ABSTRACT
Acute hepatopancreatic necrosis disease (AHPND), a high-mortality-rate shrimp disease, is caused by specific Vibrio parahaemolyticus (Vp) strains with a plasmid encoding the PirABVp toxins. As a bacterial pathogen, the invasion of AHPND-causing Vp might impose pressure on commensal microbiota in the shrimp gut, while the relationship between the pathogenesis of AHPND and the dysbiosis of gut bacterial communities remains unclear. Here we explored the temporal changes of shrimp gut microbiota in response to AHPND-causing and non-AHPND-causing Vp strains, with the non-infected controls as a baseline of the shrimp gut microbiota. The diversity and composition of bacterial communities from 168 gut samples (covering three treatments at seven time points with eight individuals per set) were investigated using 16S rRNA gene metabarcoding with high-throughput sequencing. The results showed that (i) species diversity of gut bacterial communities declined in Vp-infected shrimp, independent of the strain pathogenicity; (ii) taxonomic compositions of gut bacterial communities were significantly different between shrimp infected by AHPND-causing and non-AHPND-causing Vp strains; (iii) short-term (within 6 hours) compositional shifts in the gut microbiota were found only in AHPND-causing Vp-infected shrimp; (iv) the gut microbiota of AHPND-causing Vp-infected shrimp was enriched with genera Photobacterium and Vibrio, with a decline in Candidatus Bacilliplasma; and (v) functional predictions suggested the loss of normal metabolism due to compositional shifts in the gut microbiota. Our work reveals distinct features of community dynamics in shrimp gut microbiota, associated with pathogenic versus non-pathogenic Vibrio infections, providing a new perspective of the pathogenesis of AHPND.
IMPORTANCE
Shrimp production is continually threatened by newly emerging diseases, such as AHPND, which is caused by specific Vp strains. Previous studies on the pathogenesis of AHPND have mainly focused on the histopathology and immune responses of the host. However, more attention needs to be paid to the gut microbiota, which acts as the first barrier to pathogen colonization. In this study, we revealed that shrimp gut microbiota responded differently to pathogenic and non-pathogenic Vp strains, with bacterial genera Photobacterium and Vibrio enriched in pathogenic Vp-infected shrimp, and Candidatus Bacilliplasma enriched in non-pathogenic Vp-infected shrimp. Moreover, functional predictions suggested that changes in taxonomic compositions would further affect normal metabolic functions, emphasizing the importance of sustaining an equilibrium in the gut microbiota. Several biomarkers associated with specific microbial taxa and functional pathways were identified in our data sets, which help predict the incidence of disease outcomes.
KEYWORDS: acute hepatopancreatic necrosis disease (AHPND), gut microbiota, Litopenaeus vannamei, pathogen invasion, Vibrio parahaemolyticus
INTRODUCTION
Pacific white shrimp (Litopenaeus vannamei) is one of the most widely farmed crustacean species with a high economic value in aquaculture (1), while its production is threatened by disease outbreaks in recent years (2). Among the diseases listed by the World Organization for Animal Health (founded as the Office International des Epizooties, OIE), acute hepatopancreatic necrosis disease (AHPND) is one of major diseases that severely threaten shrimp farming in Asia and Latin America (2).
Primary pathogenic agents of AHPND are unique strains of Vibrio parahaemolyticus (Vp), a Gram-negative bacterium commonly found in aquatic habitats (3). The AHPND-causing Vp strains contain an extrachromosomal plasmid encoding specific genes for PirAVp and PirBVp toxins, while this plasmid is not found in non-AHPND-causing Vp strains (4). The AHPND-causing Vp strains initially colonize the shrimp stomach and release the binary toxins from the stomach into the hepatopancreas, inducing sloughing of tubule epithelial cells followed by shrimp mortality (5 – 7). Since the histological hallmark of AHPND is hepatopancreatic pathognomonic lesions in the absence of any causative pathogen, it has been suggested that the pathology of the AHPND-causing strain is due to secreted toxins rather than the presence of the bacteria itself (6). However, during the terminal phase of AHPND, the hepatopancreatic tubules are surrounded by hemocytic capsules as a response to secondary bacterial infections, possibly caused by a vibriosis (8). Moreover, the presence of PirABVp toxins could modulate the virulence of non-AHPND-causing Vibrio species and aggravate vibriosis (9). Although many efforts have been made, the pathogenesis of AHPND remains unclear and needs further investigation.
Aquatic crustaceans literally live in water with diverse microorganisms, making them susceptible to potential pathogens, including viruses, bacteria, fungi, and protists (10). In recent years, emerging evidence suggests that the gut microbiota plays a critical role in host physiological processes such as nutrient acquisition, immune activation, and initial defense against infection (11, 12). Gut microbiota comprises diverse commensal bacteria that provide intrinsic protection against pathogen colonization and stimulate the host immune responses, particularly during pathogen invasion (13). Once the balance of the gut microbiota is disrupted, the host may become more susceptible to the invading pathogens. In the aquaculture system, gut microbiota dysbiosis (the altered composition of bacterial communities) could significantly disrupt the stability of normal gut functions, leading to disease aggravation (14). Therefore, an understanding of the dynamics of the shrimp gut microbiota during the invasion of AHPND-causing Vp is crucial for a better understanding of the pathogenesis of AHPND.
Previous studies have mainly focused on the histopathology and immune responses of the shrimp host during AHPND infection (15). However, more attention needs to be paid to the gut microbiota, which acts as the first barrier to pathogen colonization. Some authors have shown that AHPND infection indeed has effects on shrimp gut microbiota (16 – 18). Significant differences in gut bacterial communities were detected between healthy and AHPND-infected shrimp, with an increase in some disease-specific bacterial taxa (16). Abundances of specific bacterial taxa have been reported to show a positive correlation with immune gene expressions of the host (19, 20), suggesting that the compositional changes of gut microbiota are vital in disease pathogenesis (21, 22).
For the invasion of an external microbe, competition for space and nutrients is crucial for successful colonization of gut ecosystems that already contain resident bacteria. Pathogenic bacteria have antagonistic interactions with the gut microbiota through the release of virulence factors (23, 24), while non-pathogens or probiotics might have neutral or commensal interactions with the gut microbiota (25, 26). The differences between AHPND-causing and non-AHPND-causing Vp in carbon source utilizations have been reported, implicating that they adopt distinct strategies to acquire resources in the shrimp gut (27). However, it remains unclear whether the gut microbiota responds differently to AHPND-causing and non-AHPND-causing Vp strains. Therefore, in this study, we aimed to characterize the dynamic responses of the gut microbiota during the invasion by both AHPND-causing and non-AHPND-causing Vp strains, as a reference for the pathogenesis of AHPND. We hypothesized that the colonization of both AHPND-causing and non-AHPND-causing Vp strains would result in changes in the shrimp gut microbiota, while these changes might differ in terms of diversity and composition.
Herein, we investigated the temporal changes of the gut microbiota during the infection with AHPND-causing and non-AHPND-causing Vp strains, with non-infected controls as a baseline of the shrimp gut microbiota. We collected a total of 168 shrimp gut samples, focusing on the early infection stage (0, 3, 6, 12, 24, 48, and 72 hours post immersion) to closely observe the responses of gut bacterial communities to the colonization of a bacterial pathogen in the shrimp stomach. We employed high-throughput sequencing of the 16S ribosomal RNA gene to characterize the dynamics of shrimp gut microbiota. We compared the diversity and composition of gut bacterial communities in shrimp infected with either AHPND-causing or non-AHPND-causing Vp strains to explore differential community responses mediated by bacterial interactions. Exploring dynamic community patterns of the gut microbiota during the course of the disease would provide further insights into the pathogenesis of AHPND.
RESULTS
Time-varying pattern of AHPND virulence in shrimp
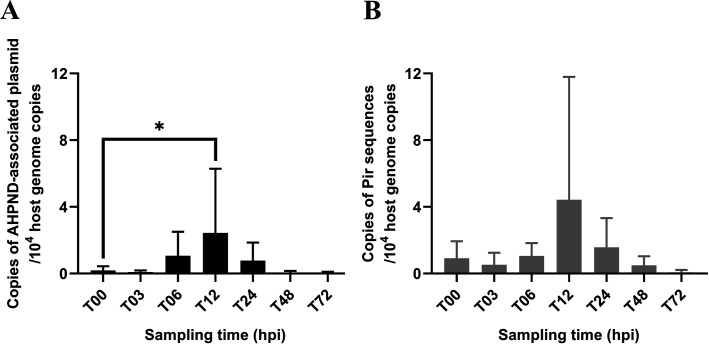
The real-time PCR results showed that for shrimp in the 5HP-infected group, the copy numbers of the AHPND plasmid and toxin gene were highest at T12 (Fig. 1). The absence of AHPND-associated gene fragments was confirmed by PCR amplification for the S02-infected and tryptic soy broth (TSB)-treated groups. The mortality rate of shrimp in the 5HP-infected group was higher than that of shrimp in the S02-infected and TSB-treated groups (Fig. S1). Specifically, approximately 11% of mortality was observed between T06 and T24 in the 5HP-infected group, whereas very few shrimp individuals died in the S02-infected and TSB-treated groups. The majority of deaths in the 5HP-infected group occurred at T12, which was consistent with the presence of elevated levels of AHPND-associated genes at T12 (Fig. 1).
FIG 1.
AHPND detection in 5HP-infected shrimp. The relative copies of AHPND-associated plasmid (A) and toxin gene (B) of pathogenic Vp, detected in shrimp individuals of the 5HP-infected group collected at different time points. The relative copies of AHPND plasmid and toxin gene were normalized against the shrimp genome copies. The sample size at each time point was n = 8, except for T72 (n = 5). Shrimp collected at T12 contained significantly higher numbers of AHPND-associated plasmid copies than those collected at T00. Statistical significance was calculated based on one-way analysis of variance with Dunnett’s test. *: p < 0.05.
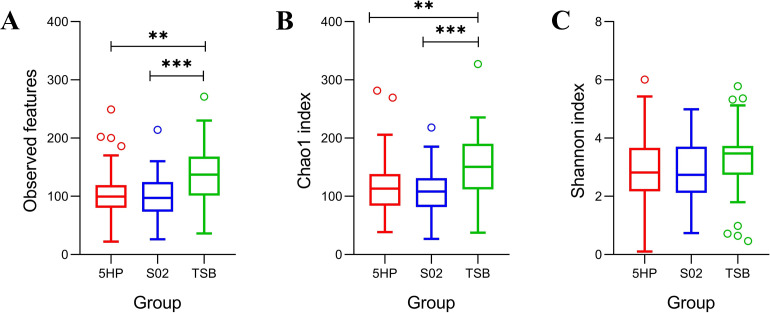
Differences in α-diversity of gut microbiota
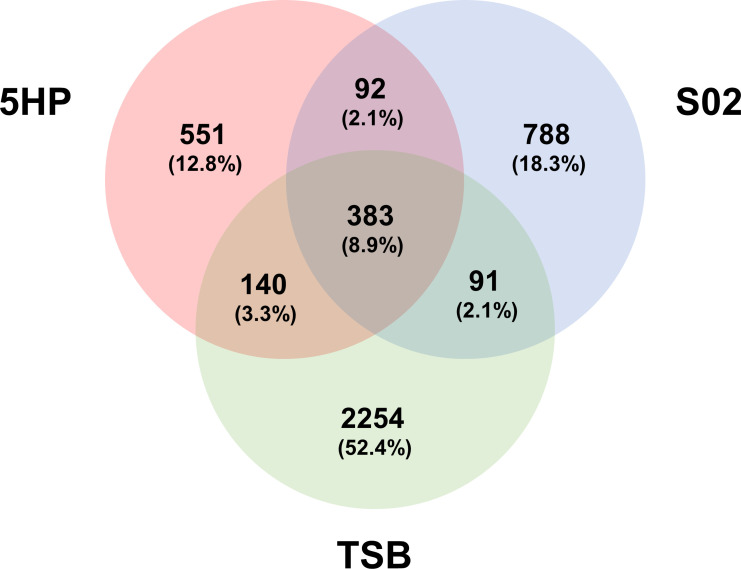
Regarding species diversity of the shrimp gut microbiota, the bacterial communities of the 5HP-infected and S02-infected groups showed lower α-diversity values compared to the TSB-treated group, with significant differences in observed features and Chao1 indices but not in Shannon (Fig. 2). However, considering the temporal variation, the α-diversity values in the 5HP-infected and S02-infected groups showed no significant changes at different time points (Fig. S2). Focusing on the differences in richness among the three groups, the Venn diagram showed that 383 amplicon sequence variants (ASVs) were common to all groups, while each group contained its own unique ASVs, with 551 unique ASVs in the 5HP-infected group, 788 in the S02-infected group, and 2,254 in the TSB-treated group (Fig. 3). These results indicated that the invasion of both Vp 5HP and S02 strains would reduce the number of bacterial species (i.e., richness) in the shrimp gut microbiota.
FIG 2.
Differences in species diversity of gut microbiota among three experimental groups. Gut microbiota α-diversity was determined using three diversity indices, including (A) observed features, (B) Chao1, and (C) Shannon. Different colors in boxplots indicate gut microbiota samples from three distinct experimental groups: red indicates 5HP-infected (n = 56); blue indicates S02-infected (n = 54); and green indicates TSB-treated (n = 55) groups. The samples of the three experimental groups included all samples from all time points. The TSB-treated group showed significantly higher α-diversity in terms of observed features and Chao1 but not Shannon. Statistical significance was calculated based on the Kruskal-Wallis test and post hoc Dunn tests. **P < 0.01, ***P < 0.001.
FIG 3.
Amplicon sequence variant (ASV) Venn diagram of gut microbiota of three groups. Venn diagram showed the overlapping patterns of ASVs among three experimental groups from all time points: the red circle indicates 5HP-infected (n = 56); the blue circle indicates S02-infected (n = 54); and the green circle indicates TSB-treated (n = 55) groups.
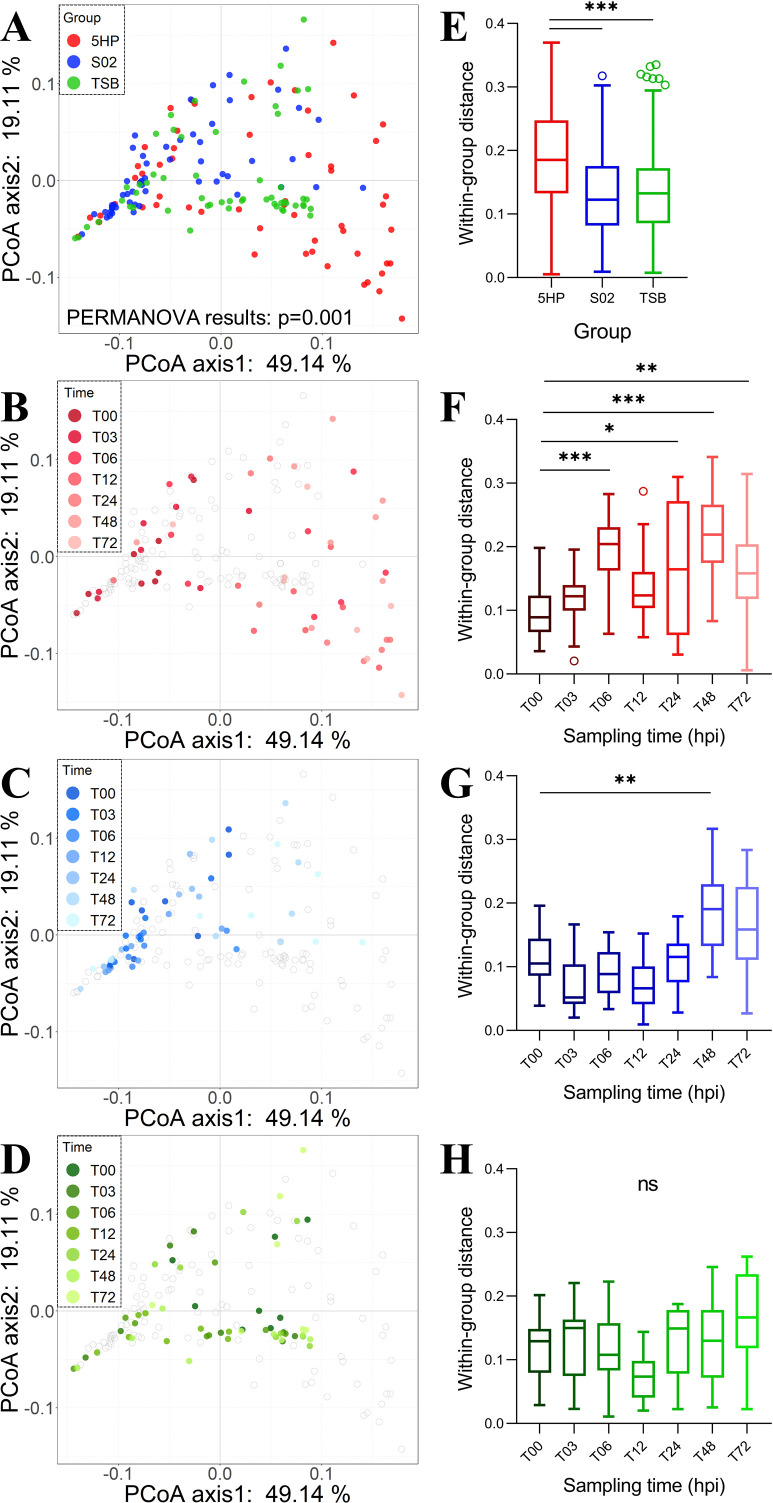
Differences in β-diversity of gut microbiota
Principal co-ordinate analysis (PCoA) based on the weighted UniFrac distances was conducted to illustrate the differences in community composition of gut microbiota (Fig. 4). The permutational multivariate analysis of variance (PERMANOVA) test confirmed the unique compositional characteristics of gut bacterial communities in each group (Fig. 4A; Table S1). In terms of the within-group mean distances, both the S02-infected and TSB-treated groups were significantly lower than the 5HP-infected group, but there was no significant difference in the gut microbiota between S02-infected and TSB-treated groups (Fig. 4E). Specifically, the 5HP-infected group contained samples with relatively high dispersion and exhibited unique taxonomic compositions that differed from the other two groups. Furthermore, while samples in all three groups showed a trend toward dispersion over time (Fig. 4B through D), only the 5HP-infected group had high compositional shifts in the gut microbiota within 6 hours (Fig. 4F through H). Specifically, in the 5HP-infected group, the within-group variation increased significantly from T06 (Fig. 4F), and at T06, the taxonomic composition began to differ significantly from the initial point T00 (Fig. S3A). By contrast, the gut microbiota in the S02-infected group showed higher variation at later T48 and T72 (Fig. 4G; Fig. S3B), and the gut microbiota in the TSB-treated group maintained a constant structure at different time points (Fig. 4H; Fig. S3C).
FIG 4.
Changes in community composition of gut microbiota among three experimental groups across seven sampling time points. Similarity and dissimilarity in bacterial compositions of gut microbiota were determined by principal co-ordinate analysis (PCoA) based on the weighted UniFrac distances for three experimental groups (5HP-infected, S02-infected, and TSB-treated) or distinct sampling times (T00 to T72) within the group: (A) all data points from three groups, (B) 5HP-infected group, (C) S02-infected group, and (D) TSB-treated group. Boxplots (E–H) showed the variation of within-group pairwise distances (based on the weighted UniFrac) corresponding to colored groups in panels A–D. In PCoA, the numbers of the co-ordinates refer to the variance explained by the axes, and the statistical significance of the differences between the three groups was tested using PERMANOVA test (P = 0.001). In boxplots, statistical significance was calculated based on the Kruskal-Wallis test and post hoc Dunn tests. In panels F–H, only the statistical results in comparison to T00 are marked. *P < 0.05, **P < 0.01, ***P < 0.001.
The random forest analysis confirmed that the gut microbiota of different experimental groups could be well classified and predicted, with an accuracy of 100% and 95.9% for the training (Fig. S4A) and validation (Fig. S4B) data sets, respectively. For the validation data set (Fig. S4B), the 5HP-infected group showed perfect performance in both sensitivity and accuracy (100%), whereas the fitting performance of the S02-infected group was relatively lower with an accuracy of 89.5%, and the sensitivity of the TSB-treated group was relatively lower with an accuracy of 86.7%. These results supported the distinctiveness of the 5HP-infected group and the relative similarity between the S02-infected and TSB-treated groups as observed in the PCoA (Fig. 4A).
Taxonomic abundance patterns of shrimp gut microbiota
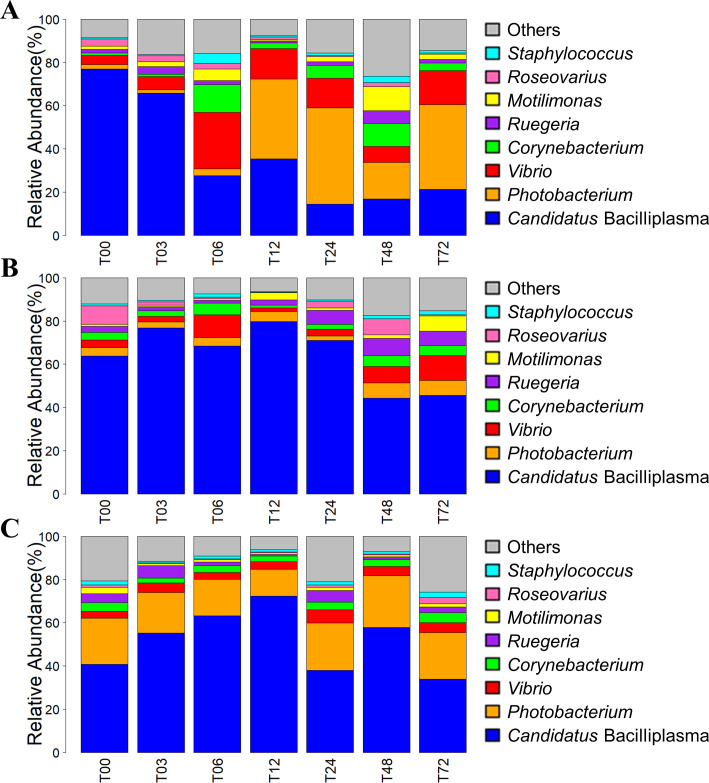
As revealed by sequence-based taxonomic annotation, eight bacterial genera had an average abundance greater than 1% across all gut samples (Fig. 5). Candidatus Bacilliplasma maintained its predominance in the S02-infected and TSB-treated groups at all sampling times (Fig. 5B and C), whereas Vibrio and Photobacterium were enriched in the 5HP-infected group, with high abundances from T06 to T72 (Fig. 5A).
FIG 5.
Differences in taxonomic composition of gut microbiota among three experimental groups across seven sampling time points. Bacterial taxonomic composition (at genus level) of shrimp gut microbiota in three experimental groups: (A) 5HP-infected, (B) S02-infected, and (C) TSB-treated groups is shown. Eight dominant genera (with the average abundance greater than 1%) are shown as their relative abundances, while the rest were grouped as “others.”
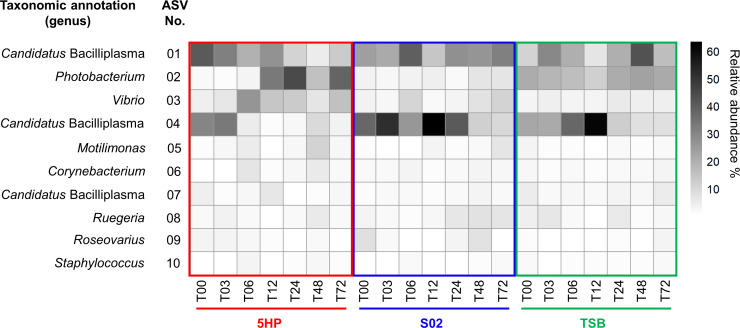
Furthermore, we focused on the temporal dynamics of the top 10 dominant ASVs to identify the main contributors to compositional changes over time (Fig. 6). Actually, the top four ASVs cumulatively accounted for ~70% of the total abundance, being good representation. The ASV-01 belonging to Candidatus Bacilliplasma existed in a generally rich abundance in all groups and showed weak temporal variation. In contrast, the ASV-04 also belonging to Candidatus Bacilliplasma showed a low abundance at T12 in the 5HP-infected group, compared to a high abundance at T12 in the other two groups. The ASV-02 belonging to Photobacterium and the ASV-03 belonging to Vibrio was particularly increased in the 5HP-infected group, with the highest abundance at T24 and T06, respectively. Importantly, the ASV-03 abundance dynamics might represent the temporal colonization of the Vibrio pathogen, as the representative sequence of the ASV-03 was 100% identical to the AHPND-causing Vp 5HP strain.
FIG 6.
The relative abundances of the top dominant ASVs. Relative abundances of 10 ASVs (with the average abundance greater than 1%) are shown on average for each experimental group at seven time points. The vertical axis shows the ASV number corresponding to the numbers in Tables S5, S6, and S7. The changes in the color intensity indicate the relative abundance of respective ASVs.
Through 10-fold cross-validation, the random forest classification identified 33 top-ranking important microbial ASV markers for classifying experimental groups (Fig. S5). Of the top 33 ASVs contributing to the accuracy of the random forest classification model, the dominant ASVs (with average relative abundance >1%, marked as ASV-01 to 10) were all included. These 33 ASVs were mainly from bacterial genera Candidatus Bacilliplasma (seven ASVs) and Corynebacterium (four ASVs) (Fig. S5).
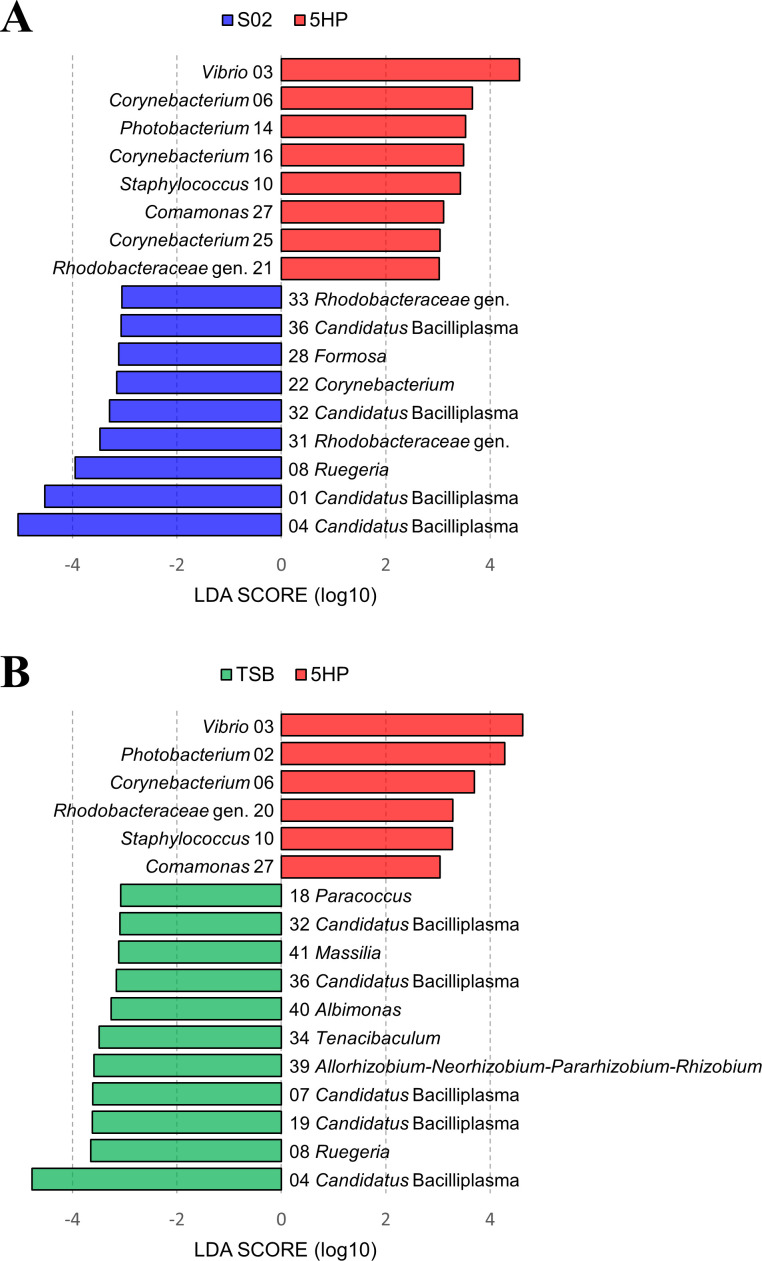
Linear discriminant analysis effect size (LEfSe) analysis revealed that several ASVs showed significant differences in the abundance levels between the 5HP-infected group and the other two groups, respectively [linear discriminant analysis (LDA) >3.0, all P < 0.05; Fig. 7A and B]. Importantly, the top eight ASVs (with mean decrease Gini >1) identified by the random forest analysis (Fig. S5) were also detected by the LEfSe analysis, indicating their high contribution to the discrimination between experimental treatments. Vibrio ASV-03 was identified as the most specific feature in the 5HP-infected group with the highest LDA score, while Candidatus Bacilliplasma ASV-04 was the most important feature in the S02-infected and TSB-treated groups (Fig. 7A and B). Moreover, ASVs belonging to the genera Vibrio, Photobacterium, Staphylococcus, and Comamonas were significantly abundant in the 5HP-infected group, whereas ASVs of the genera Candidatus Bacilliplasma and Ruegeria were relatively dominant in the S02-infected and TSB-treated groups (Fig. 7).
FIG 7.
Linear discriminant analysis effect size analysis of the abundance patterns of bacterial ASVs. Identification of bacterial ASVs that differentiated (A) 5HP-infected (red) vs S02-infected (blue) groups or (B) 5HP-infected (red) vs TSB-treated (green) groups by LDA effect size. The ASV numbers corresponded to the numbers in Tables S5, S6, and S7. The differences were significant (P < 0.05) among classes (Kruskal-Wallis test). The threshold of the logarithmic LDA score was 3.0.
Functional analysis of shrimp gut microbiota
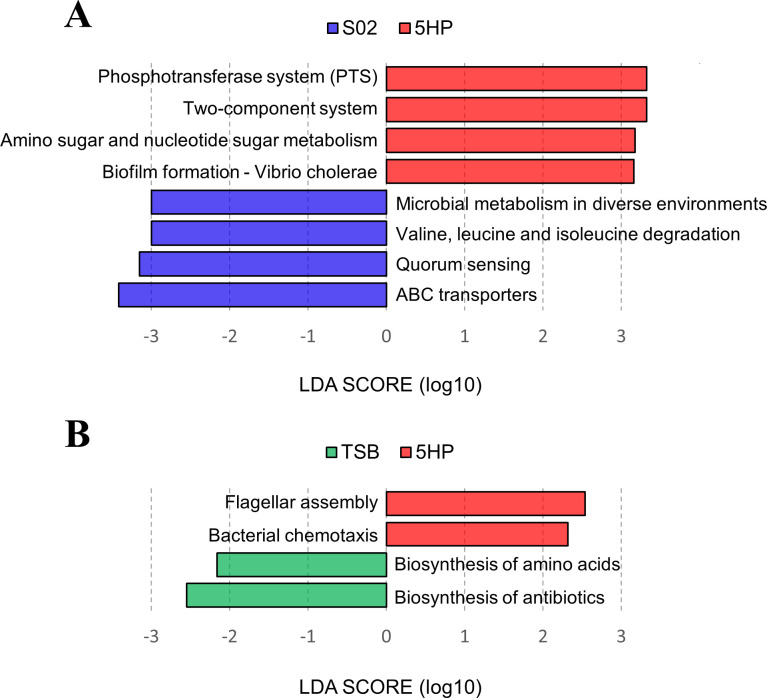
The potential functions of the shrimp gut microbiota were predicted by Tax4Fun2 through the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and the biomarkers were revealed by LEfSe. Significant differences in eight functional pathways between 5HP-infected and S02-infected groups (LDA >3.0, all P < 0.05) and differences in four functional pathways between 5HP-infected and TSB-treated groups (LDA >2.0, all P < 0.05), respectively, were reported (Fig. 8A and B). In the 5HP-infected group, pathways associated with phosphotransferase system, two-component system, amino sugar and nucleotide sugar metabolism, biofilm formation—Vibrio cholerae, flagellar assembly, and bacterial chemotaxis were enriched. In the S02-infected group, functions related to ABC transporters, quorum sensing, valine/leucine/isoleucine degradation, and microbial metabolism in diverse environments were relatively abundant. In the TSB-treated group, pathways associated with biosynthesis of antibiotics and biosynthesis of amino acids were the key features. The highly distinguishable pathways for the 5HP-infected group were mainly associated with signal transduction and cell motility, whereas those for the S02-infected and TSB-treated groups were mainly associated with metabolism (Table S2), suggesting that the shifts in taxonomic compositions of the gut microbiota would likely have an impact on metabolic functions in the gut.
FIG 8.
Linear discriminant analysis effect size analysis of functional biomarkers. Identification of functional pathways in KEGG level three that differentiated (A) 5HP-infected (red) vs S02-infected (blue) groups or (B) 5HP-infected (red) vs TSB-treated (green) groups by LDA effect size. The differences were significant (P < 0.05) among classes (Kruskal-Wallis test). The threshold of the logarithmic LDA score was 3 for panel A and 2 for panel B.
DISCUSSION
AHPND-causing Vp colonized the shrimp stomach in a short time
The real-time PCR results showed that the toxin-related genes were apparently detected from 6 to 24 hpi in the 5HP-infected group (Fig. 1), in accord with the enzyme-linked immunoassay conducted by Lai et al. (6). Similar to findings of the previous study (28), the infection of AHPND-causing Vp (the 5HP strain) induced high mortality between 6 and 24 hpi, whereas there was no apparent death caused by the infection of non-AHPND-causing Vp (the S02 strain) (Fig. S1). The detection of toxin-related genes, together with shrimp mortality, suggested that the harmful effect of the AHPND-causing Vp strain correlated with the level of toxin production (29).
However, the sequence abundance of the 5HP strain (annotated as the ASV-03) showed the highest percentage at 6 hpi (Fig. 6) rather than 12 hpi (the high toxin gene detection, Fig. 1). This inconsistency indicated that AHPND-causing Vp might generate plasmids and produce toxins only after stable colonization. Recent evidence suggests that virulence factors of Vibrio are regulated by a quorum-sensing system and are expressed only when reaching a certain cell concentration threshold or population density (30, 31), which may explain the time lag between Vp invasion and toxin release. Moreover, these pathogenic processes are consistent with the proposed model of (32) that the incipient entry of AHPND-causing Vp into the shrimp stomach leads to the dysbiosis of microbiota, as a result of direct bacterial competition or the release of signaling metabolites. Moreover, the dysbiosis of the commensal bacterial community would allow the AHPND-causing Vp to further replicate and colonize, with the release of PirABVp toxins. Later, the PirABVp toxins induce the disruption of tight junctions between stomach epithelial cells, allowing the migration of AHPND-causing Vp to the hepatopancreas. In another study (6), AHPND-causing Vp could be detected in the hepatopancreas at 12 hpi, indicating the later phase of infection (15). Overall, these findings suggest that the colonization resistance of gut microbiota in the early stage (~6 hpi) is critical to prevent or mitigate the effect of AHPND.
Shrimp gut microbiota diversity reduced after Vibrio invasion
In this study, the gut bacterial communities of the Vibrio-infected shrimp (5HP-infected and S02-infected groups) showed lower α-diversity values than those of the TSB-treated group, which reflected in species richness (Fig. 2). Specifically, the number of ASVs in the Vibrio-infected groups markedly declined for all dominant bacterial genera (Table S3). Species diversity is of great significance for promoting stability and performance in all ecosystems (33, 34), and the diversity of gut microbiota is considered to be a good indicator of host health (35, 36). The disease outbreak is often accompanied by the reduced diversity of microbiota (16, 37, 38); thus, the loss of α-diversity in the 5HP-infected group could be attributed to AHPND. Interestingly, the α-diversity values in the S02-infected group suggested that even exposure to an avirulent strain could reduce the diversity of gut microbiota (Fig. 2). It has been reported that even high concentrations of probiotics may decrease the species richness of shrimp gut microbiota (39). Indeed, exposure to an external microorganism may act as a stressful stimulus which would affect the stability of commensal bacteria (11, 40) as well as alter the relative abundance (41).
Similarly, reduced diversity in the gut microbiota of both Vibrio-infected groups (5HP-infected and S02-infected) implicated that a universal initial response of microbiota to opportunistic pathogens is independent of the virulence (42). This initial response indicated that any opportunistic pathogen surrounding the shrimp host may trigger the transition of symbiont gut microbiota and causes a change in health status. Low species diversity in the gut bacterial community would likely become more sensitive to environmental stresses and surrounding microbes (43), reinforcing the emergence of shrimp diseases (44). Our findings, along with previous studies, suggest that the disruption of the symbiotic microbiota is the primary and crucial step in opportunistic pathogen invasion. Thus, developing effective strategies to maintain and restore healthy gut microbiota could be critical for shrimp disease prevention.
Dysbiosis in shrimp gut microbiota by AHPND-causing Vp
Our study found that bacterial compositions of the gut microbiota were significantly different among healthy (TSB-treated), non-AHPND-infected (S02-infected), and AHPND-infected (5HP-infected) shrimp (Fig. 4). The AHPND-infected (5HP-infected) group was distinct from the healthy (TSB-treated) and non-AHPND-infected (S02-infected) groups, as evidenced by within-group β-distances (Fig. 4E) and random forest analysis (Fig. S4). This result is in accordance with previous report (17) where AHPND-infected shrimp showed microbiota compositions that were distinct from healthy shrimp. Our findings indicate that AHPND-causing Vp adapts unique strategies to break the colonization resistance of commensal bacteria and obtain a competitive advantage. Compared to non-AHPND-causing Vp, the presence of a pVA plasmid carrying PirABVp toxin genes along with other virulence genes (45) in the AHPND-causing Vp may serve as a characteristic feature to outcompete other microbes. In addition to the plasmid, an AHPND-causing Vp strain (13–028/A3 strain) has been reported to utilize multiple carbon sources more efficiently than non-AHPND-causing Vp (RIMD2210633 and BB22OP strains) (27). The capability to efficiently metabolize various substrates contributes to the competitive advantage of microbes (46).
Considering the temporal changes in gut microbiota, in the 5HP-infected group, the gut microbiota composition shifted and the variation increased at 6 hpi, while the microbiota in the S02-infected and TSB-treated groups maintained a relatively stable structure during the course of the experiment (Fig. 4). The minor fluctuations of microbiota in the TSB-treated group is representative of usual state in a healthy host (47 – 49). The exposure to both AHPND-causing Vp and non-AHPND-causing Vp, respectively, could perturb the community composition. However, a resilient community could recover to normal functions after a lag phase (49). The recovery process in the S02-infected group was reflected during 6 to 24 hpi by the increasing abundance of ASVs that belong to Candidatus Bacilliplasma and maintained diverse genus-level composition during 48 to 72 hpi (Fig. 5). However, in the 5HP-infected group, the perturbation by the AHPND-causing Vp would directly change the taxonomic composition of gut microbiota, exceeding the threshold of resilience.
The relative abundances of Vibrio and Photobacterium were increased in the gut microbiota of the 5HP-infected shrimp, while Candidatus Bacilliplasma maintained its predominance in the S02-infected and TSB-treated shrimp (Fig. 5). Both Vibrio and Photobacterium belong to the Vibrionaceae family. Vibrionaceae has served as the signature for AHPND diagnosis (50). The higher abundances of the Vibrionaceae family and Vibrio genus were postulated to be caused by the colonization of the AHPND-causing Vp and related to secondary Vibrio infections (51). Apart from some highly virulent strains causing the primary disease, Vibrio spp. are often considered as secondary or opportunistic pathogens in shrimp (52). Most shrimp vibrioses occur either combined with physical stresses or following primary infections by other pathogens (53). An argument is that AHPND is not a typical vibriosis infection but an acute intoxication caused by PirABVp toxins (8). However, if PirABVp toxins could modulate the virulence of non-AHPND-causing Vibrio species (9), shrimp may indeed die because of a secondary vibriosis after being weakened by PirABVp toxins. In addition, the relatively high abundance of Photobacterium may be associated with secondary luminous bacterial infections (54), as reported in pearl gentian grouper, where infection with high virulent Vibrio significantly increased the abundance of predominant Photobacterium (55). It has been suggested that the stress from the invasion of Vibrio might open a niche for Photobacterium to opportunistically occupy (56).
Candidatus Bacilliplasma, reported as a possible novel lineage of class Mollicutes (57), is often observed in the digestive tract of shrimp and has been implicated in several shrimp diseases (42, 58, 59). As a prominence in the shrimp gut, the abundance of Candidatus Bacilliplasma has been reported to decrease in shrimp subjected to microcystin-LR (a variant of microcystin with leucine and arginine) stress and White spot syndrome virus (WSSV) infection (60, 61). In our study, the ASV-04 belonging to Candidatus Bacilliplasma exhibited an inverse trend to ASV-03 belonging to Vibrio (Fig. 7), which indicated its antagonism to the opportunistic or pathogenic bacterial strains. Our finding implied that the decrease of Candidatus Bacilliplasma may indicate the diseased status of shrimp. However, by analyzing bacterial interaction networks, Chen et al. (42) suggested that varied subspecies of Candidatus Bacilliplasma interacted with the pathogenic Vibrio strains, and they either enhance or inhibit infection. Thus, the interaction dynamics between ASVs belonging to Candidatus Bacilliplasma and Vibrio should be reassessed further to understand their interaction mechanisms.
Microbiota regulates the maintenance of host gut functions
The functional pathways of shrimp gut microbiota infected by AHPND-causing Vp (5HP-infected) were mainly associated with “signal transduction” and “cell motility,” while those in non-AHPND-causing Vp (S02-infected) and control (TSB-treated) groups were mainly associated with “metabolism” (Table S2). In diseased shrimp, pathogens are extremely efficient in arranging their virulence repertoire through complex signaling transduction systems in response to the compound signals from the host or the normal gut microbiota (62). The distinguished pathways of cell motility were supposed to be enriched due to the dual flagellar systems of Vp strains adapted for locomotion under different circumstances (63). In healthy shrimp, the gut microbiota provides baseline functions associated with general metabolism, especially in the metabolism of amino acids and carbohydrates (64). It is generally considered that aquatic animals do not have all the essential enzymes to cope with their dietary challenges (65). For example, low activities of polysaccharide digestive enzymes were detected in the intestine of shrimp (66). Several bacterial strains isolated from the shrimp digestive tract have the genetic capacity to produce extracellular enzymes (67). Thus, the microbiota producing digestive enzymes was inferred to remedy the shortage (64). Specifically, the gut microbiota plays a major role in protein digestion, amino acid metabolism, lipid metabolism, and fatty acid synthesis that are vital for host health (68, 69).
Kumar et al. (70) previously suggested that the induction of genes involved in bile acid synthesis was observed in response to AHPND. Crude bile acids positively influence both biofilm formation and the secretion of PirABVp toxins (71). Elevated taurocholate concentrations were correlated with enhanced biofilm formation in AHPND-causing Vp (70), consistent with the results from our LEfSe analysis that the biofilm formation was enriched in the AHPND-causing Vp (5HP-infected) group (Fig. 8). However, bile acid and taurocholic bile salt treatments failed to induce the biofilm formation of the non-AHPND-causing Vp (S02), which may be due to the significant genomic differences between the AHPND-causing and non-AHPND-causing strains (4). Pathogens may resist the antibacterial properties of bile and may also use bile as a signal to modulate their virulence (72). Thus, the effects of bile acid constituents on pathogen biofilm formation are selective and distinct (73). The differential utilization of metabolites results in the differential effects of pathogenic and non-pathogenic bacterial invasions on the gut microbiota.
Our results, together with previous studies, indicate that the gut microbiota plays an indispensable role in host metabolic functions, highlighting the importance of maintaining a balance in the shrimp gut microbiota. We found that AHPND-causing Vp could induce dysbiosis of the shrimp microbiota, altering the metabolic functions of gut bacteria. In contrast, non-AHPND Vp was found to induce non-pathological changes in the gut microbiota, suggesting that the gut microbiota responds differently to pathogenic and non-pathogenic Vibrio parahaemolyticus.
MATERIALS AND METHODS
Experimental animals and bacterial strains
Specific pathogen-free Pacific white shrimp (Litopenaeus vannamei, with average body weight 2.0 ± 0.5 g) were obtained from the National Pingtung University of Science and Technology. Shrimp were kept in tanks containing 30 L of artificial seawater (maintaining salinity ~20 ppt, temperature ~27°C, and pH value ~8.0) for 2 days as acclimatization prior to the immersion challenge. During the experimental periods, environmental conditions of the water tanks were kept the same as described above for all groups (Table S4).
Two strains of Vp were used for this study: 5HP strain (AHPND-causing Vp) and S02 strain (non-AHPND-causing Vp) both isolated from Thailand (74). Bacterial glycerol stocks were provided by the laboratory of Prof. Han-Ching Wang in the National Cheng Kung University and preserved in 25% glycerol at −80℃ before the immersion challenge.
Experimental groups and immersion challenges
To explore the Vp infection effects on the gut microbiota, we performed three experimental treatments with (i) 5HP-infected group: immersion challenge with Vp 5HP strain, (ii) S02-infected group: immersion challenge with Vp S02 strain, and (iii) TSB-treated group: immersion in the TSB as a negative control. To detect the change in gut microbiota during the critical period of AHPND pathogenesis (32), shrimp stomach samples were collected, respectively, at seven time points (0, 3, 6, 12, 24, 48, and 72 hours post immersion; shown as T00, T03, T06, T12, T24, T48, and T72 hereafter).
Shrimp were randomly distributed into six tanks (three treatments in duplicates, n = 35/per tank). The immersion challenges were performed as described previously by Lai et al. (6) with slight modifications. To recover bacterial stocks, Vp 5HP and S02 strains were separately cultured on thiosulfate citrate bile salts sucrose agar plates. The colonies were inoculated into TSB medium with 2% NaCl and incubated at 30°C, 180 rpm, for 16 hours as starting cultures. Overnight bacterial culture was then scaled up and the cell density was adjusted to OD600 = 0.1 (approximately 107 CFU/mL) assessed by spectrophotometer. Individual bacterial inoculums (100 mL) were mixed with 900-mL seawater to adjust the concentration to 106 CFU/mL for immersion challenges. For the 5HP-infected and S02-infected groups, shrimp were immersed in the 106-CFU/mL inoculum mixture for 15 min and then transferred back to their respective tanks. To keep shrimp under the infected condition, ~300-mL inoculum mixture was added to 30-L seawater in the tank (final bacterial density 104 CFU/mL). At T00, T03, T06, T12, T24, T48, and T72, the entire stomach of each shrimp, including the contents and mucosa, was aseptically dissected and stored at −80°C until DNA extraction. The stomach was sliced into two parts: one-fifth of the stomach was used for AHPND diagnosis and the rest was kept for gut microbiome analysis (42). At each time point, four shrimp individuals were collected from each tank, giving a total of 168 samples for subsequent data analysis.
AHPND detection
The DNA for AHPND diagnosis was extracted from shrimp stomach using a DTAB/CTAB DNA extraction kit (GeneReach Biotechnology Corp, Taiwan). The DNA yield and quality were assessed by a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA). To quantitatively determine AHPND-related markers, TaqMan real-time PCR was performed on a CFX96 real-time system (Bio-Rad, USA), using an IQ REAL AHPND/EMS Quantitative System (targeting the AHPND plasmid and PirABVp gene) and an IQ REAL WSSV Quantitative System (targeting host genome) (Gene Reach Biotechnology Corp, Taiwan). A two-temperature PCR amplification protocol was applied: 40 cycles of denaturation at 93°C for 15 s and annealing/extension at 60°C for 1 min (42). The artificial DNA provided in the kit contained partial sequence fragments of the AHPND plasmid and PirABVp gene (Toxin 1), which were used as standards to construct standard curves. The relative copy number of the AHPND plasmid or toxin 1 was normalized per cell against host genome copies. One-way analysis of variance with Dunnett’s test was performed using the GraphPad Prism V.8 software for Windows (GraphPad Software, USA; https://www.graphpad.com/) to evaluate the differences in the copy numbers of AHPND-associated genes over time.
Microbiome profiling by 16S rRNA gene sequencing
The DNA for gut microbiome analysis was extracted using QIAamp PowerFecal DNA Kit (QIAGEN, German), according to the manufacturer’s instructions. The hypervariable V4 region of the 16S rRNA gene was amplified by the specific 515F/806R PCR primers (515F: GTGYCAGCMGCCGCGGTAA, 806R: GGACTACNVGGGTWTCTAAT) (75), following the steps: initial denaturing at 95°C for 3 min; 28 cycles of 95°C for 30 s, 55°C for 40 s, 72°C for 50 s; and a final extension at 72°C for 5 min. The sizes of PCR products were checked by gel electrophoresis. For the samples with only one specific band, the PCR products were purified by Agencourt AMPuer XP (Beckman Coulter, USA), while those with multiple bands were purified by QIAquick Gel Extraction Kit (QIAGEN) focusing on the band with the expected size. To build the amplicon library for high-throughput sequencing, the PCR products were bound to adapters with barcodes as follows: initial denaturing at 95°C for 3 min; 7 cycles of 95°C for 30 s, 55°C for 40 s, 72°C for 50 s; and a final extension at 72°C for 5 min. After confirming the product size and purification, the amplicon concentrations were measured by Qubit 1X dsDNA HS Assay Kit on a QubitTM Fluorometer. Pooled library containing equal DNA concentration for each sample was sequenced on the Illumina Miseq platform, with 2 × 300 bp paired-end reads (Genomics BioSci. & Tech, Taiwan).
Processing of raw sequencing data
Using QIIME2 v.2021.11 (76), paired-end FASTQ sequence reads were demultiplexed based on sample unique barcodes. The PCR primer flanks were trimmed using Cutadapt (77). Quality filtration, sequence merging, and feature abundance table were conducted by the DADA2 plugin (78). Sequence bases with low Q scores were truncated considering Q30 as the benchmark. The truncated reads were further grouped into ASVs, followed by chimera removal. The representative sequences were extracted to generate a rooted phylogenetic tree, produced by the QIIME2 phylogeny tool: align-to-tree-mafft-fast tree (79, 80). ASVs were taxonomically classified from phylum to genus levels by using the classify-sklearn with a naïve Bayes classifier trained on Silva 138 99% operational taxonomic units (OTUs) from the 515F/806R region of sequences (MD5: e05afad0fe87542704be96ff483824d4) (40, 81 – 83). ASVs that were not assigned to bacteria (i.e., unassigned, archaeal, and eukaryotic ASVs, as well as chloroplast and mitochondrial ASVs) were excluded.
The feature abundance table generated by DADA2 was rarefied to a depth of 8,800 sequence reads per sample. For equal comparison, two samples of the S02-infected group and one sample of the TSB-treated group were excluded due to insufficient reads. The final rarefied table contained a total of 165 samples.
Microbiome community analysis
To detect the changes in shrimp gut microbiota, α-diversity indices, including observed features, Chao1, and Shannon were estimated using QIIME2 (76). To determine the differences in bacterial community composition, PCoA based on weighted UniFrac distances were applied to analyze and visualize the patterns of β-diversity (84, 85). The diversity core-metrics-phylogenetic plugin in QIIME2 was used to calculate the weighted UniFrac distances, and the β-diversity was visualized through PCoA using the ggplot function from ggplot2 package (86) in R v.4.2.0 (87). Boxplots of within-group α-diversity and β-diversity were performed using the GraphPad Prism v.8 software for Windows (GraphPad Software, https://www.graphpad.com/). Kruskal-Wallis tests and post hoc Dunn tests were performed using the GraphPad Prism v.8 software for Windows (GraphPad Software, https://www.graphpad.com/) to assess differences in α-diversity values, within-group β-diversity distances, and temporal compositional variations among three experimental groups, with the significance level set at P < 0.05. Moreover, PERMANOVA was employed using QIIME2 (76) to evaluate differences in the gut microbiota among three experimental groups, with the significance level set at P < 0.05.
To test whether the gut microbiota of different experimental groups could be well classified and predicted, a random forest model was constructed with the abundances of ASVs, using package randomForest in R (88). The samples were randomized into a training data set (70%) and a validation data set (30%). The contribution of ASVs to group classification was based on the mean decrease of Gini. To identify the top-ranking important ASVs, the predictive performance was evaluated using 10-fold cross-validation with the random forest model.
To further infer the functional potential of the gut microbiota, Tax4Fun2 v.1.1.5 (89) based on the KEGG database (90) was applied for acquiring the functional composition of the gut microbiota, and the predicted functional categories of Tax4Fun2 were grouped into three levels according to the KEGG database.
To identify the taxonomic or functional biomarkers that differed significantly between experimental treatments, the LEfSe (91) was applied for biomarker discovery using the Galaxy/Hutlab tool (https://huttenhower.sph.harvard.edu/galaxy/). The significance level was set at P < 0.05 and the threshold for LDA scores was 3.0.
ACKNOWLEDGMENTS
This study was supported financially by the Ministry of Science and Technology, Taiwan (MOST 108-2611-M-006-001-MY2).
We thank the International Center for the Scientific Development of Shrimp Aquaculture, National Cheng Kung University, for providing experimental equipment and technical support.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Hsiao-Pei Lu, Email: hplu@gs.ncku.edu.tw.
Pei-Yuan Qian, Hong Kong University of Science and Technology, Hong Kong .
DATA AVAILABILITY
The raw sequencing data reported in this study has been archived in the Sequence Read Archive of the National Center for Biotechnology Information under accession number PRJNA921960.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.01180-23.
Fig. S1 to S5 and Tables S1 to S7.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. FAO . 2022. The state of world fisheries and aquaculture 2022. Sustain Action. [Google Scholar]
- 2. Tang KF, Bondad-Reantaso MG, Arthur JR, MacKinnon B, Hao B, Alday-Sanz V, Dong X, Liang Y. 2020. Shrimp acute hepatopancreatic necrosis disease strategy manual, p 1–65. In FAO fisheries and aquaculture circular. doi: 10.4060/cb2119en [DOI] [Google Scholar]
- 3. Santos HM, Tsai C-Y, Maquiling KRA, Tayo LL, Mariatulqabtiah AR, Lee C-W, Chuang KP. 2020. Diagnosis and potential treatments for acute hepatopancreatic necrosis disease (AHPND): a review. Aquac Int 28:169–185. doi: 10.1007/s10499-019-00451-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang Y-T, Chen I-T, Lee C-T, Chen C-Y, Lin S-S, Hor L-I, Tseng T-C, Huang Y-T, Sritunyalucksana K, Thitamadee S, Wang H-C, Lo C-F. 2014. Draft genome sequences of four strains of Vibrio parahaemolyticus, three of which cause early mortality syndrome/acute hepatopancreatic necrosis disease in shrimp in China and Thailand. Genome Announc 2:e00816-14. doi: 10.1128/genomeA.00816-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tran L, Nunan L, Redman RM, Mohney LL, Pantoja CR, Fitzsimmons K, Lightner DV. 2013. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis Aquat Organ 105:45–55. doi: 10.3354/dao02621 [DOI] [PubMed] [Google Scholar]
- 6. Lai H-C, Ng TH, Ando M, Lee C-T, Chen I-T, Chuang J-C, Mavichak R, Chang S-H, Yeh M-D, Chiang Y-A, Takeyama H, Hamaguchi H, Lo C-F, Aoki T, Wang H-C. 2015. Pathogenesis of acute hepatopancreatic necrosis disease (AHPND) in shrimp. Fish Shellfish Immunol 47:1006–1014. doi: 10.1016/j.fsi.2015.11.008 [DOI] [PubMed] [Google Scholar]
- 7. Prachumwat A, Taengchaiyaphum S, Mungkongwongsiri N, Aldama-Cano DJ, Flegel TW, Sritunyalucksana K. 2019. Update on early mortality syndrome/acute hepatopancreatic necrosis disease by April 2018. J World Aquacult Soc 50:5–17. doi: 10.1111/jwas.12559 [DOI] [Google Scholar]
- 8. Lozano-Olvera R, Gomez-Gil B, Bolanmejia C, Aguilar-Rendon K, Soto-Rodriguez S, Enciso-Ibarra J. 2018. Pathological, genomic and phenotypical characterization of Vibrio parahaemolyticus, causative agent of acute hepatopancreatic necrosis disease (AHPND) in Mexico. Asian Fish Sci 31S:102–111. [Google Scholar]
- 9. Tran PTN, Kumar V, Bossier P. 2020. Do acute hepatopancreatic necrosis disease-causing PirABVP toxins aggravate vibriosis? Emerg Microbes Infect 9:1919–1932. doi: 10.1080/22221751.2020.1811778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bachère E. 2003. Anti-infectious immune effectors in marine invertebrates: potential tools for disease control in larviculture. Aquaculture 227:427–438. doi: 10.1016/S0044-8486(03)00521-0 [DOI] [Google Scholar]
- 11. Zhu J, Dai W, Qiu Q, Dong C, Zhang J, Xiong J. 2016. Contrasting ecological processes and functional compositions between intestinal bacterial community in healthy and diseased shrimp. Microb Ecol 72:975–985. doi: 10.1007/s00248-016-0831-8 [DOI] [PubMed] [Google Scholar]
- 12. Fan L, Li QX. 2019. Characteristics of intestinal microbiota in the Pacific white shrimp Litopenaeus vannamei differing growth performances in the marine cultured environment. Aquaculture 505:450–461. doi: 10.1016/j.aquaculture.2019.02.075 [DOI] [Google Scholar]
- 13. Stecher B, Hardt W-D. 2011. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol 14:82–91. doi: 10.1016/j.mib.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 14. Rungrassamee W, Klanchui A, Maibunkaew S, Karoonuthaisiri N. 2016. Bacterial Dynamics in intestines of the black Tiger shrimp and the Pacific white shrimp during Vibrio harveyi exposure. J Invertebr Pathol 133:12–19. doi: 10.1016/j.jip.2015.11.004 [DOI] [PubMed] [Google Scholar]
- 15. Kumar V, Roy S, Behera BK, Bossier P, Das BK. 2021. Acute hepatopancreatic necrosis disease (AHPND): virulence, pathogenesis and mitigation strategies in shrimp aquaculture. Toxins (Basel) 13:524. doi: 10.3390/toxins13080524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cornejo-Granados F, Lopez-Zavala AA, Gallardo-Becerra L, Mendoza-Vargas A, Sánchez F, Vichido R, Brieba LG, Viana MT, Sotelo-Mundo RR, Ochoa-Leyva A. 2017. Microbiome of Pacific whiteleg shrimp reveals differential bacterial community composition between Wild, Aquacultured and AHPND/EMS outbreak conditions. Sci Rep 7:11783. doi: 10.1038/s41598-017-11805-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hossain MS, Dai J, Qiu D. 2021. Dysbiosis of the shrimp (Penaeus monodon) gut microbiome with AHPND outbreaks revealed by 16S rRNA metagenomics analysis . Aquaculture Research 52:3336–3349. doi: 10.1111/are.15178 [DOI] [Google Scholar]
- 18. Shen H, Song T, Lu J, Qiu Q, Chen J, Xiong J. 2021. Shrimp AHPND causing Vibrio anguillarum infection: quantitative diagnosis and identifying antagonistic bacteria. Mar Biotechnol (NY) 23:964–975. doi: 10.1007/s10126-021-10079-8 [DOI] [PubMed] [Google Scholar]
- 19. Liu W-C, Zhou S-H, Balasubramanian B, Zeng F-Y, Sun C-B, Pang H-Y. 2020. Dietary seaweed (Enteromorpha) polysaccharides improves growth performance involved in regulation of immune responses, intestinal morphology and microbial community in banana shrimp Fenneropenaeus merguiensis. Fish Shellfish Immunol 104:202–212. doi: 10.1016/j.fsi.2020.05.079 [DOI] [PubMed] [Google Scholar]
- 20. Shi C, Xia M, Li R, Mu C, Zhang L, Liu L, Ye Y, Wang C. 2019. Vibrio alginolyticus infection induces coupled changes of bacterial community and metabolic phenotype in the gut of swimming crab. Aquaculture 499:251–259. doi: 10.1016/j.aquaculture.2018.09.031 [DOI] [Google Scholar]
- 21. te Biesebeke R. 2018. Balancing microbial ecosystems within humans and animals to prevent medical conditions. jnfrt 1:40–40. doi: 10.30881/jnfrt.00009 [DOI] [Google Scholar]
- 22. Buttó LF, Haller D. 2016. Dysbiosis in intestinal inflammation: cause or consequence. Int J Med Microbiol 306:302–309. doi: 10.1016/j.ijmm.2016.02.010 [DOI] [PubMed] [Google Scholar]
- 23. Pickard JM, Zeng MY, Caruso R, Núñez G. 2017. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev 279:70–89. doi: 10.1111/imr.12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamada N, Chen GY, Inohara N, Núñez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14:685–690. doi: 10.1038/ni.2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Restrepo L, Domínguez-Borbor C, Bajaña L, Betancourt I, Rodríguez J, Bayot B, Reyes A. 2021. Microbial community characterization of shrimp survivors to AHPND challenge test treated with an effective shrimp probiotic (Vibrio diabolicus). Microbiome 9:88. doi: 10.1186/s40168-021-01043-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Du S, Chen W, Yao Z, Huang X, Chen C, Guo H, Zhang D. 2021. Enterococcus faecium are associated with the modification of gut microbiota and shrimp post-larvae survival. Anim Microbiome 3:88. doi: 10.1186/s42523-021-00152-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williams SL, Jensen RV, Kuhn DD, Stevens AM. 2017. Analyzing the metabolic capabilities of a Vibrio parahaemolyticus strain that causes early mortality syndrome in shrimp. Aquaculture 476:44–48. doi: 10.1016/j.aquaculture.2017.03.030 [DOI] [Google Scholar]
- 28. Ng TH, Lu C-W, Lin S-S, Chang C-C, Tran LH, Chang W-C, Lo C-F, Wang H-C. 2018. The Rho signalling pathway mediates the pathogenicity of AHPND-causing V. parahaemolyticus in shrimp. Cell Microbiol 20:e12849. doi: 10.1111/cmi.12849 [DOI] [PubMed] [Google Scholar]
- 29. Tinwongger S, Nochiri Y, Thawonsuwan J, Nozaki R, Kondo H, Awasthi SP, Hinenoya A, Yamasaki S, Hirono I. 2016. Virulence of acute hepatopancreatic necrosis disease Pir AB‐Like relies on secreted proteins not on gene copy number. J Appl Microbiol 121:1755–1765. doi: 10.1111/jam.13256 [DOI] [PubMed] [Google Scholar]
- 30. Pumkaew M, Taengchaiyaphum S, Powtongsook S, Pungrasmi W, Sritunyalucksana K. 2019. Production of acute hepatopancreatic necrosis disease toxin is affected by addition of cell‐free supernatant prepared from AI‐2‐Producing Vibrio harveyi mutant . J World Aquacult Soc 50:878–886. doi: 10.1111/jwas.12618 [DOI] [Google Scholar]
- 31. Soowannayan C, Boonmee S, Puckcharoen S, Anatamsombat T, Yatip P, Ng W-K, Thitamadee S, Tuchinda P, Munyoo B, Chabang N, Nuangsaeng B, Sonthi M, Withyachumnarnkul B. 2019. Ginger and its component shogaol inhibit Vibrio Biofilm formation in vitro and orally protect shrimp against acute hepatopancreatic necrosis disease (AHPND). Aquaculture 504:139–147. doi: 10.1016/j.aquaculture.2019.02.007 [DOI] [Google Scholar]
- 32. Kumar R, Ng TH, Wang HC. 2020. Acute hepatopancreatic necrosis disease in penaeid shrimp. Rev Aquacult 12:1867–1880. doi: 10.1111/raq.12414 [DOI] [Google Scholar]
- 33. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clément K, Doré J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker J-D, Raes J, Hansen T, MetaHIT consortium, Bork P, Wang J, Ehrlich SD, Pedersen O. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500:541–546. doi: 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 35. Li T, Li H, Gatesoupe F-J, She R, Lin Q, Yan X, Li J, Li X. 2017. “Bacterial signatures of “Red-Operculum” disease in the gut of crucian carp (Carassius auratus)”. Microb Ecol 74:510–521. doi: 10.1007/s00248-017-0967-1 [DOI] [PubMed] [Google Scholar]
- 36. Li E, Wang X, Chen K, Xu C, Qin JG, Chen L. 2017. Physiological change and nutritional requirement of Pacific white shrimp Litopenaeus vannamei at low salinity . Rev Aquacult 9:57–75. doi: 10.1111/raq.12104 [DOI] [Google Scholar]
- 37. Xiong J, Wang K, Wu J, Qiuqian L, Yang K, Qian Y, Zhang D. 2015. Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl Microbiol Biotechnol 99:6911–6919. doi: 10.1007/s00253-015-6632-z [DOI] [PubMed] [Google Scholar]
- 38. Xiong J, Zhu J, Dai W, Dong C, Qiu Q, Li C. 2017. Integrating gut microbiota immaturity and disease‐discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ Microbiol 19:1490–1501. doi: 10.1111/1462-2920.13701 [DOI] [PubMed] [Google Scholar]
- 39. Xie J-J, Liu Q, Liao S, Fang H-H, Yin P, Xie S-W, Tian L-X, Liu Y-J, Niu J. 2019. Effects of dietary mixed probiotics on growth, non-specific immunity, intestinal morphology and microbiota of juvenile Pacific white shrimp, Litopenaeus vannamei. Fish & Shellfish Immunology 90:456–465. doi: 10.1016/j.fsi.2019.04.301 [DOI] [PubMed] [Google Scholar]
- 40. Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. 2011. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav Immun 25:397–407. doi: 10.1016/j.bbi.2010.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Galley JD, Bailey MT. 2014. Impact of stressor exposure on the interplay between commensal microbiota and host inflammation. Gut Microbes 5:390–396. doi: 10.4161/gmic.28683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen W-Y, Ng TH, Wu J-H, Chen J-W, Wang H-C. 2017. Microbiome dynamics in a shrimp grow-out pond with possible outbreak of acute hepatopancreatic necrosis disease. Sci Rep 7:9395. doi: 10.1038/s41598-017-09923-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bass D, Stentiford GD, Wang H-C, Koskella B, Tyler CR. 2019. The pathobiome in animal and plant diseases. Trends in Ecology & Evolution 34:996–1008. doi: 10.1016/j.tree.2019.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yao Z, Yang K, Huang L, Huang X, Qiuqian L, Wang K, Zhang D. 2018. Disease outbreak accompanies the dispersive structure of shrimp gut bacterial community with a simple core microbiota. AMB Express 8:120. doi: 10.1186/s13568-018-0644-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ghenem L, Elhadi N, Alzahrani F, Nishibuchi M. 2017. Vibrio parahaemolyticus: a review on distribution, pathogenesis, virulence determinants and epidemiology. Saudi J Med Med Sci 5:93–103. doi: 10.4103/sjmms.sjmms_30_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rohmer L, Hocquet D, Miller SI. 2011. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol 19:341–348. doi: 10.1016/j.tim.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hsiao A, Ahmed AMS, Subramanian S, Griffin NW, Drewry LL, Petri WA, Haque R, Ahmed T, Gordon JI. 2014. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515:423–426. doi: 10.1038/nature13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwab C, Berry D, Rauch I, Rennisch I, Ramesmayer J, Hainzl E, Heider S, Decker T, Kenner L, Müller M, Strobl B, Wagner M, Schleper C, Loy A, Urich T. 2014. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J 8:1101–1114. doi: 10.1038/ismej.2013.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. 2017. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol 15:630–638. doi: 10.1038/nrmicro.2017.58 [DOI] [PubMed] [Google Scholar]
- 50. Yu W, Wu J-H, Zhang J, Yang W, Chen J, Xiong J. 2018. A meta-analysis reveals universal gut bacterial signatures for diagnosing the incidence of shrimp disease. FEMS Microbiol Ecol 94:fiy147. doi: 10.1093/femsec/fiy147 [DOI] [PubMed] [Google Scholar]
- 51. Soo TCC, Bhassu S. 2022. Biochemical indexes and gut microbiota testing as diagnostic methods for Penaeus monodon health and physiological changes during AHPND infection with food safety concerns. Food Sci Nutr 10:2694–2709. doi: 10.1002/fsn3.2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Manilal A, Sujith S, Selvin J, Shakir C, Gandhimathi R, Kiran GS. 2010. Virulence of vibrios isolated from diseased black Tiger shrimp, Penaeus monodon, Fabricius. J World Aquac Soc 41:332–343. doi: 10.1111/j.1749-7345.2010.00375.x [DOI] [Google Scholar]
- 53. Sung H-H, Hsu S-F, Chen C-K, Ting Y-Y, Chao W-L. 2001. Relationships between disease outbreak in cultured tiger shrimp (Penaeus monodon) and the composition of Vibrio communities in pond water and shrimp hepatopancreas during cultivation. Aquaculture 192:101–110. doi: 10.1016/S0044-8486(00)00458-0 [DOI] [Google Scholar]
- 54. Prayitno SB, Latchford JW. 1995. Experimental infections of crustaceans with luminous bacteria related to Photobacterium and Vibrio. effect of Salinity and pH on Infectiosity. Aquaculture 132:105–112. doi: 10.1016/0044-8486(94)00374-W [DOI] [Google Scholar]
- 55. Deng Y, Zhang Y, Chen H, Xu L, Wang Q, Feng J. 2020. Gut–liver immune response and gut microbiota profiling reveal the pathogenic mechanisms of Vibrio harveyi in pearl gentian grouper (Epinephelus lanceolatus♂× E. fuscoguttatus♀). Front Immunol 11. doi: 10.3389/fimmu.2020.607754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ringø E, Olsen RE, Jensen I, Romero J, Lauzon HL. 2014. Application of vaccines and dietary supplements in aquaculture: possibilities and challenges. Rev Fish Biol Fisheries 24:1005–1032. doi: 10.1007/s11160-014-9361-y [DOI] [Google Scholar]
- 57. Kostanjsek R, Strus J, Avgustin G. 2007. Candidatus Bacilloplasma,” a novel lineage of Mollicutes associated with the hindgut wall of the terrestrial isopod Porcellio scaber (Crustacea: Isopoda. Appl Environ Microbiol 73:5566–5573. doi: 10.1128/AEM.02468-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hou D, Huang Z, Zeng S, Liu J, Wei D, Deng X, Weng S, Yan Q, He J. 2018. Intestinal bacterial signatures of white feces syndrome in shrimp. Appl Microbiol Biotechnol 102:3701–3709. doi: 10.1007/s00253-018-8855-2 [DOI] [PubMed] [Google Scholar]
- 59. Huang Z, Zeng S, Xiong J, Hou D, Zhou R, Xing C, Wei D, Deng X, Yu L, Wang H, Deng Z, Weng S, Kriengkrai S, Ning D, Zhou J, He J. 2020. Microecological Koch’s postulates reveal that intestinal microbiota dysbiosis contributes to shrimp white feces syndrome. Microbiome 8:1–13. doi: 10.1186/s40168-020-00802-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang J, Huang Y, Xu K, Zhang X, Sun H, Fan L, Yan M. 2019. White spot syndrome virus (WSSV) infection impacts intestinal microbiota composition and function in Litopenaeus vannamei. Fish & Shellfish Immunology 84:130–137. doi: 10.1016/j.fsi.2018.09.076 [DOI] [PubMed] [Google Scholar]
- 61. Duan Y, Lu Z, Zeng S, Dan X, Mo Z, Zhang J, Li Y. 2021. Integration of intestinal microbiota and transcriptomic and metabolomic responses reveals the toxic responses of Litopenaeus vannamei to microcystin-LR. Ecotoxicol Environ Saf 228:113030. doi: 10.1016/j.ecoenv.2021.113030 [DOI] [Google Scholar]
- 62. Lustri BC, Sperandio V, Moreira CG. 2017. Bacterial chat: intestinal metabolites and signals in host-microbiota-pathogen interactions. Infect Immun 85:e00476-17. doi: 10.1128/IAI.00476-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim Y-K, McCarter LL. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus . J Bacteriol 182:3693–3704. doi: 10.1128/JB.182.13.3693-3704.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gao S, Pan L, Huang F, Song M, Tian C, Zhang M. 2019. Metagenomic insights into the structure and function of intestinal microbiota of the farmed Pacific white shrimp (Litopenaeus vannamei). Aquaculture 499:109–118. doi: 10.1016/j.aquaculture.2018.09.026 [DOI] [Google Scholar]
- 65. Ringø E, Harikrishnan R, Soltani M, Ghosh K. 2022. The effect of gut microbiota and probiotics on metabolism in fish and shrimp. Animals (Basel) 12:3016. doi: 10.3390/ani12213016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Y-B. 2007. Effect of probiotics on growth performance and digestive enzyme activity of the shrimp Penaeus vannamei. Aquaculture 269:259–264. doi: 10.1016/j.aquaculture.2007.05.035 [DOI] [Google Scholar]
- 67. Tzuc JT, Escalante DR, Rojas Herrera R, Gaxiola Cortés G, Ortiz MLA. 2014. Microbiota from Litopenaeus vannamei: digestive tract microbial community of Pacific white shrimp (Litopenaeus vannamei). Springerplus 3:280. doi: 10.1186/2193-1801-3-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dai Z-L, Wu G, Zhu W-Y. 2011. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci (Landmark Ed) 16:1768–1786. doi: 10.2741/3820 [DOI] [PubMed] [Google Scholar]
- 69. Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. 2011. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A 108 Suppl 1:4523–4530. doi: 10.1073/pnas.1006734107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kumar R, Tung T-C, Ng TH, Chang C-C, Chen Y-L, Chen Y-M, Lin S-S, Wang H-C. 2021. Metabolic alterations in shrimp stomach during acute hepatopancreatic necrosis disease and effects of Taurocholate on Vibrio parahaemolyticus. Front Microbiol 12:631468. doi: 10.3389/fmicb.2021.631468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kumar R, Ng TH, Chang CC, Tung TC, Lin SS, Lo CF, Wang HC. 2020. Bile acid and bile acid transporters are involved in the pathogenesis of acute hepatopancreatic necrosis disease in white shrimp Litopenaeus vannamei. Cell Microbiol 22:e13127. doi: 10.1111/cmi.13127 [DOI] [PubMed] [Google Scholar]
- 72. Yang M, Liu Z, Hughes C, Stern AM, Wang H, Zhong Z, Kan B, Fenical W, Zhu J. 2013. Bile salt–induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc Natl Acad Sci U S A 110:2348–2353. doi: 10.1073/pnas.1218039110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sanchez LM, Cheng AT, Warner CJA, Townsley L, Peach KC, Navarro G, Shikuma NJ, Bray WM, Riener RM, Yildiz FH, Linington RG. 2016. Biofilm formation and detachment in gram-negative pathogens is modulated by select bile acids. PLoS One 11:e0149603. doi: 10.1371/journal.pone.0149603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee C-T, Chen I-T, Yang Y-T, Ko T-P, Huang Y-T, Huang J-Y, Huang M-F, Lin S-J, Chen C-Y, Lin S-S, Lightner DV, Wang H-C, Wang A-J, Wang H-C, Hor L-I, Lo C-F. 2015. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc Natl Acad Sci U S A 112:10798–10803. doi: 10.1073/pnas.1503129112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS II, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j 17:10. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 78. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Price MN, Dehal PS, Arkin AP. 2010. Fasttree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bolyen E, Rideout JR, Dillon MR, Al-Ghalith GA, Asnicar F, Arumugam v, Alm EJ, Alexander H, Bokulich NA, Abnet C. 2018. QIIME 2: Reproducible, interactive, Scalable, and extensible Microbiome data science. PeerJ Preprints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V. 2011. Scikit-learn: Machine learning in python. J Mach Learn Res 12:2825–2830. [Google Scholar]
- 83. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–6. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dray S, Legendre P, Peres-Neto PR. 2006. Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecological Modelling 196:483–493. doi: 10.1016/j.ecolmodel.2006.02.015 [DOI] [Google Scholar]
- 85. Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi: 10.1128/AEM.01996-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wickham H, Chang W, Wickham MH.. 2016. Package ‘ggplot2’. Create elegant data visualisations using the grammar of graphics Version 2:1-189.
- 87. Team RC . 2013. R: A language and environment for statistical computing.
- 88. Liaw A, Wiener M. 2002. Classification and regression by randomForest. R news 2:18–22. [Google Scholar]
- 89. Wemheuer F, Taylor JA, Daniel R, Johnston E, Meinicke P, Thomas T, Wemheuer B. 2020. Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ Microbiome 15:11. doi: 10.1186/s40793-020-00358-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. 2023. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res 51:D587–D592. doi: 10.1093/nar/gkac963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:1–18. doi: 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5 and Tables S1 to S7.
Data Availability Statement
The raw sequencing data reported in this study has been archived in the Sequence Read Archive of the National Center for Biotechnology Information under accession number PRJNA921960.