Abstract
GR135402, a sordarin derivative, was isolated in an antifungal screening program. GR135402, sordarin, and derivatives of both compounds were evaluated for their ability to inhibit cell-free translational systems from five different pathogenic fungi (Candida albicans, Candida glabrata, Candida krusei, Candida parapsilosis, and Cryptococcus neoformans). The activity profile of GR135402 is extended to other chemical compounds derived from sordarin. Experimental results indicate that sordarin analogs exert their antifungal effects by specifically inhibiting the protein synthesis elongation cycle in yeasts but do not affect protein synthesis machinery in mammalian systems. Intrinsically resistant strains owe their resistance to differences in the molecular target of sordarins in these strains. Preliminary studies performed to elucidate the mode of action of this new class of antifungal agents have shown that the putative target of sordarins is one of the protein synthesis elongation factors.
In the last few years fungal infections have emerged as one of the major complications in immunocompromised patients (14, 23). A study of the present antifungal drugs indicates that all of them have several drawbacks, with the more important ones being resistance and toxicity (3). Therefore, new approaches to the design of novel drugs seem to be necessary. In this sense, identification of physiological processes different from those targeted by currently used antifungal agents would be of great interest.
Protein synthesis has always been considered one of the more attractive targets in the development of antimicrobial agents (9); in fact, many of the presently known antibiotics are inhibitors of bacterial protein synthesis. However, application of this idea to the field of antifungal therapy is not an easy task since selectivity is hampered due to the eukaryotic nature of fungi and therefore to the great degree of similarity between the fungal and mammalian protein synthesis machineries. Nevertheless, fungal translation has evolved as a desirable target with the discovery of a new essential factor for protein synthesis which is unique to yeasts, i.e., elongation factor 3 (EF-3) (1, 8, 20).
On the basis of this idea and taking advantage of the possibility of assaying yeast protein synthesis in vitro (5), a high-throughput screening program was established with the aim of finding selective inhibitors of the fungal protein synthesis system. As a result, compound GR135402 was isolated and characterized from a broth obtained from Graphium putredinis (13). The compound belongs to the sordarin family (10) and appeared to be a selective and potent inhibitor of Candida albicans protein synthesis. A synthetic chemical program was then initiated to produce novel analogs of the compound. In the present study it was demonstrated that the activity profile of GR135402 can be extended to other chemically synthesized sordarin derivatives. The study also aimed at ascertaining whether protein synthesis is actually the primary cellular target of this new class of antifungal agents and at elucidating the molecular basis of the intrinsic resistance in fungal species.
MATERIALS AND METHODS
Materials.
Sordarin derivatives were chemically synthesized by the Organic Synthesis Laboratories of the Research Department at Glaxo Wellcome S.A. The microorganisms used in the study (C. albicans 2005E, Candida glabrata 2375E, Candida krusei 2374E, Candida parapsilosis 2372E, and Cryptococcus neoformans 2867E) were obtained from the Glaxo Wellcome culture collection. Media for growth were from Difco (Detroit, Mich.). Radiochemicals and the rabbit reticulocyte system were from Amersham (Little Chalfont, United Kingdom). RNA-guard was from Pharmacia (Uppsala, Sweden). Yeast tRNA was from Boehringer Mannheim (Mannheim, Germany). All other chemicals were from Sigma (St. Louis, Mo.).
All solutions and buffers were prepared in 0.1% (vol/vol)-diethyl pyrocarbonate-treated water, and all procedures were performed at 4°C unless stated otherwise.
Methods. (i) Preparation of cell-free lysates.
To obtain a cell-free lysate capable of in vitro protein synthesis, the procedure described by Tuite and Plesset (22) was followed. Basically, cells were grown to the mid-logarithmic phase in yeast nitrogen base medium supplemented with 2% (wt/vol) glucose. At this point the cells were harvested by centrifugation, washed twice with lysis buffer (8.5% [wt/vol] mannitol, 30 mM HEPES-KOH [pH 7.4], 100 mM potassium acetate, 2 mM magnesium acetate, 2 mM dl-dithiothreitol), and finally, resuspended in an equal volume of this buffer supplemented with 1 mM phenylmethylsulfonyl fluoride. The cells were broken by grinding with glass beads for three cycles of 4 min each in a Vibrogen cell homogenizer (Edmund Bühler, Tübingen, Germany) refrigerated with a circulating ice-cold water bath. The lysate was decanted, centrifuged at 5,000 × g for 5 min to remove the beads and cell debris, and spun at 30,000 × g for 20 min. The resulting supernatant was aspirated off and centrifuged at 100,000 × g for 30 min, thus yielding a postpolysomal supernatant (S-100 fraction). This material was immediately frozen under liquid nitrogen and stored at −80°C until it was tested for cell-free protein synthesis activity. Alternatively, the S-100 fraction was split into ribosomes and soluble factors by centrifugation at 100,000 × g for 4 h. The postribosomal supernatant was then removed and was stored at −80°C until it was used, while the ribosomal pellet was carefully resuspended in 1/20 of the initial volume of lysis buffer, frozen under liquid nitrogen, and stored at −80°C.
(ii) Poly(U)-directed in vitro translation assay.
Poly(U)-directed in vitro protein synthesis in fungal cell-free systems was examined by measuring the level of incorporation of [14C]Phe into trichloroacetic acid (TCA)-precipitable material over 60 min. The assay was adapted from the method of Tuite and Plesset (22) and was performed in Multiscreen 96-well plates (Millipore, Bedford, Mass.). Final concentrations in the 50-μl assay volume were as follows: 20 mM HEPES-KOH (pH 7.4), 20 mM dl-dithiothreitol, 150 mM potassium acetate, 10 mM magnesium acetate, 0.38 U of RNA-guard, 100 μM GTP, 450 μM ATP, 24 mM phosphocreatine, 70 μg of creatine phosphokinase per ml, 0.5 mg of poly(U) per ml, and 0.75 μM [14C]Phe (12.5 kBq/ml). For the rabbit reticulocyte system, the instructions from the manufacturer were followed. To terminate the assay 50 μl of 1 M NaOH was added to each well and the plates were incubated at room temperature for 10 min. Afterward, 25 μl of ice-cold 50% (wt/vol) TCA was added to each well and the plates were incubated for 1 h at 4°C. The amount of synthesized poly-Phe was measured by harvesting the plate under vacuum, followed by the addition of scintillator Meltilex (Wallac, Turku, Finland) and counting in a Wallac MicroBeta scintillation counter.
(iii) Compound testing.
All compounds were dissolved in 25% (vol/vol) dimethyl sulfoxide (DMSO) and were serially diluted in this solvent. For determination of the 50% inhibitory concentrations (IC50s) in the in vitro translation assays, the final concentration of the compounds ranged from 0.005 to 2.5 μg/ml for the C. albicans, C. glabrata, and C. neoformans systems and from 0.2 to 100 μg/ml for the rabbit reticulocyte and the C. krusei and C. parapsilosis systems. In both cases the final DMSO concentration was 2.5% (vol/vol). The IC50 is defined as the compound concentration that inhibits 50% of the activity of the control. For MIC assays compound concentrations ranged from 0.001 to 125 μg/ml, while the DMSO concentration was kept constant at 1% (vol/vol) and MICs were determined according to the procedures of the National Committee for Clinical Laboratory Standards (16). The MIC is defined as the minimum concentration that causes 95% inhibition of growth with respect to the growth of the control.
(iv) Phe-tRNAPhe synthetase assay.
The Phe-tRNAPhe synthetase assay was performed in a manner similar to that described above for the in vitro translation assay except that postribosomal supernatant was used as the enzyme source, poly(U) was omitted, yeast tRNA was included at 1 mg/ml, and NaOH was not added to avoid alkaline hydrolysis of the recently formed Phe-tRNAPhe molecules.
(v) Measurement of de novo protein and RNA synthesis in whole C. albicans cells.
Methods for measuring the level of incorporation of radioactively labelled precursors into TCA-precipitable material have been described previously (19), and those methods were essentially followed. Briefly, cells were grown in yeast nitrogen base medium without amino acids, harvested at the mid-logarithmic phase, and diluted in fresh medium to 5 × 105 cells/ml. A pulse of either [35S]methionine or [3H]uridine was then added. Cells were preincubated at 37°C to initiate the incorporation of radiolabelled precursor, and then compound was added. For the protein synthesis experiments, aliquots were removed at regular intervals, TCA was added to 5% (vol/vol), and the sample was boiled for 20 min and allowed to precipitate at 4°C for 1 h. For the RNA synthesis experiments aliquots were removed and sodium dodecyl sulfate was added to 1.5% (wt/vol). After 30 min at room temperature samples were precipitated at 4°C for 1 h with 5% TCA in the presence of 10 mg of yeast tRNA per ml as a carrier. In both cases samples were finally harvested and counted by liquid scintillation.
RESULTS
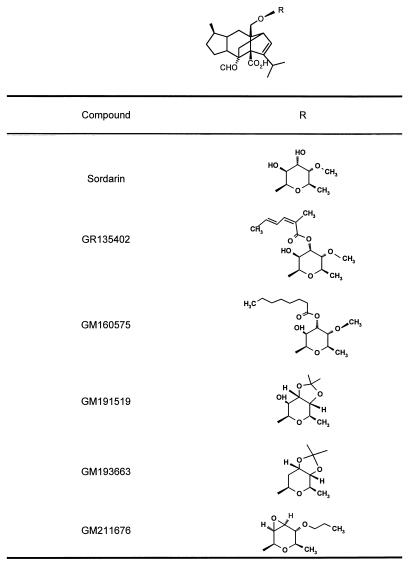
GR135402, sordarin, and several sordarin semisynthetic derivatives (structures are given in Table 1) were tested for their ability to inhibit in vitro protein synthesis and cell growth for five different species of pathogenic fungi (Table 2). Their ability to inhibit protein synthesis in mammalian systems (rabbit reticulocyte) was also tested. In terms of the IC50s, all compounds showed good activity against Candida albicans, C. glabrata, and Cryptococcus neoformans. However, C. krusei and C. parapsilosis were resistant to the actions of the sordarin compounds in both protein synthesis and cell growth assays. These compounds are derivatives that include the sordarin moiety with different kinds of substitutions. Since all of them contain the common sordarin structure as the main part of the molecule, it is reasonable to assume that they all point to the same target. However, substitutions may give rise to differences in the interactions with specific residues at the binding site of the targeted protein. These, combined with differences in the target among the tested fungal species, might explain the diversity of results presented in Table 2. On the other hand, in vitro protein synthesis in the rabbit reticulocyte system is not affected by these compounds over the range of concentrations tested (up to 100 μg/ml), showing the selective behavior of sordarins. Resistance in the cell-free translational system correlates with a lack of sensitivity in growth inhibition assays, thus suggesting that protein synthesis is the target for sordarins in the whole cell. However, the physiological significance of the assay is questionable due to the presence of artificial components: nonnatural mRNA, high ionic strength to force initiation, etc. (22). Therefore, experimental evidence is required to prove that inhibition of cell growth is actually due to the arrest of protein synthesis.
TABLE 1.
Chemical structures of the sordarin derivatives tested
TABLE 2.
Effect of sordarin derivatives on the cell-free protein synthesis assay (IC50s) and on cell growth (MICs)
| Compound |
C. albicans 2005E
|
C. glabrata 2375E
|
C. parapsilosis 2372E
|
C. krusei 2374E
|
C. neoformans 2867E
|
IC50 (μg/ml) for rabbit reticulocytes | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (μg/ml) | MIC (μg/ml) | IC50 (μg/ml) | MIC (μg/ml) | IC50 (μg/ml) | MIC (μg/ml) | IC50 (μg/ml) | MIC (μg/ml) | IC50 (μg/ml) | MIC (μg/ml) | ||
| Sordarin | 0.01 | 8.0 | 0.2 | >125 | >100 | >125 | >100 | >125 | 0.06 | >125 | >100 |
| GR135402 | 0.2 | 0.015 | 0.8 | >125 | >100 | >125 | >100 | >125 | 0.2 | 0.25 | >100 |
| GM160575 | 0.08 | <0.001 | 0.4 | >125 | >100 | >125 | >100 | >125 | 0.01 | 0.25 | >100 |
| GM191519 | <0.005 | 0.12 | 0.5 | 31.0 | 100 | >125 | 100 | >125 | 0.005 | 125 | >100 |
| GM193663 | <0.005 | <0.001 | 0.02 | 31.0 | >100 | >125 | >100 | >125 | 0.2 | 8.0 | >100 |
| GM211676 | <0.005 | 0.001 | 0.01 | 8.0 | 100 | >125 | 100 | >125 | 0.12 | 1.0 | >100 |
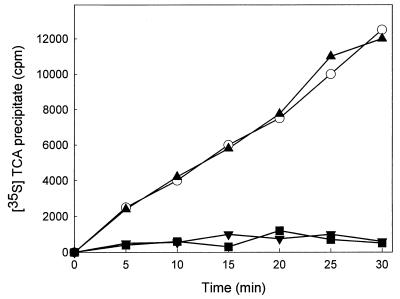
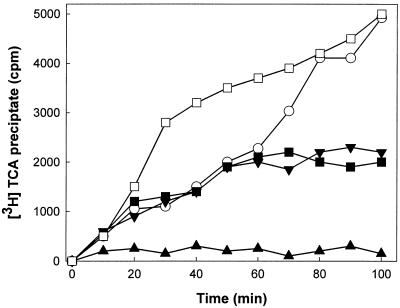
With the aim of obtaining this evidence we studied the effect of sordarins on protein and RNA synthesis de novo in intact C. albicans cells and compared this to the effects of verrucarin (a specific protein synthesis inhibitor) (2) and phenanthroline (an inhibitor of transcription) (11). The experiment was performed with GM160575 since this compound fulfilled the criteria of having good activity in the whole cell and having adequate chemical tractability. From Fig. 1 it is obvious that the compound prevented the incorporation of [35S]methionine to the same level that verrucarin does, therefore inhibiting the synthesis of new proteins. However, a clear difference between the behavior of GM160575 and phenanthroline on RNA synthesis was observed (Fig. 2). Phenanthroline caused drastic and fast inhibition, whereas GM160575 led to a short-term stimulatory effect at low concentrations. This effect, which disappeared with longer incubation times, was not seen with higher concentrations, with which inhibition of RNA synthesis was observed after 60 min due to the antifungal effect of the drug. Other investigators have already reported on the stimulation of RNA synthesis upon the addition of inhibitors of protein elongation to amino acid-starved yeast cells (17); this result thus supports the notion that sordarin derivatives exert their antifungal effect by selectively inhibiting protein synthesis. On the other hand, when the sensitivity and resistance profile of sordarin was compared to those of several known protein synthesis inhibitors whose modes of action have been elucidated (Table 3), none of the compounds had a profile identical to that for sordarin, that is, total inhibition in the C. albicans system and no inhibition in the C. krusei and C. parapsilosis systems. Similar patterns were obtained with emetine and fusidic acid; however, they showed the same potency against C. albicans as they did against C. parapsilosis. Although this result is not conclusive, it suggests that sordarin inhibits protein synthesis in a different manner than the other inhibitors do.
FIG. 1.
Effect of GM160575 on protein synthesis in intact C. albicans cells. Protein synthesis was measured as the ability to incorporate [35S]methionine into actively growing C. albicans cells. The experiments were performed as described in Materials and Methods. Time zero corresponds to the moment of compound addition. The results are the means of three independent experiments performed in duplicate. ○, control; ■, 0.2 μg of GM160575 per ml; ▴, 5 μg of phenanthroline per ml; ▾, 2.5 μg of verrucarin per ml.
FIG. 2.
Effect of GM160575 on RNA synthesis in intact C. albicans cells. RNA synthesis was measured as the ability to incorporate [3H]uridine into actively growing C. albicans cells. The experiments were performed as described in Materials and Methods. Time zero corresponds to the moment of compound addition. Results are the means of three independent experiments performed in duplicate. ○, control; ■, 0.2 μg of GM160575 per ml; □, 0.02 μg of GM160575 per ml; ▴, 5 μg of phenanthroline per ml; ▾, 2.5 μg of verrucarin per ml.
TABLE 3.
Activity profiles of several protein synthesis inhibitors in cell-free systems from sordarin-sensitive and -resistant Candida species
| Compound (concn [μg/ml]) | % Protein synthesis activity
|
Step inhibiteda | ||
|---|---|---|---|---|
| C. albicans | C. krusei | C. parapsilosis | ||
| Cycloheximide (1) | 94 | 15 | 25 | Translocation |
| Verrucarin A (1) | 5 | 8 | 7 | Peptide bond formation |
| Paromomycin (100) | 20 | 43 | 24 | Translocation, peptide bond formation |
| Anisomycin (1) | 41 | 50 | 36 | Peptide bond formation |
| Hygromycin (10) | 51 | 90 | 67 | Translocation |
| Puromycin (10) | 48 | 18 | 16 | Peptide bond formation |
| Emetine (100) | 77 | 97 | 74 | Unknown |
| Fusidic acid (100) | 77 | 100 | 80 | Translocation |
| Homoharringtonine (100) | 23 | 6 | 15 | Loading of aminoacyl-tRNA |
| Sordarin (1) | 1 | 100 | 94 | ? |
As described previously (2).
In order to gain more knowledge about the precise mechanism by which sordarins inhibit the protein synthesis elongation cycle and hence ascertain the molecular basis of intrinsic resistance in natural strains, cell-free systems from sensitive (C. albicans) or resistant (C. krusei and C. parapsilosis) species were split into ribosomes and soluble factors by centrifugation. The fractions obtained in this way were remixed to generate homologous (both fractions from the same species) or heterologous (fractions from different species) systems, and these regenerated systems proved to be active on in vitro protein synthesis. They were then used to identify whether the primary site of action of sordarins was the ribosome or a soluble factor, the rationale being that the sensitive or resistant nature of the reconstituted system is conferred by the target. Results from this experiment are summarized in Table 4. As expected, the homologous system from C. albicans was sensitive to inhibition, whereas those from C. krusei and C. parapsilosis were resistant to sordarin. However, the heterologous systems were sensitive only when soluble factors came from a sensitive species (C. albicans) and resistant when the source of such factors was a resistant species (C. krusei or C. parapsilosis). On the contrary, the ribosome source did not seem to play a role in conferring resistance or sensitivity to the heterologous systems. Since it was checked and confirmed that resistance in C. krusei and C. parapsilosis was not due to any sordarin-masking or -inactivating activity (data not shown), it can be assumed that differences in the specific target for sordarin are indeed responsible for the different sensitivities of the cell-free systems, and thus, it is concluded that the target is one of the soluble factors involved in the elongation cycle.
TABLE 4.
Effect of sordarin on protein synthesis in reconstituted systems generated with sordarin-sensitive and -resistant Candida species
| Source of soluble factors | % Remaining protein synthesis activity for the following ribosome sourcesa:
|
||
|---|---|---|---|
| C. albicans | C. krusei | C. parapsilosis | |
| C. albicans | 4 | 13 | 5 |
| C. krusei | 79 | 96 | 100 |
| C. parapsilosis | 80 | 98 | 100 |
Remaining protein synthesis activity in the presence of 0.5 μg of sordarin per ml. Each percentage is calculated with respect to the activity in the absence of sordarin for each reconstituted system.
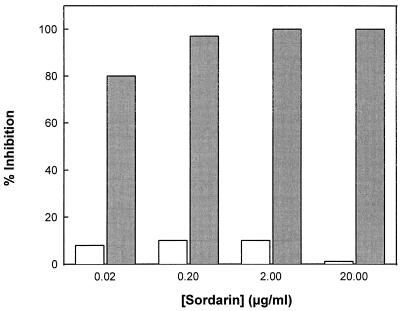
Four types of nonribosomal soluble proteins are known to be involved in the protein synthesis elongation cycle in yeasts (15): aminoacyl-tRNA synthetases and the three elongation factors EF-1, EF-2, and EF-3. Figure 3 shows evidence that sordarin has no effect on C. albicans Phe-tRNAPhe synthetase activity at the concentrations at which total inhibition of protein synthesis is achieved. The results presented above are in agreement that the sordarin target is one of the three elongation factors present in yeasts.
FIG. 3.
Effect of sordarin on Phe-tRNAPhe synthetase activity in C. albicans. The experiment was performed as described in Materials and Methods. The effect of sordarin on Phe-tRNAPhe synthetase activity (white bars) is compared with the effect of the same drug on cell-free protein synthesis activity (shaded bars).
DISCUSSION
The increasing impact of fungal infections on immunocompromised patients has intensified the need for new antifungal agents. The key to the development of these new drugs is the identification and characterization of new targets. Owing to their eukaryotic nature, fungal cells have only a restricted set of specific targets that do not overlap with those in their mammalian counterparts. In that respect most of the presently known antifungal agents point to targets which reflect clear differences between both kinds of cells: the fungal membrane (whose singularity lies in the presence of ergosterol, the function of which can be impaired either by direct interaction [with polyenes] or by inhibiting its synthesis [with azoles and allylamines]) and the cell wall (which can be damaged either by blocking the synthesis of its individual components β-(1,3)glucan [with echinocandins] and chitin [with nikkomycins] or by direct perturbation of the structure [with pradimicins]). However, different and more audacious strategies in the search for new antifungal agents could be of great value.
Although protein synthesis is an attractive target from the previous point of view, the lack of selective inhibitors so far may be due to the high degree of similarity between the fungal and the mammalian systems. Despite this similarity, sordarins have proved to be potent inhibitors of translation in fungi with an extremely high level of selectivity. All compounds inhibited in vitro translation in C. albicans, C. glabrata, and C. neoformans, but to varying degrees. Generally, there is a good correlation between inhibition of whole-cell growth and cell-free protein synthesis, and any differences may be indicative of differences in the uptake or in the intracellular stabilities of these compounds. The lack of sensitivity of C. krusei and C. parapsilosis to sordarins, however, is apparently explained by a difference in the recognition of the compound at the molecular target level. Differences in protein synthesis machinery among Candida species have not been described elsewhere. Knowledge is basically limited to C. albicans, and no information is available concerning the molecular features of protein synthesis factors from other non-C. albicans Candida species. The intriguing resistance of C. krusei and C. parapsilosis to the sordarins in comparison with the extremely high levels of potency of the sordarins against C. albicans suggests that these compounds have a highly specific binding site which may also be the basis for the greater selectivity of these compounds to inhibit the fungal but not the mammalian system from rabbit reticulocytes. Both aldehyde and carboxylic groups of the diterpene moiety have been shown to be essential for the retention of the activities of sordarin compounds (data not shown), presumably due to the participation of these groups in fine interactions with particular amino acid residues at the binding pocket of the target protein.
Experiments to measure the effects of these compounds on de novo protein and RNA synthesis in intact cells clarify the role of sordarins as specific inhibitors of protein synthesis and more precisely as inhibitors of the elongation cycle, since initiation and termination are not represented in the cell-free assay. The inhibition of [35S]methionine incorporation into newly synthesized proteins observed with the sordarins correlates with that observed with verrucarin, a potent and specific inhibitor of translation that acts on peptide bond formation (2). On the other hand, low concentrations of GM160575 stimulate the synthesis of RNA in C. albicans under conditions of amino acid starvation, whereas at higher concentrations inhibition is detected only after 60 min of exposure to the drug. This inhibition cannot be attributed to the specific arrest of RNA synthesis but is attributed more to the antifungal effect of the drug, since the inhibitory effect of phenanthroline, which acts as a transcriptional inhibitor (11), is observed immediately. It has been reported by other investigators that certain inhibitors of the elongation step in protein synthesis are able to stimulate de novo RNA synthesis not only in starved yeast cells (17, 18) but in mammalian cells as well (4, 6), and this has been associated with the specific stimulation of ongoing RNA synthesis. In addition to this, incubation of GM160575 with actively growing Saccharomyces cerevisiae cells lowers the monosome population, with a consequent increase in the polysome population (21a). All of this evidence is consistent with the freezing of the elongation cycle as the cellular mechanism of action of sordarins.
The protein synthesis elongation cycle in yeasts involves the participation of ribosomes and several nonribosomal soluble proteins, i.e., aminoacyl-tRNA synthetases and three elongation factors (EF-1, EF-2, and EF-3). The cycle proceeds through four main steps: (i) charging of tRNA with its corresponding amino acid residue in a reaction catalyzed by the corresponding aminoacyl-tRNA synthetase; (ii) occupancy of the ribosomal A site by the aminoacyl-tRNA in a process driven by EF-1α; (iii) formation of a peptide bond catalyzed by the peptidyl transferase center of the ribosome; and (iv) translocation of the nascent peptide, attached to tRNA, from the A to the P site in the ribosome, this step being conducted by EF-2. The precise role of EF-3 is still unclear, although it seems to be involved in maintaining translational accuracy as well as stimulating the function of EF-1α (12, 21). Our results indicate that the resistance or sensitivity of reconstituted heterologous systems relies on the source of the nonribosomal fraction more than on that of the ribosomes. The easiest and more reasonable explanation for this comes from the assumption that the target is directly related to this resistance-sensitivity profile. This leads us to discard the ribosome as the primary target of sordarin compounds, which, as a consequence, are not inhibitors of peptide bond formation. Moreover, because aminoacyl-tRNA synthetase activity is not affected by sordarin, we can infer that one of the three elongation factors might be the target of sordarin compounds. As shown in a further study (7), this conclusion has been confirmed and EF-2 has been identified as the primary target of sordarin derivatives in C. albicans.
ACKNOWLEDGMENTS
We are indebted to M. F. Tuite for helpful discussions. J. M. Viana is also thanked for his technical assistance, and E. Herreros is thanked for providing us with MIC data.
REFERENCES
- 1.Belfield G P, Tuite M F. Translation elongation factor 3: a fungus-specific translation factor? Mol Microbiol. 1993;9:411–418. doi: 10.1111/j.1365-2958.1993.tb01702.x. [DOI] [PubMed] [Google Scholar]
- 2.Carrasco L, Fernandez-Puentes C, Vazquez D. Antibiotics and compounds affecting translation by eukaryotic ribosomes. Specific enhancement of aminoacyl-tRNA binding by methylxanthines. Mol Cell Biochem. 1976;10:97–122. doi: 10.1007/BF01742203. [DOI] [PubMed] [Google Scholar]
- 3.Clark A M. The need for new antifungal drugs. In: Fernandes P B, editor. New approaches for antifungal drugs. Boston, Mass: Birkhäuser; 1992. pp. 1–19. [Google Scholar]
- 4.Coleclough C, Kuhn L, Lefkovits I. Regulation of mRNA abundance in activated T lymphocytes: identification of mRNA species affected by the inhibition of protein synthesis. Proc Natl Acad Sci USA. 1990;87:1753–1757. doi: 10.1073/pnas.87.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colthurst D R, Chalk P, Hayes M, Tuite M F. Efficient translation of synthetic and natural mRNAs in an mRNA-dependent cell-free system from the dimorphic fungus Candida albicans. J Gen Microbiol. 1991;137:851–857. doi: 10.1099/00221287-137-4-851. [DOI] [PubMed] [Google Scholar]
- 6.Dittman W A, Kumada T, Majerus P W. Transcription of thrombomodulin mRNA in mouse hemangioma cells is increased by cycloheximide and thrombin. Proc Natl Acad Sci USA. 1989;86:7179–7182. doi: 10.1073/pnas.86.18.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domínguez J M, Martin J J. Identification of elongation factor 2 as the essential protein targeted by sordarins in Candida albicans. Antimicrob Agents Chemother. 1998;42:2279–2283. doi: 10.1128/aac.42.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gooday G W. The potential of novel antifungal drugs for the treatment of disease in the immunocompromised host. Exp Opin Invest Drugs. 1995;4:679–691. [Google Scholar]
- 9.Hall C C, Bertasso A B, Watkins J D, Georgopapadakou N H. Screening assays for protein synthesis inhibitors. J Antibiot. 1992;45:1697–1699. doi: 10.7164/antibiotics.45.1697. [DOI] [PubMed] [Google Scholar]
- 10.Hauser D, Sigg H P. Isolierung und Abbau von Sordarin. Helv Chim Acta. 1971;54:1178–1190. doi: 10.1002/hlca.19710540427. [DOI] [PubMed] [Google Scholar]
- 11.Herruer M H, Mager W H, Raue H A, Vreken P, Wilms E, Planta R J. Mild temperature shock affects transcription of yeast ribosomal protein genes as well as the stability of their mRNAs. Nucleic Acids Res. 1988;16:7917–7929. doi: 10.1093/nar/16.16.7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kambampati R, Chakraburtty K. Functional subdomains of yeast elongation factor 3. Localization of ribosome binding domain. J Biol Chem. 1997;272:6377–6381. doi: 10.1074/jbc.272.10.6377. [DOI] [PubMed] [Google Scholar]
- 13.Kinsman O S, Chalk P A, Jackson H C, Middleton R F, Shuttleworth A, Rudd B A M, Jones C A, Noble H M, Wildman H G, Dawson M J, Stylli C, Sidebottom P J, Lamont B, Lynn S, Hayes M V. Isolation and characterisation of an antifungal antibiotic ( GR135402) with protein synthesis inhibition. J Antibiot. 1998;51:41–49. doi: 10.7164/antibiotics.51.41. [DOI] [PubMed] [Google Scholar]
- 14.Lortholary O, Dupont B. Antifungal prophylaxis during neutropenia and immunodeficiency. Clin Microbiol Rev. 1997;10:477–504. doi: 10.1128/cmr.10.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moldave K. Eukaryotic protein synthesis. Annu Rev Biochem. 1985;54:1109–1149. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts. Proposed standard M27-P. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1992. [Google Scholar]
- 17.Oliver S G, McLaughlin C S. The regulation of RNA synthesis in yeasts. I. Starvation experiments. Mol Gen Genet. 1977;154:145–153. doi: 10.1007/BF00330830. [DOI] [PubMed] [Google Scholar]
- 18.Oliver S G, Warmington J R. Transcription. In: Rose A H, Harrison J S, editors. The yeasts. 3. Metabolism and physiology of yeasts. San Diego, Calif: Academic Press, Inc.; 1991. pp. 117–160. [Google Scholar]
- 19.Oliver S G, Williamson D H. The conditions required for the induction of petite yeast mutants by fluorinated pyrimidines. Mol Gen Genet. 1976;146:261–268. doi: 10.1007/BF00701249. [DOI] [PubMed] [Google Scholar]
- 20.Skogerson L, Wakatama E. A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc Natl Acad Sci USA. 1976;73:73–76. doi: 10.1073/pnas.73.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Triana-Alonso F J, Chakraburtty K, Nierhaus K H. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J Biol Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 21a.Tuite, M. F. Personal communication.
- 22.Tuite M F, Plesset J. mRNA-dependent yeast cell-free translation systems: theory and practice. Yeast. 1986;2:35–52. doi: 10.1002/yea.320020103. [DOI] [PubMed] [Google Scholar]
- 23.Walsh T J. Invasive fungal infections: problems and challenges for developing new antifungal compounds. In: Sutcliffe J A, Georgopapadakou N H, editors. Emerging targets in antibacterial and antifungal chemotherapy. New York, N.Y: Chapman & Hall; 1992. pp. 349–373. [Google Scholar]