hu Y.; Liang W.-h.; Ou C.-q.; He J.-x.; Liu L.; Shan H.; Lei C.-l.; Hui D. S. C.; Du B.; Li L.-j.; Zeng G.; Yuen K.-Y.; Chen R.-c.; Tang C.-l.; Wang T.; Chen P.-y.; Xiang J.; Li S.-y.; Wang J.-l.; Liang Z.-j.; Peng Y.-x.; Wei L.; Liu Y.; Hu Y.-h.; Peng P.; Wang J.-m.; Liu J.-y.; Chen Z.; Li G.; Zheng Z.-j.; Qiu S.-q.; Luo J.; Ye C.-j.; Zhu S.-y.; Zhong N.-s. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J. Med. 2020, 382 (18), 1708–1720. 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri D.; Ardila A. COVID-19 Pandemic: A Neurological Perspective. Cureus 2020, 12 (4), e7889 10.7759/cureus.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L.; Jin H.; Wang M.; Hu Y.; Chen S.; He Q.; Chang J.; Hong C.; Zhou Y.; Wang D.; Miao X.; Li Y.; Hu B. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol 2020, 77 (6), 683–690. 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepburn M.; Mullaguri N.; George P.; Hantus S.; Punia V.; Bhimraj A.; Newey C. R. Acute symptomatic seizures in critically ill patients with COVID-19: is there an association?. Neurocrit Care 2021, 34 (1), 139–143. 10.1007/s12028-020-01006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbey J. E.; Fujinami R. S. Neurotropic viral infections leading to epilepsy: focus on Theiler’s murine encephalomyelitis virus. Future Virol 2011, 6 (11), 1339–1350. 10.2217/fvl.11.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhi P. Infectious causes of seizures and epilepsy in the developing world. Dev Med. Child Neurol 2011, 53 (7), 600–609. 10.1111/j.1469-8749.2011.03928.x. [DOI] [PubMed] [Google Scholar]
- Poyiadji N.; Shahin G.; Noujaim D.; Stone M.; Patel S. C.; Griffith B. COVID-19-associated acute hemorrhagic necrotizing encephalopathy: imaging features. Radiology 2020, 296 (2), E119. 10.1148/radiol.2020201187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig A. M. Neurological manifestations in COVID-19 caused by SARS-CoV-2. CNS Neurosci Ther 2020, 26 (5), 499. 10.1111/cns.13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAbee G. N.; Brosgol Y.; Pavlakis S.; Agha R.; Gaffoor M. Encephalitis Associated with COVID-19 Infection in an 11-Year-Old Child. Pediatr Neurol 2020, 109, 94. 10.1016/j.pediatrneurol.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechien J. R.; Chiesa-Estomba C. M.; De Siati D. R.; Horoi M.; Le Bon S. D.; Rodriguez A.; Dequanter D.; Blecic S.; El Afia F.; Distinguin L. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur. Arch Otorhinolaryngol 2020, 277 (8), 2251–2261. 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gane S. B.; Kelly C.; Hopkins C. Isolated sudden onset anosmia in COVID-19 infection. A novel syndrome. Rhinology 2020, 58 (3), 299–301. 10.4193/Rhin20.114. [DOI] [PubMed] [Google Scholar]
- Amer M. A.; Elsherif H. S.; Abdel-Hamid A. S.; Elzayat S. Early recovery patterns of olfactory disorders in COVID-19 patients; a clinical cohort study. Am. J. Otolaryngol 2020, 41 (6), 102725. 10.1016/j.amjoto.2020.102725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet M.; Geddes J. R.; Husain M.; Luciano S.; Harrison P. J. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry 2021, 8 (5), 416–427. 10.1016/S2215-0366(21)00084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings M. J; Baldwin M. R; Abrams D.; Jacobson S. D; Meyer B. J; Balough E. M; Aaron J. G; Claassen J.; Rabbani L. E; Hastie J.; Hochman B. R; Salazar-Schicchi J.; Yip N. H; Brodie D.; O'Donnell M. R Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet 2020, 395 (10239), 1763–1770. 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J.; Scorza F. A.; Ghosh R. COVID-19 polyradiculitis in 24 patients without SARS-CoV-2 in the cerebro-spinal fluid. J. Med. Virol 2021, 93 (1), 66. 10.1002/jmv.26121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L.; Druce J. D.; Catton M. G.; Jans D. A.; Wagstaff K. M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020, 178, 104787. 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Cao R.; Xu M.; Wang X.; Zhang H.; Hu H.; Li Y.; Hu Z.; Zhong W.; Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov 2020, 6, 16. 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Cao R.; Zhang L.; Yang X.; Liu J.; Xu M.; Shi Z.; Hu Z.; Zhong W.; Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30 (3), 269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.; Ye F.; Zhang M.; Cui C.; Huang B.; Niu P.; Liu X.; Zhao L.; Dong E.; Song C.; Zhan S.; Lu R.; Li H.; Tan W.; Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis 2020, 71 (15), 732–739. 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runfeng L.; Yunlong H.; Jicheng H.; Weiqi P.; Qinhai M.; Yongxia S.; Chufang L.; Jin Z.; Zhenhua J.; Haiming J.; Kui Z.; Shuxiang H.; Jun D.; Xiaobo L.; Xiaotao H.; Lin W.; Nanshan Z.; Zifeng Y. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol. Res. 2020, 156, 104761. 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N.; Sun Y.; Yang M.; Lu Q.; Wang J.; Xiao C.; Wang Y.; Du L.; Ji K.; Xu C.; Liu Q. Analysis of circular RNA expression profile in HEK 293T cells exposed to ionizing radiation. Dose-Response 2019, 17 (2), 1. 10.1177/1559325819837795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma-Lauer Y.; Carbajo-Lozoya J.; Hein M. Y.; Muller M. A.; Deng W.; Lei J.; Meyer B.; Kusov Y.; von Brunn B.; Bairad D. R.; Hunten S.; Drosten C.; Hermeking H.; Leonhardt H.; Mann M.; Hilgenfeld R.; von Brunn A. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (35), E5192–E5201. 10.1073/pnas.1603435113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Jiang M.; Bi H.; Chen X.; He L.; Li X.; Wu J. Conversion of female germline stem cells from neonatal and prepubertal mice into pluripotent stem cells. J. Mol. Cell Biol. 2014, 6 (2), 164–171. 10.1093/jmcb/mju004. [DOI] [PubMed] [Google Scholar]
- Zeng F.; Huang F.; Guo J.; Hu X.; Liu C.; Wang H. Emerging methods to generate artificial germ cells from stem cells. Biol. Reprod. 2015, 92 (4), 89. 10.1095/biolreprod.114.124800. [DOI] [PubMed] [Google Scholar]
- Guo J.; Wang H.; Hu X. Reprogramming and transdifferentiation shift the landscape of regenerative medicine. DNA Cell Biol. 2013, 32 (10), 565–572. 10.1089/dna.2013.2104. [DOI] [PubMed] [Google Scholar]
- Liu H.; Zeng F.; Zhang M.; Huang F.; Wang J.; Guo J.; Liu C.; Wang H. Emerging landscape of cell penetrating peptide in reprogramming and gene editing. J. Controlled Release 2016, 226, 124–137. 10.1016/j.jconrel.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Geng J.; Xia X.; Teng L.; Wang L.; Chen L.; Guo X.; Belingon B.; Li J.; Feng X.; Li X.; Shang W.; Wan Y.; Wang H. Emerging landscape of cell-penetrating peptide-mediated nucleic acid delivery and their utility in imaging, gene-editing, and RNA-sequencing. J. Controlled Release 2022, 341, 166–183. 10.1016/j.jconrel.2021.11.032. [DOI] [PubMed] [Google Scholar]
- Deng X. Y.; Wang H.; Wang T.; Fang X. T.; Zou L. L.; Li Z. Y.; Liu C. B. Non-viral methods for generating integration-free, induced pluripotent stem cells. Curr. Stem Cell Res. Ther 2015, 10 (2), 153–158. 10.2174/1574888X09666140923101914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M.; Acevedo-Cintrón J.; Jhaldiyal A.; Wang H.; Andrabi S. A.; Eacker S.; Karuppagounder S. S.; Brahmachari S.; Chen R.; Kim H.; Ko H. S.; Dawson V. L.; Dawson T. M. Defects in Mitochondrial Biogenesis Drive Mitochondrial Alterations in PARKIN-Deficient Human Dopamine Neurons. Stem Cell Reports 2020, 15 (3), 629–645. 10.1016/j.stemcr.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel A. E.; Chintawar S.; Lalic T.; Whiteley E.; Vowles J.; Giustacchini A.; Argoud K.; Sopp P.; Nakanishi M.; Bowden R.; Cowley S.; Newey S.; Akerman C.; Ponting C. P.; Cader M. Z. Assessing similarity to primary tissue and cortical layer identity in induced pluripotent stem cell-derived cortical neurons through single-cell transcriptomics. Hum. Mol. Genet. 2016, 25 (5), 989–1000. 10.1093/hmg/ddv637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F.; Pather S. R.; Huang W. K.; Zhang F.; Wong S. Z. H.; Zhou H.; Cubitt B.; Fan W.; Chen C. Z.; Xu M.; Pradhan M.; Zhang D. Y.; Zheng W.; Bang A. G.; Song H.; Carlos de la Torre J.; Ming G. L. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27 (6), 937–950. 10.1016/j.stem.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Han Y.; Nilsson-Payant B. E.; Gupta V.; Wang P.; Duan X.; Tang X.; Zhu J.; Zhao Z.; Jaffré F. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell 2020, 27 (1), 125–136. 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.-Z.; Chu H.; Han S.; Shuai H.; Deng J.; Hu Y.-f.; Gong H.-r.; Lee A. C.-Y.; Zou Z.; Yau T.; Wu W.; Hung I. F.-N.; Chan J. F.-W.; Yuen K.-Y.; Huang J.-D. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Research 2020, 30 (10), 928–931. 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S. K.; Wang S.; Smith D.; Carlin A. F.; Rana T. M. Revealing tissue-specific SARS-CoV-2 infection and host responses using human stem cell-derived lung and cerebral organoids. Stem Cell Reports 2021, 16 (3), 437–445. 10.1016/j.stemcr.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Bermejo J. A.; Kang S.; Rockwood S. J.; Simoneau C. R.; Joy D. A.; Silva A. C.; Ramadoss G. N.; Flanigan W. R.; Fozouni P.; Li H.; Chen P.-Y.; Nakamura K.; Whitman J. D.; Hanson P. J.; McManus B. M.; Ott M.; Conklin B. R.; McDevitt T. C. SARS-CoV-2 infection of human iPSC-derived cardiac cells reflects cytopathic features in hearts of patients with COVID-19. Sci. Transl Med. 2021, 13 (590), eabf7872 10.1126/scitranslmed.abf7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y. W.; Hysolli E.; Kim K. Y.; Tanaka Y.; Park I. H. Human induced pluripotent stem cells and neurodegenerative disease: prospects for novel therapies. Curr. Opin Neurol 2012, 25 (2), 125–130. 10.1097/WCO.0b013e3283518226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G.; Chiuppesi F.; Chen X.; Wang C.; Tian E.; Nguyen J.; Kha M.; Trinh D.; Zhang H.; Marchetto M. C.; Song H.; Ming G.-L.; Gage F. H.; Diamond D. J.; Wussow F.; Shi Y. Modeling Human Cytomegalovirus-Induced Microcephaly in Human iPSC-Derived Brain Organoids. Cell Rep. Med. 2020, 1 (1), 100002. 10.1016/j.xcrm.2020.100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. M.; Rana P. S. J. B.; Jaeger H. K.; O’Dowd J. M.; Balemba O. B.; Fortunato E. A. Human Cytomegalovirus Compromises Development of Cerebral Organoids. J. Virol 2019, 93 (17), 1. 10.1128/JVI.00957-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang J.; Tiwari S. K.; Lichinchi G.; Qin Y.; Patil V. S.; Eroshkin A. M.; Rana T. M. Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell 2016, 19 (2), 258–265. 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X.; Nguyen H. N.; Jacob F.; Song H.; Ming G.-l. Using brain organoids to understand Zika virus-induced microcephaly. Development 2017, 144 (6), 952–957. 10.1242/dev.140707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M. M.; Beumer J.; Van Der Vaart J.; Knoops K.; Puschhof J.; Breugem T. I.; Ravelli R. B.; Paul van Schayck J.; Mykytyn A. Z.; Duimel H. Q. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369 (6499), 50–54. 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V.; Kwon H.; Prado P.; Hagelkruys A.; Wimmer R. A.; Stahl M.; Leopoldi A.; Garreta E.; Hurtado del Pozo C.; Prosper F.; Romero J. P.; Wirnsberger G.; Zhang H.; Slutsky A. S.; Conder R.; Montserrat N.; Mirazimi A.; Penninger J. M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 2020, 181 (4), 905–913. 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A.; Müller L.; Ostermann P. N.; Gabriel E.; Abida-Islam P.; Müller-Schiffmann A.; Mariappan A.; Goureau O.; Gruell H.; Walker A. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020, 39 (20), e106230 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Li C.; Liu X.; Chiu M. C.; Zhao X.; Wang D.; Wei Y.; Lee A.; Zhang A. J.; Chu H. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020, 26 (7), 1077–1083. 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- Pellegrini L.; Albecka A.; Mallery D. L.; Kellner M. J.; Paul D.; Carter A. P.; James L. C.; Lancaster M. A. SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell 2020, 27 (6), 951–961. 10.1016/j.stem.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Boutros P. C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 2011, 12 (1), 1–7. 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E.; Zhang C.; Israelow B.; Lu-Culligan A.; Prado A. V.; Skriabine S.; Lu P.; Weizman O.-E.; Liu F.; Dai Y. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp Med. 2021, 218 (3), e20202135 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J.; Hughes C. C. Of mice and not men: differences between mouse and human immunology. J. Immunol 2004, 172 (5), 2731–2738. 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Wan Y.; Shang J.; Graham R.; Baric R. S.; Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol 2020, 94 (7), 1 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C.; Maher L.; Lee C.; Kang W. COVID-19 preclinical models: human angiotensin-converting enzyme 2 transgenic mice. Hum Genomics 2020, 14 (1), 20. 10.1186/s40246-020-00272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L.; Deng W.; Huang B.; Gao H.; Liu J.; Ren L.; Wei Q.; Yu P.; Xu Y.; Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583 (7818), 830–833. 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Jiang R.-D.; Liu M.-Q.; Chen Y.; Shan C.; Zhou Y.-W.; Shen X.-R.; Li Q.; Zhang L.; Zhu Y.; Si H.-R.; Wang Q.; Min J.; Wang X.; Zhang W.; Li B.; Zhang H.-J.; Baric R. S.; Zhou P.; Yang X.-L.; Shi Z.-L. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 2020, 182 (1), 50–58. 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P. B.; Pewe L.; Wohlford-Lenane C.; Hickey M.; Manzel L.; Shi L.; Netland J.; Jia H. P.; Halabi C.; Sigmund C. D.; Meyerholz D. K.; Kirby P.; Look D. C.; Perlman S. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol 2007, 81 (2), 813–821. 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P. B. Jr; Pewe L.; Wohlford-Lenane C.; Hickey M.; Manzel L.; Shi L.; Netland J.; Jia H. P.; Halabi C.; Sigmund C. D.; Meyerholz D. K.; Kirby P.; Look D. C.; Perlman S. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol 2007, 81 (2), 813–821. 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau G. B.; Burgess S. L.; Sturek J. M.; Donlan A. N.; Petri W. A.; Mann B. J. Evaluation of K18-hACE2Mice as a Model of SARS-CoV-2 Infection. Am. J. Trop Med. Hyg 2020, 103 (3), 1215–1219. 10.4269/ajtmh.20-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R. D.; Liu M. Q.; Chen Y.; Shan C.; Zhou Y. W.; Shen X. R.; Li Q.; Zhang L.; Zhu Y.; Si H. R.; Wang Q.; Min J.; Wang X.; Zhang W.; Li B.; Zhang H. J.; Baric R. S.; Zhou P.; Yang X. L.; Shi Z. L. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell 2020, 182 (1), 50–58. 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K. H.; Leist S. R.; Schäfer A.; Edwards C. E.; Martinez D. R.; Montgomery S. A.; West A.; Yount B. L.; Hou Y. J.; Adams L. E.; Gully K. L.; Brown A. J.; Huang E.; Bryant M. D.; Choong I. C.; Glenn J. S.; Gralinski L. E.; Sheahan T. P.; Baric R. S. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 2020, 586 (7830), 560–566. 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. Y.; Anand-Jawa V.; Chatterjee S.; Wong K. K. Immune responses to adeno-associated virus and its recombinant vectors. Gene Ther. 2003, 10 (11), 964–976. 10.1038/sj.gt.3302039. [DOI] [PubMed] [Google Scholar]
- Liu F.; Wu K.; Sun J.; Duan Z.; Quan X.; Kuang J.; Chu S.; Pang W.; Gao H.; Xu L.; Li Y.; Zhang H.; Wang X.; Luo R.; Feng X.; Schöler H. R.; Chen X.; Pei D.; Wu G.; Zheng Y.; Chen J. Rapid generation of ACE2 humanized inbred mouse model for COVID-19 with tetraploid complementation. Natl. Sci. Rev. 2021, 8 (2), 1. 10.1093/nsr/nwaa285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. H.; Chen Q.; Gu H. J.; Yang G.; Wang Y. X.; Huang X. Y.; Liu S. S.; Zhang N. N.; Li X. F.; Xiong R.; Guo Y.; Deng Y. Q.; Huang W. J.; Liu Q.; Liu Q. M.; Shen Y. L.; Zhou Y.; Yang X.; Zhao T. Y.; Fan C. F.; Zhou Y. S.; Qin C. F.; Wang Y. C. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe 2020, 28 (1), 124–133. 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.-T. K.; Huang C.; Newman P.; Wang N.; Narayanan K.; Watts D. M.; Makino S.; Packard M. M.; Zaki S. R.; Chan T.-s.; Peters C. J. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J. Virol 2007, 81 (3), 1162–1173. 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J.; Kremer S.; Merdji H.; Clere-Jehl R.; Schenck M.; Kummerlen C.; Collange O.; Boulay C.; Fafi-Kremer S.; Ohana M.; Anheim M.; Meziani F. Neurologic features in severe SARS-CoV-2 infection. N Engl J. Med. 2020, 382 (23), 2268–2270. 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K. W.; Brann D. H.; Farruggia M. C.; Bhutani S.; Pellegrino R.; Tsukahara T.; Weinreb C.; Joseph P. V.; Larson E. D.; Parma V.; Albers M. W.; Barlow L. A.; Datta S. R.; Di Pizio A. COVID-19 and the chemical senses: supporting players take center stage. Neuron 2020, 107 (2), 219–233. 10.1016/j.neuron.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.; Wong L.-Y. R.; Li K.; Verma A. K.; Ortiz M. E.; Wohlford-Lenane C.; Leidinger M. R.; Knudson C. M.; Meyerholz D. K.; McCray P. B.; Perlman S. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 2021, 589 (7843), 603–607. 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. Scientists set out to connect the dots on long COVID. Nat. Methods 2021, 18 (5), 449–453. 10.1038/s41592-021-01145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen E. B.; Orvold S. N.; Swan C. L.; Yourkowski A.; Thivierge B. M.; Francis M. E.; Ge A.; Rioux M.; Darbellay J.; Howland J. G.; Kelvin A. A. After the virus has cleared—Can preclinical models be employed for Long COVID research?. PLoS Pathog 2022, 18 (9), e1010741 10.1371/journal.ppat.1010741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J.; Chard L. S.; Wang Z.; Wang Y. Syrian hamster as an animal model for the study on infectious diseases. Front Immunol 2019, 10, 2329. 10.3389/fimmu.2019.02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butowt R.; Meunier N.; Bryche B.; von Bartheld C. S. The olfactory nerve is not a likely route to brain infection in COVID-19: a critical review of data from humans and animal models. Acta Neuropathol 2021, 141 (6), 809–822. 10.1007/s00401-021-02314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M.; Iwatsuki-Horimoto K.; Hatta M.; Loeber S.; Halfmann P. J.; Nakajima N.; Watanabe T.; Ujie M.; Takahashi K.; Ito M. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (28), 16587–16595. 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D. A.; Møller R.; Uhl S. A.; Oishi K.; Frere J.; Golynker I.; Horiuchi S.; Panis M.; Blanco-Melo D.; Sachs D.; Arkun K.; Lim J. K.; tenOever B. R. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity 2021, 54 (3), 557–570. 10.1016/j.immuni.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo G. D.; Lazarini F.; Levallois S.; Hautefort C.; Michel V.; Larrous F.; Verillaud B.; Aparicio C.; Wagner S.; Gheusi G.; Kergoat L.; Kornobis E.; Donati F.; Cokelaer T.; Hervochon R.; Madec Y.; Roze E.; Salmon D.; Bourhy H.; Lecuit M.; Lledo P. M. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl Med. 2021, 13 (596), 1 10.1126/scitranslmed.abf8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F.-W.; Zhang A. J.; Yuan S.; Poon V. K.-M.; Chan C. C.-S.; Lee A. C.-Y.; Chan W.-M.; Fan Z.; Tsoi H.-W.; Wen L. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis 2020, 71 (9), 2428–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S. F.; Yan L.-M.; Chin A. W. H.; Fung K.; Choy K.-T.; Wong A. Y. L.; Kaewpreedee P.; Perera R. A. P. M.; Poon L. L. M.; Nicholls J. M.; Peiris M.; Yen H.-L. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583 (7818), 834–838. 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y.-K.; Ali G. D.; Jia F.; Li Q.; Kelvin D.; Couch R. C.; Harrod K. S.; Hutt J. A.; Cameron C.; Weiss S. R.; Jonsson C. B. The SARS-CoV ferret model in an infection-challenge study. Virology 2008, 374 (1), 151–163. 10.1016/j.virol.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Shi J.; Deng G.; Guo J.; Zeng X.; He X.; Kong H.; Gu C.; Li X.; Liu J. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 2013, 341 (6144), 410–414. 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- Kim Y.-I.; Kim S.-G.; Kim S.-M.; Kim E.-H.; Park S.-J.; Yu K.-M.; Chang J.-H.; Kim E. J.; Lee S.; Casel M. A. B. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host & Microbe 2020, 27 (5), 704–709. 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala S. S.; Menachery V. D. The search for a COVID-19 animal model. Science 2020, 368 (6494), 942–943. 10.1126/science.abc6141. [DOI] [PubMed] [Google Scholar]
- Shi J.; Wen Z.; Zhong G.; Yang H.; Wang C.; Huang B.; Liu R.; He X.; Shuai L.; Sun Z.; Zhao Y.; Liu P.; Liang L.; Cui P.; Wang J.; Zhang X.; Guan Y.; Tan W.; Wu G.; Chen H.; Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368 (6494), 1016–1020. 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs E. C.; Reid T. J. Animals and SARS-CoV-2: Species susceptibility and viral transmission in experimental and natural conditions, and the potential implications for community transmission. Transbound Emerg Dis 2021, 68 (4), 1850–1867. 10.1111/tbed.13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P.; Qi F.; Xu Y.; Li F.; Liu P.; Liu J.; Bao L.; Deng W.; Gao H.; Xiang Z. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med. 2020, 3 (1), 93–97. 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D. K.; Singh B.; Ganatra S. R.; Gazi M.; Cole J.; Thippeshappa R.; Alfson K. J.; Clemmons E.; Gonzalez O.; Escobedo R. Responses to acute infection with SARS-CoV-2 in the lungs of rhesus macaques, baboons and marmosets. Nat. Microbiol 2021, 6 (1), 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.; Zhao Y.; Yu W.; Yang Y.; Gao J.; Wang J.; Kuang D.; Yang M.; Yang J.; Ma C. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct Target Ther 2020, 5 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L.; Deng W.; Gao H.; Xiao C.; Liu J.; Xue J.; Lv Q.; Liu J.; Yu P.; Xu Y.. Lack of reinfection in rhesus macaques infected with SARS-CoV-2. BioRxiv, 2020, 10.1101/2020.03.13.990226. [DOI] [Google Scholar]
- Rockx B.; Kuiken T.; Herfst S.; Bestebroer T.; Lamers M. M.; Oude Munnink B. B.; De Meulder D.; Van Amerongen G.; Van Den Brand J.; Okba N. M. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020, 368 (6494), 1012–1015. 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey C.; Borisevich V.; Prasad A. N.; Agans K. N.; Deer D. J.; Dobias N. S.; Heymann J. C.; Foster S. L.; Levine C. B.; Medina L.; Melody K.; Geisbert J. B.; Fenton K. A.; Geisbert T. W.; Cross R. W. Establishment of an African green monkey model for COVID-19 and protection against re-infection. Nat. Immunol 2021, 22 (1), 86–98. 10.1038/s41590-020-00835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V. J.; Feldmann F.; Williamson B. N.; van Doremalen N.; Perez-Perez L.; Schulz J.; Meade-White K.; Okumura A.; Callison J.; Brumbaugh B.; Avanzato V. A.; Rosenke R.; Hanley P. W.; Saturday G.; Scott D.; Fischer E. R.; de Wit E. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020, 585 (7824), 268–272. 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkai I.; Mayer M. G.; Hellmers L. M.; Ning B.; Huang Z.; Monjure C. J.; Coyne C.; Silvestri R.; Golden N.; Hensley K.; Chandler K.; Lehmicke G.; Bix G. J.; Maness N. J.; Russell-Lodrigue K.; Hu T. Y.; Roy C. J.; Blair R. V.; Bohm R.; Doyle-Meyers L. A.; Rappaport J.; Fischer T. Neuropathology and virus in brain of SARS-CoV-2 infected non-human primates. Nat. Commun. 2022, 13 (1), 1–13. 10.1038/s41467-022-29440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L.; Yang Y.; Yu W.; Zhao Y.; Long H.; Gao J.; Ding K.; Ma C.; Li J.; Zhao S. The olfactory route is a potential way for SARS-CoV-2 to invade the central nervous system of rhesus monkeys. Signal Transduct Target Ther 2021, 6 (1), 1–11. 10.1038/s41392-021-00591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F. S.; Stoffels P. Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV): An Unprecedented Partnership for Unprecedented Times. JAMA 2020, 323, 2455–2457. 10.1001/jama.2020.8920. [DOI] [PubMed] [Google Scholar]
- Vogel A. B.; Kanevsky I.; Che Y.; Swanson K. A.; Muik A.; Vormehr M.; Kranz L. M.; Walzer K. C.; Hein S.; Güler A. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 2021, 592 (7853), 283–289. 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- Corbett K. S.; Edwards D. K.; Leist S. R.; Abiona O. M.; Boyoglu-Barnum S.; Gillespie R. A.; Himansu S.; Schäfer A.; Ziwawo C. T.; DiPiazza A. T. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586 (7830), 567–571. 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salguero F. J.; White A. D.; Slack G. S.; Fotheringham S. A.; Bewley K. R.; Gooch K. E.; Longet S.; Humphries H. E.; Watson R. J.; Hunter L.; Ryan K. A.; Hall Y.; Sibley L.; Sarfas C.; Allen L.; Aram M.; Brunt E.; Brown P.; Buttigieg K. R.; Cavell B. E.; Cobb R.; Coombes N. S.; Darby A.; Daykin-Pont O.; Elmore M. J.; Garcia-Dorival I.; Gkolfinos K.; Godwin K. J.; Gouriet J.; Halkerston R.; Harris D. J.; Hender T.; Ho C. M. K.; Kennard C. L.; Knott D.; Leung S.; Lucas V.; Mabbutt A.; Morrison A. L.; Nelson C.; Ngabo D.; Paterson J.; Penn E. J.; Pullan S.; Taylor I.; Tipton T.; Thomas S.; Tree J. A.; Turner C.; Vamos E.; Wand N.; Wiblin N. R.; Charlton S.; Dong X.; Hallis B.; Pearson G.; Rayner E. L.; Nicholson A. G.; Funnell S. G.; Hiscox J. A.; Dennis M. J.; Gleeson F. V.; Sharpe S.; Carroll M. W. Comparison of rhesus and cynomolgus macaques as an infection model for COVID-19. Nat. Commun. 2021, 12 (1), 1260. 10.1038/s41467-021-21389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostanoski L. H.; Wegmann F.; Martinot A. J.; Loos C.; McMahan K.; Mercado N. B.; Yu J.; Chan C. N.; Bondoc S.; Starke C. E. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020, 26 (11), 1694–1700. 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostanoski L. H.; Wegmann F.; Martinot A. J.; Loos C.; McMahan K.; Mercado N. B.; Yu J.; Chan C. N.; Bondoc S.; Starke C. E.; Nekorchuk M.; Busman-Sahay K.; Piedra-Mora C.; Wrijil L. M.; Ducat S.; Custers J.; Atyeo C.; Fischinger S.; Burke J. S.; Feldman J.; Hauser B. M.; Caradonna T. M.; Bondzie E. A.; Dagotto G.; Gebre M. S.; Jacob-Dolan C.; Lin Z.; Mahrokhian S. H.; Nampanya F.; Nityanandam R.; Pessaint L.; Porto M.; Ali V.; Benetiene D.; Tevi K.; Andersen H.; Lewis M. G.; Schmidt A. G.; Lauffenburger D. A.; Alter G.; Estes J. D.; Schuitemaker H.; Zahn R.; Barouch D. H. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020, 26 (11), 1694–1700. 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F. P.; Thomas S. J.; Kitchin N.; Absalon J.; Gurtman A.; Lockhart S.; Perez J. L.; Marc G. P.; Moreira E. D.; Zerbini C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J. Med. 2020, 383, 2603–2615. 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R. K.; Paliwal V. K. Spectrum of neurological complications following COVID-19 vaccination. Neurol Sci. 2022, 43 (1), 3–40. 10.1007/s10072-021-05662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M.; Clemens S. A. C.; Madhi S. A.; Weckx L. Y.; Folegatti P. M.; Aley P. K.; Angus B.; Baillie V. L.; Barnabas S. L.; Bhorat Q. E. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397 (10269), 99–111. 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. US authorization of first COVID vaccine marks new phase in safety monitoring. Nature 2020, 588 (7838), 377–379. 10.1038/d41586-020-03542-4. [DOI] [PubMed] [Google Scholar]
- Gottlieb R. L.; Vaca C. E.; Paredes R.; Mera J.; Webb B. J.; Perez G.; Oguchi G.; Ryan P.; Nielsen B. U.; Brown M.; Hidalgo A.; Sachdeva Y.; Mittal S.; Osiyemi O.; Skarbinski J.; Juneja K.; Hyland R. H.; Osinusi A.; Chen S.; Camus G.; Abdelghany M.; Davies S.; Behenna-Renton N.; Duff F.; Marty F. M.; Katz M. J.; Ginde A. A.; Brown S. M.; Schiffer J. T.; Hill J. A. Early Remdesivir to Prevent Progression to Severe Covid-19 in Outpatients. N Engl J. Med. 2022, 386 (4), 305–315. 10.1056/NEJMoa2116846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibaldi B. T.; Wang K.; Robinson M. L.; Zeger S. L.; Bandeen-Roche K.; Wang M.-C.; Alexander G. C.; Gupta A.; Bollinger R.; Xu Y. Comparison of Time to Clinical Improvement With vs Without Remdesivir Treatment in Hospitalized Patients With COVID-19. JAMA Network Open 2021, 4 (3), e213071 10.1001/jamanetworkopen.2021.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D. J.; Gebo K. A.; Shoham S.; Bloch E. M.; Lau B.; Shenoy A. G.; Mosnaim G. S.; Gniadek T. J.; Fukuta Y.; Patel B.; Heath S. L.; Levine A. C.; Meisenberg B. R.; Spivak E. S.; Anjan S.; Huaman M. A.; Blair J. E.; Currier J. S.; Paxton J. H.; Gerber J. M.; Petrini J. R.; Broderick P. B.; Rausch W.; Cordisco M.-E.; Hammel J.; Greenblatt B.; Cluzet V. C.; Cruser D.; Oei K.; Abinante M.; Hammitt L. L.; Sutcliffe C. G.; Forthal D. N.; Zand M. S.; Cachay E. R.; Raval J. S.; Kassaye S. G.; Foster E. C.; Roth M.; Marshall C. E.; Yarava A.; Lane K.; McBee N. A.; Gawad A. L.; Karlen N.; Singh A.; Ford D. E.; Jabs D. A.; Appel L. J.; Shade D. M.; Ehrhardt S.; Baksh S. N.; Laeyendecker O.; Pekosz A.; Klein S. L.; Casadevall A.; Tobian A. A. R.; Hanley D. F. Early Outpatient Treatment for Covid-19 with Convalescent Plasma. N Engl J. Med. 2022, 386 (18), 1700–1711. 10.1056/NEJMoa2119657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA: Coronavirus (COVID-19) | Drugs. https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs (accessed August 6, 2023).
- Taquet M.; Geddes J. R.; Husain M.; Luciano S.; Harrison P. J. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry 2021, 8 (5), 416–427. 10.1016/S2215-0366(21)00084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann A.; Wu C.-H.; Hardwick M.; Baillie J. K.; Openshaw P. J. M.; Semple M. G.; Böhning D.; Pett S.; Michael B. D.; Thomas R. H.; Galea I. investigators, f. t. I. C., Fewer COVID-19 Neurological Complications with Dexamethasone and Remdesivir. Ann. Neurol. 2023, 93 (1), 88–102. 10.1002/ana.26536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin R. Baricitinib Is First Approved COVID-19 Immunomodulatory Treatment. JAMA 2022, 327 (23), 2281. 10.1001/jama.2022.9846. [DOI] [PubMed] [Google Scholar]
- Wang C.; Zhang M.; Garcia Jr G.; Tian E.; Cui Q.; Chen X.; Sun G.; Wang J.; Arumugaswami V.; Shi Y. ApoE-isoform-dependent SARS-CoV-2 neurotropism and cellular response. Cell Stem Cell 2021, 28 (2), 331–342. 10.1016/j.stem.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-H.; Liu J.-F.; Zhang Y.-L.; Dong Z. Systematic review of efficacy and safety of Lianhua Qingwen Capsules in treatment of viral influenza. Zhongguo Zhong Yao Za Zhi 2019, 44 (7), 1503–1508. 10.19540/j.cnki.cjcmm.20190102.001. [DOI] [PubMed] [Google Scholar]
- Ding Y.; Zeng L.; Li R.; Chen Q.; Zhou B.; Chen Q.; Cheng P. L.; Yutao W.; Zheng J.; Yang Z.; Zhang F. The Chinese prescription lianhuaqingwen capsule exerts anti-influenza activity through the inhibition of viral propagation and impacts immune function. BMC Complement Altern Med. 2017, 17 (1), 130. 10.1186/s12906-017-1585-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P.; Li Y.; Wan S.; Wang Y.. Observation of therapeutic effect of Lianhua Qingwen granule combined with abidor on mild new coronavirus pneumonia. Chin J. Pharm. 2020, 55, 1042−1045. [Google Scholar]
- Hu K.; Guan W.-j.; Bi Y.; Zhang W.; Li L.; Zhang B.; Liu Q.; Song Y.; Li X.; Duan Z. Efficacy and safety of Lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus disease 2019: a multicenter, prospective, randomized controlled trial. Phytomedicine 2021, 85, 153242. 10.1016/j.phymed.2020.153242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Wang H.; Chen H.; Yue Y.; Bu F.; Zhang X. Lianhua Qingwen capsule and interferon-α combined with lopinavir/ritonavir for the treatment of 30 COVID-19 patients. J. Bengbu Med. Coll 2020, 45 (2), 154–155. [Google Scholar]
- Chen D.; Wang W.; Li Y.; Wu X.; Zhou B.; Song Q. Cases of COVID-19 patients with Chinese medicine Lianhuaqingwen curative effect analysis: a multi-center retrospective study. Tianjin Journal of Tradition Chinese Medicine 2020, 37, 509–516. [Google Scholar]
- Yao K.-t.; Liu M.-y.; Li X.; Huang J.-h.; Cai H.-b. Retrospective clinical analysis on treatment of coronavirus disease 2019 with traditional Chinese medicine Lianhua Qingwen. Chinese Journal of Experimental Traditional Medical Formulae 2020, 24, 8–12. [Google Scholar]
- Zhuang J.; Dai X.; Wu Q.; Cai H.; Fu X.; Zhang W.; Chen B. A meta-analysis for Lianhua Qingwen on the treatment of Coronavirus disease 2019 (COVID-19). Complement Ther Med. 2021, 60, 102754. 10.1016/j.ctim.2021.102754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Gao Y.; Yuan Y.; Yang K.; Shi S.; Tian J.; Zhang J. Efficacy and safety of herbal medicine (Lianhuaqingwen) for treating COVID-19: A systematic review and meta-analysis. Integr Med. Res. 2021, 10 (1), 100644. 10.1016/j.imr.2020.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.; Sarker M. M. R.; Afrin S.; Richi F. T.; Zhao C.; Zhou J.-R.; Mohamed I. N. Traditional Herbal Medicines, Bioactive Metabolites, and Plant Products Against COVID-19: Update on Clinical Trials and Mechanism of Actions. Front Pharmacol 2021, 12, Review. 10.3389/fphar.2021.671498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay M.; Thomas D.; Craighead J.; Economides C.; Rosenthal J. Clinical Development Success Rates 2006–2015. Nat. Biotechnol. 2014, 32, 40–51. 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- Wang L.; Sievert D.; Clark A. E.; Lee S.; Federman H.; Gastfriend B. D.; Shusta E. V.; Palecek S. P.; Carlin A. F.; Gleeson J. G. A human three-dimensional neural-perivascular ’assembloid’ promotes astrocytic development and enables modeling of SARS-CoV-2 neuropathology. Nat. Med. 2021, 27 (9), 1600–1606. 10.1038/s41591-021-01443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D.; Lei W.; Hu S. Cardiac organoid—A promising perspective of preclinical model. Stem Cell Res. Ther 2021, 12 (1), 1–10. 10.1186/s13287-021-02340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena M.; Ning L.; King A.; Hwang B.; Jin L.; Serpooshan V.; Sloan S. A. 3D bioprinting of neural tissues. Adv. Healthc Mater. 2021, 10 (15), 2001600. 10.1002/adhm.202001600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S. I.; Sei Y. J.; Park H.-J.; Kim J.; Ryu Y.; Choi J. J.; Sung H.-J.; MacDonald T. J.; Levey A. I.; Kim Y. Microengineered human blood-brain barrier platform for understanding nanoparticle transport mechanisms. Nat. Commun. 2020, 11 (1), 1–12. 10.1038/s41467-019-13896-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A.; De C.; Abad Fernandez M.; Lenarcic E. M.; Xu Y.; Cockrell A. S.; Cleary R. A.; Johnson C. E.; Schramm N. J.; Rank L. M.; Newsome I. G.; Vincent H. A.; Sanders W.; Aguilera-Sandoval C. R.; Boone A.; Hildebrand W. H.; Dayton P. A.; Baric R. S.; Pickles R. J.; Braunstein M.; Moorman N. J.; Goonetilleke N.; Victor Garcia J. Precision mouse models with expanded tropism for human pathogens. Nat. Biotechnol. 2019, 37 (10), 1163–1173. 10.1038/s41587-019-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunney J. K.; Van Goor A.; Walker K. E.; Hailstock T.; Franklin J.; Dai C. Importance of the pig as a human biomedical model. Sci. Transl Med. 2021, 13 (621), 1 10.1126/scitranslmed.abd5758. [DOI] [PubMed] [Google Scholar]
- Montgomery R. A.; Stern J. M.; Lonze B. E.; Tatapudi V. S.; Mangiola M.; Wu M.; Weldon E.; Lawson N.; Deterville C.; Dieter R. A. Results of two cases of pig-to-human kidney xenotransplantation. N Engl J. Med. 2022, 386 (20), 1889–1898. 10.1056/NEJMoa2120238. [DOI] [PubMed] [Google Scholar]
- Reardon S. First pig-to-human heart transplant: what can scientists learn?. Nature 2022, 601 (7893), 305–306. 10.1038/d41586-022-00111-9. [DOI] [PubMed] [Google Scholar]
- Mohs R. C.; Greig N. H. Drug discovery and development: Role of basic biological research. Alzheimers Dement (N Y) 2017, 3 (4), 651–657. 10.1016/j.trci.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N. B.; Krieger K.; Khan F. M.; Huffman W.; Chang M.; Naik A.; Yongle R.; Hameed I.; Krieger K.; Girardi L. N.; Gaudino M. The current state of animal models in research: A review. Int. J. Surg 2019, 72, 9–13. 10.1016/j.ijsu.2019.10.015. [DOI] [PubMed] [Google Scholar]
- Vatansever S.; Schlessinger A.; Wacker D.; Kaniskan H. Ü.; Jin J.; Zhou M. M.; Zhang B. Artificial intelligence and machine learning-aided drug discovery in central nervous system diseases: state-of-the-arts and future directions. Med. Res. Rev. 2021, 41 (3), 1427–1473. 10.1002/med.21764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S.; Zhang D.; Wang H.; An Q.; Yu G.; Han J.; Jiang C.; Huang J. Editorial: Computational and systematic analysis of multi-omics data for drug discovery and development. Front Med. (Lausanne) 2023, 10, 1146896. 10.3389/fmed.2023.1146896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes J. M.; Yang K.; Swanson K.; Jin W.; Cubillos-Ruiz A.; Donghia N. M.; MacNair C. R.; French S.; Carfrae L. A.; Bloom-Ackermann Z.; Tran V. M.; Chiappino-Pepe A.; Badran A. H.; Andrews I. W.; Chory E. J.; Church G. M.; Brown E. D.; Jaakkola T. S.; Barzilay R.; Collins J. J. A deep learning approach to antibiotic discovery. Cell 2020, 180 (4), 688–702. 10.1016/j.cell.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Wang J.; Geng J.; Xiao L.; Wang H. Emerging Landscape of SARS-CoV-2 Variants and Detection Technologies. Mol. Diagn Ther 2023, 27 (2), 159–177. 10.1007/s40291-022-00631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B.; Yi X.; Sun Y.; Bi X.; Du J.; Zhang C.; Quan S.; Zhang F.; Sun R.; Qian L.; Ge W.; Liu W.; Liang S.; Chen H.; Zhang Y.; Li J.; Xu J.; He Z.; Chen B.; Wang J.; Yan H.; Zheng Y.; Wang D.; Zhu J.; Kong Z.; Kang Z.; Liang X.; Ding X.; Ruan G.; Xiang N.; Cai X.; Gao H.; Li L.; Li S.; Xiao Q.; Lu T.; Zhu Y.; Liu H.; Chen H.; Guo T. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182 (1), 59–72. 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X.; Qian L.; Sun R.; Huang B.; Dong X.; Xiao Q.; Zhang Q.; Lu T.; Yue L.; Chen S.; Li X.; Sun Y.; Li L.; Xu L.; Li Y.; Yang M.; Xue Z.; Liang S.; Ding X.; Yuan C.; Peng L.; Liu W.; Yi X.; Lyu M.; Xiao G.; Xu X.; Ge W.; He J.; Fan J.; Wu J.; Luo M.; Chang X.; Pan H.; Cai X.; Zhou J.; Yu J.; Gao H.; Xie M.; Wang S.; Ruan G.; Chen H.; Su H.; Mei H.; Luo D.; Zhao D.; Xu F.; Li Y.; Zhu Y.; Xia J.; Hu Y.; Guo T. Multi-organ proteomic landscape of COVID-19 autopsies. Cell 2021, 184 (3), 775–791. 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migaud M.; Gandotra S.; Chand H. S.; Gillespie M. N.; Thannickal V. J.; Langley R. J. Metabolomics to Predict Antiviral Drug Efficacy in COVID-19. Am. J. Respir. Cell Mol. Biol. 2020, 63 (3), 396–398. 10.1165/rcmb.2020-0206LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danlos F.-X.; Grajeda-Iglesias C.; Durand S.; Sauvat A.; Roumier M.; Cantin D.; Colomba E.; Rohmer J.; Pommeret F.; Baciarello G.; Willekens C.; Vasse M.; Griscelli F.; Fahrner J.-E.; Goubet A.-G.; Dubuisson A.; Derosa L.; Nirmalathasan N.; Bredel D.; Mouraud S.; Pradon C.; Stoclin A.; Rozenberg F.; Duchemin J.; Jourdi G.; Ellouze S.; Levavasseur F.; Albigès L.; Soria J.-C.; Barlesi F.; Solary E.; André F.; Pène F.; Ackerman F.; Mouthon L.; Zitvogel L.; Marabelle A.; Michot J.-M.; Fontenay M.; Kroemer G. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death & Disease 2021, 12 (3), 258. 10.1038/s41419-021-03540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz N.; Penant G.; Jardot P.; Duclos N.; La Scola B. Culture of SARS-CoV-2 in a panel of laboratory cell lines, permissivity, and differences in growth profile. Eur. J. Clin Microbiol Infect Dis 2021, 40 (3), 477–484. 10.1007/s10096-020-04106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Kruger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N.-H.; Nitsche A.; Muller M. A.; Drosten C.; Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181 (2), 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.; Chan J. F.-W.; Yuen T. T.-T.; Shuai H.; Yuan S.; Wang Y.; Hu B.; Yip C. C.-Y.; Tsang J. O.-L.; Huang X. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe 2020, 1 (1), e14 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X.; Liu Y.; Lei X.; Li P.; Mi D.; Ren L.; Guo L.; Guo R.; Chen T.; Hu J.; Xiang Z.; Mu Z.; Chen X.; Chen J.; Hu K.; Jin Q.; Wang J.; Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11 (1), 1–12. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.-H.; Chen Q.; Gu H.-J.; Yang G.; Wang Y.-X.; Huang X.-Y.; Liu S.-S.; Zhang N.-N.; Li X.-F.; Xiong R. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host & Microbe 2020, 28 (1), 124–133. 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K. H.; Leist S. R.; Schafer A.; Edwards C. E.; Martinez D. R.; Montgomery S. A.; West A.; Yount B. L.; Hou Y. J.; Adams L. E.; Gully K. L.; Brown A. J.; Huang E.; Bryant M. D.; Choong I. C.; Glenn J. S.; Gralinski L. E.; Sheahan T. P.; Baric R. S. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 2020, 586 (7830), 560–566. 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T.; Chen R.; He X.; Yuan Y.; Deng X.; Li R.; Yan H.; Yan S.; Liu J.; Zhang Y.; Zhang X.; Yu F.; Zhou M.; Ke C.; Ma X.; Zhang H. Infection of wild-type mice by SARS-CoV-2 B. 1.351 variant indicates a possible novel cross-species transmission route. Signal Transduct Target Ther 2021, 6 (1), 1–12. 10.1038/s41392-021-00848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladunni F. S.; Park J.-G.; Pino P. A.; Gonzalez O.; Akhter A.; Allué-Guardia A.; Olmo-Fontánez A.; Gautam S.; Garcia-Vilanova A.; Ye C. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020, 11 (1), 6122. 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce V. M.; Costoya J. A. SARS-CoV-2 infection in K18-ACE2 transgenic mice replicates human pulmonary disease in COVID-19. Cell Mol. Immunol 2021, 18 (3), 513–514. 10.1038/s41423-020-00616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryche B.; St Albin A.; Murri S.; Lacote S.; Pulido C.; Ar Gouilh M.; Lesellier S.; Servat A.; Wasniewski M.; Picard-Meyer E.; Monchatre-Leroy E.; Volmer R.; Rampin O.; Le Goffic R.; Marianneau P.; Meunier N. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav Immun 2020, 89, 579–586. 10.1016/j.bbi.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. J.; Lee A. C.-Y.; Chu H.; Chan J. F.-W.; Fan Z.; Li C.; Liu F.; Chen Y.; Yuan S.; Poon V. K.-M.; Chan C. C.-S.; Cai J.-P.; Wu K. L.-K.; Sridhar S.; Chan Y.-S.; Yuen K.-Y. Severe acute respiratory syndrome coronavirus 2 infects and damages the mature and immature olfactory sensory neurons of hamsters. Clin Infect Dis 2021, 73 (2), e503 10.1093/cid/ciaa995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Brand J.; Haagmans B. L.; Leijten L.; van Riel D.; Martina B. E.; Osterhaus A.; Kuiken T. Pathology of experimental SARS coronavirus infection in cats and ferrets. Vet Pathol 2008, 45 (4), 551–562. 10.1354/vp.45-4-551. [DOI] [PubMed] [Google Scholar]
- Cleary S. J.; Pitchford S. C.; Amison R. T.; Carrington R.; Robaina Cabrera C. L.; Magnen M.; Looney M. R.; Gray E.; Page C. P. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br. J. Pharmacol. 2020, 177 (21), 4851–4865. 10.1111/bph.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D. S.; Wheatley A. K.; Ramuta M. D.; Reynaldi A.; Cromer D.; Subbarao K.; O’Connor D. H.; Kent S. J.; Davenport M. P. Measuring immunity to SARS-CoV-2 infection: comparing assays and animal models. Nat. Rev. Immunol 2020, 20 (12), 727–738. 10.1038/s41577-020-00471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.; Qiao S.; Zhang G. Analysis of angiotensin-converting enzyme 2 (ACE2) from different species sheds some light on cross-species receptor usage of a novel coronavirus 2019-nCoV. J. Infect 2020, 80 (4), 469–496. 10.1016/j.jinf.2020.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W.; Bao L.; Liu J.; Xiao C.; Liu J.; Xue J.; Lv Q.; Qi F.; Gao H.; Yu P. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science 2020, 369 (6505), 818–823. 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D.; Nilsson-Payant B. E.; Liu W.-C.; Uhl S.; Hoagland D.; Møller R.; Jordan T. X.; Oishi K.; Panis M.; Sachs D.; Wang T. T.; Schwartz R. E.; Lim J. K.; Albrecht R. A.; tenOever B. R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020, 181 (5), 1036–1045. 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.; Zhao Y.; Yu W.; Yang Y.; Gao J.; Wang J.; Kuang D.; Yang M.; Yang J.; Ma C.; Xu J.; Qian X.; Li H.; Zhao S.; Li J.; Wang H.; Long H.; Zhou J.; Luo F.; Ding K.; Wu D.; Zhang Y.; Dong Y.; Liu Y.; Zheng Y.; Lin X.; Jiao L.; Zheng H.; Dai Q.; Sun Q.; Hu Y.; Ke C.; Liu H.; Peng X. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct Target Ther 2020, 5 (1), 157. 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q.; Zhou J.; He Q.; Li R. T.; Yang G.; Zhang Y.; Wu S. J.; Chen Q.; Shi J. H.; Zhang R. R.; Zhu H. M.; Qiu H. Y.; Zhang T.; Deng Y. Q.; Li X. F.; Liu J. F.; Xu P.; Yang X.; Qin C. F. SARS-CoV-2 infection in the mouse olfactory system. Cell Discovery 2021, 7 (1), 49. 10.1038/s41421-021-00290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela C.; Dowling W. E.; Funnell S. G.; Gsell P.-S.; Riveros-Balta A. X.; Albrecht R. A.; Andersen H.; Baric R. S.; Carroll M. W.; Cavaleri M. Animal models for COVID-19. Nature 2020, 586 (7830), 509–515. 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]