ABSTRACT
Carbapenem-resistant Klebsiella pneumoniae (CRKP), which harbors the bla NDM plasmid, has been reported extensively and is considered a global threat clinically. However, characterization and comparisons of bla NDM-1-carrying and bla NDM-5-harboring IncX3-type plasmids in CRKP are lacking. Here, we systematically compared the differences in the characteristics, genetic backgrounds, transferability, and fitness costs between bla NDM-1-carrying and bla NDM-5-carrying plasmids in K. pneumoniae isolates. Fifteen NDM-producing CRKP isolates were recovered from 1376 CRKP isolates between 2019 and 2021, of which 4 were positive for bla NDM-1 and 11 were positive for bla NDM-5. All strains were highly resistant to carbapenem but remained susceptible to tigecycline and colistin. Core-genome-based phylogenetic analyses revealed that these strains were not clonally related. Whole-genome sequencing showed that bla NDM-1 and bla NDM-5 were located on ~54 kb and ~46 kb IncX3-type plasmids, respectively. The backbone, genetic context, and fitness cost of the bla NDM-1-bearing plasmid were highly similar to those of the bla NDM-5-carrying plasmid, but the transferability of the bla NDM-1-positive plasmid was greater than that of the bla NDM-5-positive plasmid. In conclusion, the transmission of bla NDM-1 or bla NDM-5 is mainly disseminated by plasmids rather than clonal spread. The high transfer frequency of the IncX3 plasmid facilitates the prevalence and dissemination of NDM-KP among Enterobacteriaceae.
IMPORTANCE
The emergence of NDM-producing Klebsiella pneumoniae is a severe challenge to public health. The widespread presence of bla NDM-1 and bla NDM-5 in Enterobacteriaceae has aroused broad concern. In this study, we performed molecular characterization of bla NDM-1-carrying and bla NDM-5-harboring IncX3-type plasmids in carbapenem-resistant Klebsiella pneumoniae (CRKP) and compared their phenotypes between strains with different bla NDM subtype. Our findings highlight the importance of IncX3-type plasmids in the transfer of the bla NDM-1 and bla NDM-5 genes and demonstrate that the bla NDM-1 plasmid possesses higher transfer ability. These data will provide important insights into carbapenem resistance gene transfer via plasmids and their further spread in clinical settings.
KEYWORDS: carbapenemases, NDM-1, NDM-5, IncX3, fitness
INTRODUCTION
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is a serious global health threat (1). New Delhi metallo-β-lactamase (NDM) is an important carbapenemase with the ability to hydrolyze almost all β-lactams, even carbapenems (2). NDM-1 was first identified in K. pneumoniae in 2008 (3), and an increasing number of variants have since been reported. NDM-5 differs from NDM-1 by only two amino acid substitutions and confers elevated carbapenem and expanded-spectrum cephalosporin resistance (4).
The dissemination and prevalence of NDM-1 and NDM-5 in K. pneumoniae and other Enterobacteriaceae are largely due to the plasmid-mediated transfer of bla NDM. The bla NDM gene was reported to be located on different incompatibility typing plasmids, such as IncX3, IncFII, IncN, and IncF (5). The horizontal spread of bla NDM is mediated by plasmids in many Enterobacteriaceae species. Escherichia coli and K. pneumoniae are the major hosts of bla NDM, followed by Citrobacter freundii and Salmonella enterica (2). Current research has raised great concern for when hypervirulent K. pneumoniae strains acquire a bla NDM-carrying plasmid. The wide spread of conjugative bla NDM-carrying plasmids has contributed to the emergence and prevalence of CR-hvKp strains (6).
In this study, we compared bla NDM-1-positive plasmid and bla NDM-5-bearing plasmid in CRKP isolates to elucidate the dissemination mechanism and confirm the horizontal gene transfer of bla NDM among Enterobacteriaceae.
RESULTS
Bacterial strains and antimicrobial susceptibility
Among 15 NDM-KP isolates collected from two different hospitals (A and B), 4 bla NDM-1-producing K. pneumoniae and 11 bla NDM-5-positive K. pneumoniae were identified (Table 1). In this study, NDM-KP isolates were recovered from adult patients with age ranging from 42 to 88 and were mainly collected from sputum (40%, 6/15) and urine (26.67%, 4/15). The antimicrobial susceptibility testing results showed that all strains were resistant to carbapenem (MIC >128 mg/L), ceftazidime (MIC >128 mg/L), cefepime (MIC >128 mg/L), amoxicillin-clavulanic acid (MIC >128 mg/L), and ceftazidime-avibactam (MIC >128/4 mg/L), but most were susceptible to tigecycline (MIC 0.25–4 mg/L) and colistin (MIC 0.25–2 mg/L) (Table 2). In addition, 12 isolates were susceptible to amikacin (MIC 0.5–8 mg/L), and 3 isolates were resistant (MIC >128 mg/L). Concerning ciprofloxacin, three isolates were intermediate, and others were resistant (MIC 4- > 128 mg/L).
TABLE 1.
Clinical information of the 15 NDM-KP isolates in the study
| Strain | Hospital | Age | Sex | Specimen | Diagnosis |
|---|---|---|---|---|---|
| ZRY2400 | A | 68 | Male | Blood | Bloodstream infection, esophageal tumor |
| NB0030 | B | 56 | Female | Sputum | Pneumonia |
| NB0011 | B | 63 | Male | Sputum | Pneumonia, gastric cancer |
| NB0010 | B | 58 | Female | Blood | Bloodstream infection, acute myelogenous leukemia |
| NB1827 | B | 70 | Male | Ascites | Bile duct infection |
| ZRY3974 | A | 51 | Male | Urine | Pneumonia, hemiplegia |
| ZRY3951 | A | 72 | Male | Sputum | Bronchiectasis |
| ZRY4277 | A | 81 | Female | Sputum | Pneumonia, cerebral infarction |
| ZRY4810 | A | 51 | Female | Sputum | Pneumonia, brainstem hemorrhage |
| ZRY5867 | A | 89 | Male | Sputum | Pneumonia, Alzheimer’s disease |
| ZRY1178 | A | 58 | Female | Urine | Urinary tract infection, hemiplegia |
| ZRY9668 | A | 60 | Male | Blood | Bloodstream infection, respiratory failure |
| ZRY2312 | A | 88 | Female | Urine | Urinary tract infection, hypertension |
| ZRY4942 | A | 76 | Male | Catheter | Hemiplegia |
| NB0045 | B | 42 | Male | Urine | Urinary tract infection, cerebral hemorrhage |
TABLE 2.
Antimicrobial susceptibility of the 15 NDM-KP isolates in the study
| Strain | NDM type | ST | MIC (mg/L) a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FEP | CAZ | AMC | AK | CIP | IPM | MEM | ETP | TGC | CST | CZA | |||
| ZRY2400 | NDM-1 | 152 | >128 | >128 | >128 | >128 | 8 | >128 | >128 | >128 | 0.25 | 0.25 | >128/4 |
| NB0030 | NDM-1 | 45 | >128 | >128 | >128 | 8 | 16 | >128 | >128 | >128 | 2 | 0.5 | >128/4 |
| NB0011 | NDM-1 | 3303 | >128 | >128 | >128 | 2 | 2 | >128 | >128 | >128 | 0.25 | 0.25 | >128/4 |
| NB0010 | NDM-1 | 12 | >128 | >128 | >128 | 8 | 4 | >128 | >128 | >128 | 1 | 0.5 | >128/4 |
| NB1827 | NDM-5 | 1 | >128 | >128 | >128 | 0.5 | >128 | >128 | >128 | >128 | 1 | 0.5 | >128/4 |
| ZRY3974 | NDM-5 | 17 | >128 | >128 | >128 | 1 | >128 | >128 | >128 | >128 | 0.5 | 1 | >128/4 |
| ZRY3951 | NDM-5 | 17 | >128 | >128 | >128 | 2 | 32 | >128 | >128 | >128 | 0.5 | 2 | >128/4 |
| ZRY4277 | NDM-5 | 17 | >128 | >128 | >128 | 2 | 32 | >128 | >128 | >128 | 0.5 | 0.5 | >128/4 |
| ZRY4810 | NDM-5 | 17 | >128 | >128 | >128 | 2 | 32 | >128 | >128 | >128 | 1 | 0.5 | >128/4 |
| ZRY5867 | NDM-5 | 1326 | >128 | >128 | >128 | 2 | 4 | >128 | >128 | >128 | 0.5 | 0.25 | >128/4 |
| ZRY1178 | NDM-5 | 37 | >128 | >128 | >128 | 0.5 | 64 | >128 | >128 | >128 | 4 | 0.5 | >128/4 |
| ZRY9668 | NDM-5 | 17 | >128 | >128 | >128 | 8 | 2 | >128 | >128 | >128 | 0.25 | 1 | >128/4 |
| ZRY2312 | NDM-5 | 17 | >128 | >128 | >128 | 4 | 2 | >128 | >128 | >128 | 0.5 | 0.25 | >128/4 |
| ZRY4942 | NDM-5 | 15 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 1 | 0.25 | >128/4 |
| NB0045 | NDM-5 | 5837 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 0.25 | 0.25 | >128/4 |
| ZRY2400J53 | – b | – | 64 | >128 | >128 | 16 | ≤0.125 | 64 | 64 | >128 | ≤0.0625 | 0.25 | >128/4 |
| NB1827J53 | – | – | >128 | >128 | >128 | 8 | ≤0.125 | >128 | >128 | >128 | 0.0625 | 0.25 | >128/4 |
| J53 | – | – | ≤0.125 | ≤0.125 | 2 | 1 | ≤0.125 | 0.5 | 0.25 | ≤0.125 | ≤0.0625 | 0.25 | 0.125 |
FEP, cefepime; CAZ, ceftazidime; AMC, amoxicillin-clavulanic acid; AK, amikacin; CIP, ciprofloxacin; IPM, imipenem; MEM, meropenem; ETP, ertapenem; TGC, tigecycline; CST, colistin; CZA, ceftazidime/avibactam.
–, not applicable.
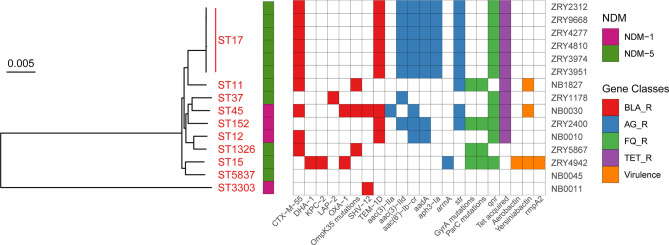
To uncover the resistance genes and virulence genes in the 15 isolates, we selected all strains for genome sequencing using the Illumina HiSeq sequencing platform. The distribution of resistance genes and virulence genes in the 15 isolates is listed in Fig. 1. Three isolates were positive for irp/ybtAEPQSTUX (encoding yersiniabactin), and only one isolate, ZRY4942, harbored iucABCD-iutA (encoding aerobactin) and rmpA2 (encoding mucoid-phenotype regulators); others were negative for these virulence genes.
Fig 1.
Core genome phylogenetic tree and gene heatmap of 15 NDM-KP strains. Red dots represent K. pneumoniae with bla NDM-1, and green dots represent K. pneumoniae with bla NDM-5. The gene heatmap contains resistance genes and virulence genes.
Ten of the 15 isolates carried the extended-spectrum beta-lactamase (ESBL) gene bla CTX-M-55, and two isolates carried bla SHV-12. In addition, majority of the isolates harbored bla TEM-1D. Of note, isolate ZRY4942 carried bla KPC-2 and bla NDM-5, but none co-carried another bla OXA-48-like carbapenemase gene.
Genetic relatedness
To investigate the phylogenetic relationship and genetic relatedness of NDM-KP isolates, multilocus sequence typing (MLST) and core genome phylogenetic tree analyses were carried out (Fig. 1). Six NDM-5-KP isolates belonged to the same sequence type, ST 17, and they were the most closely related to each other in the core genome phylogenetic alignment cluster. Others belonged to nine different sequence types (STs) (Table 2). In accordance with the MLST results, these strains displayed various clusters in the core genome phylogenetic tree.
Characterization and location of the bla NDM gene
To determine the location of the bla NDM gene, we selected two isolates, ZRY2400 and NB1827, as representatives for sequencing by Nanopore technology. In isolate ZRY2400, bla NDM-1 was located on a 54,035-bp IncX3 plasmid, pZRY2400-NDM1, with a GC content of 49.0% that was predicted to harbor 74 open reading frames (ORFs). Illumina sequence reads from other NDM-1-KP strains were mapped to pZRY2400-NDM1 with high identity and coverage, supporting the presence of a similar IncX3 plasmid in the bla NDM-1-carrying isolates (Fig. 2A).
Fig 2.
Genomic and molecular analyses of the bla NDM-1-positive plasmid pZRY2400-NDM1 and bla NDM-5-positive plasmid pNB1827-NDM5. (A) Schematic map of the pf plasmids pZRY2400-NDM1 and pNB1827-NDM5. (B) Comparative analysis of bla NDM-carrying IncX3 plasmids.
In isolate NB1827, bla NDM-5 was located on a 46,161-bp plasmid, pNB1827-NDM5, with a 46.7% GC content that belonged to the IncX3 group and did not harbor any resistance genes other than bla NDM-5. The complete sequence of pNB1827-NDM5 was predicted to harbor 66 ORFs with a 46.7% GC content. BLAST analysis showed that the plasmids with bla NDM-5 in other strains were highly homologous to pNB1827-NDM5 (Fig. 2A).
Plasmid sequence and comparative analysis
BLASTN search of GenBank showed that pZRY2400-NDM1 matched well with the bla NDM-1-carrying plasmid pA575-NDM (100% identity, 100% coverage) (GenBank accession no. NZ_MH917283.1), which is a typical bla NDM-1-positive plasmid from KP. The other plasmid pNB1827-NDM5 in our study was nearly identical to plasmid pEC463-NDM (GenBank accession no. MG545911) (99.95% identity and 100% coverage). The backbones of the bla NDM-5-positive plasmid pNB1827-NDM5 were highly similar to most of those in the bla NDM-1-carrying plasmid pZRY2400-NDM1, which indicated that integration or detachment events may have occurred during plasmid evolution.
In the ZRY2400 strain, the genetic environment of the bla NDM-1 gene (△ISAba125-IS5- bla NDM-1-ble MBL-trpF-IS26) was identical to that of pA575-NDM. In the NB1827 strain, the bla NDM-5 gene was flanked in the upstream region by △ISAba125-IS5 and downstream by ble MBL-trpF-IS26-△umuD-ISKox3, and this genetic context is the same as that of plasmid pEC463-NDM (Fig. 2B). Structural differences in the genetic elements surrounding bla NDM-1 and bla NDM-5 were observed only in the orientation of IS5.
Transmissibility and fitness cost of plasmids bearing bla NDM-1 and bla NDM-5
To explore the transferability of plasmids with bla NDM-1 and bla NDM-5, a conjugation experiment was performed. The pZRY2400-NDM1 plasmid was transferred to E. coli J53 by conjugation at a frequency of 8.7 × 10−2 (transconjugant/recipient). The conjugation frequency of pNB1827-NDM5 (recipient strain, E. coli J53) was 4.3 × 10−4. The transconjugant displayed a similar antibiotic resistance phenotype to strains ZRY2400 and NB1827, whereas the MICs of imipenem, meropenem, and cefepime of the transconjugant NB1827J53 with the bla NDM-5-carrying plasmid were all >128 mg/L, which were higher than that of the transconjugant ZRY2400J53 with the bla NDM-1-carrying plasmid (64 mg/L).
To compare the fitness of the bla NDM-harboring plasmid, growth kinetics assays were performed with strains with and without the plasmids pZRY2400-NDM1 or pNB1827-NDM5. Notably, a significant decrease was observed in the growth rate and area under the growth curve of the transconjugants ZRY2400J53 and NB1827J53 compared with the recipient E. coli J53 strain (P < 0.001) (Fig. 3). Results showed impaired growth as a result of the acquisition of plasmid pZRY2400-NDM1 or pNB1827-NDM5. However, no differences in the growth dynamics between the two transconjugants ZRY2400J53 and NB1827J53 were observed, indicating that the fitness burden of the bla NDM-1-positive plasmid and bla NDM-5-positive plasmid to the host was similar.
Fig 3.
(A) Bacterial growth curve of strains ZRY2400J53, NB1827J53, and J53. OD600, optical density at 600 nm. (B) Relative growth rates of strains ZRY2400J53, NB1827J53, and J53. (C) Area under the growth curve of strains ZRY2400J53, NB1827J53, and J53. ****, P < 0.0001.
DISCUSSION
Carbapenem resistance in K. pneumoniae is an urgent clinical problem that needs to be solved. Carbapenemase NDM remains a severe challenge and concern of many studies (2, 5). To date, carbapenemases NDM-1 and NDM-5 have been described mostly in Enterobacteriaceae isolates. However, few studies have systematically compared the differences between bla NDM-1-carrying and bla NDM-5-carrying plasmids. Hence, we compared the differences in the characteristics, genetic background, transferability, and fitness cost between the bla NDM-1-carrying plasmid and bla NDM-5-carrying plasmid in K. pneumoniae isolates obtained in the local region. Our results revealed that the IncX3-type plasmid played a vital role in the transport of bla NDM-1 and bla NDM-5 in K. pneumoniae, and the backbone, genetic context, and fitness cost of bla NDM-1 were highly similar to those of the bla NDM-5-carrying plasmid; however, the transferability of the bla NDM-1-positive plasmid was greater than that of the bla NDM-5-positive plasmid. Taken together, our findings suggest that the emerging threats of plasmid-mediated transfer and spread of bla NDM in K. pneumoniae require urgent improvements in the monitoring and prevention of NDM-KP.
Compared to NDM-1, NDM-5 has greater hydrolytic activity toward carbapenems and expanded-spectrum cephalosporins (2, 4, 7, 8). Similarly, the transconjugant acquisition of bla NDM-5-carrying plasmid in our study displayed elevated cefepime, imipenem, and meropenem MICs compared with transconjugants with bla NDM-1-positive plasmid.
Clonal spread and plasmid horizontal transmission are the two common ways to spread carbapenemase-producing K. pneumoniae. The MLST and CgMLST results in our study revealed that NDM-KP isolates were not clonally related. The NDM-KP strains in this study have a surprising diversity of STs, and they are not clonally related. Many STs, such as ST11, ST14, ST15, and ST147, are common NDM-positive K. pneumoniae clonal lineages that have been found in multiple studies, but sufficient evidence to demonstrate that these STs are high-risk clones mediating the transmission of bla NDM is lacking (9, 10).
Bla NDM-1 is mainly located on plasmids and plays a role in the dissemination and spread of resistance genes. bla NDM-1 and bla NDM-5 have been reported to be located on plasmids with many replicon types (11). The replicon types of bla NDM-1-carrying plasmids in K. pneumoniae included IncC, ColE10, IncFIA, IncFIB, IncHI1, IncHI3, IncN2, IncL/M, IncP, IncR, IncT, IncX1, IncX3, and IncY (2). For bla NDM-5-carrying plasmids, there is an equal variety of replicon types, such as IncFUA, IncFIB, Inc FIC, IncFII, IncX3, IncX4, and IncY. Of note, IncX3 appears to be the most popular replicon type of the bla NDM-carrying plasmid. To date, IncX3 plasmids carrying bla NDM-1 and bla NDM-5 have been reported worldwide. Most of the bla NDM-carrying IncX3 plasmids deposited in GenBank have been found in China and neighboring countries in East Asia (2).
The genetic environments of bla NDM-1 and bla NDM-5 in our study were nearly identical and were associated with △ISAba125 upstream and ble MBL downstream. The similar genetic environment of bla NDM-1 and the backbones of the bla NDM-positive plasmid in this study suggest that bla NDM-5 may have evolved from bla NDM-1 because another copy of IS26 was found in the bla NDM-1-harboring plasmid; one IS26-mediated deletion event might have happened, which contributed to the evolution from a bla NDM-1-harboring plasmid to a bla NDM-5-harboring plasmid (12). The IncX3 plasmid may be a major vehicle mediating the dissemination and evolution of bla NDM variants.
IncX3 plasmids appear to be a potentially successful vehicle for spreading bla NDM. In Ma’s study, the majority of wild-type strains acquiring the IncX3 plasmid showed a low fitness cost in the absence of antibiotic selection pressure (13). Strains with plasmids carrying bla NDM-1 and bla NDM-5 in our study imposed significant but similar fitness costs on their hosts. We could not compare them with other plasmids and with other replicons due to the absence of other replicons in our study. Additionally, high transfer frequency was the other characteristic of the IncX3 plasmid. The conjugation frequency of the IncX3 bla NDM-carrying plasmid was between 10−4 and 10−2, which is consistent with the findings of Liu et al. (14). The transferability of the bla NDM-1-positive plasmid was greater than that of the bla NDM-5-positive plasmid. The basic structure of the bla NDM-1-harboring plasmid and the bla NDM-5-bearing plasmid is similar, and the core genes of the backbone including plasmid replication-related genes, conjugation-related genes, and conjugation/type IV secretion system (T4SS, with 11 genes, pilX1 to pilX11) were identical. In addition, differences between the two plasmids included chaperonin GroEL, class A extended-spectrum β-lactamase SHV-12, DeoR/GlpR transcriptional regulator, and NAD(P)-dependent oxidoreductase; however, these differential genes do not affect the transmissibility of plasmids. Furthermore, more ISs were identified in the bla NDM-1-bearing plasmid, such as more copies of IS26. This may be a cause of higher transmissibility.
Conclusions
We described the molecular characterization of NDM-KP strains that belonged to various STs. Both bla NDM-1 and bla NDM-5 were located on a self-transmissible IncX3 plasmid, which could be transferred to E. coli with a low fitness cost and high transfer frequency. Moreover, the genetic environment of bla NDM-1 and the backbones of the bla NDM-positive plasmid was similar, suggesting that the IncX3 plasmid may be a major vehicle mediating the dissemination and evolution of bla NDM variants.
MATERIALS AND METHODS
Bacterial strains
A total of 15 NDM-producing Klebsiella pneumoniae (NDM-KP) strains were collected from 1,376 CRKP isolates between 2019 and 2021 in two hospitals in Zhejiang, China. The MBL carbapenemase gene bla NDM was screened by PCR using specific primers from the previous study and confirmed by Sanger sequence (15).
Antimicrobial susceptibility testing
The MICs of antibiotics were determined by the broth microdilution method as per Clinical and Laboratory Standards Institute (CLSI) recommendations. The results of colistin and tigecycline were interpreted according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints (www.eucast.org) and other antibiotics in Table 1 according to CLSI criteria (16). E. coli ATCC 25922 was used for the quality control strain.
Whole-genome sequencing and analysis
These NDM-KP isolates were genome sequenced using HiSeqTM 2000 (Illumina Inc., San Diego, USA). Reads were de novo assembled into contigs using CLC Genomics Workbench (CLC Bio 8.0). Acquired resistance genes were analyzed by ResFinder database plasmid replicons PlasmidFinder (both at https://cge.cbs.dtu.dk/) and sequence typing (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
Long-read genome sequencing and analysis
Representative isolates of NDM-1 and NDM-5-producing KP, NB1827 and ZRY2400, were selected to sequence by long-read genome sequencing by a MinION Sequencer (Nanopore; Oxford, UK). The complete genome sequence was obtained by de novo hybrid assembly of both short (Illumina) and long reads using Unicycler v0.4.3 and Pilon v1.24. Short reads of other isolates were mapped to pZRY2400-NDM1 and pNB1827-NDM5 using CGView (17).
Phylogenomic analysis of NDM-KP
The phylogenetic tree was based on the core genome which consisted of 4,143 genes defined by Panaroo (v.1.2.10) that existed in 99%–100% strains. The phylogenetic tree was constructed by RAxML-NG (v.1.0.1) based on the bootstrap analysis. We chose the autoMRE option, and the bootstrapping converged after 650 replicates (Fig. S1) (18). The virulence and antimicrobial resistance genes were annotated by Kleborate3 (v.2.2.0) with default options (19). The phylogenetic tree and gene heatmap were visualized by ggtree4 (20).
Conjugation assay
Mating experiments were performed as previously described, and E. coli J53 was used as the recipient strain as described previously (15, 21, 22). Putative transconjugants were selected on MH agar plates supplemented with ampicillin (100 mg/L) and sodium azide (300 mg/L). The transconjugants were confirmed through PCR with specific primers (NDM_F58 5′-ggcggaatggctcatcacga-3′; NDM_R344 5′-cgcaacacagcctgactttc-3′).
Growth kinetics
Growth curves for the recipients were performed in 96-well plates as described previously (23). The recipients J53/NDM-1 and J53/NDM-5 and J53 were diluted in LB broth medium in 96-well microtiter plates and incubated at 37°C for 24 h. OD600 measurements were taken hourly to construct a growth curve. Relative growth rates were measured via R script, and AUC values were calculated using GraphPad Prism software 8.0.2 (GraphPad Software, San Diego, CA, USA). Growth kinetics assays were performed in triplicate.
Statistical analysis
Relative growth rates were evaluated using Student’s t-test. Statistical significance was analyzed using GraphPad Prism 8.0.2 (GraphPad Software, San Diego, CA, USA). P < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (82172306), Zhejiang Provincial Public Projects (LGD21H190001), and the Medical and Health Research Project of Zhejiang Province, China (2022KY531), the Research and Development Program of Zhejiang Province (2023C03068), Medical and Health Technology Project of Hangzhou (A20210087).
We declare no conflicts of interest.
Contributor Information
Hua Zhou, Email: zhouhua1@zju.edu.cn.
N. Esther Babady, Memorial Sloan Kettering Cancer Center, New York, New York, USA .
ETHICS APPROVAL
This study was conducted in accordance with the Declaration of Helsinki and had been reviewed and approved by the Research Ethics Committee of the First Affiliated Hospital of Zhejiang University (IIT20210268A).
DATA AVAILABILITY
The genome sequences used in this study have been deposited in the National Center for Biotechnology Information database under BioProject PRJNA936643.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.01028-23.
The bootstrap analysis of core genome phylogenetic tree.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Wang M, Earley M, Chen L, Hanson BM, Yu Y, Liu Z, Salcedo S, Cober E, Li L, Kanj SS, Gao H, Munita JM, Ordoñez K, Weston G, Satlin MJ, Valderrama-Beltrán SL, Marimuthu K, Stryjewski ME, Komarow L, Luterbach C, Marshall SH, Rudin SD, Manca C, Paterson DL, Reyes J, Villegas MV, Evans S, Hill C, Arias R, Baum K, Fries BC, Doi Y, Patel R, Kreiswirth BN, Bonomo RA, Chambers HF, Fowler VG, Arias CA, van Duin D, Abbo LM, Anderson DJ, Arias R, Arias CA, Baum K, Bonomo RA, Chambers HF, Chen L, Chew KL, Cober E, Cross HR, De PP, Desai S, Dhar S, Di Castelnuovo V, Diaz L, Dinh AQ, Doi Y, Earley M, Eilertson B, Evans B, Evans S, Fowler Jr VG, Fries BC, Gao H, Garcia-Diaz J, Garner OB, Greenwood-Quaintance K, Hanson B, Herc E, Hill C, Jacob JT, Jiang J, Kalayjian RC, Kanj SS, Kaye KS, Kim A, Komarow L, Kreiswirth BN, Lauterbach C, Li L, Liu Z, Manca C, Marimuthu K, Marshall SH, McCarty T, Munita J, Ng OT, Oñate Gutierrez JM, Ordoñez K, Patel R, Paterson DL, Peleg A, Reyes J, Rudin SD, Salata RA, Salcedo S, Satlin MJ, Schmidt-Malan S, Smitasin N, Spencer M, Stryjewski M, Su J, Tambyah PA, Valderrama S, van Duin D, Villegas Botero MV, Wang M, Waters M, Weston G, Wong D, Wortmann G, Yang Y, Yu Y, Zhang F. 2022. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis 22:401–412. doi: 10.1016/S1473-3099(21)00399-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. 2019. NDM metallo-Β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev 32:e00115-18. doi: 10.1128/CMR.00115-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the new delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 55:5952–5954. doi: 10.1128/AAC.05108-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55:5403–5407. doi: 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang X, Sun Q, Li J, Jiang Y, Li Y, Lin J, Chen K, Chan EW-C, Zhang R, Chen S. 2022. Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg Microbes Infect 11:841–849. doi: 10.1080/22221751.2022.2049458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu W, Wang X, Qin J, Liang W, Shen Z, Gales AC. 2020. Dissemination and stability of the bla(NDM-5)-Carrying IncX3-type plasmid among multiclonal Klebsiella pneumoniae isolates. mSphere 5:e00917-20. doi: 10.1128/mSphere.00917-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khan AU, Maryam L, Zarrilli R. 2017. Structure, genetics and worldwide spread of new Delhi metallo-Β-lactamase (NDM): a threat to public health. BMC Microbiol 17:101. doi: 10.1186/s12866-017-1012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giske CG, Fröding I, Hasan CM, Turlej-Rogacka A, Toleman M, Livermore D, Woodford N, Walsh TR. 2012. Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob Agents Chemother 56:2735–2738. doi: 10.1128/AAC.06142-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, Li Y, Liao K, Chen S. 2017. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19:98–106. doi: 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo X, Chen R, Wang Q, Li C, Ge H, Qiao J, Li Y. 2022. Global prevalence, characteristics, and future prospects of IncX3 plasmids: a review. Front Microbiol 13:979558. doi: 10.3389/fmicb.2022.979558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hua X, Zhang L, Moran RA, Xu Q, Sun L, van Schaik W, Yu Y. 2020. Cointegration as a mechanism for the evolution of a KPC-producing multidrug resistance plasmid in Proteus mirabilis. Emerg Microbes Infect 9:1206–1218. doi: 10.1080/22221751.2020.1773322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma T, Fu J, Xie N, Ma S, Lei L, Zhai W, Shen Y, Sun C, Wang S, Shen Z, Wang Y, Walsh TR, Shen J. 2020. Fitness cost of bla(NDM-5)-carrying p3R-IncX3 plasmids in wild-type NDM-free Enterobacteriaceae. Microorganisms 8:377. doi: 10.3390/microorganisms8030377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Z, Wang Y, Walsh TR, Liu D, Shen Z, Zhang R, Yin W, Yao H, Li J, Shen J. 2017. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob Agents Chemother 61:e02233-16. doi: 10.1128/AAC.02233-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li X, Fu Y, Shen M, Huang D, Du X, Hu Q, Zhou Y, Wang D, Yu Y. 2018. Dissemination of blaNDM-5 Gene via an IncX3-type Plasmid among non-Clonal Escherichia coli in China. Antimicrob Resist Infect Control 7. doi: 10.1186/s13756-018-0349-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clinical and Laboratory Standards Institute . 2020. CLIS document M100. Performance standards for antimicrobial susceptibility testing. 30th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17. Grant JR, Stothard P. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–4. doi: 10.1093/nar/gkn179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. doi: 10.1038/s41467-021-24448-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu G. 2020. Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinformatics 69:e96. doi: 10.1002/cpbi.96 [DOI] [PubMed] [Google Scholar]
- 21. Liu H, Moran RA, Chen Y, Doughty EL, Hua X, Jiang Y, Xu Q, Zhang L, Blair JMA, McNally A, van Schaik W, Yu Y. 2021. Transferable Acinetobacter baumannii Plasmid pDETAB2 encodes OXA-58 and NDM-1 and represents a new class of antibiotic resistance plasmids. J Antimicrob Chemother 76:1130–1134. doi: 10.1093/jac/dkab005 [DOI] [PubMed] [Google Scholar]
- 22. Moran RA, Liu H, Doughty EL, Hua X, Cummins EA, Liveikis T, McNally A, Zhou Z, van Schaik W, Yu Y. 2022. GR13-type plasmids in Acinetobacter potentiate the accumulation and horizontal transfer of diverse accessory genes. Microb Genom 8:mgen000840. doi: 10.1099/mgen.0.000840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi Y, Hua X, Xu Q, Yang Y, Zhang L, He J, Mu X, Hu L, Leptihn S, Yu Y. 2020. Mechanism of eravacycline resistance in Acinetobacter baumannii mediated by a deletion mutation in the sensor kinase adeS, leading to elevated expression of the efflux pump AdeABC. Infect Genet Evol 80:104185. doi: 10.1016/j.meegid.2020.104185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The bootstrap analysis of core genome phylogenetic tree.
Data Availability Statement
The genome sequences used in this study have been deposited in the National Center for Biotechnology Information database under BioProject PRJNA936643.