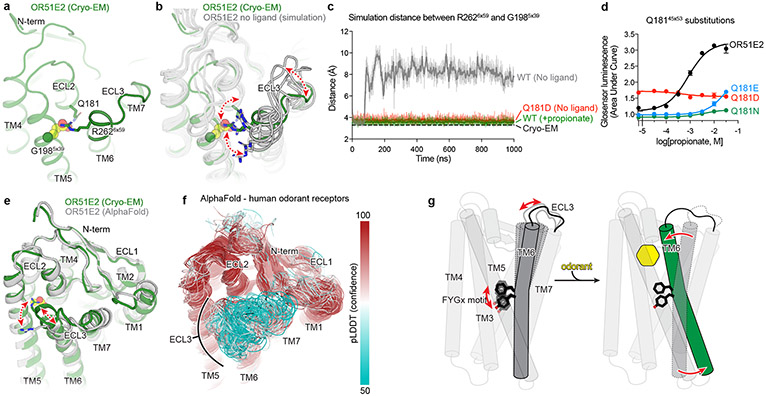
Figure 5: Structural dynamics of ECL3 in OR function.
a) Residue R2626x59 in ECL3 makes a critical contact with propionate. Residue Q18145x53 in ECL2 is highlighted. b) Molecular dynamics simulations of OR51E2 with propionate removed shows increased flexibility of R2626x59. Representative snapshots are displayed from five independent simulation replicates after 1000 ns of simulation time. c) In simulations of wild-type (WT) OR51E2 bound to propionate, the minimum distance between R2626x59 and G1985x39 heavy atoms is stable and similar to the cryo-EM structure. Simulations of WT OR51E2 without propionate (no ligand) show increased minimum distance between R2626x59 and G1985x39. In simulations of Q18145x53D mutant without propionate, the minimum distance between R2626x59 and G1985x39 is similar to WT OR51E2 bound to propionate. Minimum distance was measured between R2626x59 sidechain atoms and G1985x39 main chain atoms (excluding the hydrogens) over the course of 1000 ns MD simulation (see Extended Data Fig. 8 for simulation replicates). Thick traces represent smoothed values with an averaging window of 8 nanoseconds; thin traces represent unsmoothed values. d) Conservative mutagenesis of Q18145x53 shows that the Q18145x53D mutant is constitutively active, potentially because it substitutes a carboxylic acid in the OR51E2 binding pocket. e) Comparison of cryo-EM structure of OR51E2 with the AlphaFold2 predicted structure shows high similarity in the extracellular domain with the exception of the ECL3 region. The AlphaFold2 model shows an outward displacement of R2626x59 and ECL3 similar to simulations of apo OR51E2. f) AlphaFold2 predictions for all human odorant receptors show low confidence in the ECL3 region and high confidence in other extracellular loops. g) A model for ECL3 as a key site for odorant receptor activation.