Abstract
The 47-kbp plasmid pGFT1 from Salmonella enterica subsp. enterica serovar Dublin mediated tetracycline resistance via a tet(A) gene located on an integrated copy of a Tn1721-analogous transposon. The integration site of the transposon was located within the reading frame of a fip gene. Plasmid pGFT1 was shown to be conjugative and to be able to replicate and express tetracycline resistance in Escherichia coli.
Salmonella enterica subsp. enterica serovar Dublin (referred to herein as S. dublin) isolates are common causes of severe infections in cattle (4). Clinical S. dublin infections are characterized by fever, diarrhea, circulatory collapses, septicemia, pneumonia, or diseases of the central nervous system. Although S. dublin is considered a cattle-adapted pathogen, it may also cause severe systemic infections in humans (2, 14). Appropriate antibiotic treatment is required to control systemic forms of S. dublin infections (4). So far, very little is known about antibiotic resistance of S. dublin isolates.
In the present study, we examined 17 S. dublin isolates collected at the Veterinary Faculty of the University of Leipzig, Leipzig, Germany, from independent cases of septicemia, pneumonia, nervous disorder, or diarrhea in cattle for their resistance to ampicillin (10 μg), cefazolin (30 μg), chloramphenicol (30 μg), gentamicin (10 μg), kanamycin (30 μg), nalidixic acid (30 μg), neomycin (30 μg), streptomycin (10 μg), sulfamethoxazole (23.75 μg), tetracycline (30 μg), and trimethoprim (5 μg) by the agar diffusion assay (17). All S. dublin isolates proved to be susceptible to all of the antibiotics tested except for four isolates that exhibited resistance to tetracycline. To confirm the epidemiological unrelatedness of the isolates suggested by their history, the four tetracycline-resistant S. dublin isolates were investigated by macrorestriction analysis using the restriction enzymes XbaI, SpeI, and BlnI as previously described (6). Among the four S. dublin isolates, four XbaI patterns, three SpeI patterns, and four different BlnI patterns were observed. The patterns obtained with the same restriction enzyme differed from one another by up to five fragments (data not shown).
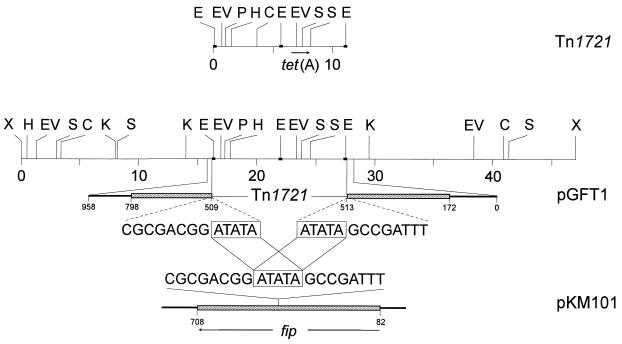
All four S. dublin isolates exhibited a common plasmid profile consisting of two plasmids of 76 and 47 kbp. The 76-kbp plasmid was identified as the S. dublin serovar-specific virulence plasmid by hybridization with a spvB/C gene probe (6). The 47-kbp plasmids of each of the four isolates mediated tetracycline resistance as confirmed by transformation (10) into Escherichia coli JM107 (Stratagene, Heidelberg, Germany). Restriction endonuclease analysis with 16 endonucleases revealed no differences in the restriction fragment patterns of each of the 47-kbp plasmids. Therefore, a common designation, pGFT1, was chosen for these 47-kbp tetracycline resistance plasmids of S. dublin. Hybridization experiments with gene probes for the five tetracycline resistance genes (tet) of classes A to E described to occur among members of the family Enterobacteriaceae (7, 8) were performed to determine the class of the tet gene carried by plasmid pGFT1. Previous studies identified tet genes of classes B and C to occur most frequently in bacteria of the genus Salmonella (5, 8). No hybridization signals were obtained with probes specific for the genes tet(B) (1.275-kbp HincII fragment of plasmid R222), tet(C) (0.929-kbp BstNI fragment of plasmid pSC101), tet(D) (3.05-kbp HindIII-PstI fragment of plasmid RA1), and tet(E) (2.5-kbp ClaI-PvuI fragment of pSL1456). These findings were confirmed by the absence of amplification products in specific PCR assays for the tet genes of classes B to E and also for those of classes G and H (3). However, the tet(A)-specific PCR assay (3) yielded the expected amplification product of 0.95 kbp. Moreover, the nonradioactively labelled (enhanced chemiluminescence system; Amersham-Buchler, Braunschweig, Germany) internal 750-bp SmaI fragment of the tet(A) gene of plasmid RP1 hybridized with a 5.5-kbp EcoRI fragment of pGFT1. Restriction mapping of plasmid pGFT1 revealed striking homology between an 11-kbp region of pGFT1 and the map of the tet(A)-carrying transposon Tn1721 (1) (Fig. 1). A 15.7-kbp KpnI fragment of pGFT1 that contained the entire Tn1721-homologous part was cloned into pBluescript II SK+ (Stratagene). This recombinant plasmid also mediated tetracycline resistance after transformation into E. coli JM107.
FIG. 1.
Comparison of the restriction maps of Tn1721 and plasmid pGFT1. Restriction endonuclease cleavage site abbreviations: C, ClaI; E, EcoRI; EV, EcoRV; H, HindIII; K, KpnI; P, PstI; S, SmaI; X, XhoI. A distance scale in kilobase pairs is indicated below the maps of Tn1721 and pGFT1. The shaded rectangles represent the fip reading frames in pGFT1 and pKM101. The repeated sequence at the integration site of the Tn1721-like transposon in pGFT1 and the corresponding sequence in the fip gene of pKM101 are marked by open rectangles. The numbering of the fip gene area refers to the numbering of the deposited nucleotide sequences of pGFT1 (no. Y11027 and Y11028) and pKM101 (no. U42978).
The nonconjugative 11.1-kbp transposon Tn1721 is a member of the Tn21 subgroup of the Tn3 family of bacterial transposons (1, 13). It carries two terminal 38-bp inverted repeats and a 38-bp internal repeat that separates the transposon into two almost identically sized parts, one of which contains the genes required for transposition and the other of which contains those for functional expression of tetracycline resistance (1, 13). EcoRI restriction sites are located in the 38-bp repeated sequences. To determine the integration site of the Tn1721-like transposon in plasmid pGFT1, EcoRI-KpnI fragments of 2 and 2.6 kbp which contained the junctions between the original plasmid and the Tn1721-analogous transposon were subcloned. Tn1721 has been reported to produce 5-bp direct repeats at its integration site (13). A first comparison of the sequences immediately upstream and downstream of the Tn1721-like transposon in pGFT1 suggested the presence of 6-bp inverted repeats. To further characterize the location of the integration site in pGFT1, the sequences of the upstream and downstream flanking regions were determined on both strands by the dideoxy chain termination method (11) and analyzed for the presence of open reading frames. The sequences of both sides of the transposon corresponded very closely to the coding sequence of a fip gene (12, 15). Only one base pair difference, T→C at position 606, which, however, did not change the amino acid sequence, was detected in the fip coding region of pGFT1 as compared to that of plasmid pKM101 (12, 15). A comparison of the fip gene sequence with the potential repeats at the integration site confirmed that there is definitely only a 5-bp repeat of dyad symetry (ATATA) in pGFT1 (Fig. 1). This 5-bp repeated sequence differed from all repeats observed so far in connection with the integration of Tn1721-like transposons (1, 13).
Conjugative broad-host-range plasmids such as the IncN plasmid pKM101 (16) have been reported to carry fip genes. The fip gene product represents a fertility inhibition protein which inhibits conjugal transfer of cohabitating IncP plasmids. The disruption of the fip reading frame by integration of a Tn1721-like transposon in pGFT1 did not affect essential biological properties of the plasmid. This observation was in accordance with the observations of fip-deficient pKM101 derivatives produced by Tn5 insertion into the fip reading frame (16). Mating experiments with the plasmid-free, kanamycin-resistant E. coli strain CSW893 obtained from the strain collection of the Institut für Tierzucht und Tierverhalten, Celle, Germany, were performed (9) and identified the plasmid pGFT1 as the conjugative plasmid. The transfer ratio of plasmid pGFT1 from S. dublin to E. coli CSW893 was calculated to be 5.4 × 10−7.
These data showed that tetracycline resistance in different isolates of S. dublin was due to the integration of a nonconjugative tet(A)-carrying transposon into a conjugative plasmid that was able to replicate and express tetracycline resistance in E. coli.
Nucleotide sequence accession number.
The sequences corresponding to part of the terminal 38-bp repeats and the adjacent flanking regions of pGFT1 have been deposited in the EMBL database under accession no. Y11027 and Y11028.
Acknowledgments
G.F. is the recipient of a Dr. Alhard von Burgsdorff scholarship.
We thank Stuart B. Levy for providing the gene probes for tet genes of classes A to E and Christiane Werckenthin for providing the kanamycin-resistant E. coli isolate CSW893.
REFERENCES
- 1.Allmeier H, Cresnar B, Greck M, Schmitt R. Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene. 1992;111:11–20. doi: 10.1016/0378-1119(92)90597-i. [DOI] [PubMed] [Google Scholar]
- 2.Fang F F, Fierer J. Human infections with Salmonella dublin. Medicine. 1991;70:198–207. doi: 10.1097/00005792-199105000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Hansen L M, Blanchard P C, Hirsh D C. Distribution of tet(H) among Pasteurella isolates from the United States and Canada. Antimicrob Agents Chemother. 1996;40:1558–1560. doi: 10.1128/aac.40.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsh D C. Salmonella. In: Biberstein E L, Zee Y C, editors. Review of veterinary microbiology. Cambridge, Mass: Blackwell Scientific Publications, Inc.; 1990. pp. 110–115. [Google Scholar]
- 5.Levy S B. Tetracycline resistance determinants are widespread. ASM News. 1988;54:418–421. [Google Scholar]
- 6.Liebisch B, Schwarz S. Evaluation and comparison of molecular techniques for epidemiological typing of Salmonella enterica subsp. enterica serovar dublin. J Clin Microbiol. 1996;34:641–646. doi: 10.1128/jcm.34.3.641-646.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMurray L, Petrucci R E, Jr, Levy S B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendez B, Tachibana C, Levy S B. Heterogeneity of tetracycline resistance determinants. Plasmid. 1980;3:99–108. doi: 10.1016/0147-619x(80)90101-8. [DOI] [PubMed] [Google Scholar]
- 9.Provence D L, Curtiss R., III . Gene transfer in gram-negative bacteria. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. 1st ed. Washington, D.C: American Society for Microbiology; 1994. pp. 317–347. [Google Scholar]
- 10.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 11.Sanger F, Nicklen S, Coulson A. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santini, J. M., and V. A. Stanisich. 1995. GenBank accession no. U42978.
- 13.Schöffl F, Arnold W, Pühler A, Altenbuchner J, Schmitt R. The tetracycline resistance transposons Tn1721 and Tn1771 have three 38-base-pair repeats and generate five-base-pair direct repeats. Mol Gen Genet. 1981;181:87–94. doi: 10.1007/BF00339010. [DOI] [PubMed] [Google Scholar]
- 14.Taylor D N, Bied J M, Munro J S, Feldman R A. Salmonella dublin infections in the United States, 1979–1980. J Infect Dis. 1982;146:322–327. doi: 10.1093/infdis/146.3.322. [DOI] [PubMed] [Google Scholar]
- 15.Winans, S. C. 1995. GenBank accession no. U43676.
- 16.Winans S C, Walker G C. Fertility inhibition of RP1 by IncN plasmid pKM101. J Bacteriol. 1985;161:425–427. doi: 10.1128/jb.161.1.425-427.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woods G L, Washington J A. Antibacterial susceptibility tests: dilution and disk diffusion methods. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 1327–1341. [Google Scholar]