Abstract
Background and Aims:
HBV infection is restricted to the liver, where it drives exhaustion of virus-specific T and B cells and pathogenesis through dysregulation of intrahepatic immunity. Our understanding of liver-specific events related to viral control and liver damage has relied almost solely on animal models, and we lack useable peripheral biomarkers to quantify intrahepatic immune activation beyond cytokine measurement. Our objective was to overcome the practical obstacles of liver sampling using fine-needle aspiration and develop an optimized workflow to comprehensively compare the blood and liver compartments within patients with chronic hepatitis B using single-cell RNA sequencing.
Approach and Results:
We developed a workflow that enabled multi-site international studies and centralized single-cell RNA sequencing. Blood and liver fine-needle aspirations were collected, and cellular and molecular captures were compared between the Seq-Well S3 picowell-based and the 10× Chromium reverse-emulsion droplet–based single-cell RNA sequencing technologies. Both technologies captured the cellular diversity of the liver, but Seq-Well S3 effectively captured neutrophils, which were absent in the 10× dataset. CD8 T cells and neutrophils displayed distinct transcriptional profiles between blood and liver. In addition, liver fine-needle aspirations captured a heterogeneous liver macrophage population. Comparison between untreated patients with chronic hepatitis B and patients treated with nucleoside analogs showed that myeloid cells were highly sensitive to environmental changes while lymphocytes displayed minimal differences.
Conclusions:
The ability to electively sample and intensively profile the immune landscape of the liver, and generate high-resolution data, will enable multi-site clinical studies to identify biomarkers for intrahepatic immune activity in HBV and beyond.
INTRODUCTION
Eight hundred million people worldwide are at risk for liver cirrhosis, including over 290 million chronically infected with HBV1,2. Chronic hepatitis B (CHB) is a highly heterogeneous disease characterized by variable viral loads and liver inflammation3. Antiviral therapies suppress viral replication but rarely lead to a cure4. Currently, there are no known peripheral blood biomarkers to monitor anti-HBV responses in the liver to predict disease progression or cure5. This is a significant knowledge gap that could be addressed through elective liver tissue sampling and next-generation single-cell RNA sequencing (scRNA-seq).
Liver tissue is typically available through surgical resections or percutaneous needle biopsies. However, these procedures are dependent on clinical need rather than elective sampling necessary to understand immune mechanisms in a dynamic disease such as CHB. Fine-needle aspirates (FNA) of the liver are collected using a 25-gauge (G; 0.51 mm outer diameter) needle, which is significantly smaller than needles used for standard liver biopsies (16-18 G; 1.65–1.27 mm outer diameter) and venipuncture (21 G; 0.82 mm outer diameter). As a result, the FNA technique poses minimal risk and discomfort to participants. The safety profile of liver FNAs enables elective liver sampling, including longitudinal clinical studies with less than 2 weeks between samplings6,7. However, compared to more invasive needle biopsies, liver FNAs yield fewer cells (<100,000 cells)8,9. Moreover, they are best used immediately onsite to provide the most accurate representation of the liver immune environment.
To capitalize on the utility of FNAs and address obstacles of site-specific variability, we developed and optimized a workflow that enables international, multi-site collection of liver FNAs and centralized scRNA-seq library generation. We developed metrics to assess FNA quality, followed by the comparison of 2 methods for scRNA-seq on the collected cells, the Seq-Well S3 and 10× Genomics 3′ v2 single-cell platforms. We further optimized a method to freeze and ship loaded Seq-Well S3 arrays, simplifying onsite processing for clinical workflows and facilitating enhanced reproducibility through centralized whole transcriptome amplification and sequencing. By analyzing matched blood and liver FNA samples collected at 4 international sites, we highlight the impact of the liver microenvironment on immune cells and examine the impact of antiviral therapy on gene expression. This optimized workflow will enable multi-site international studies of human liver biology.
METHODS
Ethical statement
Peripheral blood and liver FNAs were collected from 35 participants living with CHB at the Erasmus MC University Medical Center (Rotterdam, The Netherlands), the Toronto General Hospital (Toronto, Canada), and the Massachusetts General Hospital (Boston, USA). Three healthy volunteer blood and liver FNAs were collected at the Janssen Clinical Pharmacology Unit (Antwerp, Belgium). All participants provided written informed consent. This study was approved by institutional review boards at all sites and was conducted in accordance with both the declaration of Helsinki and Istanbul.
Collection of liver fine-needle aspirates
Paired liver FNA and blood samples were obtained to compare intrahepatic and peripheral immune profiles. To minimize variation between sites, the liver FNA procedure was standardized between participating sites and performed as follows. The position and movement of the liver during respiration were assessed using ultrasound to avoid large blood vessels. The puncture site was cleaned using chlorhexidine, and the participant was covered in sterile dressings, leaving only the puncture site exposed. Upon exhalation, a 25 G spinal needle (Braun Spinocan) was inserted intercostally into the liver parenchyma. The stylet was carefully removed, and a 10 ml syringe (BD Bioscience) was attached to the needle. Liver cells were aspirated from the parenchyma by pulling back the syringe to create negative pressure while simultaneously advancing the needle ~2.5 cm into the liver. Finally, the needle was retracted from the participant and moved to a sterile table where approximately 500 µL of cold RPMI 1640 medium without phenol red (Lonza) was aspirated into the syringe. The cell suspension was transferred to a 5 mL tube (Axygen) and placed on ice immediately. This was repeated 3 times (a total of 4 passes), using fresh needles and syringes for each pass.
Detailed methodology for sample processing and analysis is provided in the Supplemental materials, http://links.lww.com/HEP/H830
RESULTS
Fine-needle aspirates (FNAs) to assess the diversity of intrahepatic immune cells
Unlike core biopsies that cut a tissue cylinder, FNA sampling uses negative pressure and forward motion of the syringe to aspirate cells. This introduces the potential for blood contamination if the needle engages hepatic blood vessels. Because of the collection method, the cellular composition of liver FNAs lies between peripheral whole blood and core biopsies10. To ensure consistent sampling of intrahepatic tissue, it is important to objectively assess the extent of peripheral blood contamination in each sample.
We established a standardized protocol, and instructional videos, for obtaining FNA material (Supplemental video 1), followed by an immediate assessment of sample quality (Supplemental video 2). Individual FNA passes display different degrees of peripheral blood composition based on visual inspection (Figure 1A). We used flow cytometry to quantify naïve T cells in FNAs, which are typically not found in solid organs8. However, flow cytometry requires significant time to prepare, a large fraction of the collected sample, and infrastructure at different sites within a clinical trial. Therefore, we examined whether an optical density (OD) to measure red blood cell (RBC) content would deliver comparable results while minimizing sample use and processing time. We compared OD450 measurements with conventional immunological profiling on different FNA passes from individual participants (Supplemental Fig. 1, http://links.lww.com/HEP/H831). As the OD450 value increased, the CD4:CD8 ratio inverted. CD8 T cells dominated in samples with a low OD450, while there was a predominance of CD4 T cells in high OD450 samples, as it is characteristic of the blood (Figure 1B)6. The OD450 measurement was further validated by finding a significant positive correlation between OD450 and the frequency of naïve CD4 and CD8 T cells (Figures. 1C and D). Furthermore, there was a significant negative correlation between OD450 and frequencies of mucosal-associated invariant T (MAIT) cells (Figure 1E), which are compartmentalized to the liver11. These data demonstrate that RBC content measured by OD450 is a robust indicator of peripheral blood composition within FNA passes and supports the use of this simple, quantitative, sample-sparing test to account for peripheral blood content in FNA samples.
FIGURE 1.
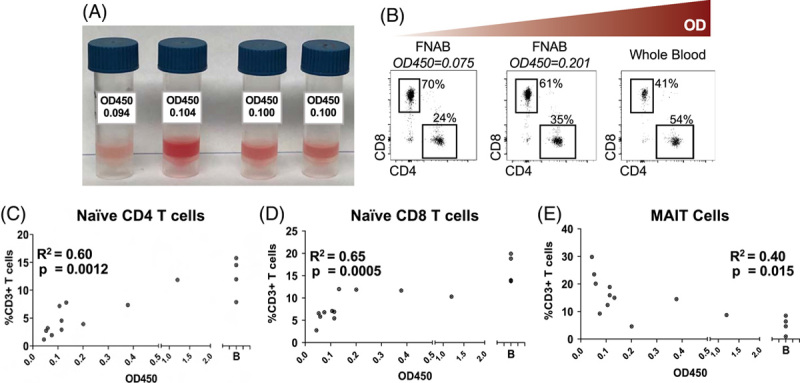
Quantification of RBC contamination of liver FNAs. (A) Images showing variable RBC contents of individual FNA passes. (B) CD4:CD8 ratio in FNA passes from a single patient with increasing OD450 values. Frequency of (C) naïve CD4 T cells (D) naïve CD8 T cells and (E) MAIT cells compared to OD450 values of each FNA pass. Analysis was done using the Pearson pairwise correlation. Abbreviation: MAIT, mucosal-associated invariant T.
Video 1.
Standard protocol and instructions for obtaining FNA material.
Video 2.
Processing FNA material for use in downstream assays.
Comparing scRNA-seq technology platforms for analysis of human liver FNAs
We next sought to identify the best means of comprehensively profiling the total cellular diversity in FNA samples. Reverse-emulsion droplet and picowell-based high-throughput scRNA-seq platforms are amenable to low-input samples. The 10× Genomics Chromium system, such as the Drop-seq and inDrops platforms, uses a reverse-emulsion microfluidic system to co-capture uniquely barcoded beads and cells12,13. Picowell-based platforms, such as Seq-Well S3 14 and BD Rhapsody15, use small wells to isolate individual cells. Both the Seq-Well S3 and 10× Genomics platforms have been successfully used to analyze peripheral blood and digested tissue16,17. However, cell capture, cell type representation, sequencing depth and quality, and the robustness of data generation across multiple sites from primary human liver tissue samples are not known.
Four FNA passes were collected from each of the 4 volunteers. The lowest OD450 passes were pooled and analyzed in parallel using Seq-Well S3 and the 10× Genomics 3′ version 2 (10×v2) platforms. After filtering low-quality cells based on a minimum of 300 genes and 500 UMIs per cell, the number of transcripts (p = 0.044), the number of genes captured per cell and cell capture from 15,000 cell inputs were significantly higher in liver FNAs using Seq-Well S3 (Supplemental Fig. S2a-c, http://links.lww.com/HEP/H832). For the peripheral blood, only the number of transcripts showed a significant difference between Seq-Well S3 and 10× Genomics (Supplemental Fig. 2d-f, http://links.lww.com/HEP/H832).
UMAP visualization of the Seq-Well S3 and 10 × 3′ v2 datasets readily identified lymphocytes and myeloid cells (Figure 2A-D). We compared cell type frequencies between the platforms by calculating the number of each cell type recovered divided by the total sequenced cells passing quality thresholds in the sample (Figure 2E). The 10× v2 captured significantly more γδ T cells whereas Seq-Well S3 captured significantly more granulocytes and was capable of efficiently capturing both blood and liver neutrophils, which were undetectable in the 10 × 3′ v2 dataset (Figure 2E). A cluster of regulatory T cells (CD3, CTLA4, IL2RB, and FoxP3) was unique to the Seq-Well S3 dataset, when compared to data obtained using the 10 × 3′ v2 kit. Few high-quality nonimmune cells, such as hepatocytes, were captured using either method.
FIGURE 2.
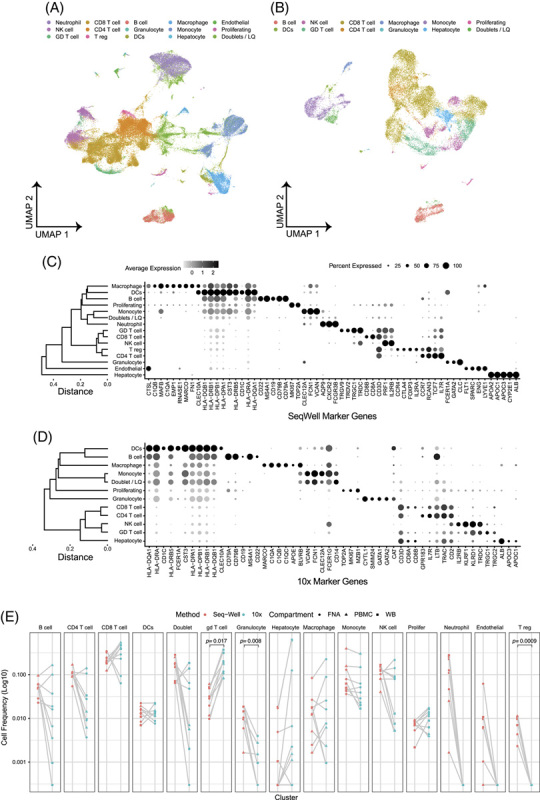
10 × 3′v2 versus Seq-Well S3 comparison. UMAPs of all data from (A) Seq-Well and (B) ×10. Dot plot of cell type marker genes for (C) Seq-Well and (D) ×10. (E) Cell type frequencies compared across matched samples (gray lines) for Seq-Well S3 (red) and ×10 (blue). Paired t test was used to test for statistical significance. Samples from FNA, PBMC, and WB are circles, triangles, and squares, respectively, n = 4 patients run in parallel, and 8 samples account for blood and liver from each patient. Abbreviation: FNA, fine-needle aspirates.
We used publicly available datasets to determine how liver sample processing impacted cell capture. We compared liver FNAs, which required minimal processing, to datasets that sorted CD45 + cells from liver biopsies18 or collected total cells after perfusion-based tissue digestion16. Each method resulted in a distinct cell capture profile (Supplemental Fig S3, http://links.lww.com/HEP/H833). Perfusion-based tissue digestion resulted in high hepatocyte, LSEC, and plasma cell capture but loss of most of the non-adherent lymphocytes. CD45 sorting from dissociated biopsies lost all hepatocytes and LSECs and dramatically reduced myeloid cell capture, presumably due to their lower CD45 expression. RBC-depleted FNAs sat in the middle, providing a good representation of lymphocytes, myeloid cells, and neutrophils while still capturing some parenchymal cells. These data support the use of liver FNAs to capture a representative cellular repertoire of the liver, with the understanding that parenchymal cells are underrepresented in FNAs.
Cryopreservation of Seq-Well S 3 arrays for centralized library generation
Based on cell type capture, we selected the picowell-based Seq-Well S3 system to collect the remaining samples for this study. We further developed a protocol that allowed for centralized processing by freezing Seq-Well S3 arrays immediately after cell loading and membrane sealing. This allowed us to capture freshly isolated cells onsite and perform library generation centrally to minimize onsite processing and batch effects.
Fresh FNA and PBMC samples from 6 participants from 3 sites were loaded onto parallel arrays up to membrane sealing. One array was processed onsite up to the reverse transcription step and the other was frozen at −80oC and shipped on dry ice to the central lab. Freezing and shipping after cell loading yielded an equivalent number of transcripts, genes per cell, and total cell recovery compared to processing onsite for both the liver FNA (Figure 3A,C,E) and peripheral blood samples (Figure 3B,D,F). Fresh and frozen arrays clustered together, without data integration, with only a maximum of 7 consensus differentially expressed genes (consensus = expressed in > 50% of comparisons) observed in CD8 T cells (Figure 3G). Arrays from individual patients showed similar cellular diversity (Figure 3H). We used the optimized workflow to characterize blood and liver FNA samples from 13 CHB participants and 3 healthy participants (Table 1, representing 4, 4, 5, and 3 samples from the 4 sites). We recovered a total of 66,446 high-quality cells, which were analyzed in a lineage-specific manner to investigate transcriptional differences between the blood and liver.
FIGURE 3.
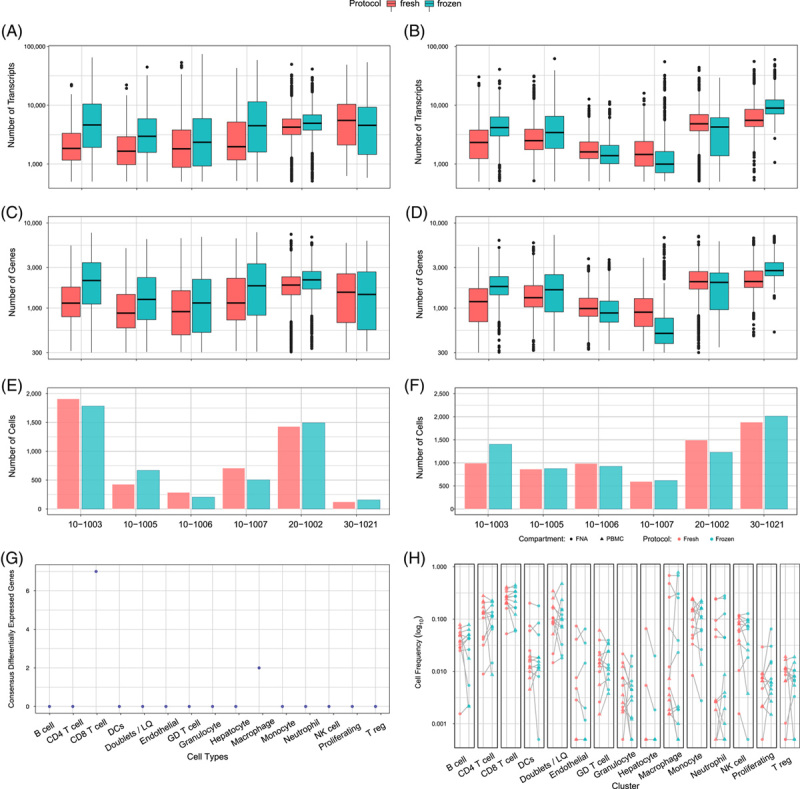
Data comparison of freshly processed versus frozen arrays. (A) Number of transcripts/cells, (B) Number of genes/cells, and (C) number of cells captured in liver FNAs from 6 different patients. (D) number of transcripts/cells (E) number of genes/cell and (F) number of cells captured in peripheral blood from 6 different patients. Paired t test was used to test for statistical significance in panels (a–). (G) Number of differentially expressed genes in matched cell types between freshly processed and frozen arrays. Differential expression was performed on a participant and compartment basis. Genes were considered consensus differentially expressed if they were significant at adjusted p values < 0.05 and the absolute value of Cohen’s d > 0.2 across at least 6 of the 12 individual and compartment combinations. (H) Comparison of cell type capture across matched samples (gray lines) for fresh (pink) and frozen (blue) arrays. Samples from FNA and PBMC are circles and triangles, respectively. Paired t test was used to test for statistical significance (n = 6 patients, blood and liver). Abbreviation: PBMC, peripheral blood mononuclear cells.
TABLE 1.
Donor used for Seq-Well scRNA-seq
| ID | Age | sex | HBV DNA (Log10 IU/ml) | ALT (U/L) | HBsAg (Log10 IU/ml) | HBeAg + /- | Treatment | Cell# FNA | Cell# blood |
|---|---|---|---|---|---|---|---|---|---|
| 10-1003 | 38 | M | < 20 | 24 | 2.1 | Neg | + | 3692 | 2396 |
| 10-1005 | 26 | M | 4.3 | 68 | 2.1 | Neg | − | 1096 | 1739 |
| 10-1006 | 51 | M | < 20 | 29 | 1.7 | Neg | + | 491 | 1910 |
| 10-1007 | 55 | M | < 20 | 28 | 2.1 | Neg | + | 1564 | 1211 |
| 20-1002 | 55 | M | 2.5 | 21 | + | Neg | − | 6247 | 3465 |
| 20-1003 | 43 | M | 3.8 | 24 | + | Neg | − | 1377 | 1063 |
| 20-3001 | 62 | F | < 20 | 26 | + | Neg | + | 2592 | 1267 |
| 20-3002 | 42 | M | < 20 | 10 | + | Neg | + | 973 | 601 |
| 30-1021 | 32 | F | 2.4 | 9 | 3.1 | Neg | − | 2074 | 5273 |
| 30-1022 | 24 | F | 1.4 | 25 | 2.1 | Neg | − | 4168 | 859 |
| 30-1023 | 29 | F | 3.7 | 40 | 3.8 | Neg | − | 3430 | 2602 |
| 30-1024 | 19 | F | 3.2 | 23 | 3.6 | Neg | − | 5012 | 1655 |
| 30-1025 | 29 | M | 2.8 | 33 | 2.4 | Neg | − | 444 | 774 |
| 22-0002 | 61 | F | N/A | 17 | N/A | N/A | N/A | 2592 | 2009 |
| 22-0004 | 64 | M | N/A | 18 | N/A | N/A | N/A | 1809 | 1864 |
| 22-0010 | 66 | M | N/A | 24 | N/A | N/A | N/A | 18 | 1680 |
Notes: + indicates qualitative HBsAg and on nucleoside analog therapy.
22-0002, 22-0004, and 22-0010 were healthy donors with no viral markers.
Subset enrichment and transcriptional adaption of lymphocytes in the liver
The composition and phenotypic profiles of liver T cells are distinct from the blood19. Subclustering data revealed 5 distinct CD8 T cell populations (Figure 4A). The differentially expressed genes discriminating each cluster are shown in Figure 4B (a full list of gene markers is available in Supplemental Table 1, http://links.lww.com/HEP/H834). The observed clusters were comparable to prototypic CD8 T cell subpopulations known from prior studies of infection and cancer20,21. Cluster 0 (“GZMB”) displayed strong expression of granzymes B (GZMB) and H (GZMH), together with CX3CR1 and other markers typically found in effector T cells20,22. Cluster 1 (“GZMK”) was dominated by expression of GzmK, the transcription factor EOMES, and chemokine receptors CCR5 and CXCR6, resembling transitional, or precursor, CD8 T cells described in liver cancer20. Cluster 2 (“NR4A2”) expressed the transcription factor NR4A2 that has been associated with dysfunctional CD8 T cells23. Cluster 3 (“CCR7 + TCF7 + ”) expressed the chemokine receptor CCR7 and transcription factor TCF7, together with LEF1 and SELL, which is characteristic of both naïve or stem-like CD8 T cells24. Cluster 4 (“MAIT”) co-expressed genes associated with MAIT cells such as KLRB1, SLC4A10, DPP4, and IL7R 25. Interestingly, the MAIT cell cluster expressed many of the GZMK cluster 1 defining genes in addition to its own characteristic gene expression signature (Figure 4C).
FIGURE 4.
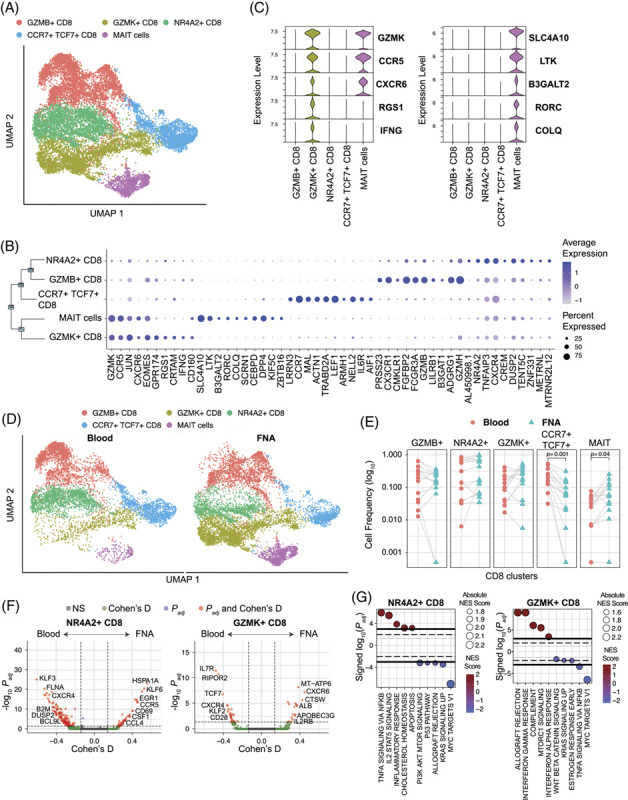
CD8 T cell composition in liver versus blood. (A) scRNA-seq UMAP for CD8 T cells colored by cluster IDs. (B) Dot plot showing the top 10 marker genes for each cluster ID. (C) Violin plot showing the top 5 marker genes for GZMK + CD8 and MAIT cells. Significance was determined using the Wilcoxon Rank sum test. (D) scRNA-seq UMAP colored by cluster and split based on tissue of origin, that is, liver and blood. (E) Comparison of cell frequencies between blood and liver within sample (connected through grey lines) for each CD8 cluster. Significance was determined using Wilcoxon Signed-Rank test with the Bonferroni correction (adjusted p-value < 0.05). (F) Volcano plots depicting differences in gene expression between blood and FNA in NR4A2 + and GZMK + CD8 T cells. The R-package MAST was used to obtain hurdle p values that were Bonferroni corrected for multiple hypothesis testing. Positive Cohen’s d value suggests higher expression in liver. Cohen’s d cutoff calculated as mean + ×2 SD of Cohen’s d values of all genes. (G) Hallmark gene sets enriched by NR4A2 + and GZMK + CD8 T cells. The normalized enrichment score was calculated based on a vector of gene-level signed statistic and false discovery rate was adjusted based on the Benjamini-Hochberg (BH) Correction. x-axis represents the signed log10 of adjusted p-value for the gene sets, and the positive value suggests enrichment in the liver. Abbreviation: MAIT, mucosal-associated invariant T.
The composition and phenotypic profiles of liver T cells are distinct from what is found in the blood19. Differences in relative T cell frequencies were especially apparent for MAIT cells, which are enriched in the liver (Figure 4D,E)11. The GZMK population was also more prevalent in the liver. In contrast, the naïve/naïve-like CCR7 TCF7 population was enriched in the blood (Figure 4E)26. While all 5 clusters were found in both tissue compartments, the UMAP visualization in Figure 4D showed that liver and blood-derived T cells occupy distinct spaces within most clusters, suggesting different transcriptional states. Detailed comparison of transcriptional signatures between blood and liver within clusters revealed profound differences in gene expression for the NR4A2 and GZMK CD8 T cell clusters (Figure 4F), representing key cellular pathways involved in the inflammatory response, IL2 and TNF-α signaling in the liver NR4A2 population and IFN-γ and mTORC1 signaling in the liver GZMK population compared to blood (Figure 4G). The overall T cell composition and clear differences in the transcriptional landscape support that liver FNAs predominantly captured liver-resident T cells.
Other major lymphocyte populations showed fewer differences when analyzed by compartment. CD4 T cells were less complex than CD8 T cells, with only 2 distinct clusters (excluding the Tregs cluster in Figure 2C) and only marginal differences with regard to the relative size of clusters or their transcriptional landscape (Supplemental Fig. 4, http://links.lww.com/HEP/H835). NK cell analysis showed enrichment of the CD56hi NK cell population in the liver FNAs, as expected, but no significant impact of the liver compartment on the transcriptional profiles (Supplemental Fig. 5, http://links.lww.com/HEP/H836). B cells showed no significant compartment-specific enrichment or transcriptional profiles (Supplemental Fig. 6, http://links.lww.com/HEP/H837).
Complex neutrophil compartment in the blood and liver
Animal studies suggest that neutrophils are recruited first in the inflammatory cascade and facilitate immune cell infiltration into the liver parenchyma, but little data are available from the patient’s liver27,28. After recognizing that the picowell-based scRNA-seq approach efficiently captured neutrophils, we changed our RBC depletion to magnetic bead–based depletion to preserve the neutrophil population. As a result, we present neutrophil data from 7 participants with matched whole blood and FNA, as the remaining 9 had PBMCs isolated by density gradient centrifugation. Subclustering of neutrophils yielded 6 subpopulations (Figure 5A). The loss of neutrophils was apparent in the PBMC samples (Figure 5B). Neutrophils shared an overlapping transcriptional profile with monocytes but were clearly distinguishable from monocytes due to the lack of VCAN and CD68 (not shown). The 5 clusters expressed known neutrophil markers CXCR2, IL8, and S100A genes29,30. Cluster 0, IL8(hi) SYAP1- neutrophils, and cluster 4, IL8(hi) SYAP1 + neutrophils expressed the highest level of IL8 (CXCL8), which is found in activated neutrophils31. Expression of SYAP1, a described target of caspases, distinguished the IL-8 + neutrophils (Figure 5C). Both cluster 3 (MME) and cluster 5 (MMP8) neutrophils expressed genes whose products are stored in secretory granules and aid in the process of neutrophil recruitment to tissues and matrix degradation32,33. However, they expressed different secreted molecules, such as MME and FCN1 or MMP8 and LCN2, in clusters 3 and 5, respectively (Figure 5C). Cluster 1, IFN-stim neutrophils, displayed a profile consistent with activation by type I interferon, with increased expression of RSAD2, IFIT1, MX1, ISG15, and OAS3. Cluster 2 (SIGLEC10) neutrophils expressed SIGLEC10, which has been described as an inhibitory receptor of other immune cells in the context of tumor escape34 (Figure 5C).
FIGURE 5.
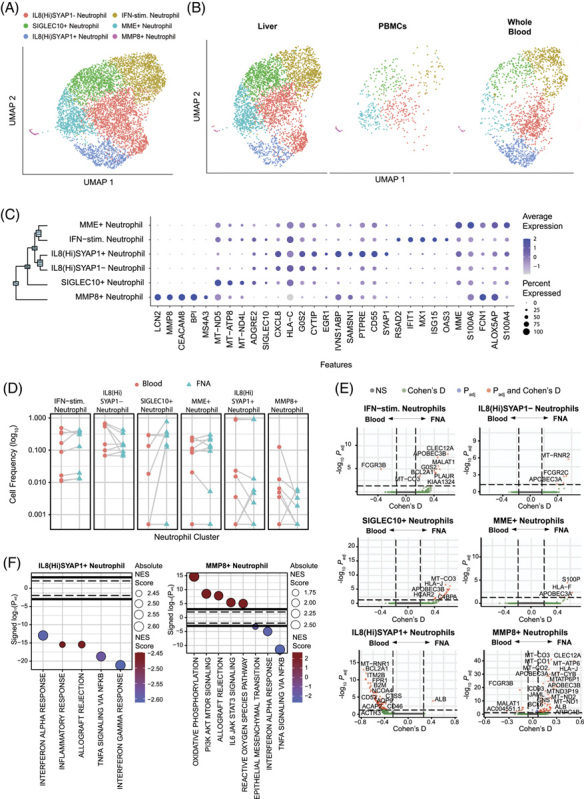
Neutrophil identification. (A) scRNA-seq UMAP for neutrophils colored by cluster IDs. (B) UMAP dimensionality reduction of neutrophils by compartment. (C) Dot plot showing top 5 marker genes for each cluster determined using the Wilcoxon Rank sum test with the Bonferroni correction (adjusted p-value < 0.05). (D) Differential frequency of neutrophil clusters between compartments. Participants with 0 cells within a cell population were excluded. Significant differences between compartments were assessed using the Wilcoxon Signed-Rank test with the Bonferroni correction (adjusted p-value < 0.05). (E) Volcano plots depicting differences in gene expression between compartments within each cluster. The R-package MAST was used to obtain hurdle p values that were Bonferroni corrected for multiple hypothesis testing. Positive Cohen’s d value suggests higher expression in the liver. (F) Hallmark gene sets enriched for clusters with differentially expressed genes between compartments. The normalized enrichment score was calculated based on a vector of gene-level signed statistic and the false discovery rate was adjusted based on the Benjamini-Hochberg (BH) correction. x-axis represents signed log10 of adjusted p-value for the gene sets, and positive value suggests enrichment in liver.
The neutrophil distribution between the blood and liver did not show significant enrichment of any cluster in either compartment (Figure 5D). However, similar to the CD8 T cells, neutrophil clusters displayed distinct liver and blood transcriptional signatures within clusters (Figure 5E). Enrichment analysis of IL8(hi)SYAP1 + neutrophils revealed significant enrichment of pathways associated with inflammation and type I and II interferons (IFNs) in the blood. MMP8 neutrophils displayed distinct signatures within the cluster with type I and II IFN signatures in the blood and pathways enriched in IL6 signaling and metabolic processes in the liver (Figure 5F). While their role in the progression of CHB is unclear, the ability to capture neutrophils in the scRNA-seq data will allow for deeper investigation and a more in-depth understanding of their role.
Liver FNAs capture macrophage diversity in the human liver
The plasticity of myeloid cells allows them to tune their functionality to specific environments. We identified a highly diverse monocyte population between the blood and liver, consisting of 10 monocyte subpopulations (Supplemental Fig. 7a, http://links.lww.com/HEP/H838). Monocyte subpopulations were not enriched in either compartment (Supplemental Fig. 7c,d, http://links.lww.com/HEP/H838), consistent with their ability to circulate and patrol tissues35. Two monocyte clusters displayed significant differentially expressed genes (DEGs) between the compartments: cluster 0, cMono(1), and cluster 1, cMono(2) (Supplemental Fig. 7e, http://links.lww.com/HEP/H838). These data demonstrate the potential diversity of short-lived monocyte populations.
Macrophages regulate the inflammatory environment in the liver. They have been associated with HBV-mediated liver inflammation and the progression of fibrosis36,37. Macrophages exist on a functional spectrum between activating and suppressing38. They are tightly bound to the endothelium, and studies that have characterized human macrophages with scRNA-seq have used collagenase to digest liver tissue16. It was unclear if we would capture them with FNAs. We noted that macrophage capture was variable among patients. Therefore, caution should be taken when comparing frequencies of adherent cells, such as macrophages, between time points or between patients.
Subclustering macrophages yielded 5 subpopulations (Figure 6A) that were restricted to the liver (Figure 6B). The macrophage clusters displayed markers shared across populations, including complement (C1QA), FCGR3A (CD16), MARCO, CTSS, and MSR1 (Figure 6C). However, we identified unique markers capable of distinguishing each population (Figure 6D). Cluster 0, TIMD4 macrophages, has been defined as comprising embryonically derived, or long-lived, tissue macrophages39,40. TIMD4 macrophages expressed LYVE1, also associated with long-lived-tissue–resident macrophages39, and CD163, a scavenger receptor associated with fibrosis in chronic hepatitis B (Figure 6E)41. Cluster 1, SLC40A1 (ferroportin) macrophages, expressed markers that indicate recent monocyte-to- macrophage differentiation, including NR1H3 (Liver X receptor alpha, LxRa) and SPIC (Figure 6E)42,43. Cluster 2, C1QA low macrophages, and cluster 3, HBB (hemoglobin) macrophages expressed the lowest level of shared macrophage markers C1QA and MARCO and may represent transient or transitional macrophage populations44 (Figure 6C). Cluster 2, C1QA low macrophages, expressed high levels of monocyte markers VCAN and LYZ, suggesting that these macrophages are in a transitional state of differentiation (Figure 6E)45. Cluster 4, HBB + macrophages, may represent a transient liver macrophage population responsible for clearance of RBCs prior to differentiating to SLC40A1 macrophages46. Cluster 4, CD9 macrophages, expressed OLR1 and LGALS3 consistent with scar-associated macrophages found in the cirrhotic liver (Figure 6E)17. These data confirm that the collection of FNAs allows the capture of adherent macrophages and identifies unique macrophage states associated with different stages of liver disease.
FIGURE 6.
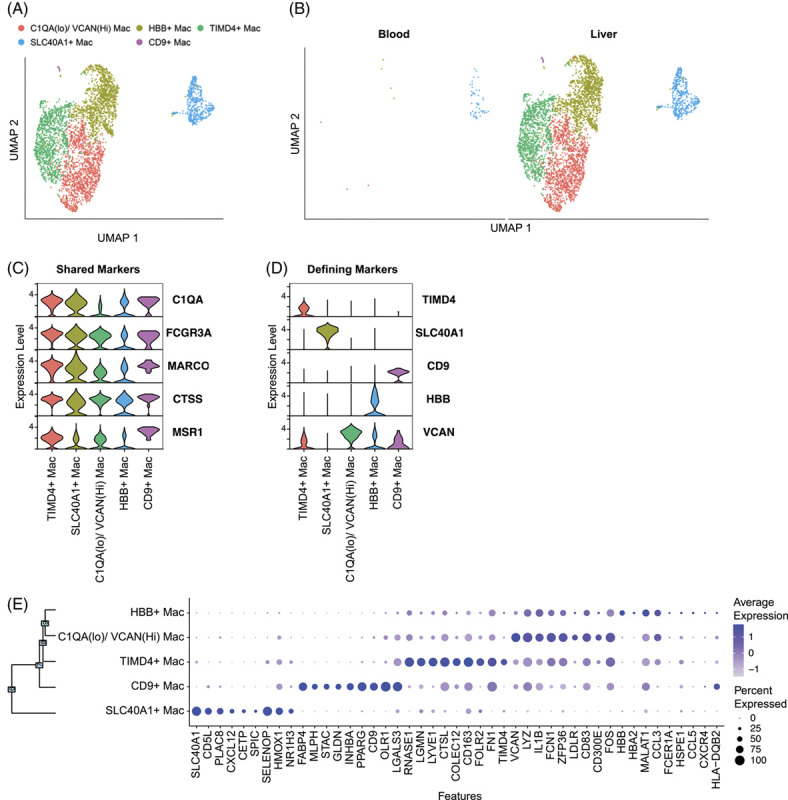
Macrophage identification in liver FNAs. (A) scRNA-seq UMAP for macrophages colored by cluster IDs. (B) UMAP dimensionality reduction of macrophages by compartment. Violin plots of (C) macrophage-shared genes and (D) unique cluster-defining genes. (E) Dot plot showing the top 9 cluster-defining genes. Significance was determined using the Wilcoxon Rank-Sum test with the Bonferroni correction (adjusted p-value < 0.05). Abbreviations: Mac, macrophages; scRNA-seq, single cell RNA sequencing.
Myeloid cells show significant differences between treated and untreated patients
Nucleoside analog treatment suppresses HBV replication and normalizes ALT. Five patients with CHB in our study were on nucleoside analog therapy at the time of FNA collection. The remaining 8 had varying HBV DNA levels, and all but one (ALT 1.7xULN) had ALT levels in the normal range (Table 1). This allowed us to investigate how the presence of HBV DNA impacts the transcriptional profile of immune cells with little to no influence by liver damage. The groups were divided based on treatment, and cell clusters were projected onto UMAP plots (Figure 7A, B). None of the clusters showed a significant difference in frequency between treated and untreated patients (Figure 7C). Macrophages were not included in the frequency comparison due to their adherent nature, but showed the greatest changes in DEGs between patient groups, followed by monocytes (Figure 7D). DEGs from monocytes and macrophages comprised numerous immune-related genes that included chemokines, cytokines, and interferon-stimulated genes (Figure 7E). In contrast, lymphocytes displayed few significant DEGs between cohorts, which were largely not immune-related and comprised of many ribosomal genes (Figure 7E). The impact of nucleoside analog therapy, particularly on liver macrophage gene expression, indicates that ongoing HBV replication induces significant changes in the liver environment despite comparable ALT levels.
FIGURE 7.
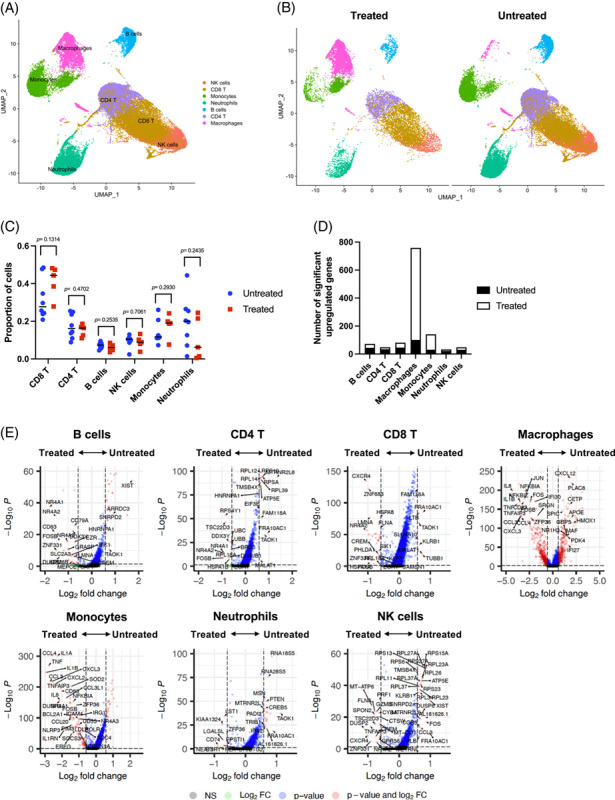
Myeloid cells are the most sensitive to treatment in patients with CHB. (A) scRNA-seq UMAP of cells from treated and untreated patients with CHB. (B) UMAP dimensionality reduction of intrahepatic cells by treatment. (C) Comparison of cell proportions between treatment. Dots represent individual donors; bars indicate mean values. The unpaired t test was used to test for statistical significance. (D) Number of differentially upregulated genes in each intrahepatic cluster between treated and untreated patients with CHB. (E) Volcano plots showing genes that are differentially expressed between treated and untreated patients with CHB. Significance was determined using the Wilcoxon rank sum test with Bonferroni correction (adjusted p-value < 0.05). Thresholds: p < 0.05 and log2 fold change ≥ 1.5. Genes upregulated in untreated patients are shown to the right-hand side of each plot. Genes upregulated in treated patients are shown on the left-hand side of each plot. Abbreviation: scRNA-seq, single cell RNA sequencing.
DISCUSSION
Liver FNAs revolutionize access to liver tissue, allowing frequent, scheduled tissue sampling to collect the most informative time points in disease progression or treatment. Our comparison of tissue processing approaches showed that RBC-depleted liver FNAs represented a balance between CD45 + immune cells and perfusion-based tissue digestion, effectively capturing lymphocytes, myeloid cells, and some parenchymal cells. The OD450 measurement was developed as a quantitative, sample-sparing approach to assess peripheral blood contamination reserves > 95% of the FNA sample for scientific investigation. Using a picowell-based technology provided the ability to load fresh cells onto the arrays locally and freeze and ship them. This allowed us to use a central laboratory for library generation and sequencing to support the international multi-site collaboration.
Elective liver sampling with FNAs allows for monitoring therapeutic interventions and opens the opportunity to re-define the classical stages of chronic hepatitis B. Clinically, the stages are based on viral load, HBeAg, and liver damage (ALT) but are given immunological names such as “immune tolerant” or “immune active”47. While such staging has been relevant for clinical management, it may obscure the effective use of novel therapies or the actual immune status in the liver. Recent studies have used microarray, or total RNA sequencing from core liver biopsies, to distinguish immunological signatures between stages but lack resolution of individual cell types and comparisons to the blood48,49. Recently, Zhang et al performed scRNA-seq on core biopsies after enzymatic digestion and CD45 + cell sorting and showed distinct compartmentalization of T cells and transcriptional profiles associated with liver damage18. In contrast, the workflow that we present using FNAs avoids the time and manipulation involved with digestion and cell sorting and was used to electively sample patients without liver damage. We took advantage of this to compare the effect of nucleoside analog therapy in patients with CHB without liver damage. While lymphocytes showed relatively minor alterations between groups, monocytes and macrophages showed significant changes between treated and untreated patients. Given that macrophages represent a primary sentinel of inflammation in the liver, the transcriptional changes suggest ongoing immunological activation in the absence of clinically evident liver damage.
Our technology comparison showed that cellular diversity between the emulsion-based and picowell-based approaches was similar for lymphocytes and myeloid cells but diverged significantly in their ability to capture granulocytes. Human neutrophil capture is a known issue with the 10 × 3′v2 data because of their low RNA and high nuclease content. Newer versions of the 10 × Genomics reagents may improve neutrophil capture, but our data suggest that picowell strategies were more effective. As a result, we identified liver-specific transcriptional profiles in neutrophil subtypes known to modulate the adaptive response in other settings50. This presents an opportunity to investigate the role of neutrophils in HBV infection, particularly in liver damage, where only existing data were generated in mouse models27,28.
We did not observe vastly different frequencies of CD4 and CD8 T cell subpopulations by compartment, as seen by Zhang et al, likely because our patient cohort did not include patients with hepatitis18. However, we found transcriptional differences between blood and liver immune cell populations in the absence of inflammation. CD8 T cells displayed the most unique transcriptional profiles between compartments. In the 5 CD8 T cell clusters that we analyzed, GZMK CD8 T cells displayed the chemokine receptors CCR5 and CXCR6, which are associated with homing to the liver51, and were the only cluster positive for the IFN-γ transcript. This suggests that a CD8 T cell population, which is likely not HBV-specific due to its size, may play a role in viral control in the HBV-infected liver and/or the potential to drive pathogenesis through induction of IFN-γ regulated chemokines CXCL-9 and CXCL-10.
B cells and NK cells did not show compartment-specific transcriptional profiles. However, it should be stated that our study was not powered to discover all DEGs and included patients with different disease profiles, including healthy donors, with a primary goal of mapping cellular diversity. Moreover, the number of cells recovered from 3 healthy donors did not allow for a robust comparison to patients’ with CHB liver FNA samples. Our analysis used stringent thresholds for identifying compartment-specific gene expression. Genes specifically expressed in the blood or liver were compared while taking the participants into account as covariates. This was necessary to avoid bias toward individual participants but resulted in lower statistical power in this small cohort. We have observed lower inter-patient variability when patients are selected by defined inclusion/exclusion criteria and more robust transcriptional changes in longitudinal samples from individual patients with CHB52.
Liver macrophages have been characterized in studies from healthy livers16, livers with cholestatic liver disease53, and cirrhosis17, all of which required tissue digestion. The FNA sampling approach captured macrophages without the need for tissue digestion, minimizing processing time to preserve in vivo transcriptional profiles. However, macrophage capture varied between patients using the FNA sampling approach. Therefore, calculations such as frequency would be prone to error, but their transcriptional profiles can be highly informative of the inflammatory environment. In a heterogeneous disease, such as CHB, we expected to find a heterogeneous macrophage population, and did so, identifying 5 different clusters. Some macrophage populations displayed markers of monocyte-to- macrophage differentiation, suggesting that datasets such as these could be used to investigate the transition of liver-infiltrating monocytes. However, because of variability in the collection, macrophage-related data should be validated in tissue sections where their transcriptional profiles can be validated by spatial transcriptomics, and their frequency and localization can be assessed by fluorescent microscopy.
We found the greatest diversity of cellular phenotypes within short-lived monocytes and neutrophils54,55. Because of their short-lived nature, they are dynamically regulated within the tissues and periphery by changing environmental cues. Therefore, these cell types may be ideal sentinels for immunomodulatory therapies that induce inflammatory cytokines. The FNA procedure, properly timed after treatment, could be used to interrogate the intrahepatic response to immunomodulation to identify more robust biomarkers related to antiviral immunity and immune activation.
Overall, establishing this workflow across 4 international collaborating sites provides a framework to compare the immunological status between the blood and liver across the different stages of CHB, where specific cohorts may be differently represented in geographical regions and facilitate deployment in future clinical studies. We are highly confident that when this approach is applied to clinical studies with defined enrollment criteria and longitudinal sampling, the ability to detect changes in transcriptional profiles between time points will be significantly more sensitive.
Supplementary Material
Footnotes
Abbreviations: CHB, chronic hepatitis B; DEG, differentially expressed gene; FNA, fine-needle aspirate; Mac, macrophage; MAIT, mucosal-associated invariant T cell; scRNAseq, single-cell RNA sequencing.
Alex S. Genshaft, Sonu Subudhi, Arlin Keo, Juan Diego Sanchez Vasquez, and Nádia ConceiC̦ão-Neto shared first authorship.
Georg M. Lauer, Andre Boonstra, Alex K. Shalek, and Adam J. Gehring shared senior authorship.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, http://www.hepjournal.com.
Contributor Information
Alex S. Genshaft, Email: alexgenshaft@gmail.com.
Sonu Subudhi, Email: ssubudhi@mgh.harvard.edu.
Arlin Keo, Email: d.keo-denboer@erasmusmc.nl.
Juan Diego Sanchez Vasquez, Email: jd.sanchezvasquez@mail.utoronto.ca.
Nádia Conceição-Neto, Email: nneto@its.jnj.com.
Deeqa Mahamed, Email: Deeqa.mahamed@fluidigm.com.
Lauke L. Boeijen, Email: l.l.boeijen-3@umcutrecht.nl.
Nadia Alatrakchi, Email: nalatrakchi@mgh.harvard.edu.
Chris Oetheimer, Email: chrisoetheimer@gmail.com.
Mike Vilme, Email: mvilme@mit.edu.
Riley Drake, Email: riled@mit.edu.
Ira Fleming, Email: irafleming@gmail.com.
Nancy Tran, Email: ntran@broadinstitute.org.
Constantine Tzouanas, Email: c.tzouanas@gmail.com.
Jasmin Joseph-Chazan, Email: jasminjoseph@fas.harvard.edu.
Martin Arreola Villanueva, Email: mav@mit.edu.
Harmen J. G. van de Werken, Email: h.vandewerken@erasmusmc.nl.
Gertine W. van Oord, Email: g.vanoord@erasmusmc.nl.
Zwier M. A. Groothuismink, Email: z.groothuismink@erasmusmc.nl.
Boris J. Beudeker, Email: b.beudeker@erasmusmc.nl.
Zgjim Osmani, Email: z.osmani@erasmusmc.nl.
Shirin Nkongolo, Email: Shirin.Nkongolo@med.uni-heidelberg.de.
Aman Mehrotra, Email: amanmehrotra89@gmail.com.
Kurt Spittaels, Email: kspittae@its.jnj.com.
Jordan Feld, Email: Jordan.Feld@uhn.ca.
Raymond T. Chung, Email: Chung.Raymond@mgh.harvard.edu.
Robert J. de Knegt, Email: r.deknegt@erasmusmc.nl.
Harry L. A. Janssen, Email: h.janssen@erasmusmc.nl.
Jeroen Aerssens, Email: jaerssen@its.jnj.com.
Jacques Bollekens, Email: jbolleke@its.jnj.com.
Nir Hacohen, Email: nhacohen@mgh.harvard.edu.
Georg M. Lauer, Email: glauer@helix.mgh.harvard.edu.
Andre Boonstra, Email: p.a.boonstra@erasmusmc.nl.
Alex K. Shalek, Email: shalek@mit.edu.
Adam J. Gehring, Email: Adam.Gehring@uhnresearch.ca.
DATA AVAILABILITY STATEMENT
The scRNA-seq data have been deposited in Gene Expression Omnibus (GEO) with accession number: GSE234241
AUTHOR CONTRIBUTIONS
Alex S. Genshaft and Adam J. Gehring wrote the manuscript. Alex S. Genshaft, Sonu Subudhi, Arlin Keo, Juan Diego Sanchez Vasquez, Nádia Conceição-Neto, Nadia Alatrakchi, Constantine Tzouanas, Jasmin Joseph-Chazan, Martin Arreola Villanueva, Harmen J. G. van de Werken, Gertine W. van Oord, Zwier M.A. Groothuismink, Boris J. Beudeker, Zgjim Osmani, Nir Hacohen, Georg M. Lauer, Andre Boonstra, and Alex K. Shalek performed data analysis. Alex S. Genshaft, Sonu Subudhi, Arlin Keo, Juan Diego Sanchez Vasquez and Adam J. Gehring wrote the manuscript. Deeqa Mahamed, Lauke L. Boeijen, Nadia Alatrakchi Riley Drake, Ira Fleming, Nancy Tran, Shirin Nkongolo, and Aman Mehrotra performed the experiments. Kurt Spittaels provided the Supplemental videos. Jordan Feld, Raymond T. Chung, Robert J. de Knegt, Harry L. A. Janssen, Jeroen Aerssens, and Jacques Bollekens, enrolled the patients. Jordan Feld, Raymond T. Chung, Robert J. de Knegt, Harry L. A. Janssen, Jeroen Aerssens, Jacques Bollekens, Nir Hacohen, Georg M. Lauer, Andre Boonstra, Alex K. Shalek and Adam J. Gehring worked on conceptual development. Jordan Feld, Raymond T. Chung, Robert J. de Knegt, Harry L. A. Janssen, Jeroen Aerssens, Jacques Bollekens, Georg M. Lauer, and Adam J, Gehring revised the paper. All authors revised and approved the manuscript.
FUNDING INFORMATION
This study was funded by Janssen Pharmaceuticals. Raymond T. Chung was supported by the Massachusetts General Hospital Research Scholars Program. Adam J. Gehring received support from The Toronto Centre for Liver Disease and Canada Foundation for Innovation. Alex K. Shalek receives funding from the NIH, Break Through Cancer, the Pew-Stewart Scholars Program, Foundation MIT, Wellcome Leap, the Bill & Melinda Gates Foundation, and the Moore Foundation.
CONFLICTS OF INTEREST
Nádia Conceição-Neto is employed by Janssen Pharmaceutica NV. Jordan Feld consults and received grants from Arbtus, AbbVie, Gilead, GlaxoSmithKline, Janssen, Roche, and Vir. He received grants from Eiger and Enanta. Raymond T. Chung received grants from AbbVie, Boehringer, Bristol Myers Squibb, Gilead, GlaxoSmithKline, Janssen, Merck, and Roche. Robert J. de Knegt consults, is on the speakers' bureau, and received grants from AbbVie, Echosens, and Gilead. He consults and received grants from Bristol Myers Squibb, Janssen, Medtronic, and Roche. He consults for Merck and Norgine. He received grants from GlaxoSmithKline and Phillips. Henry L.A. Janssen consults and received grants from Arbutus, Gilead, GlaxoSmithKline, Janssen, Merck, Roche, and Vir. He consults for Aligos, Antios, Arena, Eiger, Enyo, Regulus, VBI Vaccines, and Viroclinics. He received grants from AbbVie, Arbutus, and Bristol Myers Squibb. Jeroen Aerssens owns stock in Johnson & Johnson. He is employed by Janssen. Jacques Bollekens consults, owns stock in, and holds intellectual property rights with Johnson & Johnson. He is employed by Janssen. Nir Hacohen consults and owns stock in DangerBio. He owns stock in Biontech. Georg M. Lauer consults for and received grants from Janssen. He consults for Altimmune, GlaxoSmithKline, and Roche. Andre Boonstra received grants from Fujirebio, Gilead, GlaxoSmithKline, and Janssen. Alex K. Shalek consults, advises, and owns stock in Cellarity, Dahlia Biosciences, Honeycomb Biotechnologies, Ochre Bio, Repertoire Immune Medicines, Relation Therapeutics, and Santa Ana Bio. He consults and received grants from Janssen. He consults for FL82, Hovione, Merck, Roche, and Third Rock Ventures. He received grants from Leo Pharma A/S and Novo Nordisk. Adam J. Gehring consults and received grants from BlueJay Therapeutics, Gilead, GlaxoSmithKline, Janssen Pharmaceuticals, Roche, and Vir Biotech. He consults for Assembly, Finich Therapeutics, SQ BioTech, Viron Therapeutics, and VBI Vaccines. The remaining authors have no conflicts to report.
REFERENCES
- 1.Marcellin P, Kutala BK. Liver diseases: a major, neglected global public health problem requiring urgent actions and large-scale screening. Liver Int. 2018;38(Suppl 1):2–6. [DOI] [PubMed] [Google Scholar]
- 2.Polaris Observatory C. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383–403. [DOI] [PubMed] [Google Scholar]
- 3.Yuen MF, Chen DS, Dusheiko GM, Janssen HLA, Lau DTY, Locarnini SA, et al. Hepatitis B virus infection. Nat Rev Dis Primers. 2018;4:18035. [DOI] [PubMed] [Google Scholar]
- 4.Lok AS, Zoulim F, Dusheiko G, Ghany MG. Hepatitis B cure: From discovery to regulatory approval. Hepatology. 2017;66:1296–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Revill PA, Chisari FV, Block JM, Dandri M, Gehring AJ, Guo H, et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol Hepatol. 2019;4:545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claassen MA, de Knegt RJ, Janssen HL, Boonstra A. Retention of CD4 + CD25 + FoxP3 + regulatory T cells in the liver after therapy-induced hepatitis C virus eradication in humans. J Virol. 2011;85:5323–5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spaan M, van Oord GW, Janssen HL, de Knegt RJ, Boonstra A. Longitudinal analysis of peripheral and intrahepatic NK cells in chronic HCV patients during antiviral therapy. Antiviral Res. 2015;123:86–92. [DOI] [PubMed] [Google Scholar]
- 8.Sprengers D, van der Molen RG, Kusters JG, Kwekkeboom J, van der Laan LJ, Niesters HG, et al. Flow cytometry of fine-needle-aspiration biopsies: a new method to monitor the intrahepatic immunological environment in chronic viral hepatitis. J Viral Hepat. 2005;12:507–512. [DOI] [PubMed] [Google Scholar]
- 9.Pembroke T, Gallimore A, Godkin A. Tracking the kinetics of intrahepatic immune responses by repeated fine needle aspiration of the liver. J Immunol Methods. 2015;424:131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill US, Pallett LJ, Thomas N, Burton AR, Patel AA, Yona S, et al. Fine needle aspirates comprehensively sample intrahepatic immunity. Gut. 2019;68:1493–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jo J, Tan AT, Ussher JE, Sandalova E, Tang XZ, Tan-Garcia A, et al. Toll-like receptor 8 agonist and bacteria trigger potent activation of innate immune cells in human liver. PLoS Pathog. 2014;10:e1004210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gierahn TM, Wadsworth MH, II, Hughes TK, Bryson BD, Butler A, Satija R, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shum EY, Walczak EM, Chang C, Christina Fan H. Quantitation of mRNA Transcripts and Proteins Using the BD Rhapsody™ Single-Cell Analysis System. Adv Exp Med Biol. 2019;1129:63–79. [DOI] [PubMed] [Google Scholar]
- 16.MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Li J, Cheng Y, Meng F, Song JW, Fan X, et al. Single-cell RNA sequencing reveals intrahepatic and peripheral immune characteristics related to disease phases in HBV-infected patients. Gut. 2023;72:153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroy DC, Ciuffreda D, Cooperrider JH, Tomlinson M, Hauck GD, Aneja J, et al. Liver environment and HCV replication affect human T-cell phenotype and expression of inhibitory receptors. Gastroenterology. 2014;146:550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 2017;169:1342–1356 e1316. [DOI] [PubMed] [Google Scholar]
- 21.Andreatta M, Corria-Osorio J, Müller S, Cubas R, Coukos G, Carmona SJ. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat Commun. 2021;12:2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, et al. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity. 2016;45:1270–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, López-Moyado IF, Seo H, Lio CJ, Hempleman LJ, Sekiya T, et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature. 2019;567:530–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurtulus S, Madi A, Escobar G, Klapholz M, Nyman J, Christian E, et al. Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1(-)CD8( + ) Tumor-Infiltrating T Cells. Immunity. 2019;50:181–194 e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrot T, Gorin JB, Ponzetta A, Maleki KT, Kammann T, Emgård J, et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci Immunol. 2020;5:eabe1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weninger W, Crowley MA, Manjunath N, von Andrian UH. Migratory properties of naive, effector, and memory CD8( + ) T cells. J Exp Med. 2001;194:953–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitia G, Isogawa M, Kakimi K, Wieland SF, Chisari FV, Guidotti LG. Depletion of neutrophils blocks the recruitment of antigen-nonspecific cells into the liver without affecting the antiviral activity of hepatitis B virus-specific cytotoxic T lymphocytes. Proc Natl Acad Sci USA. 2002;99:13717–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sitia G, Isogawa M, Iannacone M, Campbell IL, Chisari FV, Guidotti LG. MMPs are required for recruitment of antigen-nonspecific mononuclear cells into the liver by CTLs. J Clin Invest. 2004;113:1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silvin A, Chapuis N, Dunsmore G, Goubet AG, Dubuisson A, Derosa L, et al. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell. 2020;182:1401–1418 e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calcagno DM, Zhang C, Toomu A, Huang K, Ninh VK, Miyamoto S, et al. SiglecF(HI) Marks Late-Stage Neutrophils of the Infarcted Heart: A Single-Cell Transcriptomic Analysis of Neutrophil Diversification. J Am Heart Assoc. 2021;10:e019019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tecchio C, Micheletti A, Cassatella MA. Neutrophil-derived cytokines: facts beyond expression. Front Immunol. 2014;5:508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Didangelos A. COVID-19 Hyperinflammation: What about Neutrophils? mSphere. 2020;5:e00367-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin M, Jackson P, Tester AM, Diaconu E, Overall CM, Blalock JE, et al. Matrix metalloproteinase-8 facilitates neutrophil migration through the corneal stromal matrix by collagen degradation and production of the chemotactic peptide Pro-Gly-Pro. Am J Pathol. 2008;173:144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin SS, Gao FH. Molecular Mechanism of Tumor Cell Immune Escape Mediated by CD24/Siglec-10. Front Immunol. 2020;11:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. 2013;39:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heymann F, Peusquens J, Ludwig-Portugall I, Kohlhepp M, Ergen C, Niemietz P, et al. Liver inflammation abrogates immunological tolerance induced by Kupffer cells. Hepatology. 2015;62:279–291. [DOI] [PubMed] [Google Scholar]
- 37.Bility MT, Cheng L, Zhang Z, Luan Y, Li F, Chi L, et al. Hepatitis B virus infection and immunopathogenesis in a humanized mouse model: induction of human-specific liver fibrosis and M2-like macrophages. PLoS Pathog. 2014;10:e1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Li S, Duan X, Yang C, Xu M, Chen L. Macrophage Phenotypes and Hepatitis B Virus Infection. J Clin Transl Hepatol. 2020;8:424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol. 2019;20:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw TN, Houston SA, Wemyss K, Bridgeman HM, Barbera TA, Zangerle-Murray T, et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J Exp Med. 2018;215:1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kazankov K, Barrera F, Møller HJ, Bibby BM, Vilstrup H, George J, et al. Soluble CD163, a macrophage activation marker, is independently associated with fibrosis in patients with chronic viral hepatitis B and C. Hepatology. 2014;60:521–530. [DOI] [PubMed] [Google Scholar]
- 42.Bonnardel J, T’Jonck W, Gaublomme D, Browaeys R, Scott CL, Martens L, et al. Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity. 2019;51:638–654 e639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai M, Troutman TD, Seidman JS, Ouyang Z, Spann NJ, Abe Y, et al. Liver-Derived Signals Sequentially Reprogram Myeloid Enhancers to Initiate and Maintain Kupffer Cell Identity. Immunity. 2019;51:655–670 e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sander J, Schmidt SV, Cirovic B, McGovern N, Papantonopoulou O, Hardt AL, et al. Cellular Differentiation of Human Monocytes Is Regulated by Time-Dependent Interleukin-4 Signaling and the Transcriptional Regulator NCOR2. Immunity. 2017;47:1051–1066 e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7:10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theurl I, Hilgendorf I, Nairz M, Tymoszuk P, Haschka D, Asshoff M, et al. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat Med. 2016;22:945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370–398. [DOI] [PubMed] [Google Scholar]
- 48.van Buuren N, Ramirez R, Turner S, Chen D, Suri V, Aggarwal A, et al. Characterization of the liver immune microenvironment in liver biopsies from patients with chronic HBV infection. JHEP Rep. 2022;4:100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montanari NR, Ramírez R, Aggarwal A, van Buuren N, Doukas M, Moon C, et al. Multi-parametric analysis of human livers reveals variation in intrahepatic inflammation across phases of chronic hepatitis B infection. J Clin Invest. 2023;133:e158903. [DOI] [PubMed] [Google Scholar]
- 50.Ponzetta A, Carriero R, Carnevale S, Barbagallo M, Molgora M, Perucchini C, et al. Neutrophils Driving Unconventional T Cells Mediate Resistance against Murine Sarcomas and Selected Human Tumors. Cell. 2019;178:346–360 e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boeijen LL, van Oord GW, Hou J, van der Heide-Mulder M, Gaggar A, Li L, et al. Gene expression profiling of human tissue-resident immune cells: Comparing blood and liver. J Leukoc Biol. 2019;105:603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nkongolo S, Mahamed D, Kuipery A, Sanchez Vasquez JD, Kim SC, Mehrotra A, et al. Longitudinal liver sampling in patients with chronic hepatitis B starting antiviral therapy reveals hepatotoxic CD8 + T cells. J Clin Invest. 2023;133:e158903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor SA, Chen SY, Gadhvi G, Feng L, Gromer KD, Abdala-Valencia H, et al. Transcriptional profiling of pediatric cholestatic livers identifies three distinct macrophage populations. PLoS ONE. 2021;16:e0244743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pillay J, den Braber I, Vrisekoop N, Kwast LM, de Boer RJ, Borghans JA, et al. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 2010;116:625–627. [DOI] [PubMed] [Google Scholar]
- 55.Patel AA, Zhang Y, Fullerton JN, Boelen L, Rongvaux A, Maini AA, et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med. 2017;214:1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The scRNA-seq data have been deposited in Gene Expression Omnibus (GEO) with accession number: GSE234241
AUTHOR CONTRIBUTIONS
Alex S. Genshaft and Adam J. Gehring wrote the manuscript. Alex S. Genshaft, Sonu Subudhi, Arlin Keo, Juan Diego Sanchez Vasquez, Nádia Conceição-Neto, Nadia Alatrakchi, Constantine Tzouanas, Jasmin Joseph-Chazan, Martin Arreola Villanueva, Harmen J. G. van de Werken, Gertine W. van Oord, Zwier M.A. Groothuismink, Boris J. Beudeker, Zgjim Osmani, Nir Hacohen, Georg M. Lauer, Andre Boonstra, and Alex K. Shalek performed data analysis. Alex S. Genshaft, Sonu Subudhi, Arlin Keo, Juan Diego Sanchez Vasquez and Adam J. Gehring wrote the manuscript. Deeqa Mahamed, Lauke L. Boeijen, Nadia Alatrakchi Riley Drake, Ira Fleming, Nancy Tran, Shirin Nkongolo, and Aman Mehrotra performed the experiments. Kurt Spittaels provided the Supplemental videos. Jordan Feld, Raymond T. Chung, Robert J. de Knegt, Harry L. A. Janssen, Jeroen Aerssens, and Jacques Bollekens, enrolled the patients. Jordan Feld, Raymond T. Chung, Robert J. de Knegt, Harry L. A. Janssen, Jeroen Aerssens, Jacques Bollekens, Nir Hacohen, Georg M. Lauer, Andre Boonstra, Alex K. Shalek and Adam J. Gehring worked on conceptual development. Jordan Feld, Raymond T. Chung, Robert J. de Knegt, Harry L. A. Janssen, Jeroen Aerssens, Jacques Bollekens, Georg M. Lauer, and Adam J, Gehring revised the paper. All authors revised and approved the manuscript.
FUNDING INFORMATION
This study was funded by Janssen Pharmaceuticals. Raymond T. Chung was supported by the Massachusetts General Hospital Research Scholars Program. Adam J. Gehring received support from The Toronto Centre for Liver Disease and Canada Foundation for Innovation. Alex K. Shalek receives funding from the NIH, Break Through Cancer, the Pew-Stewart Scholars Program, Foundation MIT, Wellcome Leap, the Bill & Melinda Gates Foundation, and the Moore Foundation.
CONFLICTS OF INTEREST
Nádia Conceição-Neto is employed by Janssen Pharmaceutica NV. Jordan Feld consults and received grants from Arbtus, AbbVie, Gilead, GlaxoSmithKline, Janssen, Roche, and Vir. He received grants from Eiger and Enanta. Raymond T. Chung received grants from AbbVie, Boehringer, Bristol Myers Squibb, Gilead, GlaxoSmithKline, Janssen, Merck, and Roche. Robert J. de Knegt consults, is on the speakers' bureau, and received grants from AbbVie, Echosens, and Gilead. He consults and received grants from Bristol Myers Squibb, Janssen, Medtronic, and Roche. He consults for Merck and Norgine. He received grants from GlaxoSmithKline and Phillips. Henry L.A. Janssen consults and received grants from Arbutus, Gilead, GlaxoSmithKline, Janssen, Merck, Roche, and Vir. He consults for Aligos, Antios, Arena, Eiger, Enyo, Regulus, VBI Vaccines, and Viroclinics. He received grants from AbbVie, Arbutus, and Bristol Myers Squibb. Jeroen Aerssens owns stock in Johnson & Johnson. He is employed by Janssen. Jacques Bollekens consults, owns stock in, and holds intellectual property rights with Johnson & Johnson. He is employed by Janssen. Nir Hacohen consults and owns stock in DangerBio. He owns stock in Biontech. Georg M. Lauer consults for and received grants from Janssen. He consults for Altimmune, GlaxoSmithKline, and Roche. Andre Boonstra received grants from Fujirebio, Gilead, GlaxoSmithKline, and Janssen. Alex K. Shalek consults, advises, and owns stock in Cellarity, Dahlia Biosciences, Honeycomb Biotechnologies, Ochre Bio, Repertoire Immune Medicines, Relation Therapeutics, and Santa Ana Bio. He consults and received grants from Janssen. He consults for FL82, Hovione, Merck, Roche, and Third Rock Ventures. He received grants from Leo Pharma A/S and Novo Nordisk. Adam J. Gehring consults and received grants from BlueJay Therapeutics, Gilead, GlaxoSmithKline, Janssen Pharmaceuticals, Roche, and Vir Biotech. He consults for Assembly, Finich Therapeutics, SQ BioTech, Viron Therapeutics, and VBI Vaccines. The remaining authors have no conflicts to report.


