Abstract
(Genome-wide Association Studies) GWAS have identified ∼42 late-onset Alzheimer’s disease (LOAD)-associated loci, each of which contains multiple single nucleotide polymorphisms (SNPs) in linkage disequilibrium (LD) and most of these SNPs are in the non-coding region of human genome. However, how these SNPs regulate risk gene expression remains unknown. In this work, by using a set of novel techniques, we identified 6 functional SNPs (fSNPs) rs9271198, rs9271200, rs9281945, rs9271243, and rs9271247 on the LOAD-associated HLA-DRB1/DQA1 locus and 42 proteins specifically binding to five of these 6 fSNPs. As a proof of evidence, we verified the allele-specific binding of GATA2 and GATA3, ELAVL1 and HNRNPA0, ILF2 and ILF3, NFIB and NFIC, as well as CUX1 to these five fSNPs, respectively. Moreover, we demonstrate that all these nine proteins regulate the expression of both HLA-DQA1 and HLA-DRB1 in human microglial cells. The contribution of HLA class II to the susceptibility of LOAD is discussed.
Keywords: functional SNPs, post-GWAS functional analysis, Late onset Alzheimer's disease (LOAD)
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder that affects ∼50 million people and that number is predicted to reach around 152 million by 2050 in the world. 1 It is an age-related disease that affects 5% of all people by age of 65, however, 50% by age of 85. According to the age at onset, AD can be classified into early‐onset AD (EOAD) and late‐onset AD (LOAD). While EOAD is mainly caused by mutations in amyloid precursor protein (APP), presenilin 1 (PSEN1) and presenilin 2 (PSEN2), LOAD is likely to be driven by a complex interplay between genetic and environmental factors. 2 Clinically, AD is characterized by severe behavioral and cognitive impairments, which often lead to memory loss in patients. 3 Pathologically, AD is characterized by two neuropathological hallmarks: that is, neurofibrillary tangles of hyperphosphorylated tau and beta-amyloid (Aβ) plaques in the brains of AD patients. 4 Although the exact etiology of AD is unknown, it is now believed that genetics plays a big role in the pathogenesis of this disease contributed to ∼70% of AD risk. 5
Genome‐wide association studies (GWAS) have identified ∼42 loci that are specifically associated with LOAD.6-13 These include multiple risk genes that are involved in immune responses such as CD33, TREM2, and HLA-DRB1/DQA1,14,15 implicating the important role of neuroinflammation in AD pathogenesis. It is currently accepted that the contribution of each of these LOAD risk genes is rather small, but a combination of multiple risk genes, which seemingly have no connections, that could form a presently uncharacterized genetic network that ultimately determines a person’s overall susceptibility to AD. Therefore, it is important to understand how each risk gene expression is regulated by the LOAD-associated fSNPs and how the regulation of all the risk genes is connected.
In humans, the human leukocyte antigen (HLA) locus encodes 6 classical transplantation HLA genes and at least 132 protein coding genes that have important roles in the regulation of the immune system as well as some other fundamental molecular and cellular processes.16,17 This ∼.7 Mb DNA segment of human genome has been associated with more than 100 different diseases, including Alzheimer’s disease, rheumatoid arthritis and multiple sclerosis.18-20 An early GWAS analysis identified one SNP rs9271192 on the HLA-DRB1/DQA1 locus associated with LOAD with an overall OR = 1.11 and P < 2.9 × 10−1221 and, later on, this association was independently validated by other two groups.22,23 However, in linkage disequilibrium (LD) with this tagged SNP (r2 > .8), there are additional 58 SNPs. All these SNPs are located in the non-coding region having no effect on the protein coding sequences. Therefore, if such a SNP is functional, it likely exerts its function as part of a regulatory element to modulate risk gene expression by recruiting regulatory proteins. However, technically to say, it is difficult to identify the non-coding functional SNPs (fSNPs) among all the disease-associated SNPs in LD and it is also difficult to understand how these non-coding fSNPs influence the susceptibility of AD. Other fSNPs could be SNPs in the coding regions (rare variants) altering protein sequences, in the microRNA changing the microRNA sequences, or on the RNA splicing sites disrupting RNA splicing.
Both HLA-DRB1 and HLA-DQA1 are HLA class II molecules that are normally expressed only on the surface of professional antigen-presenting cells (APCs) such as dendritic cells, macrophages and B cells. They are important in presenting epitopic peptides to CD4+ T cells and initiating adaptive immune responses including secretion of cytokines and generation of antibodies. 24 In the central nervous system (CNS), microglia is a brain-resident macrophage acting as the main form of immune defense within the brain. 25 While expression of HLA class II in microglia is low in homeostatic conditions in the brain, it can be rapidly upregulated in activated microglia in neurodegenerative diseases. 26 In AD, microglia can be activated by the accumulation of amyloid plagues, which was evidenced by the presence of activated microglia in pathological lesions in AD.27-29 However, how these activated microglia regulate the expression of HLA class II gene in response to amyloid deposition is large unknown.
Recently, we have developed a sequential methodology to not only identify, but also characterize disease-associated fSNPs. 30 We use: (1) Reel-seq, an electrophoresis mobility shift assay (EMSA)-based high throughput techniques to identify fSNPs; (2) SNP-specific DNA competition pulldown (SDCP)-MS, a unique DNA pulldown assay modified from Flanking restriction enhanced pulldown (FREP)-MS 31 to identify fSNP-bound regulatory proteins; and (3) Allele-imbalanced DNA pulldown (AIDP)-Wb, a Western blot analysis to validate both a fSNP and its binding protein simultaneously by detecting allele-specific protein:fSNP binding. In this study, using this approach, we identified and validate 6 noncoding fSNPs on the LOAD-associated HLA locus. We characterized nine proteins that regulate both HLA-DQA1 and HLA-DRB1 expression via binding to the five fSNPs on the LOAD-associated HLA-DRB1/DQA1 locus in human microglia cells.
Results
Identification and Validation of 6 fSNPs on the LOAD-Associated HLA-DRB1/DQA1 Locus Using Reel-seq
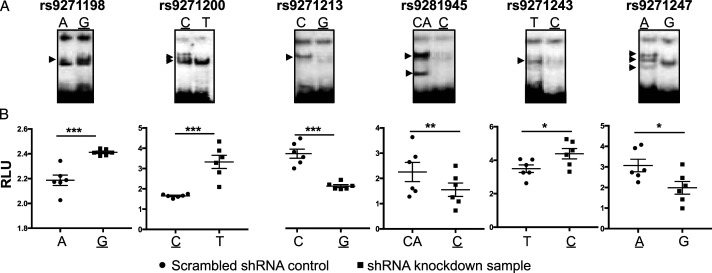
GWAS have identified an association of the HLA-DRB1/DQA1 locus with LOAD. 21 At this locus in LD with r2 > .8, there are 58 disease-associated SNPs in the non-coding region. To understand the mechanisms underlying the contribution of these disease-associated SNPs to the susceptibility of AD, by using Reel-seq, 30 we identified 6 candidate fSNPs, rs9271198, rs9271200, rs9271213, rs9271243, rs9271247, and rs9281945 with a -.05 > slope > .05 and a P-value < .05; 11 putative fSNPs with a -.05 < slope < .05 and a P-value < .05; and 42 non-fSNPs with a P-value > .05 including 4 SNPs with incomplete data from the screen (Supplemental Table 1). To demonstrate if these 6 candidate fSNPs are functional, EMSA was performed using nuclear extract (NE) isolated from microglia cell line HMC3. As the results presented in Figure 1A, all these 6 SNPs showed a pattern of allele-imbalanced gel shifting, indicating that they are functional. We also performed the same EMSA using NE isolated from human frontal cortex and similar results were observed (Supplemental Figure 1). However, the patterns of the allele-imbalanced gel shifting are slightly different from that observed in the EMSA using NE isolated from HMC3, suggesting a cell-type specificity. To further demonstrate these 6 fSNPs, a luciferase reporter assay was performed in HMC3 cells and, consistently, our results showed that each of these 6 SNPs has a significant allele-imbalanced luciferase activity between the risk and non-risk alleles (Figure 1B). Together, these data clearly confirm that all these 6 candidate fSNPs are functional.
Figure 1.
Validation of 6 candidate fSNPs on the HLA-DQA1/DRB1 locus. (A). EMSA showing allele-imbalanced gel shifting with the 6 potential fSNPs on the HLA-DRB1/DQA1 locus using NE from human microglia cell line HMC3. Arrow indicating the allele-imbalanced gel shifting. Risk alleles are underlined. Data for Western blots represents three biologically independent experiments (n = 3). (B). Luciferase reporter assay showing allele-imbalanced luciferase activity with the 6 potential fSNPs in HMC3. RLU: relative luciferase unit. Data for luciferase reporter assay represents 6 biologically independent repeats (n = 6). * P < .05; ** P < .001; and *** P, .0001.
In addition, we tested the 11 putative fSNPs and 10 SNPs randomly selected from the 42 non-fSNPs. At least 8 putative fSNPs and 4 non-fSNPs showed allele-imbalanced gel shifting (Supplemental Figure 1), indicating that they are potentially functional. This data together with the data obtained from Figure 1 also suggest a ∼83% positive recovery rate and a 40% false negative rate for this Reel-seq screen. We also performed in silico analysis on these 6 fSNPs using HaploReg 4.1, a web-based tool for epigenetic and functional annotation of genetic variants. 32 We analyzed each of the 6 SNPs on a scale of 0 to 5 to reflect the number of positive annotations for promoter and enhancer histone methylation, DNase hypersensitivity, predicted protein binding, and predicted alteration in binding motifs. As results shown in Supplemental Table 2, we scored 4 on both rs9271198 and rs9271200, 3 on rs9271213, 2 on both rs9271243 and rs9281945, and one on rs9271247, indicating that there was no obvious concordance between the Reel-seq screen and the in silico analysis using Haploreg in identifying fSNPs.
Identification of Proteins That Specifically Bind to the 6 Confirmed fSNPs at the HLA-DRB1/DQA1 Locus by SDCP-MS
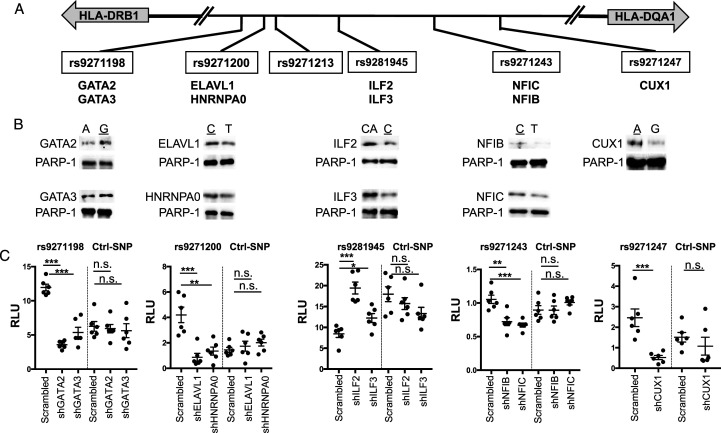
As shown in Figure 2A, all the 6 confirmed fSNPs are located in the non-coding region in between HLA-DRB1 and HLA-DQA1, which are in opposite transcriptional orientations. As non-coding fSNPs, all these SNPs may likely exert their function as part of a regulatory element by binding to regulatory proteins modulating risk gene expression.33-36 To identify the regulatory proteins that specifically bind to the 6 fSNPs, we performed SDCP-MS 30 using the alleles that bind more proteins as shown in Figure 1A. In total, 199 proteins were recovered, among which 109 were filtered for their non-specific binding to all these 6 fSNPs and 48 for binding to the 6 fSNPs without reproducibility. By removing these 157 proteins, we identified 42 unique proteins that bind to 5 fSNPs as shown in Table 1. Some of these proteins are highly specific as they only bind to one fSNP such as CUX1 to rs9271247 and ILF2 and ILF3 to rs9271243, others are relatively specific as they bind to more than one fSNPs. However, we did not identify any protein that specifically binds to rs9271213. Among these 42 proteins, 36 were reported as nuclear proteins, four cytosolic proteins and two membrane proteins as analyzed by Ingenuity Pathway Analysis (IPA, Qiagen) (Supplemental Figure 3). Also, among these 42 proteins, only 16 were previously reported as transcription factors including GATA2/3, DLX6, HOXD9, PRRX2, TEAD1, ILF2/3, DEK, ZNF638, NFIB/C/A/X, KHDRBS1, and CUX1.
Figure 2.
Characterization of nine proteins that specifically bind to the five fSNPs at the HLA-DQA1/DRB1 locus. (A). Partial genomic arrangement showing the relative location of the 6 fSNPs in between HLA-DQA1 and HLA-DRB1 and the nine proteins identified by SDCP-MS. Risk alleles are underlined. (B). AIDP-Wb showing the allele-imbalanced binding of GATA2 and GATA3 to rs9271198; ELAVL1and HNRNPA0 to rs9271200; ILF2 and ILF3 to rs9281945; NFIB and NFIC to rs9271243; and CUX1 to rs9271247. PARP-1, a double-stranded DNA end binding protein was used as an internal loading control. Data for AIDP-Wb represents three biologically independent experiments (n = 3). (C). Luciferase reporter assay demonstrating the specific binding of these nine proteins to their corresponding fSNPs by showing altered luciferase activities when all these nine proteins were downregulated by shRNA knockdown in HMC3 cells. Ctrl: negative control with an irrelevant SNP sequence. Data for luciferase reporter assay represents 6 biologically independent repeats (n = 6). * P < .05; ** P < .001; and *** P, .0001. n.s.: not significant.
Table 1.
Peptide spectrum counts showing proteins that specifically bind to the five fSNPs on the HLA-DQA1/DRB1 locus. Each SNP was performed in duplicate.
| Gene name | rs9271198 | rs9271200 | rs9281945 | rs9271243 | rs9271247 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| repeat1 | repeat2 | repeat1 | repeat2 | repeat1 | repeat2 | repeat1 | repeat2 | repeat1 | repeat2 | |
| GATA2 | 8 | 14 | 1 | 1 | ||||||
| GATA3 | 5 | 8 | ||||||||
| DLX6 | 4 | 5 | ||||||||
| HOXD9 | 2 | 2 | ||||||||
| PRRX2 | 3 | 3 | ||||||||
| TEAD1 | 2 | 1 | ||||||||
| E9PSI1 | 1 | 2 | ||||||||
| RPS9 | 2 | 1 | ||||||||
| ELAVL1 | 1 | 1 | 5 | 2 | ||||||
| HNRNPA0 | 3 | 5 | 5 | 7 | ||||||
| TCOF1 | 8 | 6 | 22 | 15 | ||||||
| PCBP1 | 2 | 2 | 6 | 1 | ||||||
| SRP14 | 2 | 2 | 5 | 5 | ||||||
| NOLC1 | 3 | 3 | 6 | 6 | ||||||
| HNRNPK | 1 | 1 | 2 | 5 | 5 | 4 | ||||
| ILF2 | 1 | 0 | 9 | 9 | ||||||
| ILF3 | 17 | 16 | ||||||||
| DHX9 | 25 | 8 | ||||||||
| TMPO | 3 | 1 | ||||||||
| HNRNPR | 3 | 2 | ||||||||
| ASH2L | 6 | 2 | ||||||||
| LRP2 | 1 | 3 | ||||||||
| POLDIP3 | 1 | 2 | 5 | 4 | 2 | 2 | ||||
| PRPF19 | 1 | 1 | 6 | 4 | 2 | 2 | ||||
| ADAR | 3 | 5 | ||||||||
| TOP2A | 6 | 5 | ||||||||
| HNRNPC | 8 | 7 | 3 | 3 | ||||||
| PTBP1 | 3 | 1 | ||||||||
| DEK | 7 | 10 | 5 | 1 | 7 | 8 | ||||
| ZNF638 | 1 | 2 | ||||||||
| MATR3 | 10 | 3 | ||||||||
| RECQL | 5 | 4 | ||||||||
| NFIB | 3 | 8 | 30 | 31 | ||||||
| NFIC | 1 | 4 | 20 | 25 | ||||||
| NFIX | 8 | 6 | ||||||||
| NFIA | 7 | 7 | ||||||||
| U2AF1 | 4 | 4 | ||||||||
| KHDRBS1 | 5 | 3 | 1 | 1 | ||||||
| SSBP1 | 2 | 3 | ||||||||
| HNRNPH1 | 1 | 2 | ||||||||
| CUX1 | 48 | 50 | ||||||||
Validation of Nine Proteins Specifically Binding to the Five fSNPs at the HLA-DRB1/DQA1 Locus by AIDP-Wb and luciferase Reporter Assay
To demonstrate that these 42 proteins identified by SDCP-MS specifically bind to their corresponding fSNPs, as a proof of evidence, we applied two assays on nine of these 42 proteins, they are GATA2 and GATA3 on rs9271198; ELAVL1 and HNRNPA0 on rs9271200; ILF2 and ILF3 on rs9281945; NFIB and NFIC on rs9271243; and CUX1 on rs9271247 as shown in Figure 2A. First, we performed AIDP-Wb(30) to detect allele-specific protein:fSNP binding instead of protein:DNA binding as both gel supershift assay and ChIP assay do. By using AIDP-Wb, we validated both GATA2 and GATA3 binding to rs9271198 with the risk allele G binding more than the non-risk allele A; ELAVL1 and HNRNPA0 to rs9271200 with the risk allele C binding more than the non-risk allele T; ILF2 and ILF3 to rs9281945 with the risk allele C binding less than the non-risk allele CA; NFIB and NFIC to rs9271243 with the risk allele C binding less than the non-risk allele T; and CUX1 to rs9271247 with the risk allele A binding more than the non-risk allele G (Figure 2B). The allele-imbalanced binding of these nine proteins to their corresponding fSNPs verifies these 9 proteins as fSNP-bound proteins, and, at the same time, it also validates these 5 SNPs as fSNPs.
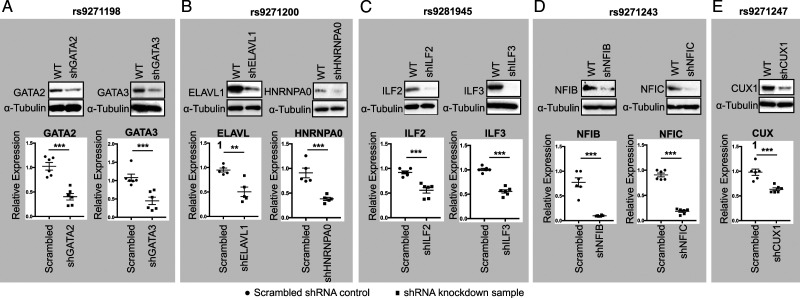
Second, we performed a luciferase reporter assay using the reporter construct used in Figure 1B that carry the risk allele for each of these 5 fSNP such as G allele from rs9271198; A from rs9271247; and C from rs9271200, rs9281945, and rs9271243. To perform this assay, we first generated HMC3 cell lines with each of these nine proteins downregulated by using a shRNA lentivirus as shown in Figure 3A-3E. We performed these luciferase reporter assays in these shRNA knockdown cells together with their scrambled shRNA controls. As can be seen in Figure 2C, knockdown of GATA2 and GATA3; ELAVL1 and HNRNPA0; NFIB and NFIC; and CUX1 results in a significant decrease in luciferase activities on the fSNP rs9271198; rs9271200; rs9271243; and rs9271247, respectively, by comparing to their corresponding controls. However, downregulation of ILF2 and ILF3 results in an increased luciferase activity on the fSNP rs9281945 (Figure 2C), suggesting that these two proteins function as transcriptional suppressors. We also performed a control luciferase reporter assay using a construct containing an irrelevant SNP sequence in the same shRNA knockdown HMC3 cells. In this case, no significant change of luciferase activities was observed (Figure 2C). These data suggest that these nine proteins are the regulatory proteins that regulate luciferase activity via interacting with the fSNPs in the reporter constructs.
Figure 3.
Western blots (Upper panel) and qPCR (Lower panel) showing shRNA knockdown of GATA2 and GATA3 (A); ELAVL1 and HNRNPA0 (B); ILF2 and ILF3 (C); NFIB and NFIC (D); and CUX1 (E) in HMC3 cells. Data for Western blots represents three biologically independent experiments (n = 3). Data for qPCR represents the combination of three biologically independent experiments (n = 3), each in duplicate. α-Tubulin: loading control for Western blot and GAPDH was used as control for qPCR. * P < .05; ** P < .001; and *** P, .0001.
Together, the data from both AIDP-Wb and luciferase reporter assay demonstrate that at least nine of the 42 proteins identified by SDCP-MS are fSNP binding proteins.
Regulation of HLA-DQA1 and HLA-DRB1 Expression by GATA2 and GATA3; ELAVL1 and HNRNPA0; ILF2 and ILF3; NFIB and NFIC; and CUX1
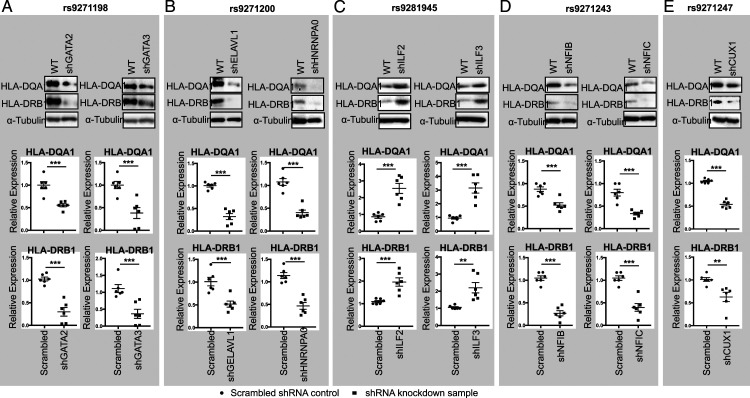
The allele-imbalanced binding of GATA2 and GATA3 to rs9271198; ELAVL1 and HNRNPA0 to rs9271200; ILF2 and ILF3 to rs9281945; NFIB and NFIC to rs9271243; and CUX1 to rs9271247 (Figure 2B), as well as the luciferase reporter assay (Figure 2C) suggest that these nine proteins could be the potential transcription regulators that control the HLA-DQA1 and HLA-DRB1 expression. To demonstrate this, using shRNA lentiviruses, we first generated polyclonal HMC3 cells that have a reduced expression on GATA2 and GATA3 (Figure 3A); ELAVL1 and HNRNPA0 (Figure 3B); ILF2 and ILF3 (Figure 3C); NFIB and NFIC (Figure 3D); and CUX1 (Figure 3E). Downregulation of these nine genes was validated on both protein (Figure 3, Upper panel) and mRNA levels (Figure 3, Lower panel). shRNA knockdown of GATA2, GATA3, ELAVL1, HNRNPA0, NFIB, NFIC, and CUX1 equivalently downregulated the expression of both HLA-DRB1 and HLA-DQA1 (Figures 4A, 4B, 4D, and 4E), however, shRNA knockdown of ILF2 and ILF3 resulted in a significant upregulation of both HLA-DRB1 and HLA-DQA1 expression (Figure 4C). In all these cases, regulation of HLA-DRB1 and HLA-DQA1 expression was evidenced on both the mRNA (Figure 4, Upper) and protein level (Figure 4, Lower), which indicates that these 9 proteins are transcriptional regulators. In addition, these data are also consistent with the luciferase reporter assays showed in Figure 2C that indicate GATA2, GATA3, ELAVL1, HNRNPA0, NFIB, NFIC, and CUX1 are transcriptional activators and ILF2 and ILF3 are transcriptional suppressors. To further confirm these data, as a proof of evidence, we also performed siRNA knockdown on five of these nine proteins: GATA2, GATA3, NFIB, NFIC, and CUX1 in human HMC3 cells with the targeting sequences in these five siRNAs different from that in the shRNA knockdown. Consistent with our shRNA knockdown assays, we observed a significant downregulation of both HLA-DRB1 and HLA-DQA1 expression in these siRNA knockdown cells detected by qPCR (Supplemental Figure 4). In addition, based on an eQTL analysis (https://gtexportal.org/home/snp/rs9271192.), in human brain, we also identified 6 additional candidate risk genes in the HLA locus associated with rs9271192 including HLA-DMA, HLA-DQA2, HLA-DQB1, HLA-DQB2, HLA-DRB5, and HLA-DRB6. To assess if these genes are also regulated by the transcriptional regulators identified by SDCP-MS, as a proof of evidence, we performed qPCR analysis on these genes in the CUX1, GATA2, and GATA3 shRNA knockdown HMC3 cells. While we cannot detect the expression of HLA-DQA2, HLA-DQB2, HLA-DRB5, and HLA-DRB6 in these HMC3 cells, a significant downregulation of both HLA-DMA and HLA-DQB1 were observed in these three shRNA knockdown HMC3 cells (Supplemental Figure 5).
Figure 4.
Western blots (Upper panel) and qPCR (Lower panel) showing the altered expression of HLA-DQA1 and HLA-DRB1 in the GATA2 and GATA3 (A); ELAVL1 and HNRNPA0 (B); ILF2 and ILF3 (C); NFIB and NFIC (D); and CUX1 (E) shRNA knockdown HMC3 cells. Data for Western blots represents three biologically independent experiments (n = 3). Data for qPCR represents the combination of three biologically independent experiments (n = 3), each in duplicate. α-Tubulin: loading control for Western blot and GAPDH was used as control for qPCR. * P < .05; ** P < .001; and *** P, .0001.
Together, our results demonstrate that GATA2, GATA3, ELAVL1, HNRNPA0, NFIB, NFIC, CUX1, and ILF2 and ILF3 are the transcription regulators that control the LOAD risk genes HLA-DRB1 and HLA-DQA1 expression at least in human HMC3 cells.
Discussion
The LOAD-associated SNP rs9271192 was showed to be significantly associated with increased levels of HLA-DRB1 in the brain of all patients with AD. 37 In this study, using Reel-seq, we identified 6 fSNPs on the LOAD-associated HLA-DRB1/DQA1 locus. To further investigate the role of these 6 fSNPs, using SDCP-MS, we identified 42 proteins that specifically bind to five of the 6 fSNPs and, as a proof of evidence, we validated nine of these proteins by showing allele-imbalanced binding to the five fSNPs by using AIDP-Wb and luciferase reporter assay. We also demonstrated these nine proteins are the HLA-DRB1/DQA1 regulators. Among these nine proteins, we observed that more binding of GATA2, GATA3, ELAVL1, HNRNPA0, NIFB, NIFC, and CUX1 to their corresponding risk alleles was correlated to an increased activation of HLA-DRB1/DQA1, suggesting that these five proteins are the activators. We also observed that less binding of ILF2 and ILF3 to the risk allele of rs9281945 activated the HLA-DRB1/DQA1 expression, indicating that these two proteins are the suppressors of HLA-DRB1/DQA1. These findings explain the mechanisms underlying the regulation of AD risk gene HLA-DRB1/DQA1 expression. Among these 9 proteins, CUX1 is the layer maker in human cerebral cortex with high expression. 38 Also, transcriptome analysis in AD patients demonstrated that both CUX1 and HLA-DQA1 were upregulated in hippocampus. 39 The same transcriptome analysis also showed that both GATA2 and GATA3 were co-expressed with HLA-DQA1 in AD hippocampus. 39 All these reports are consistent with our findings that CUX1, GATA2/3 are a transcriptional activator for HLA expression in human brain. In addition, we also investigated the possibility that Aβ activates the HLA-DRB1/DQA1 expression via modulating CUX1 as well as GATA2/3 by treating human microglia cell HMC3 with aggregated Aβ42 and no induction of CUX1 as well as GATA2/3 was observed (data not shown).
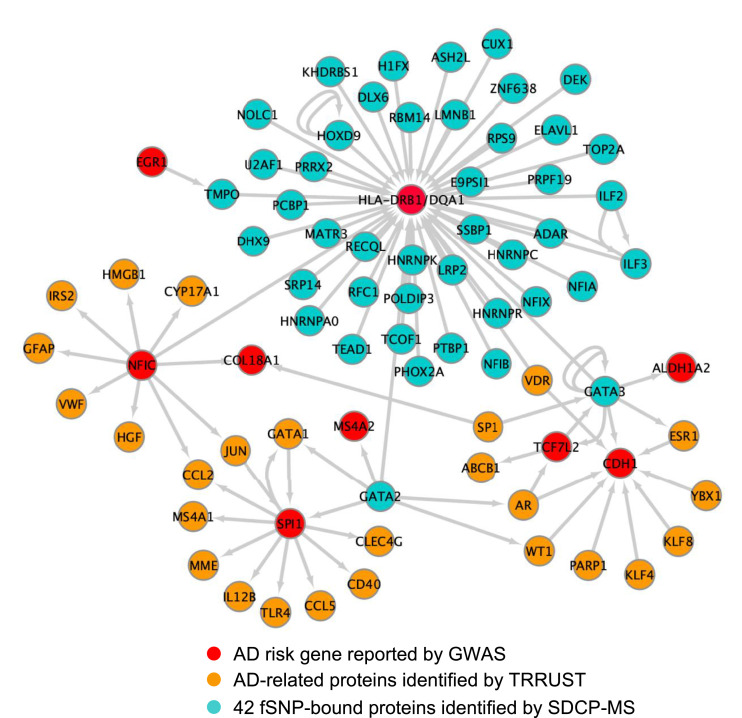
To further understand the contribution of these AD transcriptional regulators to the pathogenesis of, and the susceptibility of AD, we applied TRRUST v2, a manually curated database of human and mouse transcriptional regulatory networks, 40 to generate an HLA-DRB1/DQA1-related transcriptional regulation network (TRN) that is associated with AD as shown in Figure 5. In this TRN, we discovered additional 8 AD risk genes: SPI1 (PU.1), MS4A2, TCF7L2, CDH1, ALDH1A2, COL18A1, EGR1, and NFIC. NFIC itself is one of the 42 transcriptional regulators identified by SDCP-MS. Among these 8 AD risk genes, SPI1 (PU.1) and MS4A2 are regulated by GATA2; TCF7L2, CDH1, and ALDH1A2 by GATA3; and COL18A1 by NFIC. As previously reported, SPI1 (PU.1) is a critical transcriptional factor regulating both AD-associated genes in primary human microglia 41 and microglial inflammatory response. 42 MS4A2 is a key modulator of soluble TREM2, another LOAD risk gene. 43 CDH1, an activator subunit of APC/C (The anaphase promoting complex/cyclosome), is highly expressed in post-mitotic neurons 44 and its degradation induced by Aβ resulted in neuronal apoptosis. 45 These TRN analyses, while preliminary, are consistent with the notion that even though each fSNP may confer small individual risk by regulating one risk gene expression, functional combination of all the fSNPs via the disease-associated signal transduction and transcriptional regulation networks (STTRNs), 46 together with environmental factors, are believed to contribute significantly to the etiology of the disease.
Figure 5.
TRN analysis by TRRUST v2. Simplified diagram showing the HLA-DRB1/DQA1-related transcriptional regulation network that is associated with AD. Grey arrows indicate transcription regulation identified in this work as well as revealed by TRRUST v2.
HLA-DRB1/DQA1 belongs to class II HLA antigen that is a glycoprotein complex on the surface of professional antigen-presenting cells that displays short antigen peptides to CD4+ helper T cells activating adaptive immunity. Within the brain, class II HLA is primarily expressed on microglia, where it is generally considered a marker of activated cells. 47 As reviewed by Hopperton et al 48 (2018), around 50 studies were reported that quantitatively compared markers of class II HLA between AD and control in post mortem human brains. Even though there exits some controversy, most of the data indicate that AD pathology may stimulate upregulation of HLA class II. While these data are consistent with our findings, at the current stage, it is still not clear whether the increased expression of HLA-DRB1/DQA1 in AD is the cause, rather than a result of AD pathology. Besides, in addition to their classical function in antigen presentation, class II HLA molecules were also shown to be involved in activating signaling pathways such as PKC and TLR leading to cell death, and thereby contribute to termination of the immune response.49,50 However, whether or not the non-classical function of class II HLA molecules plays a role in the pathogenesis of AD remains undetermined.
Materials and Methods
Cell Culture
The human microglia cell line HMC3 was purchased from ATCC (cat#: CRL-3304). It is free of mycoplasma. HMC3 cells were cultured in a 1:1 mixture of DMEM and F12 Medium containing 10% FBS. All cells were cultured at 37°C in 5% CO2.
Primers and Antibodies
All primers used are listed in Supplemental Table 3. Anti-human antibodies used are listed in Supplemental Table 4 with corresponding supplier information.
Reel-seq
To identify non-coding fSNPs at the HLA-DRB1/DQA1 locus, Reel-seq was performed as previously described. 30 The Reel-seq library was built and amplified by primers seq and G3 with Accuprime Taq polymerase (Invitrogen). For screening, ∼10 μg NE isolated from HMC3 cells (buffer for control) was mixed with ∼50 ng of library DNA using the binding buffer from LightShift™ Chemiluminescent EMSA Kit (Thermo Fisher Scientific) and subsequently incubated at RT for 2 hr. The reaction was performed in triplicate with three buffer-treated controls and three NE-treated samples. All samples were resolved on a 6% TBE native gel for gel shifting. After the completion of electrophoresis, unshifted bands (libraries) from each of the controls and samples were cut and isolated. The isolated library DNA was next amplified by PCR using seq and G3 primers and the regenerated libraries were used for the next cycle of Reel-seq screen. In total, 7 cycles were performed. After the screening, as previously described, next generation sequencing was performed and analyzed with the PCR product from cycle 1, 4, and 7.30,31,51
SDCP-MS
An 84 bp DNA fragment biotinylated at the 5’ end was conjugated to streptavidin-coated Dynabeads as per manufacturer’s instructions (Invitrogen). This fragment was engineered to include a 31 bp target SNP sequence flanked by restriction enzyme cleavage sites with BamH I and EcoR I. The same fragment with a 7 bp deletion in the middle of the 31 bp SNP sequence was used as negative competitor. 15 μg magnetic beads-linked DNA was mixed with 1 mg nuclear extract from HMC3 cells in 1x binding buffer from LightShift™ Chemiluminescent EMSA Kit (Thermo Fisher Scientific) at room temperature for 2 h. After magnetic selection and wash with PBS+.05% Tween 20, DNA beads with bound proteins were digested with EcoR I at 37°C for 30 min. After another magnetic selection and wash, the DNA beads were digested with BamH I at 37°C for 45 min and the supernatant was collected. Mass spectrometry was performed to identify specific binding proteins within the complex.
EMSA
EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific) according to manufacturer’s instructions. For the probe, a 31 bp SNP fragment with the SNP centered in the middle was made by annealing two oligos. The double stranded oligos were then biotinylated using the Biotin 3' End DNA Labeling Kit (Thermo Fisher Scientific). NE was isolated from human HMC3 cells. After the DNA and NE incubation at RT for 30 min, the DNA-NE complex was resolved on 6% TBE native gel for mobility shifting. The data represents three independent biological replicates (n = 3).
Luciferase Reporter Assay
Luciferase reporter assays were performed in HMC3 cells using pGL3 luciferase reporter vector (cat#: E1751, Promega). Insert sequences are listed in Supplemental Table 3. Luciferase reporter construct DNA was transfected into HMC3 cells by FuGENE HD transfection reagent (Promega) together with the control vector pRL-TK. The luciferase activity was measured by the Dual-Glo® Luciferase Reporter Assay System (cat#: E2920, Promega). All experiments were performed according to the manufacturer’s protocol. The data represents 6 independent biological replicates (n = 6).
AIDP-Western Blots
AIDP-Wb was performed as previously described. 30 In brief, a 31 bp biotinylated SNP sequence centered with either the risk or non-risk allele was generated by annealing two biotinylated primers (IDT). Approximately 1 μg DNA was then attached to 40 μl of Dynabeads™M-280 Streptavidin. DNA-beads were mixed with ∼100 μg NE isolated from HMC3 cells at RT for 1 hour with rotation. After washing off the unbound proteins, the DNA bound proteins were eluted by sample buffer and resolved on an SDS-PAGE gel for Western blot analysis. For an internal control, the same blot was probed using an antibody directed against PARP-1. The data represents three independent biological replicates (n = 3).
RNA Interference
For shRNA stable knockdown, the shRNA vectors (pLKO.1) were purchased from Sigma (the MISSION® shRNA Library) (Supplemental Table 3). Lentiviruses were produced and shRNA knockdown was performed in HMC3 cells according to the manufacturer’s protocol (Addgene). For siRNA transient knockdown, siRNAs were purchased from Dharmacon (Horizon Discovery), all the procedures were performed following the manufacturer’s protocol.
RNA Isolation and RT-PCR
Total RNA was isolated using RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. cDNA was synthesized from 1 μg RNA after DNase I treatment using iScript™ cDNA Synthesis Kit (Bio-rad). qPCR was performed with the StepOne real-time PCR system using the power SYBR green PCR master mix (Applied Biosystems) according to the manufacturer’s protocol. The sequence-specific primers are listed in Supplemental Table 3.
Statistical Analysis
All the experimental data are presented as means ± SEM. Statistical significance was evaluated with a two-tailed unpaired Student’s t test. Data for Western blot represent three independent experiments (n = 3). Data for qPCR represent the combination of three independent samples (n = 3) with each in duplicate. Data for luciferase reporter assay represent the combination of 6 independent samples (n = 6).
Supplemental Material
Supplemental Material, sj-pdf-1-aja-10.1177_15333175221085066 for Regulation of the Late Onset alzheimer’s Disease Associated HLA-DQA1/DRB1 Expression by Xiaoyu Zhang, Meijaun Zou, Yuwei Wu, Danli Jiang, Ting Wu, Yihan Zhao, Di Wu, Jing Cui and Gang Li in American Journal of Alzheimer's Disease & Other Dementias
Acknowledgments
We thank Drs. Shihui Liu, Aditi Uday Gurkar, Xiaojun Tan and Yvonne S. Eisele for scientific discussions concerning this work.
Author Contributions: G.L. designed the study, analyzed the data, and drafted and revised the manuscript; X.Z. performed all the experiments and participated in drafting the manuscript; M.Z., D.J., T.W., Y.Z. assisted with the experiments; D.W., J.C. performed sequencing and statistical data analysis.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported partly by grants from NIH NIA R01AG056279 (G.L.).
Supplemental Material: Supplemental material for this article is available online.
ORCID iD
References
- 1.Risk Reduction of Cognitive Decline and Dementia: WHO Guidelines . Geneva2019. [PubMed]
- 2.Uddin MS, Hasana S, Hossain MF, et al. Molecular genetics of early- and late-onset Alzheimer's disease. Curr Gene Ther. 2021;21(1):43-52. doi: 10.2174/1566523220666201123112822. [DOI] [PubMed] [Google Scholar]
- 3.Fillenbaum GG, Belle G, Morris JC, et al. Consortium to establish a registry for Alzheimer's disease (CERAD): the first twenty years. Alzheimer's Dementia. 2008;4(2):96-109. doi: 10.1016/j.jalz.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wes PD, Sayed FA, Bard F, Gan L. Targeting microglia for the treatment of Alzheimer's disease. Glia. 2016;64:1-23. doi: 10.1002/glia.22988. [DOI] [PubMed] [Google Scholar]
- 5.Brouwers N, Sleegers K, Van Broeckhoven C. Molecular genetics of Alzheimer's disease: an update. Ann Med. 2008;40(8):562-583. doi: 10.1080/07853890802186905. [DOI] [PubMed] [Google Scholar]
- 6.Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414-430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832-1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert J-C, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45(12):1452-1458. doi: 10.1038/ng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43(5):436-441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreno DJ, Ruiz S, Ríos Á, et al. Association of GWAS top genes with late-onset Alzheimer's disease in colombian population. Am J Alzheimers Dis Other Demen. 2017;32(1):27-35. doi: 10.1177/1533317516679303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wightman DP, Jansen IE, Savage JE, et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer's disease. Nat Genet. 2021;53(9):1276-1282. doi: 10.1038/s41588-021-00921-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Sidorenko J, Couvy-Duchesne B, et al. Risk prediction of late-onset Alzheimer's disease implies an oligogenic architecture. Nat Commun. 2020;11(1):4799. doi: 10.1038/s41467-020-18534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seto M, Weiner RL, Dumitrescu L, Hohman TJ. Protective genes and pathways in Alzheimer's disease: moving towards precision interventions. Mol Neurodegener. 2021;16(1):29. doi: 10.1186/s13024-021-00452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giri M, Lü Y, Zhang M. Genes associated with Alzheimer's disease: an overview and current status. Clin Interv Aging. 2016;11:665-681. doi: 10.2147/cia.S105769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karch CM, Goate AM. Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77(1):43-51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54(1):15-39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 17.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54(1):15-39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann D, Wiebusch H, Marshall SE, et al. HLA class I, II & III genes in confirmed late-onset Alzheimer's disease. Neurobiol Aging. 2001;22(1):71-77. doi: 10.1016/s0197-4580(00)00180-9. [DOI] [PubMed] [Google Scholar]
- 19.van Heemst J, van der Woude D, Huizinga TW, Toes RE. HLA and rheumatoid arthritis: how do they connect? Ann Med. 2014;46(5):304-310. doi: 10.3109/07853890.2014.907097. [DOI] [PubMed] [Google Scholar]
- 20.Barcellos LF, Sawcer S, Ramsay PP, et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15(18):2813-2824. doi: 10.1093/hmg/ddl223. [DOI] [PubMed] [Google Scholar]
- 21.Lambert J-C, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45(12):1452-1458. doi: 10.1038/ng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat Genet. 2019;51(3):404-413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno-Grau S, de Rojas I, Hernández I, et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer's disease and three causality networks: The GR@ACE project. Alzheimers Dement. 2019;15(10):1333-1347. doi: 10.1016/j.jalz.2019.06.4950. [DOI] [PubMed] [Google Scholar]
- 24.Mangalam AK, Taneja V, David CS. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J Immunol. 2013;190(2):513-519. doi: 10.4049/jimmunol.1201891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YB, Nagai A, Kim SU. Cytokines, chemokines, and cytokine receptors in human microglia2002;69(1):94-103. doi: 10.1002/jnr.10253. [DOI] [PubMed] [Google Scholar]
- 26.Wyss-Coray T, Mucke L. Inflammation in neurodegenerative disease—a double-edged sword. Neuron. 2002;35(3):419-432. doi: 10.1016/S0896-6273(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 27.Bateman RJ, Xiong C, Benzinger TLS, et al. Dominantly inherited Alzheimer N. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med. 2012;367(9):795-804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro V, Sanchez-Mejias E, Jimenez S, et al. Microglia in Alzheimer's disease: activated, dysfunctional or degenerative. Front Aging Neurosci. 2018;10:140. doi: 10.3389/fnagi.2018.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGeer PL, Kawamata T, Walker DG, Akiyama H, Tooyama I, McGeer EG. Microglia in degenerative neurological disease. Glia. 1993;7(1):84-92. doi: 10.1002/glia.440070114. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, Wu D, Jiang D, et al. A sequential methodology for the rapid identification and characterization of breast cancer-associated functional SNPs. Nat Commun. 2020;11(1):3340. doi: 10.1038/s41467-020-17159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G, Martínez-Bonet M, Wu D, et al. High-throughput identification of noncoding functional SNPs via type IIS enzyme restriction. Nat Genet. 2018;50(8):1180-1188. doi: 10.1038/s41588-018-0159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877-D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sur I, Tuupanen S, Whitington T, Aaltonen LA, Taipale J. Lessons from functional analysis of genome-wide association studies. Cancer Res. 2013;73(14):4180-4184. doi: 10.1158/0008-5472.CAN-13-0789. [DOI] [PubMed] [Google Scholar]
- 34.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461(7261):199-205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190-1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dickel DE, Visel A, Pennacchio LA. Functional anatomy of distant-acting mammalian enhancers. Philos Trans R Soc Lond B Biol Sci. 2013;368(1620):20120359. doi: 10.1098/rstb.2012.0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen M, Kachadoorian M, Carrasquillo MM, et al. Late-onset Alzheimer disease risk variants mark brain regulatory loci. Neurology Genetics. 2015;1(2):e15. doi: 10.1212/nxg.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nieto M, Monuki ES, Tang H, et al. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II–IV of the cerebral cortex2004;479(2):168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- 39.Dharshini SAP, Taguchi Y-h., Gromiha MM. Investigating the energy crisis in Alzheimer disease using transcriptome study. Sci Rep. 2019;9(1):18509. doi: 10.1038/s41598-019-54782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han H, Cho J-W, Lee S, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46(D1):D380-D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rustenhoven J, Smith AM, Smyth LC, et al. PU.1 regulates Alzheimer's disease-associated genes in primary human microglia. Mol Neurodegener. 2018;13(1):44. doi: 10.1186/s13024-018-0277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pimenova AA, Herbinet M, Gupta I, et al. Alzheimer's-associated PU.1 expression levels regulate microglial inflammatory response. Neurobiol Dis. 2021;148:105217. doi: 10.1016/j.nbd.2020.105217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deming Y, Filipello F, Cignarella F, et al. The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer's disease risk. Sci Transl Med. 2019;11(505). doi: 10.1126/scitranslmed.aau2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almeida A. Regulation of APC/C-Cdh1 and its function in neuronal survival. Mol Neurobiol. 2012;46(3):547-554. doi: 10.1007/s12035-012-8309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuchsberger T, Martínez-Bellver S, Giraldo E, Teruel-Martí V, Lloret A, Viña J. Abeta induces excitotoxicity mediated by APC/C-Cdh1 depletion that can be prevented by glutaminase inhibition promoting neuronal survival. Sci Rep. 2016;6:31158. doi: 10.1038/srep31158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou M, Jiang D, Wu T, et al. Post-GWAS functional studies reveal an RA-associated CD40-induced NF-kB signal transduction and transcriptional regulation network targeted by class II HDAC inhibitors. Hum Mol Genet. 2021;30(9):823-835. doi: 10.1093/hmg/ddab032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee YB, Nagai A, Kim SU. Cytokines, chemokines, and cytokine receptors in human microglia. J Neurosci Res. 2002;69(1):94-103. doi: 10.1002/jnr.10253. [DOI] [PubMed] [Google Scholar]
- 48.Hopperton KE, Mohammad D, Trépanier MO, Giuliano V, Bazinet RP. Markers of microglia in post-mortem brain samples from patients with Alzheimer's disease: a systematic review. Mol Psychiatr. 2018;23(2):177-198. doi: 10.1038/mp.2017.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Daccak R, Mooney N, Charron D. MHC class II signaling in antigen-presenting cells. Curr Opin Immunol. 2004;16(1):108-113. doi: 10.1016/j.coi.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 50.Hassan GS, Mourad W. An unexpected role for MHC class II. Nat Immunol. 2011;12(5):375-376. doi: 10.1038/ni. [DOI] [PubMed] [Google Scholar]
- 51.Larman HB, Laserson U, Querol L, et al. PhIP-Seq characterization of autoantibodies from patients with multiple sclerosis, type 1 diabetes and rheumatoid arthritis. J Autoimmun. 2013;43:1-9. doi: 10.1016/j.jaut.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-aja-10.1177_15333175221085066 for Regulation of the Late Onset alzheimer’s Disease Associated HLA-DQA1/DRB1 Expression by Xiaoyu Zhang, Meijaun Zou, Yuwei Wu, Danli Jiang, Ting Wu, Yihan Zhao, Di Wu, Jing Cui and Gang Li in American Journal of Alzheimer's Disease & Other Dementias