ABSTRACT
Staphylococcus aureus is a highly infective Gram-positive bacterial pathogen that causes a wide range of diseases in both healthy and immunocompromised individuals. It can evade host immune defenses by expressing numerous virulence factors and toxins. Coupled with the inability of the human host to develop protective immunity against S. aureus, the emergence of antibiotic-resistant strains complicates treatment options. The non-canonical Sts phosphatases negatively regulate signaling pathways in varied immune cell types. To determine the role of the Sts proteins in regulating host responses to a Gram-positive microorganism, we investigated the response of mice lacking Sts expression to S. aureus infection. Herein, we demonstrate that Sts −/− animals are significantly resistant to lethal intravenous doses of S. aureus strain USA300. Resistance is characterized by significantly enhanced survival and accelerated bacterial clearance in multiple peripheral organs. Infected Sts −/− animals do not display increased levels of cytokines TNFα, IFNγ, and IL-6 in the spleen, liver, and kidney during the early stages of the infection, suggesting that a heightened pro-inflammatory response does not underlie the resistance phenotype. In vivo ablation of mononuclear phagocytes compromises the Sts −/− enhanced CFU clearance phenotype. Additionally, Sts −/− bone marrow-derived macrophages demonstrate significantly enhanced restriction of intracellular S. aureus following ex vivo infection. These results reveal the Sts enzymes to be critical regulators of host immunity to a virulent Gram-positive pathogen and identify them as therapeutic targets for optimizing host anti-microbial responses.
KEYWORDS: host-pathogen interactions, Staphylococcus aureus, macrophages, innate immunity
INTRODUCTION
Staphylococcus aureus is a Gram-positive facultative anaerobe that is found widely in the human microbiome, persisting either as a harmless commensal or an intermittent colonizer in over half of the world’s population (1). It can be highly infective and cause a range of diseases in healthy immune-competent individuals, including skin and soft-tissue infections, endocarditis, organ abscesses, pneumonia, and sepsis (2 – 4). S. aureus is also an opportunistic pathogen and is a leading cause of infections of immunocompromised individuals and within healthcare settings (5). Serious S. aureus infections have a mortality rate that exceeds 20%, and the number of S. aureus-attributed deaths in the USA exceeds 20,000 annually (6).
The success of S. aureus as a pathogen rests in part on its ability to evade host immune defenses through the expression of numerous virulence factors and toxins or leukocidins (4). S. aureus secreted immune evasion proteins (estimated to number between 100 and 200) target and impair diverse facets of host immunity, including cells such as T cells, neutrophils, and other phagocytes, elements of the complement cascade, and different clotting factors. Complicating efforts to control S. aureus infections, the human host does not naturally develop protective immunity (7). Further, attempts to develop an effective vaccine strategy have not been successful, despite overwhelming efforts (8, 9). Finally, the emergence and rapid spread over the past several decades of multiple S. aureus strains that are resistant to common antibiotics such as penicillin and penicillinase-resistant beta-lactams has alarmed many public health experts. In the years since methicillin-resistant S. aureus (MRSA) appeared, it has disseminated throughout the world and has led to numerous outbreaks in healthcare and community-wide settings (5). Although a number of antibiotics effective against MRSA have been developed and approved by the FDA in the last decade, a high rate of mortality associated with MRSA infections remains. The continued limitations involved in managing and resolving S. aureus infections highlight the need for additional therapeutic options.
The two Sts enzymes (Sts-1 and Sts-2) are evolutionarily conserved homologous phosphatases that have been shown to have overlapping, if not redundant, signaling functions as negative regulators of diverse cells signaling pathways (10). They are more than 50% identical at the amino acid level, with Sts-1 ubiquitously expressed in a large number of tissues and cell types and Sts-2 possessing a more limited pattern of expression that is primarily confined to cells of hematopoietic origin (11, 12). The two proteins are characterized by a distinct structure consisting of four separate domains. A prominent feature is their unique C-terminal HP phosphatase domain whose distinctive active site architecture and catalytic mechanism differentiates them from both pSer/pThr phosphatases and members of the canonical PTP phosphatase family (13). Of the two, Sts-1 is the more active enzyme in vitro (14). The two enzymes are unique among intracellular phosphatases for having two protein interaction domains not found in any other phosphatase: an N-terminal ubiquitin-association (UBA) domain and a central SH3 domain. These domains are thought to modulate Sts signaling functions (10). Finally, a cyclic nucleotide phosphodiesterase (PDE) enzyme activity associated with the region between the UBA and SH3 domains was recently identified, and a functional role for Sts-1 PDE activity in regulating signaling downstream of different surface receptors has been demonstrated (15). The modular organization of the four Sts protein domains, not found together in any other polypeptide, distinguishes Sts from all other intracellular signaling molecules and suggests they occupy a unique intracellular signaling niche.
Human and mouse Sts-1 and Sts-2 are highly conserved (98% and 83%, respectively). Interestingly, mice lacking Sts-1 and Sts-2 expression (Sts-1/2 −/− or Sts −/−) have no overt phenotypic disorders, although some immune cells derived from Sts −/− mice show heightened responsiveness in vitro (13, 16, 17). The role of the Sts proteins in regulating host-pathogen interactions is an area of active interest that has begun to be explored. Recently, Sts −/− mice were evaluated for susceptibility to bloodstream infection by the clinically relevant fungal pathogen Candida albicans. Unlike wild-type mice, in which rapid fungal proliferation in peripheral organs leads to progressive sepsis and rapid lethality, Sts −/− mice exhibit a profound resistance to infection (18). Importantly, the Sts −/− response is associated with a significant reduction in fungal burden in critical target organs (kidneys), sharply diminished levels of many inflammatory molecules beginning at 24 h post-infection, and an absence of inflammatory lesions. Although the mechanisms underlying the enhanced resistance of Sts −/− animals to C. albicans have not been definitively established, bone marrow monocytes and marrow-derived dendritic cells lacking Sts display enhanced activation of signaling molecules downstream of fungal receptor Dectin-1 and a significantly potentiated reactive oxygen species response (19). A similar in vivo resistance phenotype was observed in the response of Sts −/− mice to the Gram-negative bacterial pathogen Francisella tularensis (Live Vaccine Strain, LVS). Specifically, Sts −/− animals demonstrated significantly reduced mortality following intradermal F. t. LVS infection, with resistance accompanied by enhanced bacterial clearance in multiple peripheral organs (20). Interestingly, Sts −/− marrow monocytes, but not marrow-derived macrophages, demonstrated more effective restriction of LVS than wild-type cells, a phenotype that was dependent on heightened production of IFNγ by Sts −/− cells (21). In sum, results obtained using these two infection models indicate that Sts inactivation leads to distinct immune responses that are uniquely capable of restricting distinct pathogens.
In the present study, we sought to broaden our understanding of the role of the Sts proteins in regulating host anti-microbial immunity. We report here that animals lacking Sts expression display significantly heightened resistance to infection by the Gram-positive bacterium S. aureus. The resistance phenotype is characterized by significantly enhanced survival following lethal inoculums and more rapid bacterial clearance in multiple peripheral organs. Further, Sts −/− macrophages (BMDMs) exhibit accelerated restriction of intracellular bacteria following ex vivo infection. These results identify Sts-1 and Sts-2 as critical regulators of host antistaphylococcal immunity, including macrophage-mediated bacterial killing.
MATERIALS AND METHODS
Mice
The generation of mice containing the Sts mutations, backcrossed 10 generations onto the C57/B6 background, has been described (22). All animals, including Ccr2 −/− and Ccr2-DTR mice (23, 24) (kind gifts from E. Pamer), Sts −/− × Ccr2 −/− and Sts −/− × Ccr2-DTR mice were housed and bred in the Stony Brook University Animal Facility in accordance with Division of Laboratory Animal Resources regulations. Animal protocols followed guidelines established within the “Guide for the Care and Use of Laboratory Animals” (8th ed.) published by the National Research Council of the National Academies. Protocols were approved by the Stony Brook University Institutional Animal Care and Use Committee.
Infections
S. aureus strain USA300 (LAC) was grown overnight at 37°C in tryptic soy broth. On the morning of infection, overnight cultures were diluted 1:100 and grown to an OD600 = 0.4–0.5. Cultures were washed and resuspended in phosphate-buffered saline (PBS) to the desired CFU/mL. For survival studies, 8-week-old female mice were anesthetized with isoflurane and inoculated via the retro-orbital route with a lethal dose (2.5 × 107 CFUs) of S. aureus suspended in 0.1 mL PBS. For survival studies involving Ccr2 −/− and Ccr2-DTR animals, 8-week-old males were inoculated as described above with 7.5 × 107 CFUs S. aureus. Graphing and statistical analysis of survival after infection were carried out using a log-rank test (Mantel-Cox) with Prism (v9) software (Dotmatics, Inc.). For clodronate depletion studies, mice were administered by intravenous injection of 0.5 mg control liposomes or clodronate/liposome formulation (Encapsula NanoSciences) at −24 h and +24 h relative to intravenous infection with S. aureus (7.5 × 104 CFUs). For histology studies, 8-week-old female mice were infected as previously described with a sublethal dose of (2.5 × 106 CFUs) of S. aureus Newman suspended in 0.1 mL PBS. Mice were euthanized at Day 5 for histological and organ burden analysis.
Organ burden analysis
Organs from 8-week-old male mice infected with a sublethal dose of S. aureus (7.5 × 106 CFUs) were harvested, weighed, and homogenized in PBS. Whole blood was collected via cardiac puncture. Homogenates and blood were serially diluted, plated on tryptic soy agar, and incubated at 37°C for 1 day.
Macrophage culture
Marrow cells were cultured in Dulbecco’s modified Eagle’s medium containing 30% L929 cell supernatant, 20% fetal bovine serum (FBS), 1% non-essential amino acid (NEAA), 1% penicillin/streptomycin, and 1 mM sodium pyruvate for 5–7 days. Cell counts were determined using trypan blue staining.
Macrophage infections
For ex vivo infections, cells were harvested, counted, and seeded in triplicate at a density of 5 × 105 cells/well in a 24-well plate, in media containing 15% L929 cell supernatant, 10% FBS, 1% NEAA, and 1 mM sodium pyruvate. Gentamicin protection assays were carried out as previously described (25). Briefly, S. aureus strain USA300 (LAC) was prepared as described above and resuspended in cell culture media without antibiotics. To infect cells, culture media were replaced with medium containing bacteria. Cells were incubated at 37°C for 30 min and then washed with PBS. Media containing gentamicin (300 µg/mL) was added to the cultures for 1 h, following which it was replaced with fresh media containing 100 µg/mL gentamicin for the duration of the infection. The latter media change was designated t = 0. In some cases as noted, the initial gentamicin(+) media was replaced with gentamicin(−) media for the duration of the infection. At intervals thereafter, cells were washed with PBS and lysed with 0.01% Triton in PBS. Lysates were serially diluted, plated, and incubated at 37°C for 24 h, after which colonies were enumerated. Cell images were acquired at 40× on an EVOS M5000 microscope.
May-Gruenwald Giemsa staining
Cells were seeded on glass coverslips and infected as described above. Coverslips were fixed in methanol for 5 min, rinsed with H2O and stained with May-Gruenwald solution (EMS 26250-01) for 2 min, followed by 10% Giemsa solution (EMS 26250-02) for 15 min before air drying and mounting onto glass slides. Staining procedure was adapted from Giemsa staining protocol previously developed (26). Bacteria were counted using ImageJ software, with >300 cells analyzed from 15 different fields of view. Representative results from three infections are displayed. Images (60×) were acquired with a Zeiss Observer D1 Inverted Phase Contrast Fluorescence Microscope, Axiocam 105 color camera, and Zeiss Zen pro microscope software.
Histology
Kidneys isolated from uninfected or infected animals were fixed in buffered formalin, paraffin embedded, and sectioned at 5 µM by the Stony Brook University Histology Core. Sections were counterstained with hematoxylin and eosin (H&E). Digitized slides were produced with PrimeHisto XE Histology Slide Scanner and HistoView Software. Images (60×) were acquired with a Zeiss Observer D1 Inverted Phase Contrast Fluorescence Microscope, Axiocam 105 color camera, and Zeiss Zen pro microscope software. Images were obtained from four sections of varying depth from each mouse. Staphylococcal abscess communities (SACs) within infection foci were quantified using ImageJ Software.
Cytokine analysis
Organs were snap frozen in liquid nitrogen and homogenized in buffer (100 mM Tris pH7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.5% sodium deoxycholate) with 1× protease inhibitor cocktail (Roche). Whole blood was collected by cardiac puncture and allowed to clot at room temperature for 30 min before centrifugation to isolate serum. Clarified homogenates, serum, and cell culture supernatants were analyzed for levels of indicated cytokines using ELISA MAX Standard kits (BioLegend).
Cytotoxicity assays
Culture supernatants were collected after ex vivo bacterial infection and analyzed for the presence of lactate dehydrogenase (LDH) using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega) according to the manufacturer’s instructions. Background LDH release was quantified by evaluating supernatants from uninfected cells. Maximum LDH release was quantified by evaluating supernatants from uninfected cells that were incubated with the provided lysis buffer for 30 min. Percent LDH release was quantified by subtracting background LDH release from all sample values, dividing by maximum LDH release, and multiplying by 100.
RESULTS
Sts −/− mice are highly resistant to bloodstream S. aureus infection
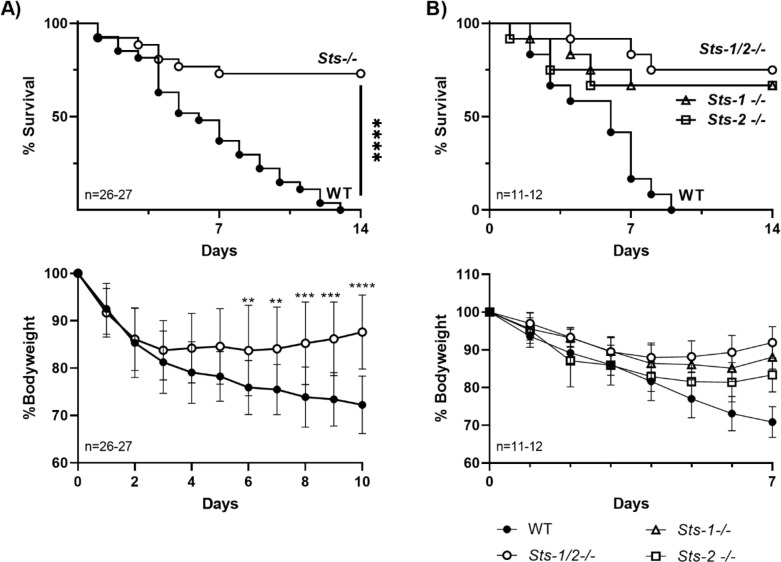
The role of the Sts enzymes in regulating host anti-microbial responses to S. aureus infection was examined by evaluating the susceptibility of Sts −/− mice to intravenous infection. Mice were infected with a lethal inoculum via the retro-orbital route and animals were monitored for 14 days. More than 50% of wild-type control mice became moribund within the first week of infection and all animals succumbed by the end of the 2-week period (Fig. 1A, top). Consistently, wild-type animals also failed to recover from the sharp weight loss that occurred 2–3 days post-infection and continued to lose weight over a 10-day period (Fig. 1A, bottom). In contrast to wild-type mice, Sts −/− mice displayed significantly enhanced survival, with only 30% of the animals in the Sts −/− cohort becoming moribund within the 14-day time frame. In addition, as a group, Sts −/− animals stopped losing weight by 4 days post-infection and subsequently began to recover from their initial weight loss. Survival studies performed with both higher and lower infectious doses displayed similar differences between wild-type versus Sts −/− mice (data not shown).
Fig 1.
Resistance of Sts −/− mice to intravenous S. aureus infection. (A). (top) Mice (females) were infected with S. aureus USA300 (2.5 × 107 CFUs) and monitored for 14 days. Sts −/− mice demonstrated significantly enhanced survival. A cumulative total of 26 mice per genotype were evaluated, in three independent experiments. ****, P < 0.0001 by log-rank Mantel-Cox test. (bottom). In contrast to wild-type mice, Sts −/− mice begin to recover from their initial weight loss by Days 3–4 post-infection. Significant differences in overall weights of the two groups appeared by Days 6–7 post-infection. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.0001 (by two-way ANOVA, and Sidak multiple-comparison test). Male mice demonstrated an identical phenotype. (B) (top) Mice with individual deletions of either Sts-1 or Sts-2 demonstrate significant survival advantage relative to wild-type mice following intravenous infection with S. aureus USA300. A total of 11–12 male mice per genotype were evaluated, in three independent experiments. P values were < 0.01 (log-rank Mantel-Cox test) for Sts-1 −/− versus WT (P = 0.0017) and Sts-2 −/− versus WT (P = 0.0043). P values for Sts-1 −/− versus Sts −/− (dKO) and Sts-2 −/− versus Sts −/− (dKO) were 0.5784 and 0.5409, respectively. (bottom) Unlike wild-type mice, Sts-1 −/− and Sts-2 −/− mice begin to recover from their initial weight loss after Day 6 post-infection (P < 0.0001 for Sts-1 −/− versus WT, P = 0.0007 for Sts-2 −/− versus WT, two-way ANOVA, and Tukey multiple-comparison test).
To investigate the individual contributions of Sts-1 and Sts-2 to the regulation of host immunity to S. aureus infection, we compared the survival of corresponding single mutant mice to wild-type and Sts −/− animals. Both Sts-1 −/− and Sts-2 −/− mice displayed enhanced survival similar to animals lacking expression of both Sts-1 and Sts-2 together (Fig. 1B). Thus, consistent with interesting functional differences between the Sts proteins, including their in vitro phosphatase activities and their patterns of tissue expression (10, 12, 14), they appear to carry out non-redundant functions in regulating the overall host response to S. aureus infection. Altogether, these results demonstrate that mice lacking either Sts-1 and Sts-2 expression individually or together are able to resist lethal systemic doses of S. aureus strain USA300, a highly pathogenic Gram-positive bacterium.
Accelerated bacterial clearance in Sts −/− mice
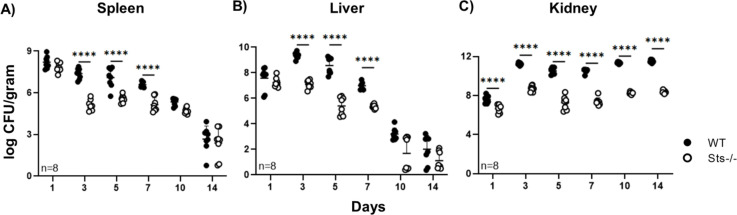
The enhanced ability of Sts −/− mice to survive a high-dose bloodstream S. aureus infection suggested they could either tolerate high bacterial burdens more effectively than wild-type mice or they were better at overcoming bacterial virulence mechanisms and clearing the infection, or a combination of both. To determine how Sts inactivation impacts the kinetics of S. aureus clearance, we infected mice and subsequently assessed bacterial load in peripheral tissues. At 4h post-infection, both wild-type and Sts −/− mice had identical CFUs in the bloodstream, liver, spleen, and kidney (Fig. S1), suggesting that initial bacterial dissemination occurs similarly in both strains. On Day 1, wild-type and Sts −/− spleen and liver also contained similarly high CFUs, ~107 CFU/g (Fig. 2A and B). However, by Day 3 and beyond, as the infection was cleared following mobilization of the host response, the bacterial load in Sts −/− spleen and liver was consistently 10- to 100-fold lower than in wild-type organs (Fig. 2A and B). Kidney CFUs were also significantly different, with Day 1 CFUs displaying close to a 1-log difference for Sts −/− versus wild-type animals and almost three orders of magnitude reduced bacterial load in Sts −/− kidneys for the remainder of the time course (Fig. 2C).
Fig 2.
Accelerated bacterial clearance in peripheral organs of Sts −/− mice. Bacterial burden in (A) spleen, (B) liver, and (C) kidneys at indicated time points following infection of 8-week-old male mice (7.5 × 106 S. aureus USA300 CFUs/mouse). Cumulative data compiled from two separate experiments with four mice per group. ****, P < 0.0005 (by two-way ANOVA and Sidak multiple-comparison test). Female mice demonstrated an identical response.
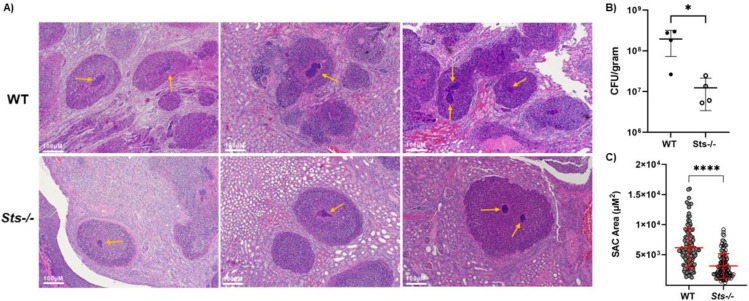
In addition to the overall CFU differences between wild-type versus Sts −/− kidneys, we were able to discern a measurable difference in the apparent sizes of the SACs that form at the center of abscess lesions in wild-type versus Sts −/− kidneys (27). SACs are formed by a dense cluster of bacterial cells and as illustrated in Fig. 3, the average cross-sectional area of numerous visible SACs that form in Day 5 infected kidneys is significantly reduced in Sts −/− kidneys relative to wild-type kidneys.
Fig 3.
Altered Staphylococcal abscess communities (SACs) within Sts −/− kidney abscesses. (A) Representative H&E-stained histological sections of wild-type and Sts −/− kidneys 5 days after intravenous infection (S. aureus strain Newman, 2.5 × 106 CFUs/mouse) of 8-week-old female mice. SACs are indicated by yellow arrows. (B) Reduced S. aureus strain Newman CFUs in Sts −/− kidneys on Day 5 p.i., relative to CFUs in wild-type kidneys. Cumulative data compiled from two separate experiments with two mice per group (inoculum = 2.5 × 106). *, P < 0.05 (Mann-Whitney t test). (C) Average cross-sectional area of SACs within Sts −/− kidneys is significantly reduced relative to the average cross-sectional area of wild-type SACs. n = 113 (wild type), 125 (Sts −/−). Cumulative data compiled from two separate experiments with four mice per group. ****, P < 0.0005 (Mann-Whitney t test).
Similar expression of key pro-inflammatory cytokines in wild-type and Sts −/− infected animals
The Sts proteins have been shown to negatively regulate the expression of diverse cytokines, including pro-inflammatory cytokines TNFα and IFNγ (12, 28). Nakane et al. identified opposing roles for tissue IFNγ and TNFα in the host response to S. aureus, with TNFα playing a positive role and IFNγ expression having detrimental effects (29, 30). Because the enhanced ability of Sts −/− mice to resist high-dose bloodstream S. aureus infection might be due to an underlying imbalance in levels of expression of critical pro-inflammatory cytokines, we evaluated levels of IFNγ, TNFα, and IL-6 in tissues of infected wild-type and Sts −/− animals. However, no significant differences in the tissue levels of the three important pro-inflammatory cytokines during early time points, except for 6.5 h p.i. liver expression of IL-6, which was significantly elevated in wild-type tissues relative to Sts −/− tissue (Fig. S2).
Mononuclear phagocytes contribute to the enhanced CFU clearance in Sts −/− animals
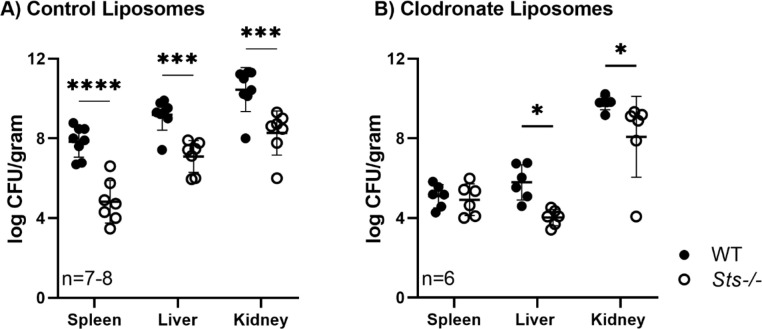
Having observed the ability of Sts −/− mice to survive high-dose S. aureus infection, we next investigated underlying cellular mechanisms. The enhanced suppression of bacterial growth evident in Sts −/− infected tissues at early time points after infection (see Fig. 2) suggests the involvement of elements of the innate immune system. Therefore, we utilized chemical-mediated cell ablation to investigate the contribution of Sts −/− mononuclear phagocytes. Clodronate treatment abrogated the enhanced CFU clearance phenotype normally observed in Day 3 Sts −/− spleens (Fig. 4), suggesting mononuclear phagocytes play a critical role in the spleen. Kidney and liver CFUs in Day 3 clodronate-treated Sts −/− animals, however, were reduced relative to clodronate-treated wild-type animals. These data implicate Sts as an important regulator of splenic mononuclear phagocyte anti-microbial responses in vivo, but suggest they also regulate the responses of additional critical innate cell types in other organs such as the liver and kidney.
Fig 4.
Ablation of mononuclear phagocytes partially abrogates Sts −/− enhanced clearance phenotype. Eight-week-old male mice were treated with control liposomes (A) or liposomes loaded with clodronate (B) prior to infection with S. aureus USA300 (7.5 × 107 or 7.5 × 104 CFUs/mouse, respectively). Cumulative data compiled from two separate experiments with three to four mice per group. *, P < 0.05; ***, P < 0.005; ****, P < 0.0001 by two-way ANOVA and Sidak multiple-comparison test.
To begin to address the identity of the phagocyte population(s) that play critical non-redundant roles in the Sts −/− enhanced resistance phenotype, we investigated the role for Ccr2+ inflammatory monocytes (IMs) (31). In animals lacking expression of the chemokine receptor Ccr2, IM recruitment to peripheral tissues following bacterial infection is impaired (32). We generated Sts −/− animals that lacked expression of Ccr2 (Sts −/− × Ccr2 −/− triple knockout animals) and evaluated their response to systemic S. aureus infection. As illustrated in Fig. S3A, Sts −/− animals lacking Ccr2 expression demonstrated enhanced survival following high-dose S. aureus infection, relative to Sts +/+ × Ccr2 −/− animals. We also evaluated Sts−/− animals that express the Ccr2-DTR transgene, within which rapid and targeted deletion of IMs occurs following administration of diphtheria toxin (DT) (24). As illustrated in Fig. S3B, Sts −/− × Ccr2-DTR mice treated with DT display significantly enhanced survival relative to Sts +/+ × Ccr2-DTR mice treated with DT. Taken together, these results suggest that Ccr2+ inflammatory monocytes do not play an essential nonredundant role in the Sts −/− enhanced survival phenotype.
Macrophages lacking Sts expression demonstrate increased resilience to S. aureus infection
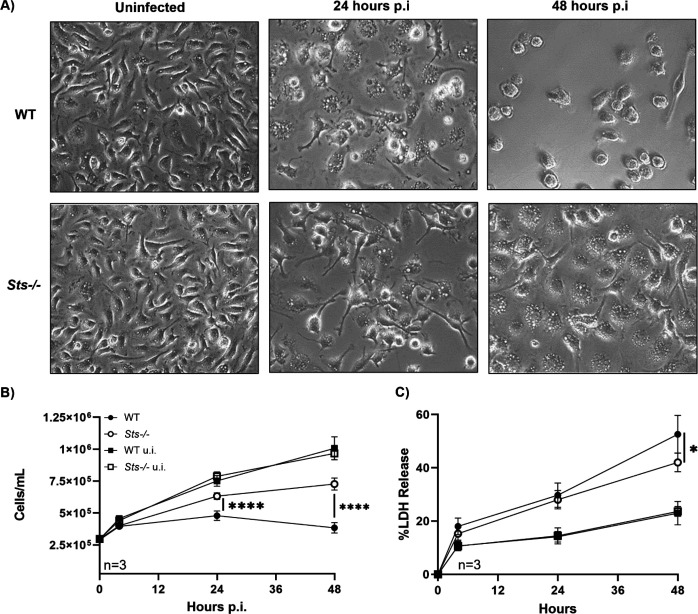
Macrophages are known to play a critical role in containing and resolving S. aureus infections (33, 34). However, despite macrophages possessing a wide array of potent anti-microbial defenses, recent studies have also demonstrated their susceptibility to S. aureus-mediated cytotoxicity. For example, S. aureus can survive and proliferate within the inhospitable interior of the macrophage phagolysosome, eventually triggering apoptosis and cell rupture (35). To investigate a potential role for Sts in regulating phagocyte anti-microbial responses, we utilized bone marrow-derived macrophages (BMDMs). Uninfected wild-type and Sts −/− BMDMs displayed no overt differences in appearance or growth rate when cultured in vitro (Fig. 5A and B). Following infection with S. aureus, however, BMDMs prepared from wild-type and Sts −/− animals responded differently. While both groups of cells took on the typical appearance of activated macrophages at 24 h post-infection (Fig. 5A, middle panels), the growth rate of infected wild-type BMDMs slowed considerably during the first 24 h p.i. and then they began to die by 24–48 h p.i. In contrast, Sts −/− BMDM cultures continued to expand during the 0- to 48-h period following infection, albeit at a lesser rate than observed for uninfected cultures (Fig. 5B). Concomitantly, levels of LDH in culture supernatants, indicative of cell damage, were also significantly lower in infected Sts −/− cultures versus infected wild-type cultures (Fig. 5C). Therefore, lack of Sts expression protects BMDMs from S. aureus infection-related cytotoxicity.
Fig 5.
Reduced S. aureus-induced cytotoxicity exhibited by Sts −/− macrophages. (A) By 48 h p.i., the increased susceptibility of wild-type cells versus Sts −/− to S. aureus-induced cytotoxicity is visible in ex vivo culture. Brightfield images (40×) depict cells before (left) and 24 h (middle) or 48 h (right) post-infection. (B) Uninfected wild-type macrophages (black squares) and Sts −/− macrophages (white squares) display identical growth rates. Infected wild-type macrophages cultures (black circles) exhibit significantly reduced cell numbers relative to infected Sts −/− cultures (white circles). (C) Infected wild-type macrophages release more LDH into culture medium than Sts −/− BMDMs (black and white circles, respectively). LDH release (%) is normalized to LDH in culture medium of lysed BMDMs (100%). (LDH levels in uninfected wild-type and Sts −/− cultures, black and white squares, respectively). Results represent the mean ± SD of three independent experiments. ****, P < 0.0001 by two-way ANOVA and Sidak multiple-comparison test.
Enhanced restriction of intracellular S. aureus by Sts −/− macrophages
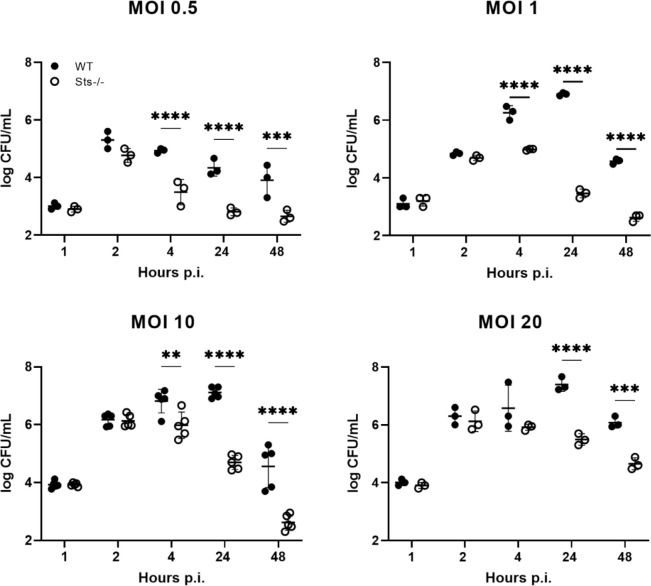
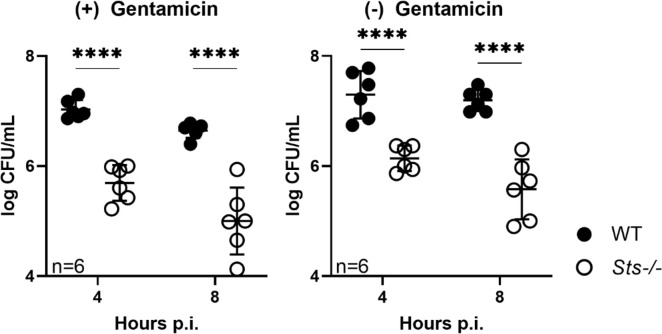
To evaluate the effects of Sts inactivation on the dynamic interactions between macrophages and S. aureus following infection, we established BMDM cultures, infected them with S. aureus at different multiplicities of infection (MOIs), and evaluated bacterial intracellular growth by plating for CFUs at regular intervals post-infection. At 1 h post-infection, similar CFUs were recovered from wild-type and Sts −/− cultures, suggesting similar levels of bacterial uptake in the two cultures (Fig. 6). At the end of the subsequent hour, the two cultures also displayed similar levels of intracellular CFUs, indicating similar bacterial proliferation in wild-type versus Sts −/− cells during the first 2 h post-infection. By 4 h post-infection, however, the number of CFUs isolated from Sts −/−−/− cells were significantly decreased relative to wild-type CFUs in all infected cultures except for those infected with MOI 20, and by 24 h post-infection Sts −/− cells displayed a 2–3 log-fold reduction in levels of intracellular S. aureus relative to wild-type cells, regardless of initial MOI (Fig. 6). The difference in intracellular CFUs between wild-type and Sts −/− cells was also maintained after 24 h (Fig. 6). Importantly, we obtained similar results when ex vivo infected macrophage cultures were not exposed to prolonged extracellular antibiotic treatment during the course of infection (Fig. 7).
Fig 6.
Enhanced restriction of S. aureus within Sts −/− bone marrow-derived macrophages. Wild-type and Sts −/− BMDMs were infected ex vivo at MOIs 0.5, 1, 10, and 20, cells were lysed at the indicated time points and bacterial CFUs enumerated. Results are representative of three to five5 independent experiments. **, P < 0.01; ***, P < 0.005; ****, P < 0.0001 (by two-way ANOVA and Sidak multiple-comparison test).
Fig 7.
Enhanced restriction of S. aureus within Sts −/− macrophages in the absence of extracellular antibiotic. BMDMs were infected ex vivo at MOI 10. Following 60 min incubation with media containing gentamicin to eliminate extracellular bacteria, cells were incubated for the indicated time points in gentamicin(+) media (left) or gentamicin(−) (right) media, lysed at the indicated time points, and bacterial CFUs enumerated.
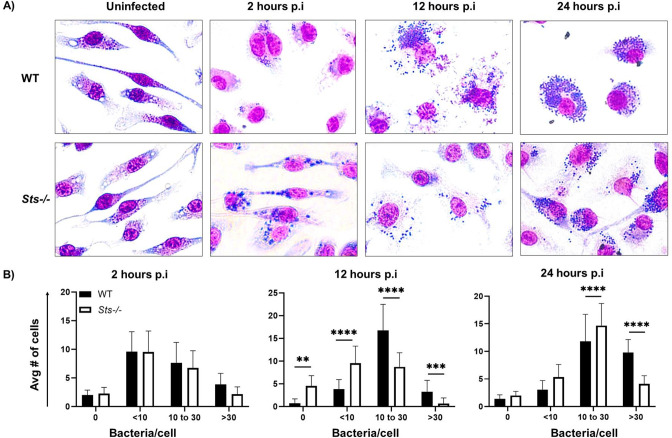
We then supplemented the bulk culture macrophage infection studies by tracking S. aureus intracellular growth within macrophages at the single-cell level. Macrophages were infected (MOI 10) and stained with Giemsa at regular intervals post-infection (Fig. 8). We then determined the distribution of cells containing different levels of intracellular bacteria at various time points post-infection. At early time points, wild-type and Sts −/− cells had a similar distribution of bacteria per cell, with the majority of cells containing less than 10 S. aureus/cell at 2 h p.i. As the infection progressed, however, we observed a significant shift in the levels of bacteria within individual cells of the different cultures. In particular, at 12 h p.i., the majority of wild-type cells contained 10–30 bacteria/cell while the majority of Sts −/− cells contained fewer than 10 bacteria/cell. Likewise, at 24 h p.i., a significantly greater proportion of wild-type cells displayed >30 bacterium/cell than Sts −/− cells (Fig. 8). Altogether, these results indicate that inactivation of the Sts proteins markedly enhances the ability of macrophages to restrict intracellular S. aureus.
Fig 8.
Single-cell analysis reveals altered distribution of S. aureus within Sts −/− BMDMs. (A) Following infection (MOI 10) of wild-type and Sts −/− BMDMs, cells were fixed and stained with May-Grunwald Giemsa dyes at the indicated time points. Representative images illustrate the increased bacterial burden of wild-type BMDMs at 12 h and 24 h post-infection relative to Sts −/− BMDMs (cytoplasm pink, cell nuclei purple, and bacteria dark blue). (B) Cells were categorized by the number of discrete intracellular bacteria (0, 10, 10–30, and <30). Approximately ~350 cells were counted from a total of 15 random fields of view, quantifying the number of cells containing different bacteria within each field. A greater distribution of wild-type cells contains 10–30 (12 h) and 30+ (24 h) bacteria per cell than observed in Sts −/− cultures. ****, P ≤ 0.0001 (by two-way ANOVA and Sidak multiple-comparison test).
We also evaluated expression levels of the pro-inflammatory cytokines IFNγ, TNFα, and IL-6 within the macrophage cultures. No significant differences in the levels of the three important pro-inflammatory cytokines within culture supernatants during early time points (Fig. S4), suggesting the accelerated decrease in CFUs within Sts −/− cells did not occur as a result of increased signaling downstream of the critical pro-inflammatory cytokines IFNγ, TNFα, and IL-6 (Fig. S4).
DISCUSSION
Infectious diseases are leading contributors to the global disease burden, causing persistently high morbidity and mortality in underdeveloped countries (36, 37). Intractable infections are also on the rise in developed nations, especially among the elderly and immunocompromised (38). In many cases, a lack of effective vaccine strategies coupled with the difficulty of the human host to develop adequate protective immunity contributes to high mortality (39 – 41). Although antibiotics continue to be the most important front-line defense against such infections, many that are in current use suffer from a number of drawbacks, including toxicity, limited bioavailability, and a narrow spectrum of activity (42). Additionally, the continual threat posed by the emergence of antibiotic resistance is of deep concern. Therefore, there is great interest in developing additional therapeutic options (43).
In recent years, we have reported that mice lacking expression of the two noncanonical Sts phosphatases, Sts-1 and Sts-2, are highly resistant to infection by two distinct pathogens, the commensal fungus C. albicans and the Gram-negative bacterium F. tularensis LVS (18, 20). These previous studies established the Sts enzymes as critical regulators of host anti-microbial immunity and highlighted that Sts inactivation could lead to enhanced protection against diverse pathogens. In the current study, we sought to broaden our understanding of the role of Sts in regulating host responses to common microbial infections by examining how Sts −/− mice respond to challenge with the virulent Gram-positive pathogen S. aureus. Our results illustrate that mice lacking Sts expression exhibit profound resistance to lethal bloodstream inoculums of S. aureus, a mouse infection model that recapitulates aspects of severe sepsis in humans. The resistance phenotype is characterized by significantly enhanced survival and accelerated bacterial clearance in multiple peripheral tissues. Given the similarities in the overall response of Sts −/− animals to C. albicans, F. tularensis, and S. aureus infections, we conclude that the Sts proteins regulate a key host pathway(s) that is important for the immunological control of diverse pathogens. Further, given Sts' established role as a negative regulator of select cell signaling pathways, we hypothesize that Sts inactivation potentiates critical anti-microbial responses within essential populations of responding cells. Importantly, we demonstrate that macrophages lacking Sts expression, when infected ex vivo, possess a significantly enhanced ability to restrict the growth of intracellular S. aureus. As macrophages play an important role in the control of S. aureus infections (33, 34), this latter observation suggests that altered macrophage responses could underlie aspects of the Sts −/− host resistance phenotype. This is supported by in vivo ablation studies in which targeted elimination of mononuclear phagocytes leads to less pronounced organ CFU differences that are normally observed between wild-type and Sts −/− animals.
Interestingly, over the course of analyzing ex vivo infection of Sts −/− phagocytes with three distinct pathogens, we have observed a degree of cell specificity that accompanies the responses to each pathogen. For example, for C. albicans, we observed that Sts −/− BMDCs but not BMDMs or BM-derived monocytes, displayed increases in growth-suppressing activity relative to corresponding wild-type cells (19). Further, in the context of infection by F. tularensis, Sts −/− BM-derived monocytes, but not BMDMs or BMDCs, demonstrated enhanced bactericidal activity that was accompanied by significantly reduced infection-induced cytotoxicity (21). Finally, for S. aureus, we observed that only Sts −/− BMDMs, but not BMDCs, displayed the resistance phenotype. The underlying origins of this cell specificity are currently unclear, although we speculate that it is related to pathogen-induced intracellular innate immune responses that are uniquely induced within each cell type.
In the context of the Sts −/− in vivo response to S. aureus infection, it is currently unclear which cellular immune components contribute to the enhanced resistance. Based on preliminary ex vivo infection studies, macrophages or macrophage-like cells are a leading cellular candidate (Fig. 5–7). For example, liver Kupffer cells (KCs) are macrophage-like cells that play an essential role in clearing bloodstream S. aureus (44, 45), and the accelerated bacterial clearance observed in Sts −/− livers could be linked to enhanced KC microbicidal functionality. To address this possibility, it will be important to compare directly the abilities of wild-type versus Sts −/− KCs to restrict intracellular S. aureus ex vivo or evaluate their in vivo functional role using the infection model described herein. It would also be instructive to examine the ex vivo antimicrobial responses of additional resident macrophage-like cell populations, including kidney resident phagocytes, alveolar macrophages, or peritoneal macrophages.
The identity of additional cell populations that contribute to the Sts −/− resistance phenotype is an important question. Intriguingly, we observed that functional targeting of Ccr2+ inflammatory monocytes through the use of the Ccr2 −/− and Ccr2-DTR mouse models did not eliminate the enhanced survival phenotype exhibited by Sts −/− animals. This suggests that inflammatory monocytes that are recruited from the bone marrow to peripheral tissues following infection do not play an essential nonredundant role in the enhanced survival of lethally infected Sts −/− animals. Regarding additional cell types that may contribute to the Sts −/− phenotype, neutrophils are an important candidate. Neutrophils are rapidly recruited to sites of infection, engulfing and killing pathogens while simultaneously expressing cytokines to support the anti-microbial activities of other immune cells. Further investigations will be necessary to determine whether they contribute to the increased resistance to bloodstream S. aureus infection that is evident in Sts −/− animals.
In a similar vein, the intracellular pathway(s) regulated by Sts that are involved in phagocyte anti-microbial responses are currently unclear. Previous studies identified non-redundant roles for Sts in negatively regulating receptor proximal signaling elements in diverse immune cell types. For example, in T cells, they target the kinase Zap-70 downstream of TCR activation (12). Similar studies illustrated a role for Sts-1 in targeting the Zap-70 homolog Syk in platelets and mast cells (16, 17). Interestingly, Syk is also activated downstream of the C-type lectin fungal receptor Dectin 1 and it is hyper-phosphorylated in Sts −/− BMDCs following stimulation with live or heat-killed C. albicans (19). The identity of the intracellular pathways regulated by Sts during the macrophage response to S. aureus infection is under active investigation.
In our current model, the Sts proteins negatively regulate a signaling pathway upstream of a critical phagocyte anti-microbial effector response that engages a wide variety of pathogens. In this model, Sts inactivation would potentiate effector activity in such a way as to shift the interactions between phagocyte and pathogen in favor of the phagocyte, thereby enabling enhanced pathogen clearance. Such a shift in activation levels are likely most impactful early during the course of an infection, prior to the generation of widespread tissue damage that can occur during an exuberant antimicrobial inflammatory response. It is intriguing to consider the possibility that targeted transient inactivation of Sts-1/2 could enhance host antimicrobial immunity and provide a therapeutic benefit.
ACKNOWLEDGMENTS
The authors also thank Laurie Levine, Jean Rooney, Sandy Scherrer, and the Stony Brook Department of Laboratory Animal Services for help with animal procedures and care, the Stony Brook Research Histology Core Lab for help with histological analysis, and Neena Kaur, Hara Seo, and JoAnn Mugavero for additional assistance. The authors would also like to thank Dominique Missiakas for S. aureus strains USA300 & Newman and for important discussions.
This work was supported by the Stony Brook University startup funds (H.K.K.) and funds from the National Institutes of Health: R01AI141592 (N.C.) and R21AI156238 (N.C.). The authors thank the Stony Brook University Department of Microbiology and Immunology for additional support. Additionally, research in this publication was supported by the Center for Biotechnology, a New York State Center for Advanced Technology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Center for Biotechnology. Additional support was provided by Stony Brook University.
We declare no financial conflict of interest.
Contributor Information
Nick Carpino, Email: nicholas.carpino@stonybrook.edu.
Victor J. Torres, St Jude Children's Research Hospital, Memphis, Tennessee, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/iai.00260-23.
Fig. S1 to S4.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Thomer L, Schneewind O, Missiakas D. 2016. Pathogenesis of Staphylococcus aureus bloodstream infections. Annu Rev Pathol Mech Dis 11:343–364. doi: 10.1146/annurev-pathol-012615-044351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Okumura CYM, Nizet V. 2014. Subterfuge and sabotage: evasion of host innate defenses by invasive gram-positive bacterial pathogens. Annu Rev Microbiol 68:439–458. doi: 10.1146/annurev-micro-092412-155711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kwiecinski JM, Horswill AR. 2020. Staphylococcus aureus bloodstream infections: pathogenesis and regulatory mechanisms. Curr Opin Microbiol 53:51–60. doi: 10.1016/j.mib.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de NWM, Kessel KPM, Strijp JAG. 2019. Immune evasion by Staphylococcus aureus. Microbiol Spectr 7. doi: 10.1128/microbiolspec.GPP3-0061-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 6. Lee AS, de Lencastre H, Garau J, Kluytmans J, Malhotra-Kumar S, Peschel A, Harbarth S. 2018. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers 4:18033. doi: 10.1038/nrdp.2018.33 [DOI] [PubMed] [Google Scholar]
- 7. Proctor RA. 2019. Immunity to Staphylococcus aureus: implications for vaccine development. Microbiol Spectr 7:GPP3–0037. doi: 10.1128/microbiolspec.GPP3-0037-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Proctor RA. 2015. Recent developments for Staphylococcus aureus vaccines: clinical and basic science challenges. Eur Cell Mater 30:315–326. doi: 10.22203/ecm.v030a22 [DOI] [PubMed] [Google Scholar]
- 9. Miller LS, Fowler VG, Shukla SK, Rose WE, Proctor RA. 2020. Development of a vaccine against Staphylococcus aureus invasive infections: evidence based on human immunity, genetics and bacterial evasion mechanisms. FEMS Microbiol Rev 44:123–153. doi: 10.1093/femsre/fuz030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsygankov AY. 2020. TULA proteins as signaling regulators. Cell Signal 65:109424. doi: 10.1016/j.cellsig.2019.109424 [DOI] [PubMed] [Google Scholar]
- 11. Carpino N, Kobayashi R, Zang H, Takahashi Y, Jou S-T, Feng J, Nakajima H, Ihle JN. 2002. Dentification, cDNA cloning, and targeted deletion of p70, a novel, ubiquitously expressed SH3 domain-containing protein. Mol Cell Biol 22:7491–7500. doi: 10.1128/MCB.22.21.7491-7500.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carpino N., Turner S, Mekala D, Takahashi Y, Zang H, Geiger TL, Doherty P, Ihle JN. 2004. Regulation of ZAP-70 activation and TCR signaling by two related proteins, Sts-1 and Sts-2. Immunity 20:37–46. doi: 10.1016/s1074-7613(03)00351-0 [DOI] [PubMed] [Google Scholar]
- 13. Rigden DJ. 2008. The histidine phosphatase superfamily: structure and function. Biochem J 409:333–348. doi: 10.1042/BJ20071097 [DOI] [PubMed] [Google Scholar]
- 14. Mikhailik A, Ford B, Keller J, Chen Y, Nassar N, Carpino N. 2007. A phosphatase activity of Sts-1 contributes to the suppression of TCR signaling. Mol Cell 27:486–497. doi: 10.1016/j.molcel.2007.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yin Y, Frank D, Zhou W, Kaur N, French JB, Carpino N. 2020. An unexpected 2-histidine phosphoesterase activity of suppressor of T cell receptor signaling protein-1 contributes to the suppression of cell signaling. J Biol Chem 295:8514–8523. doi: 10.1074/jbc.RA120.013482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Castro RO, Zhang J, Groves JR, Barbu EA, Siraganian RP. 2012. Once phosphorylated, tyrosines in carboxyl terminus of protein-tyrosine kinase Syk interact with signaling proteins, including TULA-2, a negative regulator of mast cell degranulation. J Biol Chem 287:8194–8204. doi: 10.1074/jbc.M111.326850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thomas DH, Getz TM, Newman TN, Dangelmaier CA, Carpino N, Kunapuli SP, Tsygankov AY, Daniel JL. 2010. A novel histidine tyrosine phosphatase, TULA-2, associates with Syk and negatively regulates GPVI signaling in platelets. Blood 116:2570–2578. doi: 10.1182/blood-2010-02-268136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Naseem S, Frank D, Konopka JB, Carpino N. 2015. Protection from systemic Candida albicans infection by inactivation of the Sts phosphatases. Infect Immun 83:637–645. doi: 10.1128/IAI.02789-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frank D, Naseem S, Russo GL, Li C, Parashar K, Konopka JB, Carpino N, Klein BS, Alspaugh JA. 2018. Phagocytes from mice lacking the Sts phosphatases have an enhanced antifungal response to Candida albicans. mBio 9:e00782-18. doi: 10.1128/mBio.00782-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parashar K, Kopping E, Frank D, Sampath V, Thanassi DG, Carpino N. 2017. Increased resistance to intradermal Francisella tularensis LVS infection by inactivation of the Sts phosphatases. Infect Immun 85:e00406-17. doi: 10.1128/IAI.00406-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parashar K, Carpino N. 2020. A role for the Sts phosphatases in negatively regulating IFNγ-mediated production of nitric oxide in monocytes. Immun Inflamm Dis 8:523–533. doi: 10.1002/iid3.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. San Luis B, Sondgeroth B, Nassar N, Carpino N. 2011. Sts-2 is a phosphatase that negatively regulates zeta-associated protein (ZAP)-70 and T cell receptor signaling pathways. J Biol Chem 286:15943–15954. doi: 10.1074/jbc.M110.177634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuziel WA, Morgan SJ, Dawson TC, Griffin S, Smithies O, Ley K, Maeda N. 1997. Severe reduction in leukocyte adhesion and monocyte extravasation in mice deficient in CC chemokine receptor 2. Proc Natl Acad Sci U S A 94:12053–12058. doi: 10.1073/pnas.94.22.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hohl TM, Rivera A, Lipuma L, Gallegos A, Shi C, Mack M, Pamer EG. 2009. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe 6:470–481. doi: 10.1016/j.chom.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sokolovska A, Becker CE, Stuart LM. 2012. Measurement of phagocytosis, phagosome acidification, and intracellular killing of Staphylococcus aureus. Curr Protoc Immunol Chapter 14:14. doi: 10.1002/0471142735.im1430s99 [DOI] [PubMed] [Google Scholar]
- 26. Kobayashi S, Saio M, Fujimori M, Hirato J, Oyama T, Fukuda T. 2020. Macrophages in Giemsa-stained cerbrospinal fluid specimens predict carcinomatous meningitis. Oncol Lett 20:352. doi: 10.3892/ol.2020.12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thammavongsa V, Kim HK, Missiakas D, Schneewind O. 2015. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol 13:529–543. doi: 10.1038/nrmicro3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Newman TN, Liverani E, Ivanova E, Russo GL, Carpino N, Ganea D, Safadi F, Kunapuli SP, Tsygankov AY. 2014. Members of the novel UBASH3/STS/TULA family of cellular regulators suppress T-cell-driven inflammatory responses in vivo. Immunol Cell Biol 92:837–850. doi: 10.1038/icb.2014.60 [DOI] [PubMed] [Google Scholar]
- 29. Nakane A, Okamoto M, Asano M, Kohanawa M, Minagawa T. 1995. Endogenous gamma interferon, tumor necrosis factor, and Interleukin-6 in Staphylococcus aureus infeciton in mice. Infect Immun 63:1165–1172. doi: 10.1128/iai.63.4.1165-1172.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sasaki S, Nishikawa S, Miura T, Mizuki M, Yamada K, Madarame H, Tagawa YI, Iwakura Y, Nakane A. 2000. Interleukin-4 and Interleukin-10 are involved in host resistance to Staphylococcus aureus infection through regulation of gamma interferon. Infect Immun 68:2424–2430. doi: 10.1128/IAI.68.5.2424-2430.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Serbina NV, Jia T, Hohl TM, Pamer EG. 2008. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol 26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Serbina NV, Pamer EG. 2006. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol 7:311–317. doi: 10.1038/ni1309 [DOI] [PubMed] [Google Scholar]
- 33. Flannagan RS, Heit B, Heinrichs DE. 2015. Antimicrobial mechanisms of macrophages and the immune evasion strategies of Staphylococcus aureus. Pathogens 4:826–868. doi: 10.3390/pathogens4040826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pidwill GR, Gibson JF, Cole J, Renshaw SA, Foster SJ. 2021. The role of macrophages in Stahylococcus aureus infection. Front. Immunol 11. doi: 10.3389/fimmu.2020.620339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Flannagan RS, Heit B, Heinrichs DE. 2016. Intracellular replication of Staphylococcus aureus in mature phagolysosomes in macrophages precedes host cell death, and bacterial escape and dissemination. Cell Microbiol 18:514–535. doi: 10.1111/cmi.12527 [DOI] [PubMed] [Google Scholar]
- 36. Baker RE, Mahmud AS, Miller IF, Rajeev M, Rasambainarivo F, Rice BL, Takahashi S, Tatem AJ, Wagner CE, Wang L-F, Wesolowski A, Metcalf CJE. 2022. Infectious disease in an era of global change. Nat Rev Microbiol 20:193–205. doi: 10.1038/s41579-021-00639-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brower JL. 2018. The threat and response of infectious diseases. Microb Ecol 76:19–36. doi: 10.1007/s00248-016-0806-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McGrath B, Broadhurst M, Roman C. 2020. Infectious disease considerations in immuno-compromised patients. JAAPA 33:16–25. doi: 10.1097/01.JAA.0000694948.01963.f4 [DOI] [PubMed] [Google Scholar]
- 39. Maslow JN. 2019. Challenges and solutions in the development of vaccines against emerging and neglected infectious diseases. Hum Vaccin Immunother 15:2230–2234. doi: 10.1080/21645515.2019.1661209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trovato M, Sartorius R, D’Apice L, Manco R, De Berardinis P. 2020. Viral emerging diseases: challenges in developing vaccination strategies. Front Immunol 11:2130. doi: 10.3389/fimmu.2020.02130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Excler J-L, Saville M, Berkley S, Kim JH. 2021. Vaccine development for emerging infectious diseases. Nat Med 27:591–600. doi: 10.1038/s41591-021-01301-0 [DOI] [PubMed] [Google Scholar]
- 42. Fouladkhah AC, Thompson B, Camp JS. 2020. The threat of antibiotic resistance in changing climate. Microorganisms 8:748. doi: 10.3390/microorganisms8050748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zumla A, Rao M, Wallis RS, Kaufmann SHE, Rustomjee R, Mwaba P, Vilaplana C, Yeboah-Manu D, Chakaya J, Ippolito G, Azhar E, Hoelscher M, Maeurer M, Host-Directed Therapies Network consortium . 2016. Host-directed therapies for infectious diseases: current status, recent progress, and future prospects. Lancet Infect Dis 16:e47–63. doi: 10.1016/S1473-3099(16)00078-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Surewaard BGJ, Deniset JF, Zemp FJ, Amrein M, Otto M, Conly J, Omri A, Yates RM, Kubes P. 2016. Identification and treatment of the Staphylococcus aureus reservoir in vivo. J Exp Med 213:1141–1151. doi: 10.1084/jem.2016033411032016c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pollitt EJG, Szkuta PT, Burns N, Foster SJ. 2018. Staphylococcus aureus infection dynamics. PLoS Pathog 14:e1007112. doi: 10.1371/journal.ppat.1007112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4.