ABSTRACT
Pseudomonas aeruginosa (P. aeruginosa) is one of the leading causes of chronic infections, including reinfection, relapse, and persistent infection, especially in cystic fibrosis patients. Relapse P. aeruginosa infections are more harmful because of repeated hospitalization and undertreatment of antimicrobials. However, relapse P. aeruginosa infection in China remains largely unknown. Herein, we performed a 3-year retrospective study from 2019 to 2022 in a tertiary hospital, which included 442 P. aeruginosa isolates from 196 patients. Relapse infection was identified by screening clinical records and whole-genome sequencing (WGS). We found that 31.6% (62/196) of patients had relapsed infections. The relapse incidence of carbapenem-resistant P. aeruginosa infection (51.4%) is significantly higher than that of carbapenem-susceptible P. aeruginosa infection (20.2%, P < 0.0001). These isolates were assigned to 50 distinct sequence types and sporadically distributed in phylogeny, indicating that relapsed infections were not caused by certain lineages. Fast adaptation and evolution of P. aeruginosa isolates were reflected by dynamic changes of antimicrobial resistance, gene loss and acquisition, and single-nucleotide polymorphisms during relapse episodes. Remarkably, a convergent non-synonymous mutation that occurs in a pyochelin-associated virulence gene fptA (T1056C, M252T) could be a considerable target for the diagnosis and treatment of relapse P. aeruginosa infection. These findings suggest that integrated utilization of WGS and medical records provides opportunities for improved diagnostics of relapsed infections. Continued surveillance of the genomic dynamics of relapse P. aeruginosa infection will generate further knowledge for optimizing treatment and prevention in the future.
IMPORTANCE
Pseudomonas aeruginosa is a predominant pathogen that causes various chronic infections. Relapse infections promote the adaptation and evolution of antimicrobial resistance and virulence of P. aeruginosa, which obscure evolutionary trends and complicate infection management. We observed a high incidence of relapse P. aeruginosa infection in this study. Whole-genome sequencing (WGS) revealed that relapse infections were not caused by certain lineages of P. aeruginosa isolates. Genomic dynamics of relapse P. aeruginosa among early and later stages reflected a plasticity scattered through the entire genome and fast adaptation and genomic evolution in different ways. Remarkably, a convergent evolution was found in a significant virulence gene fptA, which could be a considerable target for diagnosis and treatment. Taken together, our findings highlight the importance of longitudinal surveillance of relapse P. aeruginosa infection in China since cystic fibrosis is rare in Chinese. Integrated utilization of WGS and medical records provides opportunities for improved diagnostics of relapse infections.
KEYWORDS: Pseudomonas aeruginosa, relapse infection, prevalence, molecular epidemiology, whole-genome sequencing
INTRODUCTION
Pseudomonas aeruginosa (P. aeruginosa) is a non-fermentative and aerobic Gram-negative bacillus that is one of the leading causes of severe healthcare-associated infections, such as nosocomial pneumonia, urinary tract infections, and bloodstream infections, which results in high mortality rates and brings huge medical burdens to human society (1, 2). Carbapenems are the most effective antimicrobial agents against P. aeruginosa infections. However, the emergence and global dissemination of carbapenem-resistant P. aeruginosa (CRPA) threatened the effectiveness of antimicrobial therapy, which is identified as a global public health crisis (1, 3).
A relatively large genome (5.5–7 Mb) of P. aeruginosa encodes a wide array of virulence factors, regulatory genes, and catabolic functions, which provides the pathogenicity and adaptability to establish acute and chronic infections and against the different stressors in the environment with the facility to tailor its response (3, 4). Its tremendous ability to adapt greatly facilitates its capacity to cause chronic infections and became a dominant pathogen that causes persistent and chronic lung infections, which contributes to morbidity and mortality in patients with cystic fibrosis (CF) and burn injury particularly (3, 5).
Chronic infections could divide into three types, including (i) persistent infection that occurs when the bacteria causing the infection are not completely eliminated by treatment and continuously reproduce and induce inflammation in the body; (ii) relapse infection that occurs when a patient’s symptoms improve and the bacteria were not detected by the clinical laboratory after treatment but then return later infected by the same bacterial clone, often because the initial treatment did not completely eradicate the infection; and (iii) reinfection that occurs when a person was invaded by new clones (6 – 8). During chronic infections, P. aeruginosa isolates are facing long-term host-derived stressors and antimicrobial treatment, which are significant selective pressures for evolution and adaptation (8 – 11). Remarkably, relapse infections are more harmful since the patients could be erroneously identified as being cured which obscure evolutionary trends and complicate infection management, leading to the repeated hospitalization and undertreatment of antimicrobials (12). Previous studies have confirmed that CF patients are susceptible to establish P. aeruginosa infection which accounts for 95% of deaths in CF patients (13, 14). Notably, P. aeruginosa isolates from CF patients evolved in the phenotype and genotype which confer a fitness advantage or antimicrobial resistance, establishing persistent colonization and infection (5, 15). However, CF is the most common autosomal recessive genetic disorder among Caucasians, whereas it rarely happens in Chinese (16). Other than CF, chronic obstructive pulmonary disease (COPD), bronchiectasis, pulmonary tuberculosis, lung cancer, and interstitial lung diseases are lung-associated risk factors causing repeated lung infections (12 – 15). However, the characterizations of relapse P. aeruginosa infections in China are largely unknown.
The aim of this study was to investigate the incidence of relapse P. aeruginosa infections and reveal the genomic dynamics of these relapse P. aeruginosa isolates. We performed a 3-year retrospective study that included 196 subjects who were diagnosed with P. aeruginosa infections at the Guangzhou Provincial Hospital of Chinese Medicine from September 2019 to December 2022. The P. aeruginosa isolates were purified and subjected to whole-genome sequencing. The population structure, multilocus sequence typing (MLST), antimicrobial resistance genes (ARGs), and virulence factors were used to demonstrate the molecular epidemiology of relapse P. aeruginosa isolates. Additionally, we determined the pairwise single nucleotide polymorphism (SNP) distance and evolution of isolates from early and later stages of relapse infections with the purpose of providing information on adaptive processes that could identify genetic markers and targets of relapse P. aeruginosa infections.
MATERIALS AND METHODS
Subject inclusion and exclusion criteria
This study was approved by the Guangdong Provincial Hospital of Chinese Medicine, Guangzhou, China. The clinical isolates were routinely collected and whole-genome sequenced in the hospital. To identify the subjects with P. aeruginosa infections, we screened the clinical records of subjects from 1 September 2019 to 31 December 2022. The subjects were included if clinical samples were evidenced by a positive P. aeruginosa culture, and the patients had typical clinical infectious symptoms and abnormal laboratory markers of infections (Table S1). The patients were excluded if the clinical samples were positive for the culture of P. aeruginosa but had no infectious symptoms, which were regarded as colonization. The source of infection was determined as pneumonia, urinary tract infection, surgical site infection, intra-abdominal infection, catheter-related infection, or bacteremia as defined by the Centers for Disease Control and Prevention (17).
Identification of relapse P. aeruginosa infections
Recurrence infection was defined as the return of P. aeruginosa infections after documentation of negative cultures of clinical samples or clinical improvement after completing a course of antimicrobial therapy, according to previous studies (8, 18, 19). Therefore, all patients with recurrent episodes (≥2 episodes that occurred ≥30 days apart with clinical sample culture results negative for P. aeruginosa in the interim) were identified. Their medical records were reviewed to verify clearance (resolution of all clinical signs of infection) between recurrences and to determine the clinical characteristics of the episodes. Recurrence was further subclassified as reinfection or relapse infection: if genomes of P. aeruginosa isolates from the patient were closely related to and distributed into the same lineage of the phylogeny, the recurrence was considered as a relapse infection. Conversely, if the genomic contents are greatly different and the isolates were sporadically distributed on phylogeny, the recurrent episode was considered to be reinfection.
Isolation of P. aeruginosa strains
Clinical samples and cultured isolates were collected as part of clinical management and hospital surveillance in the Guangdong Provincial Hospital of Chinese Medicine. Clinical samples (urine, blood, sputum, and wound samples) from patients with infections were plated on Columbia Blood Agar (CBA) with 5% sheep blood (Luqiao, Beijing, China), except blood samples that were injected into blood culture bottles (BACT/ALERT FN/FA Plus) processing in the BacT/ALERT 3D System (bioMerieux Inc.). After the culture at 37°C in 24 hours or positive alarm of blood culture, if P. aeruginosa is identified as the most abundant species in clinical samples, three P. aeruginosa colonies were selected and stored. DNA was extracted using the boiling method. P. aeruginosa isolates were cultured by adding 3 mL of nutrient broth and incubating for 18–24 hours at 37°C. Subsequently, total DNA was obtained from the supernatant by boiling 1 mL of the broth at 100°C for 10–15 minutes. Species identification was first confirmed by MALDI-TOF MS (BrukerDaltonik GmbH, Bremen, Germany). Where the species could not be reliably assigned by MALDI-TOF, 16S rDNA sequencing was applied. After these processes, we randomly choose one representative P. aeruginosa isolate for the subsequent analysis including MIC testing and whole-genome sequencing.
Antimicrobial susceptibility testing
The antimicrobial susceptibilities of the P. aeruginosa isolates were determined by using the Vitek 2 Compact system (bioMérieux, France) according to the manufacturer’s instructions. The minimum inhibitory concentrations (MICs) of cefepime, ceftazidime, imipenem, meropenem, ticarcillin-clavulanate, levofloxacin, ciprofloxacin, tobramycin, and amikacin were determined, and the results were interpreted according to Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute, document M100-S29) guidelines. The MICs of colistin and polymyxin E (colistin) were determined using the broth dilution method (EUCAST breakpoints, version 9.0; CLSI M100-S29). The antibiotic susceptibilities of the P. aeruginosa isolates are provided in Table S2.
Whole-genome sequencing and data processing
A total of 163 P. aeruginosa isolates from 65 patients with relapse infections (identified using MALDI-TOF MS ± 16s rDNA sequencing) were subjected to whole-genome sequencing. The Qiagen Blood and Tissue Kit (Qiagen, Hilden, Germany) was used to extract genomic DNA for the P. aeruginosa isolate according to the manufacturer’s instructions. DNA libraries were constructed with 350-bp paired-end fragments and sequenced using an Illumina HiSeq 2000 platform (Illumina Inc., USA). The sequencing yielded >100-fold coverage sequences (~1G) of raw paired-end (150 bp) per isolate. Adaptor, unreliable reads, and low-quality bases of raw reads were trimmed using fastp with default parameters (20). SPAdes v3.13.1 with the “--careful” parameter and automatically determined k-mer values (21, 33, 55, 77) were used to perform draft genome de novo assembly (21). Contigs less than 500 bp in size were excluded for further analysis. WGS-based species identification was performed using JSpeciesWS v3.2.7 (22). Isolates not confirmed as P. aeruginosa using WGS-based species identification, and contaminated/mixed sequences, were manually excluded from subsequent analyses.
Prediction and annotation of open reading frames
Open reading frames were predicted using prodigal v2.6.3 and annotated by Prokka v1.13.3 (23). Antimicrobial resistance genes, plasmid replicons, and virulence-associated genes were identified using SRST2 v0.2 by mapping reads to ResFinder, PlasmidFinder, and VFDB database (24 – 27); meanwhile, the locations of these traits in a draft genome level were annotated using ABRicate 0.8.7 with “--minid 75 --mincov 60” parameters (https://github.com/tseemann/abricate). In silico MLST of P. aeruginosa was assigned using PubMLST (https://pubmlst.org/) against seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE). New alleles and sequence types (STs) were uploaded on the PubMLST database.
Pan-genome analysis and phylogenetic construction
Pan-genome analyses were performed using Roary (28), and a concatenated alignment of genes shared among ≥99% of all isolates (core genome) was extracted using Mafft v7.407 (29). Core genome SNPs (cgSNPs) were extracted from this concatenated alignment using SNP sites (30). To obtain high-quality cgSNPs, we filtered SNPs that are of low quality and frequency by vcftools v0.1.17 with “--minDP 5 --max-missing 0.95 --maf 0.05” parameters (31). The paired SNP distance among all isolates was calculated by snp-dists v0.7.
A maximum likelihood (ML) phylogeny for cgSNPs was reconstructed using RAxML v8.2.10 implementing a generalized time-reversible nucleotide substitution model with a Γ distribution (GTR + G) (32). Branch lengths and bootstrap supports for bipartitions were estimated using 1,000 bootstrap replicates. The population structure was assessed using cgSNPs with hierBAPS, which was run four times using maximum clustering sizes of 20, 40, 60, and 80 (33). Sequence clusters (SCs) identified using hierBAPS were labeled on the core genome phylogeny on iTOL (https://itol.embl.de/).
Statistical analysis
Statistical analysis was conducted using R software v3.5.3. Statistical significance between groups was calculated using a two-sided Wilcoxon rank sum test and a Fisher’s exact test. P values less than 0.05 were considered as statistically significant.
RESULTS
Identification of patients with relapse P. aeruginosa infections
A total of 442 non-duplicate P. aeruginosa isolates were identified in 196 patients at the Guangdong Provincial Hospital of Chinese Medicine from September 2019 to December 2022 (Table S1). Through screening of clinical records and inclusion criteria, we found that 33.2% (65/196) of patients had at least twice P. aeruginosa infections during the study period, which include relapsed infection and reinfection (Fig. 1).
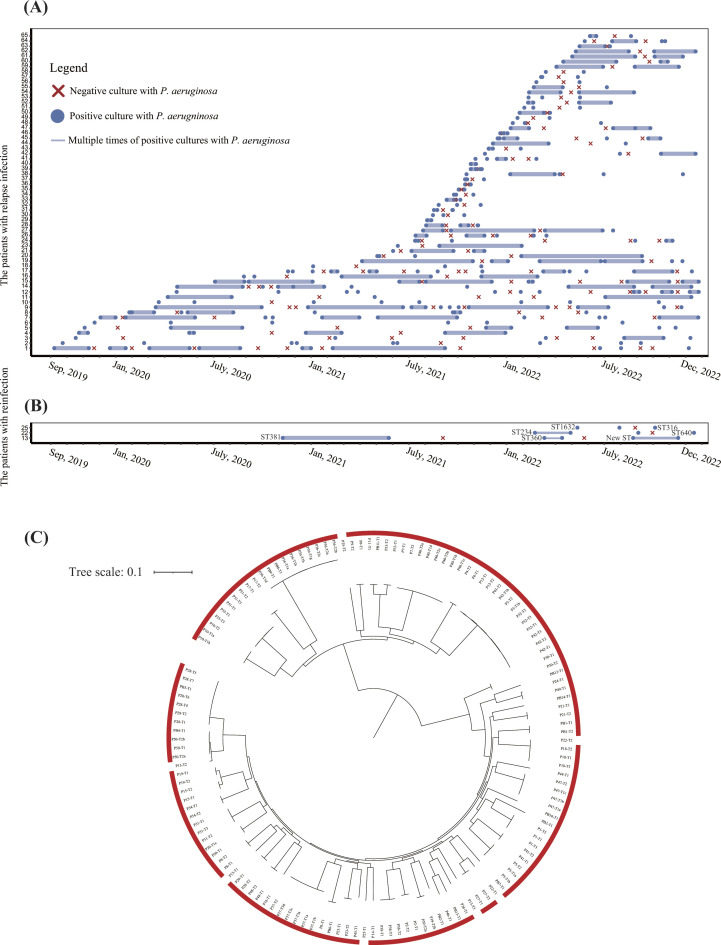
Fig 1.
Identification of relapse infection by screening of clinical data and whole-genome sequencing. (A) Timelines of relapse P. aeruginosa infection for each patient. Data are shown for isolates of 62 patients (1–62) from 1 September 2019, to 31 December 2022. The blue dots represent the positive cultures with P. aeruginosa isolates. The red forks represent negative culture with P. aeruginosa isolates. The docs connected with solid lines represent multiple times (>2) of positive cultures with P. aeruginosa isolates. (B) Timelines of reinfection of P. aeruginosa for each patient. Sequence type (ST) of the isolate was annotated beside the dot. (C) Phylogenetic tree of 163 P. aeruginosa isolates from 65 patients with multiple times of P. aeruginosa infections. The tree was constructed using core genome SNPs with maximum likelihood method. The isolates labeled with red frame indicate the relapse infection, while no frame indicates reinfection.
We hypothesize that the relapse P. aeruginosa isolates should be closely related, meaning the differences in genomes should be minor and the isolates are distributed into the same lineage on phylogeny. To precisely identify the relapse P. aeruginosa infections, we performed WGS and phylogenetic analysis for 163 isolates from 65 patients; we identified that 156 isolates from 62 patients were distributed into the same lineage, whereas seven isolates from three patients were distributed sporadically (Fig. 1C), which indicated that 62 patients (95.4%, n = 65) had relapse infections and three patients (4.6%) had reinfections (Fig. 1A and B).
Comparison of clinical characterizations among relapse and non-relapse P. aeruginosa isolates
Among our cohort, 66.8% (131/196) were male, with the median age being 67 years (interquartile range [IQR], 55–81 years). There are no differences in gender (male: 64.5% [40/62] vs 67.9% [91/134], P = 0.6388) and age distribution (≥60 years: 22.6% [14/62] vs 32.1% [43/134], P = 0.1782) between relapse and non-relapse infection groups (Table 1). Notably, 30.8% (20/65) of the patients were diagnosed with chronic lung diseases which were considered as risk factors in relapse P. aeruginosa infections (Table S1). However, only 16.9% (11//65) of patients were diagnosed with COPD, 4.6% (n = 3) with bronchiectasis, 7.7% (n = 5) with interstitial lung diseases, and 1.5% (n = 1) with lung cancer, and none was diagnosed with CF disease, indicating that no certain chronic lung disease could be a direct indicator.
TABLE 1.
Clinical characteristics of 196 patients infected by P. aeruginosa a
| Characteristics | Relapse infection (N = 62) | Non-relapse infection (N = 134) | P value |
|---|---|---|---|
| Gender | |||
| Male | 64.5% (40) | 67.9% (91) | 0.639 |
| Female | 35.5% (22) | 32.1% (43) | |
| Age | |||
| ≥60 years | 22.6% (14) | 32.1% (43) | 0.173 |
| <60 years | 77.4% (48) | 67.9% (91) | |
| Clinical department | |||
| Pulmonology | 29.0% (18) | 26.1% (35) | 0.669 |
| Rehabilitation | 17.7% (11) | 13.4% (18) | 0.429 |
| ICU | 12.9% (8) | 8.2% (11) | 0.302 |
| Neurology | 12.9% (8) | 7.5% (10) | 0.220 |
| Cardiology | 9.7% (6) | 5.2% (7) | 0.392 b |
| Neurosurgery | 8.1% (5) | 5.2% (7) | 0.652 b |
| Others | 9.7% (6) | 34.3% (46) | 0.0003 |
| Specimen source | |||
| Sputum | 64.5%(40) | 54.5% (73) | 0.186 |
| Alveolar lavage fluid | 21% (13) | 5.2% (7) | 0.001 |
| Wound | 6.5% (4) | 12.7% (17) | 0.189 |
| Urine | 4.8% (3) | 20.9% (28) | 0.004 |
| Blood | 3.2% (2) | 1.5% (2) | 0.799 |
| Others | 0% (0) | 5.2% (7) | 0.156 |
| Clinical diagnosis | |||
| Pneumonia | 91.9% (57) | 68.6% (92) | 0.0003 |
| UTI | 8.1% (5) | 22.4% (30) | 0.0158 |
| Wound infection | 0% (0) | 3.0% (4) | 0.1222 |
| BSI | 0% (0) | 3.0% (4) | 0.1222 |
| Pneumonia and BSI | 0% (0) | 1.5% (2) | 0.4981 |
| Others | 0% (0) | 1.5% (2) | 0.4981 |
Data are presented as percentage of patients with the corresponding number in parentheses. UTI, urinary tract infection.
Statistical analysis with Fisher’s exact test. BSI, bloodstream infection.
Of the patients, 27.0% (53/196) were from the pulmonology department, followed by the rehabilitation department (14.8%, n = 29), ICU (9.7%, n = 19), neurology department (9.2%, n = 18), and other 16 departments (39.3%, n = 77). There is no difference in department distributions between patients with relapse and non-relapse infections (P > 0.05; Table 1).
The P. aeruginosa isolates from 57.7% (n = 113) of patients were collected from sputum, followed by urine (15.8%, n = 31), wound (10.7%, n = 21), and bronchoalveolar lavage fluid (10.2%, n = 20). The P. aeruginosa isolates (n = 6) were found in both sputum and blood in two patients and in both sputum and wound in one patient, which caused systematic infections. We found that the relapse P. aeruginosa isolates were mainly collected from sputum (64.5%, n = 40) and bronchoalveolar lavage fluid (21.0%, n = 13).
Most patients (77.0%, 151/196) had lung infections, followed by urinary tract infections (17.9%, n = 35), bloodstream infection (3.1%, n = 6), and wound infections (2.0%, n = 4). The incidence of relapse infection in patients with lung infection (39.1%, 59/151) was significantly higher than that of urinary tract infections (14.3%, 5/35, P = 0.0054), which indicated that the relapse P. aeruginosa infections usually happened in the lungs.
Antimicrobial resistance of relapse P. aeruginosa isolates
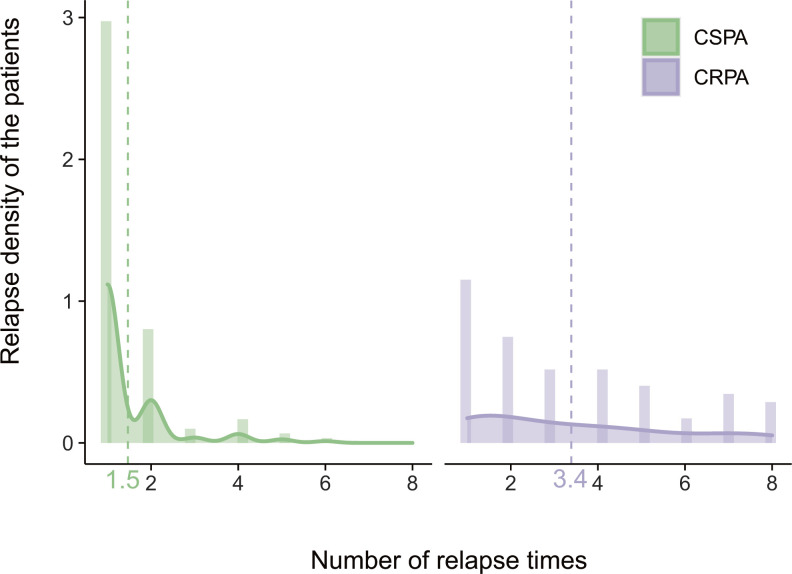
Among 442 isolates collected from 196 patients, 41.0% (n = 181) of isolates were resistant to ticarcillin/clavulanate, followed by imipenem (38.5%, n = 170), meropenem (28.7%, n = 127), and levofloxacin (26.7%, n = 118) (Table 2). Through antimicrobial susceptibility testing (AST), we found that 41.0% (181/442) of P. aeruginosa isolates were resistant to carbapenems. Among 196 patients included in this study, 36.7% of patients (n = 72) were infected with CRPA isolates, whereas 63.3% of patients (n = 124) were infected with CSPA isolates. Remarkably, we observed that the incidence of relapse infection of CRPA isolates (51.4%, 37/72) was significantly higher than that of CSPA (20.2%, 25/124) isolates (P < 0.0001, Fig. 2), which could be attributed to the incomplete eradication since CRPA is commonly resistant to most of the antimicrobials (34).
TABLE 2.
Antimicrobial susceptibility of 442 Pseudomonas aeruginosa isolates a
| Antimicrobials | Antimicrobial susceptibility (N = 442) | ||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| Ticarcillin-clavulanate | 37.3% (165) | 21.7% (96) | 41% (181) |
| Cefepime | 0.72 (318) | 20.8% (92) | 7.2% (32) |
| Ceftazidime | 74.4% (329) | 5.2% (23) | 20.4% (90) |
| Amikacin | 93.2% (412) | 1.8% (8) | 5% (22) |
| Tobramycin | 91.4% (404) | 1.1% (5) | 7.5% (33) |
| Meropenem | 61.8% (273) | 9.5% (42) | 28.7% (127) |
| Imipenem | 0.61 (270) | 0.5% (2) | 38.5% (170) |
| Ciprofloxacin | 0.79 (349) | 10.6% (47) | 10.4% (46) |
| Levofloxacin | 63.8% (282) | 9.5% (42) | 26.7% (118) |
| Colistin | 95.9% (424) | – | 4.1% (18) |
The numbers in the table refer to the percentages of isolates showing susceptibility, intermediate susceptibility, and resistance to each antimicrobial. The numbers in parentheses indicate the number of isolates. –, not applicable.
Fig 2.
Density of relapse times for CRPA and CSPA isolates The x-axis represents the number of relapse times, while the y-axis represents the relapse density of the patients. Vertical dotted line represents the mean value of relapse times. CRPA, carbapenem-resistant P. aeruginosa; CSPA, carbapenem-susceptible P. aeruginosa.
During the different relapse episodes, the antimicrobial resistance spectrum of P. aeruginosa isolates changed in 17 patients (27.0%, N = 63). Decreased susceptibility of antimicrobials was observed in 15 patients, including imipenem (n = 13), meropenem (n = 13), ticarcillin-clavulanate (n = 8), cefoperazone/sulbactam (n = 5), ceftazidime (n = 5), and amikacin (n = 2), whereas increased susceptibility of ciprofloxacin and levofloxacin was observed in one patient and imipenem and meropenem in another one (Table S2). These phenotypical changes could be ascribed to antimicrobial therapy during relapse episodes, reflecting a fast adaptation of P. aeruginosa facing antimicrobial selective pressures.
Virulence-associated genes, antimicrobial resistance genes, and plasmid of relapse P. aeruginosa isolates
Among 156 isolates from patients with relapse P. aeruginosa infection, a total of 60 kinds of virulence-associated genes and 2 kinds of plasmid replicons were identified against VFDB and PlasmidFinder databases (Tables S4 and S5). The average number of virulence-associated genes for each isolate is 48.4 (standard deviation [SD]: ±3.7). A total of 45 isolates harbored IncFIA plasmid, and 12 isolates harbored IncQ plasmid. No VFs and plasmid replicons were lost or acquired among different time points of infections in all patients.
A total of 25 ARGs were detected among relapse P. aeruginosa isolates (Table S3). The average number of ARGs for each isolate is 5.7 (SD: ±1.1). We found that 41 patients had the same ARGs among different time points of infections. Notably, the P. aeruginosa isolates of nine patients lost ARGs at the later stages of infections, such as aph(3')-IIa (n = 5) and aac (3)-IV (n = 5), while P. aeruginosa isolates of 10 patients acquired new ARGs at a later stage, such as catA1 (n = 7) and aac (3)-IV (n = 5). Acquisition and loss of the entire genes are important for evolution and adaptation during infection, which is commonly connected with horizontal transfer of mobile genes and transposons that can rapidly introduce large genomic and phenotypic changes, conferring antimicrobial resistance, virulence, or fitness advantage (12, 13, 35).
Genomic characterizations of relapse P. aeruginosa isolates
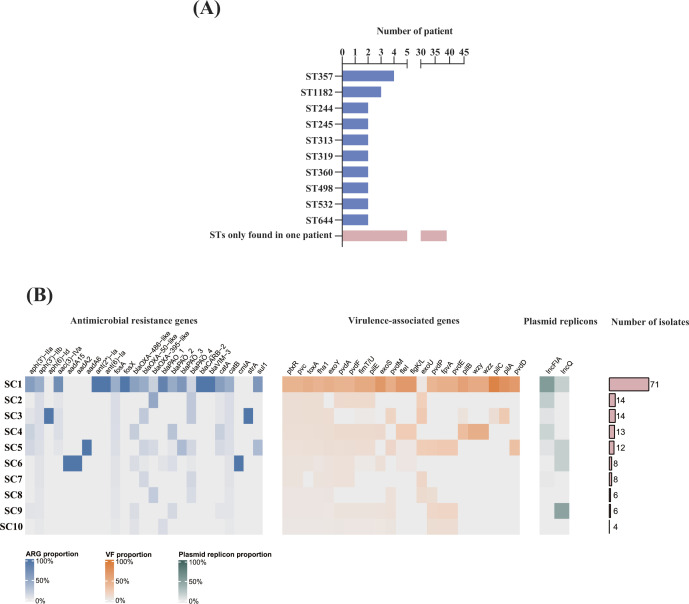
These relapse P. aeruginosa isolates were assigned into 50 distinct STs, of which 8 new STs were identified among 16 isolates (Fig. 4; Table S6). Notably, our results showed that four patients were infected with ST357 isolates and three patients with ST1182 isolates, while 40 STs were detected in only one patient and 8 STs in two patients, which indicated that the relapse infections were not caused by certain lineages of P. aeruginosa.
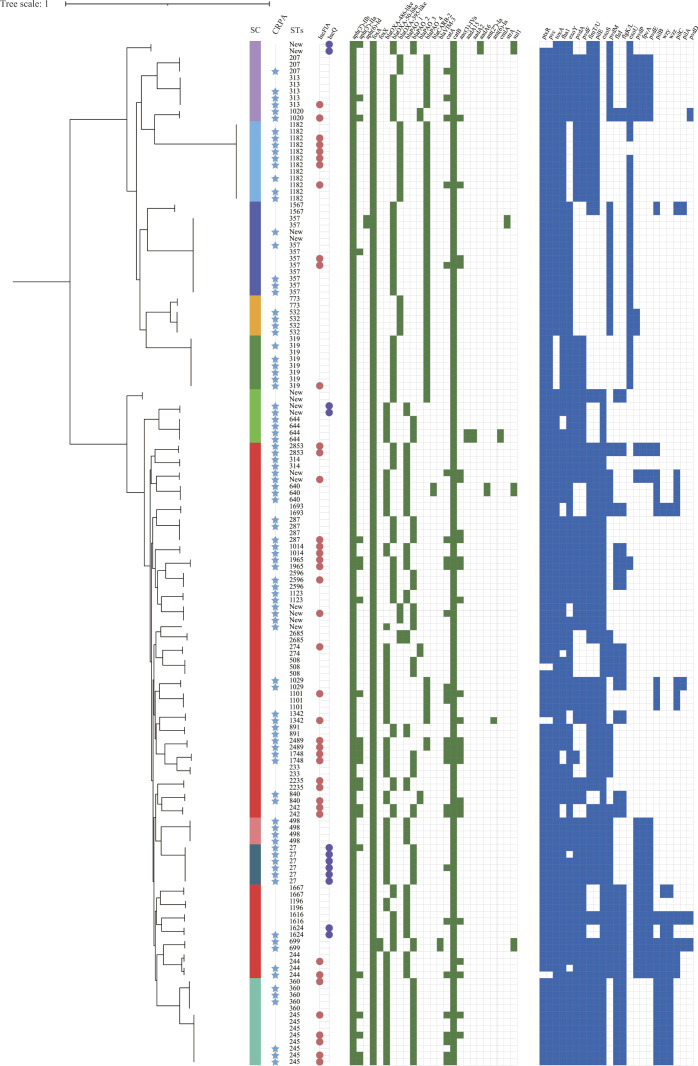
Pan-genome analysis of 156 relapse P. aeruginosa isolates from 62 patients identified 4,785 core genes representing a 4.5-Mb alignment in ≥99% of genomes. hierBAPS analysis of the relapse P. aeruginosa population structure based on 59,631 cgSNPs identified 10 sequence clusters (SCs), of which SC7 was polyphyletic in the ML phylogeny and bifurcated as two distinct monophyletic clades (Fig. 3). SC7 was the most prevalent cluster containing 46.8% (73/156) of isolates (Fig. 4). We observed that the relapse P. aeruginosa isolates were distributed into different branches, and the distribution of ARGs, VFs, and plasmid replicons was scattered, indicating no lineage specificity in relapse P. aeruginosa isolates (Fig. 3).
Fig 3.
Phylogenetic tree and genomic characteristics of P. aeruginosa isolates from relapse infections. The ML phylogenetic tree was constructed by cgSNPs of 156 P. aeruginosa isolates from 62 patients with relapse P. aeruginosa infections. The sequence clusters (SCs), CRPA, MLST, and plasmid replicon were labeled by a color strip, star, text, and color dot. In the heatmap, green blocks denote the presence of an ARG, while blue blocks denote the presence of a virulence factor.
Fig 4.
MLST, ARGs, virulence-associated genes, and plasmid replicons for each sequence cluster based on phylogeny. (A) The distribution of MLST. The pink bar represents the number of patients of which ST is only found in one patient. (B) The proportion heatmap of ARGs, VFs, and plasmid replicons for each sequence cluster. The proportions of ARGs, virulence-associated genes, and plasmid replicons were represented by blue, orange, and green heatmaps, respectively.
Pairwise genetic distance and evolution of relapse P. aeruginosa isolates
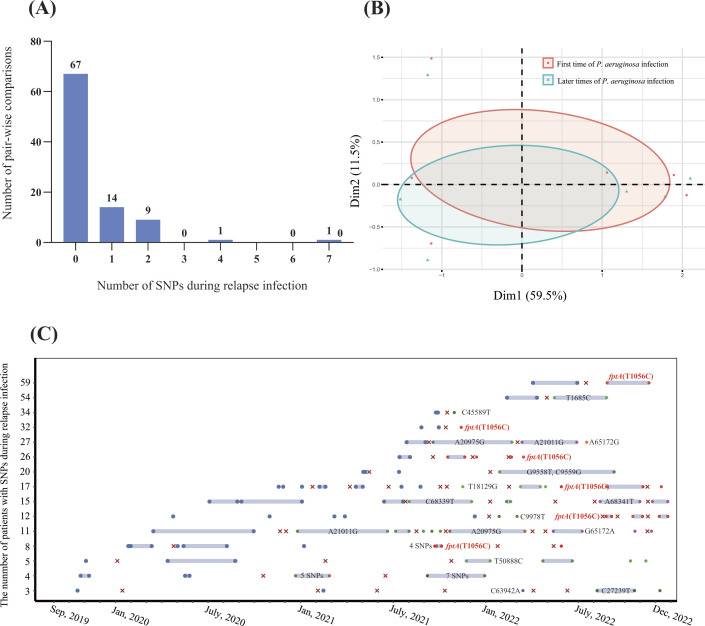
Previous studies demonstrated that micro-evolution and adaptation could happen in P. aeruginosa isolate under the selective pressure of antimicrobials and host immune systems, which could promote virulence and antimicrobial resistance of P. aeruginosa isolates (5, 12 – 15) . Therefore, we performed a genomic comparative analysis to reveal the evolution of relapse P. aeruginosa isolates. The results of pairwise SNP distance of P. aeruginosa isolates for each patient showed that SNPs of the isolates from 15 patients occurred during early and subsequent stages of relapse infections (ranging from one to seven SNPs), whereas 47 patients occurred with no SNP among different time points (Fig. 5A). These SNPs were located on 25 sites of P. aeruginosa chromosome, and 24 of them are detected in only one patient.
Fig 5.
Dynamic of SNPs in P. aeruginosa genome during the episodes of relapse infection (A) Number of SNPs in P. aeruginosa core genome among pair-wise comparisons during relapse infection of 62 patients. (B) PCA analysis of cgSNPs among first time and later times of relapse P. aeruginosa isolates. (C) Timelines for each patient with SNPs during the episodes of relapse infections. Data are shown for isolates of 15 patients from 1 September 2019, to 31 December 2022. The blue dots represent the positive cultures with P. aeruginosa isolates. The blue dots represent the genotype of P. aeruginosa isolate for each patient. The red dots labeled with red text represent SNP found in the fptA gene (T1056C). The green, purple, and orange dots represent the SNPs found in other SNP sites.
Principal component analysis (PCA) showed that P. aeruginosa isolates among the first episode and later episodes were divided into two parts, which may reveal that there is a convergent evolution during the episodes of relapse infections (Fig. 5B). Remarkably, we found an SNP, which causes a non-synonymous mutation in the fptA gene (T1056C, M252T), was detected in relapse P. aeruginosa of six patients (Fig. 5C). FptA, a TonB-dependent transporter, promotes the utilization of iron that permits the high-affinity binding and transport of Fe(III)-pyochelin complex across the outer membrane, which is associated with the virulence of P. aeruginosa (36, 37). The mutation of the fptA gene has also been detected in patients with long-term P. aeruginosa infections, which indicated that convergent microevolution occurs in relapse P. aeruginosa isolates (38).
DISCUSSION
Relapse P. aeruginosa infection is responsible for repeated admissions and adverse outcomes of patients. In European and American countries, relapse P. aeruginosa infection commonly happened in CF patients. However, the prevalence and characteristics of relapse P. aeruginosa infections in China remain unknown since CF is a rare disease in China (16). In this study, we performed a 3-year retrospective study to investigate the epidemiological and genomic characteristics of P. aeruginosa isolates causing relapse infections in a tertiary hospital in China.
Whole-genome sequencing is a precise method for recognition among relapse infection, persistent infection, and reinfection of a certain pathogen (6 – 8). Our results showed a high accuracy (95.4%, 62/65) in identifying relapse infections by screening clinical records using the standards in this study. However, the distinguishment between relapse and persistent infections relied on both the genotyping by WGS and clinical symptoms. In many previous studies, relapse and persistent infections were collectively analyzed since the clinical symptoms during the treatment periods were ignored, which underestimated the influences of entirely different dynamics of pathogen virulence and the host’s immune status (10, 11, 14, 38 – 40).
Our results revealed that the incidence of relapse P. aeruginosa infection in this study (31.6%, 62/196) is high but relatively lower than the results from European and American countries, which could be attributable to the reason that CF patients were not found in our cohort, which is an important risk factor for colonization and infection of P. aeruginosa (13, 41). Relapse P. aeruginosa infections are commonly detected in the lungs (91.9%, 57/62), indicating that the specific host lung environment favors P. aeruginosa colonization and persistence. Notably, 30.8% (20/65) of the patients were diagnosed with chronic lung diseases, but no direct indicator was found. Besides, we found that most of the patients are all elderly and with prolonged hospitalization which are significant risk factors for P. aeruginosa colonization (42). Therefore, we propose that decolonization could be an efficient therapy for the treatment and prophylaxis of relapse P. aeruginosa infection.
We revealed that the incidence of relapse in CRPA isolates (51.4%, 37/72) is significantly higher than that in CSPA isolates (20.2%, 25/124, P < 0.0001), which could be attributed to the reason that CRPA is commonly resistant to most of the antimicrobials and hard to eradicate completely (34). Decreased susceptibility of several antimicrobials was observed in 27% (7/63) of patients in the late stage of relapse, including carbapenems, β-lactams, and aminoglycosides. Besides, we found that two aminoglycoside resistance genes (aph(3')-IIa and aac(3)-IV) underwent dynamic events of loss or acquisition in the late stage of relapse infection. As previously confirmed, the acquisition of ARGs could confer resistance, whereas the loss of ARGs could confer a fitness advantage, which is important for evolution and adaptation during infection (13, 35). However, only one-half of patients (4/10) have received aminoglycoside treatment, which may imply that phenotypical and genotypical dynamics of antimicrobial resistance reflect fast adaptation and genomic evolution not just in antimicrobial resistance but also in changing fitness, resulting in incomplete eradication of P. aeruginosa isolates or increased bacterial fitness by lost genes with a burden.
The identification of pathoadaptive genes, in which mutations accumulate with higher frequencies than expected statistically during relapse infection, gave hope to finding genetic markers that might predict the potential to establish chronic infection in the clinic. In our study, the results showed that relapse infections are not caused by specific lineages of P. aeruginosa isolates since these isolates are assigned to distinct STs and sporadically distributed in the phylogenetic tree. Therefore, we proposed that the relapse infection could impute to non-lineage-associated factors, such as genetic evolutions of horizontal gene transfer, mutation, or relapse-associated functional virulence-associated genes. To test our hypothesis, we additionally analyzed the genetic evolution at different times of relapse P. aeruginosa isolates. We found several mutations of cgSNPs in the later time of relapse infections, reflecting a dramatic genetic evolution during relapse infections. Notably, we found a convergent evolution of a non-synonymous mutation in the fptA gene (T1056C, M252T). FptA is a TonB-dependent transporter that permits the high-affinity binding and transport of Fe(III)-pyochelin complex, driving the import of the bacteriocin across the outer membrane and promoting the virulence of P. aeruginosa, especially in the pulmonary and urinary tract infections (43, 44). Consistently, another study has found that the fptA gene was hypermutated in P. aeruginosa isolates collected from prolonged ICU inpatients; even the epidemiological and genomic information of these isolates were entirely different from the isolates in this study (38). Since the host lung microbiome is able to guard against colonization and infection of pathogens by competing for the nutrient metal iron (40), the P. aeruginosa is warranted to increase its metal iron acquisition function to overcome the iron limitation which could be a selective pressure facilitating the microevolution of fptA gene, thus in favor of the fitness and adaptation of P. aeruginosa isolates in relapse and prolonged infections. Previous studies have indicated that FptA confers increased susceptibility to siderophore-drug conjugates and is considered a novel target for treating multidrug-resistant P. aeruginosa (37). According to the above evidence, we speculate that FptA could be a considerable target for diagnostics, decolonization, and treatment of relapse and prolonged infections by P. aeruginosa isolates.
Our study has several limitations. First, we included the patients with P. aeruginosa infections in a single clinical center, and our epidemiological findings may not be generalizable. We realize that more wide-ranging studies involving the surveillance and genomic investigations are needed to clarify the prevalence and genomic characterizations of relapse P. aeruginosa infection since CF is a rare disease in China. Secondly, we are not able to trace the clinical records of patients with relapse infections who were hospitalized in other medical centers, which could underestimate the incidence of relapse P. aeruginosa infections. Thirdly, we were not able to consider the influence of selective pressure from the host immune environment on the convergent evolution of relapse P. aeruginosa isolates. Above two limitations were restricted by the retrospective study design. A prospective cohort study and the collection of comprehensive clinical samples would supply strong evidence to address these gaps. Finally, we were not able to assess the causality between our genomic findings and relapse infection based on an epidemiological study. Further metatranscriptomics and experimental evidence would be helpful to address this noteworthy question and is part of our future work.
Despite these limitations, our data provide a comprehensive understanding of the high prevalence and genomic dynamics of relapse P. aeruginosa isolates across different stages. Integrated utilization of WGS and medical records provides opportunities for improved diagnostics of relapse infections. Relapse P. aeruginosa infections were not caused by a certain lineage of isolates but could be attributable to the convergent evolution of the siderophore-encoding gene. Continued surveillance of the genomic dynamics of relapse P. aeruginosa infection will generate further knowledge for optimizing the treatment and prevention of relapse infection in the future, and our approach could potentially be used as a template for monitoring and diagnosing relapse infection of such prolonged infection by pathogens more widely.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant numbers 82302598, 82072345, 82172329, and 82202590), Guangdong Basic and Applied Research Foundation (grant number 2022A1515111171), China Postdoctoral Science Foundation (grant numbers 2022M720922 and 2023T160150), Guangdong Provincial Hospital of Chinese Medicine (grant numbers YN2022QN11 and 10812), Guangzhou Basic and Applied Foundation (grant number 2023A04J0456), and Military Logistics Research Fund Project (CLB21J018).
C.C., B.H., and C.S. designed the study. C.S. did the literature search and wrote the manuscript, which was reviewed and edited by C.C. and B.H. C.S., J.Z., and Y.X. performed the experiments and produced the tables and figures. C.S., J.Z., and Y.X. contributed to data analysis and interpretation. All authors contributed to sample collection and data collection. All authors reviewed, revised, and approved the final submission.
We declare no competing interests.
Contributor Information
Cong Shen, Email: shencong456@163.com.
Bin Huang, Email: huangb3@mail.sysu.edu.cn.
Cha Chen, Email: chencha906@163.com.
Benjamin M. Liu, Children's National Hospital, George Washington University, Washington, DC, USA
ETHICS APPROVAL
Ethical approval was sought and given by the Ethics Committee of Guangdong Provincial Hospital of Chinese Medicine (approval number ZE2023-077-01). Individual consent forms were translated into Mandarin Chinese, and consent was obtained for all patients. Patients were individually contacted, either face-to-face or by phone, and consent sought to use these samples/data for this study described herein. All participants had the right to withdraw from the study at any stage.
DATA AVAILABILITY
The genome assemblies of P. aeruginosa reported in this study have been deposited in the NCBI GenBank genomic DNA database under BioProject accession number PRJNA974176.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.05312-22.
Legends for Tables S1 to S6.
Tables S1 to S6.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Hu Y-Y, Cao J-M, Yang Q, Chen S, Lv H-Y, Zhou H-W, Wu Z, Zhang R. 2019. Risk factors for carbapenem-resistant Pseudomonas aeruginosa, zhejiang province, China. Emerg Infect Dis 25:1861–1867. doi: 10.3201/eid2510.181699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jurado-Martín I, Sainz-Mejías M, McClean S. 2021. Pseudomonas aeruginosa: an audacious pathogen with an adaptable arsenal of virulence factors. Int J Mol Sci 22:3128. doi: 10.3390/ijms22063128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Silby MW, Winstanley C, Godfrey SAC, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x [DOI] [PubMed] [Google Scholar]
- 5. Vanderwoude J, Fleming D, Azimi S, Trivedi U, Rumbaugh KP, Diggle SP. 2020. The evolution of virulence in Pseudomonas aeruginosa during chronic wound infection. Proc Biol Sci 287:20202272. doi: 10.1098/rspb.2020.2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Howden BP, Giulieri SG, Wong Fok Lung T, Baines SL, Sharkey LK, Lee JYH, Hachani A, Monk IR, Stinear TP. 2023. Staphylococcus aureus host interactions and adaptation. Nat Rev Microbiol 21:380–395. doi: 10.1038/s41579-023-00852-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hill PWS, Moldoveanu AL, Sargen M, Ronneau S, Glegola-Madejska I, Beetham C, Fisher RA, Helaine S. 2021. The vulnerable versatility of salmonella antibiotic persisters during infection. Cell Host Microbe 29:1757–1773. doi: 10.1016/j.chom.2021.10.002 [DOI] [PubMed] [Google Scholar]
- 8. Cho J, Cunningham S, Pu M, Lennon RJ, Dens Higano J, Jeraldo P, Sampathkumar P, Shannon S, Kashyap PC, Patel R. 2021. Clostridioides difficile whole-genome sequencing differentiates relapse with the same strain from reinfection with a new strain. Clin Infect Dis 72:806–813. doi: 10.1093/cid/ciaa159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stapels DAC, Hill PWS, Westermann AJ, Fisher RA, Thurston TL, Saliba A-E, Blommestein I, Vogel J, Helaine S. 2018. Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 362:1156–1160. doi: 10.1126/science.aat7148 [DOI] [PubMed] [Google Scholar]
- 10. Winstanley C, O’Brien S, Brockhurst MA. 2016. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol 24:327–337. doi: 10.1016/j.tim.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hogardt M, Heesemann J. 2010. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int J Med Microbiol 300:557–562. doi: 10.1016/j.ijmm.2010.08.008 [DOI] [PubMed] [Google Scholar]
- 12. Bartell JA, Sommer LM, Haagensen JAJ, Loch A, Espinosa R, Molin S, Johansen HK. 2019. Evolutionary highways to persistent bacterial infection. Nat Commun 10:629. doi: 10.1038/s41467-019-08504-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rossi E, La Rosa R, Bartell JA, Marvig RL, Haagensen JAJ, Sommer LM, Molin S, Johansen HK. 2021. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol 19:331–342. doi: 10.1038/s41579-020-00477-5 [DOI] [PubMed] [Google Scholar]
- 14. Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907 [DOI] [PubMed] [Google Scholar]
- 15. Yang L, Jelsbak L, Marvig RL, Damkiær S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Høiby N, Sommer MOA, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A 108:7481–7486. doi: 10.1073/pnas.1018249108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang T, Tian X, Xu KF. 2020. Cystic fibrosis: a rare disease emerging in China. Sci China Life Sci 63:1082–1084. doi: 10.1007/s11427-020-1620-x [DOI] [PubMed] [Google Scholar]
- 17. Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. 1988. CDC definitions for nosocomial infections, 1988. Am J Infect Control 16:128–140. doi: 10.1016/0196-6553(88)90053-3 [DOI] [PubMed] [Google Scholar]
- 18. Baran J, Riederer KM, Ramanathan J, Khatib R. 2001. Recurrent vancomycin-resistant enterococcus bacteremia: prevalence, predisposing factors, and strain relatedness. Clin Infect Dis 32:1381–1383. doi: 10.1086/319996 [DOI] [PubMed] [Google Scholar]
- 19. Chang F-Y, Peacock JE, Musher DM, Triplett P, MacDonald BB, Mylotte JM, O’Donnell A, Wagener MM, Yu VL. 2003. Staphylococcus aureus bacteremia: recurrence and the impact of antibiotic treatment in a prospective multicenter study. Medicine (Baltimore) 82:333–339. doi: 10.1097/01.md.0000091184.93122.09 [DOI] [PubMed] [Google Scholar]
- 20. Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 24. Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kutilova I, Medvecky M, Leekitcharoenphon P, Munk P, Masarikova M, Davidova-Gerzova L, Jamborova I, Bortolaia V, Pamp SJ, Dolejska M. 2021. Extended-spectrum beta-lactamase-producing Escherichia coli and antimicrobial resistance in municipal and hospital wastewaters in czech republic: culture-based and metagenomic approaches. Environ Res 193:110487. doi: 10.1016/j.envres.2020.110487 [DOI] [PubMed] [Google Scholar]
- 26. Carattoli A, Hasman H. 2020. Plasmidfinder and in Silico pMLST: Identification and typing of plasmid replicons in Whole-Genome Sequencing (WGS). Methods Mol Biol 2075:285–294. doi: 10.1007/978-1-4939-9877-7_20 [DOI] [PubMed] [Google Scholar]
- 27. Liu B, Zheng D, Zhou S, Chen L, Yang J. 2022. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res 50:D912–D917. doi: 10.1093/nar/gkab1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Page Andrew J, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rozewicki J, Li S, Katoh K, Standley DM. 2021. Analysis of protein intermolecular interactions with MAFFT-DASH. Methods Mol Biol 2231:163–177. doi: 10.1007/978-1-0716-1036-7_11 [DOI] [PubMed] [Google Scholar]
- 30. Page A.J, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, Harris SR. 2016. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom 2:e000056. doi: 10.1099/mgen.0.000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group . 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RaXML-NG: a fast, Scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455. doi: 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. 2018. RhierBAPS: an R implementation of the population clustering algorithm hierBAPS. Wellcome Open Res 3:93. doi: 10.12688/wellcomeopenres.14694.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tenover FC, Nicolau DP, Gill CM. 2022. Carbapenemase-producing Pseudomonas aeruginosa - an emerging challenge. Emerg Microbes Infect 11:811–814. doi: 10.1080/22221751.2022.2048972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gabrielaite M, Johansen HK, Molin S, Nielsen FC, Marvig RL. 2020. Gene loss and acquisition in lineages of Pseudomonas aeruginosa evolving in cystic fibrosis patient airways. mBio 11:e02359-20. doi: 10.1128/mBio.02359-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manzoor S, Ahmed A, Moin ST. 2021. Iron coordination to pyochelin siderophore influences dynamics of FptA receptor from Pseudomonas aeruginosa: a molecular dynamics simulation study. Biometals 34:1099–1119. doi: 10.1007/s10534-021-00332-x [DOI] [PubMed] [Google Scholar]
- 37. Luscher A, Moynié L, Auguste PS, Bumann D, Mazza L, Pletzer D, Naismith JH, Köhler T. 2018. TonB-dependent receptor repertoire of Pseudomonas aeruginosa for uptake of siderophore-drug conjugates. Antimicrob Agents Chemother 62:e00097-18. doi: 10.1128/AAC.00097-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu H, Yang L, Chen Q, Song H, Bo X, Guo J, Li P, Ni M, Goldberg JB. 2022. Time series genomics of Pseudomonas aeruginosa reveals the emergence of a hypermutator phenotype and within-host evolution in clinical inpatients. Microbiol Spectr 10. doi: 10.1128/spectrum.00057-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lucca F, Guarnieri M, Ros M, Muffato G, Rigoli R, Da Dalt L. 2018. Antibiotic resistance evolution of Pseudomonas aeruginosa in cystic fibrosis patients (2010-2013). Clin Respir J 12:2189–2196. doi: 10.1111/crj.12787 [DOI] [PubMed] [Google Scholar]
- 40. Marvig RL, Damkiær S, Khademi SMH, Markussen TM, Molin S, Jelsbak L. 2014. Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. mBio 5:e00966-14. doi: 10.1128/mBio.00966-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bell SC, Mall MA, Gutierrez H, Macek M, Madge S, Davies JC, Burgel P-R, Tullis E, Castaños C, Castellani C, Byrnes CA, Cathcart F, Chotirmall SH, Cosgriff R, Eichler I, Fajac I, Goss CH, Drevinek P, Farrell PM, Gravelle AM, Havermans T, Mayer-Hamblett N, Kashirskaya N, Kerem E, Mathew JL, McKone EF, Naehrlich L, Nasr SZ, Oates GR, O’Neill C, Pypops U, Raraigh KS, Rowe SM, Southern KW, Sivam S, Stephenson AL, Zampoli M, Ratjen F. 2020. The future of cystic fibrosis care: a global perspective. Lancet Respir Med 8:65–124. doi: 10.1016/S2213-2600(19)30337-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu Y, Qing Y, Chen J, Liu C, Lu J, Wang Q, Zhen S, Zhou H, Huang L, Zhang R. 2021. Prevalence, risk factors, and molecular epidemiology of intestinal carbapenem-resistant Pseudomonas aeruginosa. Microbiol Spectr 9:e0134421. doi: 10.1128/Spectrum.01344-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hare NJ, Soe CZ, Rose B, Harbour C, Codd R, Manos J, Cordwell SJ. 2012. Proteomics of Pseudomonas aeruginosa Australian epidemic strain 1 (AES-1) cultured under conditions mimicking the cystic fibrosis lung reveals increased iron acquisition via the siderophore pyochelin. J Proteome Res 11:776–795. doi: 10.1021/pr200659h [DOI] [PubMed] [Google Scholar]
- 44. Snopkova K, Dufkova K, Klimesova P, Vanerkova M, Ruzicka F, Hola V. 2020. Prevalence of bacteriocins and their co-association with virulence factors within Pseudomonas aeruginosa catheter isolates. Int J Med Microbiol 310:151454. doi: 10.1016/j.ijmm.2020.151454 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends for Tables S1 to S6.
Tables S1 to S6.
Data Availability Statement
The genome assemblies of P. aeruginosa reported in this study have been deposited in the NCBI GenBank genomic DNA database under BioProject accession number PRJNA974176.