Abstract
The inebriation of wild African elephants from eating the ripened and rotting fruit of the marula tree is a persistent myth in Southern Africa. However, the yeasts responsible for alcoholic fermentation to intoxicate the elephants remain poorly documented. In this study, we considered Botswana, a country with the world's largest population of wild elephants, and where the marula tree is indigenous, abundant and protected, to assess the occurrence and biodiversity of yeasts with a potential to ferment and subsequently inebriate the wild elephants. We collected marula fruits from over a stretch of 800 km in Botswana and isolated 106 yeast strains representing 24 yeast species. Over 93% of these isolates, typically known to ferment simple sugars and produce ethanol comprising of high ethanol producers belonging to Saccharomyces, Brettanomyces, and Pichia, and intermediate ethanol producers Wickerhamomyces, Zygotorulaspora, Candida, Hanseniaspora, and Kluyveromyces. Fermentation of marula juice revealed convincing fermentative and aromatic bouquet credentials to suggest the potential to influence foraging behaviour and inebriate elephants in nature. There is insufficient evidence to refute the aforementioned myth. This work serves as the first work towards understanding the biodiversity marula associated yeasts to debunk the myth or approve the facts.
Keywords: yeast biodiversity, fermentation, African elephants, intoxication myth, fermenting marula fruits
A microbial perspective of the marula fruit and elephant intoxication myth, delving into the biodiversity of marula associated yeasts.
Introduction
The persistent myth of the inebriated African wild elephants (Loxodonta africana) after consumption of marula fruits (Sclerocarya birrea subsp caffra) baffled the tourists visiting Africa's game reserves and protected areas. The scientific community has not been spared either, for many decades. This probably follows our excessive fascination of the effects of ethanol on animal behaviours as a species with long evolutionary relationship with euphoric ethanolic beverages (Carrigan et al. 2015, Dudley and Maro 2021). Perhaps the gigantic body size of the elephants, their preferential foraging of marula fruits as well as the natural disperser of the seeds of the tree and the possible presence of unknown amounts of naturally occurring alcohol are central of the myth (Lewis 1987). However, empirical evidence to suggest that the alcohol in the naturally fermenting fruits could inebriate elephants, to substantiate the myth, remains elusive.
The marula tree is a deciduous tree belonging to the Anacardiaceae family indigenous to Southern African miombo woodlands, although it is also found in Sudano–Sahelian range of West Africa as well as the savanna woodlands of Eastern Africa and Madagascar (Velempini and Ketlhoilwe 2022). The tree bears thousands of succulent edible (up to 100 000 fruits per tree), very juicy filled flesh (up to 8 mL per fruit) and sugar-rich plum-sized fruits (up to 16° Brix) (Shackleton 2002, Mkwezalamba et al. 2015, Phiri et al. 2022). Traditionally, the sugar-sweet marula fruits are spontaneously fermented to produce an alcoholic beverage known as morula in Botswana, mukumbi in Zimbabwe (Mugochi et al. 1999) and mokhope or ubuganu in South Africa (Krige 1937, Cunningham 1990, Maluleke 2019). The production of traditional alcoholic drinks, dating back to the Neolithic period, is thought to be due to the evolutionary ripened fruit-eating behaviour of primates and subsequently our closest ancestors (Guerra-Doce and Theory 2015). It is notable that the renowned African Marula Cream Liqueur™, whose appealing label features an enormous elephant head and the fruit, is incidentally made from the fruits of this tree (Van Wyk 2011). These fruits most likely contain yeasts as agents of microbial decay characterised by alcoholic fermentation (Lewis 1987). Aerobic and anaerobic fermentation of abundant and fermentable fruit sugars is highly likely to yield the euphoric and intoxicating substance, ethanol, among other alcohols, as well as other volatile compounds, which could attract and inebriate elephants. This could be the most probable reason to substantiate the myth, although some speculations such as change of behaviour due to a mere finding of the special fruit and or intoxication after ingesting poisonous marula tree bark inhabiting beetle pupae have been brought forward (Goosen et al. 1985).The intoxication of such a gigantic animal weighing between 1800 and 6300 kg from naturally occuring alcohol remains a major concern to debunk or approve of the myth (Langman et al. 1995, Morris et al. 2006). Morris et al. (2006) were not in agreement with inebriation myth on the basis that the elephant´s body size is too large to be affected by amounts of ethanol in fermenting fruits cited as very low. The authors based their argument on the insufficient amounts of ethanol accumulated by fermenting marula fruits. The occurrence of yeasts species responsible for fermentations (Mpofu et al. 2008, Phiri et al. 2022) has been described rather inconclusively to warrant the inebriation of elephants. There are several anecdotal reports of frugivorous and nectarivorous animals being inebriated of naturally occurring alcohol. The Swedish elk is intoxicated from rotting and ripening apples (Cooke 2018) some birds have been reported to lose coordination and ability to fly and even fatally inebriated by fermented fruits and sap (Dennis 1987). Several marula fruit feeding animals including warthogs, baboons and giraffes have been reportedly intoxicated after consuming the fermented marula fruits (Dudley 2014). However, the elephant inebriation myth remains the most interesting of all times and a priority for research.
As a first step to potentially debunk the inebriation myth, we investigated the occurrence and biodiversity of fermenting yeasts associated with marula fruits. We took advantage of the ecological and geographical uniqueness of Botswana; a country with the world´s largest population of elephants (about 130 000 out of 500 000 in the whole world) (Azeem et al. 2020) and a country where the marula fruit tree is indigenous and abundant from 1.6 trees per hectare in arid regions (size over 3.9 million ha) to 23 trees per hectare in the Okavango Delta (size close to 2 million ha) (Neuenschwander et al. 2002, Wynberg et al. 2002, Batisani and Yarnal 2010). A remarkable diversity of yeasts with fermentative abilities was found in marula fruit samples collected countrywide (over a stretch of 800 km) from elephant-inhabited pristine and protected game reserves. Selected subsets of these yeasts were then investigated for their ability to ferment the marula fruit juice. We also assessed for their ability to produce possible aromatic compounds, which are thought to influence the foraging decisions in elephants. We then discussed the two attributes of the marula-associated yeasts to either substantiate the inebriation myth or otherwise.
Materials and methods
Sample collection
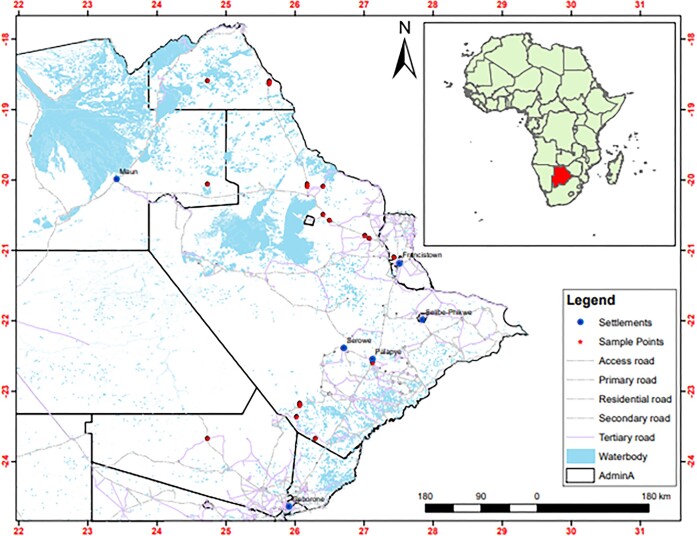
We collected 74 samples of marula (Sclerocarya birrea) wild fruits from 21 different locations on a stretch of over 800 km in Botswana (Fig. 1, Table 1). The fruits were collected over a two-year marula fruit ripening period. Ripened fruits as well as those with insect lacerations were aseptically collected and put in sealed sterile zipper-lock plastic bags and stored in a chilled cooler box before transporting to the laboratory. Upon arrival, samples were stored at 4 ºC until they could be processed.
Figure 1.
Map of Botswana depicting the points where marula fruits were collected. A total of 21 localities (red dots) within a stretch of 800 km where 106 strains described in this study were isolated are shown. GPS coordinates of the locations of the sampling points used to draw this map are available in the supplementary material (Table S1).
Table 1.
Species identification of yeasts isolated from marula fruits (Sequence comparison was done in February 2023).
| Strain ID | Nearest species match | Accession number | Percent Match | Query cover | Number of nucleotides in sequences |
|---|---|---|---|---|---|
| Z2iii | Clavispora lusitaniae | KP131863.1 | 100.00% | 99% | 318 |
| Z1i | Clavispora lusitaniae | LC413208.1 | 100.00% | 100% | 312 |
| Z17i | Candida albicans | ON851010.1 | 100.00% | 100% | 323 |
| Z15iii | Cyberlindnera mississippiensis | GQ340433.1 | 100.00% | 100% | 550 |
| Y0299 | Pichia kudriavzevii | MN310532.1 | 100.00% | 100% | 467 |
| Y0296 | Pichia kudriavzevii | MN913464.1 | 100.00% | 99% | 470 |
| Y0295 | Pichia manshurica | KM368827.1 | 100.00% | 100% | 424 |
| Y0293 | Pichia manshurica | KJ810825.1 | 100.00% | 95% | 426 |
| Y0292 | Pichia sporocuriosa | EU315763.1 | 100.00% | 99% | 470 |
| Y0291 | Pichia kudriavzevii | MK373022.1 | 100.00% | 97% | 472 |
| Y0289 | Pichia kudriavzevii | MN310532.1 | 100.00% | 100% | 469 |
| Y0288 | Pichia kudriavzevii | MN913464.1 | 100.00% | 99% | 473 |
| Y0287 | Pichia kudriavzevii | MN310532.1 | 100.00% | 99% | 473 |
| Y0286 | Saccharomyces cerevisiae YJM681 | CP006454.1 | 100.00% | 100% | 797 |
| Y0284 | Pichia manshurica | OM523901.1 | 100.00% | 100% | 420 |
| Y0283 | Hanseniaspora guilliermondii | FJ491945.1 | 100.00% | 99% | 686 |
| Y0282 | Pichia kudriavzevii | MN913464.1 | 100.00% | 98% | 464 |
| Y0281 | Pichia kudriavzevii | MN913464.1 | 100.00% | 98% | 473 |
| Y0280 | Pichia manshurica | OM523901.1 | 100.00% | 97% | 437 |
| Y0279 | Pichia manshurica | OM523901.1 | 100.00% | 100% | 418 |
| Y0278 | Pichia kudriavzevii | MH263646.1 | 100.00% | 99% | 471 |
| Y0277 | Hanseniaspora guilliermondii | KY103518.1 | 100.00% | 99% | 712 |
| Y0276 | Pichia manshurica | KJ810825.1 | 100.00% | 98% | 427 |
| Y0275 | Pichia kudriavzevii | KY104590.1 | 100.00% | 99% | 472 |
| Y0274 | Pichia manshurica | KM368827.1 | 100.00% | 94% | 429 |
| Y0273 | Pichia sporocuriosa | EU315763.1 | 100.00% | 99% | 471 |
| Y0272 | Pichia kudriavzevii | KP675519.1 | 100.00% | 97% | 472 |
| Y0271 | Starmera stellimalicola | NR_155825.1 | 100.00% | 99% | 473 |
| Y0270 | Hanseniaspora guilliermondii | KY103518.1 | 100.00% | 99% | 699 |
| Y0268 | Hanseniaspora opuntiae | MH934975.1 | 100.00% | 95% | 319 |
| Y0265 | Pichia manshurica | OM523901.1 | 100.00% | 100% | 419 |
| Y0263 | Pichia manshurica | OM523901.1 | 100.00% | 100% | 422 |
| Y0262 | Kluyveromyces marxianus | CP067319.1 | 100.00% | 92% | 686 |
| Y0261 | Pichia manshurica | FM199959.1 | 99.85% | 99% | 424 |
| Y0260 | Pichia sp. AUMC 7766 | JQ425352.1 | 99.83% | 100% | 463 |
| Y0256 | Pichia manshurica | OM523901.1 | 99.83% | 100% | 425 |
| Y0255 | Zygosaccharomyces bailii | KP132936.1 | 99.83% | 100% | 495 |
| Y0254 | Pichia manshurica | OM523901.1 | 99.82% | 94% | 745 |
| Y0253 | Pichia kudriavzevii | MN310532.1 | 99.79% | 100% | 472 |
| Y0252 | Pichia kudriavzevii | MN913464.1 | 99.79% | 100% | 453 |
| Y0247 | Pichia manshurica | FM199959.1 | 99.79% | 99% | 421 |
| Y0246 | Saccharomyces cerevisiae | KY104996.1 | 99.78% | 100% | 534 |
| Y0245 | Pichia kudriavzevii | MN913464.1 | 99.78% | 99% | 473 |
| Y0244 | Pichia kudriavzevii | MG183700.1 | 99.78% | 100% | 462 |
| Y0243 | Pichia manshurica | KM368827.1 | 99.75% | 94% | 431 |
| Y0241 | Pichia manshurica | KM368827.1 | 99.74% | 98% | 412 |
| Y0240 | Zygosaccharomyces bailii | KY076624.1 | 99.73% | 100% | 453 |
| Y0239 | Hanseniaspora guilliermondii | KY103523.1 | 99.72% | 99% | 688 |
| Y0238 | Saccharomyces cerevisiae YJM1419 | CP006415.1 | 99.71% | 99% | 601 |
| Y0237 | Pichia kudriavzevii | MN861069.1 | 99.71% | 99% | 466 |
| Y0235 | Saccharomyces cerevisiae YJM1419 | CP006415.1 | 99.65% | 100% | 633 |
| Y0231 | Saccharomyces cerevisiae YJM1419 | CP006415.1 | 99.62% | 99% | 796 |
| Y0230 | Pichia kudriavzevii | MN861069.1 | 99.57% | 99% | 461 |
| Y0228 | Pichia manshurica | MW045578.1 | 99.53% | 97% | 421 |
| Y0227 | Pichia manshurica | OM523901.1 | 99.52% | 100% | 425 |
| Y0226 | Pichia manshurica | OM523901.1 | 99.52% | 99% | 425 |
| Y0225 | Zygotorulaspora sp. | MN721359.1 | 99.49% | 99% | 563 |
| Y0224 | Pichia manshurica | KM368827.1 | 99.42% | 96% | 429 |
| Y0220 | Pichia kudriavzevii | MG183700.1 | 99.42% | 100% | 452 |
| W9i | Wickerhamomyces anomalus | AY231612.1 | 99.37% | 100% | 580 |
| W6iii | Naganishia randhawai | MT542688.1 | 99.37% | 99% | 588 |
| W21iii | Clavispora lusitaniae | KP131863.1 | 99.35% | 99% | 337 |
| W20ii | Candida albicans | KM036428.1 | 99.30% | 99% | 324 |
| W19 | Wickerhamomyces anomalus | MT321266.1 | 99.29% | 100% | 572 |
| W18 | Wickerhamomyces anomalus | MH545921.1 | 99.29% | 99% | 568 |
| W16ii | Papiliotrema laurentii | MN660253.1 | 99.19% | 100% | 491 |
| W14iii | Cyberlindnera fabianii | KU961975.1 | 99.15% | 100% | 578 |
| W12 | Wickerhamomyces anomalus | MT321266.1 | 99.06% | 100% | 574 |
| W11i | Candida albicans | ON851010.1 | 99.05% | 99% | 341 |
| W10 | Wickerhamomyces anomalus | MT321266.1 | 99.05% | 100% | 555 |
| SN221 | Pichia kudriavzevii | MN913464.1 | 98.74% | 100% | 462 |
| S82 | Saccharomyces cerevisiae | KU535591.1 | 98.70% | 100% | 899 |
| S64 | Pichia kudriavzevii | MK298061.1 | 98.56% | 99% | 461 |
| S63 | Pichia kudriavzevii | MT234392.1 | 98.55% | 100% | 453 |
| S62 | Pichia kudriavzevii | MN913464.1 | 98.52% | 100% | 461 |
| S61 | Pichia kudriavzevii | MG183700.1 | 98.50% | 100% | 462 |
| S6 | Brettanomyces bruxellensis | KY103313.1 | 98.49% | 99% | 430 |
| S41 | Pichia kudriavzevii | MN310532.1 | 98.49% | 100% | 465 |
| S31 | Pichia kudriavzevii | MG183700.1 | 98.29% a | 99% | 464 |
| S2B3 | Saccharomyces cerevisiae | KY105010.1 | 98.12% a | 99% | 797 |
| S2B2 | Pichia sp. | MG757432.1 | 97.57% a | 100% | 470 |
| S2B1 | Pichia kudriavzevii | MN310532.1 | 97.57% a | 100% | 464 |
| S22 | Saccharomyces cerevisiae | MK680912.1 | 97.12% a | 100% | 338 |
| S2 | Zygotorulaspora sp. | MN721359.1 | 96.28% a | 100% | 563 |
| S164 | Saccharomyces cerevisiae | KY109257.1 | 96.10% a | 100% | 787 |
| S163 | Saccharomyces cerevisiae | OP562387.1 | 96.00% a | 100% | 781 |
| S141 | Pichia kudriavzevii | MN310532.1 | 95.99% a | 100% | 465 |
| S12 | Pichia kudriavzevii | OK073656.1 | 95.73% a | 99% | 464 |
| OK5 | Papiliotrema flavescens | FN428902.1 | 95.56% a | 99% | 490 |
| N71 | Pichia kudriavzevii | MN913464.1 | 95.49% a | 100% | 467 |
| N61 | Saccharomycodes ludwigii | KY105242.1 | 95.30% a | 100% | 644 |
| N31 | Pichia kudriavzevii | MT102789.1 | 95.25% a | 99% | 466 |
| N2B1 | Pichia kudriavzevii | OP418395.1 | 95.06% a | 99% | 452 |
| N273 | Pichia kudriavzevii | MK587457.1 | 94.90% a | 99% | 471 |
| N25 | Pichia kudriavzevii | KP675519.1 | 94.45% a | 100% | 466 |
| N242 | Pichia kudriavzevii | MK298061.1 | 93.17% a | 99% | 461 |
| N241 | Saccharomyces cerevisiae | KT175188.1 | 93.13% a | 99% | 757 |
| N231 | Pichia sp. | MF662390.1 | 92.92% a | 100% | 462 |
| N191 | Pichia kudriavzevii | KY104590.1 | 91.93% a | 99% | 467 |
| N181 | Pichia kudriavzevii | MN310532.1 | 91.74% a | 97% | 478 |
| N172 | Pichia sporocuriosa | EU315763.1 | 89.18% a | 97% | 460 |
| N161 | Saccharomyces cerevisiae | MK973014.1 | 88.87% a | 100% | 467 |
| N14 | Saccharomyces cerevisiae | KY105078.1 | 86.78% a | 100% | 614 |
| N1 | Pichia kudriavzevii | MN310532.1 | 86.47% a | 100% | 457 |
| 0K10 | Meyerozyma caribbica | NR_149348.1 | 86.14% a | 99% | 563 |
The identities are below the internal transcribed spacer barcoding threshold of yeasts, which is 98.41% (Vu et al. 2016).
Yeast isolation
The inner marula fruit mesocarp and endocarp were finely cut into small pieces using a sterile scalpel and homogenized using a pestle and mortar. One gram of each of the homogenates was transferred to 2 mL microcentrifuge tubes. An aliquot of 1 mL sterile distilled water was then added to each tube and further homogenized using a vortex (Stuart, London, UK). The homogenates were transferred into test tubes containing 2 mL of modified YPD (1% yeast extract, 2% peptone and 2% glucose at a pH of 4) supplemented with a cocktail of antibiotics (ampicillin, streptomycin and tetracycline at 20 µg/mL of each) to inhibit bacterial growth. The test tubes were sealed with parafilm and incubated at 30ºC in a shaking incubator (KS 3000, Thermo Fischer Scientific, Waltham, MA, USA) set at 180 rpm for 24 h. After incubation, an aliquot of 100 µL of the fermentation broth was ten-fold serially diluted (10˗1 to 10˗5) and 100 µL of the dilutions was spread plated onto YPD agar plates. The agar plates were incubated for 2–3 days at 30 ºC. From each sample, representative non-filamentous yeast-like circular colonies were picked based on different morphologies and verified using a compound microscope (Carl Zeiss, Jena, Germany). The colonies were purified by multiple streaking and cryopreserved at ‒80ºC in 25% (v/v) glycerol.
Molecular identification of yeasts
Putative identification of yeasts isolates was carried out by extracting genomic DNA, amplifying the ITS1-5.8S-ITS4 rRNA gene, sequencing and comparing the resultant sequences using NCBI databases. In brief, genomic DNA was extracted using a cell lysis solution containing 200 Mm LiOAc 1% SDS (Lõoke et al. 2011) and amplified with universal primers, ITS1 (5′˗TCCGTAGGTGAACCTGCGG˗3′) and ITS4 (5′˗TCCTCCGCTTATTGATATGC˗3′) (White et al. 1990) using the Applied Biosystems Proflex Thermal cycler (Thermo Fisher, Marsiling, Singapore). The PCR program was run as follows: initial denaturation at 98ºC for 5 min; 45 cycles of denaturation (98ºC for 45 s), primer annealing (54ºC for 1 min), extension (72ºC for 1 min), a final elongation step (72ºC for 7 min).Four Saccharomyces reference strains: Ale yeast (Saccharomyces cerevisiae, strain T58, fermentis, France), Baker's yeast (Saccharomyces cerevisiae, Gold Star, South Africa), Lager yeast (Saccharomyces pastorianus, Lallemand Brewing, Austria), CBS 8340 (Saccharomyces cerevisiae) were also included. The amplicons were sequenced at Inqaba Biotechnological Industries (Pty) Ltd using the Sanger Sequencing method. The sequences were then quality trimmed using BioEdit ver.7.2 (http://www.bioedit.com). Species identification was carried out by comparing with those in the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/blast).
Phylogenetic analysis marula-associated yeasts
Phylogenetic relatedness among marula-associated yeasts, was determined using a Molecular Evolutionary Genetics Analysis software (MEGA X ver.10.2.6) (Kumar et al. 2018). In brief, the ITS1-5.8S-ITS4 sequences were aligned using multiple sequence comparison by log expectation (MUSCLE) (Edgar 2004) inbuilt in the MEGA X software. Evolutionary history was inferred using the Maximum Likelihood method based on the Kimura 2-parameter model (Ranneby 1984). Support was estimated with a setting of 1000 bootstrap replicates. CBS356 strain was used an out-group.
In-silico PCR-restriction fragment length polymorphisms (RFLP) of marula yeasts
An in silico restriction fragment length polymorphism (PCR-RFLP) was used to determine genetic diversity among closely related marula yeasts isolates using the ITS1-5.8S-ITS4 amplicon sequences from the section above. The SnapGene® viewer software ver.4.2.11 (http://www.snapgene.com) was used to view restriction fragment profiles simulated at 4% agarose gel with TBE buffer settings after digestion with 4 restriction enzymes: Hae111, HinfI, CfoI, and BsiEI simultaneously. Biozym Quantitas (25 to 500 bp) was used as a molecular weight marker to estimate the restriction fragment lengths. The amplicon sequences of the 4 reference strains (noted in section 2.2) were used in the simulations together with yeasts from marula fruits based on trimmed consensus sequence regions.
Characterization of inebriation potential of selected marula-associated isolates.
We selected a subset of 29 out of 109 isolates representing 9 species with a generally regarded as safe (GRAS) status (due to our laboratory limitations of working with risk-to-high risk microorganisms). We assessed their fermentative capacity using marula juice as well as their ability to produce aromatic volatile compounds that are a possible important factor that influences foraging of marula fruits by elephants. The fermentative capacity of the isolates was compared to 2 commercial brewing yeasts (Ale yeast (Saccharomyces cerevisiae, strain T58, Fermentis, France and a Lager yeast (Saccharomyces pastorianus, Lallemand Brewing, Austria), one baker's yeast (sometimes used for traditional beer brewing) (Saccharomyces cerevisiae, Gold Star, South Africa) and one laboratory yeast strain Y706 (Saccharomyces cerevisiae CBS 8340). Pure cultures were inoculated in 2 mL YPD broth in 15 mL centrifuge tubes followed by incubation at 30 ºC for 18 h on a rotary shaker set to 180 rpm. After incubation, cells were pelleted by centrifuging at 2000 x g for 2 min before discarding the supernatant. We then washed the cells by suspending the pelleted cells in 5 mL sterile distilled water. This was followed by brief vortexing before centrifuging again under the same conditions. The supernatant was discarded and the washing procedure was repeated twice before cells were used for fermentation assays.
Fermentation of marula fruit juice
Marula juice was extracted from ripened fruits by piercing through its leathery skin using sterile pipette tips. The fruits were pressed by hand and the juice was collected into a 1000 mL Erlenmeyer flask. The juice was diluted with sterile distilled water at a ratio of 1:1 to reduce the viscosity of the juice as reported by Fundira et al. (2002). The freshly pressed and diluted marula juice was stored frozen at −20°C until further analyses. A volume of 5 mL marula juice in 15 mL conical centrifuge tubes was inoculated with pre-grown cells to a final concentration of OD600nm = 1. The tubes were tightly closed and sealed with parafilm and incubated for 2 weeks at 30°C without agitation. After 2 weeks, the fermented broth was centrifuged at 8000 x g for 5 min and sterile filtered through 2.2 µm filters before storing at ‒20°C for further analyses. The accumulated ethanol was quantified using an enzymatic assay kit (K-ETOH 08–18, Megazyme, Ireland) according to the manufacturer's recommendations. The 4 reference strains were included and analyzed simultaneously. The experiment was carried out in triplicates and repeated for a minimum of three times.
Volatile organic compounds analysis
The fermented marula juice from above was also used for analyses of volatile organic compounds. One mL distilled water, 500 µL of NaCl, 400 µL of the sample was added in a 20 mL headspace vial. We added 2.13 mg/L of 2-octanol (dissolved in ethanol) as an internal standard. A Trace GC Ultra gas chromatography cojoined to a TSQ Quantum XLS version mass spectrometer (Thermo Scientific, Milan, Italy) with a joined PAL combi˗xt autosampler (CTC, Zwingen, Switzerland) was used to analyse the organic compounds. A Solid Phase Microextraction (DVB/CAR/PDMS) (Germany) fiber of 2 cm was used for extraction. The compounds were desorbed from the filter and analyzed using the VF-wax GC capillary column (30 m length, 0.25 mm inner diameter, 0.25 µm thick film). The gas chromatograph was set to split-less mode (5 min) at 250°C. Helium gas (5.5 grade) was used as carrier gas at a constant flow of 1.2 mL min−1. The GC oven temperature was initially set at 40°C for 4 min and increased to 250°C (6°C min−1) with a final hold (5 min). Total run-time was 44 min. The results were analyzed in triplicates using R package ver.1.0.12 ‘pheatmap’ software (Kolde 2019) to generate a heatmap.
Statistical analysis
The ethanol produced by the isolates was analyzed using STATISTICA ver.13.2 (StatSoft Inc., Oklahoma, USA). One-way analysis of variance (ANOVA) was used for comparison of means and the Tukey's Post˗Hoc test (95% confidence interval) used to compare multiple paired means.
Results and discussion
Ripe and rotting marula fruits harbor diverse fermenting yeast species
We successfully isolated a total of 106 yeast strains from 75 marula fruits collected from 21 locations in Botswana stretching over 800 km (Fig. 1 and Table 1).
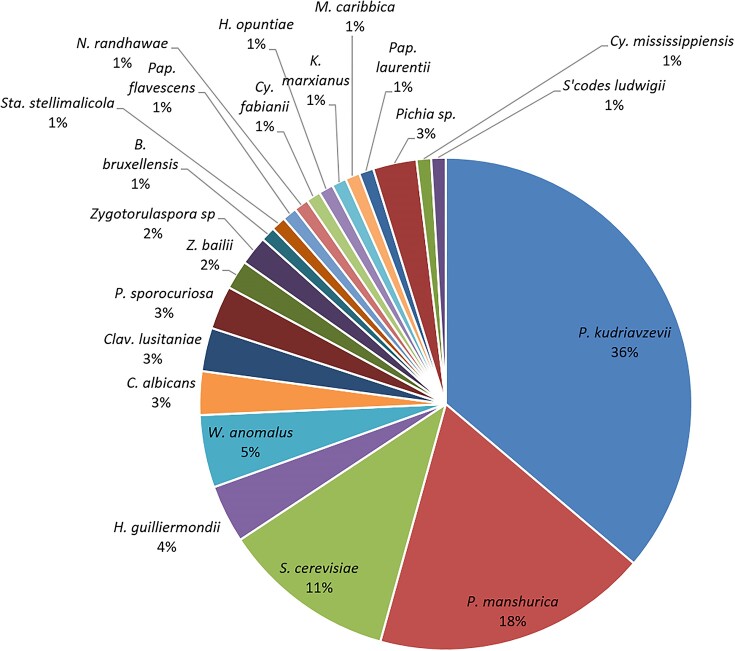
The morphologically Ascomycetous-like smooth or rough surface and white to cream colony colour (data not shown) belonged to 24 yeast species (Fig. 2). Among the 24 yeast species, the most abundant isolates belonged to the Pichia genus (56%) (Fig. 2, Table S2) largely represented by P. kudriavzevii (36%) and P. manshurica (18%).
Figure 2.
The distribution of yeast species of marula isolates. The yeast isolates were obtained from 75 marula fruits collected from 21 localities within a stretch of 800 km (Fig. 1, Table S2).
The high frequency and occurrence of Pichia spp. among the yeast isolates is not surprising as many researchers have reported the predominance of Pichia yeasts in various niches such as fruits, tree barks, the soil and wine (Bhadra et al. 2008, Zhao et al. 2021). The fermentation credentials of members of the Pichia genus is well described in literature (Amaya-Delgado et al. 2013, Holt et al. 2018, Zhang et al. 2021, Scansani et al. 2022). The most abundant members, P. kudriavzevii are described as ethanologenic yeasts with formidable stress tolerance described in cocoa bean and bioethanol fermentations (Daniel et al. 2009, Mukherjee et al. 2017). This genus is reported to efficiently ferment both hexose and pentose sugars, which could be important in increasing the ethanol titers especially from latter carbon sources, which mostly cannot be fermented by a variety of conventional yeasts (Fonseca and Gonwa-Grauslund 2007, Kumar et al. 2009).
The second most common genus was the Saccharomyces genus represented by the Saccharomyces cerevisiae species (with high frequency of 12%). The findings reveal that this species whose fermentative prowess and domestication due to ability to ferment was the third most abundant yeast species. Although S. cerevisiae is known to be found in very low populations in sugar-rich niches such as vineyards and grapes (Fleet 1993, Taylor et al. 2014, Goddard and Greig 2015) the species prevails in fermentation outcompeting many other species with their ability to ferment, leading to higher ethanol concentrations after fermentation (Bauer and Pretorius 2000). Abundance of this species in this study can be attributed to the isolation procedure, which involved a fermentation enrichment stage.
The isolation of Saccharomyces cerevisiae, an ‘industrial workhorse’ with broad alcoholic fermentation applications in baking, winery and brewing (Capece et al. 2018, Gallone et al. 2018) suggests alcoholic fermentation of the marula fruit to intoxicate elephants could be substantiated. One of the distinguishing characteristics of yeasts inhabiting sugar rich niches is the ability to ferment sugars and produce a competitor intoxicating substance, such as ethanol (Dashko et al. 2014). This fermentation process, an ecosystem engineering strategy, generates heat and large amounts of carbon dioxide, which further inhibits heat sensitive and annihilates competitors respectively (Piškur et al. 2006, Goddard and Greig 2015). Yeasts belonging to the two genera (Saccharomyces and Pichia) are well known for their prodigious alcoholic fermentation abilities and could be enlightening in our quest to substantiate the inebriation myth.
To further suggest that there are yeasts with additional fermentative ability to increase the ethanol titers in the fermenting fruits, the results reveal that there are other genera ranked third in abundance such as Hanseniaspora (4%) and Wickerhamomyces (4%). It could be argued that the fermentative ability of these non-Saccharomyces yeasts is documented to be at a lower efficiency when compared to Saccharomyces yeasts (Fleet et al. 1984, Querol et al. 1990, Zhou et al. 2017), but their contribution to the total ethanol titers in the fermenting fruit cannot be neglected. Wickerhamomyces spp. have recently been reported to be applicable as alternative baker's yeast (Zhou et al. 2017, Semumu et al. 2021) suggesting their fermentative capabilities are higher than previously thought.
On the other hand, Hanseniaspora has been reported as one of the most abundant yeast genera on various fruits and musts (Spencer et al. 1992, Vadkertiová et al. 2012). Yeasts of this genus have also been reported to be responsible for spontaneous fermentation of fruit juices (Cadez and Smith 2011). Their presence could significantly; in co-fermentation with Saccharomyces spp. and Pichia spp. elevate the final ethanol concentrations important in understanding the basis of the myth. Additionally, our results document the presence of other well-known fermenting yeasts although at lower frequencies such as Brettanomyces bruxellensis, Candida spp, Hanseniaspora opuntiae, and Kluyveromyces marxianus, among others evident in Fig. 2. Brettanomyces bruxellensis is a wine and beer yeast, whose ability to produce ethanol is comparable to that of S. cerevisiae. The two yeasts are Crabtree positive yeast, well-known for their production of very high concentrations of ethanol (Galafassi et al. 2011). The phenomenon, also known as the Crabtree effect (Pronk et al. 1996), despite its energetic inefficiency when compared to aerobic respiration (Goddard and Greig 2015) together with production of other products of fermentation such as heat and CO2 (Goddard 2008) and a fast consumption of sugars (Dashko et al. 2014) allows yeasts to make, accumulate and consume ethanol in the presence of oxygen (Lin et al. 2012, Tronchoni et al. 2022). This species has a key role in spontaneous beer fermentations (Colomer et al. 2020, Motlhanka et al. 2020) and biofuels (Schifferdecker et al. 2014), therefore, its presence in marula fruits could further elevate the concentrations of ethanol accumulated in the rotting fruits.
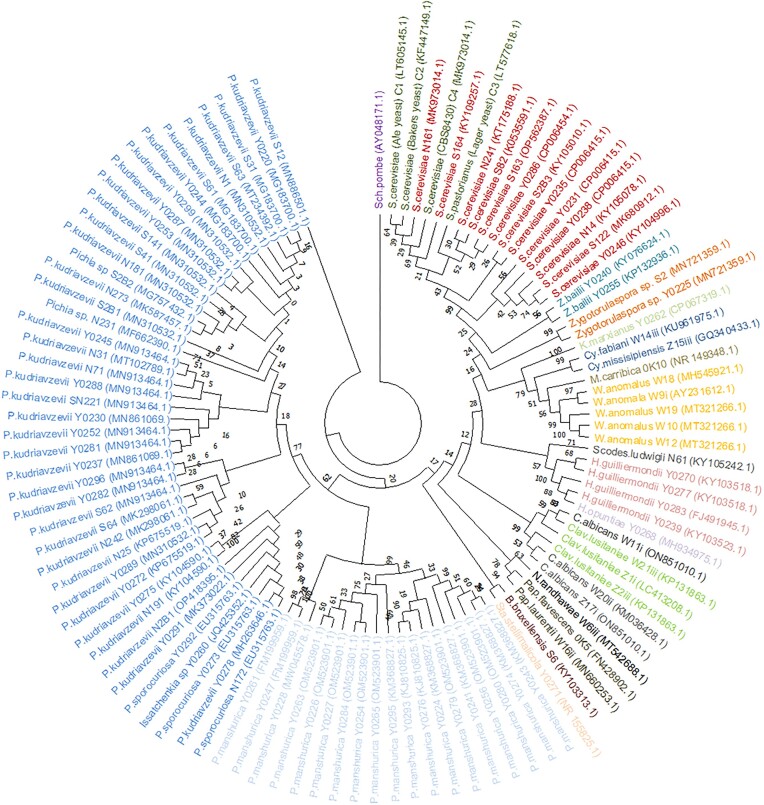
Pre-whole genome duplication (WGD) yeasts typically producing intermediate amounts of ethanol such as Zygotorulaspora spp. and Zygosaccharomyces bailii were also found within the marula fruits niche. The isolation of non- or poor-fermenting yeasts such as Candida spp, Meyerozyma carribica, Cryptococcus and basidiomycetous yeasts such as Papiliotrema laurentii yeasts is normal if the species can prevail even in a fermentation-engineered niche (Zhou 2015). Fermentation credentials among yeasts are more pronounced in yeasts with a phylogenetic proximity to the Saccharomyces spp. as they appear in the phylogenetic tree (Fig. 3). However, other yeasts that evolved to ferment independent of the Saccharomyces yeasts such as the Dekkera/Bruxellensis lineages are well less related to the Saccharomyces yeasts (Rozpędowska et al. 2011).
Figure 3.
Phylogenetic tree depicting Saccharomyces and non-Saccharomyces yeast strains isolated from marula fruits. The phylogenetic tree was constructed using Maximum likelihood analysis and Kimura 2-parameter set at 1000 bootstrap replicates based on the ITS1-5.8S-ITS4 region. Isolates that belong to the same species are highlighted using the same colour.
Alcoholic fermentation in naturally occurring fruits has been cited to be an ecosystems engineering strategy by yeasts (Goddard 2008) to annihilate and outcompete other microorganisms in ephemeral sugar-rich fruits and sap niches characteristic of autumn when fruits ripen (Dashko et al. 2014, Zhou et al. 2017). The colonization of flowers, tree sap and rotten fruits by fermentative yeasts bears probable link to yeasts being responsible for intoxicating animals with fruits and sap diets. Literature further suggests that many animals are inebriated by naturally occurring alcohol albeit at different levels due to variation in abilities to metabolize ethanol (Janiak et al. 2020). The isolation of yeasts from sugar-rich fruits as niches harbouring diverse yeasts is well documented (Conant and Wolfe 2007, Becher et al. 2012, Dashko et al. 2014, Camargo et al. 2018)). The isolation of yeasts from the ancient spontaneous winemaking and traditional brewing is irrefutable evidence of fermenting yeasts. When marula fruits are gathered and crushed as in the traditional African marula wine processing steps, they spontaneously ferment to produce an alcoholic beverage, where yeasts are known to play a major role in this process (Motlhanka et al. 2020, Phiri et al. 2022). The 106 isolates suggest that yeasts from the Saccharomycotina complex with a diverse phylogenetic background dominate marula fruit niches (Fig. 3). In agreement to our studies, colleagues from South Africa, Zimbabwe, Namibia and Swaziland (Shackleton 2002) have also documented evidence of the occurrence of phylogenetically diverse fermenting yeasts belonging to the genera we presented here. A few more genera such as the Metschnikowia and Lachancea have been documented. This indicates that ripened marula fruits are fermented by mixed cultures of yeasts with wide ranging abilities comparable to the currently used industrial yeasts.
In-silico PCR-restriction fragment length polymorphisms (RFLP) reveals genetic diversity within isolates of the same species
We sought to analyze the intra-species genetic variation of marula isolates, which showed a significantly high percentage of similarity of their ITS1-5.8S-ITS4 region. In-Silico PCR-RFLP restriction patterns are comparable to restriction patterns obtained in vitro (Raspor et al. 2007). The precision of discrimination of PCR-RFLP has been considered to be parallel to sequencing analysis and therefore as an alternative method for species identification and delimitation (Pham et al. 2011, da Fonseca Meireles et al. 2022) and the better option in instances where rapid validation or identification is needed considering its simplicity, speed, high reproducibility and high throughput (Raspor et al. 2007). While da Fonseca Meireles et al. (2022) used one restriction enzyme to differentiate yeasts of different genera and species, here we further increase the precision by using multiple restriction enzyme to differentiate within strains .Single restriction enzymes often have poor resolution (similar restriction profiles) in differentiating closely similar sequences of strains, we further revealed that use of a combination of different restriction enzymes was sufficient to resolve the genetic differences.
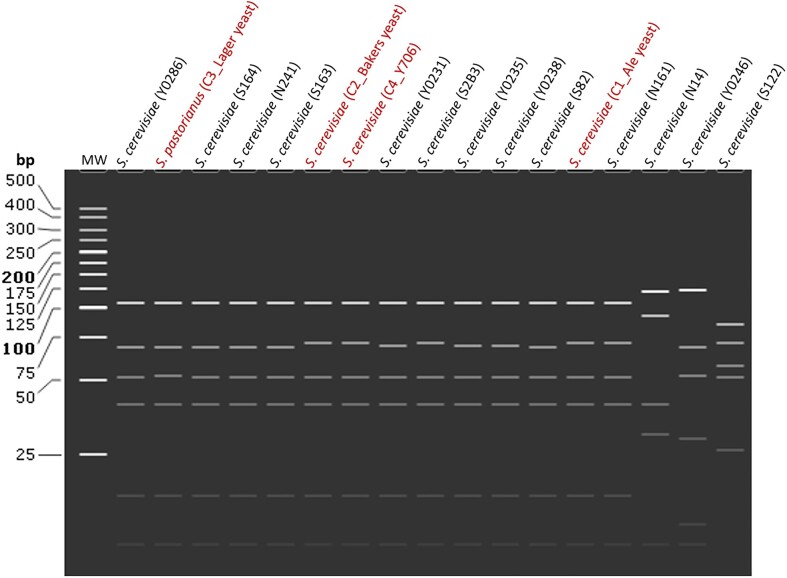
The restriction fragment pattern results show that most of the S. cerevisiae isolates were genetically distinct from each other (see the exact single base pair differences on Table S3) and from the control yeasts (Fig. 4, Supplementary Materials, Table S3). An exception was observed where the restriction fragments profiles of S. cerevisiae (S163) and S. cerevisiae (Y0231) were similar. In addition the fragment profiles of S. cerevisiae (N241) were similar to that of the baker's yeast reference strain. The high degree of genetic variation among isolates belonging to the Saccharomyces genus could suggest differences in fermentation physiology of these yeasts (Pham et al. 2011, Gibson et al. 2017). Subsequently, there is a possibility to increase the titers of ethanol to intoxicate elephants, which further validate the myth (Fig. 4).
Figure 4.
Restriction fragment patterns of the marula isolates. In-silico PCR-RFLP was simulated using Snapgene software to generate fragments from restriction digestion using 4 enzymes: Hae111, Cfo1, Hinfl, BsiEI on trimmed consensus sequences for isolated Saccharomyces yeasts. The restriction fragment list is shown in supplementary materials (Table S3A).
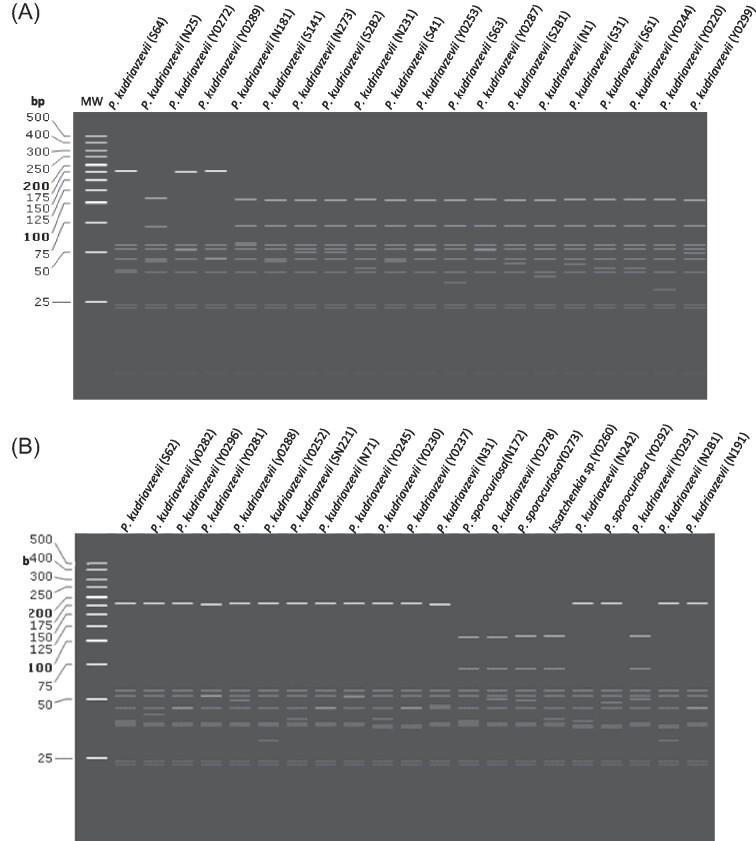
On the other hand, restriction fragment patterns of non-Saccharomyces yeasts presented the highest interspecies strain similarity (Figs. 5 and 6). Yeast species isolated at a low frequency such as H. guilliermondii and W. anomalus had the least genetic variation in comparison to the other isolates (Fig. 6). From the 4 H. guilliermondii isolates, only 1 strain (Y0239) was genetically distinct with 2/8 (25%) variable fragment sizes. The fragment patterns of the H. opuntiae strain Y0268 and S'codes ludwigii strain N61 investigated in this study, shows only 4 and 6 fragments respectively, since the sequences were not able to be cleaved by some restriction enzymes. All 4 W. anomalus isolates were indistinguishable.
Figure 5.
(A&B): Restriction fragment patterns of the marula isolates. In-silico PCR-RFLP was simulated using Snap Gene to generate fragments from restriction digestion using 4 enzymes: Hae111, Cfo1, Hinfl, BsiEI on trimmed consensus sequences for isolated non-Saccharomyces yeasts. The restriction fragment list is shown in supplementary materials (Table S3).
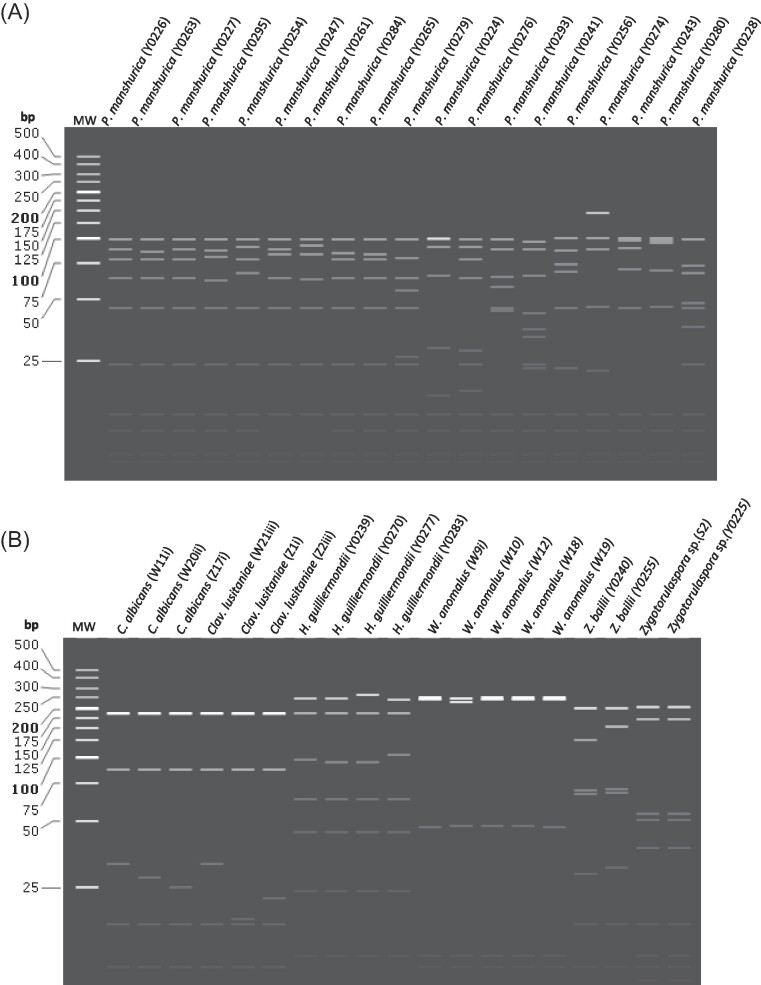
Figure 6.
(A&B): Restriction fragment patterns of the marula isolates. In-silico PCR-RFLP was simulated using Snapgene to generate fragments from restriction digestion using 4 enzymes: Hae111, Cfo1, Hinfl, BsiEI on trimmed consensus sequences for isolated non-Saccharomyces yeasts. The restriction fragment list is shown in supplementary materials (Table S3A).
Out of the 19 P. manshurica isolates, only 2 (10.5%) (Y0226 and Y0227) had the same restriction fragment profile. The C. albicans isolates showed 2/3 (66.7%) strain similarities while P. sporocuriosa, Cryptococcus spp., P. laurentii, and M. caribbica isolates, had different fragment patterns. Pichia kudriavzevii as the most predominant species, presented clusters of isolates with identical profiles: cluster 1 (SN221, Y0244, and S61); cluster 2 (S62, S64 and N242); cluster 3 (N31, and Y0289); cluster 4 (Y0287, Y0272, Y0262, Y0253, Y0296, Y0281, S2B2), cluster 5 (Y0291, and Y0278), cluster 6 (Y0275 and Y0288), cluster 7 (N71, N25, and S141). The remaining 61% of P. kudriavzevii isolates had unique fragment patterns (Fig. 5).
Marula-associated yeasts ferment marula juice: suggestive of inebriation potential
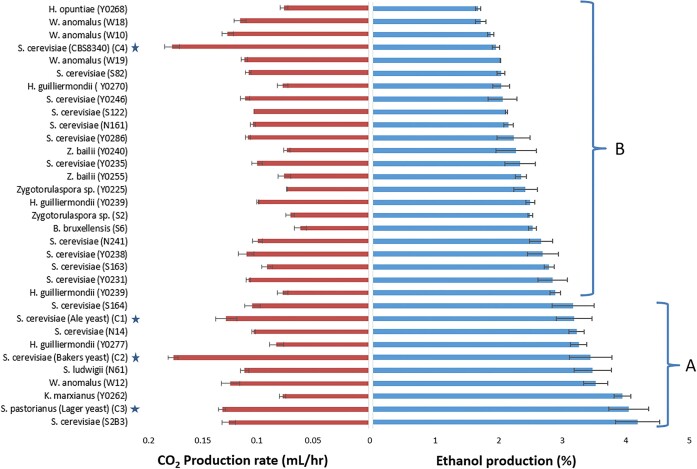
One indispensable trait of yeasts associated with the fermentation of marula fruits and intoxication myth, should be the ability to ferment the sugars found in marula juice and accumulate ethanol. The fermentative capacity of a subset of isolates and the capability to accumulate ethanol using marula juice was investigated. Our results suggest that there is both intra- and inter-species variation among the 29 isolates (compared to the average of reference strains) in terms of the amount of ethanol produced (Fig. 7).
Figure 7.
Fermentation capacity based on CO2 production rate and amount of ethanol produced (%) by the isolates and the reference strains when fermented in marula juice. Ethanol production and fermentation capacity of the isolates relative to the ethanol produced by 4 control yeasts (blue star); S. cerevisiae, Ale yeast (C1); S. cerevisiae, Baker's yeast (C2); S. pastorianus, Lager yeast (C3) and S. cerevisiae, CBS8340 (C4); averaged and represented by blue stars. Group A shows isolates with ethanol production higher than 3% while Group B shows isolates with ethanol production lower than 3%. However, the carbon dioxide production rate was highly variable among the isolates (see attached Table S4 and 5 in Supplementary Material).
The isolates were divided into 2 groups, based on the amounts of ethanol they accumulated in comparison to amounts produced by commercial brewing industrial strains. Group A comprised of 7 Saccharomyces and non-Saccharomyces isolates that produced similar ethanol concentrations (on average 3.55% (v/v) to the one produced by the averaged control yeast while Group B yeasts produced significantly lower ethanol concentrations (on average 2.26% (v/v)) when compared to the averaged control yeast. The ethanol accumulation of the average of control yeasts was also compared to individual reference strains and the results suggest that the yeasts could be grouped based on their potential ethanol production (Fig. 7). The most paramount question on the intoxication of elephants’ myth is how much ethanol is produced when marula fruits are fermented to be able to inebriate elephants. Although these findings do not provide evidence of the inebriation potential, there is not enough evidence to debunk the myth.
A previous study of marula fermentations reported ethanol production within the range of 2%–5% (v/v) (Hiwilepo-van Hal et al. 2014). These ethanol concentrations are in agreement with our findings. Although there is limited information on the amount of ethanol required to intoxicate an elephant. On average, a 3000 kg elephant would require about 10–27 L of 7% (v/v) to intoxicate it (Morris et al. 2006). It is noteworthy that this is entirely correct as it is just an assumption based on human physiology and may not be the case with that of elephants. If the above assertion holds, consuming about double the amount presented i.e. 20–54 L since the average amount of ethanol produced by all isolates was 2.673%, would intoxicate such an elephant. This is a very practical amount of fermented juice at a single instance, considering that an adult elephant can consume about 300 kg of vegetation (Laws 1970, Stephenson and Ntiamoa-Baidu 2010). Although elephants preferentially feed on fruits (White et al. 1993), they do not exclusively feed on fruits, foraging on just half of the possible daily feeds (150 200 kg) this could be over 11 000 fruits if each fruit weigh about 18 g on average as described by Tapiwa (2019). Each fruit contains on average 3–8 mL of fermentable juice (results based on our observations when we prepared the juices but not shown), there could be 33 L–56 L of juice available for fermentation. The relative amounts of sugars in a single fruit also determine the amount of possible ethanol. The average sugar content has been recorded to be up to 16° Brix depending on the season and environment (Suárez et al. 2012, Phiri et al. 2022). The question is, are there enough marula fruits for a single elephant to forage on huge numbers of fruits to yield enough juice and volume of ethanol? There is a huge density of marula trees in Botswana ranging from 1.6 trees per hectare in arid regions (covering over 3.9 million ha) to 23 trees per hectare in the Okavango Delta (covering close to 2 million ha) (Neuenschwander et al. 2002, Wynberg et al. 2002, Batisani and Yarnal 2010). Therefore the number of available fruits per elephant is highly unlikely to be limiting. On average a single marula plant can produce about 1400 kg of fruits (i.e. about 78 000 of fruits at 18–30 g each) (Venter and Venter 1996, Botelle et al. 2002, Hiwilepo-van Hal et al. 2013, Tapiwa 2019). Our results show that our marula isolates produced from 1.67% to 4.19% ethanol, which is suggestively close enough to the ascertained required amounts to intoxicate an elephant. The highest ethanol production reported in this study was 4.2 ± 0.34% (v/v) by using a single culture of S. cerevisiae strain S2B3. However, wild marula yeasts ferment the fruits as mixed cultures that may result in higher titers of ethanol produced in spontaneous fermentations of the marula juice. Some early studies suggested ethanol production of as high as 7% (v/v) per marula fruit (Eriksson and Nummi 1983, Dudley 2000). Although domesticated commercial brewing yeasts produce high yields of ethanol using brewing wort (with mostly maltose as the abundant sugar), (Morris et al. 2006, Gibson et al. 2017), they were not the best fermenters in non-starchy marula juice (80 g/L sucrose, 17 g/L glucose and 17 g/L fructose (Phiri et al. 2022). The 4 Saccharomyces reference strains used: ale yeast (Saccharomyces cerevisiae, strain T58, fermentis, France), baker's yeast (Saccharomyces cerevisiae, Gold Star, South Africa), lager yeast (Saccharomyces pastorianus, Lallemand Brewing, Austria), CBS8340 (Saccharomyces cerevisiae) produced 3.19 ± 0.28, 2.35 ± 0.15, 3.45 ± 0.33, and 4.05 ± 0.32% (v/v) of ethanol, respectively. This study suggests that marula fruits can contain a significant amount of ethanol, but if this amount is sufficient enough to inebriate elephants remains elusive.
In addition to the possible amount of ethanol produced when yeasts spontaneously ferment marula juice when they are attached to the plant or when they rot after abscission, fermentation may continue in the stomach of the elephant. The resident time food takes in the elephant's gut, which is reported as at 36–48 hours (Morris et al. 2006, Viljoen 2013) could be one factor that increase the final ethanol titers. Another suggested possibility to support this myth, is that upon ingestion of the fruit, the elephants do not crush all the fruit thus continued fermentation could persist in the elephants’ gut. Other than the amounts of ethanol in fermented marula fruits, and the body size of the elephant, there is another factor that could be of importance in substantiating the myth i.e. the inability of elephants to metabolize ethanol efficiently when compared to human beings. Recent studies suggest that class IV gene alcohol dehydrogenase gene (ADH7), a gene involved in the breakdown of ethanol, in both African and Asian elephants is non-functional (Janiak et al. 2020). In addition, human beings have a mutation (a gene inactivating stop codon) on the ADH7 gene, which makes them breakdown ethanol about 40 times faster than most primates (Morris et al. 2006, Carrigan et al. 2015). Therefore, there is a possibility that even lower amounts of ethanol than known could inebriate elephants when compared to human beings if the inability to detoxify themselves of ethanol is important. Therefore the inebriation myth requires a multi-dimensional approach and may not be debunked by assuming the size of the body logic, the amount of ethanol compared to the amount that is known to intoxicate human beings. Although other possible ethanol breakdown pathways not involving the ADH7 gene may exist in elephants, there isn't enough evidence thus far to reject the inebriation myth. Genetic polymorphisms of alcohol metabolizing enzymes have also been suggested on some organisms such as treeshrews (Wiens et al. 2008). These organisms have evolved to increase the amount of alcohol intake without being inebriated. It's apparent that evolutionary solutions to alcohol inebriation are varied among organism in a wide phylogenetic history. Even among members of the same species, some organisms tend to be inefficient in dealing with challenges, for example members of the Asian population have an aldehyde dehydrogenase ALDH variant gene which produces a nonfunctional enzyme (Agarwal 2001). Therefore, in order to disprove the myth, more research is needed. For example, it would be beneficial to examine the effects of known alcohol concentrations on elephant intoxication, compare the results of these studies between two genetically distinct elephant populations, inoculate marula fruit with various types of yeasts to study potential intoxication, and conduct other studies.
Marula yeasts produce wide aromatic bouquets: a possible attractant for elephant foraging
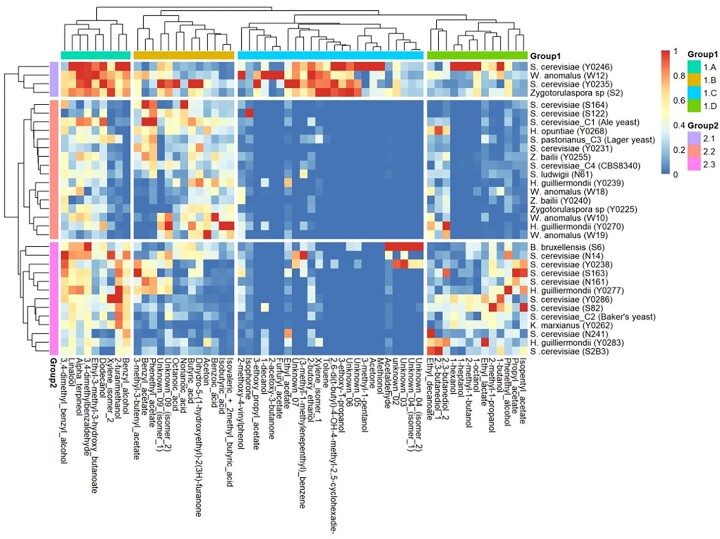
Other than the effect of the euphoric substance, elephants are thought to be drawn to the aromatic bouquets produced by ripe marula fruits (Nevo et al. 2020). The fermentation of sugars, often mistaken as the microbial rotting of fruits, is known to produce a variety of aromatic metabolites. If the aromatic profiles are important in elephants’ foraging behaviours whose olfactory system is well-developed than most animals (von Dürckheim et al. 2018), then it was necessary to characterise the aroma compounds produced by different marula-associated yeast isolates. Our findings suggest that the aroma profiles of marula juice fermented by selected marula-associated isolates were very diverse, clustering into four groups (Fig. 8). A large proportion of the yeasts (Group 2.2) were primarily distinguished by the production of moderate organic acids such as isovaleric, isobutyric, and nonanoic acids. Does the contribution of these notes to bittering and sourness in beverages (Thompson Witrick et al. 2017) matter in the preference for foraging in elephants? Although no empirical studies have explained the effects of such acids in foraging, it is likely that these notes underlie foraging preference in elephants.
Figure 8.
Hierarchical clustering of heatmap representing volatile aromatic compounds produced by 29 isolates after fermentation of marula juice. Varying degrees of red show relatively high volatile compound concentration, while varying degrees of blue show relatively low volatile compound concentrations (Table S8).
Based on the clustering algorithm, there are four main groups of the volatile aromatic compounds (Group 1) and three main groups of isolates (Group 2). The volatile organic compounds in Group 1D (green) comprise of a larger proportion of higher alcohols and relatively less of esters. Such a product will have a solvent-like aroma and a fruity or floral contribution from esters. They were mostly produced by isolates in Group 2.3 (pink) closely associated with the baker's yeast strain. On the other hand, Group 1C (blue) aromas had high concentrations of acetate esters and other solvents like acetone and toluene. Typically such high concentrations of acetate may be responsible for pleasant aromas such as apple, pear, strawberry, or floral notes like rose or violet in wines (Gutiérrez et al. 2018). These were mostly produced by isolates in Group 2.1 (purple), which had four isolates belonging to three different genera, Saccharomyces, Wickerhamomyces, and Zygotorulaspora. The volatile organic compounds in Group 1B (gold) show high concentrations of organic acids and acetate esters, while Group 1A (bright teal) shows an even distribution of diverse compounds including alcohols, esters and terpenes. The isolates in Group 2.2 (peach) dominated the Group 1B, and this is where we had the largest proportion of the yeast strains including the ale and lager yeasts. Although the Group 1A had all the yeasts featured, the highest concentrations were among Group 2.1 isolates.
Overally, the isolates produced complex aroma profile including higher alcohols, esters, acids, ketones, terpenes, aldehydes, and furans which impart diverse flavors at particular thresholds. If all these aromatic notes are the most important in elephant foraging, then our isolates exhibit the best credentials to attract elephants. Furthermore, several studies show that moderate concentrations of higher alcohols produced by group D and F of yeast contribute to the desired warm mouth-feel tone found in most beers, along with green herbal aromas due to acetaldehyde production (Callejo et al. 2019, Viejo et al. 2019, Einfalt and Technology 2021). All groups of isolates produce esters at varying concentrations, and some produce aldehydes, alcohols, along with some unknown aromatic compounds which altogether impart fresh floral and fruity aromas. These metabolites have been proposed to act as signals that animals use to find ripe fruits in monkeys and bats (Hodgkison et al. 2013, Nevo et al. 2015). A behavioral and chemical assay study by (Nevo et al. 2020) suggest that elephants use marula fruit aroma profiles to choose fruits with highest sugar content. Some volatile aromatic compounds produced from the fermentation of marula fruits resulted in burnt plastic and horse sweat associated aromas, a Dekk/Bretts characteristic (Vanbeneden et al. 2008, Lentz 2018, Callejo et al. 2019, Motlhanka et al. 2020).
Although primary metabolites were initially thought to be the main signals with a direct correlation with the sugar levels of the fruits (Dudley 2004), recent research has also revealed the significance of secondary metabolites, mainly esters as signals for the nutritional sugar content and quality of the fruits. Elephants have a high preference for high sugar contents marula fruits (Nevo et al. 2020) which could be directly correlated with the ethanol levels: high sugar containing marula fruits are likely to produce high ethanol titers (Dominy 2004) but negatively correlated with the concentration of ethyl acetates (Nevo et al. 2020). This could possibly account for the inebriation of elephants after ingesting marula fruits: in search of high sugar content in marula fruits, elephants are more directed towards fermented fruits with more ethanol.
Conclusion
The inebriation of elephants is a persistent myth that has baffled humankind. To debunk or approve of the myth, studies on the presence of fermentative yeast species to account for sufficient ethanol to inebriate elephants are important. Our work suggests that there is a high diversity of fermentative yeasts resident on the marula tree fruits whose fermentative capacity could be responsible for the inebriation of elephants. The yeasts were dominated by members of the Saccharomycetaceae family whose elevated fermentative capacity is in agreement with our findings. Although the inebriation of elephants is dependent upon many other factors such as the amounts of ethanol per given fruit and the ability to efficiently metabolise ethanol, the fermentative capacity of yeasts is an important trail towards understanding inebriation of elephants from ingestion of marula fruits. In addition, our study revealed that marula-associated isolates produce varying amounts of aromatic chemicals, which could be essential in establishing the foraging behaviour of elephants towards the potentially inebriating and fermented fruits. However, more research is needed to explore the inebriation potential of all the diverse non-Saccharomyces and Saccharomyces yeasts in controlled mixed culture fermentations and the ability of the elephants to match the fermented juice to inebriating levels.
Supplementary Material
Acknowledgments
This is a short text to acknowledge the contributions of specific colleagues, institutions, or agencies that aided the efforts of the authors. The authors would like to thank MarGarcia‐Aloy from the Metabolomics Unit, Food Quality and Nutrition Department, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige in Italy for assisting with the heatmap presentations.
Contributor Information
Tawanda Proceed Makopa, Department of Biological Sciences and Biotechnology, Botswana International University of Science and Technology, Plot 10071, Boseja, Palapye, Botswana, 00267.
Gorata Modikwe, Department of Biological Sciences and Biotechnology, Botswana International University of Science and Technology, Plot 10071, Boseja, Palapye, Botswana, 00267.
Urska Vrhovsek, Food Quality and Nutrition, Research and Innovation Centre, Fondazione Edmund Mach, San Michelle All'Adige, Via E. Mach, 1, Italy, 38010.
Cesare Lotti, Food Quality and Nutrition, Research and Innovation Centre, Fondazione Edmund Mach, San Michelle All'Adige, Via E. Mach, 1, Italy, 38010.
José Paulo Sampaio, UCIBIO, Departamento de Ciencias da Vida, Faculdade de Ciencias e Tecnologia, Universidade Nova de Lisboa, Caparica, Portugal , 2829-516.
Nerve Zhou, Department of Biological Sciences and Biotechnology, Botswana International University of Science and Technology, Plot 10071, Boseja, Palapye, Botswana, 00267.
Author contributions
TPM: drafted, wrote and revised the manuscript
GM: Isolated the yeasts, analysed their fermentative credentials, reviewed the manuscript
UV: Critically revised the manuscript
CL: Analysed the aroma profiles
JPS: Critically revised the manuscript
NZ: conception, design, analysis and interpretation of data and wrote the manuscript
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
The research work was funded by Botswana International University of Science and Technology (BIUST).
References
- Agarwal DP. Genetic polymorphisms of alcohol metabolizing enzymes. Pathol Biol (Paris). 2001;49:703–9. [DOI] [PubMed] [Google Scholar]
- Amaya-Delgado L, Herrera-López E, Arrizon J et al. Performance evaluation of Pichia kluyveri, Kluyveromyces marxianus and Saccharomyces cerevisiae in industrial tequila fermentation. World J Microbiol Biotechnol. 2013;29:875–81. [DOI] [PubMed] [Google Scholar]
- Azeem S, Bengis R, Van Aarde R et al. Mass die-off of African elephants in Botswana: pathogen, poison or a perfect storm?. S Afr J Wildl Res. 2020;50:149–56. [Google Scholar]
- Batisani N, Yarnal B. Rainfall variability and trends in semi-arid Botswana: implications for climate change adaptation policy. Appl Geogr. 2010;30:483–9. [Google Scholar]
- Bauer F, Pretorius IS. Yeast stress response and fermentation efficiency: how to survive the making of wine. S Afr J Enol Vitic. 2000;21:27–51. [Google Scholar]
- Becher PG, Flick G, Rozpędowska E et al. Yeast, not fruit volatiles mediate Drosophila melanogaster attraction, oviposition and development. Funct Ecol. 2012;26:822–8. [Google Scholar]
- Bhadra B, Rao RS, Singh PK et al. Yeasts and yeast-like fungi associated with tree bark: diversity and identification of yeasts producing extracellular endoxylanases. Curr Microbiol. 2008;56:489–94. [DOI] [PubMed] [Google Scholar]
- Botelle A, du Plessis P, Pate K et al. A survey of marula fruit yields in North-Central Namibia. CRIAA SA-DC. 2002.
- Cadez N, Smith MT. Hanseniaspora Zikes (1912). The Yeasts, 2011; pp. 421–34. Elsevier. [Google Scholar]
- Callejo M, Navas JG, Alba R et al. Wort fermentation and beer conditioning with selected non-Saccharomyces yeasts in craft beers. Eur Food Res Technol. 2019;245:1229–38. [Google Scholar]
- Camargo JZ, Nascimento VM, Stefanello I et al. Biochemical evaluation, molecular characterization and identification of novel yeast strains isolated from Brazilian savannah fruits, chicken litter and a sugar and alcohol mill with biotechnological potential for biofuel and food industries. Biocatal Agric Biotechnol. 2018;16:390–9. [Google Scholar]
- Capece A, Romaniello R, Pietrafesa A et al. Use of Saccharomyces cerevisiae var. Boulardii in co-fermentations with S. cerevisiae for the production of craft beers with potential healthy value-added. Int J Food Microbiol. 2018;284:22–30. [DOI] [PubMed] [Google Scholar]
- Carrigan MA, Uryasev O, Frye CB et al. Hominids adapted to metabolize ethanol long before human-directed fermentation. Proc Natl Acad Sci. 2015;112:458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer MS, Chailyan A, Fennessy RT et al. Assessing population diversity of brettanomyces yeast species and identification of strains for brewing applications. Front Microbiol. 2020;11:637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol. 2007;3:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke L. The Truth about Animals: Stoned Sloths, Lovelorn Hippos, and Other Tales from the Wild Side of Wildlife. New York, USA: Basic Books, 2018. [Google Scholar]
- Cunningham AB. The regional distribution, marketing and economic value of the palm wine trade in the ingwavuma district Natal, South Africa. S Afr J Bot. 1990;56:191–8. [Google Scholar]
- da Fonseca Meireles S, dos Santos SF, Rafael MS et al. Yeasts from the nests of two Amazonian stingless bees: screening and PCR-RFLP molecular analysis. Symbiosis. 2022;87:153–63. [Google Scholar]
- Daniel H-M, Vrancken G, Takrama JF et al. Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Res. 2009;9:774–83. [DOI] [PubMed] [Google Scholar]
- Dashko S, Zhou N, Compagno C et al. Why, when, and how did yeast evolve alcoholic fermentation?. FEMS Yeast Res. 2014;14:826–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis JJBW. If you drink, don't fly: fermented fruit and sap can inebriate birds. Bird World. 1987;1:15–9. [Google Scholar]
- Dominy NJ. Fruits, fingers, and fermentation: The sensory cues available to foraging primates. Integr Comp Biol. 2004;44:295–303. [DOI] [PubMed] [Google Scholar]
- Dudley R, Maro A. Human evolution and dietary ethanol. Nutrients. 2021;13:2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley R. Ethanol, Fruit Ripening, and the Historical Origins of Human Alcoholism in Primate Frugivory. Integr Comp Biol. 2004;44:315–23. [DOI] [PubMed] [Google Scholar]
- Dudley R. Evolutionary origins of human alcoholism in primate frugivory. Q Rev Biol. 2000;75:3–15. [DOI] [PubMed] [Google Scholar]
- Dudley R. The Drunken Monkey: Why We Drink and Abuse Alcohol. Berkeley, CA, USA: Univ of California Press, 2014. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einfalt D. Barley-sorghum craft beer production with Saccharomyces cerevisiae, Torulaspora delbrueckii and Metschnikowia pulcherrima yeast strains. Eur Food Res Technol. 2021;247:385–93. [Google Scholar]
- Eriksson K, Nummi H. Alcohol accumulation from ingested berries and alcohol metabolism in passerine birds. Ornis Fenn. 1983;60:2–9. [Google Scholar]
- Fleet G, Lafon-Lafourcade S, Rib reau-Gayon P. Evolution of yeasts and lactic acid bacteria during fermentation and storage of Bordeaux wines. Appl Environ Microbiol. 1984;48:1034–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet G. Yeast-growth during fermentation. Wine Microbiology and Biotechnology. 1993;27–54.
- Fonseca H-HBKK, Gonwa-Grauslund CS-MI. MF, 2007. Towards industrial pentose–fermenting yeast strains. Appl Microbiol Biotechnol. 2007;74:937. [DOI] [PubMed] [Google Scholar]
- Fundira M, Blom M, Pretorius I et al. Selection of yeast starter culture strains for the production of marula fruit wines and distillates. J Agric Food Chem. 2002;50:1535–42. [DOI] [PubMed] [Google Scholar]
- Galafassi S, Merico A, Pizza F et al. Dekkera/Brettanomyces yeasts for ethanol production from renewable sources under oxygen-limited and low-pH conditions. J Ind Microbiol Biotechnol. 2011;38:1079–88. [DOI] [PubMed] [Google Scholar]
- Gallone B, Mertens S, Gordon JL et al. Origins, evolution, domestication and diversity of Saccharomyces beer yeasts. Curr Opin Biotechnol. 2018;49:148–55. [DOI] [PubMed] [Google Scholar]
- Gibson B, Geertman J-M, Hittinger C et al. New yeasts—New brews: modern approaches to brewing yeast design and development. FEMS Yeast Res. 2017;17:fox038. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Greig D. Saccharomyces cerevisiae: a nomadic yeast with no niche?. FEMS Yeast Res. 2015;15:fov009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard MR. Quantifying the complexities of Saccharomyces cerevisiae's ecosystem engineering via fermentation. Ecology. 2008;89:2077–82. [DOI] [PubMed] [Google Scholar]
- Goosen H, Holtzhausen L, Van W et al. The marula is tamed. South African Panorama. 1985;30:21–5. [Google Scholar]
- Guerra-Doce E. The origins of inebriation: archaeological evidence of the consumption of fermented beverages and drugs in prehistoric Eurasia. J Archaeol Method Theory. 2015;22:751–82. [Google Scholar]
- Gutiérrez A, Boekhout T, Gojkovic Z et al. Evaluation of non-saccharomyces yeasts in the fermentation of wine, beer and cider for the development of new beverages. J Inst Brew. 2018;124:389–402. [Google Scholar]
- Hiwilepo-van Hal P, Bille PG, Verkerk R et al. A review of the proximate composition and nutritional value of Marula (Sclerocarya birrea subsp. caffra). Phytochem Rev. 2014;13:881–92. [Google Scholar]
- Hiwilepo-van Hal P, Robben J, Verkerk R et al. Optimising the juice yield and quality of marula fruit (Sclerocarya birrea subsp. Caffra) with pectolytic enzymes by a response surface method. Processing of Marula (Sclerocarya Birrea Subsp Caffra) Fruits: A Case Study on health-promoting Compounds in Marula Pulp. 2013;51:49–51. [Google Scholar]
- Hodgkison R, Ayasse M, Häberlein C et al. Fruit bats and bat fruits: the evolution of fruit scent in relation to the foraging behaviour of bats in the New and Old World tropics. Funct Ecol. 2013;27:1075–84. [Google Scholar]
- Holt S, Mukherjee V, Lievens B et al. Bioflavoring by non-conventional yeasts in sequential beer fermentations. Food Microbiol. 2018;72:55–66. [DOI] [PubMed] [Google Scholar]
- Janiak MC, Pinto SL, Duytschaever G et al. Genetic evidence of widespread variation in ethanol metabolism among mammals: revisiting the ‘myth' of natural intoxication. Biol Lett. 2020;16:20200070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. pheatmap: Pretty Heatmaps. R package version 1.0. 12. 2019.
- Krige EJ. Note on the Phalaborwa and their Morula Complex. Bantu Stud. 1937;11:357–66. [Google Scholar]
- Kumar A, Singh L, Ghosh S. Bioconversion of lignocellulosic fraction of water-hyacinth (Eichhornia crassipes) hemicellulose acid hydrolysate to ethanol by Pichia stipitis. Bioresour Technol. 2009;100:3293–7. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M et al. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langman VA,Roberts TJ,Black J et al. Moving cheaply: energetics of walking in the African elephant. J Exp Biol. 1995;3:629–32. [DOI] [PubMed] [Google Scholar]
- Laws RM. Elephants as agents of habitat and landscape change in east africa. Oikos. 1970;1–15. [Google Scholar]
- Lentz MR. The impact of simple phenolic compounds on beer aroma and flavor. Fermentation. 2018;4:20. [Google Scholar]
- Lewis DM. Fruiting patterns, seed germination, and distribution of Sclerocarya caffra in an elephant-inhabited woodland. Biotropica. 1987;19:50–6. [Google Scholar]
- Lin Y, Zhang W, Li C et al. Factors affecting ethanol fermentation using Saccharomyces cerevisiae BY4742. Biomass Bioenergy. 2012;47:395–401. [Google Scholar]
- Lõoke M, Kristjuhan K, Kristjuhan AJB. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques. 2011;50:325–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluleke E. Characterisation of the microorganisms and determination of the chemical constituents of Marula brews during fermentation. Thesis. 2019.
- Mkwezalamba I, Munthali CR, Missanjo E. Phenotypic variation in fruit morphology among provenances of Sclerocarya birrea (A. Rich.) Hochst. Intern J Forestry Res. 2015;2015:1–8. [Google Scholar]
- Morris S, Humphreys D, Reynolds D. Myth, marula, and elephant: an assessment of voluntary ethanol intoxication of the African elephant (Loxodonta africana) following feeding on the fruit of the marula tree (Sclerocarya birrea). Physiol Biochem Zool. 2006;79:363–9. [DOI] [PubMed] [Google Scholar]
- Motlhanka K, Lebani K, Boekhout T et al. Fermentative microbes of khadi, a traditional alcoholic beverage of botswana. Fermentation. 2020;6:51. [Google Scholar]
- Mpofu A, Kock J, Pretorious E et al. Identification of yeasts isolated from mukumbi, a Zimbabwean traditional wine. J Sustain Dev Afr. 2008;10:88–102. [Google Scholar]
- Mugochi T, Parawira W, Mpofu A et al. Survival of some species of Salmonella and Shigella in mukumbi, a traditional Zimbabwean wine. Int J Food Sci Nutr. 1999;50:451–5. [DOI] [PubMed] [Google Scholar]
- Mukherjee V, Radecka D, Aerts G et al. Phenotypic landscape of non-conventional yeast species for different stress tolerance traits desirable in bioethanol fermentation. Biotechnol Biofuels. 2017;10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenschwander AL, Crawford MM, Ringrose S. Monitoring of Seasonal Flooding in the Okavango Delta Using EO-1 Data. Vol. 6:2002; pp. 3124–6. IEEE. [Google Scholar]
- Nevo O, Garri RO, Salazar LTH et al. Chemical recognition of fruit ripeness in spider monkeys (Ateles geoffroyi). Sci Rep. 2015;5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo O, Schmitt MH, Ayasse M et al. Sweet tooth: elephants detect fruit sugar levels based on scent alone. Ecol Evolut. 2020;10:11399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T, Wimalasena T, Box W et al. Evaluation of ITS PCR and RFLP for differentiation and identification of brewing yeast and brewery ‘wild'yeast contaminants. J Inst Brew. 2011;117:556–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phiri A, La Grange D, Moganedi KJB. Microbial and chemical dynamics during marula wine fermentation. Beverages. 2022;8:50. [Google Scholar]
- Piškur J, Rozpędowska E, Polakova S et al. How did Saccharomyces evolve to become a good brewer?. Trends Genet. 2006;22:183–6. [DOI] [PubMed] [Google Scholar]
- Pronk JT, Yde Steensma H, Van Dijken JP. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–33. [DOI] [PubMed] [Google Scholar]
- Querol A, Jiménez M, Huerta T. Microbiological and enological parameters during fermentation of musts from poor and normal grape-harvests in the region of Alicante (Spain). J Food Science. 1990;55:1603–6. [Google Scholar]
- Ranneby B. The maximum spacing method. An estimation method related to the maximum likelihood method. Scand J Stat. 1984;93–112.
- Raspor P, Zupan J, Čadež N. Validation of yeast identification by in silico RFLP. J Rapid Method Autom Microbiol. 2007;15:267–81. [Google Scholar]
- Rozpędowska E, Hellborg L, Ishchuk OP et al. Parallel evolution of the make–accumulate–consume strategy in Saccharomyces and Dekkera yeasts. Nat Commun. 2011;2:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scansani S, van Wyk N, Nader KB et al. The film-forming pichia spp. In a winemaker's toolbox: a simple isolation procedure and their performance in a mixed-culture fermentation of Vitis vinifera L. cv. Gewürztraminer must. Int J Food Microbiol. 2022;365:109549. [DOI] [PubMed] [Google Scholar]
- Schifferdecker AJ, Dashko S, Ishchuk OP et al. The wine and beer yeast Dekkera bruxellensis. Yeast. 2014;31:323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semumu T, Gamero A, Boekhout T et al. Evolutionary engineering of Wickerhamomyces subpelliculosus and Kazachstania gamospora for baking. 2021. [DOI] [PubMed]
- Shackleton S. The informal marula beer traders of Bushbuckridge, Limpopo Province, South Africa. Rhodes University, South Africa. 2002. [Google Scholar]
- Spencer DM, Spencer J, De Figueroa L et al. Yeasts associated with rotting citrus fruits in Tucumán, Argentina. Mycol Res. 1992;96:891–2. [Google Scholar]
- Stephenson PJ, Ntiamoa-Baidu YJO. Conservation planning for a widespread, threatened species: WWF and the African elephant Loxodonta africana. Oryx. 2010;44:194–204. [Google Scholar]
- Suárez C, Beckett K, Adel S et al. Investigation of the marula fruit ripening process: correlation between quality aspects and local knowledge of Marula fruit. Agro Food Ind Hi-Tech. 2012;23:20–2. [Google Scholar]
- Tapiwa KA. Harvesting and utilization of Marula (Sclerocarya birrea) by smallholder farmers: a review. JOJ Wildlife and Biodiversity. 2019;1:76–9. [Google Scholar]
- Taylor MW, Tsai P, Anfang N et al. Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ Microbiol. 2014;16:2848–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson Witrick K, Duncan SE, Hurley KE et al. Acid and volatiles of commercially-available lambic beers. Beverages. 2017;3:51. [Google Scholar]
- Tronchoni J, Gonzalez R, Guindal AM et al. Exploring the suitability of Saccharomyces cerevisiae strains for winemaking under aerobic conditions. Food Microbiol. 2022;101:103893. [DOI] [PubMed] [Google Scholar]
- Vadkertiová R, Molnárová J, Vránová D et al. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can J Microbiol. 2012;58:1344–52. [DOI] [PubMed] [Google Scholar]
- Van Wyk B-E. The potential of South African plants in the development of new food and beverage products. S Afr J Bot. 2011;77:857–68. [Google Scholar]
- Vanbeneden N, Gils F, Delvaux F et al. Formation of 4-vinyl and 4-ethyl derivatives from hydroxycinnamic acids: occurrence of volatile phenolic flavour compounds in beer and distribution of Pad1-activity among brewing yeasts. Food Chem. 2008;107:221–30. [Google Scholar]
- Velempini K, Ketlhoilwe. MJ. Experiences with wild pedagogies in teacher education in Botswana. Canadian J Environ Educat. 2022;25:155–74. [Google Scholar]
- Venter F, Venter J-A. Making the Most of Indigenous Trees. South Africa: Briza publications, 1996. [Google Scholar]
- Viejo CG, Fuentes S, Torrico DD et al. Chemical characterization of aromas in beer and their effect on consumers liking. Food Chem. 2019;293:479–85. [DOI] [PubMed] [Google Scholar]
- Viljoen S. Elephant Fruit: The Dispersal Attributes of Balanites maughamii. Thesis, South Africa: University of Cape Town, 2013. [Google Scholar]
- von Dürckheim KE, Hoffman LC, Leslie A et al. African elephants (Loxodonta africana) display remarkable olfactory acuity in human scent matching to sample performance. Appl Anim Behav Sci. 2018;200:123–9. [Google Scholar]
- Vu D, Groenewald M, Szöke S et al. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud Mycol. 2016;85:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White LJ, Tutin CE, Fernandez M. Group composition and diet of forest elephants, Loxodonta africana cyclotis Matschie 1900, in the Lopé Reserve, Gabon. African J Ecol. 1993;31:181–99. [Google Scholar]
- White TJ, Bruns T, Lee S et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. 1990;18:315–22. [Google Scholar]
- Wiens F, Zitzmann A, Lachance M-A et al. Chronic intake of fermented floral nectar by wild treeshrews. Proc Natl Acad Sci USA. 2008;105:10426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynberg R, Cribbins J, Leakey R et al. Knowledge on Sclerocarya birrea subsp. Caffra with emphasis on its importance as a non-timber forest product in South and southern Africa: a summary: Part 2: commercial use, tenure and policy, domestication, intellectual property rights and benefit-sharing. South Afrn Forestry J. 2002;2002:67–78. [Google Scholar]
- Zhang H, Wang L, Tan Y et al. Effect of Pichia on shaping the fermentation microbial community of sauce-flavor Baijiu. Int J Food Microbiol. 2021;336:108898. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sun Q, Zhu S et al. Biodiversity of non-Saccharomyces yeasts associated with spontaneous fermentation of Cabernet Sauvignon wines from Shangri-La wine region. China. 2021;11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Schifferdecker AJ, Gamero A et al. Kazachstania gamospora and Wickerhamomyces subpelliculosus: two alternative baker's yeasts in the modern bakery. Int J Food Microbiol. 2017;250:45–58. [DOI] [PubMed] [Google Scholar]
- Zhou N. Carbon Metabolism in Non-conventional Yeasts: Biodiversity, Origins of Aerobic Fermentation and Industrial Applications. Sweden: Department of Biology, Lund University, 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.