Abstract
PURPOSE
The investigation of multiple molecular targets with next-generation sequencing (NGS) has entered clinical practice in oncology, yielding to a paradigm shift from the histology-centric approach to the mutational model for personalized treatment. Accordingly, most of the drugs recently approved in oncology are coupled to specific biomarkers. One potential tool for implementing the mutational model of precision oncology in daily practice is represented by the Molecular Tumor Board (MTB), a multidisciplinary team whereby molecular pathologists, biologists, bioinformaticians, geneticists, medical oncologists, and pharmacists cooperate to generate, interpret, and match molecular data with personalized treatments.
PATIENTS AND METHODS
Since May 2020, the institutional MTB set at Fondazione IRCCS Istituto Nazionale Tumori of Milan met weekly via teleconference to discuss molecular data and potential therapeutic options for patients with advanced/metastatic solid tumors.
RESULTS
Up to October 2021, among 1,996 patients evaluated, we identified >10,000 variants, 43.2% of which were functionally relevant (pathogenic or likely pathogenic). On the basis of functionally relevant variants, 711 patients (35.6%) were potentially eligible to targeted therapy according to European Society of Medical Oncology Scale for Clinical Actionability of Molecular Targets tiers, and 9.4% received a personalized treatment. Overall, larger NGS panels (containing >50 genes) significantly outperformed small panels (up to 50 genes) in detecting actionable gene targets across different tumor types.
CONCLUSION
Our real-world data provide evidence that MTB is a valuable tool for matching NGS data with targeted treatments, eventually implementing precision oncology in clinical practice.
#MolecularTumorBoard is key to optimize #PrecisionOncology: from #NextGenerationSequencing to new therapies, MTB increases the chance of a match. Here, the real-world study from referral Center @IstTumori #JCOPO @JCOPO_ASCO.
INTRODUCTION
In the past 2 decades, the molecular characterization of tumors has become an essential tool to guide patients' treatment. Recent evidence suggests that up to 37% of patients with cancer harbor at least one potentially druggable genetic alteration.1 Accordingly, in 2021, over a third of new Food and Drug Administration (FDA) drug registrations (6/17) in oncology were represented by compounds targeted to specific DNA-based biomarkers.2,3 Along this line, European Society for Medical Oncology (ESMO) guidelines recommend routine use of next-generation sequencing (NGS) in patients with advanced nonsquamous non–small-cell lung cancer (NSCLC), colorectal cancer (CRC, if not implying increased costs over single-gene testing), prostate cancer, ovarian cancer (OC), cholangiocarcinoma (CCA),4 and, more recently, metastatic and/or recurrent thyroid carcinoma (TC) and salivary gland carcinoma.5,6 In addition, ESMO recommends clinical research centers to extend multigene NGS testing to other cancer types, including breast cancer (BC), pancreatic cancer (PC), and hepatocellular carcinoma,4 in the context of molecular screening programs to inform clinical researchers and to promote access to innovative drugs. These circumstances, along with a steady decrease in the costs of NGS analyses, led to a widespread diffusion of NGS procedures, which produce huge volumes of molecular data to be swiftly translated into clinically meaningful information.7-9 To this aim, several institutions recently set multidisciplinary teams (MDTs) where molecular pathologists, biologists, bioinformaticians, geneticists, clinical oncologists, and pharmacists cooperate to match molecular data with targeted treatments. This multidisciplinary model commonly falls under the definition of Molecular Tumor Board (MTB), as originally described by Kurzrock’s group in 2014.10 Ever since, MTBs have been proposed as valuable tools in dissecting the complex interaction between molecular data and patient management, implementing molecular test application, data interpretation, and access to therapies.11 Nevertheless, data are still immature relative to the analytical procedures and clinical utility of MTBs in daily clinical practice, with most of the reports focusing on clinical trials or specific cancer settings. Here, we present the structure, composition, procedure, and clinical results of an institutional MTB operating in a cancer center in Italy, focusing on its composition and workflow, type and output of NGS panels used, as well as procedures for variant annotation, definition of actionability, and therapeutic recommendations.
CONTEXT
Key Objective
Molecular Tumor Boards (MTBs) are candidate to represent essential tools for the management of next-generation sequencing (NGS) data for targeted therapies in precision oncology. In 2020, an institutional MTB has been set at the National Cancer Institute of Milan for the weekly discussion of potential therapeutic options for patients with advanced/metastatic solid tumor, on the basis of the mutational model.
Knowledge Generated
The current study reports the composition, workflow, and results of a real-world MTB: in the first 18 months of activity, our MTB indicated the potential eligibility to targeted therapy in 35.6% of almost 2,000 patients, 9.4% of whom ultimately received a personalized treatment. Furthermore, comprehensive genomic profiling was found to outperform the use of small panels in detecting variants potentially actionable in clinical trials.
Relevance
Collectively, these data point toward a central role of MTB in mastering NGS testing and targeted therapy management in clinical practice.
PATIENTS AND METHODS
The prospectively collected mono-institutional cohort described in this study includes consecutive patients with cancer whose tumor specimens were molecularly characterized by NGS at the Pathology Department of the Fondazione IRCCS Istituto Nazionale Tumori (INT) of Milan, and discussed by the institutional MTB, between May 2020 and October 2021. NGS testing was carried out, after informed consent collection, in the following patient settings: (1) meeting the above mentioned ESMO criteria4; (2) with metastatic/advanced disease progressing on standard therapeutic options independent of ESMO criteria; (3) potentially eligible in prospective clinical studies requiring NGS characterization; and (4) included in institutional screening programs (NSCLC, melanoma, and gastrointestinal stromal tumors), independent of tumor stage. NGS testing in liquid biopsy was performed at diagnosis whenever tissue analysis was unsafe or in patients relapsing upon therapies with tyrosine kinase inhibitors. NGS was performed in patients with 0-2 performance status.
Baseline demographic, clinical, and molecular data from the institutional electronic health record (EHR) were collected and stored in a dedicated database based on the institutional RedCap platform interface (Data Supplement [Methods]). The study was approved by the INT Institutional Ethics Committee (code INT-277/20) and was carried out in accordance with the Declaration of Helsinki.
MTB Composition and Workflow
The core of the MTB was composed of five molecular pathologists, six biologists, two bioinformaticians, two geneticists, five oncologists, one pharmacologist, and one data scientist. The institutional MDTs were involved in MTB activities through formally designed delegates who discussed cases with the MTB core team and reported to the patients. The MTB workflow included (1) selection of patients eligible for molecular testing; (2) identification of suitable and cost-effective gene panels meeting the requirements of the treating oncologist; (3) interpretation of variant biological and clinical significance; (4) identification of the optimal treatment for the individual patient; (5) identification of patients eligible for genetic counseling; (6) creation of a formal report enclosed into the EHR; and (7) prospective annotation of outcome data. The MTB core team and delegates from MDTs met weekly via videoconference (average meeting duration of 2 hours). Usually, the MTB discussion was carried out within 10 working days from the NGS report.
Further details regarding NGS methodologic procedures, gene variant functional and clinical annotation, and therapeutic recommendations can be found in the Data Supplement (Methods).
RESULTS
Study Population
Between May 2020 and October 2021, tumor samples and/or liquid biopsies from 1996 patients were molecularly characterized through 2,517 NGS tests. Median patient age at the time of NGS analysis was 64.2 years (ranging from 1 month to 91.9 years). Specifically, NGS tests were performed on tumor samples collected during standard diagnostic procedures: tumors from 1,963 patients (98.3%) were characterized on archival formalin-fixed paraffin-embedded blocks, including 1,078 (55%) primary tumors and 885 (45%) metastatic samples, while in 50 patients (2.5%), tumor genomic characterization was performed by liquid biopsy (17 patients were tested on both tumor tissue and plasma). The most common primary site was lung (26%), followed by colon-rectum (14.1%) and female reproductive system (12.3%; Fig 1A; Appendix Table A1).
FIG 1.

(A) Distribution of patients included in the study cohort by primary tumor localization. (B) Prevalence of the NGS assays used in the study cohort. Most (77.4%) tests were performed in house, and the remaining (22.6%) were outsourced and profiled by FoundationOne CDx (550 cases) or OncotypeMAP (two cases). AFP-Lun, Archer FusionPlex Lung panel; AFP-Sar, Archer FusionPlex Sarcoma panel; BRCA, Oncomine BRCA Research Assay; CHP, Ion AmpliSeq Cancer Hotspot Panel v2; FOneCDX-CGP, FoundationOne CDX; LKB1 custom, custom lung LKB1 v.2 panel; OCAplusDNA, Oncomine Comprehensive Assay Plus, DNA.
Most patients (1,533; 76.8%) were characterized by a single molecular test, while multiple panels were applied in 463 patients. In most of these patients (446 of 463), a combination of two panels for the detection of DNA mutations and RNA aberrations was used. Figure 1B shows the gene panels used in NGS analysis and their prevalence.
NGS Results
In the whole study cohort, after polymorphisms and synonymous variant filtering, a total of 10,475 variants (mean, 6.49; median, 2; range, 1-219 mutations per case) were identified in 1,612 patients (80.7%). Three hundred eighty-five patients had no reportable alterations (wild-type, 19.3%). In particular, 43.2% of the variants were classified as functionally relevant (39.9% pathogenic and 3.3% likely pathogenic), while 56.8% were classified as neutral (55.7% variant of unknown significance, 0.4% benign, and 0.7% likely benign), leading to 1,525 (76.4%) patients with at least one pathogenic or likely pathogenic variant (Data Supplement [Fig A1]). The output in Figure 2 depicts the landscape of all the pathogenic variants found in our data set and their specific alteration.
FIG 2.
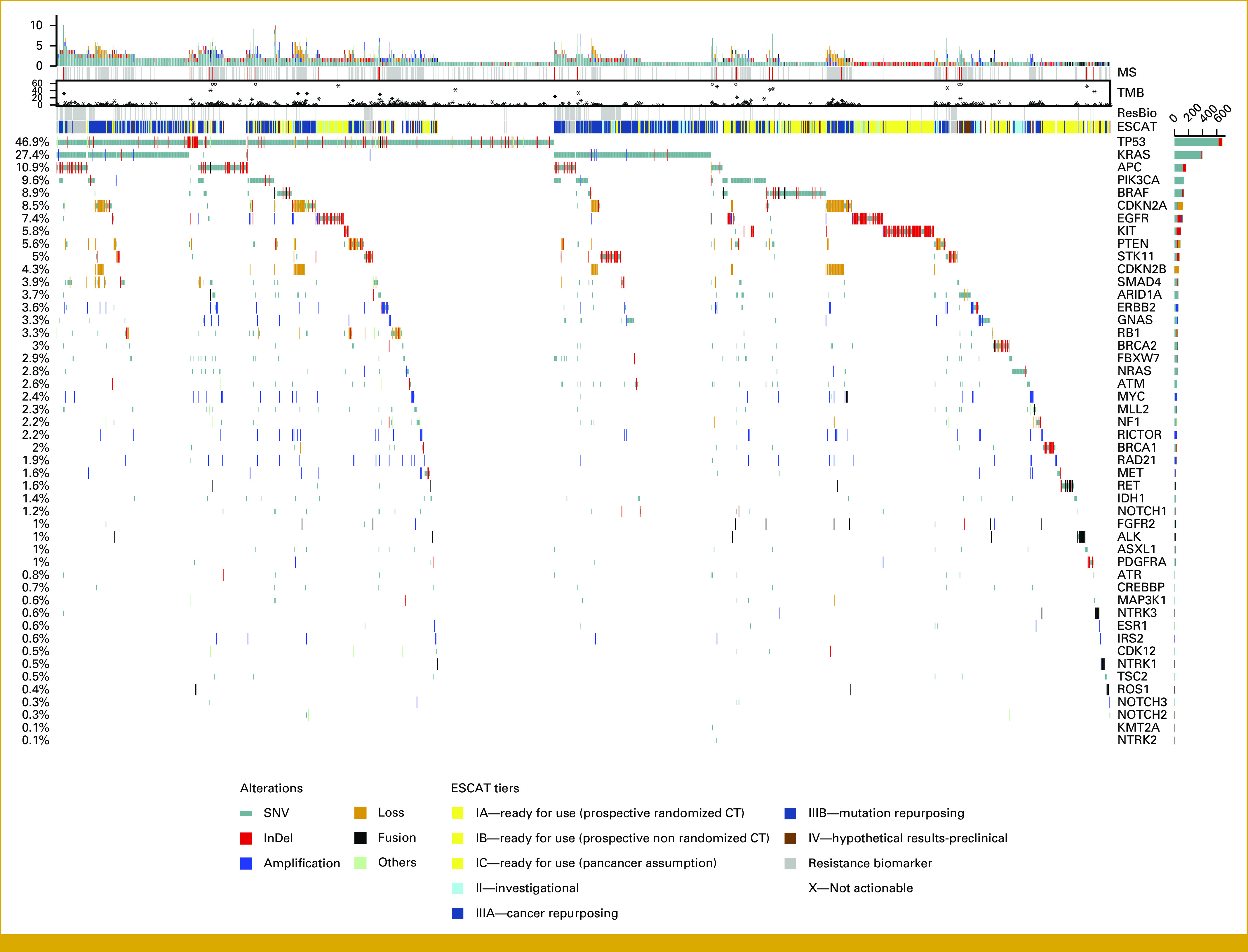
Oncoprint plot visualizing pathogenic genomic alterations in patients of the study cohort (in columns), including SNVs, indels, CNVs, and fusions, ranked according to their prevalence (right bars). MSI status, TMB, whenever available (upper panel), and the level of clinical actionability according to ESCAT classification (right panel) are also provided. As expected, TP53 and KRAS mutations were largely represented, with a prevalence of 47% and 27.1%, respectively. Among 566 (28.4%) patients evaluated with CGP panels (FoundationOne CDx, Oncomine Comprehensive Assay Plus, and OncotypeMap), we were able to obtain TMB and MSI status in 493 and 482 cases, respectively. High MSI was detected in 27 patients (5.6%), with the highest prevalence in patients with gastroesophageal carcinoma (10 of 64 patients, 15.6%) and in patients with CRC (11 of 75 patients, 14.7%). TMB ranged from 0 to 252 mutations/Mbp (median, 3.78). Sixty-three (12.8%) patients had a TMB >10 mutation/Mbp, including a case with a POLE mutation and a TMB of 252 mutation/Mbp. Highest percentages of TMB >10 were detected in patients with neuroendocrine carcinoma (5/19; 26.3%), CRC (16/75; 21.3%), gastroesophageal carcinoma (11/61; 18%), and breast carcinoma (5/32; 15.6%). Overall, 149 gene fusions (80) or rearrangements (69) were detected in 135 patients of 1,014 patients (13.3%) tested with Archer lung (29 cases of 296 tests; 9.8%), Archer sarcoma (8/29; 27.5%), FoundationOne CDx (103/550; 18.7%), and Oncomine Comprehensive Assay RNA Plus (9/151; 5.9%) panels, for a total amount of 1,026 tests. Of the 135 patients bearing gene fusions/rearrangements, 56 (41.4%, corresponding to 5.4% of the whole series) had actionable alterations, and targeted treatment was administered to 22 of them (24%, 2.1%), while eight patients were lost at follow-up. CGP, comprehensive genomic profiling; CNVs, copy number variations; CRC, colorectal cancer; ESCAT, European Society of Medical Oncology Scale for Clinical Actionability of Molecular Targets; MSI, microsatellite instability; SNVs, single-nucleotide variations; TMB, tumor mutational burden.
Definition of Clinical Actionability
Pathogenic variants were stratified into European Society of Medical Oncology Scale for Clinical Actionability of Molecular Targets (ESCAT) classes. ESCAT I-IV actionable alterations were found in 57.8% (1,154/1,996) of the study population. Table 1 summarizes the prevalence of patients harboring actionable alterations according to ESCAT scale. Of note, 21.7% (433) and 7.1% (141) of patients harbored an ESCAT Tier I or a Tier II mutation, respectively, with a cumulative prevalence of 28.8% (574 patients). As expected, gene variants with strong evidence of clinical actionability were associated with higher rates of treatment recommendation and therapy initiation.
TABLE 1.
Prevalence of ESCAT Classes, Drug Recommendation, and Targeted Treatment in Patients Discussed by the MTB

When stratified by tumor types, patients with gastro-intestinal stromal tumors (GIST), BC, TC, lung adenocarcinoma, OC, and melanoma showed the highest prevalence of putatively druggable targets, in line with the high prevalence of KIT (GIST), PIK3CA (BC), BRAF (melanoma and TC) and RET (TC), EGFR, ALK and ROS1 (lung adenocarcinoma), and BRCA1 and BRCA2 (BC and OC) alterations. Figure 3A summarizes the distribution of ESCAT classes across the different tumor types.
FIG 3.

(A) ESCAT classification of pathogenic variants across different tumor types. The high prevalence of targetable genes found in SCLC and carcinosarcoma (ie, malignant mixed mullerian tumors [2/4, including one ALK fusion and one p.L858R EGFR mutation; and 1/4, MSI-high, respectively]) was reasonably due to the small number of patients evaluated. (B) Venn plot describing the prevalences of gene alterations, MTB recommendation, and therapeutic interventions in the whole cohort. Pathogenic or likely pathogenic: patients harboring at least one functionally significant gene variant; actionable: patients with at least one ESCAT 1-4 alteration; eligible: patients for whom MTB gave a therapeutic recommendation; actioned: patients actually receiving the recommended therapy. CT, computed tomography; ESCAT, European Society of Medical Oncology Scale for Clinical Actionability of Molecular Targets; GIST, gastro-intestinal stromal tumor; MTB, Molecular Tumor Board; NET, neuro-endocrine tumor; NOS, not otherwise specified; NSCLC, non–small-cell lung cancer; SCLC, small-cell lung cancer; VUS, variant of unknown significance.
Treatment Assignment
After MTB discussion, 711 patients (35.6%) were considered as potentially eligible to a targeted therapy. Of these, the MTB gave an indication for a standard-of-care (SOC) treatment (ie, drugs approved by the Italian drug agency Agenzia Italiana del FArmaco (AIFA) and reimbursed by the national/regional health care system [NHS]) in 241 (33.9%) patients, for an off-label drug (not approved or not yet reimbursed by NHS) in 170 patients (23.9%), and for enrollment in a clinical trial in 333 (46.8%) patients; 33 patients with gene alterations classified as ESCAT scale II-IV received more than one indication for potential non-SOC treatments. SOC was more frequently indicated for patients with NSCLC (81; 15.9%), GIST (79; 78.2%), OC (38; 15.7%), and melanoma (33; 13.6%); off-label treatment was more frequently suggested for patients with CRC (24.2%, including combinations with BRAF inhibitors), NSCLC (15.4%, including RET, KRAS G12C, and MET inhibitors), and CCA (7.1%, including IDH1, FGFR inhibitors, and PARP inhibitors in BRCAmut patients). Finally, enrollment into clinical trials was more frequently recommended for patients with NSCLC (27.3%, poziotinib, tepotinib, and RET inhibitors), CRC (13.5%, including immunotherapy for microsatellite instability [MSI]-high and tumor mutational burden [TMB]-high patients, as well as human epidermal growth factor receptor 2 [HER2] inhibitors), melanoma (6.9%, including pan-RAF and MEK inhibitors), BC (6%, including PI3K and PARP inhibitors), and CCA (5.4%, including BRAF, HER2, and MAT2A inhibitors). Median time from MTB discussion to therapy administration, evaluated in 127 of 178 patients (71.3%), was 18 days.
Of the 711 patients eligible for targeted therapies, 118 were lost at follow-up (mainly patients treated in other institutions). Of the remaining 593 patients for whom follow-up data were available, 178 (30%, corresponding to 9.4% of the whole cohort, excluding patients lost at follow-up) received a personalized treatment, including 77 (42.1% of the 178 cases) patients with a SOC recommendation (5.1% of the whole cohort), 63 (35.4%) patients with an off-label therapy recommendation (3.2% of the whole cohort), and 38 (21.3%) patients eligible for enrollment in a clinical trial (1.9% of the whole cohort; Fig 3B).
Of the 415 patients who did not receive personalized treatments after MTB discussion, 21.7% could not be addressed to actively recruiting trials, 15% were patients with early-stage tumors, 40.1% were receiving standard therapy when NGS data were produced and discussed in the MTB, and 3.2% had poor performance status (1.9%) or died before therapy (1.3%; Data Supplement [Figs A2A and A2B]).
Higher rates of druggable alterations and treatment recommendations occurred in patients with GISTs (82.18% and 21.78%, respectively), NSCLC (42.9% and 14.2%, respectively), and melanoma (45.9% and 8.2%, respectively). In rarer tumor types, we found high rates of actionability among medullary TCs (RET mutations in 10/11 patients, leading to second-line targeted therapy in two cases) and papillary TC (potentially actionable targets in 10/18 cases, with five patients starting dabrafenib/trametinib combo within a compassionate use program and one patient with an NTRK3-ETV6 fusion starting entrectinib).
More than 10% of patients with MSI-H, or EGFR, KIT, BRAF, or RET gene alterations received personalized treatments. Conversely, despite the high prevalence of cases carrying a KRAS mutation, only a minority could have access to targeted therapies (namely, patients with KRAS G12C-mutated NSCLC; Fig 4).
FIG 4.

The top-20 actioned genes in our data set. For any gene, (A) absolute and (B) relative values of pathogenic, actionable (eligibility to targeted therapy), and actioned (patients actually receiving a targeted therapy) variants are shown.
Actionability According to ESCAT Classification
We also analyzed the data set stratified according to ESCAT. In detail, among 434 patients with at least one ESCAT scale I alteration, 77 patients (17.7%) received personalized SOC therapy and eight additional patients were waiting for therapy initiation. The causes of exclusion of the 349 remaining patients are detailed in the Data Supplement [Fig A2B]. It is worth noting that for 181 ESCAT scale I patients who did not receive a SOC personalized therapy (181/349; 51.9%), the matched drug was not approved by the Italian regulatory agency at the time of MTB discussion. For 23 of these patients (12.7%), mainly patients with CRC treated with cetuximab encorafenib combo (19/23; 82.6%), MTB obtained the access to therapy through off-label indications.
Among the 291 patients bearing at least one actionable ESCAT II-IV scale target, the MTB indicated the potential utility of off-label drugs or the enrollment in a clinical trial for 55 and 230, respectively. Figure A3 in the Data Supplement shows the prevalence of drugs suggested according to variant ESCAT classification across different tumor types.
Panel Usage
Then, we analyzed the impact of NGS panels stratified into large or small according to the number of genes evaluated (>50 genes and ≤50 genes, respectively). Notably, in the whole patient cohort, large panels were able to identify actionable targets at significantly higher frequencies than small panels. Specifically, large and small panels detected actionable variants in 270/566 (47.7%) and 545/1,951 (27.9%) of patients evaluated (P < 10–6), respectively. Large panels were able to identify a significantly higher prevalence of actionable alterations in almost all the tumor types, including CRC (P < 1 × 10–7), PC (P < 1 × 10–6), gastric (P = 2.5 × 10–3), and ovarian (P = .023) carcinomas. As shown in Figure 5, we found a statistically significant benefit deriving from large panel usage. Likewise, large panels identified a significantly higher prevalence of actionable alterations in most of the tumor types analyzed, with an overall absolute increase of detection of 12.5% (37.1% v 24.6%; P = 1.38 × 10–8; Fig 5, left chart).
FIG 5.
Prevalence of pathogenic (blue), actionable (eligibility to targeted therapy; red), and actioned (patients actually receiving a targeted therapy; teal) variants in the commonest tumor types included in our cohort according to the extent of the NGS panel used. Specifically, FoundationOne CDx (324 genes), Oncomine Comprehensive Plus Assays (501 genes investigated for DNA alterations, and 49 for RNA fusions), and OncotypeMAP panel (290 genes), which are able to detect gene mutations, CNVs, gene fusions, MSI, and TMB, were defined as large panels; however, the Ion AmpliSeq Cancer Hotspot Panel v2 (50 genes), the Oncomine BRCA Research Assay (two BRCA genes), the LKB1 v.2 panel (seven genes), the GIST panel (14 genes), the FusionPlex Sarcoma panel (26 genes), and the FusionPlex Lung panel (14 genes) were classified as small panels. In patients with lung adenocarcinoma, for an unbiased evaluation of the putative added value of large panels over standard diagnostic procedures, the output of large panels was compared with that of small DNA panels (Hotspot and LKB1.v2 panels) plus RNA panels (Archer FusionPlex Lung and Oncomine Comprehensive Assay RNA Plus, which assess ALK, ROS1, and RET fusions). P values refer to the prevalence of actionable targets in large panels compared with small panels. *P < .05 in actioned targets in large versus small panel. aFor an unbiased evaluation, in NSCLC only cases with available paired DNA and RNA tests in small panels have been included in the chart. BC, breast cancer; CCA, cholangiocarcinoma; CNVs, copy number variations; CRC, colorectal carcinoma; GEC, gastroesophageal adenocarcinoma; GIST, gastro-intestinal stromal tumor; MM, malignant melanoma; MSI, microsatellite instability; NEN, neuroendocrine neoplasm (including 21 gastroenteropancreatic tumors, 14 lung tumors, and 10 rarer tumors from different districts); NGS, next-generation sequencing; NSCLC, non–small-cell lung cancer; OC, ovarian carcinoma; PC, pancreatic carcinoma; TMB, tumor mutational burden.
Overall, the prevalence of patients starting a targeted therapy was not significantly different using large or small panels, with 50 (8.8%) and 142 (7.3%) actioned molecular alterations identified, respectively (P = .46). Nonetheless, in patients with CRC, there was a significant increase in the prescription of immune checkpoint inhibitors when using large panels (P < 1 × 10–4), mostly because of their capability to capture MSI status. A clear trend, although not statistically significant, was also observed in gastric cancers (mainly because of immunotherapy administration in MSI-H and high TMB patients).
Of interest, large panels led patients to treatment with non-SOC therapies more frequently: among 566 cases profiled with large panels, 15 and 35 patients were treated within clinical trials or with off-label therapies (2.6% and 6.2% of patients profiled, respectively), in comparison with 27 and 29 patients, respectively, among the 1,951 cases profiled with small panels (1.4% and 1.5%, respectively; P < 10–6). Appendix Table A2 summarizes the distribution of the actioned targets stratified according to the NGS panel.
DISCUSSION
Herein, we report procedures and results of the first 18 months of activity of the MTB operating at INT. To the best of our knowledge, this study represents the largest mono-institutional case series reporting the activity of an MTB platform in Italy.
It has been proposed that MTB should be activated only in specific circumstances, including the finding of unusual mutational landscapes, the exhaustion of standard therapeutic regimens, or for clinical trial enrollment.12 Consistently, several studies reported the outcome of monothematic MTBs, which limited data analysis and discussion to patients affected by specific tumor types.13 Our MTB routinely discusses all NGS-profiled consecutive patients, with a real-world approach that leads to unique and realistic portrait of targeted therapy accessibility in an unselected population.
In our series, MTB gave an indication for personalized treatments for 35.6% of the patients evaluated, with 9.4% receiving target therapy. This is in line with data from the Profiler trial, which found actionable genomic aberrations in 27% of the 2,579 patients enrolled, with 6% receiving a targeted therapy.14 In the experience of the Institute Curie, 10% of the patients discussed within the MTB were enrolled in clinical trials with matched therapy.15 The Johns Hopkins' MTB recommended genomically matched therapy in 43% of 155 selected patients, resulting in treatment administration in 15% (11 off-label and 13 clinical trials).16
The application of ESCAT classification to all pathogenic and likely pathogenic variants in our series allows us analyzing its robustness and enforceability in the workflow of a real-world MTB. In the Aurora trial, at least one ESCAT I-II alteration was identified for 51% of patients with BC. These data are in line with the results from our cohort of patients with BC, that includes 22/68 (32.4%) and 36/68 (52.9%) patients with ESCAT I or ESCAT I-II alterations, respectively, mainly represented by PIK3CA, BRCA1, BRCA2, and ERBB2 alterations.
In our case series, as expected, patients with alteration(s) classified in ESCAT I frequently received therapeutic recommendation and a matched treatment more frequently (96.7% and 36%, respectively) than patients with II-IV alterations. Consistently, a recent study on simulated data indicated high concordance rates in treatment recommendations across 10 different Japanese MTBs mainly in tumor types where SOC therapies offer consolidate solutions.17 Other groups already demonstrated the predictive role of ESCAT classification in tumor type–specific case series. In a large cohort of 327 patients with CCA, 184 patients (56%) had an actionable mutation (ESCAT I-IV), 50 of whom received a matched therapy with significant benefit in terms of OS. Interestingly, patients with ESCAT I-II alterations showed longer progression-free survival (PFS) than patients with ESCAT III-IV.18 Results from the SAFIR02-BREAST showed that NGS-driven targeted therapies improved PFS in patients with ESCAT I/II variants (hazard ratio [HR], 0.41; P < .001), while no improvements were observed in the targeted therapies arm (unadjusted HR, 1.15) for ESCAT III-IV.19
The main limitation of our study is the lack, to date, of data regarding outcome and, specifically, survival of our patients: for the time being, survival data are still immature and incomplete, and we are prospectively collecting the clinical outcome of an expanded cohort including roughly 4,000 patients in our institutional database, planning to address this topic in a subsequent publication.
Large NGS panels captured a significantly higher number of actionable variants than small panels across most of the tumor types analyzed, leading to increased treatment recommendations. Unfortunately, this finding did not translate into a significant increase in personalized treatments (8.8% v 7.3%, for large v small panels, respectively), with the exception of patients with CRC in which large panels provided data on MSI and ERBB2 amplifications (Appendix Table A2). Different from small panels, designed for capturing the variants most frequently actioned using personalized SOC therapy, large panels usually identify higher prevalence of ESCAT III-IV alterations targetable only in clinical trials or using off-label drugs. It has also to be underlined that several drugs validated by FDA are still under consideration in Italy (ie, immunotherapy for TMBhigh), further hampering the treatment of patients with actionable variants. These data highlight the urgent need to speed and harmonize the drug validation process across European countries, as well as ease off-label treatments and recruitment within national clinical trials. Furthermore, establishing the cost-effectiveness of comprehensive genomic profiling is pivotal, since its implementation has a number of nontrivial infrastructural, organizational, and scientific implications for pathology laboratories. The access to active off-label drugs and clinical trials represents the bottleneck for personalized treatment and would strongly benefit from a closer relationship between institutions, regulatory agencies, and stakeholders. Our experience significantly sustains a structured governance, in the perspective of strengthening a robust platform capable of implementing existing clinical and diagnostic data collections; improving the process of integration, updating, and interdisciplinary sharing; and optimizing the quality of existing prospective data sources and their availability for secondary analysis, innovating sharing of anonymous data and information among institutions, private companies, and the academia.
APPENDIX
TABLE A1.
Number and Prevalence of Tumor Types in the Study Cohort

TABLE A2.
Actioned Targets in Different Tumor Types
Andrea Vingiani
Honoraria: Roche, Lilly
Silvia Damian
Employment: Biofarma Group
Leadership: Biofarma Group
Research Funding: Basilea Pharmaceutical (Inst), Incyte (Inst), Novartis (Inst), Nerviano Medical Sciences (Inst), Roche/Genentech (Inst), Pfizer (Inst)
Claudia Proto
Consulting or Advisory Role: AstraZeneca, Roche, Bristol Myers Squibb, Janssen, MSD
Research Funding: Pfizer (Inst), Roche (Inst), AstraZeneca (Inst), Daiichi Sankyo (Inst), MSD (Inst), Celgene (Inst), Spectrum Pharmaceuticals (Inst), Janssen (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Roche
Monica Niger
Honoraria: Incyte, Servier, Sandoz, Medpoint SRL, Accademia Nazionale Di Medicina (ACCMED)
Consulting or Advisory Role: Basilea Pharmaceutical, EMD Serono, MSD/AstraZeneca, Servier, Incyte, Taiho Pharmaceutical
Travel, Accommodations, Expenses: AstraZeneca
Marta Brambilla
Speakers' Bureau: AstraZeneca
Travel, Accommodations, Expenses: Lilly
Claudio Vernieri
Consulting or Advisory Role: Daiichi Sankyo/Astra Zeneca, Novartis, Pfizer
Speakers' Bureau: Novartis, Istituto Gentili, Lilly, Accademia Nazionale Di Medicina (ACCMED)
Research Funding: Roche
Claudio Jommi
Honoraria: Amgen, AstraZeneca, Bristol Myers Squibb, Dephaforum, Gilead Sciences, Incyte, Market Access Provider, MSD, Roche, Sanofi, Takeda, Alira Health, CSL Behring
Consulting or Advisory Role: Amgen, AstraZeneca, Bristol Myers Squibb, Celgene, Dephaforum, Gilead Sciences, Incyte, Market Access Provider, MSD, Roche, Sanofi, Takeda, Alira Health
Research Funding: AbbVie, Amgen, AstraZeneca, Bayer, Bristol Myers Squibb, Boehringer Ingelheim, Celgene, Fondazione Smith Kline, Janssen Cilag, MSD, Novartis, Roche, Pfizer, Sandoz, Sanofi, Takeda, Teva
Vito Ladisa
Employment: Roche
Leadership: MSD (Inst)
Honoraria: Takeda
Consulting or Advisory Role: Gilead Sciences
Travel, Accommodations, Expenses: Sanofi
Filippo De Braud
Honoraria: Roche, Pfizer, BMS, Merck, MSD, Servier, Sanofi, Amgen Astellas BioPharma, Incyte
Consulting or Advisory Role: Roche, Incyte, EMD Serono, Bristol Myers Squibb, Nerviano Medical Sciences, Sanofi, Novartis Italy, Nerviano Medical Sciences, Menarini, AstraZeneca, Pierre Fabre
Research Funding: Novartis (Inst), Roche (Inst), Merck Serono (Inst), Pfizer (Inst), Servier (Inst), Philogen (Inst), Loxo (Inst), Tesaro (Inst), Nerviano Medical Sciences (Inst), Kymab (Inst), Bristol Myers Squibb/Medarex, Merck KGaA, Ignyta, MedImmune, Exelixis, Bayer Health, Daiichi Sankyo Europe GmbH, Incyte, Basilea Pharmaceutical, Janssen Oncology
Giancarlo Pruneri
Honoraria: Novartis, Roche, Genomic Health
Consulting or Advisory Role: ADS Biotec
Research Funding: Roche, Roche Molecular Diagnostics
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part at ESMO Congress 2022, Paris, France, September 9-13, 2022, by Prof. Giancarlo Pruneri, in the oral presentation titled Defining the role of the pathologist in the era of Next Generation Sequencing.
SUPPORT
Supported by Fondazione IRCCS Istituto Nazionale dei Tumori institutional funds. This research was also funded by Italian Ministry of Health “Ricerca Corrente” funds.
A.V., L.A., and M.D. contributed equally to this work. F.D.B., and G.P. contributed equally to this work.
DATA SHARING STATEMENT
The data analyzed during the current study are available from the corresponding author on reasonable request. The underlying code for this study is not publicly available but may be made available to qualified researchers on reasonable request to the corresponding author.
AUTHOR CONTRIBUTIONS
Conception and design: Andrea Vingiani, Luca Agnelli, Matteo Duca, Daniele Lorenzini, Silvia Damian, Giovanni Apolone, Filippo De Braud, Giancarlo Pruneri
Provision of study materials or patients: Matteo Duca, Silvia Damian, Monica Niger, Claudia Proto, Elena Colombo, Salvatore Lopez, Vito Ladisa
Collection and assembly of data: Andrea Vingiani, Luca Agnelli, Matteo Duca, Daniele Lorenzini, Silvia Damian, Claudia Proto, Monica Niger, Federico Nichetti, Elena Tamborini, Alberta Piccolo, Marta Brambilla, Elena Colombo, Salvatore Lopez, Claudio Vernieri, Elena Conca, Fabio Bozzi, Marta Angelini, Andrea Devecchi, Rebecca Salvatori, Vito Ladisa
Data analysis and interpretation: Andrea Vingiani, Luca Agnelli, Matteo Duca, Daniele Lorenzini, Silvia Damian, Claudia Proto, Monica Niger, Federico Nichetti, Elena Tamborini, Federica Perrone, Siranoush Manoukian, Jacopo Azzollini, Elena Colombo, Francesca Marra, Elena Conca, Adele Busico, Iolanda Capone, Valentina De Micheli, Anna Baggi, Silvia Pasini, Claudio Jommi, Giancarlo Pruneri
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Andrea Vingiani
Honoraria: Roche, Lilly
Silvia Damian
Employment: Biofarma Group
Leadership: Biofarma Group
Research Funding: Basilea Pharmaceutical (Inst), Incyte (Inst), Novartis (Inst), Nerviano Medical Sciences (Inst), Roche/Genentech (Inst), Pfizer (Inst)
Claudia Proto
Consulting or Advisory Role: AstraZeneca, Roche, Bristol Myers Squibb, Janssen, MSD
Research Funding: Pfizer (Inst), Roche (Inst), AstraZeneca (Inst), Daiichi Sankyo (Inst), MSD (Inst), Celgene (Inst), Spectrum Pharmaceuticals (Inst), Janssen (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Roche
Monica Niger
Honoraria: Incyte, Servier, Sandoz, Medpoint SRL, Accademia Nazionale Di Medicina (ACCMED)
Consulting or Advisory Role: Basilea Pharmaceutical, EMD Serono, MSD/AstraZeneca, Servier, Incyte, Taiho Pharmaceutical
Travel, Accommodations, Expenses: AstraZeneca
Marta Brambilla
Speakers' Bureau: AstraZeneca
Travel, Accommodations, Expenses: Lilly
Claudio Vernieri
Consulting or Advisory Role: Daiichi Sankyo/Astra Zeneca, Novartis, Pfizer
Speakers' Bureau: Novartis, Istituto Gentili, Lilly, Accademia Nazionale Di Medicina (ACCMED)
Research Funding: Roche
Claudio Jommi
Honoraria: Amgen, AstraZeneca, Bristol Myers Squibb, Dephaforum, Gilead Sciences, Incyte, Market Access Provider, MSD, Roche, Sanofi, Takeda, Alira Health, CSL Behring
Consulting or Advisory Role: Amgen, AstraZeneca, Bristol Myers Squibb, Celgene, Dephaforum, Gilead Sciences, Incyte, Market Access Provider, MSD, Roche, Sanofi, Takeda, Alira Health
Research Funding: AbbVie, Amgen, AstraZeneca, Bayer, Bristol Myers Squibb, Boehringer Ingelheim, Celgene, Fondazione Smith Kline, Janssen Cilag, MSD, Novartis, Roche, Pfizer, Sandoz, Sanofi, Takeda, Teva
Vito Ladisa
Employment: Roche
Leadership: MSD (Inst)
Honoraria: Takeda
Consulting or Advisory Role: Gilead Sciences
Travel, Accommodations, Expenses: Sanofi
Filippo De Braud
Honoraria: Roche, Pfizer, BMS, Merck, MSD, Servier, Sanofi, Amgen Astellas BioPharma, Incyte
Consulting or Advisory Role: Roche, Incyte, EMD Serono, Bristol Myers Squibb, Nerviano Medical Sciences, Sanofi, Novartis Italy, Nerviano Medical Sciences, Menarini, AstraZeneca, Pierre Fabre
Research Funding: Novartis (Inst), Roche (Inst), Merck Serono (Inst), Pfizer (Inst), Servier (Inst), Philogen (Inst), Loxo (Inst), Tesaro (Inst), Nerviano Medical Sciences (Inst), Kymab (Inst), Bristol Myers Squibb/Medarex, Merck KGaA, Ignyta, MedImmune, Exelixis, Bayer Health, Daiichi Sankyo Europe GmbH, Incyte, Basilea Pharmaceutical, Janssen Oncology
Giancarlo Pruneri
Honoraria: Novartis, Roche, Genomic Health
Consulting or Advisory Role: ADS Biotec
Research Funding: Roche, Roche Molecular Diagnostics
No other potential conflicts of interest were reported.
REFERENCES
- 1. Bedard PL, Hyman DM, Davids MS, et al. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet. 2020;395:1078–1088. doi: 10.1016/S0140-6736(20)30164-1. [DOI] [PubMed] [Google Scholar]
- 2.2021. https://www.iqvia.com/insights/the-iqvia-institute/reports/global-oncology-trends-2021 IQVIA Global Oncology Trends 2021.
- 3.2021. https://www.ema.europa.eu/en/annual-report/2021/index.html EMA annual report 2021.
- 4. Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:1491–1505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 5. Filetti S, Durante C, Hartl DM, et al. ESMO Clinical Practice Guideline update on the use of systemic therapy in advanced thyroid cancer. Ann Oncol. 2022;33:674–684. doi: 10.1016/j.annonc.2022.04.009. [DOI] [PubMed] [Google Scholar]
- 6. van Herpen C, Vander Poorten V, Skalova A, et al. Salivary gland cancer: ESMO–European Reference Network on Rare Adult Solid Cancers (EURACAN) Clinical Practice Guideline for diagnosis, treatment and follow-up. ESMO Open. 2022;7:100602. doi: 10.1016/j.esmoop.2022.100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marino P, Touzani R, Perrier L, et al. Cost of cancer diagnosis using next-generation sequencing targeted gene panels in routine practice: A nationwide French study. Eur J Hum Genet. 2018;26:314–323. doi: 10.1038/s41431-017-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pruneri G, De Braud F, Sapino A, et al. Next-generation sequencing in clinical practice: Is it a cost-saving alternative to a single-gene testing approach? Pharmacoecon Open. 2021;5:285–298. doi: 10.1007/s41669-020-00249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Nimwegen KJ, van Soest RA, Veltman JA, et al. Is the $1000 genome as near as we think? A cost analysis of next-generation sequencing. Clin Chem. 2016;62:1458–1464. doi: 10.1373/clinchem.2016.258632. [DOI] [PubMed] [Google Scholar]
- 10. Schwaederle M, Parker BA, Schwab RB, et al. Molecular Tumor Board: The University of California-San Diego Moores Cancer Center experience. Oncologist. 2014;19:631–636. doi: 10.1634/theoncologist.2013-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tamborero D, Dienstmann R, Rachid MH, et al. The Molecular Tumor Board Portal supports clinical decisions and automated reporting for precision oncology. Nat Cancer. 2022;3:251–261. doi: 10.1038/s43018-022-00332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. VanderWalde A, Grothey A, Vaena D, et al. Establishment of a Molecular Tumor Board (MTB) and uptake of recommendations in a community setting. J Pers Med. 2020;10:252. doi: 10.3390/jpm10040252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Larson KL, Huang B, Weiss HL, et al. Clinical outcomes of Molecular Tumor Boards: A systematic review. JCO Precis Oncol. 2021 doi: 10.1200/PO.20.00495. 10.1200/PO.20.00495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tredan O, Wang Q, Pissaloux D, et al. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: Analysis from the ProfiLER trial. Ann Oncol. 2019;30:757–765. doi: 10.1093/annonc/mdz080. [DOI] [PubMed] [Google Scholar]
- 15. Basse C, Morel C, Alt M, et al. Relevance of a Molecular Tumour Board (MTB) for patients' enrolment in clinical trials: Experience of the Institut Curie. ESMO Open. 2018;3:e000339. doi: 10.1136/esmoopen-2018-000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dalton WB, Forde PM, Kang H, et al. Personalized medicine in the oncology clinic: Implementation and outcomes of the Johns Hopkins Molecular Tumor Board. JCO Precis Oncol. 2017 doi: 10.1200/PO.16.00046. 10.1200/PO.16.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Naito Y, Sunami K, Kage H, et al. Concordance between recommendations from multidisciplinary Molecular Tumor Boards and central consensus for cancer treatment in Japan. JAMA Netw Open. 2022;5:e2245081. doi: 10.1001/jamanetworkopen.2022.45081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Verdaguer H, Sauri T, Acosta DA, et al. ESMO scale for clinical actionability of molecular targets driving targeted treatment in patients with cholangiocarcinoma. Clin Cancer Res. 2022;28:1662–1671. doi: 10.1158/1078-0432.CCR-21-2384. [DOI] [PubMed] [Google Scholar]
- 19. Andre F, Filleron T, Kamal M, et al. Genomics to select treatment for patients with metastatic breast cancer. Nature. 2022;610:343–348. doi: 10.1038/s41586-022-05068-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data analyzed during the current study are available from the corresponding author on reasonable request. The underlying code for this study is not publicly available but may be made available to qualified researchers on reasonable request to the corresponding author.




