ABSTRACT
In plant-pathogen interactions, oxidative bursts are crucial for plants to defend themselves against pathogen infections. Rapid production and accumulation of reactive oxygen species kill pathogens directly and cause local cell death, preventing pathogens from spreading to adjacent cells. Meanwhile, the pathogens have developed several mechanisms to tolerate oxidative stress and successfully colonize plant tissues. In this study, we investigated the mechanisms responsible for resistance to oxidative stress by analyzing the transcriptomes of six oxidative stress-sensitive strains of the plant pathogenic fungus Fusarium graminearum. Weighted gene co-expression network analysis identified several pathways related to oxidative stress responses, including the DNA repair system, autophagy, and ubiquitin-mediated proteolysis. We also identified hub genes with high intramodular connectivity in key modules and generated deletion or conditional suppression mutants. Phenotypic characterization of those mutants showed that the deletion of FgHGG4, FgHGG10, and FgHGG13 caused sensitivity to oxidative stress, and further investigation on those genes revealed that transcriptional elongation and DNA damage responses play roles in oxidative stress response and pathogenicity. The suppression of FgHGL7 also led to hypersensitivity to oxidative stress, and we demonstrated that FgHGL7 plays a crucial role in heme biosynthesis and is essential for peroxidase activity. This study increases the understanding of the adaptive mechanisms to cope with oxidative stress in plant pathogenic fungi.
IMPORTANCE
Fungal pathogens have evolved various mechanisms to overcome host-derived stresses for successful infection. Oxidative stress is a representative defense system induced by the host plant, and fungi have complex response systems to cope with it. Fusarium graminearum is one of the devastating plant pathogenic fungi, and understanding its pathosystem is crucial for disease control. In this study, we investigated adaptive mechanisms for coping with oxidative stress at the transcriptome level using oxidative stress-sensitive strains. In addition, by introducing genetic modification technique such as CRISPR-Cas9 and the conditional gene expression system, we identified pathways/genes required for resistance to oxidative stress and also for virulence. Overall, this study advances the understanding of the oxidative stress response and related mechanisms in plant pathogenic fungi.
KEYWORDS: oxidative stress response, Fusarium graminearum, DNA damage response, autophagy, ubiquitin-proteasome pathway, heme biosynthesis
INTRODUCTION
Reactive oxygen species (ROS) are by-products generated during aerobic metabolism (1, 2). In general, ROS exist as superoxide radicals, hydrogen peroxide (H2O2), and hydroxyl radicals, and they function as signaling molecules through the oxidation of signal proteins or by themselves in the cell (3, 4). However, when cellular ROS accumulate to a high level, they can cause protein oxidation, lipid peroxidation, and/or DNA damage, all of which can lead to cell death (5 – 7).
Plants have evolved redox-related signals to deal with various biotic and abiotic stressors (8, 9). In plant-pathogen interactions, plants utilize ROS as signal molecules to regulate immune responses and physiologically inhibit pathogen infection. Plants recognize pathogen-associated molecular patterns or damage-associated molecular patterns through pattern recognition receptors in the cell membrane that then trigger pattern-triggered immunity (PTI) (10, 11). During PTI, rapid production of H2O2 and superoxide anions is concentrated in plant cells via NADPH-oxidase, peroxidases, and other ROS sources in a process generally known as an oxidative burst (12 – 14). An oxidative burst causing local cell death at the infection site can kill pathogens directly and inhibit the invasion of pathogens to the adjacent cell (15, 16).
Pathogens, meanwhile, have evolved several mechanisms to overcome oxidative bursts in host plants. ROS scavenging mechanisms are generally divided into enzymatic and non-enzymatic systems, and those two systems respond in a coordinated manner to oxidative stress (17, 18). Peroxidase, catalase, and superoxide dismutase (SOD) are representative enzymatic antioxidants, and glutathione and ascorbic acid are non-enzymatic antioxidants. These ROS-detoxifying systems have been studied thoroughly in plant pathogenic fungi. For example, peroxidases have been identified, and their roles in oxidative stress resistance have been characterized in Fusarium graminearum and Magnaporthe oryzae (19, 20). Functional analyses have revealed the role of SOD in oxidative stress resistance and virulence in Sclerotinia sclerotiorum, Botrytis cinerea, and Verticillium dahliae (21 – 23). The transcription factors such as Ap1, Skn7, and Atf1 regulating these antioxidant systems have also been extensively investigated, and it has been confirmed that their role in oxidative stress response is highly conserved in plant pathogenic fungi (24 – 26).
In addition to these antioxidant systems, mechanisms for recovering oxidative damage are essential for survival under oxidative stress conditions. DNA damage and protein oxidation are representative oxidative damage. DNA damage responses that identify and repair damaged DNA have been investigated in plant pathogenic fungi, including Ustilago maydis, V. dahliae, and F. graminearum (27 – 30). Also, proteolysis system to eliminate the damaged protein, including the ubiquitin-proteasome system (UPS), has been studied in M. oryzae and Fusarium oxysporum (31 – 33). However, a genome-wide correlation between these mechanisms and oxidative stress is required, and further identification of the mechanisms that contribute to coping with oxidative stress is still needed.
F. graminearum is one of the most destructive plant pathogenic fungi, causing Fusarium head blight (FHB) in important cereal crops worldwide (34, 35). Infection of F. graminearum, which can lead to severe yield losses and the accumulation of mycotoxins on grains, is an important cause of food shortages and a serious threat to public health (36). To control FHB, it is essential to understand how the fungus overcomes plant defense mechanisms and invades host plant cell. In this study, we aimed to identify the mechanisms/genes involved in responding to oxidative stress through a combined transcriptomic and physiological approach, and to explore the role of these mechanisms in the oxidative stress response and pathogenicity in F. graminearum.
RESULTS AND DISCUSSION
Weighted gene co-expression network analysis and identification of co-expression modules
We aimed to identify the oxidative stress response or oxidative stress-related mechanisms based on a comprehensive transcriptome analysis. We hypothesized that the expression of core genes involved in oxidative stress resistance would be dramatically changed in stress-sensitive strains than that of the wild type. In F. graminearum, eight transcription factor mutants displayed hypersensitivity to oxidative stress (20). We cultured these mutants on complete media (CM) with or without 10 mM H2O2 supplementation, and six mutants exhibiting dramatic growth defects compared with the wild-type strain were selected for further RNA-seq analyses: Δzif1, Δfgc2h010, Δfgzc236, Δfgzc086, Δfgap1, and Δfgbzip007 (Fig. 1A). The inhibition rates of Δzif1 and Δfgbzip007 were approximately twofold higher than that of the wild-type strain, and other strains showed significantly altered growth under oxidative stress conditions compared with the wild type (Fig. 1B). These six mutants were defined as “sensitive strains” in this study.
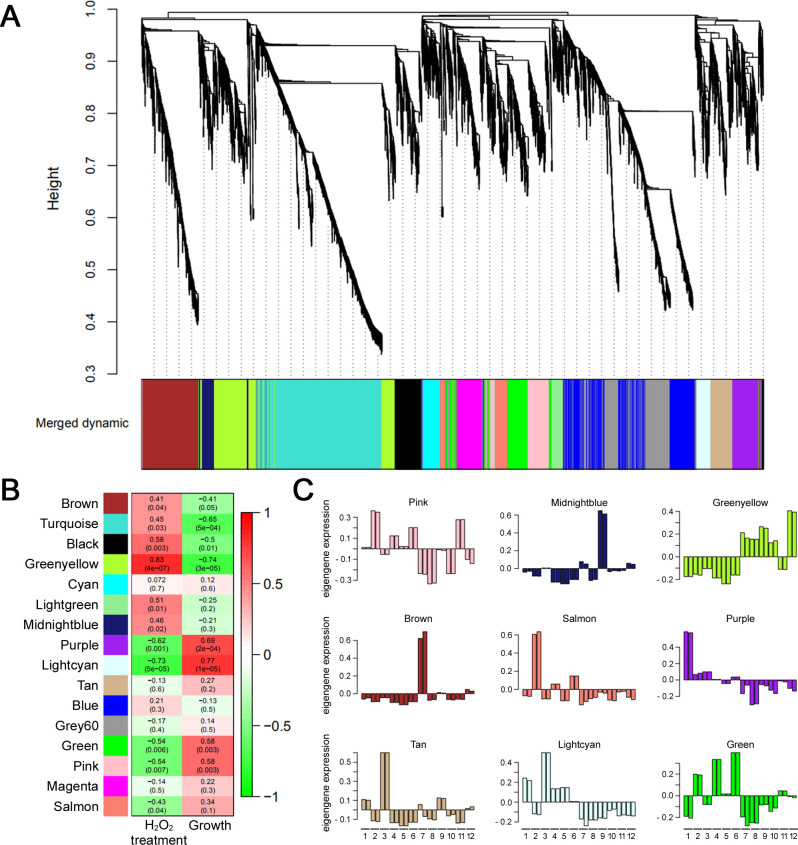
Fig 1.
Phenotypic and transcriptome analyses of oxidative stress-sensitive strains. (A) Mycelial growth of six oxidative stress-sensitive mutants under oxidative stress. Each strain was inoculated on CM and CM supplemented with 10 mM H2O2. Photographs were taken 5 days after inoculation. (B) Statistical analysis of mycelial growth inhibition under oxidative stress. Error bars represent standard deviations. Asterisks represent significant differences from the wild type (*P < 0.05; ***P < 0.001; t-test). (C) A clustering dendrogram of samples with trait heatmap. The 24 samples were clustered based on mRNA expression data. The color band underneath the tree indicates the value of treated H2O2 concentration and colony diameter. Linear gradation colors from white to red represent the values of each trait corresponding to the sample.
To reveal the mechanism related to oxidative stress response, we analyzed the transcriptomes of the six strains using RNA-seq data and conducted a weighted gene co-expression network analysis (WGCNA). All 24 samples were clustered, and samples derived from strains treated with H2O2 were clustered together, except those of Δfgbzip007 and Δfgzc086 (Fig. 1C). This clustering suggests that changes in gene expression for each sample are affected more by oxidative stress than by differences among genotypes.
Radial growth of mutants and H2O2 treatment were used as phenotypic traits for WGCNA analysis, and total gene profiles were divided into 16 modules based on transcription patterns (Fig. 2A). Among the 16 modules, three were closely correlated with H2O2 treatment and growth rate (P ≤ 0.001): Greenyellow, Lightcyan, and Purple (Fig. 2B). Gene expression patterns of the Greenyellow module were positively correlated with H2O2 treatment and negatively correlated with growth rate, which indicates that the Greenyellow module includes genes that increase expression when radial growth of the strains decreases due to oxidative stress. Conversely, the gene expression patterns of the Lightcyan and Purple modules were negatively correlated with H2O2 treatment and positively correlated with growth. The Lightcyan and Purple modules include genes for which expression decreases when radial growth of the strain decreases due to oxidative stress. The eigengene expression in each sample indicated that the Greenyellow, Lightcyan, and Purple modules were closely correlated with oxidative stress (Fig. 2C). These results imply that genes of each module can be functionally related and co-regulated by H2O2 treatment because genes with strongly correlated expression levels tend to share similar biological functions (37).
Fig 2.
Weighted gene co-expression network analysis. (A) Clustering dendrogram of genes. The genes were clustered based on the dissimilarity of the topological overlap. (B) Module-trait relationships. Each cell contains the corresponding correlation value and P-value. (C) Eigengene expression patterns in nine modules. 1-Δzif1, 2-Δfgbzip007, 3-Δfgc2h010, 4-Δfgzc236, 5-Δfgzc086, 6-Δfgap1, 7-Δzif1_H2O2, 8-Δfgzip007_H2O2, 9-Δfgc2h010_H2O2, 10-Δfgzc236_H2O2, 11-Δfgzc086_H2O2, 12-Δfgap1_H2O2.
Functional enrichment analysis of genes in the high-correlation modules
To explore the biological process of genes included in each module, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed for the three modules (Table 1). Among those genes, 245, 46, and 121 in the Greenyellow, Lightcyan, and Purple modules, respectively, had known functions. Genes in the Greenyellow module were mainly enriched in the “biosynthesis of amino acid pathway” (fgr01230), followed by “nucleotide excision repair” (fgr03420), “RNA transport” (fgr03013), and “protein processing in endoplasmic reticulum” (fgr04141). In the Lightcyan module, gene functions were enriched in the “metabolic pathway” (fgr01100) and “biosynthesis of secondary metabolites” (fgr01110). In the Purple module, genes were enriched in “metabolic pathways” (fgr01100), “oxidative phosphorylation” (fgr00190), the “MAPK signaling pathway” (fgr04011), and “Tricarboxylic acid cycle” (fgr00020) (Table 1).
TABLE 1.
KEGG pathway enrichment analysis of genes in co-expression modules
| Module | KEGG ID | Pathway | Count | P-value |
|---|---|---|---|---|
| Greenyellow | fgr03420 | Nucleotide excision repair | 16 | 2.75E−07 |
| fgr03440 | Homologous recombination | 9 | 2.49E−06 | |
| fgr03450 | Non-homologous end-joining | 5 | 4.13E−04 | |
| fgr03022 | Basal transcription factors | 10 | 7.21E−04 | |
| fgr00400 | Phenylalanine, tyrosine, and tryptophan biosynthesis | 9 | 7.90E−04 | |
| fgr03410 | Base excision repair | 8 | 8.08E−04 | |
| fgr01230 | Biosynthesis of amino acids | 25 | 1.04E−03 | |
| fgr04141 | Protein processing in endoplasmic reticulum | 15 | 9.63E−03 | |
| fgr03013 | RNA transport | 16 | 1.04E−02 | |
| fgr03430 | Mismatch repair | 6 | 1.39E−02 | |
| fgr02010 | ABC transporters | 8 | 1.59E−02 | |
| fgr04136 | Autophagy | 6 | 1.75E−02 | |
| fgr03050 | Proteasome | 8 | 1.89E−02 | |
| fgr04120 | Ubiquitin-mediated proteolysis | 10 | 3.33E−02 | |
| fgr04138 | Autophagy | 12 | 3.36E−02 | |
| Lightcyan | fgr01100 | Metabolic pathways | 39 | 4.95E−07 |
| fgr00260 | Glycine, serine, and threonine metabolism | 5 | 6.17E−03 | |
| fgr00600 | Sphingolipid metabolism | 3 | 1.31E−02 | |
| fgr00430 | Taurine and hypotaurine metabolism | 2 | 1.70E−02 | |
| fgr00100 | Steroid biosynthesis | 3 | 1.75E−02 | |
| fgr00230 | Purine metabolism | 4 | 2.31E−02 | |
| fgr01110 | Biosynthesis of secondary metabolites | 15 | 2.71E−02 | |
| fgr00250 | Alanine, aspartate, and glutamate metabolism | 3 | 4.50E−02 | |
| Purple | fgr00190 | Oxidative phosphorylation | 21 | 1.21E−09 |
| fgr04011 | MAPK a signaling pathway | 13 | 9.82E−06 | |
| fgr01100 | Metabolic pathways | 82 | 2.16E−05 | |
| fgr00020 | Tricarboxylic acid cycle | 7 | 8.67E−04 | |
| fgr01040 | Biosynthesis of unsaturated fatty acids | 4 | 2.84E−03 | |
| fgr01212 | Fatty acid metabolism | 7 | 5.63E−03 | |
| fgr04139 | Mitophagy | 5 | 2.09E−02 | |
| fgr04933 | AGE-RAGE signaling pathway in diabetic complications | 3 | 2.85E−02 | |
| fgr04113 | Meiosis | 7 | 4.42E−02 |
MAPK: mitogen activated protein kinase
Gene ontology (GO) enrichment analysis was also performed on the three gene modules (Table S1). In the biological process category, genes in the Greenyellow module were enriched in “regulation of transcription, DNA-templated” (GO:0006355), “DNA repair” (GO:0006281), “DNA recombination” (GO:0006310), and “ubiquitin-dependent protein catabolic process” (GO:0006511). In the Lightcyan and Purple modules, genes were mainly enriched in the “oxidation-reduction process” (GO:0055114) and “protein phosphorylation” (GO:0006468), respectively.
KEGG and GO functional enrichment analysis revealed that DNA damage response and protein degradation mechanisms were notably enriched in the gene group of the Greenyellow module. These mechanisms have been reported to having essential roles for protecting the cell against ROS-derived damage. DNA damage response is an important cellular process in the repair of oxidative damage in DNA (38, 39). Autophagy and the ubiquitin-proteasome system are the primary intracellular protein degradation mechanisms and play a role in the removal of proteins damaged by oxidative stress (40 – 42). Collectively, these results indicate that the DNA damage repair system, autophagy, and ubiquitin-proteasome system are upregulated to repair the damage caused by oxidative stress in F. graminearum. Furthermore, the identification of these mechanisms suggests that a transcriptomic analysis approach using stress-sensitive strains is effective for further investigating adaptive mechanisms against oxidative stress.
Identification of hub genes and construction of hub gene deletion mutants
We identified the top 30 genes with high intramodular connectivity as potential “hub” genes in each module to obtain insight into the regulatory response to oxidative stress. Among the hub genes, 16 had a KEGG orthology annotation in the Greenyellow module, and seven and eight genes were included in the Lightcyan and Purple modules, respectively (Table 2). We then constructed knock-out mutants for each gene to elucidate which genes are involved in oxidative stress response. We conducted fungal transformation using a homologous recombination method and confirmed gene deletions with diagnostic polymerase chain reaction (PCR) amplification followed by Southern blot hybridization (Fig. S3). Among the 31 hub genes, 26 were successfully deleted using a conventional fungal transformation method.
TABLE 2.
Hub genes with KEGG annotation in each module
| Module | Gene name | Locus ID | KO a identifier | Definition | Reference |
|---|---|---|---|---|---|
| Greenyellow | FgHGG1 | FGSG_01035 | K03515 | DNA repair protein REV1 | This study |
| FgHGG2 | FGSG_01132 | K08991 | Crossover junction endonuclease MUS81 | This study | |
| FgHGG3 | FGSG_01932 | K01760 | Cysteine-S-conjugate beta-lyase | This study | |
| FgHGG4 | FGSG_04274 | K11375 | Elongator complex protein 4 | This study | |
| FgHGG5 | FGSG_05904 | K15083 | DNA repair protein RAD16 | This study | |
| FgHGG6 | FGSG_05905 | K15082 | DNA repair protein RAD7 | This study | |
| FgHGG7 | FGSG_06098 | K01940 | Argininosuccinate synthase | This study | |
| FgHGG8 | FGSG_06510 | K08329 | Autophagy-related protein 17 | This study | |
| GzNF002 | FGSG_07076 | K12236 | Transcriptional repressor NF-X1 | (43) | |
| FgHGG10 | FGSG_07962 | K10875 | DNA repair and recombination protein RAD54 and RAD54-like protein | This study | |
| FgHGG11 | FGSG_08405 | K03657 | DNA helicase II/ATP-dependent DNA helicase PcrA | This study | |
| FgHGG12 | FGSG_08785 | K13431 | Signal recognition particle receptor subunit alpha | This study | |
| FgHGG13 | FGSG_10158 | K10873 | DNA repair and recombination protein RAD52 | This study | |
| FgHGG14 | FGSG_10352 | K05864 | Peptidyl-prolyl isomerase D | This study | |
| FgHGG15 | FGSG_11781 | K14284 | Nuclear RNA export factor | This study | |
| FgHGG16 | FGSG_12711 | K10884 | ATP-dependent DNA helicase 2 subunit 1 | This study | |
| Lightcyan | FgHGL1 | FGSG_01672 | K18278 | Pyrimidine precursor biosynthesis enzyme | This study |
| FgERG3A | FGSG_02502 | K00227 | Delta7-sterol 5-desaturase | (44) | |
| FgHGL3 | FGSG_02978 | K08139 | MFS transporter, SP family, sugar:H + symporter | This study | |
| FgHGL4 | FGSG_04458 | K05916 | Nitric oxide dioxygenase | This study | |
| FgHGL5 | FGSG_09373 | K18561 | FAD-dependent fumarate reductase | This study | |
| FgHGL6 | FGSG_09845 | K13076 | Sphingolipid 8-(E)-desaturase | This study | |
| FgHGL7 | FGSG_10739 | K00228 | Coproporphyrinogen III oxidase | This study | |
| Purple | FgHGP1 | FGSG_00866 | K15728 | Phosphatidate phosphatase LPIN | This study |
| FgHGP2 | FGSG_03816 | K01053 | Gluconolactonase | This study | |
| FgHGP3 | FGSG_05321 | K00667 | Fatty acid synthase subunit alpha, fungi type | This study | |
| FgHGP4 | FGSG_05322 | K00668 | Fatty acid synthase subunit beta, fungi type | This study | |
| Δ12-desaturase | FGSG_05784 | K10256 | Omega-6 fatty acid desaturase/acyl-lipid omega-6 desaturase (delta-12 desaturase) | (45) | |
| FgHGP6 | FGSG_06736 | K17775 | Mitochondrial distribution and morphology protein 34 | This study | |
| GzC2H050 | FGSG_07310 | K11215 | Transcription factor STE12 | (43) | |
| FgHGP8 | FGSG_09012 | K00236 | Succinate dehydrogenase (ubiquinone) cytochrome b560 subunit | This study |
KO: KEGG orthology
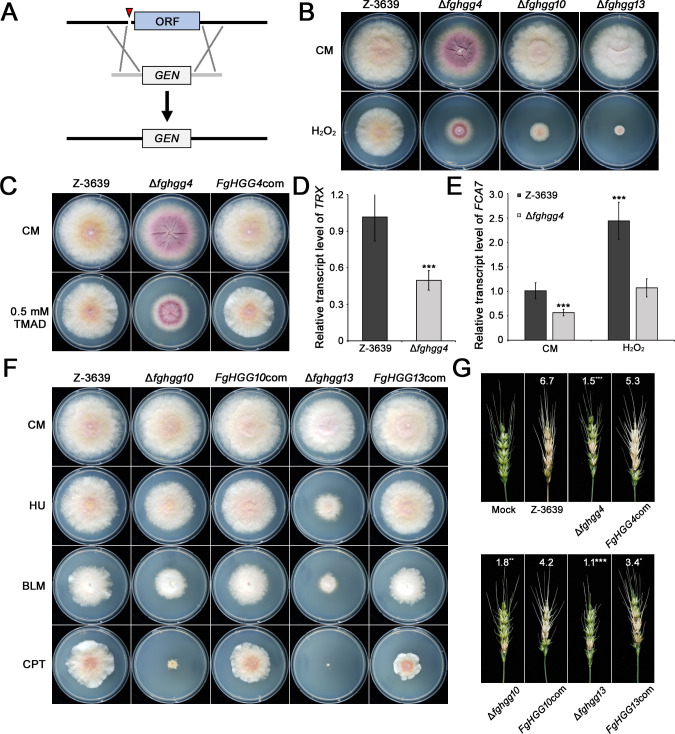
We failed to delete FgHGG4, FgHGG12, FgHGL7, FgHGP3, and FgHGP6 in three independent attempts. Among those genes, FgHGG12 and FgHGL7 are orthologs of the Saccharomyces cerevisiae genes SRP101 and HEM13, respectively, which are essential in yeast (46). Except for those essential genes, we introduced CRISPR/Cas9 system to fungal transformation for the deletion of the genes. CRISPR/Cas9, a genome-editing technology derived from a bacterial adaptive immune system, enables targeted precise editing (47, 48). Several studies have reported that the genomic nick induced by Cas9 increases homologous recombination repair efficiency (49 – 51), and CRISPR/Cas9 has been applied previously to Fusarium species (52 – 54). To increase gene deletion efficiency for FgHGG4, FgHGP3, and FgHGP6, fungal transformation was conducted with preassembled Cas9 ribonucleoproteins, and the nick site was near the start codon (Fig. 3A). Finally, we successfully obtained Δfghgg4 deletion mutants. A total of 27 hub gene mutants were cultured under oxidative stress conditions, and three mutants, Δfghgg4, Δfghgg10, and Δfghgg13, showed increased sensitivity to oxidative stress compared with the wild-type strain (Fig. 3B).
Fig 3.
Redox homeostasis and the DNA repair system are important for oxidative stress resistance. (A) An overview of the strategy for the fungal transformation using the CRISPR/Cas9 system. The red triangle indicates the cleavage site induced by the Cas9 protein. GEN, geneticin resistance gene cassette. (B) Vegetative growth of hub gene deletion mutants under oxidative stress conditions. Each strain was cultured on CM and CM supplemented with 10 mM H2O2. Photographs were taken 5 days after inoculation. (C) Vegetative growth of the fungal strains on CM with or without diamide (0.5 mM). Photographs were taken 5 days after inoculation. (D) Relative transcript levels of thioredoxin (TRX) in the wild-type and Δfghgg4 deletion strains. The transcript levels of the gene were analyzed by quantitative real-time PCR amplification (qRT-PCR), and CYP1 was used as a housekeeping gene. Asterisks represent significant differences from the wild type (***P < 0.001; t-test). (E) Transcript levels of FCA7 in the wild-type and Δfghgg4 deletion strains. Total RNA was extracted from each strain grown for 30 min in CM and CM supplemented with 5 mM H2O2. The transcripts of FCA7 were analyzed by qRT-PCR, and the CYP1 was used as a housekeeping gene (***P < 0.001; t-test). (F) Sensitivity to DNA-damaging agents. Pictures were taken 5 days after inoculation on CM and CM treated with various DNA-damaging agents (10 mM hydroxyurea, HU; 10 µg/mL bleomycin, BLM; 0.2 mM camptothecin, CPT). (G) Virulence of the wild-type and hub gene mutant strains. Pictures were taken 2 weeks after inoculation. The average of infected spikes was used as a disease index and denoted with white letter. Asterisks represent significant differences from the wild type (*P < 0.05; **P < 0.01; ***P < 0.001; t-test).
The role of FgHGG4, FgHGG10, and FgHGG13 in oxidative stress response
FgHGG4 is predicted to be an ortholog of ELP4 in S. cerevisiae, which is a subunit of the elongator complex protein. The elongator complex is composed of six subunits, named Elongator complex proteins 1 to 6 ( ELP1-6), and a holo-elongator is involved in transcriptional elongation by associating with an RNA polymerase II holoenzyme (55). In Schizosaccharomyces pombe, the elongator complex is important for oxidative stress resistance by altering the translation level of oxidative stress-related transcription factor Atf1 and Pcr1 via tRNA modification (56). The ELP complex is also known to be involved in the elongation of genes combating thiol oxidation in S. cerevisiae (57). To determine the role of FgHgg4 proteins in redox regulatory systems of F. graminearum, Δfghgg4 deletion mutants were cultured on CM supplemented with a thiol-specific oxidant, diamide. The deletion mutant exhibited growth defects compared with the wild-type and complementation strains (Fig. 3C). Transcription of the thioredoxin gene (TRX) was decreased by approximately 50% in Δfghgg4 deletion mutants compared with the wild type (Fig. 3D). A previous study confirmed that Elp3, one of the core subunits of the elongator complex, plays an important role in oxidative stress response in F. graminearum (58). In elp3 deletion mutants, several peroxidases genes, including FCA7, were downregulated or unchanged compared with those of the wild-type strain under oxidative stress conditions. We determined the transcript levels of FCA7, the major peroxidase in F. graminearum, and quantitative real-time (qRT)-PCR results indicated that FCA7 was downregulated in Δfghgg4 deletion mutants compared with the wild-type strain (Fig. 3E). The transcript level of FCA7 in Δfghgg4 deletion mutants was increased under oxidative stress conditions, but was still 2.5-fold lower than that of the wild-type strain. These results indicate that the elongator complex is involved in redox homeostasis and peroxidase regulation, and deletion of its subunit caused increased sensitivity to oxidative stress.
FgHGG10 and FgHGG13 are the respective ortholog genes of S. cerevisiae RAD54 and RAD52, which are involved in the repair of DNA double-strand breaks (59, 60). To confirm the role of FgHGG10 and FgHGG13 in DNA repair in F. graminearum, deletion mutants were cultured on media supplemented with various DNA-damaging agents. Δfghgg10 and Δfghgg13 exhibited altered sensitivity to all DNA damaged agents compared with the wild-type strain (Fig. 3F). Radial growth of Δfghgg10 and Δfghgg13 was markedly reduced on CM supplemented with camptothecin compared with the wild-type strain, and Δfghgg13 showed more severe growth defects compared with Δfghgg10. The complemented strains of Δfghgg10 and Δfghgg13 restored growth in corresponding deletion mutants under DNA-damaging conditions. These results indicate that FgHGG10 and FgHGG13 play important roles in DNA repair when exposed to oxidative stress.
Furthermore, we investigated the role of FgHGG4, FgHGG10, and FgHGG13 in asexual reproduction in F. graminearum. Compared with the wild type, the Δfghgg10 and Δfghgg13 mutants showed no altered phenotypes in conidiation. However, the deletion of FgHGG4 resulted in increased conidial formation (Fig. S1A). Also, the conidia of Δfghgg4 were longer than that of the wild type (Fig. S1B and C). To determine the sexual reproduction, we cultured the wild type and those mutants on carrot agar media. The Δfghgg4 mutant produced perithecia normally. In contrast, no perithecia were formed by Δfghgg10 and Δfghgg13 mutants (Fig. S1D). These results support that the mechanisms that function importantly in responding to stress are also required for normal reproduction in fungi.
To evaluate the virulence of the three hub gene deletion mutants, conidial suspension of each strain was injected into the middle of the flowering wheat head. Whereas the wild-type strain caused normal blight symptoms on the wheat head, Δfghgg4, Δfghgg10, and Δfghgg13 showed markedly reduced virulence (Fig. 3G). To colonize a plant’s cells, fungi must endure host-derived oxidative bursts. Deletion of each gene caused a vulnerability to oxidative stress, which appeared to lead to a failure to overcome plant defenses. These results imply that the mechanisms associated with the oxidative stress response are also directly related to pathogenicity.
Moreover, we constructed a series of double-deletion mutants of FgHGG4, FgHGG10, and FgHGG13 to identify whether they function simultaneously. The mutant construction was confirmed by PCR (Fig. S2A), and the sensitivity to oxidative stress was examined. Compared with the Δfghgg4 mutants, Δfghgg4Δfghgg10 and Δfghgg4Δfghgg13 strains showed increased sensitivity to oxidative stress (Fig. S2B and C). These results suggest that FgHgg4-mediated mechanisms function in oxidative stress resistance independently of the DNA repair system. Additionally, the growth inhibition rate of the Δfghgg10Δfghgg13 is not significantly different from that of the Δfghgg13 mutant, indicating that FgHgg13 has a role in oxidative stress response overlapping FgHgg10 functions.
Sensitivity of essential gene mutants to oxidative stress
We failed to delete FgHGG12, FgHGL7, FgHGP3, and FgHGP6 using homologous recombination among 31 hub genes. These genes are considered essential to fungal viability, and essential genes can be novel targets for disease control. We therefore introduced conditional gene expression to those genes and confirmed the phenotype. In F. graminearum, zearalenone-inducible promoter (P ZEAR) was used for conditional gene expression and β-estradiol can substitute for zearalenone (61, 62). The promoter of each gene was replaced with P ZEAR and the replacement was confirmed by Southern blot hybridization (Fig. S3). The promoters of FgHGL7, FgHGP3, and FgHGP6 were successfully replaced with P ZEAR, and we observed vegetative growth of the generated mutants, both with and without supplementation of β-estradiol. Each mutant, P ZEAR -FgHGL7 in particular, exhibited severe growth defects on CM, while vegetative growth of the mutants recovered when they were grown on CM supplemented with β-estradiol (Fig. 4). To confirm the sensitivity of each strain to oxidative stress, all mutants were cultured in CM and treated with and without 10 mM H2O2. Repression or induction of FgHGP3 and FgHGP6 did not affect sensitivity to oxidative stress. In contrast, repression of FgHGL7 resulted in hypersensitivity to oxidative stress, and partial induction of FgHGL7 gene expression through β-estradiol treatment did not restore growth defects of P ZEAR -FgHGL7. These results suggest that FgHGL7 is essential for fungal viability, and that full expression of FgHGL7 is required for growth under oxidative stress.
Fig 4.

Oxidative stress sensitivity of promoter replacement mutants. Fungal strains were cultured on CM and CM supplemented with 10 mM H2O2, and mycelial growth was observed in the absence and presence of β-estradiol. Pictures were taken 5 days after inoculation.
Heme biosynthesis pathway is important for resistance to oxidative stress
To confirm whether repression of FgHGL7 genes caused hypersensitivity to oxidative stress, we inoculated PZEAR-FgHGL7 mutants on CM supplemented with 2 mM H2O2 and observed mycelial growth at various concentrations of β-estradiol. As the concentration of β-estradiol increased, the inhibition rate of PZEAR-FgHGL7 decreased under oxidative stress (Fig. 5A and B), suggesting that expression of FgHGL7 is positively correlated with resistance to oxidative stress in F. graminearum.
Fig 5.
Heme biosynthesis is required for oxidative stress resistance. (A) Mycelial growth of the wild-type strain and PZEAR-FgHGL7 under oxidative stress conditions. Each strain was inoculated on CM and CM supplemented with 2 mM H2O2, and mycelial growth was observed at different concentrations of β-estradiol. Photographs were taken 5 days after inoculation. (B) Statistical analysis of mycelial growth inhibition under oxidative stress. (C) High-performance liquid chromatography profiling of hemin production by the wild-type strain and conditional FgHGL7 expression mutants with and without addition of β-estradiol. The gray line represents hemin. (D) Peroxidase activity of the fungal strains. Asterisks represent significant differences from the wild type (*P < 0.05; **P < 0.01; ***P < 0.001; t-test).
FgHGL7 is predicted to encode coproporphyrinogen III oxidase, an enzyme that has a role in the biosynthetic pathway of heme, converting coproporphyrinogen III into protoporphyrinogen IX through oxidative decarboxylation (63, 64). It was reported that coproporphyrinogen III oxidase is involved in the regulation of intracellular heme levels in Aspergillus niger (65). To identify the effect of suppressing FgHGL7 on heme biosynthesis in F. graminearum, the production of hemin/heme was determined in both wild-type and PZEAR-FgHGL7 mutant strains (Fig. 5C). In the wild-type strain, hemin production peaked at a retention time of 28.957 min, whereas significant reduction of peak areas was observed in PZEAR-FgHGL7 mutants. Rather, a high-performance liquid chromatography (HPLC) profile of PZEAR-FgHGL7 mutants showed two prominent peaks at retention times of 25.935 and 28.120 min, indicating the presence of heme intermediates. When β-estradiol was treated, the profiles of PZEAR-FgHGL7 mutants showed an increase in the area of the peak corresponding to hemin, indicating partial restoration of heme biosynthesis.
Several studies have reported that heme-containing peroxidases or catalases have an important role in the oxidative stress response (66 – 68). Especially, it was confirmed that the heme-containing peroxidase (Fca6, Fca7, and Fpx1) has an important role in oxidative stress response, affecting total peroxidase activity in F. graminearum (20). Based on previous studies, we aimed to investigate whether the defect in heme biosynthesis reduces the activity of peroxidases. We determined the activities of peroxidase in the wild-type and PZEAR-FgHGL7 mutant strains, and it revealed that the peroxidase activity of the PZEAR-FgHGL7 mutants was remarkably decreased compared with that of the wild-type strain (Fig. 5D). Treatment of β-estradiol slightly increased peroxidase activity in PZEAR-FgHGL7, but the levels were still lower than those of the wild type, suggesting that the partial restoration of gene expression of FgHGL7 through β-estradiol treatment was insufficient to fully restore its function. These findings indicate that FgHGL7 plays a role in heme biosynthesis, and that disruption of heme biosynthesis due to FgHGL7 suppression affects the heme-containing peroxidase activity.
Oxidative stress resistance mechanism in F. graminearum
To successfully infect a host plant, fungal pathogens must tolerate and combat the oxidative stress induced by the host’s immune system (69). Various mechanisms are activated to protect the cell from host-derived ROS. In this study, we attempted to specify those mechanisms by analyzing the gene expression profiles of oxidative stress-sensitive strains in F. graminearum. WGCNA methods identified the gene groups most closely correlated with oxidative stress, and functional enrichment analysis of the key modules revealed that several mechanisms are upregulated against oxidative stress: DNA damage response, autophagy, and ubiquitin-proteasome system.
Oxidative damage in DNA is repaired by the DNA damage response system, which prevents replication of defective DNA (38). DNA repair system and oxidative stress response appear to be closely associated. According to phenotypes described in FgTFPD (43), 4 of the 16 transcription factor mutants associated with DNA damage response in F. graminearum are sensitive to oxidative stress (70). Additionally, phenotypic analysis of the hub gene mutants revealed that the FgHGG10- and FgHGG13-encoding yeast orthologs Rad54 and Rad52, respectively, play a role in oxidative stress response, and these findings indicate that DNA repair system is essential for tolerance under oxidative stress.
Autophagy and ubiquitin-mediated proteasome systems contribute to the elimination of damaged cellular components (71). Autophagy has a role in the removal of damaged cytosolic components, and ubiquitin-mediated proteasomes selectively decompose damaged proteins (42). The ubiquitin-proteasome system and the 26S proteasome, in particular, are easily damaged by oxidative stress, but H2O2 triggers the disassembly of 26S proteasomes into 19S particles and 20S catalytic cores, and a 20S core can actively recognize and degrade oxidized protein (72). Autophagy and the UPS have been investigated in plant pathogenic fungi, and several studies have revealed that autophagy and UPS-related components, including peroxin and ubiquitin-like protein, are required for oxidative stress resistance (73 – 77). Collectively, as protein oxidations caused by oxidative stress lead to cellular dysfunction (78, 79), those mechanisms are essential to tolerating oxidative stress by controlling protein quality.
In this study, we identified core genes of each module and used them as tools to reveal the mechanisms related to oxidative stress response. Phenotypic analysis under the oxidative stress for 27 hub gene mutants identified FgHGG4, a subunit of the elongator complex, along with FgHGG10 and FgHGG13. The elongator complex has a role in transcriptional elongation and is involved in wobble modification of tRNA (80, 81). In yeast, the chemical modification of wobble base uridines of tRNA reportedly affects the translational regulation of genes involved in oxidative stress responses (56). It has also been reported that Elp3, the other subunit of the elongator complex, has a role in the oxidative stress response in F. graminearum (58). However, the role of the elongator complex subunits in oxidative stress response varies among fungal species (82 – 84), and further studies are required on the specific regulatory mechanisms of the elongator complex in the oxidative stress response in F. graminearum.
Among the hub genes, four were identified as essential for fungal viability. We determined that FgHGL7 is necessary for resistance to oxidative stress. Functional characterization of FgHGL7 revealed that heme biosynthesis is required for an effective response to oxidative stress. Heme is an iron-containing tetrapyrrole ring with an essential role in diverse biological functions, serving as the prosthetic group of hemeproteins (85 – 87). In F. graminearum, 31 putative peroxidases were identified, 23 of which contain heme (20). Multiple studies have reported the role of peroxidases in ROS detoxification and oxidative stress responses (88 – 90). Cytochromes P450s (CYPs) are also representative heme-containing proteins, and act as monooxygenases that catalyze the oxidation of cellular substrates (91, 92). However, none of the CYP mutants reportedly display an altered phenotype under oxidative stress in F. graminearum (93). These indicate that heme is required for resistance to oxidative stress as a component of heme-containing peroxidase.
Collectively, a combined transcriptomic and physiological analysis identified the several mechanisms required for oxidative stress adaptation in F. graminearum including “DNA repair,” “protein degradation mechanisms,” “transcriptional elongation,” and “heme biosynthesis.” Furthermore, this study confirms the significance of oxidative stress resistance for pathogenicity and provides a comprehensive understanding of the mechanisms to cope with oxidative stress in plant pathogenic fungi.
MATERIALS AND METHODS
Fungal strains and growth conditions
The F. graminearum Z-3639 wild-type strain and the mutants used in this study were stored as mycelia in 20% glycerol at −80°C. All media were prepared as described in The Fusarium Laboratory Manual (94). Fungal strains were grown at 25°C.
Nucleic acid manipulation and Southern blotting
Fungal genomic DNA was extracted with a cetyltrimethylammonium bromide protocol according to The Fusarium Laboratory Manual (94). Restriction endonuclease digestion, agarose gel electrophoresis, and gel blotting were performed following standard protocols (95). A North2South Biotin Random Prime Labeling Kit and a Chemiluminescent Hybridization and Detection Kit (Thermo Scientific, USA) were used for Southern blot hybridization. Total RNA was extracted using an Easy-Spin Total RNA Extraction Kit (Intron Biotech, Korea).
RNA-seq and WGCNA analysis
Conidia suspensions were cultured in liquid CM at 25°C for 24 h with shaking. Fresh mycelia were shifted to liquid CM with or without 5 mM H2O2 and cultured for 30 min. Total RNA was extracted from harvested mycelia, and RNA-seq was performed using an Illumina HiSeq 2000 system.
Adapter sequences of reads derived from RNA-seq were trimmed in the Cutadapt program (96). Unreliable low-quality reads were removed from the raw reads using the FASTX-Toolkit if the average quality within 50% of the read sequences falls below a Phred score threshold of 28. Filtered clean reads were then mapped to reference genome sequences downloaded from the National Center for Biotechnology Information using the Burrows-Wheeler Aligner (BWA) program (97). Mapped reads per gene were counted by featureCounts tool v1.5.0 (98) and normalized using the reads per kilobase per million mapped reads (RPKM) method (99).
Highly co-expressed gene modules were inferred from total genes according to WGCNA tutorials (100, 101). A matrix of pairwise Pearson correlations between all genes was calculated and transformed into a pairwise adjacency matrix by a soft power threshold (β) derived from the pickSoftThreshold algorithm. The β of 12 powers satisfied the minimum value required to generate a scale-free topology network (a R 2 value ≥0.8). The pairwise adjacency matrix was re-transformed into a topological overlap measure to capture a co-expression relationship between genes in the network. Modules as groups of co-expressed genes were defined using the cutreeDynamic algorithm (deepSplit = 4) with a minimum module size of 200. MEDissThres was set to 0.7 to merge similar modules. The closely correlated modules with traits were selected by calculating the relevance between traits and a module eigengene for further analysis. Potential hub genes as highly interconnected genes within the module were estimated with a connectivity weight threshold of 0.2 in the co-expression network.
KEGG and GO enrichment analyses were conducted to explore the biological significance of the genes in each module. KEGG pathway data were downloaded from the KEGG database (http://www.genome.jp/kegg) and GO terms were obtained from the UniProt database (https://www.uniprot.org). The statistical significance of enrichment was determined using a hypergeometric distribution (P < 0.05).
Genetic manipulations and fungal transformations
For gene deletion, 5´- and 3´- flanking regions of the open reading frame (ORF) were amplified from the genomic DNA of an F. graminearum wild-type strain. A geneticin resistance gene cassette (GEN) was amplified from a plasmid pII99. Three amplicons were fused, and final PCR products were created by double-joint (DJ) PCR methods (102). To complement the Δfghgg10 and Δfghgg13 deletion mutants (FgHGG10com, FgHGG13com), the construct that included a native promoter region of the gene, the ORF, and the 3´- flanking region of the ORF was amplified. A PCR product including the green fluorescence protein and a hygromycin resistance cassette (GFP-HYG) was amplified from the plasmid pIGPAPA. The final PCR products were transformed into the deletion strains.
For FgHGG4 and other putative essential gene deletions, a CRISPR-Cas9–mediated transformation was conducted to increase deletion efficiency (52). Briefly, 5´- flanking of the ORF was amplified with primers containing the optimal cleavage site. 3´- Flankings of the ORF and GEN cassette were amplified, and the final PCR products were constructed as described earlier. Before fungal transformation, synthesized sgRNA (Macrogen, Korea) and purchased Cas9 protein (Thermo Scientific, USA) were preassembled and added into protoplasts with the resulting amplicons.
For complementation of the Δfghgg4, the ORF of FgHGG4 and its putative promoter region (approximately 1 kb upstream of the transcription start site) were amplified. The PCR fragments were co-transformed with a pDL2/XhoI (103) linearized plasmid into yeast strain PJ69-4A. The resulting plasmid was extracted using Zymoprep Yeast Plasmid Miniprep II (Zymo Research, USA) and transformed into an Escherichia coli DH10B strain. After verification of the sequence, a recombinant plasmid was extracted and linearized for fungal transformation.
To generate a conditional gene expression mutant (PZEAR mutants), the promoter regions of the target genes were replaced with PZEAR . The hygromycin resistance gene (HYG)-PZEAR cassette was amplified from the PZEAR-FgHSP90 strains (104). The 5´- and 3´- flanking regions of the target genes were amplified and fused with HYG-PZEAR cassette using DJ PCR methods (102). After amplifying with the nested primers, the final amplicons were used for transformation.
Each final PCR product or linearized plasmid was transformed into protoplasts using polyethylene glycol-mediated transformation, and all mutations were confirmed with Southern blot hybridization analysis (Fig. S3). Primers used in this study were synthesized by an oligonucleotide synthesis facility (Bioneer, Korea) and listed in Table S2.
Quantitative real-time PCR
First-strand cDNA was synthesized from total RNA using SuperScript III reverse transcriptase (Invitrogen, USA) following the manufacturer’s instructions. Quantitative real-time PCR amplification was conducted with iTaq Universal SYBR Green Supermix (Bio-Rad, USA) and a CFX real-time PCR system (Bio-Rad). PCR amplification was performed three times with three biological replicates, and the relative transcript levels of the target gene were calculated as described previously (105). The primers used for qRT-PCR are listed in Table S2.
Virulence tests
Virulence tests were conducted using the wheat cultivar Eunpamil as previously described (106). Each fungal strain was cultured in 25 mL of carboxymethylcellulose (CMC) at 25°C for 5 days. Harvested conidia were resuspended in 0.01% Tween 20 solution. A 10 µL suspension of conidia (106 conidia/mL) was injected into the middle of the spikelet. The inoculated wheat heads were enveloped with a plastic bag to create humid conditions for 3 days. After an additional 11 days, the symptoms were observed and the number of infected spikelets was measured. Virulence tests were conducted with 10 replicate inoculations for each fungal strain.
Asexual and sexual reproduction assays
For the conidiation assay, 10 mycelial plugs of each strain were inoculated in 30 mL CMC media for 5 days. Conidia were counted with a hemocytometer. The experiment was repeated three times.
For the examination of sexual reproduction, each strain was grown on carrot agar media for 5 days, and aerial hyphae were removed with 400 µL of a 2.5% Tween 60 solution to induce sexual reproduction. The resulting media were incubated under near-UV light (Sankyo Denki, Tokyo, Japan), and the perithecia formation was observed 7 days after induction.
Heme quantification using HPLC
Heme quantification was conducted as described previously (65, 107, 108). Briefly, frozen mycelia were ground into fine powder with liquid nitrogen. Samples were homogenized in phosphate-buffered saline, and protein contents were determined by the Bradford method (109). A portion of extract was mixed with an equal volume of acetone and concentrated HCl (97.5/2.5; vol/vol). After centrifuging, the supernatant was analyzed by reversed-phase HPLC with an ultraviolet detector.
The analysis was carried out using a Shimadzu UFLC HPLC system (Shimadzu, Japan). To prepare solvent A, 0.1 M ammonium phosphate solution (pH adjusted to 3.5 with phosphoric acid) was filtered with a 0.45-µm membrane filter (Millipore, USA). The filtered solution was mixed with methanol (56:44; vol/vol), and the pH of the solution was adjusted to 3.4 with phosphoric acid. Pure methanol was used for solvent B. Samples were eluted on a C18 HPLC column (Shimadzu, Japan) at 35°C at a flow rate of 1.5 mL/min. In gradient elution, the composition of solvent B was changed from 30% to 100% for 15 min. The spectra of the samples were monitored at 405 nm.
Estimation of peroxidase enzyme activity
Total peroxidase activity assay was performed using a QuantiChrom peroxidase assay (BioAssay Systems, USA) as previously described (20). Crude proteins were extracted, and the concentration was estimated with a Bradford assay. Further steps followed the manufacturer’s instructions.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (2021R1C1C1004200). All authors certify that they have no affiliations or involvement with any organization or entity with a financial or non-financial interest in the subject matter or materials discussed in this manuscript.
Contributor Information
Young-Su Seo, Email: yseo2011@pusan.ac.kr.
Hokyoung Son, Email: hogongi7@snu.ac.kr.
Giuseppe Ianiri, Universita degli Studi del Molise, Campobasso, Italy .
DATA AVAILABILITY
The RNA-seq data have been deposited in the NCBI database under the BioProject accession number PRJNA937405.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.01485-23.
Conidiation and sexual reproduction of Δfghgg4, Δfghgg10, and Δfghgg13. Sensitivity of double deletion mutants to oxidative stress. Confirmation of the genetic manipulation by southern blot analyses.
GO enrichment analysis for the module genes.
Primers used in this study.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Aguirre J, Ríos-Momberg M, Hewitt D, Hansberg W. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol 13:111–118. doi: 10.1016/j.tim.2005.01.007 [DOI] [PubMed] [Google Scholar]
- 2. Mittler R. 2017. ROS are good. Trends Plant Sci 22:11–19. doi: 10.1016/j.tplants.2016.08.002 [DOI] [PubMed] [Google Scholar]
- 3. Zhang J, Wang X, Vikash V, Ye Q, Wu D, Liu Y, Dong W. 2016. ROS and ROS-mediated cellular signaling. Oxid Med Cell Longev 2016:4350965. doi: 10.1155/2016/4350965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. 2011. ROS signaling: the new wave? Trends Plant Sci 16:300–309. doi: 10.1016/j.tplants.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 5. Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. 2012. Oxidative stress and antioxidant defense. World Allergy Organ J 5:9–19. doi: 10.1097/WOX.0b013e3182439613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu D, Cederbaum AI. 2003. Alcohol, oxidative stress, and free radical damage. Alcohol Res Health 27:277–284. [PMC free article] [PubMed] [Google Scholar]
- 7. Betteridge DJ. 2000. What is oxidative stress? Metabolism 49:3–8. doi: 10.1016/s0026-0495(00)80077-3 [DOI] [PubMed] [Google Scholar]
- 8. Foyer CH, Noctor G. 2013. Redox signaling in plants. Antioxid Redox Signal 18:2087–2090. doi: 10.1089/ars.2013.5278 [DOI] [PubMed] [Google Scholar]
- 9. Rao MV, Lee H, Creelman RA, Mullet JE, Davis KR. 2000. Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12:1633–1646. doi: 10.1105/tpc.12.9.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jones JDG, Dangl JL. 2006. The plant immune system. Nature 444:323–329. doi: 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 11. Yuan M, Ngou BPM, Ding P, Xin X-F. 2021. PTI-ETI crosstalk: an integrative view of plant immunity. Curr Opin Plant Biol 62:102030. doi: 10.1016/j.pbi.2021.102030 [DOI] [PubMed] [Google Scholar]
- 12. Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, Bolwell GP. 2012. The apoplastic oxidative burst peroxidase in arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24:275–287. doi: 10.1105/tpc.111.093039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bigeard J, Colcombet J, Hirt H. 2015. Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8:521–539. doi: 10.1016/j.molp.2014.12.022 [DOI] [PubMed] [Google Scholar]
- 14. Kadota Y, Shirasu K, Zipfel C. 2015. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol 56:1472–1480. doi: 10.1093/pcp/pcv063 [DOI] [PubMed] [Google Scholar]
- 15. Singh Y, Nair AM, Verma PK. 2021. Surviving the odds: from perception to survival of fungal phytopathogens under host-generated oxidative burst. Plant Commun 2:100142. doi: 10.1016/j.xplc.2021.100142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Minibayeva F, Beckett RP, Kranner I. 2015. Roles of apoplastic peroxidases in plant response to wounding. Phytochemistry 112:122–129. doi: 10.1016/j.phytochem.2014.06.008 [DOI] [PubMed] [Google Scholar]
- 17. Pisoschi AM, Pop A. 2015. The role of antioxidants in the chemistry of oxidative stress: a review. Eur J Med Chem 97:55–74. doi: 10.1016/j.ejmech.2015.04.040 [DOI] [PubMed] [Google Scholar]
- 18. Irato P, Santovito G. 2021. Enzymatic and non-enzymatic molecules with antioxidant function. Antioxidants (Basel) 10:579. doi: 10.3390/antiox10040579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mir AA, Park S-Y, Abu Sadat M, Kim S, Choi J, Jeon J, Lee Y-H. 2015. Systematic characterization of the peroxidase gene family provides new insights into fungal pathogenicity in magnaporthe oryzae. Sci Rep 5:11831. doi: 10.1038/srep11831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee Y, Son H, Shin JY, Choi GJ, Lee Y-W. 2018. Genome-wide functional characterization of putative peroxidases in the head blight fungus Fusarium graminearum. Mol Plant Pathol 19:715–730. doi: 10.1111/mpp.12557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Veluchamy S, Williams B, Kim K, Dickman MB. 2012. The CuZn superoxide dismutase from Sclerotinia sclerotiorum is involved with oxidative stress tolerance, virulence, and oxalate production. Physiol Mol Plant Pathol 78:14–23. doi: 10.1016/j.pmpp.2011.12.005 [DOI] [Google Scholar]
- 22. López-Cruz J, Óscar C-S, Emma F-C, Pilar G-A, Carmen G-B. 2017. Absence of Cu-Zn superoxide dismutase BCSOD1 reduces Botrytis cinerea virulence in arabidopsis and tomato plants, revealing interplay among reactive oxygen species, callose and signalling pathways. Mol Plant Pathol 18:16–31. doi: 10.1111/mpp.12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tian L, Li J, Huang C, Zhang D, Xu Y, Yang X, Song J, Wang D, Qiu N, Short DPG, Inderbitzin P, Subbarao KV, Chen J, Dai X. 2021. Cu/Zn superoxide dismutase (VdSOD1) mediates reactive oxygen species detoxification and modulates virulence in Verticillium dahliae. Mol Plant Pathol 22:1092–1108. doi: 10.1111/mpp.13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang C, Zhang S, Zhang Q, Tao Y, Wang C, Xu J-R. 2015. FgSKN7 and FgATF1 have overlapping functions in ascosporogenesis, pathogenesis and stress responses in Fusarium graminearum. Environ Microbiol 17:1245–1260. doi: 10.1111/1462-2920.12561 [DOI] [PubMed] [Google Scholar]
- 25. Moye-Rowley WS. 2003. Regulation of the transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryot Cell 2:381–389. doi: 10.1128/EC.2.3.381-389.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fassler JS, West AH. 2011. Fungal Skn7 stress responses and their relationship to virulence. Eukaryot Cell 10:156–167. doi: 10.1128/EC.00245-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pérez-Martín J. 2009. DNA-damage response in the basidiomycete fungus Ustilago maydis relies in a sole Chk1-like kinase. DNA Repair (Amst) 8:720–731. doi: 10.1016/j.dnarep.2009.01.023 [DOI] [PubMed] [Google Scholar]
- 28. Wang S, Wu X-M, Liu C-H, Shang J-Y, Gao F, Guo H-S. 2020. Verticillium dahliae chromatin remodeling facilitates the DNA damage repair in response to plant ROS stress. PLoS Pathog 16:e1008481. doi: 10.1371/journal.ppat.1008481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jian Y, Chen X, Sun K, Liu Z, Cheng D, Cao J, Liu J, Cheng X, Wu L, Zhang F, Luo Y, Hahn M, Ma Z, Yin Y. 2023. SUMOylation regulates pre-mRNA splicing to overcome DNA damage in fungi. New Phytol 237:2298–2315. doi: 10.1111/nph.18692 [DOI] [PubMed] [Google Scholar]
- 30. Min K, Son H, Lim JY, Choi GJ, Kim J-C, Harris SD, Lee Y-W. 2014. Transcription factor RFX1 is crucial for maintenance of genome integrity in Fusarium graminearum. Eukaryot Cell 13:427–436. doi: 10.1128/EC.00293-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chondrogianni N, Petropoulos I, Grimm S, Georgila K, Catalgol B, Friguet B, Grune T, Gonos ES. 2014. Protein damage, repair and proteolysis. Mol Aspects Med 35:1–71. doi: 10.1016/j.mam.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 32. Asif N, Lin F, Li L, Zhu X, Nawaz S. 2022. Regulation of autophagy machinery in Magnaporthe oryzae. Int J Mol Sci 23:8366. doi: 10.3390/ijms23158366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duyvesteijn RGE, van Wijk R, Boer Y, Rep M, Cornelissen BJC, Haring MA. 2005. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato. Mol Microbiol 57:1051–1063. doi: 10.1111/j.1365-2958.2005.04751.x [DOI] [PubMed] [Google Scholar]
- 34. Goswami RS, Kistler HC. 2004. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol 5:515–525. doi: 10.1111/j.1364-3703.2004.00252.x [DOI] [PubMed] [Google Scholar]
- 35. Sutton JC. 1982. Epidemiology of wheat head blight and maize ear rot caused by Fusarium graminearum. Can J Plant Pathol 4:195–209. doi: 10.1080/07060668209501326 [DOI] [Google Scholar]
- 36. Desjardins AE, Proctor RH. 2007. Molecular biology of Fusarium mycotoxins. Int J Food Microbiol 119:47–50. doi: 10.1016/j.ijfoodmicro.2007.07.024 [DOI] [PubMed] [Google Scholar]
- 37. Saelens W, Cannoodt R, Saeys Y. 2018. A comprehensive evaluation of module detection methods for gene expression data. Nat Commun 9:1090. doi: 10.1038/s41467-018-03424-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barzilai A, Yamamoto K-I. 2004. DNA damage responses to oxidative stress. DNA Repair 3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 39. Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. 2019. ROS and the DNA damage response in cancer. Redox Biol 25:101084. doi: 10.1016/j.redox.2018.101084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shang F, Taylor A. 2011. Ubiquitin–proteasome pathway and cellular responses to oxidative stress. Free Radic Biol Med 51:5–16. doi: 10.1016/j.freeradbiomed.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kriegenburg F, Poulsen EG, Koch A, Krüger E, Hartmann-Petersen R. 2011. Redox control of the ubiquitin-proteasome system: from molecular mechanisms to functional significance. Antioxid Redox Signal 15:2265–2299. doi: 10.1089/ars.2010.3590 [DOI] [PubMed] [Google Scholar]
- 42. Qin Z-H. 2019. Autophagy and ubiquitin-proteasome system. Singapore: . doi: 10.1007/978-981-15-0602-4 [DOI] [Google Scholar]
- 43. Son H, Seo Y-S, Min K, Park AR, Lee J, Jin J-M, Lin Y, Cao P, Hong S-Y, Kim E-K, Lee S-H, Cho A, Lee S, Kim M-G, Kim Y, Kim J-E, Kim J-C, Choi GJ, Yun S-H, Lim JY, Kim M, Lee Y-H, Choi Y-D, Lee Y-W. 2011. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog 7:e1002310. doi: 10.1371/journal.ppat.1002310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yun Y, Yin D, Dawood DH, Liu X, Chen Y, Ma Z. 2014. Functional characterization of FgERG3 and FgERG5 associated with ergosterol biosynthesis, vegetative differentiation and virulence of Fusarium graminearum. Fungal Genet Biol 68:60–70. doi: 10.1016/j.fgb.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 45. Damude HG, Zhang H, Farrall L, Ripp KG, Tomb J-F, Hollerbach D, Yadav NS. 2006. Identification of Bifunctional Delta12/Omega3 fatty acid Desaturases for improving the ratio of Omega3 to Omega6 fatty acids in Microbes and plants. Proc Natl Acad Sci U S A 103:9446–9451. doi: 10.1073/pnas.0511079103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian K-D, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang C, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- 47. Hryhorowicz M, Lipiński D, Zeyland J, Słomski R. 2017. CRISPR/Cas9 immune system as a tool for genome engineering. Arch Immunol Ther Exp 65:233–240. doi: 10.1007/s00005-016-0427-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR Cas9 for genome engineering. Cell 157:1262–1278. doi: 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33:543–548. doi: 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]
- 50. Rong Z, Zhu S, Xu Y, Fu X. 2014. Homologous recombination in human embryonic stem cells using CRISPR/Cas9 nickase and a long DNA donor template. Protein Cell 5:258–260. doi: 10.1007/s13238-014-0032-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakajima K, Zhou Y, Tomita A, Hirade Y, Gurumurthy CB, Nakada S. 2018. Precise and efficient nucleotide substitution near genomic nick via noncanonical homology-directed repair. Genome Res 28:223–230. doi: 10.1101/gr.226027.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee N, Park J, Kim J-E, Shin J, Min K, Son H. 2022. Genome editing using Preassembled CRISPR-Cas9 Ribonucleoprotein complexes in Fusarium Graminearum. PLoS One 17:e0268855. doi: 10.1371/journal.pone.0268855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ferrara M, Haidukowski M, Logrieco AF, Leslie JF, Mulè G. 2019. A CRISPR Cas9 system for genome editing of Fusarium proliferatum. Sci Rep 9. doi: 10.1038/s41598-019-56270-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Q, Cobine PA, Coleman JJ. 2018. Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes. Fungal Genet Biol 117:21–29. doi: 10.1016/j.fgb.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Krogan NJ, Greenblatt JF. 2001. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol Cell Biol 21:8203–8212. doi: 10.1128/MCB.21.23.8203-8212.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fernández-Vázquez J, Vargas-Pérez I, Sansó M, Buhne K, Carmona M, Paulo E, Hermand D, Rodríguez-Gabriel M, Ayté J, Leidel S, Hidalgo E, Madhani HD. 2019. Modification of tRNALysUUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet 9:e1003647. doi: 10.1371/journal.pgen.1003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci U S A 101:6564–6569. doi: 10.1073/pnas.0305888101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee Y, Min K, Son H, Park AR, Kim J-C, Choi GJ, Lee Y-W. 2014. ELP3 is involved in sexual and asexual development, virulence, and the oxidative stress response in Fusarium graminearum. MPMI 27:1344–1355. doi: 10.1094/MPMI-05-14-0145-R [DOI] [PubMed] [Google Scholar]
- 59. McIlwraith MJ, West SC. 2008. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol Cell 29:510–516. doi: 10.1016/j.molcel.2007.11.037 [DOI] [PubMed] [Google Scholar]
- 60. Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, Shinohara A. 1997. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147:1545–1556. doi: 10.1093/genetics/147.4.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee J, Son H, Lee S, Park AR, Lee Y-W. 2010. Development of a conditional gene expression system using a zearalenone-inducible promoter for the ascomycete fungus Gibberella zeae. Appl Environ Microbiol 76:3089–3096. doi: 10.1128/AEM.02999-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee J-K, Son H, Lee Y-W. 2011. Estrogenic compounds compatible with a conditional gene expression system for the phytopathogenic fungus Fusarium graminearum. Plant Pathol J 27:349–353. doi: 10.5423/PPJ.2011.27.4.349 [DOI] [Google Scholar]
- 63. Franken ACW, Lokman BC, Ram AFJ, Punt PJ, van den Hondel CAMJJ, de Weert S. 2011. Heme biosynthesis and its regulation: towards understanding and improvement of heme biosynthesis in filamentous fungi. Appl Microbiol Biotechnol 91:447–460. doi: 10.1007/s00253-011-3391-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Phillips JD. 2019. Heme biosynthesis and the Porphyrias. Mol Genet Metab 128:164–177. doi: 10.1128/AEM.02999-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Franken ACW, Werner ER, Haas H, Lokman BC, van den Hondel CAMJJ, Ram AFJ, de Weert S, Punt PJ. 2013. The role of coproporphyrinogen III oxidase and ferrochelatase genes in heme biosynthesis and regulation in Aspergillus niger. Appl Microbiol Biotechnol 97:9773–9785. doi: 10.1007/s00253-013-5274-2 [DOI] [PubMed] [Google Scholar]
- 66. Zámocký M, Kamlárová A, Maresch D, Chovanová K, Harichová J, Furtmüller PG. 2020. Hybrid heme peroxidases from rice blast fungus Magnaporthe oryzae involved in defence against oxidative stress. Antioxidants 9:655. doi: 10.3390/antiox9080655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gebicka L, Krych-Madej J. 2019. The role of catalases in the prevention/promotion of oxidative stress. J Inorg Biochem 197:110699. doi: 10.1016/j.jinorgbio.2019.110699 [DOI] [PubMed] [Google Scholar]
- 68. Komalapriya C, Kaloriti D, Tillmann AT, Yin Z, Herrero-de-Dios C, Jacobsen MD, Belmonte RC, Cameron G, Haynes K, Grebogi C, de Moura APS, Gow NAR, Thiel M, Quinn J, Brown AJP, Romano MC. 2015. Integrative model of oxidative stress adaptation in the fungal pathogen Candida albicans. PLoS One 10:e0137750. doi: 10.1371/journal.pone.0137750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Baxter A, Mittler R, Suzuki N. 2014. ROS as key players in plant stress signalling. J Exp Bot 65:1229–1240. doi: 10.1093/jxb/ert375 [DOI] [PubMed] [Google Scholar]
- 70. Son H, Fu M, Lee Y, Lim JY, Min K, Kim J-C, Choi GJ, Lee Y-W. 2016. A novel transcription factor gene FHS1 is involved in the DNA damage response in Fusarium graminearum. Sci Rep 6:21572. doi: 10.1038/srep21572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kiffin R, Bandyopadhyay U, Cuervo AM. 2006. Oxidative stress and autophagy. Antioxid Redox Signal 8:152–162. doi: 10.1089/ars.2006.8.152 [DOI] [PubMed] [Google Scholar]
- 72. Wang X, Yen J, Kaiser P, Huang L. 2010. Regulation of the 26S proteasome complex during oxidative stress. Sci Signal 3:ra88. doi: 10.1126/scisignal.2001232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang B, Cui G, Chang C, Wang Y, Zhang H, Chen B, Deng Y, Jiang Z. 2019. The autophagy gene ATG8 affects morphogenesis and oxidative stress tolerance in Sporisorium scitamineum. J Integr Agric 18:1024–1034. doi: 10.1016/S2095-3119(18)62109-4 [DOI] [Google Scholar]
- 74. Na L, Sen L, Zhou S-Y, Wang C-X, Ren W-C, Li B-H. 2022. Involvement of the autophagy-related gene BdATG8 in development and pathogenicity in Botryosphaeria dothidea. J Integr Agric 21:2319–2328. doi: 10.1016/S2095-3119(21)63863-7 [DOI] [Google Scholar]
- 75. Lee KH, Gumilang A, Fu T, Kang SW, Kim KS. 2022. The autophagy protein CsATG8 is involved in asexual development and virulence in the pepper anthracnose fungus Colletotrichum scovillei. Mycobiology 50:467–474. doi: 10.1080/12298093.2022.2148393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang L, Cai X, Xing J, Liu C, Hendy A, Chen X-L. 2019. URM1-mediated Ubiquitin-like modification is required for oxidative stress adaptation during infection of the rice blast fungus. Front Microbiol 10:2039. doi: 10.3389/fmicb.2019.02039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang L, Wang L, Liang Y, Yu J. 2019. FgPEX4 is involved in development, pathogenicity, and cell wall integrity in Fusarium graminearum. Curr Genet 65:747–758. doi: 10.1007/s00294-018-0925-6 [DOI] [PubMed] [Google Scholar]
- 78. Davies KJ. 2001. Degradation of oxidized proteins by the 20s proteasome. Biochimie 83:301–310. doi: 10.1016/s0300-9084(01)01250-0 [DOI] [PubMed] [Google Scholar]
- 79. Rezayian M, Niknam V, Ebrahimzadeh H. 2019. Oxidative damage and antioxidative system in algae. Toxicol Rep 6:1309–1313. doi: 10.1016/j.toxrep.2019.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dauden MI, Kosinski J, Kolaj‐Robin O, Desfosses A, Ori A, Faux C, Hoffmann NA, Onuma OF, Breunig KD, Beck M, Sachse C, Séraphin B, Glatt S, Müller CW. 2017. Architecture of the yeast elongator complex. EMBO Reports 18:264–279. doi: 10.15252/embr.201643353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Karlsborn T, Tükenmez H, Mahmud AKMF, Xu F, Xu H, Byström AS. 2014. Elongator, a conserved complex required for wobble uridine modifications in eukaryotes. RNA Biol 11:1519–1528. doi: 10.4161/15476286.2014.992276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shao W, Lv C, Zhang Y, Wang J, Chen C. 2017. Involvement of BcElp4 in vegetative development, various environmental stress response and virulence of Botrytis cinerea. Microb Biotechnol 10:886–895. doi: 10.1111/1751-7915.12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang L, Chen S, Qi M, Cao X, Liang N, Li Q, Tang W, Lu G, Zhou J, Yu W, Wang Z, Zheng H. 2021. The putative elongator complex protein Elp3 is involved in asexual development and pathogenicity by regulating autophagy in the rice blast fungus. J Integr Agric 20:2944–2956. doi: 10.1016/S2095-3119(20)63493-1 [DOI] [Google Scholar]
- 84. Zhang Y, Wang Y, Fan J, Zhu G, Lu L, Andrianopoulos A. 2022. Aspergillus fumigatus elongator complex subunit 3 affects hyphal growth, adhesion and virulence through wobble uridine tRNA modification. PLoS Pathog 18:e1010976. doi: 10.1371/journal.ppat.1010976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Heinemann IU, Jahn M, Jahn D. 2008. The biochemistry of heme biosynthesis. Arch Biochem Biophys 474:238–251. doi: 10.1016/j.abb.2008.02.015 [DOI] [PubMed] [Google Scholar]
- 86. Ponka P. 1999. Cell biology of heme. Am J Med Sci 318:241–256. doi: 10.1097/00000441-199910000-00004 [DOI] [PubMed] [Google Scholar]
- 87. Dutt S, Hamza I, Bartnikas TB. 2022. Molecular mechanisms of iron and heme metabolism. Annu Rev Nutr 42:311–335. doi: 10.1146/annurev-nutr-062320-112625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tanabe S, Ishii-Minami N, Saitoh K-I, Otake Y, Kaku H, Shibuya N, Nishizawa Y, Minami E. 2011. The role of catalase-peroxidase secreted by Magnaporthe oryzae during early infection of rice cells. Mol Plant Microbe Interact 24:163–171. doi: 10.1094/MPMI-07-10-0175 [DOI] [PubMed] [Google Scholar]
- 89. Paris S, Wysong D, Debeaupuis J-P, Shibuya K, Philippe B, Diamond RD, Latgé J-P. 2003. Catalases of Aspergillus fumigatus.. Infect Immun 71:3551–3562. doi: 10.1128/IAI.71.6.3551-3562.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Angelova MB, Pashova SB, Spasova BK, Vassilev SV, Slokoska LS. 2005. Oxidative stress response of filamentous fungi induced by hydrogen peroxide and paraquat. Mycol Res 109:150–158. doi: 10.1017/s0953756204001352 [DOI] [PubMed] [Google Scholar]
- 91. Shin J, Kim J-E, Lee Y-W, Son H. 2018. Fungal cytochrome P450s and the P450 complement (cypome) of Fusarium graminearum. Toxins 10:112. doi: 10.3390/toxins10030112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Meunier B, de Visser SP, Shaik S. 2004. Mechanism of oxidation reactions catalyzed by cytochrome P450 enzymes. Chem Rev 104:3947–3980. doi: 10.1021/cr020443g [DOI] [PubMed] [Google Scholar]
- 93. Shin JY, Bui D-C, Lee Y, Nam H, Jung S, Fang M, Kim J-C, Lee T, Kim H, Choi GJ, Son H, Lee Y-W. 2017. Functional characterization of cytochrome P450 monooxygenases in the cereal head blight fungus Fusarium graminearum. Environ Microbiol 19:2053–2067. doi: 10.1111/1462-2920.13730 [DOI] [PubMed] [Google Scholar]
- 94. Leslie JF, Summerell BA. 2006. The fusarium laboratory manual. Blackwell publishing. doi: 10.1002/9780470278376 [DOI] [Google Scholar]
- 95. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. doi: 10.1111/1462-2920.13730 [DOI] [Google Scholar]
- 96. Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. J Comput Biol 17:10–12. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 97. Li H, Durbin R. 2009. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to Genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 99. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 100. Zhang B, Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4. doi: 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- 101. Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:1–13. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yu J-H, Hamari Z, Han K-H, Seo J-A, Reyes-Domínguez Y, Scazzocchio C. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genetics and Biology 41:973–981. doi: 10.1016/j.fgb.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 103. Zhou X, Li G, Xu J-R. 2011. Efficient approaches for generating GFP fusion and epitope-tagging constructs in filamentous fungi. Humana Press, Totowa, NJ. doi: 10.1007/978-1-61779-040-9_15 [DOI] [PubMed] [Google Scholar]
- 104. Bui D-C, Lee Y, Lim JY, Fu M, Kim J-C, Choi GJ, Son H, Lee Y-W. 2016. Heat shock protein 90 is required for sexual and asexual development, virulence, and heat shock response in Fusarium graminearum. Sci Rep 6:28154. doi: 10.1038/srep28154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔδCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 106. Son H, Lee J, Park AR, Lee Y-W. 2011. ATP citrate lyase is required for normal sexual and asexual development in Gibberella zeae. Fungal Genet Biol 48:408–417. doi: 10.1016/j.fgb.2011.01.002 [DOI] [PubMed] [Google Scholar]
- 107. Bonkovsky HL, Wood SG, Howell SK, Sinclair PR, Lincoln B, Healey JF, Sinclair JF. 1986. High-performance liquid chromatographic separation and quantitation of tetrapyrroles from biological materials. Anal Biochem 155:56–64. doi: 10.1016/0003-2697(86)90224-1 [DOI] [PubMed] [Google Scholar]
- 108. Chung D, Barker BM, Carey CC, Merriman B, Werner ER, Lechner BE, Dhingra S, Cheng C, Xu W, Blosser SJ, Morohashi K, Mazurie A, Mitchell TK, Haas H, Mitchell AP, Cramer RA, Doering TL. 2014. ChIP-Seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog 10:e1004487. doi: 10.1371/journal.ppat.1004487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conidiation and sexual reproduction of Δfghgg4, Δfghgg10, and Δfghgg13. Sensitivity of double deletion mutants to oxidative stress. Confirmation of the genetic manipulation by southern blot analyses.
GO enrichment analysis for the module genes.
Primers used in this study.
Data Availability Statement
The RNA-seq data have been deposited in the NCBI database under the BioProject accession number PRJNA937405.