Abstract
We determined the nucleotide sequence between 1,903 and 3,097 bp of pbp1a, which encodes the transpeptidase domain of PBP 1A, from clinical isolates of penicillin-resistant Streptococcus pneumoniae (PRSP) serotypes 19 (n = 8), 6 (n = 9), 23 (n = 6), and 14 (n = 2) and two penicillin-susceptible S. pneumoniae (PSSP) isolates. These serotyped PRSP strains were isolated predominantly in Japan from 1993 through 1997. The 25 resistant strains were classified into five groups on the basis of the extent of sequence differences. Strains in groups I (n = 5; serotype 6), II (n = 3; serotype 19), and III (n = 12; different serotypes) had sequences highly homologous to the sequence of pbp1a of PRSP strains from South Africa, Spain, and the United States. The group IV strain (n = 1; serotype 14) had unique deletions from or insertions in the sequences. The sequences of group V strains (n = 4; serotypes 6 and 23) had relatively few differences from the sequences of the PSSP strains. For strains (n = 18) for which the threonine at codon 371 (Thr-371) in a conserved STMK motif of PBP 1A was substituted with an alanine or a serine (in addition to having altered pbp2x and pbp2b genes), penicillin MICs were ≧1.0 μg/ml. The PBPs 1A of these strains showed decreased affinities for [3H]benzylpenicillin and slightly faster mobilities on sodium dodecyl sulfate-polyacrylamide gels. In contrast, for strains (n = 4) without a substitution at Thr-371 in PBP 1A but with mutations in both pbp2x and pbp2b, penicillin MICs were 0.125 to 0.25 μg/ml, and the affinities of their PBPs 1A were similar to that of PSSP PBPs 1A. Furthermore, for the Thr-371-substituted strains (n = 3) with altered pbp2x genes but native pbp2b genes, penicillin MICs were 0.125 to 0.25 μg/ml. These results suggest that amino acid substitution of Thr-371 contributes to the development of penicillin resistance in PRSP strains with altered pbp2x and pbp2b genes.
Severe infections due to penicillin-resistant Streptococcus pneumoniae (PRSP) have become a serious problem worldwide. The resistance of S. pneumoniae to β-lactams involves decreased affinities of penicillin-binding proteins (PBPs) resulting from alterations in PBPs (12, 21) due to the horizontal transfer of DNA blocks from oral streptococci (5, 6, 9, 16, 18). Among the PBPs usually found in S. pneumoniae, PBPs 1A, 2B, and 2X are the most important in the development of resistance (14, 18, 20, 26). Dowson et al. (5, 6, 9) performed a genetic analysis of PRSP to and found that the pbp2b alteration occurs by gene transfer. Hakenbeck et al. (12) examined the PBPs of PRSP and found multiple changes in PBPs, especially in PBP 1A and PBP 2B and the appearance of PBP 2X but no change in PBP 3. The production of PBP 2X and PBP 1A in PRSP resistant to broad-spectrum cephalosporins was reported by Muñoz et al.
The nucleotide sequences of pbp1a (18, 19), pbp2x (15, 16), and pbp2b (6–8) in penicillin-susceptible S. pneumoniae (PSSP) and PRSP have already been determined. Dowson et al. (6) classified the pbp2b of PRSP into classes A and B on the basis of mutation patterns. We previously analyzed part of pbp2b, which encodes the PBP 2B transpeptidase region in PRSP strains isolated in Japan (29). On the basis of the sequencing results, we applied PCR with primers designed to detect class A and B pbp2b genes, and we have reported that PRSP strains with class B alterations are common in Japan (25).
To study the relationship between PBP alterations in S. pneumoniae and β-lactam susceptibility, we designed primers that detect three genes: pbp1a, pbp2x, and pbp2b (26). Corresponding to the diversities in the PBP gene from PRSP, DNA amplifications occur specifically only in the unaltered genes of susceptible strains but do not occur in altered genes from a resistant strain. Our results indicated that the PBP gene most related to resistance differed depending on the type of β-lactam (26). Resistance to penicillins is affected to the same degree by altered pbp1a and pbp2b genes, while resistance to cephems is mainly affected by altered pbp1a and pbp2x genes, and resistance to carbapenems and penem is affected by altered pbp1a and pbp2b genes (20, 26). We have also reported that simultaneous alterations of the pbp1a, pbp2x, and pbp2b genes resulted in the highest level of resistance compared with the levels of resistance when only two genes, pbp2x and pbp2b, pbp1a and pbp2b, and pbp1a and pbp2x, were altered (26).
The open reading frame of pbp1a in PSSP encodes a 719-amino-acid protein with an estimated molecular mass of 79.8 kDa (19). PBP 1A is suggested to be a bifunctional enzyme with transglycosylase and transpeptidase activities, as deduced from its high degree of homology to Escherichia coli PBP 1A. DNA sequences have been determined for some PRSP strains (serotype 6, 14, or 23) isolated in South Africa, Spain (13), and the United States (3). The pbp1a genes reportedly contain four types of resistance blocks (13). As with pbp2b and pbp2x, pbp1a has been shown to be a mosaic gene that contains blocks of DNA from other sources encoding resistance. Among PRSP strains isolated in Japan, serotype 19 is the most frequently found, followed by serotypes 6 and 23. Serotype 14 is very rare (24). In this report we describe the results of genetic studies of pbp1a, along with those of pbp2x and pbp2b, from clinical PRSP isolates with different serotypes. We also investigated the nucleotide sequence between 1,903 and 3,097 bp of pbp1a and found that substitution of Thr-371 with alanine or serine predominated. The substitution of Thr-371 is associated with the level of resistance to penicillin, with PRSP strains having simultaneous alterations in pbp2x and pbp2b.
MATERIALS AND METHODS
Bacterial strains.
PRSP (n = 25) and PSSP (n = 2) strains isolated from cerebrospinal fluid (CSF), blood, nasopharynx, and sputum were used in this study. Clinical isolates were collected between 1993 and 1997 by the Japanese Working Group for PRSP (24). Details about these strains are presented in Table 1. All strains were grown at 37°C for 18 h on 5% sheep blood agar medium (Nippon Becton Dickinson, Co., Ltd., Tokyo, Japan) in a humidified atmosphere supplemented with 5% CO2.
TABLE 1.
Classification based on pbp1a sequence differences and properties of S. pneumoniae strains
| Group | Strain | Serotype | MIC (μg/ml)a
|
Substitution at STMKb | PCR resultc
|
Clinical material | ||
|---|---|---|---|---|---|---|---|---|
| Pc | Ctx | pbp2x | pbp2b | |||||
| PSSP | 1/H23 | 14 | 0.031 | 0.063 | –––– | + | − | Nasopharynx |
| 10/Z19 | 6 | 0.031 | 0.016 | –––– | − | − | CSF | |
| PRSP | ||||||||
| I | 5/H31 | 6 | 2 | 0.5 | –A–– | + | + | Nasopharynx |
| 8/Z2 | 6 | 1 | 0.5 | –A–– | + | + | CSF | |
| 11/Z20 | 6 | 1 | 0.5 | –A–– | + | + | CSF | |
| 12/Z21 | 6 | 2 | 1 | –A–– | + | + | CSF | |
| 22/HA5 | 6 | 0.25 | 0.25 | –––– | + | + | Phalynx | |
| II | 17/Z46 | 19 | 4 | 1 | –A–– | + | + | CSF |
| 20/B98 | 19 | 4 | 1 | –A–– | + | + | Blood | |
| 27/SHA3 | 19 | 4 | 4 | –A–– | + | + | Sputum | |
| III | 18/B43 | 6 | 4 | 1 | –S–– | + | + | Blood |
| 19/B90 | 6 | 0.25 | 1 | –S–– | + | − | Blood | |
| 28/H0 | 14 | 1 | 0.5 | –S–– | + | + | Nasopharynx | |
| 2/H26 | 19 | 2 | 0.5 | –S–– | + | + | Nasopharynx | |
| 3/H28 | 19 | 2 | 1 | –S–– | + | + | Nasopharynx | |
| 6/KU126 | 19 | 2 | 1 | –S–– | + | + | Nasopharynx | |
| 15/Z12 | 19 | 0.125 | 0.5 | –S–– | + | − | CSF | |
| 16/Z13 | 19 | 4 | 1 | –S–– | + | + | CSF | |
| 4/H29 | 23 | 2 | 0.5 | –S–– | + | + | Nasopharynx | |
| 13/Z34 | 23 | 2 | 1 | –S–– | + | + | CSF | |
| 21/B99 | 23 | 2 | 1 | –S–– | + | + | Blood | |
| 7/KK133 | 23 | 2 | 4 | –S–– | + | + | Nasopharynx | |
| IV | 14/Z42 | 14 | 0.125 | 0.25 | –S–– | + | − | CSF |
| V | 9/Z17 | 6 | 1 | 0.5 | –S–– | + | + | CSF |
| 23/HSB21 | 6 | 0.125 | 0.125 | –––– | + | + | Sputum | |
| 24/TJ25 | 23 | 0.25 | 0.25 | –––– | + | + | Nasopharynx | |
| 26/TJ29 | 23 | 0.25 | 0.25 | –––– | + | + | Nasopharynx | |
Pc, penicillin; Ctx, cefotaxime.
Only amino acid residues differing from the STMK sequence at positions 370 to 373 in PBP 1A are shown.
Altered PBP genes were detected by PCR as described previously (24). +, altered; −, not altered.
Antibiotic susceptibility and serotyping.
The susceptibilities of S. pneumoniae to penicillin and cefotaxime were determined with cation-adjusted Mueller Hinton agar (Eiken Co., Ltd., Tokyo, Japan) supplemented with 5% defibrinated sheep blood by the agar plate dilution method. Penicillin and cefotaxime were provided by Banyu Pharmaceutical Co., Ltd., and Japan Luther Co., Ltd., Tokyo, Japan, respectively. Isolates were serotyped by the capsular swelling technique with antisera purchased from Statens Serum Institute (Copenhagen, Denmark).
PBP assay.
S. pneumoniae PBPs were labeled as described previously (28). Whole cells (109 cells per ml) suspended in 50 mM phosphate buffer (pH 7.0) were incubated with [3H]benzylpenicillin for 10 min at 30°C; this was followed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and fluorography. For competition assays, cells were detected after preincubation with twofold-diluted nonradioactive benzylpenicillin for 10 min, followed by incubation with [3H]benzylpenicillin for a further 10 min. The relative densities of the PBPs were measured with a dual-wavelength flying-spot scanner (CS9000; Shimadzu Co., Kyoto, Japan) and were compared with the density of the PBP in the control sample with no antibiotics before exposure to the radiolabeled benzylpenicillin.
pbp1a sequencing.
The 1.3-kb DNA fragment encoding the PBP 1A transpeptidase domain was amplified from the chromosomal DNA of S. pneumoniae by PCR as reported previously (25). One colony of S. pneumoniae grown on blood agar was picked and placed in 20 μl of lysis solution, and the mixture was incubated at 60°C for 10 min and then at 90°C for 5 min. Afterward, 1 μl of the lysate was added to 100 μl of a PCR solution (10 μl of 10× PCR buffer, 8 μl of a mixture of deoxynucleoside triphosphates at a concentration of 2.5 mM each, 2.5 U of Taq polymerase [Takara Biomedicals, Kyoto, Japan], 10 pmol of the sense primer [5′-T1827GGGATGGATGTTTACACAAATG1849], and 10 pmol of the reverse primer [5′-T3098GTGCTGGTTGATGAGGATTCTG3079]). PCR conditions were as follows: 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The PCR product was electrophoresed on a 1.2% agarose gel to confirm the presence of the product and was then purified with a Chroma Spin-100 Column (Clontech Laboratories Inc., Palo Alto, Calif.) to prepare a sequencing template.
The sequencing reaction was conducted with a fmol DNA Cycle Sequencing System Kit (Promega Co., Madison, Wis.) and 5′ fluorescein isothiocyanate-labeled primers.
The following primers were used for sequencing: 5′-A1859AGCTCAAAAACATCTGTGGG1880, 5′-A2380GTAGTGAAAAAATGGCTGCTG2400, 5′-C2046TGGGGTTCTGCTATGAAACC2067, 5′-T2160ACTCCACTCTACAACTGG G2179, 5′-T2881TCGTATTTAAAAATGGTGCTCG2903, 5′-G2399CAGCCATTTTTTCACTACTTG2377, 5′-G2677TAGCTCCAGATGAAATGTTTG2699, 5′- A2392TGGCTGCTGCTTATGCTGCC2412, and 5′-C2702CAACAAACATTTCATCTGGAGC2680.
The reaction mixtures were placed in a thermal cycler and were denatured at 95°C for 2 min; they were then subjected to 30 cycles at 95°C for 30 s, 48°C for 30 s, and 72°C for 30 s. The nucleotide sequences were determined with an SQ-3000 Hitachi fluorescence DNA sequencer (Hitachi Electronics Engineering Co., Ltd., Tokyo, Japan).
PCR primers for studies with pbp2x and pbp2b.
We previously reported on the use of the primers described above for the detection of altered pbp2x and pbp2b genes of S. pneumoniae isolates. The primers were designed from the nucleotide sequences of various strains isolated in Japan (26). These primers specifically amplified parts of each gene of only susceptible strains. These parts of each gene were positioned in blocks of highly diverged sequences in the mosaic PBP genes of PRSP. The two set of primers were as follows: 5′-C1003CAGGTTCCACTATGAAAGTG1023 and 5′-C1294ATCCGTCAAACCGAAACGG1275 for pbp2x and 5′-C1636AATCTAGAGTCTGCTATGGA1656 and 5′-G1712GTCAATTCCTGTCGCAGTA1693 for pbp2b.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the following accession numbers: AB006868 (strain 1/H23), AB006869 (strains 2/H26, 4/H29, 13/Z34, 18/B43, 19/B90, 21/B99, and 28/H0), AB006870 (strains 3/H28, 6/KU126, 15/Z12, and 16/Z13), AB006871 (strains 5/H31, 8/Z2, 11/Z20, and 12/Z21), AB006872 (strain 7/KK133), AB006873 (strain 9/Z17), AB006874 (strain 10/Z19), AB006875 (strain 14/Z42), AB006876 (strains 17/Z46, 20/B98, and 27/SHA3), AB006877 (strain 22/HA5), AB006878 (strain 23/HSB21), and AB006879 (strains 24/TJ25 and 26/TJ29).
RESULTS
Nucleotide and deduced amino acid sequences of pbp1a.
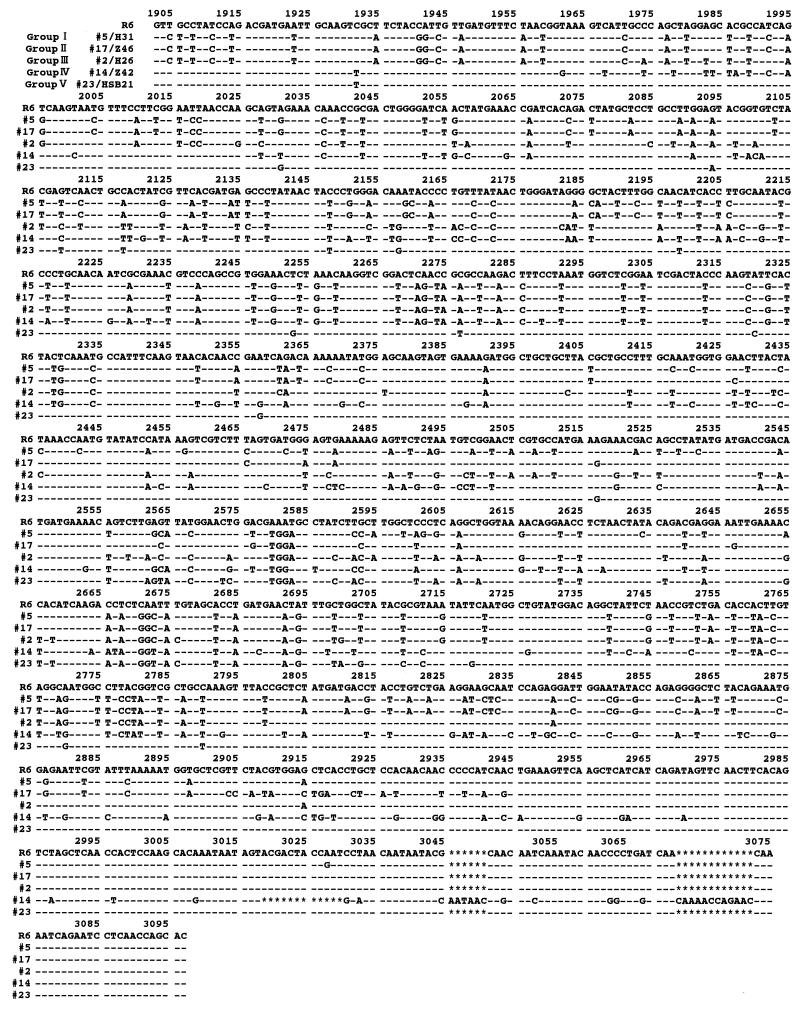
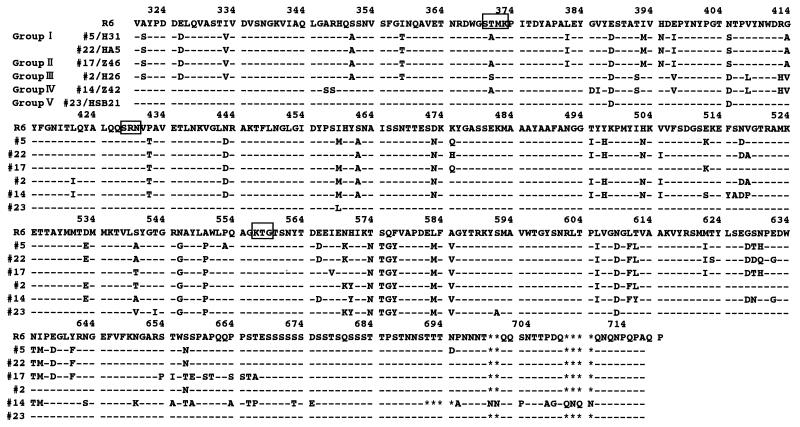
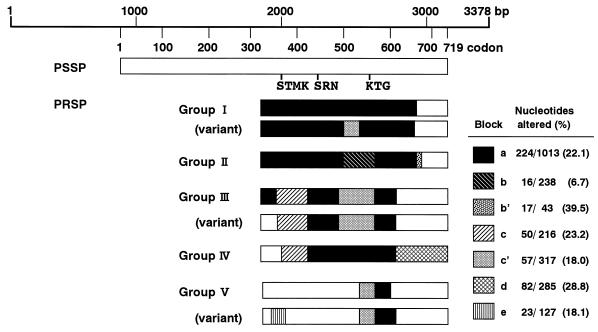
The nucleotide sequences of pbp1a between 1,903 and 3,097 bp encoding the transpeptidase region of PBP 1A in 25 PRSP and 2 PSSP strains were determined by the direct sequencing method. The nucleotide and deduced amino acid sequences of representative strains are aligned in Fig. 1 and 2, respectively, along with the previously determined sequence of a susceptible strain (strain R6) (19). The regions of the nucleotide sequence in PRSP differing by 6.7% or more from those in PSSP R6 are referred to as “resistance blocks.” Figure 3 provides a schematic illustration of the mosaic structures of pbp1a.
FIG. 1.
Nucleotide sequences of part of the pbp1a genes from representative PRSP strains. The sequence of the pbp1a gene of PSSP R6 is shown at the top. Numbering is based on published data for strain R6 (19). Only nucleotides differing from those in the R6 sequence are shown. Asterisks indicate the deletion of nucleotides.
FIG. 2.
Deduced amino acid sequence of part of PBP 1A of the strains listed in Fig. 1. Only amino acids differing from the R6 amino acid sequence are shown. Boxes indicate conserved amino acid sequences.
FIG. 3.
Mosaic structure of pbp1a genes from PRSP. Regions of the sequence coding for conserved amino acid sequences are marked. Shading indicates blocks where the sequences differ from that for the PSSP R6 strain (white area) by 6% or more, i.e., blocks a to e. Groups are as follows: group I, strains 5/H31, 8/Z2, 11/Z20, 12/Z21, and variant 22/HA5; group II, strains 17/Z46, 20/B98, and 27/SHA3; group III, strains 2/H26, 4/H29, 13/Z34, 18/B43, 19/B90, 21/B99, 28/H0, 3/H28, 6/KU126, 15/Z12, 16/Z13, and variant 7/KK133; group IV, strains 14/Z42; group V, strains 23/HSB21, 24/TJ25, 26/TJ29, and variant 9/Z17.
The sequences of two PSSP strains (strains 1/H23 and 10/Z19) differed from that of strain R6 by only five and eight nucleotides, respectively, resulting in only two and three amino acid substitutions, respectively.
Twenty-five PRSP strains were classified into five groups on the basis of differences in the mosaic pbp1a sequence (Table 1).
Group I consisted of five strains (strains 5/H31, 8/Z2, 11/Z20, 12/Z21, and 22/HA5), all of which were serotype 6. The penicillin and cefotaxime MICs for all strains except strain 22/HA5 were 1 to 2 and 0.5 to 1 μg/ml, respectively. The DNA sequences of four of the strains (all group I strains except strain 22/HA5) were highly homologous to that of the serotype 6B strains reported by Martin et al. (18), differing by only one to five nucleotides. The sequence from 1,904 to 2919 bp in these strains was named resistance block a. In this block, Thr-371 in the conserved amino acid sequence STMK (codons 370 to 373), which is considered the β-lactam binding site, was replaced by Ala in four of the strains (all group I strains except strain 22/HA5). The entire transpeptidase region of strain 22/HA5 resembled that in group I strains, differing by 8% of its nucleotides and 3% of its amino acids, but the substitution of Thr-371 was not found. The DNA sequence between 2,418 and 2,637 bp contained a part of the sequence (block c′) of group III strains. No difference in the nucleotide sequences of the pbp1a genes of strains 5/H31, 8/Z2, 11/Z20, and 12/Z21 was detected.
Group II consisted of three strains (strains 17/Z46, 20/B98, and 27/SHA3) in which parts of resistance block a were replaced by resistance blocks b (2,418 to 2,655 bp) and b′ (2,904 to 2,944 bp). Their nucleotide sequences were identical, and all three strains were serotype 19. The strains were almost identical, with 99.7% nucleotide sequence homology and 100% amino acid sequence homology to serotype 23F strains, the sequences of which were reported by Martin et al. (18). All strains showed an amino acid substitution of Thr-371 to Ala in the conserved amino acid sequence STMK.
Group III consisted of 12 strains (strains 2/H26, 3/H28, 4/H29, 6/KU126, 7/KK133, 13/Z34, 15/Z12, 16/Z13, 18/B43, 19/B90, 21/B99, and 28/H0). The DNA sequence between 1,996 and 2,211 bp was named block c, and that between 2,349 and 2,665 bp was named block c′. Parts of block a were replaced by blocks c and c′ in 11 strains but not in strain 7/KK133. The nucleotide sequences of these strains were highly homologous to the sequences of two PRSP strains isolated in the United States (3). Strain 7/KK133 was thought to be a variant of this group; the sequence between 1,904 and 1,995 bp was identical to that of PSSP R6, while the downstream region was the same as those of group III strains. In all group III strains, the Thr-371 in the STMK motif was changed to Ser. Between seven strains (strains 2/H26, 4/H29, 13/Z34, 18/B43, 19/B90, 21/B99, and 28/H0) and four strains (strains 3/H28, 6/KU126, 15/Z12, and 16/Z13), there were only two nucleotide differences in the downstream region, far from the KTG motif. Group III strains showed variable serotypes: serotypes 6 (n = 2), 14 (n = 1), 19 (n = 5), and 23 (n = 4).
Group IV consisted of strain 14/Z42 (serotype 14), which had alterations in the DNA sequence within the region upstream from STMK, but these alterations resulted in only two amino acid substitutions. Thr-371 was replaced by Ser. The sequence between 2,037 and 2,211 bp was similar to that of resistance block c, and that between 2,211 and 2,790 bp was similar to that of block a. A unique region named block d occurred between 2,791 and 3,075 bp, and this region was not observed in any other group. The most downstream region in particular showed a deletion of 12 nucleotides between 3,019 and 3,030 bp, insertions of 6 nucleotides between 3,045 and 3,046 bp, and insertions of 12 nucleotides between 3,072 and 3,073 bp.
Group V consisted of four strains (strains 9/Z17, 23/HSB21, 24/TJ25, and 26/TJ29) of serotypes 6 and 23. Group V strains had fewer mutations than the strains in the four other groups. Compared to PSSP R6, these strains had 19 to 21 amino acid changes that were concentrated in the region from 2,566 to 2,799 bp. In strain 9/Z17, the sequence in the region that includes the STMK motif was replaced by block e (1,932 to 2,058 bp). A Thr-371-to-Ser substitution was detected only in strain 9/Z17, and was not detected in the remaining three strains.
Thr-371 substitution and penicillin resistance.
Table 1 presents the 25 PRSP strains as classified into five groups and the MICs of penicillin and cefotaxime for the strains. Table 1 also indicates whether substitutions at the conserved STMK motif, together with the altered pbp2x and pbp2b genes, are present.
Strains with altered pbp2x and pbp2b genes and in which Thr-371 was substituted by Ala or Ser in the PBP 1A STMK motif (n = 18) were penicillin resistant (MICs, ≧1.0 μg/ml). For four strains (strains 22/HA5, 23/HSB21, 24/TJ25, and 26/TJ29) with alterations in both the pbp2x and pbp2b genes but not in the pbp1a STMK motif, penicillin MICs were 0.125 to 0.25 μg/ml. For strains 14/Z42, 15/Z12, and 19/B90, which had unaltered pbp2b genes but altered pbp2x genes and a Thr-371-to-Ser substitution in PBP 1A, penicillin MICs were 0.125 to 0.25 μg/ml.
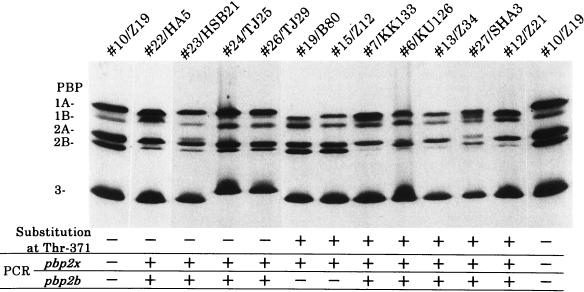
PBP 1A affinity to penicillin.
We assayed PBP profiles using [3H]benzylpenicillin (Fig. 4). The PBPs 1A of four strains without a substitution at Thr-371 were shown to have the same affinity as the PBP 1A of PSSP 10/Z19, while PBP 1A of 22/HA5 showed slightly faster mobility on SDS-polyacrylamide gels, probably because of its many amino acid substitutions typical of group I strains. PBPs 1A from all strains with a Thr-371-to-Ser or a Thr-371-to-Ala substitution showed decreased affinities for [3H]benzylpenicillin, and most had faster mobilities on SDS-polyacrylamide gels.
FIG. 4.
PBP profiles of S. pneumoniae strains. Whole cells (109 cells per ml) suspended in 50 mM phosphate buffer (pH 7.0) were labeled with [3H]benzylpenicillin for 10 min at 30°C and were then subjected to SDS-polyacrylamide gel electrophoresis and fluorography (25). The results of PCR detection of altered pbp2x and pbp2b genes (+, altered; −, not altered) and substitution of Thr-371 in PBP 1A are listed below the gel. Penicillin MICs are as follows: for 10/Z19, 0.031 μg/ml; for 22/HA5, 0.25 μg/ml; for 23/HSB21, 0.125 μg/ml; for 24/TJ25, 0.25 μg/ml; for 26/TJ29, 0.25 μg/ml; for 19/B80, 0.25 μg/ml; for 15/Z12, 0.125 μg/ml; for 7/KK133, 2.0 μg/ml; for 6/KU126, 2.0 μg/ml; for 13/Z34, 2.0 μg/ml; for 27/SHA3, 4.0 μg/ml; for 12/Z21, 2.0 μg/ml. “2A” is 2A/2X.
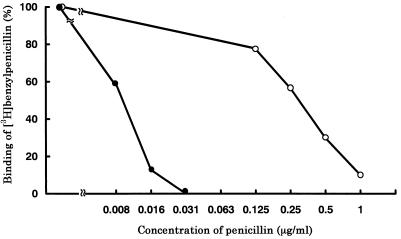
Comparison of the affinity kinetics of the PBPs 1A of strains 22/HA5 and 5/H31 showed that the PBP 1A of strain 22/HA5, which did not have a substitution at Thr-371, had a higher affinity for penicillin at a significantly lower penicillin concentration than that for the PBP 1A of strain 5/H31, which did have a substitution at Thr-371 (Fig. 5).
FIG. 5.
Competition assays of penicillin for PBPs 1A of strains 22/HA5 (•) and 5/H31 (○). Strains 22/HA5 without a substitution at Thr-371 (MIC, 0.25 μg/ml) and 5/H31 with a substitution at Thr-371 (MIC, 2.0 μg/ml) were compared. PBPs were detected with [3H]benzylpenicillin after preincubation of the cells with various concentrations of penicillin. Relative PBP density was calculated against the PBP density for the control sample that was not exposed to penicillin.
DISCUSSION
PSSP strains usually have six PBPs: high-molecular-mass PBPs 1A, 1B, 2A, 2X, and 2B and low-molecular-mass PBP 3. High-molecular-mass PBPs are supposed to have transpeptidase activity and may contain a transglycosylase region, based on homology to Escherichia coli PBPs. The pbp1a, pbp2x, and pbp2b genes are involved in penicillin resistance in S. pneumoniae (11–13, 17). The addition of an altered pbp1a to altered pbp2x or pbp2b and to altered pbp2x and pbp2b has been reported to increase the level of penicillin resistance (20, 26). The DNA sequences of these three PBP genes in PRSP have already been determined (6, 7, 16, 18, 23, 29). The genes conferring resistance in PRSP have a mosaic structure, in which parts of genes are replaced by genes from other sources, probably via gene transfer (2, 5, 9, 16, 18).
Conserved amino acid sequences SXXK with active-site serine, SXN, and K(H)T(S)G motifs are important to the transpeptidase activity and are believed to be located at the active-site cavity. If one or more amino acids in conserved motifs or adjacent amino acids are substituted by other amino acids, β-lactams may not be able to bind efficiently to the PBP, resulting in the development of resistance.
In this study we demonstrated that strains with a Thr-371 substitution in a conserved STMK motif in PBP 1A, in addition to altered pbp2x and pbp2b genes, had high levels of penicillin resistance (MICs, ≧1 μg/ml). For strains with a Thr-371 substitution in PBP 1A and an alteration in the pbp2x gene but with no pbp2b gene alteration, penicillin MICs were 0.125 to 0.25 μg/ml. On the other hand, for strains with altered pbp2x and pbp2b genes but no Thr-371 substitution, penicillin MICs were 0.25 μg/ml. All strains of PRSP examined had pbp2x alterations.
The penicillin-binding domain in the PBPs is defined as a polypeptide stretch that starts 60 residues or fewer upstream of the Ser of the conserved SXXK motif and that terminates 60 residues or fewer downstream of the Lys (His) of the conserved K(H)T(S)G motif (15). The PBPs 1A of all strains with a Thr-371 substitution had a decreased affinity for penicillin, and the PBPs 1A of four strains without this substitution had normal affinities for penicillin; the penicillin MICs for the strains were 0.125 to 0.25 μg/ml. These results support the assumption that the substitution plays an important role in the development of penicillin resistance. However, six amino acid differences other than Thr-371 are observed in the penicillin-binding domain of PBPs 1A in strains 5/H31 and 22/HA5 strains; thus, it is possible that various substitutions other than Thr-371 in the transpeptidase region of PBP 1A may be involved in penicillin resistance. Garcia-Bustos and Tomasz (10) have shown that PRSP strains produce cell walls with profoundly altered chemical compositions. It is possible that a substitution of Thr-371 adjacent to the active-site serine may change the three-dimensional structure of the transpeptidase domain and alter enzymatic activity for peptidoglycan synthesis. However, this possibility will have to be clarified in future investigations.
In Japan, PRSP strains were first isolated in CSF from children with meningitis in 1988, and the proportion of S. pneumoniae strains that are resistant to penicillin has recently increased to 40% (27). The nucleotide sequences of pbp1a genes from strains in groups I and II were highly homologous to those of certain PRSP strains isolated in South Africa and Spain between 1978 and 1988. The sequences of the pbp1a genes from strains in group III were highly homologous to those from PRSP strains isolated in the United States in 1991. This suggests that PRSP strains of similar origins may have been introduced in Japan and gradually spread. The variability of the mosaic structures in pbp1a also suggests that the recombination event mediated by gene transfer has occurred frequently in response to antibacterial pressure.
PBP genes have been reported to be easily transformed in vitro (1, 14, 22), and horizontal transfer of capsular biosynthesis genes has occurred (4). We recognized different serotypes for strains whose pbp1a gene sequences were identical (group III and IV strains) and found that the pbp1a gene sequences of members of group II of serotype 19 were almost identical to those of some serotype 23F strains previously described in South Africa and Spain. Our findings may suggest the probability of the horizontal transfer of either PBP genes or capsular genes. In conclusion, under the pressure of frequent antibiotic usage, alterations in three genes, pbp2x, pbp2b, and pbp1a, of PRSP have occurred. The highly resistant PRSP strains had pbp1a alterations that resulted in a Thr-371 substitution to alanine or serine, in addition to the simultaneous alterations in the pbp2x and pbp2b genes.
REFERENCES
- 1.Barcus V A, Ghanekar K, Yeo M, Coffey T J, Dowson C G. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol Lett. 1995;126:299–304. doi: 10.1111/j.1574-6968.1995.tb07433.x. [DOI] [PubMed] [Google Scholar]
- 2.Coffey T J, Berron S, Daniels M, Garcia-Leoni M E, Cercenado E, Bouza E, Fenoll A, Spratt B G. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F, and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 3.Coffey T J, Daniels M, McDougal L K, Dowson C G, Tenover F C, Spratt B G. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 1995;39:1306–1313. doi: 10.1128/aac.39.6.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 5.Dowson C G, Coffey T J, Kell C, Whiley R A. Evolution of penicillin resistance in Streptococcus pneumoniae; the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol Microbiol. 1993;9:635–643. doi: 10.1111/j.1365-2958.1993.tb01723.x. [DOI] [PubMed] [Google Scholar]
- 6.Dowson C G, Hutchison A, Brannigan J A, George R C, Hansman D, Liñares J, Tomasz A, Smith J M, Spratt B G. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowson C G, Hutchison A, Spratt B G. Extensive re-modelling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 8.Dowson C G, Hutchison A, Spratt B G. Nucleotide sequence of the penicillin-binding protein 2B gene of Streptococcus pneumoniae strain R6. Nucleic Acids Res. 1989;17:7518. doi: 10.1093/nar/17.18.7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowson C G, Hutchison A, Woodford N, Johnson A P, George R C, Spratt B G. Penicillin-resistant streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1990;87:5858–5862. doi: 10.1073/pnas.87.15.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Bustos J, Tomasz A. A biological price of antibiotic resistance: major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc Natl Acad Sci USA. 1990;87:5415–5419. doi: 10.1073/pnas.87.14.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grebe T, Hakenbeck R. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob Agents Chemother. 1996;40:829–834. doi: 10.1128/aac.40.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakenbeck R, Tarpay M, Tomasz A. Multiple changes of penicillin-binding proteins in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980;17:364–371. doi: 10.1128/aac.17.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jabes D, Nachman S, Tomasz A. Penicillin-binding protein families: evidence for the clonal nature of penicillin resistance in clinical isolates of pneumococci. J Infect Dis. 1989;159:16–25. doi: 10.1093/infdis/159.1.16. [DOI] [PubMed] [Google Scholar]
- 14.Kell C M, Sharma U K, Dowson C G, Town C, Balganesh T S, Spratt B G. Deletion analysis of the essentiality of penicillin-binding proteins 1A, 2B, 2X of Streptococcus pneumoniae. FEMS Microbiol Lett. 1993;106:171–175. doi: 10.1111/j.1574-6968.1993.tb05954.x. [DOI] [PubMed] [Google Scholar]
- 15.Laible G, Hakenbeck R, Sicard M A, Joris B, Ghuysen J-M. Nucleotide sequences of the pbpX genes encoding the penicillin-binding proteins 2X from Streptococcus pneumoniae R6 and a cefotaxime-resistant mutant, C506. Mol Microbiol. 1989;3:1337–1348. doi: 10.1111/j.1365-2958.1989.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 16.Laible G, Spratt B G, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 17.Markiewicz Z, Tomasz A. Variation in penicillin-binding protein patterns of penicillin-resistant clinical isolates of pneumococci. J Clin Microbiol. 1989;27:405–410. doi: 10.1128/jcm.27.3.405-410.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin C, Sibold C, Hakenbeck R. Relatedness of penicillin-binding protein 1a genes from different clones of penicillin-resistant Streptococcus pneumoniae isolated in South Africa and Spain. EMBO J. 1992;11:3831–3836. doi: 10.1002/j.1460-2075.1992.tb05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin C, Thomas B, Hakenbeck R. Nucleotide sequences of genes encoding penicillin-binding proteins from Streptococcus pneumoniae and Streptococcus oralis with high homology to Escherichia coli penicillin-binding proteins 1A and 1B. J Bacteriol. 1992;174:4517–4523. doi: 10.1128/jb.174.13.4517-4523.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muñoz R, Dowson C G, Daniels M, Coffey T J, Martin C, Hakenbeck R, Spratt B G. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1992;6:2461–2465. doi: 10.1111/j.1365-2958.1992.tb01422.x. [DOI] [PubMed] [Google Scholar]
- 21.Percheson P B, Bryan L E. Penicillin-binding components of penicillin-susceptible and resistant strains of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980;18:390–396. doi: 10.1128/aac.18.3.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sibold C, Henrichsen J, Konig A, Martin C, Chalkley L, Hakenbeck R. Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol Microbiol. 1994;12:1013–1023. doi: 10.1111/j.1365-2958.1994.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 23.Smith A M, Klugman K P. Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1995;39:859–867. doi: 10.1128/aac.39.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubukata K, Asahi Y, Okuzumi K, Konno M the Working Group for Penicillin-Resistant S. pneumoniae. Incidence of penicillin-resistant Streptococcus pneumoniae in Japan, 1993–1995. J Infect Chemother. 1996;1:177–184. doi: 10.1007/BF02350645. [DOI] [PubMed] [Google Scholar]
- 25.Ubukata K, Asahi Y, Yamane A, Konno M. Combinational detection of autolysin and penicillin-binding protein 2B genes of Streptococcus pneumoniae by PCR. J Clin Microbiol. 1996;34:592–596. doi: 10.1128/jcm.34.3.592-596.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ubukata K, Muraki T, Igarashi A, Asahi Y, Konno M. Identification of penicillin and other β-lactam resistance in Streptococcus pneumoniae by polymerase chain reaction. J Infect Chemother. 1997;3:190–197. doi: 10.1007/BF02490033. [DOI] [PubMed] [Google Scholar]
- 27.Ubukata K, Muraki T, Igarashi A, Asahi Y, Konno M the Working Group for Penicillin-Resistant S. pneumoniae. In vitro evaluation of the activity of β-lactams, new quinolones, and other antimicrobial agents against Streptococcus pneumoniae. J Infect Chemother. 1996;2:213–221. doi: 10.1007/BF02355118. [DOI] [PubMed] [Google Scholar]
- 28.Williamson R, Hakenbeck R, Tomasz A. In vitro interaction of β-lactam antibiotics with the penicillin-binding proteins of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980;18:629–637. doi: 10.1128/aac.18.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamane A, Nakano H, Asahi Y, Ubukata K, Konno M. Directly repeated insertion of 9-nucleotide sequence detected in penicillin-binding protein 2B gene of penicillin-resistant Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:1257–1259. doi: 10.1128/aac.40.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]