ABSTRACT
Gemmatimonadota is a diverse bacterial phylum commonly found in environments such as soils, rhizospheres, fresh waters, and sediments. So far, the phylum contains just six cultured species (five of them sequenced), which limits our understanding of their diversity and metabolism. Therefore, we analyzed over 400 metagenome-assembled genomes (MAGs) and 5 culture-derived genomes representing Gemmatimonadota from various aquatic environments, hydrothermal vents, sediments, soils, and host-associated (with marine sponges and coral) species. The principal coordinate analysis based on the presence/absence of genes in Gemmatimonadota genomes and phylogenomic analysis documented that marine and host-associated Gemmatimonadota were the most distant from freshwater and wastewater species. A smaller genome size and coding sequences (CDS) number reduction were observed in marine MAGs, pointing to an oligotrophic environmental adaptation. Several metabolic pathways are restricted to specific environments. For example, genes for anoxygenic phototrophy were found only in freshwater, wastewater, and soda lake sediment genomes. There were several genomes from soda lake sediments and wastewater containing type IC/ID ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). Various genomes from wastewater harbored bacterial type II RuBisCO, whereas RuBisCO-like protein was found in genomes from fresh waters, soil, host-associated, and marine sediments. Gemmatimonadota does not contain nitrogen fixation genes; however, the nosZ gene, involved in the reduction of N2O, was present in genomes from most environments, missing only in marine water and host-associated Gemmatimonadota. The presented data suggest that Gemmatimonadota evolved as an organotrophic species relying on aerobic respiration and then remodeled its genome inventory when adapting to particular environments.
IMPORTANCE
Gemmatimonadota is a rarely studied bacterial phylum consisting of a handful of cultured species. Recent culture-independent studies documented that these organisms are distributed in many environments, including soil, marine, fresh, and waste waters. However, due to the lack of cultured species, information about their metabolic potential and environmental role is scarce. Therefore, we collected Gemmatimonadota metagenome-assembled genomes (MAGs) from different habitats and performed a systematic analysis of their genomic characteristics and metabolic potential. Our results show how Gemmatimonadota have adapted their genomes to different environments.
KEYWORDS: Gemmatimonadota, gemmatimonadetes, anoxygenic phototrophs, MAGs, metagenome, RuBisCO
INTRODUCTION
The bacterial phylum Gemmatimonadota was established in 2003 when the type species, Gemmatimonas aurantiaca, was isolated from a wastewater treatment plant (1). Since then, only five more species have been described. “Gemmatirosa kalamazoonesis,” Roseisolibacter agri, and Longimicrobium terrae were isolated from various soils (2 – 4), while Gemmatimonas phototrophica and Gemmatimonas groenlandica originated from fresh waters (5, 6). Due to the low number of cultured species, our understanding of the metabolic properties of Gemmatimonadota is very limited. All isolates grow on liquid organic carbon media under aerobic or semi-aerobic conditions (7, 8). In addition, two cultured freshwater species are facultative photoheterotrophs. They perform anoxygenic phototrophy and can supplement their metabolism with light energy harvested using bacteriochlorophyll (BChl)-a-containing photosystems; however, they require a supply of organic substrate for growth (5, 9, 10). Photoheterotrophic Gemmatimonadota, similar to Proteobacteria, have their photosynthesis genes organized in the photosynthesis gene cluster, containing bch and crt genes encoding enzymes of bacteriochlorophyll and carotenoid synthesis, puf and puh operons encoding the subunits of reaction centers and light-harvesting complexes, and various regulatory genes (6, 9, 11, 12).
Metagenomic analyses have documented that Gemmatimonadota is present in a wide range of environments (13, 14). They are one of the most abundant phyla in soils, representing on average 2% of 16S rRNA gene sequences (13, 15, 16), and are relatively common in fresh waters, where they typically constitute 1% of bacteria but may contribute even up to 9% of the bacterial community (7, 12, 17, 18). Gemmatimonadota were found in soda lake sediments, where they represented ≥1% of 16S rRNA gene sequences (19). Only minimum numbers have been registered in the marine water column (20), and typically in marine environments, they are found associated with sponges (21, 22), deep-sea hydrothermal vents (23, 24), or sediments (25, 26), where they represent up to 2.4% of the total bacterial 16S rRNA reads (27).
Previously, we documented a high diversity of photoheterotrophic Gemmatimonadota in freshwater lakes (12). Interestingly, metagenome-assembled genomes (MAGs) containing genes for both anoxygenic photosynthesis and carbon fixation were identified in soda lake sediments (28, 29). However, there is only limited information about Gemmatimonadota inhabiting other environments, such as soils or marine waters. Therefore, we analyzed all publicly available MAGs (up until 3 May 2021) affiliated with Gemmatimonadota to get a global picture of their metabolic functions, patterns, and genomic differences across multiple environments. In addition, we assembled 16 MAGs from four Spanish freshwater reservoirs (Tables S1 and S2). We focused on key metabolic pathways, such as carbon assimilation, nitrogen and sulfur cycles, and photoheterotrophic capability, to define the potential roles of Gemmatimonadota in nutrient cycling and to decipher the specific differences in their physiology based on the environment from which they originate.
RESULTS AND DISCUSSION
Basic characteristics of the Gemmatimonadota genomes
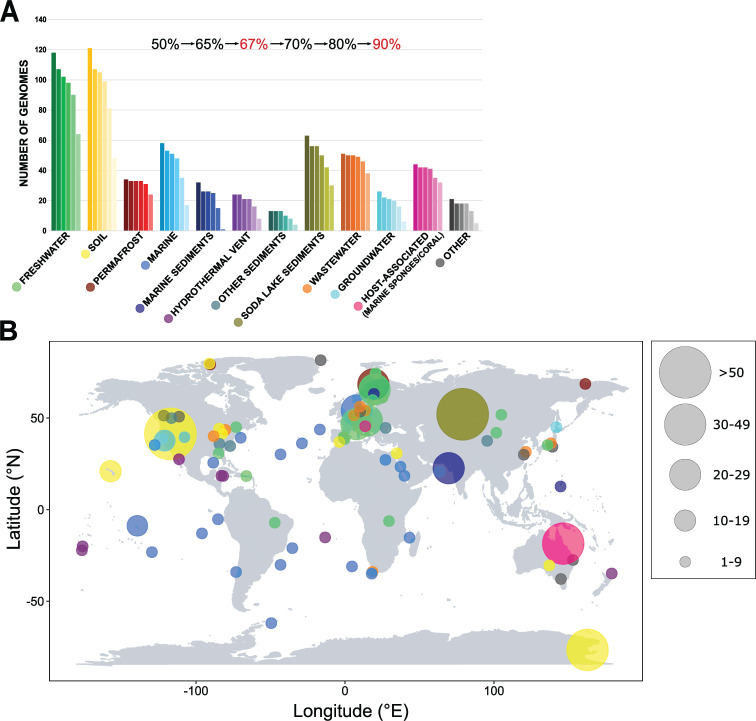
Gemmatimonadota MAGs were classified based on their environmental origin in 12 different categories (Fig. 1A and B). The numbers of dereplicated genomes within each category were as follows: fresh waters 91, soils 90, wastewaters 49, soda lake sediments 46, marine waters 42, host-associated (i.e., associated with marine sponges and coral) 25, permafrost 22, marine sediments 12, hydrothermal vents 18, groundwater 21, and other sediments 13. The final category “Other” consisted of 13 genomes from varying environments and was not included in most analyses (unless stated otherwise). Genomes from all environments varied largely in size (1.68–7.77 Mbp), with an average of 3.59 Mbp and 2,744 coding sequences (CDS) (Fig. S1A and B). The smallest genomes (1.68–3.01 Mbp) with the lowest number of genes and higher homogeneity were those from marine waters. Genomes from potentially more nutrient-rich environments, like soils, soda lake sediments, marine sediments, and wastewaters, had larger sizes as well as a higher number of CDS (Fig. S2A). This is consistent with previous studies documenting that nutrient limitation affects genome size, GC content, or coding density (30 – 32). The average coding density was 92.9% (84%–97%), and despite the high variability, it was on average higher in MAGs from fresh waters and soils than in those from marine waters, marine sediments, or wastewaters (Fig. S1C). The average median intergenic distance was 35.45 bp. Even though marine genomes are in general smaller, they have on average longer intergenic spacers than freshwater, soil, or wastewater genomes (Fig. S1D and S2B). Genomes from soda lake sediments and marine sediments have both larger sizes and longer median intergenic spacers. Lengths of intergenic spacers vary substantially among bacteria (32) and often contain regulatory elements with key functions (33).
FIG 1.
Distribution of Gemmatimonadota in different environments. (A) Bar plot showing the number of Gemmatimonadota genomes present in the NCBI database, including newly assembled freshwater MAGs, divided based on the environment of origin and completeness. Each bar represents the number of genomes with different completeness levels, from MAGs with more than >50% up to >90% completeness. The completeness levels used in subsequent analyses are marked in red. (B) Map showing where the MAGs used in our analyses originated from. Environments are color coded, and the size of the circle represents the number of MAGs obtained from the specific location.
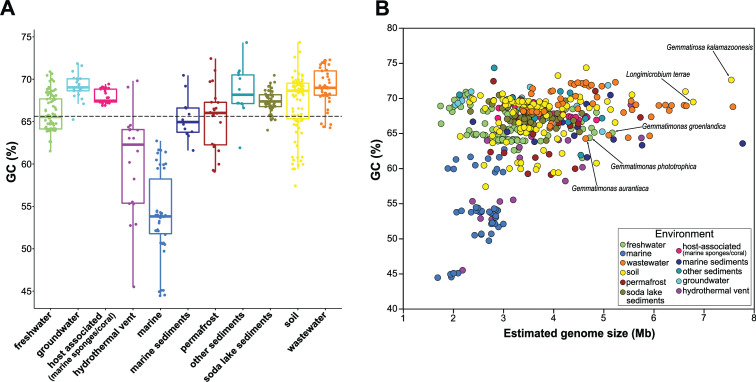
The GC content in the studied genomes ranged from 44.4% to 74.4%, with an average of 65.6% (Fig. 2A and B). Marine genomes and several others from hydrothermal vents had the lowest GC content (range 44.4%–62.7% and 45.5%–69.8%, respectively). The GC content of bacterial communities is known to be influenced by the environment (34), and low GC content among marine bacteria is a common phenomenon (35) interpreted as an adaptation to low nitrogen (36) or a result of evolutionary history (37). It must be noted that the distribution of the GC content in marine genomes was trimodal (44%–45%, 49%–54%, and 59%–62.7%), indicating an additional sub-environmental division of genomes from the same origin, possibly depending on parameters such as the water depth or water nutrient concentration. However, the associated metadata in the NCBI did not contain enough details to fully explain this pattern. The influence of environment on the GC content of bacterial communities can be observed even in closely related species, which in different environments show significant differences in GC content (34). Freshwater genomes were also smaller but had a higher GC content than marine genomes (Fig. 2A). Gemmatimonadota genomes from other environments like soil, permafrost, or wastewater varied in genome sizes and had on average a higher GC content than marine MAGs, a trait common for bacteria living in more nutrient-rich environments (38). This, combined with their larger genomes and higher number of CDS, indicates their higher metabolic potential and advantages under different environmental conditions.
FIG 2.
(A) Distribution of GC content (%) of Gemmatimonadota genomes based on their environmental origin. (B) A comparison of estimated genome size and GC content (%) of Gemmatimonadota genomes from different environments. Genomes are color-coded based on their environment. Labels depict the cultured Gemmatimonadota species.
Gemmatimonadota habitat-related core and accessory gene analysis
We explored the main shared (core) and flexible (accessory) genomes among Gemmatimonadota MAGs with >90% completeness and <10% contamination across multiple origins. Generally, the size of the habitat-dependent core and flexible genome of Gemmatimonadota differed between environments and ranged from a lower average of 2,677 genes in the marine environment (10 genomes), 2,868 in the freshwater environment (29 genomes), to the highest average of 4,659 genes in the wastewater environment (13 genomes) (Table S3), indicating how contrasting environments differentially shape their gene inventories. Larger genomes found in wastewater may encode a wider variety of enzymes for utilization in an environment often enriched with nutrients (38, 39). The size of the shared genes (strict core and soft core) also varied (Fig. S3), while accessory genes formed by the shell (40) and cloud (41) categories represented more than 50% of the flexible/accessory genome in all environments except marine waters, showing high variability in the gene inventories among the members of the phylum.
Multi-environment principal coordinate and phylogenomic analyses
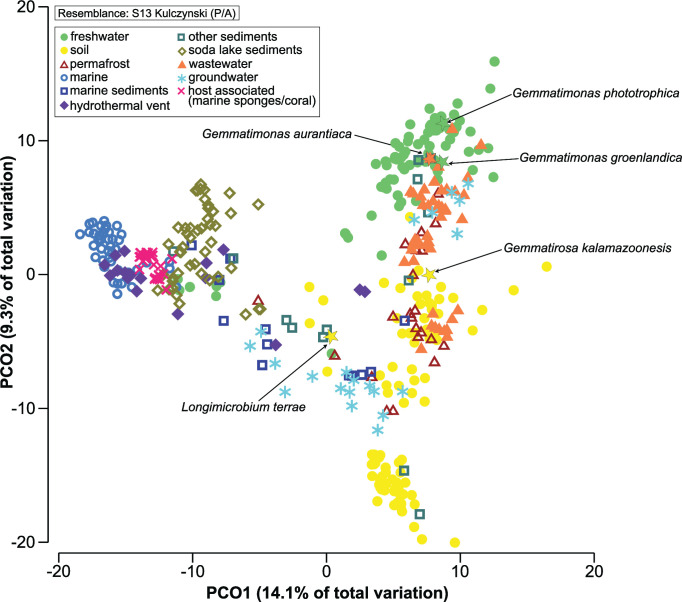
The similarity among genomes was studied using a principal coordinate analysis (PCoA) based on the presence or absence of genes (Table S4). The genomes clustered based on their environmental origin (Fig. 3), indicating their differential adaptation to specific environments. Permutational multivariate dispersion (PERMDISP) analysis documented a significant difference in heterogeneity levels (PERMDISP, P < 0.05) between various environments (Table S5), and significant differences in gene presence/absence were detected for Gemmatimonadota from all environments (PERMANOVA, P < 0.001) except marine sediments and other sediments (PERMANOVA, P = 0.019). Similarity percentage (SIMPER) analysis based on Bray-Curtis similarity showed host-associated (70.6%) and marine waters (69.9%) MAGs to be the most similar among them, while those from other sediments (56.2%), marine sediments (59.2%), and groundwater (59.7%) were the least similar (Table S5). In the comparison between different environments, marine MAGs were more like other marine-related environments such as hydrothermal vents or host-associated (with marine sponges and coral) (average dissimilarity of 37.79% and 40.21%, respectively), while freshwater MAGs were more similar to wastewater, groundwater, and permafrost MAGs (<41% of average dissimilarity). The highest dissimilarities (>47%) were seen between soil vs marine and host-associated (with marine sponges and coral) MAGs and between marine vs wastewater MAGs.
FIG 3.
PCoA using Kulczynski resemblance matrix based on SEED presence/absence of the genes in Gemmatimonadota genomes, showing grouping of genomes based on their environment. The legend in the upper left corner shows that environments are color coded and a different symbol is assigned to each environment. Cultured Gemmatimonadota species are labeled and shown with a star symbol.
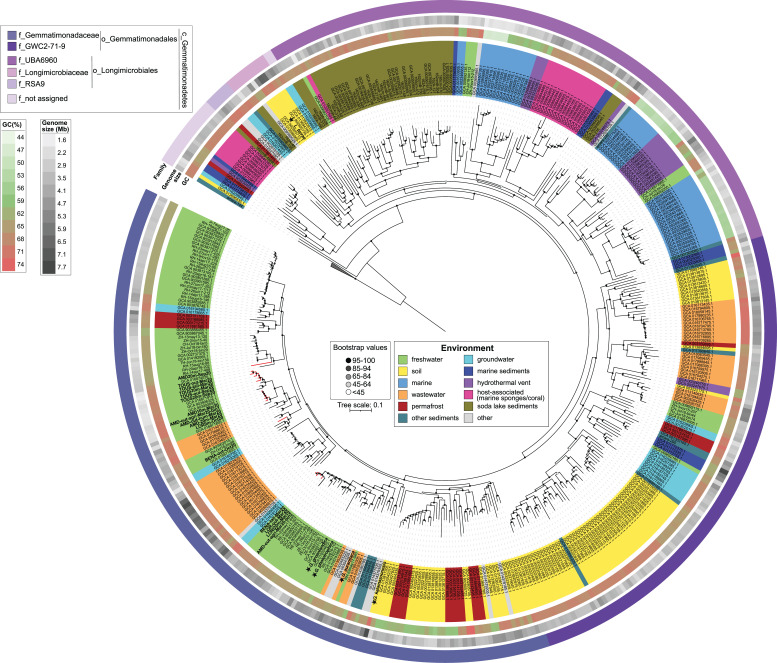
Similar patterns were found in the phylogenomic analysis (Fig. 4), albeit these genomes did not cluster exclusively according to the environment of origin. Most of the MAGs obtained belonged to two families inside the order Gemmatimonadales. The family Gemmatimonadaceae encompassed most of the MAGs from fresh, waste, and groundwater, along with genomes from permafrost and soil (Fig. S4), and cultured species G. phototrophica, G. groenlandica, G. aurantiaca, and G. kalamazoonesis. This family also contained all the MAGs from Spanish reservoirs reconstructed in this study. Eleven of them formed a clade related to MAGs from the hypolimnion of several Swiss lakes and Římov Reservoir (Czech Republic). The remaining four clustered together with a previously assigned group, Pg2 (12), which consists of freshwater phototrophic Gemmatimonadota from the epilimnion of Lake Zurich (Switzerland) and Římov Reservoir. The second family GWC2-71-9 mostly contained genomes from the soil, wastewater, permafrost, groundwater, and other sediments, with only a small number of genomes from freshwater lakes and marine sediments (Fig. S4).
FIG 4.
Phylogenomic tree of Gemmatimonadota genomes based on 400 universally conserved and most ubiquitous proteins present in the PhyloPhlAn database (42, 43). The collapsed branch represents an outgroup consisting of three genomes from the bacterial phylum Fibrobacterota (GCA_900142455.1 Hallerella intestinalis, GCA_900217845.1 Fibrobacter elongatus, GCA_000146505.1 Fibrobacter succinogenes). The strength of support for internal nodes is shown through gray-scale-colored circles (center legend). All genomes are color-coded based on their environmental origin (center legend). The following annotations, starting from innermost to outermost indicate GC content (%), estimated genome size (Mb), and family level classification. The legend for each outer circle is represented in the upper left corner. Details on all genomes can be found in Tables S1 and S2.
The second largest group was formed by MAGs belonging to the order Longimicrobiales, which was established based on the soil bacterium Longimicrobium terrae (4). This order mostly contains marine water, marine sediments, hydrothermal vents, soda lake sediments, and host-associated (with marine sponges and coral) genomes, together with several genomes from the hypolimnion of deep freshwater lakes. This is in line with our previous observations that Gemmatimonadota from deep freshwater lakes are related to those from marine environments or environments like soil and sediments (12). Host-associated MAGs (marine sponges and corals) were part of two different families (Longimicrobiales and a not assigned family), and while they are closely related to marine water genomes, the differences in the gene repertoire between these two environments were significant (PERMANOVA, P < 0.0001), and they represent real symbionts of marine sponges and corals (44, 45).
Main metabolic pathways across the Gemmatimonadota phylum
The core metabolism of the Gemmatimonadota phylum was reported recently (13). Still, both PCoA and phylogenomic analysis showed that their gene inventories vary depending on their origin, presumably due to adaptation to the specific conditions and selection pressure in any particular habitat. To study this further, we looked for the main metabolic commonalities and uniqueness associated with each environment. To do so, we individually inspected genomes from all environments to reconstruct a metabolic model of the Gemmatimonadota phylum with pathways present or absent for each environmental specialist (Fig. 5; Fig. S5; Fig. 6; Table S6).
FIG 5.
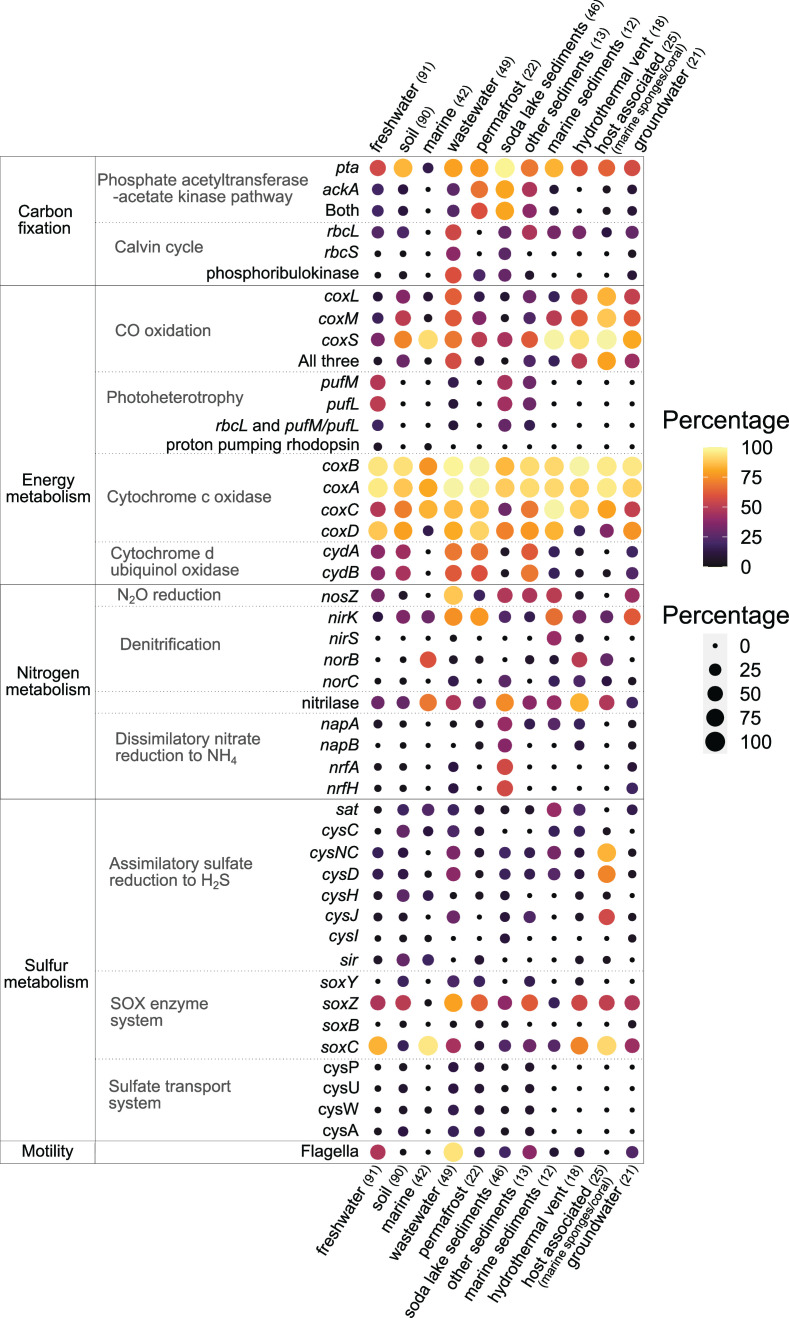
Bubble plot showing the percentages of key genes involved in specific pathways present in Gemmatimonadota genomes from different environments. Dot color and size indicate the percentage of each gene in any given environment, with the darkest color and smallest size of the dot marking the absence of said gene in that environment. The number of MAGs in each environment is labeled in parenthesis. Details about genes presence/absence can be found in Table S6.
FIG 6.
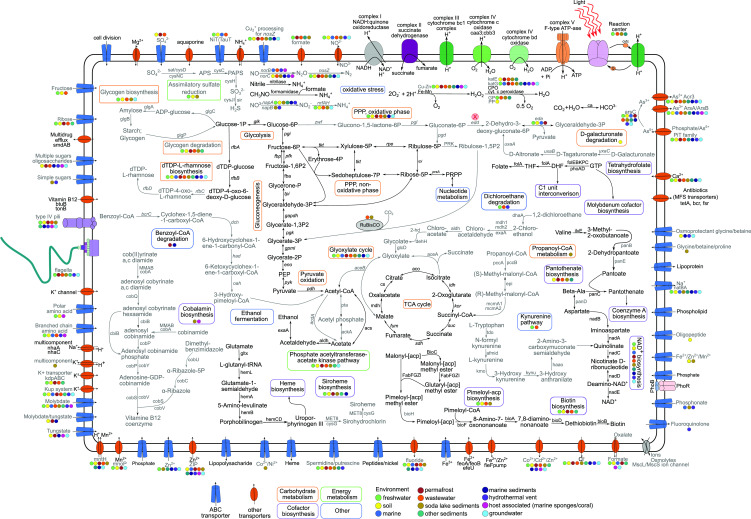
A metabolic reconstruction of Gemmatimonadota showing some of the key pathways. Four different colored rectangles depict the names of pathways and metabolic processes. Pathways labeled with black are present in all Gemmatimonadota genomes, while those labeled with gray are only found in Gemmatimonadota genomes from certain environments. Color-coded circles representing different environments of origin indicate that the said gene/pathway/transporter was present in that environment (shown if at least two genomes showed presence). Details of genomes can be found in Table S6. Abbreviations for compounds: PEP, phosphoenolpyruvate; PPP, pentose phosphate pathway; PRPP, 5-phosphoribosyl 1-pyrophosphate; CPO, chloroperoxidase; GP, glutathione peroxidase; PP, porphyrinogen peroxidase; THF, tetrahydrofolate; DHF, dihydrofolate; GTP, guanosine 5′-triphosphate; APS, adenylyl sulfate; PAPS, 3′-phosphoadenylyl sulfate; NAD+, nicotinamide adenine dinucleotide.
Basic energy metabolism
Gemmatimonadota from all environments contained basic genes for respiratory metabolism such as NADH:quinone oxidoreductase, cytochrome c oxidase, F-type ATPase, subunits of succinate dehydrogenase involved in oxidative electron transfer chains, or enzymes of heme biosynthesis (Fig. 6). Cytochrome bd ubiquinol oxidase (encoded by cydAB genes) with high affinity for oxygen (46) was present in MAGs from most environments, except for marine water and host-associated ones. The host-associated genomes also lacked succinate dehydrogenase cytochrome b subunit (sdhC), while in marine water MAGs, it was present only in one genome. Genes encoding fumarate reductase, a key enzyme in anaerobic respiration that catalyzes the reduction of fumarate to succinate, were found in host-associated (60%), soda lake sediment (36.9%), hydrothermal vent (33.3%), marine sediment (8.3%), groundwater (14.3%), and marine water (7.14%) MAGs.
Gemmatimonadota also contained genes necessary for central carbohydrate metabolism, including glycolysis (Embden-Meyerhof pathway), gluconeogenesis, tricarboxylic acid cycle, coenzyme A biosynthesis, the aerobic route of oxidation of pyruvate to acetyl-CoA via pyruvate dehydrogenase, and the biosynthesis of phosphoribosyl diphosphate, which is needed to produce purines and pyrimidines (Fig. S5). The ED (Entner-Doudoroff) pathway to obtain pyruvate without glycolysis was reported as less common in Gemmatimonadota (13), and similarly, we saw that the key enzyme for this pathway, 2-dehydro-3-deoxyphosphogluconate aldolase (eda), was present in MAGs from soil and permafrost (52.2% [47 genomes] and 27.3% [6 genomes], respectively) and almost absent in MAGs from marine, fresh, and wastewaters; other sediments; and marine sediments, where it was found only in three genomes at most. Moreover, phosphogluconate dehydratase (edd), which catalyzes another key step in this pathway, was absent in MAGs from all environments. Alternatively, the ED pathway could be supplied through the degradation of D-galacturonate (13), which can be an important carbon source for microorganisms. The pathway for degradation of D-galacturonate was present in soil Gemmatimonadota (key enzymes were present from 18.8% to 54.4% of MAGs, with a complete pathway in 16.6% of MAGs) (Fig. S5).
Gemmatimonadota from all environments encoded representative genes for the non-oxidative phase of the pentose phosphate pathway, while both key enzymes for the oxidative phase (zwf, PGD) were predominately found in permafrost (72.7%), soil (47.7%), and marine sediment genomes (41.6%). A common feature was also the presence of genes encoding the biosynthesis of dTDP-L-rhamnose, an important cell wall component, except for the host-associated MAGs, which lacked two key enzymes (rfbC and rfbD). As these MAGs live in a symbiotic association, it is likely that they do not require these enzymes, as the same pathway seems to be also missing in Alphaproteobacteria associated with marine sponges (47).
Several metabolic pathways were only common in Gemmatimonadota from specific environments. For example, key enzymes of the glyoxylate cycle (isocitrate lyase [aceA] and malate synthase [aceB]) were found in soil genomes (52.2%), other sediments (30.7%), groundwater (28.5%), permafrost (22.7%), wastewater (18.4%), marine sediments (16.6%), marine waters (7.1%), soda lake sediments (4.3%), and fresh waters (3.3%). Bacteria harboring this pathway can assimilate acetate in the absence of complex substrates (48, 49). Moreover, while some of the genes encoding the pathway for conversion of propionyl-CoA to succinyl-CoA occurred in all environments, all key genes (Fig. S5) were present only in MAGs from soda lake sediments (21.7%).
Furthermore, MAGs from all environments showed the potential to degrade polysaccharides. The gene encoding endoglucanase (cellulase) was common among MAGs from all environments, while xylanase (endo-1,4-beta-xylanase, xynA) was present in soil (22.2%), permafrost (9.1%), and sporadically in freshwater (7.7%) and wastewater MAGs (2.04%). Chitinase (chiC) was present in MAGs from wastewater (28.6%), permafrost (27.3%), marine sediments (25%), marine water (23.8%), soil (16.6%), as well as in several genomes of other sediments (7.7%) and fresh waters (8.8%). Additionally, MAGs from permafrost (59.1%), soil (58.8%), groundwater (47.6%), fresh waters (43.9%), hydrothermal vent (16.6%), and wastewater (6.1%) had chitin disaccharide deacetylase (chbG), which is suggested to catalyze the deacetylation of chitin, making it an easily degradable substrate (50).
One of the main storage molecules that helps bacteria survive periods when nutrients or energy sources are scarce is glycogen (51 – 54). The enzymes for its biosynthesis (1,4-alpha-glucan branching enzyme, glucose-1-phosphate adenylyltransferase, and glycogen synthase) were present in freshwater (21.9%), wastewater (18.3%), soil (11.1%), three groundwater, and soda lake sediment genomes. Similarly, the complete pathway for glycogen degradation was mostly found in wastewater (46.9%), other sediment (30.7%), permafrost (18.2%), freshwater (16.5%), and soil (8.8%) genomes. The ability of some bacteria to accumulate glycogen as an energy reserve (51) allows them to quickly activate their metabolism when nutrient availability increases, providing a competitive advantage in nutrient-fluctuating environments.
Carbon fixation strategies
Gemmatimonadota from several different environments contained genes encoding the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO, rbcL), sometimes in two or three copies. There are three forms of RuBisCO (type I, II, and III) that catalyze the carboxylation and oxygenation of ribulose 1,5-bisphosphate (55). The most widespread type I was reported earlier in six Gemmatimonadota MAGs reconstructed from soda lake sediments (28, 29), which were included in this analysis. Genes encoding type I RuBisCO, phylogenetically identified as type IC/ID (Fig. S6), were present in soda lake sediments (28.3%), wastewater (36.7%), groundwater (4.7%), soil (3.3%), freshwater (1.1%), and one MAG from a glacier (Other). Most of these MAGs also contained genes encoding the small subunit of RuBisCO (rbcS) and phosphoribulokinase (Fig. 5). None of the MAGs contained the proteobacterial, α-cyanobacterial (IA), or β-cyanobacterial form (IB) of RuBisCO (56).
Bacterial type II RuBisCO, which is less efficient in discriminating between CO2 and O2 and adapted to environments with low oxygen concentrations (55, 57), was present in wastewater MAGs (36.7%). Type II is commonly found in Proteobacteria (58) and organisms that also have type I (55), as is the case for some of the wastewater MAGs (12.2%), which had both types. In addition, 72 MAGs contained the so-called type IV rbcL gene, which is probably not involved in carbon fixation (55, 59, 60).
Key genes encoding the phosphate acetyltransferase-acetate kinase pathway for carbon fixation, in which acetate produced from acetyl-CoA can be used as a carbon source or electron donor, were found in soda lake sediment (80.4%), permafrost (59.1%), other sediment (38.4%), wastewater (24.5%), freshwater (19.8%), and soil MAGs (11.1%). In the freshwater environment, the presence of the two key genes was observed in two photoheterotrophic species as well as in four MAGs from Spanish reservoirs.
Carbon monoxide (CO), an atmospheric trace gas, can be an alternative energy source for some organoheterotrophic bacteria during organic carbon starvation, enhancing their survival (61 – 63). The gene encoding the large subunit of carbon monoxide dehydrogenase (coxL) is highly abundant in soils where it can facilitate atmospheric CO removal and was reported to be present in soil Gemmatimonadota (61, 64). In this study, we found the coxL gene as well as the genes for small (coxS) and medium (coxM) subunits of carbon monoxide dehydrogenase in host-associated (80%), wastewater (57.1%), hydrothermal vent (50%), groundwater (42.8%), soil (30%), other sediment (23.1%), marine sediments (16.6%), permafrost (9.1%), and freshwater (5.5%) MAGs.
Phototrophy
Many freshwater Gemmatimonadota are aerobic anoxygenic phototrophic (AAP) species (6, 11, 12). A common marker gene for AAP bacteria is the pufM gene, which encodes the M subunit of the bacterial photosynthetic reaction center (65). Here, we identified genes encoding type II photosynthetic reaction centers (pufM and/or pufL) in 51.6% of all freshwater MAGs (Fig. 5), as well as in MAGs from soda lakes (47.8%), other sediments (30.7%), wastewater (16.3%), and group Other (glacier [3 MAGs], biofilm [1 MAG]), while they were absent from all other environments. Interestingly, 28.3% of phototrophic MAGs from soda lake sediments and 8.2% from wastewater also contained type I rbcL, indicating that these species may have the potential for photoautotrophic growth (28, 29).
Many aquatic microorganisms harvest light energy using proton-pumping rhodopsins (66, 67). However, among Gemmatimonadota, this system is very rare. We found genes encoding green- or blue-light-absorbing proteorhodopsins only in five genomes, which originated from deeper layers of freshwater lakes Baikal, Constance, Zurich, and Biwa. A xanthorhodopsin gene was identified in one marine and two glacier MAGs (category Other). One of the MAGs from the glacier also contained genes for BChl-a-based photoheterotrophy, indicating the potential for dual phototrophy (68).
Nitrogen cycle
Nitrogen metabolism in Gemmatimonadota is relatively simple. No nitrogen fixation genes were found in any of the analyzed MAGs, which means that Gemmatimonadota must rely on combined nitrogen sources such as ammonium or amino acids. The gene encoding high-affinity ammonium transporter (Amt), a preferred nitrogen source for microbial growth, was present in MAGs from all environments, as well as the gene encoding nitrilase that hydrolyzes nitriles to ammonia. Branched-chain amino acid transporters were a common feature for host-associated, marine, hydrothermal vent, and wastewater genomes, while spermidine/putrescine or nitrate-nitrite/taurine transporters were more common in wastewater, freshwater, and soil genomes (Fig. 6).
The complete denitrification pathway was not identified in any of the analyzed MAGs. However, the nitrous oxide reductase (nosZ) gene was found in genomes from all environments except marine water and host-associated (Fig. 5). This enzyme catalyzes the final step of denitrification (69 – 72) but is also considered an independent respiratory reaction since it is often found in organisms lacking other genes for denitrification, such as nirK, nirS, and nor (73). In Gemmatimonadota MAGs, the nirK gene (NO−-forming nitrite reductase) was found in all environments; however, the nirS gene was only present in marine sediments and single genomes from hydrothermal vents and wastewater. Gemmatimonadota nosZ genes seem to be one of the most abundant in soil environments (74 – 76), and their high presence in other environments points to their potentially important role in reducing the N2O. Both G. aurantiaca and G. kalamazoonesis have been suggested to use N2O as a substitute for O2 to survive temporary anoxia during transitions between oxic and anoxic states, which can be common in soil or wastewater environments (71, 77). Furthermore, 59.5% of marine, 44.4% of hydrothermal vent, and 28% of host-related MAGs did not have nosZ but contained norB (nitric oxide reductase subunit B), which converts nitric oxide to N2O and could point to their genetic potential to produce N2O. The presence of this gene in host-associated Gemmatimonadota suggests their potential role in nitrogen cycling as part of the marine sponge microbiome (44). Finally, genes for dissimilatory nitrate reduction to ammonia (napAB and nrfAH) were common in soda lake sediments (28), probably due to the anaerobic conditions that can occur in these habitats.
Sulfur cycle
Regarding the sulfur cycle, the distribution of genes encoding enzymes involved in assimilatory sulfate reduction to H2S (sat, cysC, cysNC, cysD, cysH, cysJ, cysI, and sulfite reductase) was patchy (Fig. 5). In this pathway, sulfate is reduced to H2S, which is then incorporated into cysteine, which can be subsequently used for the synthesis of other sulfur-containing molecules (78, 79). The complete pathway was found in the highest numbers in soil (18.9%) and soda lake sediments (10.8%). In fresh water, hydrothermal vents, permafrost, and host-associated environments, it was only present in up to three genomes. Genes encoding for the sulfate transport system, which enables sulfate-sulfur assimilation, were mostly found in wastewater MAGs, with a lower occurrence in soil and permafrost MAGs. Furthermore, the complete sox enzyme system, involved in thiosulfate oxidation to SO4 2−, was not found in any Gemmatimonadota genomes, although they contained some genes (soxZ, soxY, soxC, or soxB), depending on the environment.
Phosphate
Phosphate is one of the main biogenic elements required for the biosynthesis of nucleic acids and lipids. Due to its low natural availability, it is the limiting nutrient in many natural environments. The main route for its uptake in Gemmatimonadota from all environments was the high-affinity phosphate transport system (pstSCAB), and they could regulate its acquisition through the PhoR-PhoB two-component system. Additionally, marine (19%), two MAGs from soda lake sediments, and hydrothermal vents had an uptake system for phosphonate (phnCDE), a good source of phosphorus under phosphate starvation (80). During phosphorus starvation, many bacteria can produce alkaline phosphatases (phoA, phoX, phoD), which catalyze hydrolysis of phosphoesters (81, 82). PhoA was found in Gemmatimonadota from all environments, with the lowest numbers in host-associated MAGs (8%) and the highest in permafrost MAGs (68.2%). In contrast, phoX was present in up to two MAGs in marine sediment and fresh waters and was generally found in lower numbers in all environments except for host-associated (92%), marine water (57.1%), and wastewater (55.1%). Finally, phoD was highly present in freshwater (92.3%), wastewater (69.4%), and hydrothermal vent (66.6%) MAGs. The polyphosphate kinase gene used for the accumulation of polyphosphate was present in all Gemmatimonadota MAGs except marine. The presence of all these genes, which are crucial during phosphorus limitation, as well as the high-affinity phosphate transport system, indicates that Gemmatimonadota has different strategies to cope with phosphorus limitations.
Protection against oxidative stress
Gemmatimonadota is composed of mostly aerobic organisms that depend on aerobic respiration. Therefore, their genomes encode many proteins involved in the protection from oxidative damage and stress that is associated with an aerobic lifestyle (Fig. 6). [Fe-Mn] and [Cu-Zn] families of superoxide dismutases were present in MAGs from all environments, except for marine and hydrothermal vents, where the [Cu-Zn] family was not found. Cytochrome c peroxidase also occurred in all environments, while glutathione peroxidase was found in high numbers in wastewater (53.1%) and fresh water (38.4%), and in other environments was present only a in a few representatives or not at all. Catalase peroxidase katG was present in all MAGs except soil, while catalase katE was found in soda lake sediments (39.1%), other sediments (23%), wastewater (22.4%), permafrost (18.2%), soils (12.2%), and only one freshwater bacterium, G. groenlandica (6). From other types of peroxidases, chloroperoxidase occurred in all environments, while porphyrinogen peroxidase was only present in two MAGs from soil and one from soda lakes and marine sediments.
So far, all cultured Gemmatimonadota contain large amounts of carotenoids. These pigments protect cells from excess light as well as against reactive oxygen species, and in AAP species, they can act as additional light-harvesting pigments (11, 83). Gemmatimonadota MAGs from wastewaters (85.7%, 67.3%), fresh waters (58.2%, 63.7%), other sediments (69.2%, 76.9%), soda lake sediments (39.13%, 91.3%), and soils (24.4%, 21.11%) contained genes encoding the initial parts for carotenoid biosynthesis, phytoene synthase (crtB) and phytoene dehydrogenase (crtI), respectively. They were almost absent in marine, permafrost, marine sediment, hydrothermal vent, and groundwater genomes, where they were found in up to two genomes. Host-associated genomes contained the crtI gene (24%), but crtB was present only in one genome. Other carotenoid biosynthesis genes found were β-carotene ketolase (crtO), present in all environments; lycopene beta-cyclase (crtY), found in a small number of soil and freshwater MAGs; up to three genomes of wastewaters, permafrost, marine water, marine sediments, and soda lake sediments; and β-carotene 3-hydroxylase (crtZ), found only in several freshwater and wastewater genomes.
Cofactors and vitamins
Gemmatimonadota cultures require a mixture of vitamins like biotin (vitamin B7), folic acid (vitamin B9), nicotinic acid (vitamin B3), pantothenic acid (vitamin B5), and cobalamin (vitamin B12) for growth (1 – 5). All analyzed MAGs contained the complete pathway for molybdenum cofactor synthesis. Molybdenum is a cofactor in numerous enzymes in prokaryotic and eukaryotic organisms (84). Folate biosynthesis could be inferred as complete in all environments, given that marine, host-associated, wastewater, and other sediment MAGs that lack one of the key enzymes (folA) have the gene encoding thyX, suggested to function as folA (85). Furthermore, MAGs from most environments encode genes involved in pantothenate biosynthesis, a precursor of coenzyme A, an essential molecule in metabolism. The exceptions were marine, freshwater, and host-associated MAGs, which lacked one of the key enzymes (panD). Genes encoding the biosynthesis of biotin, an essential cofactor of enzymes involved in fatty acid synthesis or amino acid metabolism (86), were present mostly in freshwater, wastewater, permafrost, other sediment, and several soil MAGs. In other environments, several genes for biotin biosynthesis were missing. Genes for NAD+ biosynthesis, an important metabolite and cofactor involved in nucleotide synthesis, were only sporadically present in host-associated MAGs, probably due to their incompleteness. Another biosynthetic pathway, the kynurenine pathway that leads to quinolinate, a precursor of NAD (87), was present in freshwater and wastewater MAGs. Soda lake sediment and host-associated MAGs had several genes involved in the late steps of cobalamin biosynthesis from cobyrinate a,c-diamide; however, genes involved in both aerobic and anaerobic cobalamin pathways were not found. This suggests that these genes may be used in the salvage pathway as a more effective way for obtaining cobalamin since the btuB transporter and tonB protein, which function together in cobalamin transport (88), were present in Gemmatimonadota.
Other genes
Genes for flagellar assembly were present in almost all wastewater (93.8%) and nearly half of freshwater (47.3%) MAGs (Fig. 5). This included five of the Gemmatimonadota MAGs from Spanish reservoirs and many limnic and planktonic MAGs from Římov Reservoir (12). Additionally, the presence of flagella was already shown for both freshwater cultures G. phototrophica and G. groenlandica (8). Smaller numbers were found in other sediments (38.4%), groundwater (23.8%), soda lake sediments (19.6%), and permafrost (13.6%). Host-associated and marine MAGs did not contain flagellar genes, while in soil, hydrothermal vents, and marine sediments, they only occurred in one or two genomes, respectively. Additionally, genomes from all environments but marine had genes encoding type IV pili (Fig. 6).
Some Gemmatimonadota also display different enzymes for degrading alkanes, which they may potentially use as a source of carbon and energy (89). Alkanes are naturally found in environments from sources like decaying microorganisms, algae, or plants but are also present in high content in crude oil, which can be a contaminant for the environment (89). The gene encoding alkanesulfonate monooxygenase (alkB), which degrades short alkanes (90), was found in the highest numbers in soil (30%), wastewater (18.4%), freshwater (14.3%), and marine MAGs (14.3%). Alkane-1-monooxygenase (alkM), used in the degradation of longer alkanes (90), was less common and was missing from most environments except marine (45.2%), host-associated (36%), and freshwater (8.8%) MAGs. The presence of these genes could suggest a potentially ecologically relevant role in the biodegradation of hydrocarbons in Gemmatimonadota. This potential for biodegradation is also evident in Gemmatimonadota MAGs from permafrost and marine sediments, which have genes involved in the degradation of benzoyl-CoA, a central intermediate of synthetic aromatic compounds (91). Moreover, Gemmatimonadota from wastewater, fresh water, hydrothermal vents, and one genome from marine sediments seems to be able to degrade 1,2- dichloroethane (13), an industrially produced pollutant in aquatic environments (92). Furthermore, Gemmatimonadota seems to utilize glycolate, converting it to glyoxylate, as they have genes encoding glycolate oxidase, a protein complex that consists of three subunits D, E, and F (glcDEF). This could explain the previous observation of a close association of limnic Gemmatimonadota with phytoplankton in freshwater environments (12), since glycolate is one of the most common cyanobacterial and algal exudates that can be utilized by bacteria (93 – 95). Additionally, the glc operon contains malate synthase G (glcB) that further converts glyoxylate to malate, which is then used for energy production in the TCA cycle (96).
Several different antimicrobial compounds and multidrug transport systems were present in all Gemmatimonadota, while importers and efflux systems of ions like Fe2+, Mg2+, Mn2+, Zn2+, and other heavy metals were present in different environments (Fig. 6). The Na+/H+ antiporter system to remove Na+ from cells, as well as Kch voltage-gated K+ channels, important in all prokaryotes for maintaining cellular homeostasis (31, 97), were present in all Gemmatimonadota. Trk-type, fast but low-affinity K+ transporter (97) predominated in marine water, marine sediment, and hydrothermal vent genomes, while K+ channels and the K+/H+ antiporter system predominated in MAGs from soda lake sediments, freshwater, soil, permafrost, and wastewater. To deal with hypo-osmotic stress, Gemmatimonadota from all environments had aquaporins, water channels that ease the water stress by enabling fast water efflux (98), and two types of mechanosensitive channels, MscL and/or MscS, which also helped cells return to normal, isotonic size (99).
Conclusions
We have explored a large data set of Gemmatimonadota MAGs to characterize their metabolic potential in different environments. Phylogenomics and gene content analyses indicated that Gemmatimonadota have diverse and flexible metabolisms and the ability to adapt to different conditions. A common feature of all MAGs was aerobic organoheterotrophy, but many pathways were specific to some environments. For instance, photoheterotrophy and motility (flagella) were more prevalent in freshwaters, soda lakes, and wastewaters, whereas CO oxidation was more common in soils, marine sediments, hydrothermal vents, host-associated, and groundwater. Differences between environments could also be observed in their genomes’ sizes and GC content. The size and GC content of marine MAGs were the lowest, which is a common adaptation of marine bacteria to oligotrophic conditions. Moreover, Gemmatimonadota exhibit different strategies for survival under phosphorus limitation, some of which are present in all genomes, and some, like the uptake system for phosphonate, are more common in marine and hydrothermal vent genomes, or different alkaline phosphatases like phoD or phoX are more common in host-associated, freshwater, or wastewater genomes. Gemmatimonadota are unable to fix nitrogen; however, a potential environmental role in the reduction of N2O is highlighted by the presence of nosZ genes in all Gemmatimonadota except for marine, host-associated, and hydrothermal vents, in which the presence of norB could suggest that they may rather produce N2O. Finally, pathways for the degradation of synthetic solvents and aromatic compounds found in some Gemmatimonadota point to their potential role in biodegradation in the environment, and the ability to utilize glycolate indicates a potential symbiotic relationship with phytoplankton.
MATERIALS AND METHODS
Sampling, sequencing, and assembly
Samples collected from four Spanish freshwater reservoirs (Amadorio, Tous, Benageber, and Loriguilla) (56) were reused in this analysis. All four monomictic reservoirs are located in the semi-arid eastern region of Spain, close to the Mediterranean Sea. Briefly, for each reservoir, samples were taken in two campaigns (March—winter mixing period and September/October—summer stratification period in 2020) at three locations: the dam (epilimnion, deep chlorophyll maximum-DCM, and hypolimnion in summer; and epilimnion and hypolimnion in winter), the outlet of the reservoir, and tailwaters (0.5 m). In all cases, water samples were filtered through a series of 20, 5, and 0.22-µm filters, and DNA was extracted as described in reference (56). DNA extracted from 0.22-µm filters was sequenced with Illumina NovaSeq.
Metagenomes were assembled with IDBA-UD (100), which resulted in approximately 5,000 contigs >5 kb per metagenome, which were used for further binning. Binning was conducted using METABAT2 (101), and we obtained a total of 16 MAGs ascribed to the Gemmatimonadota phylum (Table S1). A quality check of 16 MAGs was made using the CheckM v1.1.3 package (102). All MAGs had <5% contamination, while completeness ranged from the lowest 67.79% to 100%.
Analyzed data set
The obtained MAGs from the Spanish reservoirs were expanded with five cultured representatives and all publicly available MAGs of Gemmatimonadota from NCBI (downloaded on 3 May 2021) together with previously published freshwater Gemmatimonadota (12). All genomes (731) (Table S2) were checked for completeness and contamination using the CheckM package (102), and 68 MAGs with completeness below 50% and/or contamination above 10% were removed from further analysis. Moreover, basic metadata such as the environmental origin, assembly size, estimated genome size, GC content, median intergenic spacer, and coding density were obtained for each genome. Environmental origin and assembly size were collected from NCBI; GC content, median intergenic spacer, and coding density were calculated using the in-house pipeline; and estimated genome size was calculated based on the formula: (total sequence length/completeness) × (100 − contamination). All of these were taxonomically classified with the Genome Taxonomy Database (GTDB-Tk) with default settings (103). This identified 57 ambiguous genomes, which were re-classified as Latescibacterota and several other closely related bacterial phyla (e.g., Eisenbacteria, Krumholzibacteriota). These genomes were also removed from all subsequent analyses. Finally, in order to avoid bias and reduce redundancy, the remaining genomes were dereplicated using dRep v 2.3.3 (104), with parameters: -comp 50 -pa 0.99 -sa 0.995. The final data set consisted of 442 MAGs (completeness >67%, a value chosen because it represents 2/3 of the genome) that were divided based on their environmental origin into 12 different categories: freshwater, soil, marine water, marine sediment, hydrothermal vent, permafrost, soda lake sediments, other sediments, host-associated (with marine sponges and coral), wastewater, groundwater, and Other. The latter category (Other) included MAGs from glaciers (6), biofilm (1), bioreactors (2), fossil (1), compost (1), hot springs (2), and an unknown metagenome (1) (Table S2). Since the environments in this category were too diverse to be considered together, the MAGs were excluded from all of the analyses except for phylogenomics of Gemmatimonadota genomes, the RuBisCO tree, and phototrophy in metabolic analysis. Coding density plots showing comparison of estimated genome size with percent GC, number of CDS, and median intergenic spacers (bp) were performed for all genomes that had >67% completeness, excluding the category Other. The graphs were plotted using SigmaPlot v.14.0 and Rstudio v.3.6.1 (package ggplot2). The map in Fig. 1 was made in R Studio v.3.6.1 (package Maps), using data from Natural Earth, supported by NACIS (North American Cartographic Information Society) and free for use. All the graphs were edited in Inkscape v.1.0.
Analysis of core and accessory genes of Gemmatimonadota from different habitats
The analysis of core and accessory genes from Gemmatimonadota genomes was done using the GET_HOMOLOGUES package based on diamond blastp and OMCL algorithms with default parameters (105). Only environments where MAGs/cultured genomes had completeness above 90% were analyzed. These included the following categories: soil (34), freshwater (29), soda lake sediments (20), host-associated (14), wastewater (13), marine (10), and permafrost (9). To avoid bias due to redundant genomes and variability in the completeness of similar MAGs, genomes with ≥98% average nucleotide identity in the same environment were excluded. This analysis must be treated with caution due to the varying level of completeness of the analyzed MAGs regardless of the environment where they originated and the differing numbers of MAGs used for each environment. The average number of core, soft core, shell, and cloud genes was calculated for each of these environmental groups (Table S3). Core genes were defined as being present in all considered genomes of the analyzed environment, and soft core genes were defined as being present in 95% of them. The shell category comprises moderately conserved genes present in <90% of compared genomes. Finally, cloud genes are rare genes present in only one or two genomes (105).
PCoA/clustering plots and phylogenies
A principal coordinate ordination analysis with SEED (106) gene presence/absence (Table S4) was conducted for all genomes (excluding the category Other), with completeness higher than 67%. Briefly, a Kulczynski resemblance matrix based on SEED presence/absence gene values was obtained, and the derived triangular matrix was used to obtain a clustering and PCoA analysis where all genomes were distributed accordingly. Additionally, SIMPER analysis was done with the same SEED presence/absence gene values (Table S4) using Bray-Curtis (Table S5). Differences in dispersion of genes were tested by performing an analysis of PERMDISP (107) that includes pairwise comparisons of environments. To test for significant differences between environments, a PERMANOVA was performed using 9,999 permutations. All the calculations were conducted with PRIMER7 software (Primer Ltd., Lutton, UK), and the obtained graph was further edited in Inkscape v.1.0.
Phylogenomic analysis of Gemmatimonadota genomes was done with the PhyloPhlAn 3.0 tool (42, 108). Three genomes from the bacterial phylum Fibrobacterota were used as an outgroup (GCA_900142455.1 Hallerella intestinalis, GCA_900217845.1 Fibrobacter elongatus, GCA_000146505.1 F. succinogenes). PhyloPhlAN uses USEARCH (109) to screen for the presence of 400 universally conserved and ubiquitous proteins (found in the PhyloPhlAn database). The alignments of proteins against the built-in database were done using MUSCLE (110), concatenated, and used to generate a maximum-likelihood tree with RAxML (111). The tree was visualized in iTOL (112) and edited using Inkscape v.1.0.
A RuBisCO tree was constructed with representative sequences of the large subunit (rbcL/cbbL genes) from various types, including type IA, IB, IC, ID, II, intermediary II/III, III, IV, and archaeal types. This RuBisCO data set was aligned in Geneious Prime (version 2022.2.2) using MAFFT alignment (113, 114) (n = 508). Sequences that were not obtained from Gemmatimonadota MAGs were downloaded from UniProt (115) or obtained from previous studies (19, 56, 116, 117). A maximum-likelihood phylogenetic tree was calculated using IQ-TREE (118), with the LG + F + I + G4 substitution model chosen as the best-fitting model by ModelFinder according to the Bayesian Information Criterion (BIC) (119), and 1,000 ultrafast bootstrap replicates. The tree was visualized in iTOL (112) and edited using Inkscape v.1.0.
Metabolic analysis
This analysis was conducted on genomes with >67% completeness. Gene predictions were performed with PROKKA (120) and diamond (v0.9.14.115), and blastp was used to search versus the KEGG/SEED databases (Table S4). Metabolic features of MAGs were also analyzed with the RAST annotation pipeline database (106) and through BlastKOALA (121), which allowed us to obtain KO identifiers (K numbers) for orthologous genes present in all MAGs (Table S6). Metabolic pathways were then inferred from KEGG (121) and SEED (106) and manually examined for completeness. The percentages of the presence of key genes and pathways were calculated for each environment (Table S6). Plots showing the percentage of the presence of specific metabolic pathways were done in Rstudio (package bubbleplot) and edited in Inkscape 1.0. In Fig. S5, the genomes that had the majority of the genes (>60%) related to flagella were considered to have them present. The figure of metabolic reconstruction was done in Inkscape 1.0, following Table S6.
ACKNOWLEDGMENTS
The authors thank Alastair T. Gardiner for language corrections.
This work was supported by project PhotoGemm+, no. 19-28778X from the Czech Science Foundation, awarded to M.K. I.M. was additionally supported by an Erasmus + traineeship grant from the University of South Bohemia, Česke Budejovice (No. 16/2020-2021/PRF). P.J.C.-Y. was supported by an APOSTD/2019/009 Post-Doctoral Fellowship from the Generalitat Valenciana and is currently supported by a Post-Doctoral Fellowship from the Fundación Alfonso Martín Escudero, Spain. This work was supported by grants “VIREVO” CGL2016-76273-P (MCI/AEI/FEDER, EU) (cofounded with FEDER funds) to F.R.-V. and CLIMAWET-CONS (PID2019-104742RB-I00) to A.C., both from the Spanish Ministerio de Ciencia e Innovación and Agencia Estatal de Investigación, as well as “HIDRAS3” PROMETEO/2019/009 from the Generalitat Valenciana granted to both researchers. F.R.-V. was also the beneficiary of the 5top100-program of the Ministry for Science and Education of Russia.
P.J.C.-Y., A.C., I.M., and M.K. conceived the study. P.J.C.-Y., A.P., and A.C. performed the sampling campaigns from which metagenomes were derived. I.M., P.J.C.-Y., C.V.-A., and K.P. analyzed the sequence data. I.M. and C.V.-A. prepared the figures. F.R.-V., A.C., and M.K. provided the funding. I.M., M.K., and P.J.C.-Y. wrote the manuscript. All authors read, provided comments, and approved the manuscript.
The authors declare that there are no competing interests.
Contributor Information
Izabela Mujakić, Email: mujakic@alga.cz.
Michal Koblížek, Email: koblizek@alga.cz.
Sandi Orlic, Institut Ruder Boskovic, Zagreb, Croatia .
DATA AVAILABILITY
All data derived from this work are publicly available in NCBI-GenBank databases. All 16 MAGs assembled in this study have been deposited in the NCBI-GenBank database under Bioproject number PRJNA721863, biosample numbers SAMN32886101-SAMN32886116, and GenBank accession numbers JARIER000000000-JARIFG000000000. All these genomes were derived from metagenomic data sets from Spanish lakes and reservoirs that were previously deposited under Bioproject number PRJNA721863 and SRA numbers SRR15198238- SRR15198275.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.01112-23.
Fig. S1 to S6.
Basic characteristics of newly assembled Gemmatimonadota MAGs.
Basic characteristics of all Gemmatimonadota genomes.
Percentages of habitat-related core and accessory genes.
Presence/absence of genes in Gemmatimonadota based on SEED.
SIMPER analysis of similarity and dissimilarity between Gemmatimonadota genomes.
Genes and metabolic pathways found in Gemmatimonadota genomes from different environments.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Zhang H, Sekiguchi Y, Hanada S, Hugenholtz P, Kim H, Kamagata Y, Nakamura K. 2003. Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum gemmatimonadetes phyl. nov. Int J Syst Evol Microbiol 53:1155–1163. doi: 10.1099/ijs.0.02520-0 [DOI] [PubMed] [Google Scholar]
- 2. DeBruyn JM, Fawaz MN, Peacock AD, Dunlap JR, Nixon LT, Cooper KE, Radosevich M. 2013. Gemmatirosa kalamazoonesis gen. nov., sp. nov., a member of the rarely-cultivated bacterial phylum gemmatimonadetes. J Gen Appl Microbiol 59:305–312. doi: 10.2323/jgam.59.305 [DOI] [PubMed] [Google Scholar]
- 3. Pascual J, Foesel BU, Geppert A, Huber KJ, Boedeker C, Luckner M, Wanner G, Overmann J. 2018. Roseisolibacter agri gen. nov. sp. nov., a novel slow-growing member of the under-represented phylum gemmatimonadetes. Int J Syst Evol Microbiol 68:1028–1036. doi: 10.1099/ijsem.0.002619 [DOI] [PubMed] [Google Scholar]
- 4. Pascual J, García-López M, Bills GF, Genilloud O. 2016. Longimicrobium terrae gen. nov. sp. nov., an oligotrophic bacterium of the under-represented phylum gemmatimonadetes isolated through a system of miniaturized diffusion chambers. Int J Syst Evol Microbiol 66:1976–1985. doi: 10.1099/ijsem.0.000974 [DOI] [PubMed] [Google Scholar]
- 5. Zeng Y, Selyanin V, Lukeš M, Dean J, Kaftan D, Feng F, Koblížek M. 2015. Characterization of the microaerophilic, bacteriochlorophyll a-containing bacterium Gemmatimonas phototrophica sp. nov., and emended descriptions of the genus Gemmatimonas and Gemmatimonas aurantiaca. Int J Syst Evol Microbiol 65:2410–2419. doi: 10.1099/ijs.0.000272 [DOI] [PubMed] [Google Scholar]
- 6. Zeng Y, Wu N, Madsen AM, Chen X, Gardiner AT, Koblížek M. 2020. Gemmatimonas groenlandica sp. nov. is an aerobic anoxygenic phototroph in the phylum gemmatimonadetes. Front Microbiol 11:606612. doi: 10.3389/fmicb.2020.606612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mujakić I, Piwosz K, Koblížek M. 2022. Phylum gemmatimonadota and its role in the environment. Microorganisms 10:151. doi: 10.3390/microorganisms10010151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shivaramu S, Tomasch J, Kopejtka K, Nupur N, Saini MK, Bokhari SNH, Küpper H, Koblížek M. 2022. The influence of calcium on the growth morphology and gene regulation in gemmatimonas phototrophica. Microorganisms 11:27. doi: 10.3390/microorganisms11010027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koblížek M, Dachev M, Bína D, Piwosz K, Kaftan D. 2020. Utilization of light energy in phototrophic gemmatimonadetes. J Photochem Photobiol B Biol 213:112085. doi: 10.1016/j.jphotobiol.2020.112085 [DOI] [PubMed] [Google Scholar]
- 10. Qian P, Gardiner AT, Šímová I, Naydenova K, Croll TI, Jackson PJ, Kloz M, Čubáková P, Kuzma M, Zeng Y, Castro-hartmann P, Van KB, Goldie KN, Kaftan D, Hrouzek P, Hájek J, Agirre J, Siebert CA, Bína D, Sader K, Stahlberg H, Sobotka R, Russo CJ, Polívka T, Hunter CN, Koblížek M. 2022. 2.4-Å structure of the double-ring Gemmatimonas phototrophica photosystem Sci Adv 8:1–11. doi: 10.1126/sciadv.abk3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeng Y, Feng F, Medová H, Dean J, Koblížek M. 2014. Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum gemmatimonadetes. Proc Natl Acad Sci U S A 111:7795–7800. doi: 10.1073/pnas.1400295111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mujakić I, Andrei A-Ş, Shabarova T, Fecskeová LK, Salcher MM, Piwosz K, Ghai R, Koblížek M. 2021. Common presence of phototrophic gemmatimonadota in temperate freshwater lakes. mSystems 6:e01241-20. doi: 10.1128/mSystems.01241-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng X, Dai X, Zhu Y, Yang J, Jiang H, Dong H, Huang L, Hallam SJ. 2022. (Meta)genomic analysis reveals diverse energy conservation strategies employed by globally distributed gemmatimonadota. mSystems 7:e0022822. doi: 10.1128/msystems.00228-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zeng Y, Baumbach J, Barbosa EGV, Azevedo V, Zhang C, Koblížek M. 2016. Metagenomic evidence for the presence of phototrophic gemmatimonadetes bacteria in diverse environments. Environ Microbiol Rep 8:139–149. doi: 10.1111/1758-2229.12363 [DOI] [PubMed] [Google Scholar]
- 15. Janssen PH. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N. 2018. A global atlas of the dominant bacteria found in soil. Science 359:320–325. doi: 10.1126/science.aap9516 [DOI] [PubMed] [Google Scholar]
- 17. Morrison JM, Baker KD, Zamor RM, Nikolai S, Elshahed MS, Youssef NH, Aziz RK. 2017. Spatiotemporal analysis of microbial community dynamics during seasonal stratification events in a freshwater lake (Grand Lake, OK, USA). PLoS ONE 12:e0177488. doi: 10.1371/journal.pone.0177488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Villena-Alemany C, Mujakić I, Porcal P, Koblížek M, Piwosz K. 2023. Diversity dynamics of aerobic anoxygenic phototrophic bacteria in a freshwater Lake. Environ Microbiol Rep 15:60–71. doi: 10.1111/1758-2229.13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vavourakis CD, Andrei A-S, Mehrshad M, Ghai R, Sorokin DY, Muyzer G. 2018. A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments. Microbiome 6:168. doi: 10.1186/s40168-018-0548-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nunoura T, Hirai M, Yoshida-Takashima Y, Nishizawa M, Kawagucci S, Yokokawa T, Miyazaki J, Koide O, Makita H, Takaki Y, Sunamura M, Takai K. 2016. Distribution and niche separation of planktonic microbial communities in the water columns from the surface to the hadal waters of the Japan trench under the Eutrophic ocean. Front Microbiol 7:1261. doi: 10.3389/fmicb.2016.01261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thiel V, Neulinger SC, Staufenberger T, Schmaljohann R, Imhoff JF. 2007. Spatial distribution of sponge-associated bacteria in the Mediterranean sponge tethya aurantium. FEMS Microbiol Ecol 59:47–63. doi: 10.1111/j.1574-6941.2006.00217.x [DOI] [PubMed] [Google Scholar]
- 22. Slaby BM, Hackl T, Horn H, Bayer K, Hentschel U. 2017. Metagenomic binning of a marine sponge microbiome reveals unity in defense but metabolic specialization. ISME J 11:2465–2478. doi: 10.1038/ismej.2017.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kato S, Nakawake M, Kita J, Yamanaka T, Utsumi M, Okamura K, Ishibashi J-I, Ohkuma M, Yamagishi A. 2013. Characteristics of microbial communities in crustal fluids in a deep-sea hydrothermal field of the suiyo seamount. Front Microbiol 4:85. doi: 10.3389/fmicb.2013.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nunoura T, Nishizawa M, Hirai M, Shimamura S, Harnvoravongchai P, Koide O, Morono Y, Fukui T, Inagaki F, Miyazaki J, Takaki Y, Takai K. 2018. Microbial diversity in sediments from the bottom of the challenger deep, the mariana trench. Microbes Environ 33:186–194. doi: 10.1264/jsme2.ME17194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cui G, Li J, Gao Z, Wang Y. 2019. Spatial variations of microbial communities in abyssal and hadal sediments across the challenger deep. PeerJ 7:e6961. doi: 10.7717/peerj.6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peoples LM, Grammatopoulou E, Pombrol M, Xu X, Osuntokun O, Blanton J, Allen EE, Nunnally CC, Drazen JC, Mayor DJ, Bartlett DH. 2019. Microbial community diversity within sediments from two geographically separated hadal trenches. Front Microbiol 10:347. doi: 10.3389/fmicb.2019.00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vipindas PV, Mujeeb RKM, Jabir T, Thasneem TR, Mohamed Hatha AA. 2020. Diversity of sediment bacterial communities in the south eastern Arabian sea. Reg Stud Mar Sci 35:101153. doi: 10.1016/j.rsma.2020.101153 [DOI] [Google Scholar]
- 28. Vavourakis CD, Mehrshad M, Balkema C, van Hall R, Andrei A-Ş, Ghai R, Sorokin DY, Muyzer G. 2019. Metagenomes and metatranscriptomes shed new light on the microbial-mediated sulfur cycle in a siberian soda Lake. BMC Biol 17:69. doi: 10.1186/s12915-019-0688-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zorz JK, Sharp C, Kleiner M, Gordon PMK, Pon RT, Dong X, Strous M. 2019. A shared core microbiome in soda lakes separated by large distances. Nat Commun 10:4230. doi: 10.1038/s41467-019-12195-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Giovannoni SJ, Cameron Thrash J, Temperton B. 2014. Implications of streamlining theory for microbial ecology. ISME J 8:1553–1565. doi: 10.1038/ismej.2014.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salcher MM, Schaefle D, Kaspar M, Neuenschwander SM, Ghai R. 2019. Evolution in action: habitat transition from sediment to the pelagial leads to genome streamlining in methylophilaceae. ISME J 13:2764–2777. doi: 10.1038/s41396-019-0471-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappé MS, Short JM, Carrington JC, Mathur EJ. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245. doi: 10.1126/science.1114057 [DOI] [PubMed] [Google Scholar]
- 33. Thorpe HA, Bayliss SC, Hurst LD, Feil EJ. 2017. Comparative analyses of selection operating on nontranslated intergenic regions of diverse bacterial species. Genetics 206:363–376. doi: 10.1534/genetics.116.195784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Foerstner KU, von Mering C, Hooper SD, Bork P. 2005. Environments shape the nucleotide composition of genomes. EMBO Rep 6:1208–1213. doi: 10.1038/sj.embor.7400538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM, Luo H, Wright JJ, Landry ZC, Hanson NW, Thompson BP, Poulton NJ, Schwientek P, Acinas SG, Giovannoni SJ, Moran MA, Hallam SJ, Cavicchioli R, Woyke T, Stepanauskas R. 2013. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci U S A 110:11463–11468. doi: 10.1073/pnas.1304246110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McEwan CEA, Gatherer D, McEwan NR. 1998. Nitrogen-fixing aerobic bacteria have higher genomic GC content than non-fixing species within the same genus. Hereditas 128:173–178. doi: 10.1111/j.1601-5223.1998.00173.x [DOI] [PubMed] [Google Scholar]
- 37. Mann S, Chen YPP. 2010. Bacterial genomic G + C composition-eliciting environmental adaptation. Genomics 95:7–15. doi: 10.1016/j.ygeno.2009.09.002 [DOI] [PubMed] [Google Scholar]
- 38. Konstantinidis KT, Tiedje JM. 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci U S A 101:3160–3165. doi: 10.1073/pnas.0308653100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Numberger D, Ganzert L, Zoccarato L, Mühldorfer K, Sauer S, Grossart HP, Greenwood AD. 2019. Characterization of bacterial communities in wastewater with enhanced taxonomic resolution by full-length 16S rRNA sequencing. Sci Rep 9:9673. doi: 10.1038/s41598-019-46015-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaas RS, Friis C, Ussery DW, Aarestrup FM. 2012. Estimating variation within the genes and Inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 13:577. doi: 10.1186/1471-2164-13-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wolf YI, Makarova KS, Yutin N, Koonin EV. 2012. Updated clusters of orthologous genes for archaea: a complex ancestor of the archaea and the byways of horizontal gene transfer. Biol Direct 7:1–15. doi: 10.1186/1745-6150-7-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Segata N, Börnigen D, Morgan XC, Huttenhower C. 2013. Phylophlan is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4:2304. doi: 10.1038/ncomms3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rashighi M, Harris JE. 2017. Phylophlan is a new method for improved phylogenetic and taxonomic placement of microbes. Physiol Behav 176:139–148.28363838 [Google Scholar]
- 44. Engelberts JP, Robbins SJ, de Goeij JM, Aranda M, Bell SC, Webster NS. 2020. Characterization of a sponge microbiome using an integrative genome-centric approach. ISME J 14:1100–1110. doi: 10.1038/s41396-020-0591-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robbins SJ, Singleton CM, Chan CX, Messer LF, Geers AU, Ying H, Baker A, Bell SC, Morrow KM, Ragan MA, Miller DJ, Forêt S, Voolstra CR, Tyson GW, Bourne DG, ReFuGe2020 Consortium . 2019. A genomic view of the reef-building coral porites lutea and its microbial symbionts. Nat Microbiol 4:2090–2100. doi: 10.1038/s41564-019-0532-4 [DOI] [PubMed] [Google Scholar]
- 46. Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta 1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Engelberts JP, Robbins SJ, Herbold CW, Moeller FU, Jehmlich N, Laffy PW, Wagner M, Webster NS. 2023. Metabolic reconstruction of the near complete microbiome of the model sponge Ianthella basta. Environ Microbiol 25:646–660. doi: 10.1111/1462-2920.16302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kornberg HL. 1966. The role and control of the glyoxylate cycle in Escherichia coli. Biochem J 99:1–11. doi: 10.1042/bj0990001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang KH, Tang YJ, Blankenship RE. 2011. Carbon metabolic pathways in phototrophic bacteria and their broader evolutionary implications. Front Microbiol 2:165. doi: 10.3389/fmicb.2011.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao Y, Park RD, Muzzarelli RAA. 2010. Chitin deacetylases: properties and applications. Mar Drugs 8:24–46. doi: 10.3390/md8010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wilson WA, Roach PJ, Montero M, Baroja-Fernández E, Muñoz FJ, Eydallin G, Viale AM, Pozueta-Romero J. 2010. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev 34:952–985. doi: 10.1111/j.1574-6976.2010.00220.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He S, Stevens SLR, Chan L-K, Bertilsson S, Glavina Del Rio T, Tringe SG, Malmstrom RR, McMahon KD. 2017. Ecophysiology of freshwater verrucomicrobia inferred from. mSphere 2:e00277-17. doi: 10.1128/mSphere.00277-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang L, Wise MJ. 2011. Glycogen with short average chain length enhances bacterial durability. Naturwissenschaften 98:719–729. doi: 10.1007/s00114-011-0832-x [DOI] [PubMed] [Google Scholar]
- 54. Wang L, Liu Q, Hu J, Asenso J, Wise MJ, Wu X, Ma C, Chen X, Yang J, Tang D. 2018. Structure and evolution of glycogen branching enzyme N-termini from bacteria. Front Microbiol 9:3354. doi: 10.3389/fmicb.2018.03354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. 2008. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc Lond B Biol Sci 363:2629–2640. doi: 10.1098/rstb.2008.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cabello-Yeves PJ, Scanlan DJ, Callieri C, Picazo A, Schallenberg L, Huber P, Roda-Garcia JJ, Bartosiewicz M, Belykh OI, Tikhonova IV, Torcello-Requena A, De Prado PM, Millard AD, Camacho A, Rodriguez-Valera F, Puxty RJ. 2022. α-cyanobacteria possessing form IA RuBisCO globally dominate aquatic habitats. ISME J 16:2421–2432. doi: 10.1038/s41396-022-01282-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Alfreider A, Tartarotti B. 2019. Spatiotemporal dynamics of different CO2 fixation strategies used by prokaryotes in a dimictic lake. Sci Rep 9:15068. doi: 10.1038/s41598-019-51584-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. 2008. Distinct form I, II, III, and IV rubisco proteins from the three kingdoms of life provide clues about rubisco evolution and structure/function relationships. J Exp Bot 59:1515–1524. doi: 10.1093/jxb/erm361 [DOI] [PubMed] [Google Scholar]
- 59. Hanson TE, Tabita FR. 2001. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc Natl Acad Sci U S A 98:4397–4402. doi: 10.1073/pnas.081610398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ashida H, Saito Y, Kojima C, Kobayashi K, Ogasawara N, Yokota A. 2003. A functional link between RuBisCO-like protein of Bacillus and photosynthetic RuBisCO. Science 302:286–290. doi: 10.1126/science.1086997 [DOI] [PubMed] [Google Scholar]
- 61. Cordero PRF, Bayly K, Man Leung P, Huang C, Islam ZF, Schittenhelm RB, King GM, Greening C. 2019. Atmospheric carbon monoxide oxidation is a widespread mechanism supporting microbial survival. ISME J 13:2868–2881. doi: 10.1038/s41396-019-0479-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Islam ZF, Cordero PRF, Feng J, Chen YJ, Bay SK, Jirapanjawat T, Gleadow RM, Carere CR, Stott MB, Chiri E, Greening C. 2019. Two chloroflexi classes independently evolved the ability to persist on atmospheric hydrogen and carbon monoxide. ISME J 13:1801–1813. doi: 10.1038/s41396-019-0393-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. King GM, Weber CF. 2007. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol 5:107–118. doi: 10.1038/nrmicro1595 [DOI] [PubMed] [Google Scholar]
- 64. Meier DV, Imminger S, Gillor O, Woebken D. 2021. Distribution of mixotrophy and desiccation survival mechanisms across microbial genomes in an arid biological soil crust community. mSystems 6:e00786-20. doi: 10.1128/mSystems.00786-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Koblížek M. 2015. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol Rev 39:854–870. doi: 10.1093/femsre/fuv032 [DOI] [PubMed] [Google Scholar]
- 66. Olson DK, Yoshizawa S, Boeuf D, Iwasaki W, DeLong EF. 2018. Proteorhodopsin variability and distribution in the north pacific subtropical gyre. ISME J 12:1047–1060. doi: 10.1038/s41396-018-0074-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. DeLong EF, Béjà O. 2010. The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol 8:e1000359. doi: 10.1371/journal.pbio.1000359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zeng Y, Chen X, Madsen AM, Zervas A, Nielsen TK, Andrei A-S, Lund-Hansen LC, Liu Y, Hansen LH. 2020. Potential rhodopsin- and bacteriochlorophyll-based dual phototrophy in a high arctic glacier. mBio 11:e02641-20. doi: 10.1128/mBio.02641-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sanford RA, Wagner DD, Wu Q, Chee-Sanford JC, Thomas SH, Cruz-García C, Rodríguez G, Massol-Deyá A, Krishnani KK, Ritalahti KM, Nissen S, Konstantinidis KT, Löffler FE. 2012. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc Natl Acad Sci U S A 109:19709–19714. doi: 10.1073/pnas.1211238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yoon S, Nissen S, Park D, Sanford RA, Löffler FE, Kostka JE. 2016. Clade I NosZ from those harboring clade II NosZ.. Appl Environ Microbiol 82:3793–3800. doi: 10.1128/AEM.00409-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chee-Sanford J, Tian D, Sanford R. 2019. Consumption of N2O and other N-cycle intermediates by Gemmatimonas aurantiaca strain T-27. Microbiology 165:1345–1354. doi: 10.1099/mic.0.000847 [DOI] [PubMed] [Google Scholar]
- 72. Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Graf DRH, Jones CM, Hallin S, de Crécy-Lagard V. 2014. Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS ONE 9:e114118. doi: 10.1371/journal.pone.0114118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Orellana LH, Rodriguez-R LM, Higgins S, Chee-Sanford JC, Sanford RA, Ritalahti KM, Löffler FE, Konstantinidis KT. 2014. Detecting nitrous oxide reductase (nosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. mBio 5:e01193-14. doi: 10.1128/mBio.01193-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jones CM, Graf DRH, Bru D, Philippot L, Hallin S. 2013. The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J 7:417–426. doi: 10.1038/ismej.2012.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jones CM, Spor A, Brennan FP, Breuil MC, Bru D, Lemanceau P, Griffiths B, Hallin S, Philippot L. 2014. Recently identified microbial guild mediates soil N2O sink capacity. Nature Clim Change 4:801–805. doi: 10.1038/nclimate2301 [DOI] [Google Scholar]
- 77. Park D, Kim H, Yoon S. 2017. Nitrous oxide reduction by an obligate. Appl Environ Microbiol 83:1–12. doi: 10.1128/AEM.00502-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Leustek T, Martin MN, Bick JA, Davies JP. 2000. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Biol 51:141–165. doi: 10.1146/annurev.arplant.51.1.141 [DOI] [PubMed] [Google Scholar]
- 79. Grein F, Ramos AR, Venceslau SS, Pereira IAC. 2013. Unifying concepts in anaerobic respiration: insights from dissimilatory sulfur metabolism. Biochim Biophys Acta - Bioenerg 1827:145–160. doi: 10.1016/j.bbabio.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 80. Villarreal-Chiu JF, Quinn JP, McGrath JW. 2012. The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front Microbiol 3:19. doi: 10.3389/fmicb.2012.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Santos-Beneit F. 2015. The pho regulon: a huge regulatory network in bacteria. Front Microbiol 6:402. doi: 10.3389/fmicb.2015.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Luo H, Benner R, Long RA, Hu J. 2009. Subcellular localization of marine bacterial alkaline phosphatases. Proc Natl Acad Sci U S A 106:21219–21223. doi: 10.1073/pnas.0907586106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yurkov V, Csotonyi JT. 2009. Purple Phototrophic Bact Adv Photosynth Respir, p 31–55. In Hunt CN, Daldal F, Thurnauer MC, Beatty JT(ed), New light on aerobic Anoxygenic Phototrophs. Springer, Dordrecht. doi: 10.1007/978-1-4020-8815-5 [DOI] [Google Scholar]
- 84. Mendel RR. 2013. The molybdenum cofactor. J Biol Chem 288:13165–13172. doi: 10.1074/jbc.R113.455311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. 2002. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 297:105–107. doi: 10.1126/science.1072113 [DOI] [PubMed] [Google Scholar]
- 86. Tong L. 2013. Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci 70:863–891. doi: 10.1007/s00018-012-1096-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kurnasov O, Goral V, Colabroy K, Gerdes S, Anantha S, Osterman A, Begley TP. 2003. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem Biol 10:1195–1204. doi: 10.1016/j.chembiol.2003.11.011 [DOI] [PubMed] [Google Scholar]
- 88. Cadieux N, Kadner RJ. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc Natl Acad Sci U S A 96:10673–10678. doi: 10.1073/pnas.96.19.10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rojo F. 2009. Degradation of alkanes by bacteria. Environ Microbiol 11:2477–2490. doi: 10.1111/j.1462-2920.2009.01948.x [DOI] [PubMed] [Google Scholar]
- 90. Appolinario LR, Tschoeke D, Paixão RVS, Venas T, Calegario G, Leomil L, Silva BS, Thompson CC, Thompson FL. 2019. Metagenomics sheds light on the metabolic repertoire of oil-biodegrading microbes of the south Atlantic ocean. Environ Pollut 249:295–304. doi: 10.1016/j.envpol.2019.03.007 [DOI] [PubMed] [Google Scholar]
- 91. Harwood CS, Burchhardt G, Herrmann H, Fuchs G. 1998. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev 22:439–458. doi: 10.1111/j.1574-6976.1998.tb00380.x [DOI] [Google Scholar]
- 92. Dinglasan-Panlilio MJ, Dworatzek S, Mabury S, Edwards E. 2006. Microbial oxidation of 1,2-dichloroethane under anoxic conditions with nitrate as electron acceptor in mixed and pure cultures. FEMS Microbiol Ecol 56:355–364. doi: 10.1111/j.1574-6941.2006.00077.x [DOI] [PubMed] [Google Scholar]
- 93. Paver SF, Kent AD. 2010. Temporal patterns in glycolate-utilizing bacterial community composition correlate with phytoplankton population dynamics in humic lakes. Microb Ecol 60:406–418. doi: 10.1007/s00248-010-9722-6 [DOI] [PubMed] [Google Scholar]
- 94. Smith DJ, Kharbush JJ, Kersten RD, Dick GJ, Glass JB. 2022. Uptake of phytoplankton-derived carbon and cobalamins by novel acidobacteria genera in microcystis blooms inferred from metagenomic and metatranscriptomic evidence. Appl Environ Microbiol 88:e0180321. doi: 10.1128/aem.01803-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yao S, Lyu S, An Y, Lu J, Gjermansen C, Schramm A. 2019. Microalgae–bacteria symbiosis in microalgal growth and biofuel production: a review. J Appl Microbiol 126:359–368. doi: 10.1111/jam.14095 [DOI] [PubMed] [Google Scholar]
- 96. Pellicer MT, Badía J, Aguilar J, Baldomà L. 1996. Glc locus of Escherichia coli: characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J Bacteriol 178:2051–2059. doi: 10.1128/jb.178.7.2051-2059.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Chiriac MC, Haber M, Salcher MM. 2023. Adaptive genetic traits in pelagic freshwater microbes. Environ Microbiol 25:606–641. doi: 10.1111/1462-2920.16313 [DOI] [PubMed] [Google Scholar]
- 98. Sleator RD, Hill C. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x [DOI] [PubMed] [Google Scholar]
- 99. Pivetti CD, Yen M-R, Miller S, Busch W, Tseng Y-H, Booth IR, Saier MH. 2003. Two families of mechanosensitive channel proteins. Microbiol Mol Biol Rev 67:66–85. doi: 10.1128/MMBR.67.1.66-85.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174 [DOI] [PubMed] [Google Scholar]
- 101. Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH, Hancock J. 2020. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Olm MR, Brown CT, Brooks B, Banfield JF. 2017. DRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J 11:2864–2868. doi: 10.1038/ismej.2017.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. doi: 10.1128/AEM.02411-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Anderson MJ. 2006. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x [DOI] [PubMed] [Google Scholar]
- 108. Asnicar F, Thomas AM, Beghini F, Mengoni C, Manara S, Manghi P, Zhu Q, Bolzan M, Cumbo F, May U, Sanders JG, Zolfo M, Kopylova E, Pasolli E, Knight R, Mirarab S, Huttenhower C, Segata N. 2020. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using phylophlan 3.0. Nat Commun 11:1–10. doi: 10.1038/s41467-020-16366-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 110. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 112. Letunic I, Bork P. 2021. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Bateman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, Bursteinas B, Bye-A-Jee H, Coetzee R, Cukura A, Silva A, Denny P, Dogan T, Ebenezer TG, Fan J, Castro LG, Garmiri P, Georghiou G, Gonzales L, Hatton-Ellis E, Hussein A, Ignatchenko A, Insana G, Ishtiaq R, Jokinen P, Joshi V, Jyothi D, Lock A, Lopez R, Luciani A, Luo J, Lussi Y, MacDougall A, Madeira F, Mahmoudy M, Menchi M, Mishra A, Moulang K, Nightingale A, Oliveira CS, Pundir S, Qi G, Raj S, Rice D, Lopez MR, Saidi R, Sampson J, Sawford T, Speretta E, Turner E, Tyagi N, Vasudev P, Volynkin V, Warner K, Watkins X, Zaru R, Zellner H, Bridge A, Poux S, Redaschi N, Aimo L, Argoud-Puy G, Auchincloss A, Axelsen K, Bansal P, Baratin D, Blatter MC, Bolleman J, Boutet E, Breuza L, Casals-Casas C, Castro E, Echioukh KC, Coudert E, Cuche B, Doche M, Dornevil D, Estreicher A, Famiglietti ML, Feuermann M, Gasteiger E, Gehant S, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Hyka-Nouspikel N, Jungo F, Keller G, Kerhornou A, Lara V, Le Mercier P, Lieberherr D, Lombardot T, Martin X, Masson P, Morgat A, Neto TB, Paesano S, Pedruzzi I, Pilbout S, Pourcel L, Pozzato M, Pruess M, Rivoire C, Sigrist C, Sonesson K, Stutz A, Sundaram S, Tognolli M, Verbregue L, Wu CH, Arighi CN, Arminski L, Chen C, Chen Y, Garavelli JS, Huang H, Laiho K, McGarvey P, Natale DA, Vinayaka CR, Wang Q, Wang Y, Yeh LS, Ruch P, Teodoro D. 2021. Uniprot: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. doi: 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. 2007. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev 71:576–599. doi: 10.1128/MMBR.00015-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wrighton KC, Castelle CJ, Varaljay VA, Satagopan S, Brown CT, Wilkins MJ, Thomas BC, Sharon I, Williams KH, Tabita FR, Banfield JF. 2016. RubisCO of a nucleoside pathway known from archaea is found in diverse uncultivated phyla in bacteria. ISME J 10:2702–2714. doi: 10.1038/ismej.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. doi: 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 121. Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6.
Basic characteristics of newly assembled Gemmatimonadota MAGs.
Basic characteristics of all Gemmatimonadota genomes.
Percentages of habitat-related core and accessory genes.
Presence/absence of genes in Gemmatimonadota based on SEED.
SIMPER analysis of similarity and dissimilarity between Gemmatimonadota genomes.
Genes and metabolic pathways found in Gemmatimonadota genomes from different environments.
Data Availability Statement
All data derived from this work are publicly available in NCBI-GenBank databases. All 16 MAGs assembled in this study have been deposited in the NCBI-GenBank database under Bioproject number PRJNA721863, biosample numbers SAMN32886101-SAMN32886116, and GenBank accession numbers JARIER000000000-JARIFG000000000. All these genomes were derived from metagenomic data sets from Spanish lakes and reservoirs that were previously deposited under Bioproject number PRJNA721863 and SRA numbers SRR15198238- SRR15198275.