Abstract
The lung is the primary organ for the metastasis of osteosarcoma. Although the application of neoadjuvant chemotherapy and surgery has remarkably improved the survival rate of patients with osteosarcoma, prognosis is still poor for those patients with metastasis. In this study, we performed further bioinformatics analysis on single-cell RNA sequencing (scRNA-seq) data published before, containing 75,317 cells from two osteosarcoma lung metastasis and five normal lung tissues. First, we classified 17 clusters, including macrophages, T cells, endothelial cells, and so on, indicating highly intratumoral heterogeneity in osteosarcoma lung metastasis. Next, we found macrophages in osteosarcoma lung metastasis did not have significant M1 or M2 polarizations. Then, we identified that T cells occupied the most abundant among all cell clusters, and found CD8+ T cells exhibited a low expression level of immune checkpoints in osteosarcoma lung metastasis. What is more, we identified C2_Malignant cells, and found CD63 might play vital roles in determining the infiltration of T cells and malignant cells in conventional-type osteosarcoma lung metastasis. Finally, we unveiled C1_Therapeutic cluster, a subcluster of malignant cells, was sensitive to oxfendazole and mevastatin, and the potential hydrogen-bond position and binding energy of oxfendazole-KIAA0907 and mevastatin-KIAA0907 were unveiled, respectively. Our results highlighted the power of scRNA-seq technique in identifying the complex tumor microenvironment of osteosarcoma lung metastasis, making it possible to devise precision therapeutic approaches.
Keywords: osteosarcoma, lung metastasis, single-cell RNA sequencing, intratumoral heterogeneity, immune microenvironment
Impact Statement
Hitherto, the prognosis of patients with lung metastasis of osteosarcoma is still poor, despite the application of neoadjuvant chemotherapy and surgery. Therefore, it is urgently required to explore the intratumoral heterogeneity of osteosarcoma lung metastasis. In this study, we performed single-cell RNA sequencing analysis to two osteosarcoma lung metastasis and five normal lung tissues. Our result showed that a highly intratumoral heterogeneity existed in lung metastasis of osteosarcoma. We also found macrophages did not show significant M1 or M2 polarizations in osteosarcoma lung metastasis. In addition, T cells occupied the most abundant among all cell clusters, and CD8+ T cells exhibited a low expression level of immune checkpoints in osteosarcoma lung metastasis. Moreover, we found CD63 might play vital roles in determining the infiltration of T cells and malignant cells, which could be a therapeutic target. Our results make it possible to devise precision therapeutic approaches in the future.
Introduction
Osteosarcoma is a highly aggressive malignant tumor of the bone that frequently occurs in children and in young adults it tends to metastasize. 1 Despite the remarkable progress experienced in surgical resection associated with neoadjuvant and adjuvant chemotherapy for osteosarcoma over the past several decades, patients with metastatic disease still retain a poor prognosis. 2 In cases of localized osteosarcoma, patients can expect a 5-year survival rate of 60–70%, whereas those with metastatic involvement have a dismal 5-year survival rate of 20–30%.3,4 The lungs are the primary metastatic sites for osteosarcoma, and lung metastases are negatively correlated with clinical outcomes. 5 However, the pathogenesis of osteosarcoma lung metastasis remains poorly understood. Therefore, the prognosis of patients should be improved by seeking out efficient approaches to discover potential metastatic-related molecules and identifying its underlying mechanism of lung metastasis in osteosarcoma.
Tumor metastasis is driven by the accumulation of intrinsic alterations in malignant cells and requires the interaction of malignant cells and their microenvironment.6,7 The tumor microenvironment is a complex and continuously evolving entity,8,9 and is correlated to the induction of proliferation, inhibition of apoptosis and immune system, and drug resistance. 10 In addition to malignant cells, fibroblasts, and endothelial cells, the tumor microenvironment comprises immune cells, including macrophages and T cells. Increasing lines of evidence suggest that cross-talk between malignant cells and immune cells could facilitate tumor growth and metastasis when present in the tumor microenvironment. 8 Macrophages, one of the major components of the tumor microenvironment, play a critical role in facilitating tumor cell motility, 11 and mediating the disease chemoresistance by producing pro-tumorigenic cytokines and reducing the cytolytic function of CD8+ T cells.12,13 Wei et al. 14 demonstrated that macrophages induce an epithelial-mesenchymal transition program to enhance colorectal cancer migration and invasion. Yang et al. 15 illustrated that macrophages in breast cancer might induce drug resistance through IL-10/STAT3/bcl-2 signaling pathway. In addition to macrophages, CD8+ T cells, critical members of the adaptive immune system, also play key roles in tumor. 16 Studies have revealed that CD8+ T cells can enhance antitumor outcomes in breast cancer, 9 glioblastoma, 17 and cervical cancer, 18 but a state of CD8+ T cell dysfunction or exhaustion is also commonly observed in melanoma and epithelial ovarian cancer.19,20 However, the exact mechanism of immune cells affecting the disease prognosis and therapeutic outcomes in osteosarcoma, especially in osteosarcoma lung metastasis remain to be elucidated. Therefore, exploring the underlying interactions of immune and malignant cells is important for further understanding the metastatic process in osteosarcoma lung metastasis.
Previous studies widely used the population-level mRNA profiling to reveal the transcriptome of tissues. However, the traditional bulk RNA sequencing can only represent the average expression levels of all cells, and elucidating the heterogeneity among different subpopulations at a single-cell level is not feasible. Nevertheless, single-cell RNA-sequencing (scRNA-seq) could provide a new perspective into the heterogeneity of different subpopulations and unveil the differentiation trajectory.21,22
In this study, we have investigated the tumor microenvironments of conventional-type osteosarcoma lung metastasis, chondroblastic-type osteosarcoma lung metastasis, and normal lung tissues via scRNA-seq analysis and revealed the heterogeneity of macrophages, T cells, and endothelial cells. Next, we have exhibited the spatial information and main connection target between malignant and T cells in the tumor environment. Moreover, we have investigated the presence of therapeutic clusters of malignant cells that are sensitive to chemotherapy drugs and unveiled the potential hydrogen-bond position and binding energy between drugs and protein-binding sites. These results will contribute to the understanding of the intratumoral heterogeneity in osteosarcoma lung metastasis and provide a novel therapeutic strategy.
Materials and methods
scRNA-seq data
The scRNA-seq data of osteosarcoma lung metastasis tissues 23 were obtained from the GEO Database (https://www.ncbi.nlm.nih.gov/geo/, accession code: GSE152048) and the scRNA-seq data of normal lung tissues from the Human Cell Atlas Data Coordination Platform and NCBI BIOPROJECT (https://www.tissuestabilitycellatlas.org/, accession code: PRJEB31843). 24 It is worth noting that the experiment of obtaining normal lung tissues was approved by the NRES Committee of East of England-Cambridge South (15/EE/0152) with informed consent from the donor families. More detailed information about participants could be obtained from these two previous studies. In addition, the mRNA expression profiling dataset used to perform the prognostic analysis of KIAA0907 was also acquired from the GEO Database with an accession code GSE21257.
Quality control and cell type identification
A total of seven tissues, including five normal lung tissues and two different types of osteosarcoma lung metastasis tissues (conventional and chondroblastic types), were contained in our study. The Seurat package (https://satijalab.org/seurat/, version 3.2.1) in the R software (version 3.6.3) was utilized in the process of quality control. Single cells with the number of genes less than and more than 5000 and the percent of mitochondrion more than 10% were regarded as low-quality cells and directly filtered out. The Harmony function was applied to eliminate the batch effects among all the patients. Cell clusters were recognized by the FindClusters function (resolution = 1.1) and visualized using the uniform manifold approximation and projection (UMAP) analysis.
Marker genes of cell types
The assignment of cell types was conducted according to the canonical marker genes reported previously (Supplementary Table 1).
Copy number variation analysis
The copy number variation (CNV) analysis was applied utilizing the inferCNV package (https://github.com/broadinstitute/inferCNV), an R package (version 1.2.1), which can be used to visualize CNV in cells.25–27 Parameters were set as follows: cutoff = 0.1, denoise = TRUE, cluster_by_groups = TRUE, and HMM = TRUE. Unmentioned parameters are set to default values.
Pseudotime trajectory analysis
We performed the trajectory analysis to reveal the changes in T cell subpopulations using the Monocle 2 package (version 2.14.0), an R package designed for single-cell trajectories. 28 In addition, six genes (i.e. GZMA, GZMB, GZMM, GZMK, and JUND) that played important roles during the maturation of T cells were subjected to the pseudotime trajectory analysis.
Functional annotation analysis
Differently expressed genes (DEGs) were screened by the DESeq2 package. The cutoff threshold values were used as follows: adj. P value < 0.05 and fold change (FC) > 1.0. The gene set variation analysis (GSVA) was utilized to identify the gene sets enriched by DEGs. 29 These DEGs were also loaded into the clusterProfiler for the gene ontology (GO) analysis. 30
Cellular spatial organization mapper analysis
The cellular spatial organization mapper (CSOmap) analysis was applied to analyze the spatial information of cells as previously described.31,32 Briefly, we first generated the affinity matrix among cells by integrating hundreds of ligand–receptor pairs. The affinity matrix was subsequently embedded into a three-dimensional (3D) space. Then, the contributions of ligand–receptor pairs were calculated. Finally, the most contributions of receptor genes in malignant cells were selected for computer overexpression and knockdown to observe the changes in the spatial information.
M1 and M2 signature gene analyses
M1 and M2 signature gene analyses were conducted in accordance with previous research. 33 Briefly, the normalized weighted mean expression of M1 and M2 signature genes curated from the literature (Table 1) were determined using the AddModuleScore of Seurat.
Table 1.
M1/M2 signature genes of macrophages.
| Types of macrophages | Signature genes |
|---|---|
| M1 | IL1B, IL1A, TNF, IL6, CXCL9, CXCL10, IL12A, IL12B, IL23A, FCGR1A, FCGR1B, FCGR1C, CCR7, IL8, CCL5, HLA-DRA, IRF5, IRF1 |
| M2 | IL10, CD163, MARCO, MRC1, MSR1, ARG1, STAB1, TGM2, MMP7, MMP9, MMP19, TGFB1, TGFB2, TGFB3, VEGFA, FN1, CCL4, CCL22, CCL17, CCL18, IL4R, IL7R, IRF4 |
Beyondcell workflow and therapeutic clusters
The Beyondcell analysis was performed to identify the drug vulnerabilities in scRNA-seq data in accordance with a previous report (https://doi.org/10.1101/2021.04.08.438954). All codes and full documentation are available online at https://gitlab.com/bu_cnio/Beyondcell. All custom scripts for processing and analyzing the data are available upon reasonable request.
Molecular docking analysis
To unveil the potential hydrogen-bond position and binding energy between drugs and protein-binding sites, we conducted the molecular docking analysis. 34 Briefly, we downloaded the 3D structures of proteins from the RCSBPDB database (https://www.rcsb.org). Then, PyMol (version 2.4.1) was utilized to remove heteroatoms, water molecules and small molecules of proteins. We further downloaded the 3D structures of oxfendazole (sig_2649) and mevastatin (sig_8633) from the ZINC database (https://zinc.docking.org). Subsequently, sensitive proteins and drugs above-mentioned were subjected to operations by applying the AutoDock software (version 4.2.6), and the active docking sites and calculate the grid point energy were identified by AutoGrid. Finally, PyMol was applied for visual analysis.
Prognostic analysis
Based on the GSE21257 datasets, prognostic analysis of KIAA0907 was performed. P < 0.05 were used as the screening criterion.
Code availability
The R code used in the analysis of the scRNA-seq data is available on “https://github.com/hemingweigxnn/R-code-used-for-single-cell-RNA-Seq-analysis-of-osteosarcoma-lung-metastasis-data.”
Statistical analysis
Data were presented as mean ± standard deviation (SD). Statistical analysis and figures were generated via the GraphPad Prism (version 8.0.2). We utilized t-test to analyze the data if the data passed the normality test; otherwise, the nonparametric Mann–Whitney test was applied. P < 0.05 was considered as statistically significant.
Results
Cellular proportion analysis
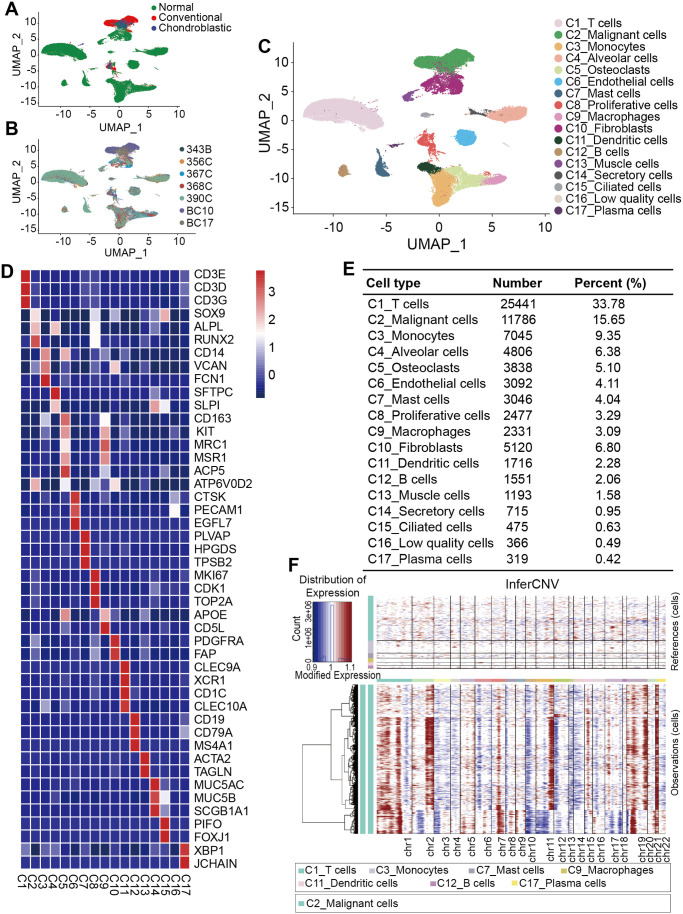
scRNA-seq analysis was performed on scRNA-seq data of the osteosarcoma lung metastasis tissues and normal lung tissues to investigate the cellular components. After the initial quality filtering, 75,317 cells, including 18,297 cells originated from osteosarcoma lung metastasis and 57,020 from normal lungs (Figure 1(A) and (B)), were available for further detection. We classified the samples into 17 clusters by unsupervised clustering analysis (Figure 1(C)), indicating high intratumoral heterogeneity. The major cell types as follows were annotated on the basis of the canonical cellular markers cited from previous studies (Figure 1(D) and (E)). C1_T cells, which accounted for 33.78% among all the cell types with a number of 25,441, were shown to highly express marker genes, including CD3D, CD3E, and CD3G. We further annotated C2_Malignant cells, which accounted for 15.65% among all the cell types with a number of 11,786, based on the marker genes involved in chondrocytes and osteoblastic differentiation, that is, SOX9, ALPL, and RUNX2. Interestingly, we found that C5_Osteoclasts expressed the marker genes of C9_Macrophages, that is, CD163, MRC, and MSR1, and conversely C9_Macrophages also expressed the marker genes of C5_Osteoclasts, that is, ACP5 and ATP6V0D2, indicating that osteoclasts and macrophages share common progenitor cells. This result was consistent with the previous research. 35 Furthermore, we defined a new cell type, named the C14_Secretory cells according to the expression of secretory mucins, that is, MUC5AC, MUC5B, and SCGB1A1. Of note, there was a group of cells expressing no specific marker genes, we therefore defined it as C16_Low quality cells. In addition, a large-scale chromosomal CNV analysis was performed to identify malignant cells (Figure 1(F)). Our results showed that the genomic regions of chromosomes 1p, 2q, 7, 11q, 15, 19, and 21 were frequently increased in malignant cells, and 3, 4, 6, 10p, 12p, 13, and 14 regions were frequently decreased.
Figure 1.
Cellular atlas of osteosarcoma lung metastasis tissues and normal lung tissues. (A–C) UMAP plots for 75,317 cells showing the (A) sample origin, (B) corresponding patients, and (C) major associated cell types. (D) Heat map of marker genes in each cell type. (E) Number and percentage of identified each cell type. (F) Hierarchical heatmap showing large-scale CNVs in malignant cells.
Heterogeneity of macrophages in osteosarcoma lung metastasis
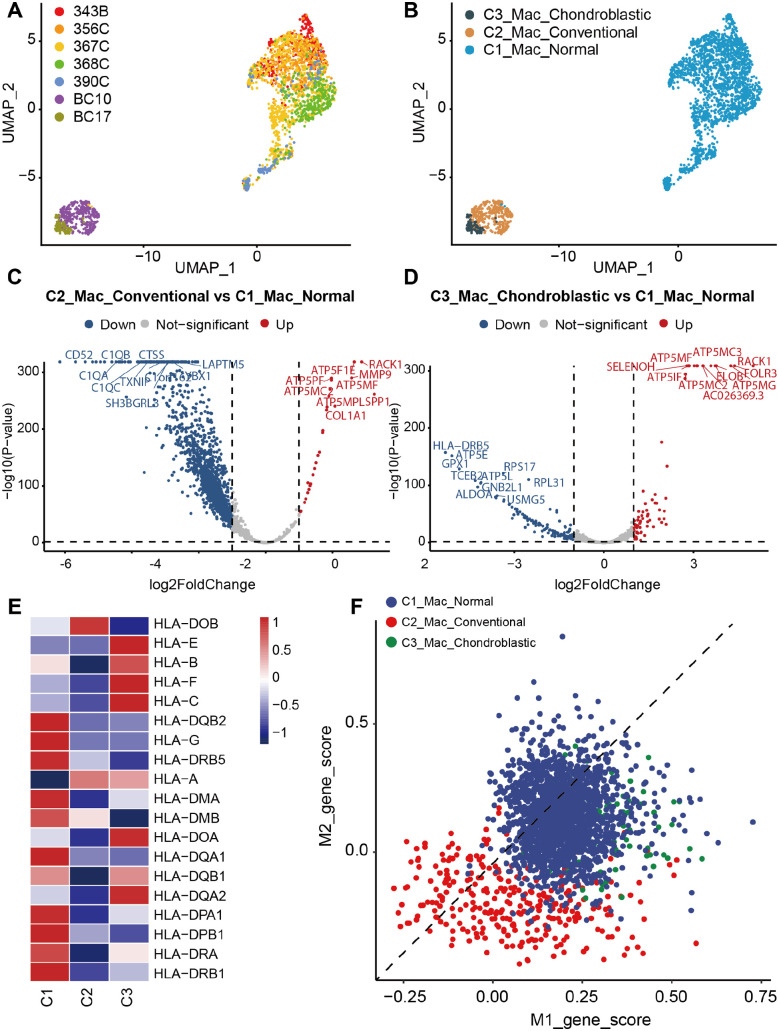
Macrophages have been widely implicated to be one of the critical abundant components of the tumor microenvironment, involving in tumor stimulating and suppressing.36,37 Three subclusters of macrophages were identified on the basis of the origin of tissues, named as C1_Mac_Normal, C2_Mac_Conventional, and C3_Mac_Chondroblastic, indicating the macrophages from these three types of lung tissues (Figure 2(A) and (B)). In addition, four subclusters of macrophages were further subdivided by their specific markers, including C1_Mac, C2_MMP9+ Mac, C3_CCL5+ Mac, and C4_SMIM25+ Mac (Supplemental Figure S1A and B). The DESeq2 package was conducted to screen DEGs from expression profiles. A total of 2019 DEGs, including 30 upregulated and 1989 downregulated DEGs, in the C2_Mac_Conventional were considered differentially expressed compared with those in the C1_Mac_Normal (Figure 2(C)). A total of 206 DEGs, including 72 upregulated and 134 downregulated DEGs, were differentially expressed between C3_Mac_Chondroblastic and C1_Mac_Normal (Figure 2(D)). As shown in Figure 2(C) and (D), the C3_Mac_Chondroblastic versus C1_Mac_Normal had a lower number of downregulated DEGs and a higher number of upregulated DEGs than C2_Mac_Conventional versus C1_Mac_Normal.
Figure 2.
Comparison of different types of macrophages (Mac) between osteosarcoma metastatic lung tissues and normal lung tissues. UMAP plots showing (A) the sample origin and (B) histopathology classification of macrophages. (C) Volcano plot showing DEGs between C2_Mac_Conventional and C1_Mac_Normal. (D) Volcano plot showing DEGs between C3_Mac_Chondroblastic and C1_Mac_Normal. (E) Heatmap of antigen-presenting related genes among C1_Mac_Normal, C2_Mac_Conventional, and C3_Mac_Chondroblastic. (F) Scatter plots showing the M1 and M2 signature genes in each cell derived from C1_Mac_Normal, C2_Mac_Conventional, and C3_Mac_Chondroblastic.
To further understand the roles of the DEGs identified, the upregulated and downregulated DEGs were annotated GO terms based on biological process, cellular component, and molecular function in this study, respectively. In C2_Mac_Conventional versus C1_Mac_Normal, the GO analysis of upregulated DEGs showed that the biological process was enriched in mitochondrial membrane organization and proton transmembrane transport; the cellular component seemed to be enriched in the inner mitochondrial protein complex, mitochondrial membrane part, mitochondrial protein complex, and mitochondrial inner membrane; and the molecular function was enriched in the proton transmembrane transporter activity (Supplemental Figure S2A). The GO analysis of downregulated DEGs showed that the biological process was enriched in neutrophil activation, neutrophil-mediated immunity, neutrophil degranulation, and neutrophil activation in immune response; the cellular component was enriched in focal adhesion, cell–substrate adherens junction, and cell–substrate junction; and the molecular function seemed to be enriched in cell adhesion molecule binding, cadherin binding, and structural constituent of ribosome (Supplemental Figure S2B).
In upregulated DEGs in C3_Mac_Chondroblastic versus C1_Mac_Normal, the biological process was enriched in leukocyte migration, response to lipopolysaccharide, and response to molecule of bacterial origin; the main cellular component seemed to be enriched in the cytoplasmic vesicle lumen, inner mitochondrial membrane protein complex, and mitochondrial proton-transporting ATP synthase complex; and the molecular function was enriched in receptor–ligand activity, G protein-coupled receptor binding, and proton transmembrane transporter activity (Supplemental Figure S2C). In terms of downregulated DEGs, the biological process was enriched in mRNA catabolic process, RNA catabolic process, and translational initiation; the cellular component was enriched in cytosolic ribosome, ribosomal subunit, and cytosolic part; and the molecular function was predominantly enriched in structural constituent of ribosome and cytokine activity (Supplemental Figure S2D).
Furthermore, we explored related antigen-presenting genes in these three subpopulations of macrophages. The C1_Mac_Normal expressed the most genes compared with C2_Mac_Conventional and C3_Mac_Chondroblastic, whereas the C2_Mac_Conventional expressed the least genes (Figure 2(E)). In the present study, we also detected the M1/M2 polarization of macrophages. The results showed that macrophages in osteosarcoma lung metastasis and normal lung tissues showed no exclusive M1 or M2 signature (Figure 2(F)).
Classification of T cells in osteosarcoma lung metastasis
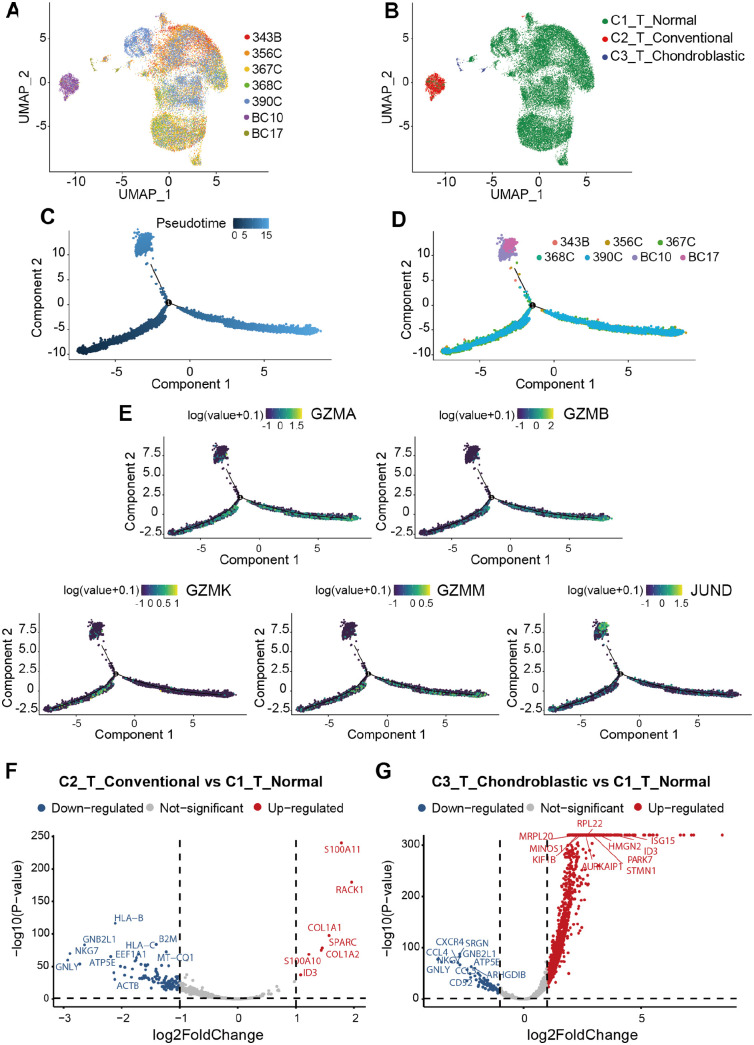
T cells are one of the major immune cells that play an essential role in regulating the response to chemotherapy and improving clinical outcomes of various cancers, including liver, 38 ovarian, 39 and breast cancers. 40 Three subclusters of T cells were defined on the basis of the origin of tissues and named as C1_T_Normal, C2_T_Conventional, and C3_T_Chondroblastic to distinguish T cells in osteosarcoma lung metastasis tissues and normal lung tissues (Figure 3(A) and (B)). Also, three subclusters of T cells, including C1_NKT cells, C2_T cells, and C3_CD8+ T cells, were further subdivided according to their specific markers (Supplemental Figure S1C-D). The cell trajectory analysis T cells showed that the T cells in BC10 and BC17 were located in the terminal position of the developmental trajectory (Figure 3(C) and (D)). It indicated that partial C1_T_Normal might differentiate into C2_T_Conventional and C3_T_Chondroblastic. The cell trajectory analyses of marker genes that play an important role in T cells were also performed in this study. Our results showed that JUND was found to be predominantly expressed in BC17, which was located at the terminal position of pseudotime trajectory with a significantly high expression level. The expression levels of GZMA, GZMB, GZMM, and GZMK were nearly equal along the pseudotime trajectory (Figure 3(E)).
Figure 3.
Comparison of T cells in different types of osteosarcoma lung metastatic tissues and normal lung tissues. UMAP plots showing (A) the sample origin and (B) histopathology classification of T cells. (C,D) The Monocle 2 trajectory plot showing the pseudotime curve (up) and dynamics (down) of T cells. (E) Pseudotime trajectories of marker genes expression (i.e. JUND, GZMA, GZMB, GZMK, and GZMM). (F) Volcano plot showing DEGs between C2_T_Conventional and C1_T_Normal. (G) Volcano plot showing DEGs between C3_T_Chondroblastic and C1_T_Normal.
The DESeq2 package was also conducted to screen DEGs from expression profiles. A total of 127 DEGs in C2_T_Conventional, including seven upregulated and 120 downregulated DEGs, were considered differentially expressed compared with those in C1_T_Normal (Figure 3(F)). A total of 1653 DEGs, including 1564 upregulated and 89 downregulated DEGs, were differentially expressed between C3_T_Chondroblastic and C1_T_Normal (Figure 3(G)). These results indicated that the number of upregulated DEGs in C2_T_Conventional versus C1_T_Normal were less than that in C3_T_Chondroblastic versus C1_T_Normal.
To further understand the different roles of DEGs in T cells, the upregulated and downregulated DEGs were also annotated GO terms based on biological process, molecular function, and cellular component. In upregulated DEGs of C2_T_Conventional versus C1_T_Normal, the biological process was enriched in response to antibiotic, response to acid chemical, and ossification; the cellular component was predominantly enriched in collagen-containing extracellular matrix; and the molecular function was enriched in extracellular matrix structural consistent and platelet-derived growth factor binding (Supplemental Figure S2E). In downregulated DEGs, the biological process was in phagocytosis, mRNA catabolic process, and response to interferon-gamma; the cellular component was in focal adhesion, cell–substrate adherens junction, and cell–substrate junction; and the molecular function was antigen binding and structural consistent of ribosome (Supplemental Figure S2F).
In upregulated DEGs in C3_T_Chondroblastic versus C1_T_Normal, the biological process was enriched in RNA catabolic process, mRNA catabolic process, and translational initiation; the cellular component was enriched in mitochondrial inner membrane, mitochondrial protein complex, and ribosome; and the molecular function was in the structural constituent of ribosome and cadherin binding (Supplemental Figure S2G). In downregulated DEGs, the biological process was in the positive regulation of leukocyte activation, positive regulation of cell activation, T cell activation, and regulation of lymphocyte activation; the cellular component was predominantly enriched in external side of plasma membrane; and the cellular component was in cytokine activity, antigen binding, and cytokine receptor binding (Supplemental Figure S2H).
Endothelial cells in osteosarcoma lung metastasis
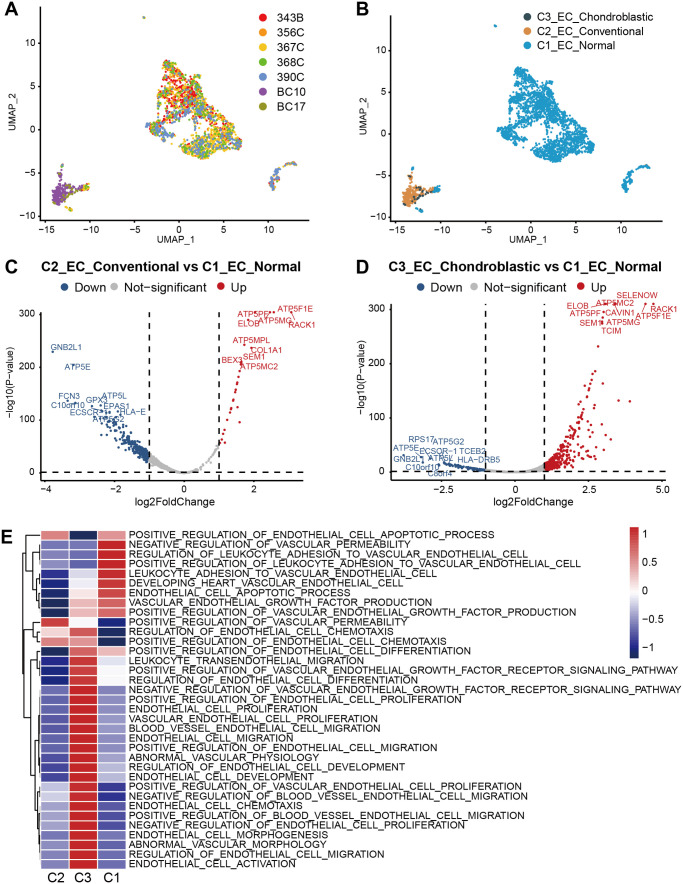
It has been demonstrated that endothelial cells promote tumor progression and chemo-resistance. 41 Thus, to identify the function of endothelial cells in our study, we detected 3092 endothelial cells. We re-clustered three subclusters of endothelial cells on the basis of the origin of tissues, named as C1_EC_Normal, C2_EC_Conventional, and C3_EC_Chondroblastic (Figure 4(A) and (B)). Meanwhile, endothelial cells were also subdivided into three subclusters according to their specific markers, including C1_ZFEP36+ ECs, C1_TMEM+ ECs, and C1_RACK1+ ECs (Supplemental Figure S1E-F). The DESeq2 package was conducted to screen DEGs from expression profiles. A total of 333 DEGs, including 30 upregulated and 303 downregulated DEGs, were considered differentially expressed in C2_EC_Conventional versus C1_EC_Normal (Figure 4(C)). A total of 511 DEGs, including 421 upregulated and 90 downregulated DEGs, were differentially expressed in C3_EC_Chondroblastic versus C1_EC_Normal (Figure 4(D)). Our results showed that the C2_EC_Conventional versus C1_EC_Normal had lower number of upregulated DEGs and higher number of downregulated DEGs than C3_EC_Chondroblastic versus C1_EC_Normal. GSVA was conducted to distinguish different cell types (Figure 4(E)). Results revealed that the C2_EC_Conventional was associated with the positive regulation of vascular permeability. The C2_EC_Conventional was abundantly associated with differentiation, proliferation, migration, and chemotaxis. The C1_EC_Normal was associated with the negative regulation of vascular permeability, which was the opposite of the main function of C2_EC_Conventional. Notably, the function of C1_EC_Normal was hardly the same as that of C3_EC_Chondroblastic. These results confirmed various functions with endothelial cells of different types of osteosarcoma lung metastasis.
Figure 4.
Comparison of different types of endothelial cells between osteosarcoma metastatic lung tissues and normal lung tissues. UMAP plots showing (A) sample origin and (B) histopathology classification. (C) Volcano plot showing DEGs between C2_EC_Conventional and C1_EC_Normal. (D) Volcano plot showing DEGs between C3_EC_Chondroblastic and C1_EC_Normal. (E) GSVA showing the functions of C1_EC_Normal, C2_EC_Conventional, and C3_EC_Chondroblastic.
The GO analysis was conducted to further explore the biological functions of the upregulated/downregulated DEGs in endothelial cells. In the GO analysis of upregulated DEGs in the C2_EC_Conventional versus C1_EC_Normal, the enrichment of GO functional significance was covered. For the biological process, the GO term of most DEGs was mitochondrial membrane organization. For the cellular component, the GO terms of most DEGs were mitochondrial proton-transporting ATP synthase complex, inner mitochondrial membrane protein complex, mitochondrial membrane part, mitochondrial protein complex, and mitochondrial inner membrane. For the molecular function, the GO terms of most DEGs were proton transmembrane transporter activity and ATPase activity (Supplemental Figure S3A). In terms of the downregulated DEGs in the C2_EC_Conventional versus C1_EC_Normal, the biological process was enriched in mRNA and RNA catabolic processes; the cellular component was enriched in focal adhesion, cell–substrate adherens junction, and cell–substrate junction; and the molecular function was enriched in cell adhesion molecule binding and cadherin binding (Supplemental Figure S3B).
In upregulated DEGs of C3_EC_Chondroblastic versus C1_EC_Normal, the biological process was enriched in extracellular matrix organization, extracellular structure organization, and cell–substrate adhesion; the cellular component was enriched in focal adhesion, cell–substrate adhesion junction, and cell–substrate junction; and the molecular function was enriched in cell adhesion molecule binding, cadherin binding, and actin binding (Supplemental Figure S3C). In terms of downregulated DEGs, the biological process was enriched in response to molecules of bacterial origin and response to lipopolysaccharide; the cellular component was enriched in cytosolic ribosome, clathrin-coated vesicle, and ribosomal subunit; and the molecular function was enriched in receptor–ligand and cytokine activities (Supplemental Figure S3D).
CD8+ T cells in osteosarcoma lung metastasis
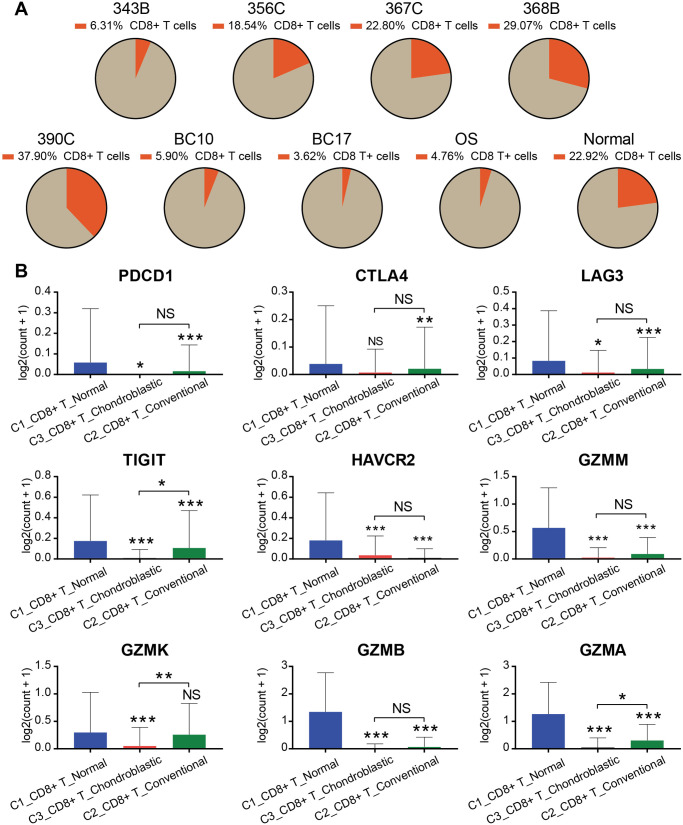
CD8+ T cells constitute an important branch of adaptive immunity, contribute to the clearance of intracellular pathogens, and provide long-term protection. 42 Herein, we extracted and analyzed the proportion of CD8+ T cells in each case (Figure 5(A)). The proportion of CD8+ T cells in normal lung tissues was significantly higher than that in lung tissue with metastatic osteosarcoma. Furthermore, we measured the expression of genes related to the function of T cells in different types of lung tissues (Figure 5(B)). The immune checkpoints (i.e. PDCD1, CTLA4, LAG3, TIGIT, and HAVCR2),43–47 which were associated with the exhaustion of T cells, were highly expressed in CD8+ C1_T_Normal but hardly expressed in CD8+ C2_T_Conventional and CD8+ C3_T_Chondroblastic. Moreover, the expression levels of cytotoxicity-related genes (i.e. GZMA, GZMB, GZMK, and GZMM) of T cells in CD8+ C1_T_Normal were higher than those in CD8+ C2_T_Conventional and CD8+ C3_T_Chondroblastic.
Figure 5.
Overview of CD8+ T cells derived from osteosarcoma metastatic lung tissues and normal lung tissues. (A) Pie chart showing the proportion of CD8+ T cells in different patients. (B) Bar chart demonstrating the expression levels of exhaustion (i.e. PDCD1, CTLA4, LAG3, TIGIT, and HAVCR2) and cytotoxicity-related (i.e. GZMA, GZMB, GZMK, and GZMM) genes of CD8+ T cells.
Critical role of TIMP1-CD63 interaction in osteosarcoma lung metastasis revealed by the CSOmap analysis
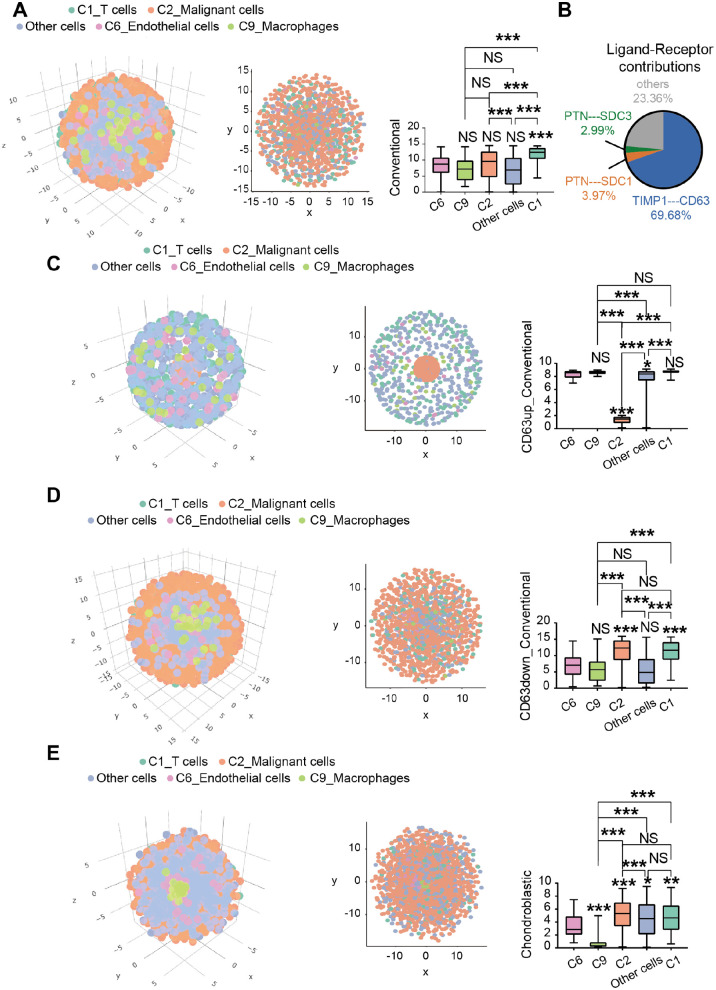
To obtain the spatial information of infiltrating cells and explore the infiltration of T cells in the tumor environment, we performed the CSOmap analysis on conventional-type osteosarcoma lung metastasis (Figure 6(A)). We identified the interactions between TIMP1 and CD63 which contributed 69.68% to the spatial reconstruction of malignant cells (Figure 6(B)). CD63, a type of membrane glycoprotein, has been reported to be involved in cell proliferation and metastasis in melanoma. 48 Through the computer simulation of upregulation of CD63 in conventional-type osteosarcoma lung metastasis, malignant cells were found to have a closer spatial structure, suggesting that malignant cells were in a state of aggregation at that situation (Figure 6(C)). When the CD63 was downregulated, malignant cells had a loose spatial structure (Figure 6(D)), suggesting that malignant cells were in a state of dispersion. Our results showed that the receptor gene CD63 might play a vital role in determining the spatial information of malignant cells in conventional-type osteosarcoma lung metastasis and act as one of the important targets for the treatment of conventional-type osteosarcoma lung metastasis. However, no significant difference was observed between malignant and T cells in chondroblastic-type osteosarcoma lung metastasis in our data (Figure 6(E)).
Figure 6.
CSOmap revealing the spatial information of malignant cells and T cells, endothelial cells, macrophages, and other cells. (A) Coordinate figure (left) and cross-section views (middle) showing the spatial information of malignant cells in conventional-type osteosarcoma lung metastasis, and histogram (right) showing the distance of cells-selected to the center of the three-dimensional axes. (B) The contribution of all ligand–receptor pairs to interactions between malignant cells and T cells, endothelial cells, macrophages, and other cells. (C) After upregulating the expression of CD63, the coordinate figure (left) and cross-section views (right) showing the spatial information of malignant cells, and the histogram (right) showing the distance of cells-selected to the center of the three-dimensional axes. (D) Coordinate figure (left) showing the global and cross-section (middle) views of spatial information of malignant cells after the downregulation of CD63, and histogram (right) showing the distance of cells-selected to the center of the three-dimensional axes. (E) Coordinate figure (left) showing the spatial information and cross-section views (middle) of malignant cells in chondroblastic-type osteosarcoma lung metastasis, and histogram (right) showing the distance of cells selected to the center of the three-dimensional axes (*P < 0.05, **P < 0.01, ***P < 0.001).
Characterization of single-cell variability in drug response in malignant cells by Beyondcell analysis
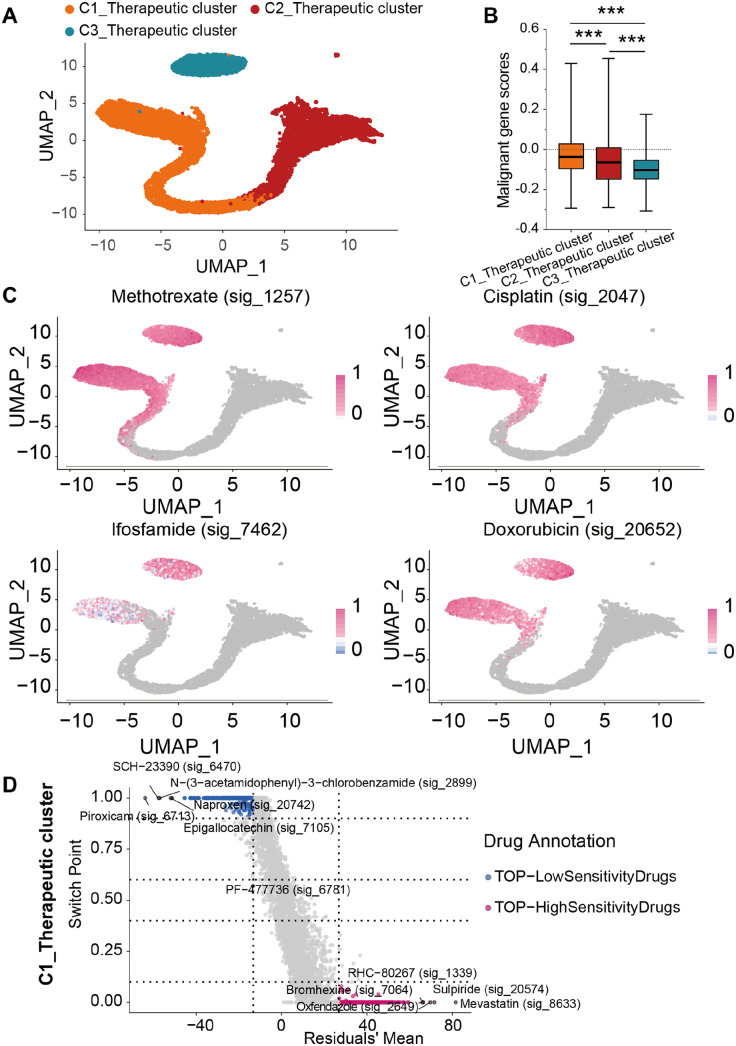
In this part, we performed the Beyondcell analysis to identify the drug-sensitive cellular populations of malignant cells that exhibited high drug resistance in osteosarcoma. Drug-sensitive cellular populations were divided into three subpopulations: C1_Therapeutic, C2_Therapeutic, and C3_Therapeutic clusters (Figure 7(A)). We obtained genes previously reported to be associated with survival of osteosarcoma (Supplementary Table 2), and then we performed a survival analysis combined with TARGET-OS dataset to obtain 18 genes with poor prognosis (Supplemental Figure S4) and 49 genes with good prognosis (Supplemental Figure S5_1-3). We then calculated the malignant gene scores of these three subpopulations in accordance with the genes associated with prognosis based on AddMouduleScore function (Figure 7(B)). Among these three subpopulations, the C1_Therapeutic cluster had the highest score. To estimate the sensitivity of traditional chemotherapy drugs to the three subpopulations, we further utilized the Beyondcell analysis and found that traditional chemotherapy drugs (i.e. methotrexate, cisplatin, ifosfamide, and doxorubicin) were sensitive to part C1_Therapeutic and C3_Therapeutic clusters, instead of the C2_Therapeutic cluster (Figure 7(C)). In addition, to determine novel drugs that were sensitive to the C1_Therapeutic cluster, we performed the Beyondcell score analysis of drugs and found five drugs with the highest sensitivity, including RHC-80267 (sig_1339), oxfendazole (sig_2649), bromhexine (sig_7064), mevastatin (sig_8633), and sulpiride (sig_20574, Figure 7(D)).
Figure 7.
Therapeutic landscapes of malignant cells. (A) UMAP plots showing the Beyondcell characterization of single-cell variability in drug response in malignant cells. (B) Malignant gene scores of C1_Therapeutic, C2_Therapeutic, and C3_Therapeutic clusters. (C) UMAP plots showing predicted sensitivity of malignant cells in response to traditional chemotherapy drugs (i.e. methotrexate, cisplatin, ifosfamide, and doxorubicin). (D) Square plots showing the differential sensitivity of Beyondcell score of drugs to C1_Therapeutic cluster.
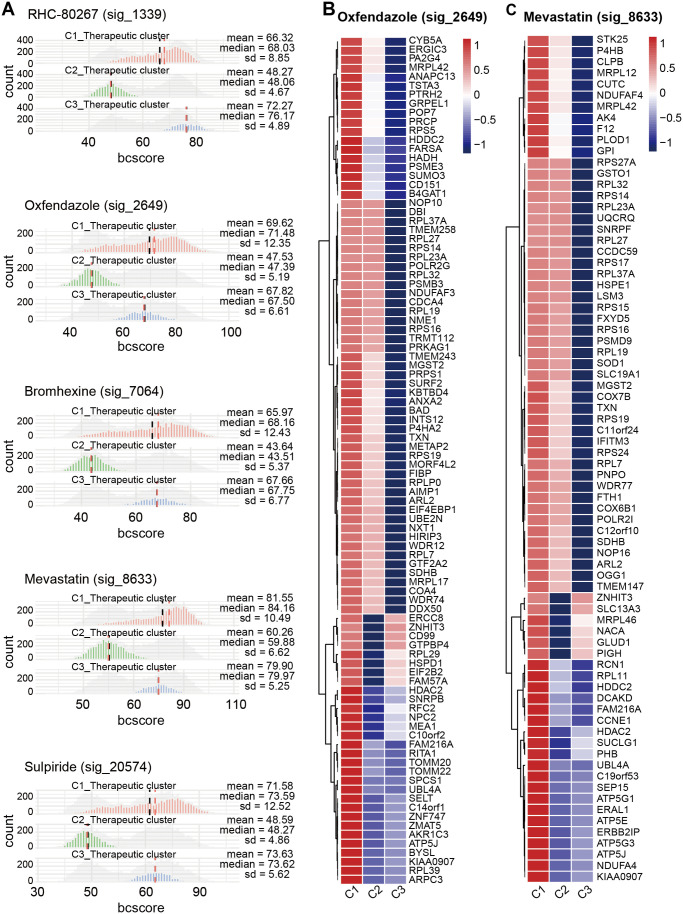
In addition, we determined the drug sensitivity signature collection scores of these five novel drugs to C1_Therapeutic, C2_Therapeutic, and C3_Therapeutic clusters and found that oxfendazole (sig_2649) and mevastatin (sig_8633) targeted the C1_Therapeutic cluster well (Figure 8(A)). Moreover, we explored the target genes of oxfendazole (sig_2649) and mevastatin (sig_8633) in these three therapeutic clusters and found that the target genes of these two drugs were enriched in the C1_Therapeutic cluster (Figure 8(B) and (C)).
Figure 8.
Drug sensitivity signature collection scores of therapeutic landscapes of malignant cells. (A) Histograms representing the Beyondcell scores of RHC-80267 (sig_1339), oxfendazole (sig_2649), bromhexine (sig_7064), mevastatin (sig_8633), and sulpiride (sig_20574) in each therapeutic cluster. Heatmap showing that downregulated genes caused by (B) oxfendazole (sig_2649) and (C) mevastatin (sig_8633) are concentrated in C1_Therapeutic cluster.
Molecular docking analysis
After identifying the target genes of oxfendazole (sig_2649) and mevastatin (sig_8633), a total of 21 shared genes were found, including “ARL2,” “ATP5J,” “FAM216A,” “HDAC2,” “HDDC2,” “KIAA0907,” “MGST2,” “MRPL42,” “RPL19,” “RPL23A,” “RPL27,” “RPL32,” “RPL37A,” “RPL7,” “RPS14,” “RPS16,” “RPS19,” “SDHB,” “TXN,” “UBL4A,” “ZNHIT3.” Survival analysis was then applied to screen out shared genes with prognostic value in the GSE21257 dataset. Eventually, KIAA0907 were considered to be the significant prognostic gene (Supplemental Figure S6A), indicating that high expression of KIAA0907 was strongly related to worse prognosis.
Subsequently, we performed the molecular docking analysis to detect the hydrogen-bond position and binding energy between oxfendazole (sig_2649) and KIAA0907, and between mevastatin (sig_8633) and KIAA0907, respectively. Our results showed that oxfendazole (sig_2649) could regulate the activities of KIAA0907 by forming a relatively stable conformation via hydrogen bonding (SER-39, HIS-43; Supplemental Figure S6B). Meanwhile, mevastatin (sig_8633) could regulate the activities of KIAA0907 by forming a relatively stable conformation through hydrogen bonding (SER-59, LEU-54, Supplemental Figure S6C). More details are listed in Table 2.
Table 2.
The hydrogen-bond position and binding energy.
| Drugs | PDB entry | Binding energy | H-bond | Target gene |
|---|---|---|---|---|
| Oxfendazole | 2YQR(KIAA0907) | –6.2 kcal/mol | SER39; HIS43 | KIAA0907 |
| Mevastatin | 2YQR(KIAA0907) | –5.9 kcal/mol | SER59; LEU54 | KIAA0907 |
PDB: protein data bank; 2YQR is the solution structure of the KH domain in KIAA0907 protein, namely 2YQR = KIAA0907.
Discussion
The lungs are the primary metastatic sites for osteosarcoma, and lung metastases are thought to be associated with poor patient outcomes. 5 Even with improvements in therapy over the past 40 years, reducing the high mortality rate of osteosarcoma, especially osteosarcoma lung metastasis, remains a challenge. Understanding the cell composition and molecular characteristics of osteosarcoma lung metastasis can contribute to solving this problem. Exploring the cell composition and molecular characteristics in detail has become possible with the first scRNA-seq method developed by Tang et al. 21 in 2009. Thus, we dissected the cellular heterogeneity of the osteosarcoma lung metastasis using the scRNA-seq data, making an important step toward understanding the intratumoral heterogeneity of osteosarcoma lung metastasis.
Macrophages, one of the main kinds of immune cells that are widely present in the body, are used to maintain homeostasis and resist pathogen invasion, 49 and commonly act as potent immune effector cells that can lead to anti- and pro-tumor functions in different settings.50,51 Macrophages are simplified into two categories (i.e. M1 classically activated macrophages and M2 alternatively activated macrophages) depending on the stimulus. 52 M1 macrophages may promote a proinflammatory Th1 response and participate in antitumor immunity, whereas M2 macrophages play a critical role in stimulating Th2 responses and promoting tumor progression. 53 It has been reported that macrophages in tumors are phenotypically similar to anti-inflammatory M2-polarized macrophages. 54 To understand the polarization of macrophages in osteosarcoma lung metastasis, we normalized the weighted mean expression of M1 and M2 signature genes. Results showed that the macrophages in osteosarcoma lung metastasis did not have significant distinct M1 or M2 polarizations, indicating that macrophages possess no polarized features and have various intermediate polarized states between the M1 and M2 states.
T cells are key mediators of tumor destruction, and their specificity for tumor-expressed antigens is important. However, other T cell-intrinsic qualities, such as durability, longevity, and functionality, also play important roles in determining the efficacy of immunotherapy. 55 In our data, C1_T cells, which had the highest proportion (33.78%) among all cells, were further re-clustered into three subtypes, including C1_T_Normal, C2_T_Conventional, and C3_T_Chondroblastic. These three subtypes of T cells followed a differentiation trajectory starting from partial of C1_T_Normal and ending with C2_T_Conventional, C3_T_Chondroblastic, and another partial of C1_T_Normal. Interestingly, we found that the expression levels of GZMA, GZMB, GZMK, and GZMM hardly increased along the pseudotime trajectory, but JUND showed relatively high expression levels in C2_T_Conventional and C3_T_Chondroblastic. GZMA, GZMB, GZMK, and GZMM are members of the protease family in the cytosolic granules of cytotoxic T and NK cells, which activate caspase-independent pyroptosis when delivered into the target cell through the immunological synapse. JUND is a core component of activator protein-1 family, involving in a substantial number of cell activities, such as proliferation, survival, and tumorigenesis.56,57 These results suggested that the T cells of osteosarcoma lung metastasis had a poor function of tumor destruction. To confirm this hypothesis, we further performed GO analysis on upregulated/downregulated DEGs in the T cells of C2_T_Conventional versus C1_T_Normal and C3_T_Chondroblastic versus C1_T_Normal. Our results showed that upregulated DEGs in C2_T_Conventional versus C1_T_Normal were predominantly enriched in response to antibiotic and acid chemical that were hardly associated with the function of T cells, the upregulated DEGs in C3_T_Chondroblastic versus C1_T_Normal, either. However, downregulated DEGs in C2_T_Conventional versus C1_T_Normal were predominantly enriched in phagocytosis and cell killing, and downregulated DEGs in C3_T_Chondroblastic versus C1_T_Normal were predominantly enriched in T cell activation, leukocyte chemotaxis, and cell killing. These results suggested that T cells in osteosarcoma lung metastasis exhibited a low function in killing tumor cells.
To detect the reason for these phenomena, we further analyzed the CD8+ T cells, one main component of T cells that play the antitumor function.58,59 The high infiltration of CD8+ T cells is reported to be associated with favorable prognoses. However, it is suggested that the proportion of CD8+ T cells in patients with osteosarcoma lung metastasis were far lower than that in normal people in our study. Notably, the proportion of CD8+ T cells was evidently distinct across different patients, suggesting the tissue heterogeneity of the immune microenvironment. Moreover, the immune checkpoints (i.e. PDCD1, CTLA4, LAG3, TIGIT, and HAVCR2) and cytotoxicity-related genes (i.e. GZMA, GZMB, GZMK, and GZMM) were highly expressed in C1_T_Normal but hardly expressed in CD8+ C2_T_Conventional or CD8+ C3_T_Chondroblastic. Therefore, CD8+ T cells of osteosarcoma lung metastasis had low numbers and were not suitable for immune checkpoint inhibitors due to the low expression genes of immune checkpoints, and this finding was in accordance with a previous study. 60
C2_Malignant cells, which were predominantly derived from the patient who was diagnosed with chondroblastic-type osteosarcoma lung metastasis (BC10), highly expressed SOX9, ALPL, and RUNX2. SOX9 and RUNX2, members of the transcription factor family, encoded the specific markers for chondrocyte differentiation and osteoblast maturation, respectively. ALPL is reported to be an important factor for bone homeostasis and skeletal mineralization. 61 Therefore, these cluster cells were named as C2_Malignant cells in our study.
It has been reported that the tumors infiltrated with abundant T cells usually showed significantly better prognosis than those not. 62 Thus, we detected the spatial information between C2_Malignant cells and T cells in conventional-type osteosarcoma lung metastasis through the CSOmap analysis. Our results showed that T cells had poor infiltration into C2_Malignant cells. This result might explain the reason why outcomes were limited when treating conventional-type osteosarcoma lung metastasis by using the chimeric antigen receptor T cell therapy. We further analyzed the ligand–receptor pairs that contributed to the spatial organization, finding the TIMP1–CD63 pair which contributed 69.68%. We also simulated the distance between C1_T cells and C2_Malignant cells after upregulating or downregulating the expression of CD63, respectively. We found that the distance between C1_T cells and C2_Malignant cells increased after upregulating the expression of CD63 (P < 0.001), whereas the distance between the two cells narrowed (P > 0.05) when the expression of CD63 was downregulated. These findings might suggest that CD63 might act as a therapeutic target for conventional-type osteosarcoma lung metastasis, which should be further addressed in our next study.
In addition to exploring the infiltration between C2_Malignant cells and T cells, we re-clustered C2_Malignant cells into three subtypes on the basis of the drug sensitivity by using the Beyondcell analysis. Among these three subgroups, we found that unlike the C2_Therapeutic cluster, the partial of C1_Therapeutic cluster and the majority of C3_Therapeutic cluster showed a relatively high drug sensitivity to traditional chemotherapy drugs (i.e. methotrexate, cisplatin, ifosfamide, and doxorubicin). These findings might explain the reason why drug resistance occurred in the treatment of osteosarcoma lung metastasis.
Furthermore, we performed the Beyondcell analysis to the current scRNA-seq data combined with drug database and found five new drugs sensitive to all three therapeutic clusters, including RHC-80267, oxfendazole, bromhexine, mevastatin, and sulpiride. We performed the Beyondcell analysis scoring and found that the C1_Therapeutic cluster showed a significant drug sensitivity to oxfendazole and mevastatin, which aroused our interest in further research. Therefore, we listed the target genes related to the effect of oxfendazole and mevastatin on the C1_Therapeutic cluster. Results showed that the most downregulated genes were found in the C1_Therapeutic cluster and suggested that the treatment of lung metastatic osteosarcoma with oxfendazole and mevastatin might be effective.
Finally, after identifying KIAA0907, a gene considered to have significant prognostic value in osteosarcoma, as the shared target gene of oxfendazole (sig_2649) and mevastatin (sig_8633), we performed molecular docking analysis and analyzed the hydrogen-bond position and binding energy between oxfendazole-KIAA0907 and mevastatin-KIAA0907, respectively. Our results indicated that oxfendazole and mevastatin could bind well to KIAA0907.
However, there were some limitations to our study. First, the number of patient samples was quite low, only two samples. Second, findings were not validated by experiments in vivo or in vitro. On one hand, the data we analyzed were mined from the online database, leading to the lack of matched tissues for further validation. On the other hand, we were unable to obtain appropriate clinical samples for validation due to the lack of clinical cases. Although these obstacles existed actually, our study provided brand-new and reliable strategies for disease analyses. The mainstream methods, such as inferCNV analysis for visualizing CNVs in cells, CSOmap analysis for analyzing the spatial information of cells, Beyondcell analysis for recognizing the drug sensitivity of malignant cells, and molecular docking analysis for predicting the molecular interactions were integrated for the first time in our study. It is of great importance to future analysis of other tumors and diseases, not only osteosarcoma. And once the number of patient samples increase in the publicly available genomic data, one day these methods will prove to be more valuable.
Collectively, we found the changes in the cytotoxicity and immune checkpoint gene expression of CD8+ T cells in osteosarcoma lung metastasis based on the scRNA-seq algorithm-based analysis in our study. In addition, we successfully recapitulated the cellular spatial organization of malignant and T cells in the tumor microenvironment, and revealed the molecular determinants of cellular interactions. Moreover, we identified the presence of therapeutic clusters of malignant cells that were sensitive to chemotherapy drugs and unveiled the potential hydrogen-bond position and binding energy between drugs and protein binding site by molecular docking analysis. Our results highlighted the power of scRNA-seq in identifying the complex tumor microenvironment of osteosarcoma lung metastasis, making it possible to devise precision therapeutic approaches.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702231171900 for A new insight of immunosuppressive microenvironment in osteosarcoma lung metastasis by Mingwei He, Xiaohong Jiang, Jifeng Miao, Wenyu Feng, Tianyu Xie, Shijie Liao, Zhaojie Qin, Haijun Tang, Chengsen Lin, Boxiang Li, Jiake Xu, Yun Liu, Zengnan Mo and Qingjun Wei in Experimental Biology and Medicine
Acknowledgments
We are grateful for the support of the Guangxi Key Laboratory of Genomics and Personalized Medicine.
Footnotes
Authors’ Contributions: QW, and ZM designed and supervised the research activity planning. MH, XJ, and JM drafted the manuscript. MH, WYF and HT finished the bioinformatics analysis. TX, and XJ performed the molecular docking analysis. CL, and ZQ helped and collected the data. SL, BL, JX, and YL revised the manuscript. All authors contributed to the manuscript and approved the submitted version.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by the Guangxi Key R&D Program (GuiKe AB22035014), the National Natural Science Foundation of China (Grant Nos 81960768 and 82260814), the Natural Science Foundation of Guangxi Province (Grant No. 2020GXNSFAA259088), the Youth Science and Technology Project of the First Affiliated Hospital of Guangxi Medical University (Grant No. 201903038), and the “Medical Excellence Award” Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University.
ORCID iDs: Jiake Xu  https://orcid.org/0000-0001-6712-5084
https://orcid.org/0000-0001-6712-5084
Qingjun Wei  https://orcid.org/0000-0003-0570-4085
https://orcid.org/0000-0003-0570-4085
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Moriarity BS, Otto GM, Rahrmann EP, Rathe SK, Wolf NK, Weg MT, Manlove LA, LaRue RS, Temiz NA, Molyneux SD, Choi K, Holly KJ, Sarver AL, Scott MC, Forster CL, Modiano JF, Khanna C, Hewitt SM, Khokha R, Yang Y, Gorlick R, Dyer MA, Largaespada DA. A Sleeping Beauty forward genetic screen identifies new genes and pathways driving osteosarcoma development and metastasis. Nat Genet 2015;47:615–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lamora A, Mullard M, Amiaud J, Brion R, Heymann D, Redini F, Verrecchia F. Anticancer activity of halofuginone in a preclinical model of osteosarcoma: inhibition of tumor growth and lung metastases. Oncotarget 2015;6:14413–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Picci P. Osteosarcoma (osteogenic sarcoma). Orphanet J Rare Dis 2007;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goguet-Surmenian E, Richard-Fiardo P, Guillemot E, Benchetrit M, Gomez-Brouchet A, Buzzo P, Karimdjee-Soilihi B, Alemanno P, Michiels JF, Schmid-Alliana A, Schmid-Antomarchi H. CXCR7-mediated progression of osteosarcoma in the lungs. Br J Cancer 2013; 109:1579–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aljubran AH, Griffin A, Pintilie M, Blackstein M. Osteosarcoma in adolescents and adults: survival analysis with and without lung metastases. Ann Oncol 2009;20:1136–41 [DOI] [PubMed] [Google Scholar]
- 6. Smith HA, Kang Y. The metastasis-promoting roles of tumor-associated immune cells. J Mol Med 2013;91:411–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li R, Zhou R, Wang H, Li W, Pan M, Yao X, Zhan W, Yang S, Xu L, Ding Y, Zhao L. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ 2019;26:2447–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res 2019;79:4557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robinson BD, Sica GL, Liu YF, Rohan TE, Gertler FB, Condeelis JS, Jones JG. Tumor microenvironment of metastasis in human breast carcinoma: a potential prognostic marker linked to hematogenous dissemination. Clin Cancer Res 2009;15:2433–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deepak KGK, Vempati R, Nagaraju GP, Dasari VR, S N, Rao DN, Malla RR. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol Res 2020;153:104683. [DOI] [PubMed] [Google Scholar]
- 11. Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006;124:263–6 [DOI] [PubMed] [Google Scholar]
- 12. Hui L, Chen Y. Tumor microenvironment: sanctuary of the devil. Cancer Lett 2015;368:7–13 [DOI] [PubMed] [Google Scholar]
- 13. Liu Z, Xie Y, Xiong Y, Liu S, Qiu C, Zhu Z, Mao H, Yu M, Wang X. TLR 7/8 agonist reverses oxaliplatin resistance in colorectal cancer via directing the myeloid-derived suppressor cells to tumoricidal M1-macrophages. Cancer Lett 2020;469:173–85 [DOI] [PubMed] [Google Scholar]
- 14. Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, Liu Q, Dou R, Xiong B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Molecular Cancer 2019;18:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang C, He L, He P, Liu Y, Wang W, He Y, Du Y, Gao F. Increased drug resistance in breast cancer by tumor-associated macrophages through IL-10/STAT3/bcl-2 signaling pathway. Med Oncol 2015;32:352. [DOI] [PubMed] [Google Scholar]
- 16. Kurachi M. CD8(+) T cell exhaustion. Semin Immunopathol 2019;41: 327–37 [DOI] [PubMed] [Google Scholar]
- 17. Kmiecik J, Poli A, Brons NH, Waha A, Eide GE, Enger PO, Zimmer J, Chekenya M. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. J Neuroimmunol 2013;264:71–83 [DOI] [PubMed] [Google Scholar]
- 18. Piersma SJ, Jordanova ES, van Poelgeest MI, Kwappenberg KM, van der Hulst JM, Drijfhout JW, Melief CJ, Kenter GG, Fleuren GJ, Offringa R, van der Burg SH. High number of intraepithelial CD8+ tumor-infiltrating lymphocytes is associated with the absence of lymph node metastases in patients with large early-stage cervical cancer. Cancer Res 2007;67:354–61 [DOI] [PubMed] [Google Scholar]
- 19. Baitsch L, Baumgaertner P, Devêvre E, Raghav SK, Legat A, Barba L, Wieckowski S, Bouzourene H, Deplancke B, Romero P, Rufer N, Speiser DE. Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J Clin Invest 2011;121:2350–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leem G, Park J, Jeon M, Kim ES, Kim SW, Lee YJ, Choi SJ, Choi B, Park S, Ju YS, Jung I, Kim S, Shin EC, Lee JY, Park SH. 4-1BB co-stimulation further enhances anti-PD-1-mediated reinvigoration of exhausted CD39(+) CD8 T cells from primary and metastatic sites of epithelial ovarian cancers. J Immunother Cancer 2020;8:e001650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 2009;6:377–82 [DOI] [PubMed] [Google Scholar]
- 22. Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 2018;18:35–45 [DOI] [PubMed] [Google Scholar]
- 23. Zhou Y, Yang D, Yang Q, Lv X, Huang W, Zhou Z, Wang Y, Zhang Z, Yuan T, Ding X, Tang L, Zhang J, Yin J, Huang Y, Yu W, Wang Y, Zhou C, Su Y, He A, Sun Y, Shen Z, Qian B, Meng W, Fei J, Yao Y, Pan X, Chen P, Hu H. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat Commun 2020;11:6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Madissoon E, Wilbrey-Clark A, Miragaia RJ, Saeb-Parsy K, Mahbubani KT, Georgakopoulos N, Harding P, Polanski K, Huang N, Nowicki-Osuch K, Fitzgerald RC, Loudon KW, Ferdinand JR, Clatworthy MR, Tsingene A, van Dongen S, Dabrowska M, Patel M, Stubbington MJT, Teichmann SA, Stegle O, Meyer KB. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biology 2019;21:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y, Feng W, Dai Y, Bao M, Yuan Z, He M, Qin Z, Liao S, He J, Huang Q, Yu Z, Zeng Y, Guo B, Huang R, Yang R, Jiang Y, Liao J, Xiao Z, Zhan X, Lin C, Xu J, Ye Y, Ma J, Wei Q, Mo Z. Single-Cell Transcriptomics reveals the complexity of the tumor microenvironment of treatment-naive osteosarcoma. Front Oncol 2021;11:709210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H, Liu L, Huang D, Jiang J, Cui GS, Yang Y, Wang W, Guo D, Dai M, Guo J, Zhang T, Liao Q, Liu Y, Zhao YL, Han DL, Zhao Y, Yang YG, Wu W. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 2019;29:725–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu Z, Lu W, Su C, Lv Y, Ye Y, Guo B, Liu D, Yan H, Mi H, Li T, Zhang Q, Cheng J, Mo Z. Single-cell RNA-seq identification of the cellular molecular characteristics of sporadic bilateral clear cell renal cell carcinoma. Front Oncol 2021;11:659251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017;14:979–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ren X, Zhong G, Zhang Q, Zhang L, Sun Y, Zhang Z. Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly. Cell Res 2020;30:763–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feng W, He M, Jiang X, Liu H, Xie T, Qin Z, Huang Q, Liao S, Lin C, He J, Xu J, Ma J, Liu Y, Wei Q. Single-cell RNA sequencing reveals the migration of osteoclasts in giant cell tumor of bone. Front Oncol 2021; 11:715552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He D, Wang D, Lu P, Yang N, Xue Z, Zhu X, Zhang P, Fan G. Single-cell RNA sequencing reveals heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations. Oncogene 2021;40:355–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bortolato A, Fanton M, Mason JS, Moro S. Molecular docking methodologies. Methods Mol Biol 2013;924:339–60 [DOI] [PubMed] [Google Scholar]
- 35. Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature 2003;423:337–42 [DOI] [PubMed] [Google Scholar]
- 36. Dazzi F. Cancer makes new friends with old tricks. Blood 2013;122:1093–4 [DOI] [PubMed] [Google Scholar]
- 37. Arneth B. Tumor microenvironment. Medicina 2019;56:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, Liu Z, Dong M, Hu X, Ouyang W, Peng J, Zhang Z. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017;169:1342–56 [DOI] [PubMed] [Google Scholar]
- 39. Santoiemma PP, Powell DJ., Jr. Tumor infiltrating lymphocytes in ovarian cancer. Cancer Biol Ther 2015;16:807–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stanton SE, Disis ML. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer 2016;4:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Minami K, Ueda N, Ishimoto K, Tsujiuchi T. LPA5-mediated signaling induced by endothelial cells and anticancer drug regulates cellular functions of osteosarcoma cells. Exp Cell Res 2020;388:111813. [DOI] [PubMed] [Google Scholar]
- 42. Mittrücker HW, Visekruna A, Huber M. Heterogeneity in the differentiation and function of CD8(+) T cells. Arch Immunol Ther Exp 2014;62:449–58 [DOI] [PubMed] [Google Scholar]
- 43. Fourcade J, Sun Z, Benallaoua M, Guillaume P, Luescher IF, Sander C, Kirkwood JM, Kuchroo V, Zarour HM. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J Exp Med 2010;207:2175–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, Thompson CB, Bluestone JA. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1994;1:405–13 [DOI] [PubMed] [Google Scholar]
- 45. Matsuzaki J, Gnjatic S, Mhawech-Fauceglia P, Beck A, Miller A, Tsuji T, Eppolito C, Qian F, Lele S, Shrikant P, Old LJ, Odunsi K. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci U S A 2010;107:7875–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harjunpää H, Guillerey C. TIGIT as an emerging immune checkpoint. Clin Exp Immunol 2020;200:108–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yan W, Liu X, Ma H, Zhang H, Song X, Gao L, Liang X, Ma C. Tim-3 fosters HCC development by enhancing TGF-beta-mediated alternative activation of macrophages. Gut 2015;64:1593–604 [DOI] [PubMed] [Google Scholar]
- 48. Kudo K, Yoneda A, Sakiyama D, Kojima K, Miyaji T, Yamazaki M, Yaita S, Hyodo T, Satow R, Fukami K. Cell surface CD63 increased by up-regulated polylactosamine modification sensitizes human melanoma cells to the BRAF inhibitor PLX4032. FASEB J 2019;33:3851–69 [DOI] [PubMed] [Google Scholar]
- 49. Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol 2013;14:986–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv Drug Deliv Rev 2017;114:206–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Anderson NR, Minutolo NG, Gill S, Klichinsky M. Macrophage-based approaches for cancer immunotherapy. Cancer Res 2021;81:1201–8 [DOI] [PubMed] [Google Scholar]
- 52. Alves AM, Diel LF, Lamers ML. Macrophages and prognosis of oral squamous cell carcinoma: a systematic review. J Oral Pathol Med 2018;47:460–7 [DOI] [PubMed] [Google Scholar]
- 53. Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol 2014;5:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Myers KV, Amend SR, Pienta KJ. Targeting Tyro3, Axl and MerTK (TAM receptors): implications for macrophages in the tumor microenvironment. Molecular Cancer 2019;18:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kishton RJ, Sukumar M, Restifo NP. Metabolic regulation of T cell longevity and function in tumor immunotherapy. Cell Metab 2017;26:94–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hernandez JM, Floyd DH, Weilbaecher KN, Green PL, Boris-Lawrie K. Multiple facets of junD gene expression are atypical among AP-1 family members. Oncogene 2008;27:4757–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peng Y, Chen Y, Chen S, Wang J, Jiang C, Hou W, Xu C. JUND-dependent up-regulation of HMOX1 is associated with cisplatin resistance in muscle-invasive bladder cancer. J Biochem 2020;168:73–82 [DOI] [PubMed] [Google Scholar]
- 58. Butler NS, Nolz JC, Harty JT. Immunologic considerations for generating memory CD8 T cells through vaccination. Cell Microbiol 2011;13:925–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nolz JC. Molecular mechanisms of CD8(+) T cell trafficking and localization. Cell Mol Life Sci 2015;72:2461–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu CC, Beird HC, Andrew Livingston J, Advani S, Mitra A, Cao S, Reuben A, Ingram D, Wang WL, Ju Z, Hong Leung C, Lin H, Zheng Y, Roszik J, Wang W, Patel S, Benjamin RS, Somaiah N, Conley AP, Mills GB, Hwu P, Gorlick R, Lazar A, Daw NC, Lewis V, Futreal PA. Immuno-genomic landscape of osteosarcoma. Nat Commun 2020;11:1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rauch F, Bardai G, Rockman-Greenberg C. ALPL mutations in adults with rheumatologic disorders and low serum alkaline phosphatase activity. J Bone Miner Metab 2019;37:893–9 [DOI] [PubMed] [Google Scholar]
- 62. Kuroda H, Jamiyan T, Yamaguchi R, Kakumoto A, Abe A, Harada O, Masunaga A. Tumor-infiltrating B cells and T cells correlate with postoperative prognosis in triple-negative carcinoma of the breast. BMC Cancer 2021;21:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702231171900 for A new insight of immunosuppressive microenvironment in osteosarcoma lung metastasis by Mingwei He, Xiaohong Jiang, Jifeng Miao, Wenyu Feng, Tianyu Xie, Shijie Liao, Zhaojie Qin, Haijun Tang, Chengsen Lin, Boxiang Li, Jiake Xu, Yun Liu, Zengnan Mo and Qingjun Wei in Experimental Biology and Medicine