ABSTRACT
Parkinson’s disease (PD) is a common neurodegenerative disease in middle-aged and elderly people, and many studies have confirmed that the disorder of gut microbiota is involved in the pathophysiological process of PD. However, the molecular mechanism of gut microbiota in regulating the pathogenesis of PD is still lacking. In this study, to investigate the impact of PD-associated gut microbiota on host transcriptome, we established various PD models with fecal microbiota transplantation (FMT) in the model organism Drosophila followed by integrative data analysis of microbiome and transcriptome. We first constructed rotenone-induced PD models in Drosophila followed by FMT in different groups. Microbial analysis by 16S rDNA sequencing showed that gut microbiota from PD Drosophila could affect bacterial structure of normal Drosophila, and gut microbiota from normal Drosophila could affect bacterial structure of PD Drosophila. Transcriptome analysis revealed that PD-associated gut microbiota influenced expression patterns of genes enriched in neuroactive ligand-receptor interaction, lysosome, and diverse metabolic pathways. Importantly, to verify our findings, we transplanted Drosophila with fecal samples from clinical PD patients. Compared to the control, Drosophila transplanted with fecal samples from PD patients had reduced microbiota Acetobacter and Lactobacillus, and differentially expressed genes enriched in diverse metabolic pathways. In summary, our results reveal the influence of PD-associated gut microbiota on host gene expression, and this study can help better understand the link between gut microbiota and PD pathogenesis through gut-brain axis.
IMPORTANCE
Gut microbiota plays important roles in regulating host gene expression and physiology through complex mechanisms. Recently, it has been suggested that disorder of gut microbiota is involved in the pathophysiological process of Parkinson’s disease (PD). However, the molecular mechanism of gut microbiota in regulating the pathogenesis of PD is still lacking. In this study, to investigate the impact of PD-associated gut microbiota on host transcriptome, we established various PD models with fecal microbiota transplantation in the model organism Drosophila followed by integrative data analysis of microbiome and transcriptome. We also verified our findings by transplanting Drosophila with fecal samples from clinical PD patients. Our results demonstrated that PD-associated gut microbiota can induce differentially expressed genes enriched in diverse metabolic pathways. This study can help better understand the link between gut microbiota and PD pathogenesis through gut-brain axis.
KEYWORDS: Parkinson’s disease, gut microbiota, Drosophila melanogaster, fecal microbiota transplantation, transcriptome
INTRODUCTION
The human gut has tens of trillions of microorganisms, including more than 1,000 species of bacteria. The number of genes of the microorganisms were 100 times larger than the human genome, and a balanced gut microbiota is critical for maintaining general health (1). More importantly, studies in the past 15 years have demonstrated gut microbiota as one of the key regulators of brain function, and have led to the appreciation of the importance of microbiota-gut-brain axis (2). Mechanistically, the highly bidirectional communication between the brain and the gut can be markedly influenced by the microbiota through integrated immunological, neuroendocrine, and neurological processes. At the molecular level, gut microbiota and its relevant metabolites interact with the host via a series of biochemical and functional pathways, thereby affecting host homeostasis and health (3).
Parkinson’s disease (PD) is the second most common neurodegenerative disease. Worldwide, about 3 million people suffer from the debilitating symptoms of PD involving motor deficits such as akinesia, muscular rigidity, tremor, slowness of movement, difficulty in walking, and gait. Also, PD is a multifactorial disease with less than 10% of cases being related to hereditary, therefore the pathogenesis of PD has a strong environmental component (4, 5). Recently, the gut microbiota-brain axis is gaining increasing attention by researchers who investigate the biological and physiological basis of psychiatric, neurodevelopmental, age-related, and neurodegenerative disorders (6). Current evidence from different clinical studies has revealed that gut microbiota composition is altered in PD patients and is related to clinical phenotypes (7), suggesting that gut microbiota plays a crucial role in PD. However, the pathogenesis of gut microbiota-mediated PD is still unclear and the microbiota-based clinical treatments are also limited. Fecal microbiota transplantation (FMT) is the most effective method to reconstruct the gut microbiota of PD patients (8). Although current research on FMT in the treatment of PD is not in-depth, FMT has been considered as a possible intervention for gut microbiota disturbance in the treatment of PD.
Fruit flies Drosophila are capable of performing complex motor behaviors such as walking, climbing, and flying, and the brains of fruit flies are functionally similar to humans (9). Basic cellular processes and many genes as well as signaling pathways are conserved between fruit flies and humans (10). The availability of genetic tools, the rapid growth and reproduction, the low cost, and the convenience to maintain in the laboratory make Drosophila an ideal model system to address biological questions relevant to human health (11, 12). The establishment of a toxin-based PD model in fruit flies has made an important contribution to our understanding of the disease (13 – 15). In such a scenario, Drosophila has emerged as a valuable model for studying mechanisms of human neurodegenerative diseases including PD. In this study, we constructed a rotenone-induced PD model in Drosophila followed by fecal microbiota transplantation, and investigated the impact of gut microbiota on host transcriptome by 16S rDNA sequencing and RNA sequencing. Our findings were verified by transplanting Drosophila with fecal samples from clinical PD patients. Our findings demonstrate the influence of PD-associated microbiota on host transcriptome in Drosophila and provide new insights into the mechanism of microbiota colonization in PD at the molecular level.
RESULTS
Rotenone-induced Parkinson’s disease models in Drosophila and FMT experiments
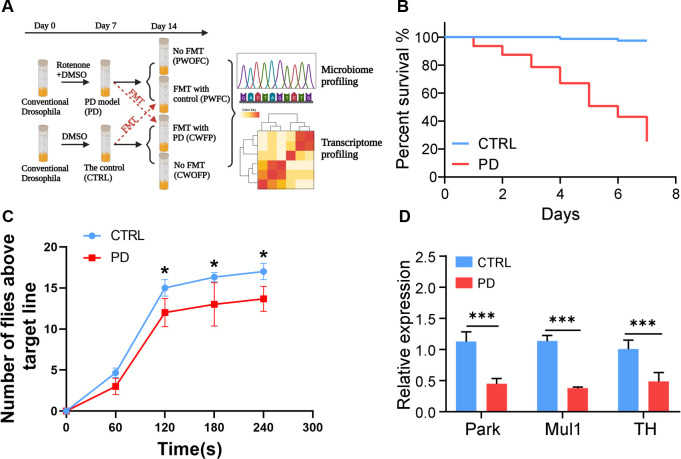
To dissect the relationship between the gut microbiota and PD, we first constructed the PD models by administering rotenone to Drosophila melanogaster for 7 days to obtain two groups of fly samples including Parkinson’s Drosophila (PD) and control Drosophila (CTRL). PD models were validated through phenotypic observation and molecular testing, then we performed cross-colonization experiments (Fig. 1A). The PD flies and control flies were subjected with or without FMT for another 7 days. After cross-colonization experiments (day 14), we harvested flies from each treatment group, including control Drosophila with FMT from Parkinson’s Drosophila (CWFP), control Drosophila without FMT from Parkinson’s Drosophila (CWOFP), Parkinson’s Drosophila with FMT from control Drosophila (PWFC), and Parkinson’s Drosophila without FMT from control Drosophila (PWOFC).
Fig 1.
Phenotypic analysis of the control and rotenone-treated Drosophila and validation of the models. (A) Experimental design of this study to show different groups of models, including control Drosophila (CTRL), Parkinson’s Drosophila (PD), control Drosophila with FMT from Parkinson’s Drosophila (CWFP), control Drosophila without FMT from Parkinson’s Drosophila (CWOFP), Parkinson’s Drosophila with FMT from control Drosophila (PWFC), and Parkinson’s Drosophila without FMT from control Drosophila (PWOFC). (B) Survival curves of CTRL and PD flies within 7 days (n = 80), significant differences were determined by log-rank test (P < 0.0001). (C) Negative geotaxis assays to show locomotor deficits of flies exposed to rotenone. Three replicates (20 flies in each replicate) were included in each timepoint and average numbers of flies were used for statistical analysis. (D) Relative expression of PD-related marker genes in CTRL and PD flies. Flies (male to female as 1:1) were used in the survival and locomotion assay. Significant differences are determined by the two-way ANOVA followed by Sidak’s post-hoc test. *P < 0.05, ***P < 0.001.
The survival curves were significantly different between the PD and CTRL groups (Log-rank test, P < 0.0001). CTRL Drosophila had a longer lifespan than PD Drosophila (Fig. 1B). Moreover, PD flies exhibited significant motor deficits. In the negative geotaxis assays, PD flies seemed not to be able to coordinate the legs and tended to stay at the bottom of the column. Statistical analysis showed that the locomotor activity of PD flies was significantly affected (Fig. 1C). Rotenone is a neurotoxin that damages mitochondria, Mul1 and Park are two major E3 ubiquitin ligases controlling the stability of mitochondria (16). Also, PD is a neuro-degenerative disease caused by the severe deficiency of Dopamine (DA) in the substantia nigra of striatum. Tyrosine hydroxylase (TH) is a key enzyme in the DA biosynthesis pathway (17). Quantitative RT-qPCR results showed that the expression levels of these marker genes in PD Drosophila were significantly lower than those in CTRL Drosophila (Fig. 1D). Thus, our results demonstrated that rotenone-induced PD flies could develop typical symptoms with marker genes changed, suggesting that our PD models were successfully constructed.
The effect of PD-associated gut microbiota on microbial composition of Drosophila
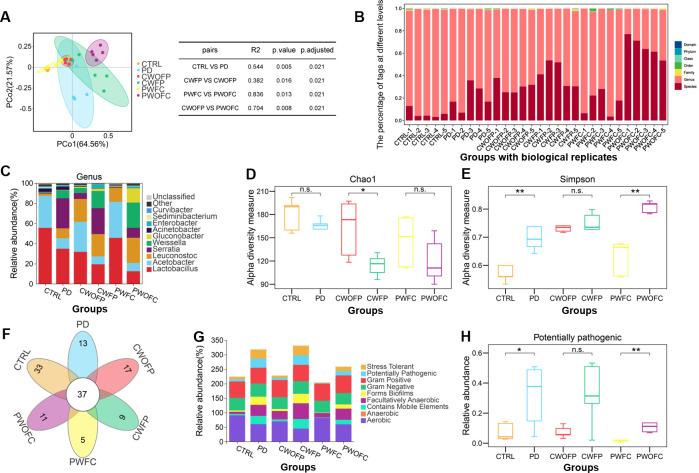
After the phenotypic confirmation of PD model and cross-colonization experiments, we performed microbial composition analysis using 16S rDNA sequencing of samples from six groups, and compared the changes of the microbiome composition between each group. Principal coordinate analysis (PCoA) of species abundance based on operational taxonomic unit (OTU) showed significant separation of samples from six groups of Drosophila, including the comparisons of CTRL vs PD, CWFP vs CWOFP, PWFC vs PWOFC, and CWOFP vs PWOFC (Fig. 2A). In order to better illustrate the effect of FMT on the gut microbiota of CTRL and PD groups, the microbiome profile of each sample was represented by a tags sequence. The global percent of tags were different at the genus and species levels for comparisons of CTRL vs PD or CWFP vs CWOFP or PWFC vs PWOFC (Fig. 2B). We counted the top 10 abundant microbial compositions in each group using stacked bar charts (Fig. 2C; Table S3). Compared to the CTRL group, PD groups had reduced Lactobacillus and Acetobacter. In addition, we found that Serratia and Weissella were mainly distributed in PD-associated groups, including PD group, CWFP group, and PWOFC group (Fig. 2C). We also presented the proportion of species in each group at the level of phylum, class, order, family, and species, respectively (Fig. S1). Together, the comparisons of CWFP vs CWOFP and PWFC vs PWOFC indicated that FMT significantly affected the global microbial composition in CTRL and PD Drosophila.
Fig 2.
FMT significantly changed the composition of gut microbiota in each group Drosophila. (A) Analysis of β-diversity of gut microbiota in Drosophila. PCoA was used to calculate the distance between samples, and the clusters between groups were tested with PERMANOVA. Each point represented a sample, n = 5. (B) According to the species annotation information of OTUs/ASVs, the number of tags sequences of each sample at each classification level was counted. The vertical axis represented the percentage of each sample in the sequence at each classification level. (C) Gut microbiota analysis of bacterial structure in six groups at the genus level. (D) Chao1 α-diversity index of grouped data (n = 5). (E) Simpson α-diversity index of grouped data (n = 5). (F) Species Venn diagrams of six groups at the genus level. (G) The phenotypic distribution of each group analyzed by BugBase. (H) Relative abundance of pathogenic microbiota (n = 5). Control Drosophila (CTRL) and Parkinson’s Drosophila (PD) are flies before cross-colonization experiments (day 7). Control Drosophila with FMT from Parkinson’s Drosophila (CWFP), control Drosophila without FMT from Parkinson’s Drosophila (CWOFP), Parkinson’s Drosophila with FMT from control Drosophila (PWFC), and Parkinson’s Drosophila without FMT from control Drosophila (PWOFC) are flies after cross-colonization experiments (day 14). Significant differences are determined by the Wilcoxon test. *P < 0.05, **P < 0.01.
To confirm the effect of FMT on microbial diversity in CTRL and PD Drosophila, we analyzed the α-diversity of gut microbiota of six groups in-depth. Dilution curves of α-diversity indexes indicated that our microbial sequencing data were reliable (Fig. S2). Further analysis on α-diversity indexes and rank abundance curves showed that species diversity was different in the comparisons of CWFP vs CWOFP or PWFC vs PWOFC (Fig. 2D and E; Fig. S3). Venn diagram showed that there were 37 shared species in the six groups at the genus level (Fig. 2F), and the CTRL group had the highest number of unique species (Fig. 2F; Fig. S4A). Importantly, FMT with gut microbiota from PD group increased the abundance of Serratia and Weissella in CWFP group, and FMT with gut microbiota from CTRL group increased the abundance of Acetobacter and Lactobacillus in PWFC group (Fig. 2C; Fig. S4B). The taxa that most likely explain the differences between groups were defined by LEfSe, which showed that Lactobacillaceae, Acetobacteraceae, and Alphaproteobacteria were significantly enriched in CTRL group, while Serratia, Enterobacteriaceae, and Weissella exhibited relatively higher abundance in the PD group (Fig. S4C). BugBase analysis of phenotype abundance showed that PD group has significantly higher abundance of pathogenic microbiota than CTRL group (Fig. 2G and H). Phenotypic abundance in CWFP group was similar to PD group, while PWFC group has significantly lower abundance of pathogenic microbiota than PWOFC group, suggesting that FMT experiments were successful and had obviously influenced the phenotype abundance in CTRL and PD Drosophila.
The impact of PD-associated microbiota on Drosophila transcriptome
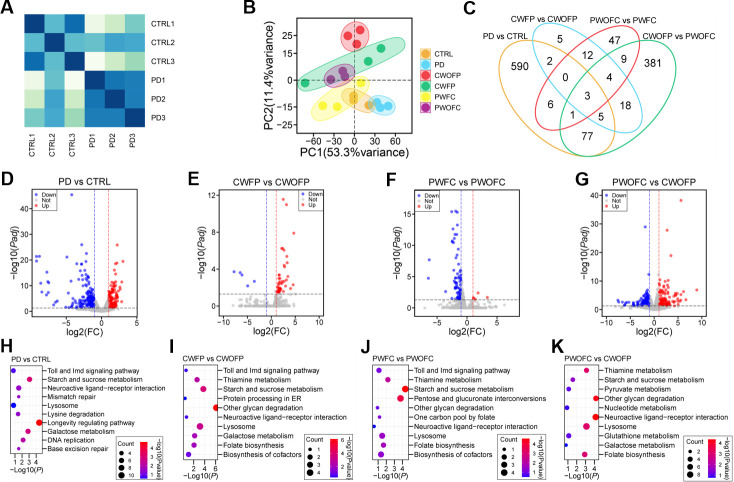
After determining the effects of FMT with PD-associated microbiota on different models, we further performed RNA sequencing using mRNAs from six groups of flies. We first drew the heatmap according to Pearson correlation coefficient of gene expression, and the results showed that the expression levels of transcripts were significantly different in the CTRL group compared with the PD group (Fig. 3A). Principal component analyses (PCAs) of transcriptome data of six groups were performed, and the results suggested that there were separation between the groups (Fig. 3B). We analyzed the differentially expressed transcripts (DETs) of six groups, and found 684 DETs between CTRL group and PD group, 49 DETs between CWFP group and CWOFP group, 82 DETs between PWFC group and PWOFC group, and 498 DETs between PWOFC group and CWOFP group (Fig. 3C; Table S2). Among these DETs, we further analyzed the upregulated and downregulated transcripts between different groups (Fig. 3D through G).
Fig 3.
RNA sequencing analysis of Drosophila samples before and after FMT experiments. (A) Pearson correlation heatmap of RNA-seq data from CTRL and PD groups. (B) PCA of RNA-seq data showed the separation of six groups, n = 3. (C) Venn diagram showed the numbers of DETs between four pairs of comparisons. (D) Volcano plot showed DETs between CTRL group and PD group. (E) Volcano plot showed DETs between CWFP group and CWOFP group. (F) Volcano plot showed DETs between PWFC group and PWOFC group. (G) Volcano plot showed DETs between PWOFC group and CWOFP group. (H) Enrichment analysis of KEGG pathway between CTRL group and PD group. (I) Enrichment analysis of KEGG pathway between CWFP group and CWOFP group. (J) Enrichment analysis of KEGG pathway between PWFC group and PWOFC group. (K) Enrichment analysis of KEGG pathway between PWOFC group and CWOFP group. The x-axis represented enrichment factors and the y-axis represented different pathways of biological processes.
KEGG analysis was performed to identify signaling pathways enriched for DETs with or without FMT experiments, and the results showed that various signaling pathways were enriched for different groups (Fig. 3H through K). Importantly, neuroactive ligand-receptor interaction, lysosome, and diverse metabolic pathways were enriched for four groups of comparisons (PD vs CTRL, CWFP vs CWOFP, PWFC vs PWOFC, and PWOFC vs CWOFP). Since all compared Drosophila were grown at the same development stage and treated with or without FMT, the observed differences in enriched pathways were proposed to derive from PD-associated microbiota and these common pathways may be associated with PD pathogenesis regulated by gut microbiota.
Correlations analysis of functional pathways influenced by PD-associated gut microbiota
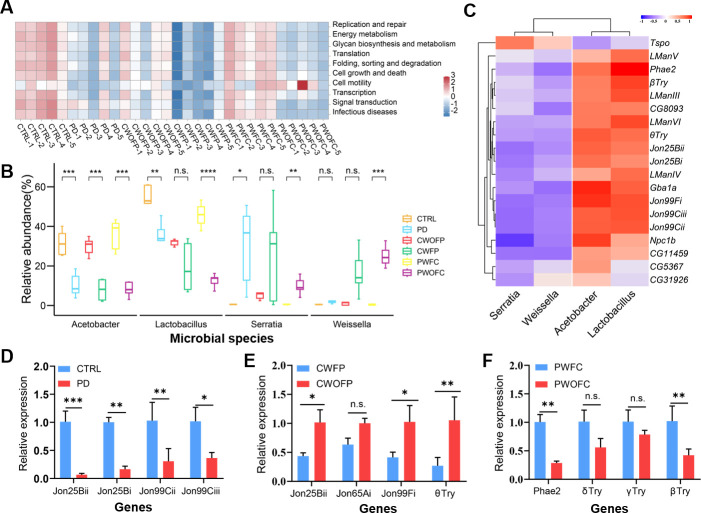
We predicted functional pathways across microbial samples using PICRUSt2, and found that various pathways were decreased in PD group than CTRL group, such as replication and repair, energy metabolism, cell growth and death, glycan biosynthesis and metabolism, etc. After FMT, the abundance of these pathways was lower in CWFP group than CWOFP group. Similarly, the abundance of these pathways was lower in PWOFC than PWFC group (Fig. 4A). Statistical analysis was performed on species at genus level among the six groups, and we found that the abundance of Lactobacillus, Acetobacter, Serratia, and Weissella was significantly different in most comparisons, including PD vs CTRL, CWFP vs CWOFP, and PWFC vs PWOFC (Fig. 4B). For example, the abundance of Acetobacter was lower in PD group than CTRL group. After cross-colonization FMT experiments (without rotenone in the medium), the abundance of Acetobacter in CWFP group became lower than CWOFP group, and the abundance of Acetobacter in PWFC group became higher than PWOFC group.
Fig 4.
Correlations analysis of functional pathways influenced by PD-associated gut microbiota and RT-qPCR validation of genes. (A) Functional prediction heatmap with PICRUSt2. Vertical axis indicated functional classification, horizontal axis indicated samples, and the color indicated the richness of the function. (B) Indicator species analysis at genus level of six groups. (C) Correlation analysis of the four gut microbes with DETs, which are representative genes from pathways of “neuroactive ligand-receptor interaction” and “lysosome” in Fig. 3. (D) Relative expression of transcripts encoding neuroactive ligand receptor interactions in CTRL group and PD group. (E) Relative expression of transcripts encoding neuroactive ligand-receptor interaction in CWFP group and CWOFP group. (F) Relative expression of transcripts encoding neuroactive ligand-receptor interactions in PWFC group and PWOFC group. Significant differences were determined by the unpaired Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
We performed correlations analysis between the four gut microbes and DETs in six groups, and the results showed that most of the DETs were positively correlated with Acetobacter and Lactobacillus, and negatively correlated with Serratia and Weissella (Fig. 4C). Moreover, we used Cytoscape software to analyze the correlation of DETs from neuroactive ligand-receptor interaction and lysosome pathways with the four gut microbes (Fig. S5), and the results showed that the DETs were correlated with the four gut microbes (Acetobacter, Lactobacillus, Serratia, and Weissella).
To verify our RNA-seq data, we performed RT-qPCR experiments and examined some DETs in neuroactive ligand-receptor interaction pathway, which are the most significantly enriched pathway between the PD and the CTRL group. The results showed that the expression levels of Jon25Bi, Jon25Bii, Jon99Cii, and Jon99Ciii were significantly downregulated by rotenone exposure (Fig. 4D). FMT significantly reduced the expression of Jon25Bii, Jon99Fi, and θTry in the CWFP group compared to the CWOFP group (Fig. 4E). The genes of βTry and Phae2 were significantly upregulated in PWFC group compared to the PWOFC group (Fig. 4F). Overall, RT-qPCR results of these DETs were generally similar to their expression patterns in RNA-seq data, indicating the validity and reliability of our RNA-seq data sets. Together, the results suggested that FMT with PD-associated gut microbiota could affect gene expression in Drosophila.
PD-associated gut microbiota from clinical patients influences microbial structure and diversity in Drosophila
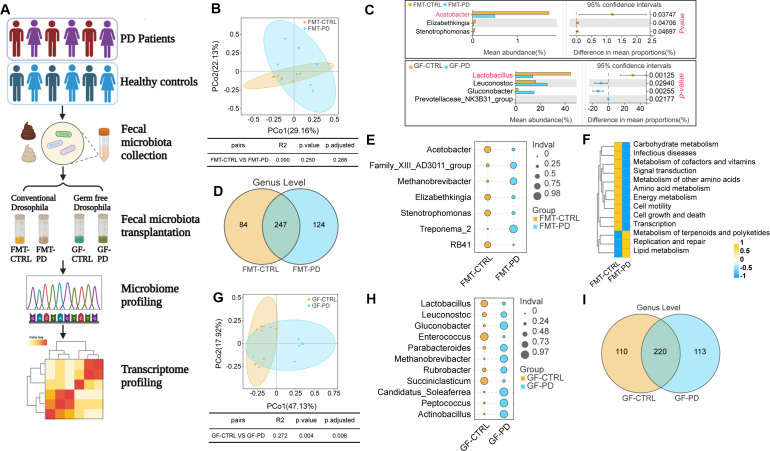
To further confirm whether gut microbiota-mediated gene expression changes derived from rotenone-induced PD model can be resembled by clinical PD-associated gut microbiota, we collected fecal samples from clinical PD patients for FMT experiments. The fecal samples were collected from six PD patients and six healthy controls with male to female ratio as 1:1. The fecal samples with gut microbiota were mixed as fecal suspensions and firstly colonized into conventional Drosophila. The effects of gut microbiota from PD patients on Drosophila were evaluated by 16S rDNA sequencing and transcriptome sequencing (Fig. 5A).
Fig 5.
Microbial structure and diversity in Drosophila colonized with fecal samples from clinical patients. (A) Schematics diagram showing experimental design of this study. (B) Analysis of β-diversity of gut microbiota in FMT-CTRL and FMT-PD groups from conventional Drosophila. PCoA was used to calculate the distance between samples, and the clusters between groups were tested with PERMANOVA. Each point represents a sample, n = 7 or 8. (C) Welch’s t-test showing differences in species abundance for conventional Drosophila and germ-free Drosophila, *P < 0.05. (D) Venn diagram indicating the numbers of species in two groups. (E) Indicator value (Indval) of species in each group at the genus level. (F) Functional prediction heatmap with PICRUSt2. (G) Analysis of β-diversity of gut microbiota in GF-CTRL and GF-PD groups from germ-free Drosophila. PCoA was used to calculate the distance between each sample. Each point represents a sample, n = 8. (H) Indicator value (Indval) of species in each group at the genus level. (I) Venn diagram indicating the numbers of species in two groups.
The clusters of FMT-CTRL Drosophila and FMT-PD Drosophila appeared to be sparse by PCoA (Fig. 5B). However, the most significantly different genus was Acetobacter (Fig. 5C), which was also the different genus in a rotenone-induced PD model. Venn diagram was made to define the core microbiome at the genus level, and we identified 124 species specifically in FMT-PD Drosophila (Fig. 5D). We calculated the indicator value of each species in each group at the genus level and confirmed that Acetobacter was one of the indicator species between FMT-CTRL Drosophila and FMT-PD Drosophila (Fig. 5E). We analyzed functional pathways with PICRUSt2, and found that various metabolism pathways were different between FMT-CTRL Drosophila and FMT-PD Drosophila (Fig. 5F). The microbial composition profiles for PD patients and healthy controls before FMT were also analyzed (Fig. S6), and the indicator values showed that Acetobacter was one of the indicator species. The alpha diversity of microbial composition for patient samples was insignificant, which may be due to the limited number of clinical samples.
The fecal samples from PD patients and healthy controls were also colonized into germ-free Drosophila followed by 16S rDNA sequencing (Fig. 5G). The indicator values of each species in each group at the genus level showed that Lactobacillus was one of the indicator species between GF-CTRL Drosophila and GF-PD Drosophila (Fig. 5H). In germ-free Drosophila, we identified 113 species specifically in GF-PD Drosophila (Fig. 5I). Together, our FMT experiments with PD patients in conventional Drosophila and germ-free Drosophila support our conclusion that Acetobacter and Lactobacillus are negatively associated with PD.
PD-associated gut microbiota from clinical patients influences Drosophila metabolic signaling pathways
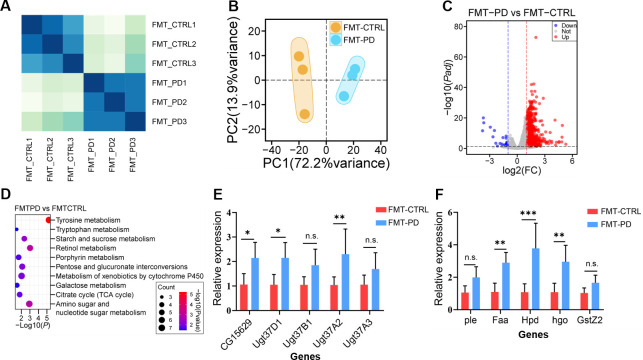
Next, we investigated the effects of PD-associated gut microbiota on host gene expression. We first analyzed the expression patterns between FMT-CTRL Drosophila and FMT-PD Drosophila groups by drawing the heatmap according to gene expression profiles. The results showed that the expression patterns were significantly different between FMT-CTRL Drosophila and FMT-PD Drosophila (Fig. 6A). PCA of RNA-seq data showed that three biological replicates from two groups were well clustered, while samples from FMT-CTRL Drosophila and FMT-PD Drosophila were clearly separated (Fig. 6B). Among these DETs, 858 transcripts were upregulated and 27 transcripts were downregulated when comparing FMT-CTRL Drosophila to FMT-PD Drosophila (Fig. 6C).
Fig 6.
Analysis of Drosophila transcriptome transplanted with PD-associated gut microbiota from clinical patients. (A) Pearson correlation heatmap of RNA-seq data from FMT-PD group and FMT-CTRL group, n = 3. (B) PCA of RNA-seq data from FMT-PD group and FMT-CTRL group, n = 3. (C) Volcano plot showing differentially expressed genes of Drosophila transplanted with PD-associated gut microbiota. (D) Enrichment analysis of the KEGG pathway of differentially expressed genes. (E) Relative expression level of genes associated with tyrosine metabolism in RT-qPCR. (F) Relative expression level of genes associated with retinol metabolism in RT-qPCR. Significant differences are determined by the unpaired Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
KEGG analysis was performed and the results revealed that differentially expressed transcripts were enriched in metabolism pathways, including tyrosine metabolism, retinol metabolism, starch and sucrose metabolism, galactose metabolism, and porphyrin metabolism (Fig. 6D). To verify the results of RNA-seq data, we selected differentially expressed genes enriched for tyrosine metabolism (Fig. 6E) and retinol metabolism (Fig. 6F) for RT-qPCR validations. The results showed that 6 out of the 10 upregulated transcripts in RNA-seq data were also upregulated in RT-qPCR experiments, indicating the validity and reliability of our RNA-seq data sets. Collectively, our Drosophila FMT experiments with fecal samples from clinical PD patients and rotenone-induced PD models revealed that PD-associated gut microbiota can influence the expression of genes enriched in metabolic signaling pathways.
DISCUSSION
Multiple studies have also proven that the pathogenesis of the chronic PD is closely related to gut microbiota dysbiosis (18 – 20). Better understanding of the interaction between gut microbiota and PD may provide novel insights into PD pathogenesis and help develop new therapeutic strategies. Drosophila melanogaster, one of many species of fruit fly, has been used as an optimal research model in the field of genetic engineering. In recent years, Drosophila has gained attention as an experimental animal for gut microbiota research in neurodegenerative diseases (21). In this study, we established PD Drosophila models by rotenone and investigated the role of gut microbiota in the pathogenesis of PD. The result showed that PD flies exhibited reduced lifespan and climbing and flight defects, meanwhile the expression levels of marker genes in PD Drosophila were significantly lower than those in CTRL Drosophila, which was consistent with the previous report. Based on 16S rDNA sequencing, we found that the species diversity in the PD group was much lower than that of the CTRL group. Our results demonstrated that the Drosophila PD model was successfully constructed in our study.
FMT with gut microbiota has been emerging as a promising therapy in various diseases (22, 23). In addition, one case report suggested that FMT can protect the gastrointestinal dysfunctions and motor disorders in a PD patient (24). To further assess the effects of FMT treatment on PD, we treated Drosophila with FMT and found that the gut microbiota of flies could be clearly separated after FMT. We counted the top 10 abundant bacteria at the genus level, and we found that Lactobacillus is the most widely distributed gut microbiota followed by Acetobacter. It has been demonstrated that the gut microbiota can regulate many biological processes by influencing gene expression in insects (25, 26). Therefore, we performed transcriptomic analysis by RNA-seq to further identify the genes involved in biological processes influenced by gut microbiota. We found that the expression levels of transcripts were significantly different in the CTRL group compared with the PD group. It is noteworthy that DETs were enriched in neuroactive ligand-receptor interaction, lysosome, and diverse metabolic pathways in all pairs of comparisons (PD vs CTRL, CWFP vs CWOFP, and PWFC vs PWOFC). Notably, all comparisons generally showed a consensus of difference in microbes and gene expression. Our results from rotenone-induced PD models were supported by FMT experiments with fecal samples from clinical PD patients. In Drosophila transplanted with fecal samples from PD patients, Acetobacter and Lactobacillus were associated with differentially expressed genes enriched in diverse metabolic pathways. Importantly, we found that starch/sucrose/galactose metabolism pathways were enriched in both rotenone-induced PD models and FMT with clinical samples. Since rotenone was not included in the fly medium when we performed cross-colonization FMT, so presumably the observed differences in microbes and gene expression from comparisons of CWFP vs CWOFP or PWFC vs PWOFC were mainly from the effect of FMT experiments. However, the side effect from rotenone in our study could not be completely excluded and should be considered in future work, since rotenone-induced PD models and FMT with clinical samples still had many different functional pathways. Taken together, our FMT experiments with gut microbiota from both rotenone-induced PD samples or clinical patients samples suggested that PD-associated gut microbiota can impact host gene expression in Drosophila.
As previously indicated, the simplicity of fruit flies provides a prominent advantage for FMT (27, 28), and the simple gut microbiota of Drosophila can help us understand the impact of gut microbiota on host physiology. Although Drosophila has orthologs for about 75% of human disease-related genes and the fly’s nervous system shares basic structure and functional features with humans (29), there are still differences in gut microbiota compositions and gene expression patterns between Drosophila and humans. In our study, we found that many gut microbiota species identified from human fecal samples were hard to be colonized in Drosophila. In both rotenone-induced PD Drosophila and Drosophila colonized with PD patient samples, the abundance of the two gut bacteria Lactobacillus and Acetobacter were obviously changed. In addition, we found that conventional Drosophila and germ-free Drosophila have different altered gut microbiota after FMT experiments. While the results from different contexts suggested that both Acetobacter and Lactobacillus were associated with PD pathogenesis, the genera Serratia and Weissella could also be related to PD pathogenesis. For instance, the genus Serratia belongs to the family Enterobacteriaceae and is now recognized as an important pathogen capable of causing many infections (30). Whether the genera Serratia and Weissella are involved into the pathogenesis of PD requires further investigations. In addition, our experiments included the same numbers of male and female Drosophila. Since male and female flies have different mechanisms for physiological processes (31, 32) and female flies are larger than male flies, the results of microbiome and transcriptome may have gender bias. RNA transcript abundance results could be biased toward female flies, and qPCR/RNA-seq experiments using single gender of flies are warranted to separate the effects of male and female flies. In the future, individual strain colonization should be carried out in a specific gender of Drosophila to explore the mechanism of gut microbiota on PD. Future work should also include more controls to rule out the side effects from other factors, such as non-FMT Drosophila controls when using clinical samples for colonization.
In summary, our results revealed the effect of gut microbiota on PD Drosophila by FMT and suggested that reduced Acetobacter and Lactobacillus were associated with PD pathogenesis. Transcriptome data analysis demonstrated that PD-associated gut microbiota could affect Drosophila gene expression. Taken together, our findings provide fundamental information for the impact of gut microbiota on transcriptomes in PD revealed by FMT. We anticipate this study can facilitate the understanding of the mechanism underlying PD treatment with FMT in clinical practice.
MATERIALS AND METHODS
Construction of rotenone-induced PD model in Drosophila
Drosophila melanogaster w1118 was reared at 25℃ under 12-hour light/12-hour dark cycles on yeast-glucose medium (1 L water, 100 g yeast, 100 g glucose, 1.2% agar, and 0.1% potassium sorbate) (33, 34). To construct PD model, we first dissolved rotenone (C23H22O6, #83–79-4, Dingguo Biotechnology Co., Ltd.) in DMSO as a stock solution (25 mM). Adult flies (day 7 post-eclosion) were cultured in a yeast-glucose-based medium supplemented with 70 µL of rotenone stock solution or DMSO for 7 days according to previous literature (16). For the control, Drosophila was cultured in a yeast-glucose-based medium containing only DMSO (#60313ES60, Yeasen). Each bottle contains the same number of male and female flies. Rotenone-containing food was freshly prepared, and the food should be changed every 2 days to ensure the efficacy.
Survival and locomotion assays
For the phenotypic experiments, adult flies were transferred to yeast-glucose medium with either DMSO (CTRL flies) or 250 µM rotenone dissolved in DMSO (PD flies) for 7 days. In order to validate the PD Drosophila model, 80 flies subdivided into 4 replicates (20 flies in each replicate) and were used for PD and CTRL, respectively. For the evaluation of the drug feeding effect, the surviving number of Drosophila was recorded every day. We determined locomotor ability with a negative geotaxis assay. Flies (male to female as 1:1) were anesthetized and placed in a vertical plastic column (length, 25 cm; diameter, 1.5 cm). After 30 minutes recovery from CO2 exposure, flies were gently tapped to the bottom of the column. Each 1 minute interval, the number of flies that climbed across a target line was recorded (35). Three replicates (20 flies in each replicate) were included in each timepoint and average numbers of flies were used for statistical analysis.
Fecal microbiota transplantation experiments
For FMT experiments with fecal samples from rotenone-induced PD models in Drosophila, we first obtained rotenone-induced PD flies or CTRL flies as in the above method. After 7 days, Drosophila were anesthetized with CO2, then surface disinfected with 70% ethanol, and washed with sterile PBS (#C0221A, Beyotime) three times. PD flies or CTRL flies were ground separately in 500 µL PBS solution with steel balls to obtain gut content. Each gut content was derived from PD flies or CTRL flies with equal numbers of males and females to avoid sex bias (10 flies per replicate). For cross-colonization experiments, PD flies were then fed for another 7 days with medium containing 50 µL of gut content from CTRL flies. Similarly, CTRL flies were fed for another 7 days with medium containing 50 µL of gut content from PD flies. For other groups of Drosophila without FMT, the flies were continuously reared in yeast-glucose medium for another 7 days. For FMT with fecal samples from clinical PD patients, stools were taken from −80℃ and thawed in water bath at 37℃. Fecal samples from individual PD patients or normal control were then pooled and homogenized in sterile PBS with a final concentration of 200 mg feces/2 mL. Pooled samples were centrifuged at 12,000 rpm for 2 minutes, and fecal suspension were collected in sterile centrifuge tubes. FMT with patient microbiome was colonized into conventional Drosophila and germ-free Drosophila. For FMT experiments in Drosophila, about 200 µL of fecal suspension was added to Drosophila medium (without rotenone). For FMT in germ-free Drosophila, we first prepared germ-free Drosophila as previous described (36), and FMT procedures were the same as the conventional Drosophila. Twenty flies (male to female as 1:1) were collected on day 7 post-eclosion for 16S rDNA sequencing and RNA sequencing.
16S rDNA sequencing
High-throughput 16S rDNA-seq was performed in Gene Denovo Biotechnology Co. Ltd. in Guangzhou. Microbial DNA from Drosophila was extracted using HiPure Stool DNA Kit (#D314103, Magen, Guangzhou, China) according to manufacturer’s protocol. The 16S rDNA target region of the ribosomal RNA gene was amplified by PCR (95℃ for 5 minutes, followed by 30 cycles at 95℃ for 1 minute, 60℃ for 1 minute, and 72℃ for 1 minute and a final extension at 72℃ for 7 minutes). The primers corresponding to V3–V4 regions of 16S rRNA gene were used for PCR: 341F (CCTACGGGNGGCWGCAG) and 806R (GGACTACHVGGGTATCTAAT). PCR reactions were performed in triplicate 50 µL mixture containing 50 ng of template DNA using SMRTbell Template Library Preparation Kit (PacBio, Menlo Park, USA). Amplicons were extracted from 2% agarose gels and purified using AxyPrep DNA Gel Extraction Kit (#AP-GX-250, Axygen Biosciences) according to manufacturer’s instructions and quantified using ABI StepOnePlus Real-Time PCR System (Life Technologies). Purified amplicons were pooled in equimolar and paired-end sequenced (PE250) on an Illumina platform according to the standard protocols.
Gut microbiome data analysis
For microbiome data analysis, the representative OTU sequences or ASV sequences were classified into organisms by a naïve Bayesian model using RDP classifier (37) (version 2.2) based on SILVA database (38) (version 132) or UNITE database (39) (version 8.0) or ITS2 database (40) (version update_2015), with the confidence threshold value of 0.8. Venn analysis was performed in R project Venn Diagram package (41) (version 1.6.16) and UpSet plot was performed in R project UpSetR package (42) (version 1.3.3) to identify unique and common species or OTUs or ASVs. Biomarker features in each group were screened by Lefse software (43) (version 1.0). OTU/ASV rarefaction curve and rank abundance curves were plotted in R project ggplot2 package (version 2.2.1). The KEGG pathway analysis of the OTUs/ASV was inferred using PICRUSt2 (44) (version 2.1.4). Microbiome composition data were analyzed using PCoA, and the clusters were tested with PERMANOVA. The software ggplot2 was used in the geom_mark_ellipse() function inside ggforce to differentiate each group of samples by drawing ellipses and colors according to the group. Based on the level of OTUs, differences in Bray-Curtis distances between groups were calculated using the R language vegan package. The pairwise Adonis test was performed based on the Bray-Curtis distance through the R language pairwiseAdonis package to determine the significance of the difference between the two groups. Chao1, Simpson (1 - ∑(Pi)2), and all other alpha diversity indexes were calculated in QIIME. OTU rarefaction curve and rank abundance curves were plotted in QIIME, and multivariate statistical techniques were calculated and plotted in the R project.
Microbiome phenotypes were classified using BugBase (https://github.com/knights-lab/BugBase), which was based on the Greengenes database using OUT sequences. The mapped microorganisms were divided into different categories, including Gram positive, Gram negative, biofilm forming, pathogenic, mobile element containing, aerobic, anaerobic, facultatively anaerobic, and oxidativestress tolerant. There are some overlaps among different categories, so the total proportion with all categories could be over 100%. For example, the total proportion will be 100% if only “Gram positive” and “Gram negative” categories are summarized, while Bugbase includes more than Gram positive and Gram negative categories.
For indicator species analysis, the numbers of OUT sequences were used to obtain differential OUTs in R project edgeR package, indicspecies and labdsv packages in R project were used to calculate the indicator value of each species. The indicator values were finally tested with cross-validation and presented as bubble plots.
RNA sequencing
After total RNA was extracted, the enriched mRNAs by Oligo(dT) beads were fragmented using fragmentation buffer and reverse transcribed into cDNA with random primers using the NEBNext Ultra RNA Library Prep Kit for Illumina (#E7530, NEB) following manufacturer’s instructions. For first-strand cDNA synthesis, the fragmented and primed mRNA was reversed into cDNA using ProtoScript II Reverse Transcriptase (#M0368S, NEB) in a 20 µL reaction with the procedure of 10 minutes at 25℃ and 15 minutes at 42℃ followed by 15 minutes at 70℃. For second-strand cDNA synthesis, the Second Strand Synthesis Enzyme Mix was added to the First-Strand Synthesis reaction to react at 16℃ for 1 hour in an 80 µL reaction. Then, the cDNA fragments were purified with QiaQuick PCR Extraction Kit (#28104, Qiagen), end-repaired, poly(A) added and ligated to Illumina sequencing adapters. The ligation products were size-selected by agarose gel electrophoresis, PCR-amplified, and sequenced using Illumina HiSeq2500 by Genedenovo Biotechnology Co, Ltd (Guangzhou, China).
Transcriptome data analysis
For data processing, quality control of sequencing data were assessed with FastQC. Adaptor sequences were trimmed by cutadapt (45) with at least 30 nt remaining length, and clean reads were mapped to the dm6 Drosophila reference genome using HISAT2 (46). FeatureCounts (47) was used to count the number of reads that mapped to a gene. Gene expression level was calculated by R package DESeq2 (48). Hierarchical clustering and principal component analysis were used to visualize the effect of different groups. The software ggplot2 was used in the geom_mark_ellipse() function inside ggforce to differentiate each group of samples by drawing ellipses and colors according to the group. Differentially expressed genes between two groups were identified by false discovery rate [adjusted P-value < 0.05 and log2(fold change) >1]. In order to obtain biological annotation of differentially expressed genes, gene ontology analysis was performed by clusterProfiler (49). For the correlation analysis of transcriptome and microbiome, the average value of three biological replicates corresponding to gene expression in each group was used for transcriptome data, and the average of the relative abundance of five biological replicates at the genus level in each group was used for microbiome data. Based on the above data for each group, the Spearman coefficient correlation analysis was performed using the corr.test function of the psych package in the R project.
RNA preparation and RT-qPCR analysis
To validate the expression level of DETs from RNA-seq, RNAs were extracted from different batch of Drosophila adults in 1 mL Trizol with sterilized steel balls. The RNA samples were dissolved in an appropriate amount of DEPC water. NanoDrop 2000 spectrophotometer (ThermoFisher) was used to quantify the concentration and purity of total RNA at a wavelength of 260 nm. The Hifair II First Strand cDNA Synthesis Super Mix for qPCR (#11123ES60, Yeasen) was used to synthesize cDNA. Three biological replicates were used for quantitative reverse transcription PCR analysis using HieffSYBR Mixture (#11201ES08, Yeasen). The relative mRNA level of gene expression was measured with Rp49 as internal control by calculating the values of ∆CtGene/∆CtRp49 and analyzed by the 2-∆∆Ct method. Primer sequences used for RT-qPCR analysis in this study are listed in Table S1.
Statistics
Statistical analyses were performed using GraphPad Prism (version 8.0), and the log-rank test was applied in order to plot the survival curves. Student’s t-test was used to evaluate the difference between the climbing assay and qPCR quantitative results. P < 0.05 was considered as significant difference.
ACKNOWLEDGMENTS
The authors would like to thank all members from the Wang lab for technical assistance and insightful discussion.
This work was supported by the National Natural Science Foundation of China (32070615, 81902093), Guangdong Provincial Natural Science Foundation (2021A1515010823 and 2022 A1515010569), Guangzhou Science and Technology Project (202002030100 to X.W.; 202201011809 to H.Z.), Panyu District Science and Technology Plan Medical and Health Project (2020-Z04-071), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme for X.W.
X.W. and H.Z. conceived and designed the experiments. X.L., M.Y., and F.Z. performed the experiments. M.Y., R.L., and X.L. analyzed the data. X.L., H.Z., and X.W. wrote the manuscript. All authors read and approved the final manuscript.
The authors declare no competing interests.
Contributor Information
Haibing Zhu, Email: zhb2@163.com.
Xiaoyun Wang, Email: wangxy@scnu.edu.cn.
John M. Chaston, Brigham Young University, Provo, Utah, USA
DATA AVAILABILITY
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. The 16S rRNA sequencing data in this study have been deposited at NCBI Sequence Read Archive under BioProject accession number PRJNA914678. The RNA sequencing data in this study have been deposited at NCBI GEO database under accession number GSE221760.
ETHICS APPROVAL
The animal protocol for this study was reviewed and approved by the Institutional Animal Care and Use Committee of South China Normal University (protocol code SCNU-SLS-2021–015). For clinical samples, written informed consent was obtained from all patients, and the research protocol was approved by the Ethical Committee of Guangzhou Panyu Central Hospital (Guangzhou, China).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.00176-23.
Fig. S1 to S6.
Table S1.
Table S2.
Table S3.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Hill-Burns EM, Debelius JW, Morton JT, Wissemann WT, Lewis MR, Wallen ZD, Peddada SD, Factor SA, Molho E, Zabetian CP, Knight R, Payami H. 2017. Parkinson's disease and Parkinson's disease medications have distinct signatures of the gut microbiome. Mov Disord 32:739–749. doi: 10.1002/mds.26942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cryan JF, O’Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, Codagnone MG, Cussotto S, Fulling C, Golubeva AV, Guzzetta KE, Jaggar M, Long-Smith CM, Lyte JM, Martin JA, Molinero-Perez A, Moloney G, Morelli E, Morillas E, O’Connor R, Cruz-Pereira JS, Peterson VL, Rea K, Ritz NL, Sherwin E, Spichak S, Teichman EM, van de Wouw M, Ventura-Silva AP, Wallace-Fitzsimons SE, Hyland N, Clarke G, Dinan TG. 2019. The microbiota-gut-brain axis. Physiol Rev 99:1877–2013. doi: 10.1152/physrev.00018.2018 [DOI] [PubMed] [Google Scholar]
- 3. Caputi V, Giron MC. 2018. Microbiome-gut-brain axis and toll-like receptors in Parkinson’s disease. Int J Mol Sci 19:19. doi: 10.3390/ijms19061689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet M-F, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK. 2016. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell 167:1469–1480. doi: 10.1016/j.cell.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parashar A, Udayabanu M. 2017. Gut microbiota: implications in Parkinson’s disease. Parkinsonism Relat Disord 38:1–7. doi: 10.1016/j.parkreldis.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi: 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scheperjans F, Aho V, Pereira PAB, Koskinen K, Paulin L, Pekkonen E, Haapaniemi E, Kaakkola S, Eerola-Rautio J, Pohja M, Kinnunen E, Murros K, Auvinen P. 2015. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov Disord 30:350–358. doi: 10.1002/mds.26069 [DOI] [PubMed] [Google Scholar]
- 8. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, Adler K, Dick-Necula D, Raskin S, Bloch N, Rotin D, Anafi L, Avivi C, Melnichenko J, Steinberg-Silman Y, Mamtani R, Harati H, Asher N, Shapira-Frommer R, Brosh-Nissimov T, Eshet Y, Ben-Simon S, Ziv O, Khan MAW, Amit M, Ajami NJ, Barshack I, Schachter J, Wargo JA, Koren O, Markel G, Boursi B. 2021. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 371:602–609. doi: 10.1126/science.abb5920 [DOI] [PubMed] [Google Scholar]
- 9. Fortini ME, Bonini NM. 2000. Modeling human neurodegenerative diseases in Drosophila: on a wing and a prayer. Trends Genet 16:161–167. doi: 10.1016/s0168-9525(99)01939-3 [DOI] [PubMed] [Google Scholar]
- 10. Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. 2001. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res 11:1114–1125. doi: 10.1101/gr.169101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bilen J, Bonini NM. 2005. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet 39:153–171. doi: 10.1146/annurev.genet.39.110304.095804 [DOI] [PubMed] [Google Scholar]
- 12. Hirth F. 2010. Drosophila melanogaster in the study of human neurodegeneration. CNS Neurol Disord Drug Targets 9:504–523. doi: 10.2174/187152710791556104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dawson TM, Ko HS, Dawson VL. 2010. Genetic animal models of Parkinson’s disease. Neuron 66:646–661. doi: 10.1016/j.neuron.2010.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whitworth AJ, Wes PD, Pallanck LJ. 2006. Drosophila models pioneer a new approach to drug discovery for Parkinson's disease. Drug Discov Today 11:119–126. doi: 10.1016/S1359-6446(05)03693-7 [DOI] [PubMed] [Google Scholar]
- 15. Kong Y, Wang L, Jiang B. 2021. The role of gut microbiota in aging and aging related neurodegenerative disorders: insights from Drosophila model. Life (Basel) 11:855. doi: 10.3390/life11080855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doktór B, Damulewicz M, Pyza E. 2019. Overexpression of mitochondrial ligases reverses rotenone-induced effects in a Drosophila model of Parkinson's disease. Front Neurosci 13:94. doi: 10.3389/fnins.2019.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagatsu T, Nagatsu I. 2016. Tyrosine hydroxylase (TH), its cofactor tetrahydrobiopterin (BH4), other catecholamine-related enzymes, and their human genes in relation to the drug and gene therapies of Parkinson's disease (PD): historical overview and future prospects. J Neural Transm (Vienna) 123:1255–1278. doi: 10.1007/s00702-016-1596-4 [DOI] [PubMed] [Google Scholar]
- 18. Dodiya HB, Forsyth CB, Voigt RM, Engen PA, Patel J, Shaikh M, Green SJ, Naqib A, Roy A, Kordower JH, Pahan K, Shannon KM, Keshavarzian A. 2020. Chronic stress-induced gut dysfunction exacerbates Parkinson's disease phenotype and pathology in a rotenone-induced mouse model of Parkinson's disease. Neurobiol Dis 135:104352. doi: 10.1016/j.nbd.2018.12.012 [DOI] [PubMed] [Google Scholar]
- 19. Bhattarai Y, Si J, Pu M, Ross OA, McLean PJ, Till L, Moor W, Grover M, Kandimalla KK, Margolis KG, Farrugia G, Kashyap PC. 2021. Role of gut microbiota in regulating gastrointestinal dysfunction and motor symptoms in a mouse model of Parkinson's disease. Gut Microbes 13:1866974. doi: 10.1080/19490976.2020.1866974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao Z, Ning J, Bao X-Q, Shang M, Ma J, Li G, Zhang D. 2021. Fecal microbiota transplantation protects rotenone-induced Parkinson's disease mice via suppressing inflammation mediated by the lipopolysaccharide-TLR4 signaling pathway through the microbiota-gut-brain axis. Microbiome 9:226. doi: 10.1186/s40168-021-01107-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kitani-Morii F, Friedland RP, Yoshida H, Mizuno T. 2021. Drosophila as a model for microbiota studies of neurodegeneration. J Alzheimers Dis 84:479–490. doi: 10.3233/JAD-215031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Borody TJ, Eslick GD, Clancy RL. 2019. Fecal microbiota transplantation as a new therapy: from Clostridioides difficile infection to inflammatory bowel disease, irritable bowel syndrome, and colon cancer. Curr Opin Pharmacol 49:43–51. doi: 10.1016/j.coph.2019.04.017 [DOI] [PubMed] [Google Scholar]
- 23. Antushevich H. 2020. Fecal microbiota transplantation in disease therapy. Clin Chim Acta 503:90–98. doi: 10.1016/j.cca.2019.12.010 [DOI] [PubMed] [Google Scholar]
- 24. Huang H, Xu H, Luo Q, He J, Li M, Chen H, Tang W, Nie Y, Zhou Y. 2019. Fecal microbiota transplantation to treat Parkinson's disease with constipation: a case report. Medicine (Baltimore) 98:e16163. doi: 10.1097/MD.0000000000016163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Broderick NA, Buchon N, Lemaitre B. 2014. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio 5:e01117–14. doi: 10.1128/mBio.01117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu L, Qi S, Xue X, Niu X, Wu L. 2020. Nitenpyram disturbs gut microbiota and influences metabolic homeostasis and immunity in honey bee (Apis mellifera L.). Environ Pollut 258:113671. doi: 10.1016/j.envpol.2019.113671 [DOI] [PubMed] [Google Scholar]
- 27. Matheoud D, Cannon T, Voisin A, Penttinen A-M, Ramet L, Fahmy AM, Ducrot C, Laplante A, Bourque M-J, Zhu L, Cayrol R, Le Campion A, McBride HM, Gruenheid S, Trudeau L-E, Desjardins M. 2019. Intestinal infection triggers Parkinson's disease-like symptoms in Pink1(-/-) mice. Nature 571:565–569. doi: 10.1038/s41586-019-1405-y [DOI] [PubMed] [Google Scholar]
- 28. Xu Y, Xie M, Xue J, Xiang L, Li Y, Xiao J, Xiao G, Wang H-L. 2020. EGCG ameliorates neuronal and behavioral defects by remodeling gut microbiota and TotM expression in Drosophila models of Parkinson's disease. FASEB J 34:5931–5950. doi: 10.1096/fj.201903125RR [DOI] [PubMed] [Google Scholar]
- 29. Aryal B, Lee Y. 2019. Disease model organism for Parkinson disease: Drosophila melanogaster. BMB Rep 52:250–258. doi: 10.5483/BMBRep.2019.52.4.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mahlen SD. 2011. Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24:755–791. doi: 10.1128/CMR.00017-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fromont C, Adair KL, Douglas AE. 2019. Correlation and causation between the microbiome, Wolbachia and host functional traits in natural populations of drosophilid flies. Mol Ecol 28:1826–1841. doi: 10.1111/mec.15041 [DOI] [PubMed] [Google Scholar]
- 32. Buck SA, Steinkellner T, Aslanoglou D, Villeneuve M, Bhatte SH, Childers VC, Rubin SA, De Miranda BR, O’Leary EI, Neureiter EG, Fogle KJ, Palladino MJ, Logan RW, Glausier JR, Fish KN, Lewis DA, Greenamyre JT, McCabe BD, Cheetham CEJ, Hnasko TS, Freyberg Z. 2021. Vesicular glutamate transporter modulates sex differences in dopamine neuron vulnerability to age-related neurodegeneration. Aging Cell 20:e13365. doi: 10.1111/acel.13365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wong AC-N, Dobson AJ, Douglas AE. 2014. Gut microbiota dictates the metabolic response of Drosophila to diet. J Exp Biol 217:1894–1901. doi: 10.1242/jeb.101725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walters AW, Hughes RC, Call TB, Walker CJ, Wilcox H, Petersen SC, Rudman SM, Newell PD, Douglas AE, Schmidt PS, Chaston JM. 2020. The microbiota influences the Drosophila melanogaster life history strategy. Mol Ecol 29:639–653. doi: 10.1111/mec.15344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Coulom H, Birman S. 2004. Chronic exposure to rotenone models sporadic Parkinson's disease in Drosophila melanogaster. J Neurosci 24:10993–10998. doi: 10.1523/JNEUROSCI.2993-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou F, Liu B, Liu X, Li Y, Wang L, Huang J, Luo G, Wang X. 2021. The impact of microbiome and microbiota-derived sodium butyrate on Drosophila transcriptome and metabolome revealed by multi-omics analysis. Metabolites 11:298. doi: 10.3390/metabo11050298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K. 2019. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264. doi: 10.1093/nar/gky1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ankenbrand MJ, Keller A, Wolf M, Schultz J, Förster F. 2015. ITS2 database V: twice as much. Mol Biol Evol 32:3030–3032. doi: 10.1093/molbev/msv174 [DOI] [PubMed] [Google Scholar]
- 41. Chen H, Boutros PC. 2011. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12:35. doi: 10.1186/1471-2105-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Conway JR, Lex A, Gehlenborg N, Hancock J. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33:2938–2940. doi: 10.1093/bioinformatics/btx364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kechin A, Boyarskikh U, Kel A, Filipenko M. 2017. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol 24:1138–1143. doi: 10.1089/cmb.2017.0096 [DOI] [PubMed] [Google Scholar]
- 46. Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 48. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6.
Table S1.
Table S2.
Table S3.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. The 16S rRNA sequencing data in this study have been deposited at NCBI Sequence Read Archive under BioProject accession number PRJNA914678. The RNA sequencing data in this study have been deposited at NCBI GEO database under accession number GSE221760.