Abstract
PURPOSE
There is a paucity of consistent data concerning genetic mutations in Brazilian patients with lung cancer. The aim of this study was to retrospectively analyze epidermal growth factor receptor (EGFR) mutations detected in a real-world scenario using a large cohort of Brazilian patients with non–small-cell lung cancer (NSCLC).
MATERIALS AND METHODS
This was a cross-sectional, observational, descriptive study on the basis of a database of EGFR molecular analysis from tumor samples of patients with a confirmatory histopathological diagnosis of primary lung cancer. Specimens were collected from 2013 to 2017 and were tested using cobas, next-generation sequencing, and Sanger sequencing platforms.
RESULTS
A total of 7,413 tumor specimens were tested. The patients were predominantly women with a median age of 67.0 years. Patients with at least one mutation represented 24.2% of the total sample. Among the positive patients, the majority had just one mutation, but two or more simultaneous mutations were observed in 1.52% of patients. Exon 19 deletion was the most prevalent alteration in the sample (12.8%), followed by exon 21 L858R (6.9%) and exon 20 insertion (1.6%). All others were considered uncommon mutations and were observed in 18.5% of all mutated patients and 4.0% of the total sample (2.3%-18.7% depending on the sequencing method).
CONCLUSION
This study examined the prevalence of EGFR mutations in Brazilian patients with NSCLC using different technologies, suggesting that the type of method used, directed or nondirected against specific mutations, influences the analysis, particularly for uncommon mutations, which will be missed by mutation-specific approaches such as cobas testing. Our estimates are the largest in Latin America and are consistent with previous reports from other parts of the world. Besides the variability in methods described here as technology incorporation advances in a nonhomogeneous manner, it is probably like the real-world clinical setting Brazilian oncologists face in their daily practice.
INTRODUCTION
With estimated 2.3 million incident cases and 1.8 million deaths in 2020, lung cancer is the leading cause of cancer deaths worldwide.1 In Brazil, approximately 38,200 lung cancer–related deaths were reported in 2020.2 Non–small-cell lung cancer (NSCLC) is the most common subtype, accounting for approximately 80%-90% of all cases.3 The therapeutic arsenal for NSCLC has faced significant changes since the introduction of targeted therapies, and the identification of actionable genetic alterations has led to the integration of molecular testing for treatment planning.4
CONTEXT
Key Objective
There is a paucity of consistent data concerning genetic mutations in Brazilian patients with lung cancer. The aim of this study was to retrospectively analyze epidermal growth factor receptor (EGFR) mutations detected in a real-world scenario using a large cohort of Brazilian patients with non–small-cell lung cancer (NSCLC). To our knowledge, this is the largest analysis of EGFR mutational status in Brazilian patients with NSCLC using multiple platforms.
Knowledge Generated
Of the 7,413 NSCLC samples successfully tested for EGFR, 1,797 carried somatic EGFR mutations, representing an average positive EGFR detection rate of 24.24%. Specimens were tested using cobas, next-generation sequencing, and Sanger sequencing platforms. Uncommon mutations affected 2.3%-18.7% of patients depending on the sequencing method.
Relevance
Besides the variability in methods described here as technology incorporation advances in a non-homogeneous manner, it is probably similar to the real-world clinical setting Brazilian oncologists face in their daily practice.
Epidermal growth factor receptor (EGFR) mutations are the most frequent actionable genetic alteration in NSCLC.5,6 EGFR mutations can be detected in 15% of adenocarcinoma subtypes of White patients in North America and Europe, in approximately 25% of Brazilian and Latin American patients, or in a much higher incidence, 40%-50% of the same subtype of East Asian NSCLC.7 The two classical EGFR mutations are exon 19 deletions (19 del) and L858R substitution in exon 21, and both can predict tumor response to EGFR tyrosine kinase inhibitors (EGFR-TKIs) in NSCLC. Other uncommon EGFR mutations were also associated with resistance (eg, exon 20 insertions) or sensitivity (eg, exon 19 insertion, p.L861Q in exon 21, p.G719X in exon 18, and p.S768I in exon 20) to EGFR-TKIs, and their reported frequency is higher in Brazil (18.0%)8 than in Asia (11.9%)9 but similar to some European countries (18.2%).10,11 Although there is no question about randomized controlled trials being the cornerstone of the best medical knowledge, there is also a strong need to narrow the gap between conventional clinical trial data and the real world, especially in some specific populations. Real-world studies are emerging in medical oncology as a useful tool to collect data from daily clinical practice, thus driving clinical choices in special patient populations. Given the speed of new insights and the increasing number of new treatment options for small subgroups of patients, the advantages of real-world studies are even clearer in lung cancer.12
After the scientific recognition of molecular testing as a key component of decision making in lung cancer management, guidelines have clearly stated that patients should be tested for EGFR mutations early in their therapeutic journey.13 On the basis of the relevance of EGFR testing in patients with NSCLC, this study retrospectively reviewed EGFR molecular testing results, including both classical and uncommon mutations, in 7,413 Brazilian patients with NSCLC whose tumor tissues were analyzed at a single testing center.
MATERIALS AND METHODS
Study Design
From January 2013 to August 2017, 7,413 formalin-fixed paraffin-embedded (FFPE) tissues of patients with NSCLC from 27 Brazilian states were tested for EGFR mutations. EGFR testing results were retrospectively reviewed in this cross-sectional study. The age of patients ranged from 24 to 97 years (median, 67 years), and there were 53.7% of female patients and 46.3% of male patients. All EGFR tests were performed at a single testing center and were requested by the attending physician for therapeutic purposes.
Molecular Testing
To detect mutations in tumor tissues, DNA was extracted from frozen and FFPE tissues collected by biopsy or surgical excision. Before EGFR testing, tumor cell content (TCC) of the specimens was assessed by an experienced pathologist. Only specimens with >5% TCC by microscopic inspection were included in these analyses. Detection of EGFR mutations from exons 18 to 21 was performed using real-time polymerase chain reaction (PCR) with specific hydrolysis probes for the detection of mutations of interest (cobas EGFR Mutation Test v2, Roche Pharma, Basel, Switzerland), direct DNA sequencing by next-generation sequencing (NGS) using the Ion Personal Genome Machine system, Sanger method, or pyrosequencing. The cobas test identified a restricted variety of EGFR mutations, including deletions in exon 19, insertions in exon 20, L858R, G719X, S768I, L861Q, and T790M, while the Sanger method was designed to amplify amplicons containing EGFR exons 18, 19, 20, and 21. Pyrosequencing of EGFR exons 18, 19, 20, and 21 was performed using a commercial EGFR Pyro Kit (Qiagen, Germantown, MD). PCR amplification was performed using 80-120 ng of genomic DNA according to the manufacturer's instructions. The PCR products were verified on 1% agarose gels using SYBR Safe DNA Gel Stain (Invitrogen, Carlsbad, CA). Template preparation and sequencing were performed using PyroMark Gold Q24 reagents in a PyroMark Q24 device following the manufacturer's instructions. Mutations were detected using the PyroMark Q24 software and default analysis parameters recommended by the manufacturer (Qiagen, Germantown, MD). A somatic mutation was present when the variant allele was detected at a frequency of >5%.14
Regarding NGS methodology, EGFR mutations were identified using the Oncomine Assay (comprising the DNA Oncomine Focus Assay [Thermo Fisher Scientific, Walthan, MS] and RNA Oncomine Fusions assay [Thermo Fisher Scientific, Walthan, MS]) hot spot panel. The library preparation followed the manufacturer's instructions using a total of 10 ng input DNA and/or RNA per sample (minimum 0.83 ng/μL sample DNA concentration). A maximum of seven DNA samples were prepared per run (six samples if both DNA and RNA analyses were required) on an Ion 318 v2 chip (Catalog No. 4488150; Thermo Fisher Scientific, Walthan, MS). The DNA panel identified hot spot mutations in AKT1, ALK, AR, BRAF, CDK4, CTNNB1, DDR2, EGFR, ERBB2, ERBB3, ERBB4, ESR1, FGFR2, FGFR3, GNA11, GNAQ, HRAS, IDH1, IDH2, JAK1, JAK2, JAK3, KIT, KRAS, MAP2K1, MAP2K2, MET, MTOR, NRAS, PDGFRA, PIK3CA, RAF1, RET, ROS1, and SMO. Analysis was performed using Ion Torrent Suite Browser version 5.0 and Ion Reporter version 5.0. The Torrent Suite. The Coverage Analysis plugin was applied to all data and used to assess amplicon coverage for regions of interest. Variants were identified using the Ion Reporter filter chain 5% Oncomine Variants (5.0). A cutoff corresponding to 500X coverage was applied for all analyses.15
The number of samples tested increased over the years, with 8% (596/7,413) of the samples tested in 2013, 11.2% (828/7,413) in 2014, 26.5% (1,966/7,413) in 2015, 31.5% (2.335/7.413) in 2016, and 22.8% (1.688/7.413) in August 2017 (incomplete year). For the percentages of NSCLC specimens tested using different platforms, 72.8% (5,395/7,413) of the samples were tested using real-time quantitative polymerase chain reaction, 21.2% (1,574/7,413) by NGS, 5.7% (424/7,413) by Sanger sequencing, and 0.3% (20/7,413) by pyrosequencing. EGFR genotyping using NGS was launched in 2016, representing 10.1% of all tests in 2016, while 79.3% of samples were tested using NGS in 2017 (data not shown).
Statistical Analysis
The prevalence of genetic variants of interest was expressed as a percentage of the total number of cases examined and the total number of patients with mutations. Potential differences in the prevalence of specific genetic variants between the subgroups of patients were estimated using the chi-square and Fisher's method. Multivariate analysis was performed to examine the potential association between EGFR status and exposure variables using multiple binary logistic regression models. All tests were performed assuming a P value of .05 as the limit for statistical significance using IBM SPSS Statistics 22 (IBM, New York, NY).
Ethical Conduct
The study was performed in accordance with ethical principles consistent with the Declaration of Helsinki, international council for harmonization of technical requirements for pharmaceuticals for human use good clinical practices, and the applicable national legislation on Noninterventional Studies and/or Observational Studies. Once the study analyzed secondary epidemiological anonymized data from a private database that compiled the results of laboratory tests performed on patients previously diagnosed with lung cancer, informed consent was not obtained. No patient was addressed directly, and no information that allowed the identification of any subject was collected.
RESULTS
EGFR Total Mutation Rates According to Different Methods
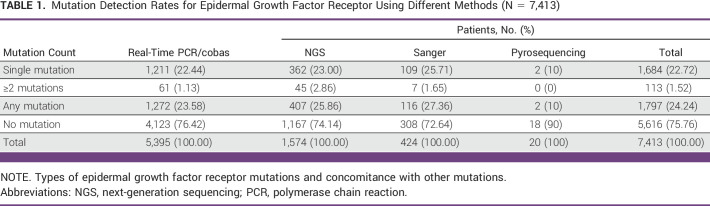
Of the 7,413 NSCLC samples successfully tested for EGFR, 1,797 carried somatic EGFR mutations, representing an average positive EGFR detection rate of 24.24%, including 23.58% (1,272/5,395) tested using real-time PCR, 25.86% (407/1,574) tested using NGS, 27.36% (116/424) tested using Sanger sequencing, and 10% (2/20) tested using pyrosequencing. Figure 1 depicts the distribution of each mutation by method (cobas, NGS, and Sanger sequencing).
FIG 1.
Distribution of each mutation by method. NGS, next-generation sequencing; PCR, polymerase chain reaction.
Molecular Testing Patterns
Among the 1,797 patients with EGFR mutations, 94% (1,684/1,797) harbored a single mutation, including 1,380 (77%) cases carrying classical mutations (19 del or L858R). Complex mutations were observed in 1.5% of the cases (Table 1). The most common complex mutations occurred in the presence of 19 del + 20 T790M mutations (n = 41, 2.3% of mutated patients), 21 L858R + 20 T790M (n = 22, 1.2% of mutated patients), and 20 S768X + 18 G719X (n = 16, 0.9% of mutated patients; data not shown).
TABLE 1.
Mutation Detection Rates for Epidermal Growth Factor Receptor Using Different Methods (N = 7,413)
Figure 2 shows the distribution of the specific EGFR mutations in this study (N = 7,413). Exon 19 del was the most prevalent alteration in the sample (12.8%), followed by exon 21 L858R (6.9%), and exon 20 insertion (1.6%). All others were considered uncommon mutations, which were present in 4.0% of the total sample, with great variation in the specific prevalence depending on the method used (Fig 2A). Complex mutations were found in 1.5% of cases, and their types are shown in Figure 2B.
FIG 2.
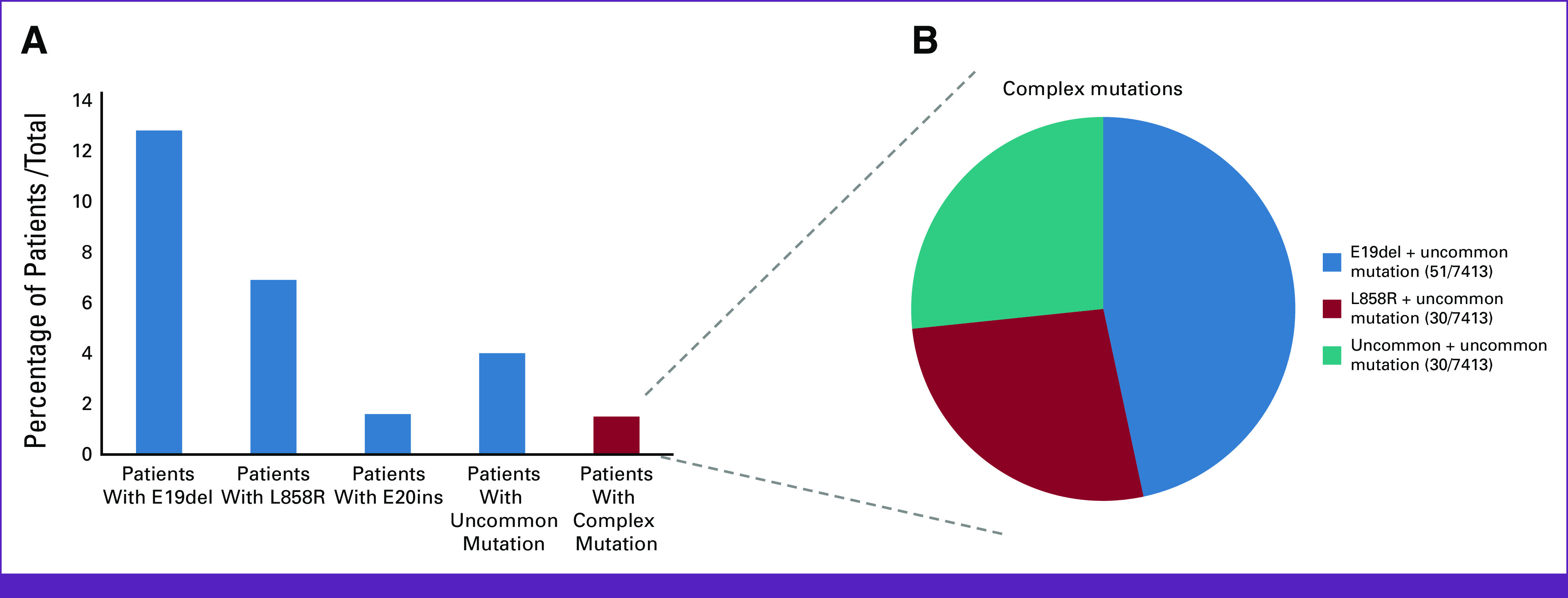
An overview of EGFR alterations present in 7,413 cases according to the mutation type. (A) A graph with the percentages of the most prevalent EGFR alterations corresponding to patients with exon 19 deletions (12.8%), patients with L858R (6.9%), patients with exon 20 insertions (1.6%), patients with uncommon mutations (4%), and patients with complex mutations (1.5%). (B) The prevalence of complex mutations in the total sample. There were no concomitant mutations with exon 20 insertions. EGFR, epidermal growth factor receptor.
Figures 3 and 4 depict the distribution of each mutation by method (cobas, NGS, and Sanger) considering only single mutations and both single and multiple mutations, respectively. Sequencing yielded 1,611 single EGFR somatic mutations and 1,911 single and multiple EGFR mutations. As expected, the proportion of each uncommon mutation depended on the testing method. Single uncommon mutations represented approximately 5.0%, 14.0%, and 66.0% of the mutated samples tested using cobas, NGS, and Sanger sequencing, respectively. When single and multiple mutations were analyzed, uncommon mutations accounted for 10.4%, 23.6%, and 69.1% with cobas, NGS, and Sanger sequencing, respectively (Figs 3 and 4). Uncommon mutations involved exon 18 for cobas and exon 21 for NGS and Sanger sequencing for single mutations (Fig 3) and exon 20 for cobas and NGS and exon 21 for Sanger sequencing, when both multiple and single mutations were analyzed together (Fig 4). The most frequent single uncommon mutations (regardless of the method used) were exon 20 T790M, exon 18 G719X, and exon 21 L861X.
FIG 3.

Distribution of EGFR mutations identified in the sample by method (single mutations only). EGFR, epidermal growth factor receptor; NGS, next-generation sequencing.
FIG 4.

Distribution of EGFR mutations identified in the sample by method (single and multiple mutations). EGFR, epidermal growth factor receptor; NGS, next-generation sequencing.
DISCUSSION
To our knowledge, this study is the largest analysis of EGFR mutational status in Brazilian patients with NSCLC using multiple platforms, which could be highly valuable in population analysis and future clinical management.
Ethnicity and geography contribute to the different frequencies of EGFR mutations in NSCLC. In this regard, our positive EGFR detection rate of 24.2% was lower than that found in the Asian population, higher than in Europe and North America, but comparable with previous Brazilian studies.16-18 The frequency of EGFR exon 19 del (12.8%) was slightly higher than that in other populations but was also in line with further Brazilian data.16,18 One important point to be considered is that our cohort was represented by a majority of female patients, and as a bias, it might increase the prevalence of EGFR mutations.
The vast majority of our results were analyzed using cobas (73%), followed by NGS (21%), Sanger sequencing (6%), and pyrosequencing (0.3%). Sanger and NGS methods showed higher positive rates for EGFR detection, and these methods are known for being able to detect novel mutations, allowing full examination of exons 18, 19, 20, and 21. By contrast, cobas real-time PCR is a mutation-specific analytical platform. Owing to the large sample size and availability of both directed and nondirected genotyping platforms, this study allowed the detection of both common and uncommon EGFR mutations in the Brazilian NSCLC population.
EGFR status is considered essential to determine therapeutic pathways for patients with NSCLC; thus, sensitive reliable molecular testing methods are needed to support treatment decision making.13,19,20 Studies have addressed the validity and comparability of different sequencing platforms, suggesting that Sanger, NGS, cobas, and pyrosequencing are comparable but present specificities that need to be considered for method selection and interpretation of results.5,21 Mutation-directed sequencing platforms such as cobas are faster and easier to use, whereas platforms with a large sequencing capacity (such as Sanger and NGS) screen for multiple targets in the same assay, providing broader genetic information. Giardina et al conducted a validation study (n = 113) comparing NGS, Sanger sequencing, pyrosequencing, competitive allele-specific TaqMan PCR (PCR), and cobas. The authors observed that NGS was concordant with other methods in 94.7% of cases but was able to detect 113 mutations originally missed by conventional platforms: 23.0% with known clinical relevance, 32.7% with potential clinical relevance, and 44.2% of unknown clinical meaning.21 In this analysis, we observed that both NGS and Sanger sequencing detected more EGFR mutations than cobas, which was the most frequently used method.
In the era of personalized medicine, research addressing the impact of genetic biomarkers on survival and response is rapidly growing, as well as the use of larger sequencing capacity platforms in clinical practice, highlighting the need to better understand the clinical impact of distinct mutations. While the classical mutations in exon 19 del and exon 21 L858R account for almost 90% of all EGFR mutations,22 post hoc analysis from EGFR-TKI randomized clinical trials and observational studies have observed a frequency of uncommon mutations of approximately 10%.9,10,22-26 In our sample, 4.0% of the patients presented with rarer mutations, regardless of the sequencing platform. The probable reason for this discrepancy is that most patients in our sample were tested with cobas, reducing the ability to comprehensively identify rarer mutations owing to the characteristics of this method. When NGS and Sanger-tested patients were analyzed, the frequencies of uncommon mutations were 6.1% and 18.7%, respectively, which is consistent with previous reports.
It is well established in the literature that patients with advanced NSCLC with activating EGFR mutations treated with EGFR-TKIs present longer progression-free survival compared with chemotherapy-treated patients and nonmutated patient.27-31 However, little is known about the interaction between uncommon mutations and patient responses to EGFR-TKIs.5 Considering the low frequency of these mutations (4.0% of all samples) and the high number of different point mutations (>30), it is difficult to attribute a clinical meaning to each of them. Despite this, it is worth mentioning that the most frequent single uncommon mutations in the sample were previously associated with impact on EGFR-TKIs response (T790M, G719X, and L861X).9,22,32 Most data on uncommon mutations and their association with response and survival were derived from EGFR-TKI trials or retrospective studies, with small sample sizes and conflicting results.33 In our sample, the single more frequent uncommon mutation was exon 20 T790M, the most well-described genetic feature associated with EGFR-TKI secondary resistance in patients with NSCLC. The exon 18 G719X was the second most frequent uncommon mutation in our sample. Previous reports have provided conflicting results about its association with response to TKIs, but the small sample size and discrepancies in findings indicate that further studies are needed on the topic.9,22,32 For patients presenting with exon 21 L861X (third most frequent uncommon mutation in our sample), progression-free survival and response rates seem similar to classical mutations, indicating that this uncommon mutation is potentially TKI-sensitive, although it has a milder effect than classical EGFR alterations.22
Another consistent subclass was represented by a group of complex mutations, which corresponded to 1.51% of our patients. However, data specifically related to complex EGFR mutations are lacking. The vast majority are derived from retrospective series and are highly heterogeneous, as the identification of compound mutations is dependent on the molecular testing methods adopted.34 In Asian reports, the incidence of EGFR complex mutations ranges from 4% to 6.7%,35 whereas in the White population, approximately 5%-7% compound EGFR mutations among EGFR-positive patients have EGFR mutations.9 Again, the use of different testing methods, with different limits of detection and reference ranges, has a significant impact on the extreme variability of complex EGFR rates.
This study also presents limitations, mostly related to its cross-sectional approach, reduced availability of clinical features, smoking status, and the absence of information on treatment selection and outcomes, which could enhance our understanding of EGFR mutation profiles in Brazil. Another concern that should be highlighted is the probable selection bias of our tested patients, represented by a majority female sex and unknown exposure to smoking. The variability in genetic sequencing techniques with different limits of detection, such as sensitivity and coverage, the difference between the frequency of the use of each test, the small number of patients examined with pyrosequencing, which is the group with a lower frequency of EGFR mutation, the much higher rate of uncommon mutation with the Sanger method, and the fact that most patients were tested using a mutation-directed method (cobas) caused the study to be heterogeneous, could affect the results and also require further consideration. Future research could focus on the impact of the distinct mutations on EGFR-TKI treatment response and survival among Brazilian patients, contributing to the growing knowledge about the clinical significance of each alteration, including the rare types. Despite these limitations, our analysis provides a comprehensive overview of the EGFR mutation status in the Brazilian population. It is worth mentioning that the variability in the methods described here is probably similar to the real-world clinical setting that Brazilian oncologists face in their daily practice; thus, the data obtained are highly representative. In comparison with other Brazilian trials, our study is the largest and unique, with these large references of uncommon and complex EGFR mutations.
In summary, our study examined the prevalence of EGFR mutations in Brazilian patients according to different commonly used molecular methods of mutation analysis. EGFR mutations were observed in 24.2% of samples, and uncommon mutations affected 2.3%-18.7% of patients depending on the sequencing method. Data regarding the impact of these mutations on treatment outcomes are scarce. This gap needs to be addressed in further studies to improve the ability of prescribers to offer more adequate tests and therapeutic options for patients with NSCLC.
ACKNOWLEDGMENT
We thank the investigators and all patients who participated in this study and their families. We also thank Alessandro Ferreira from Progenetica and Instituto Hermes Pardini, where the molecular data were generated.
Tatiane Montella
Consulting or Advisory Role: MSD Oncology, Sanofi, BMS Brazil, Takeda, Janssen
Speakers' Bureau: MSD Oncology, AstraZeneca, Sanofi, Roche, Takeda, Janssen Oncology
Travel, Accommodations, Expenses: Janssen Oncology, MSD Oncology
Mauro Zukin
Honoraria: Janssen Oncology, AstraZeneca Brasil, Roche, Lilly, AstraZeneca, Wyeth Brazil, Novartis Brazil
Consulting or Advisory Role: Janssen Oncology Panama, MSD, AstraZeneca
Speakers' Bureau: Roche, AstraZeneca, Janssen Oncology
Travel, Accommodations, Expenses: Mundipharma, MSD Oncology, Roche, BMS Brazil, MSD Oncology
Vladmir Claudio Cordeiro de Lima
Consulting or Advisory Role: BMS Brazil, AstraZeneca, MSD, Lilly, Pfizer, Janssen, Daiichi Sankyo, Amgen
Speakers' Bureau: BMS Brazil, MSD, Lilly, Novartis, AstraZeneca/Daiichi Sankyo
Research Funding: BMS (Inst)
Travel, Accommodations, Expenses: MSD, AstraZeneca, Boehringer Ingelheim, BMS Brazil
Clarissa Baldotto
Honoraria: AstraZeneca, MSD Oncology Brazil, Roche Oncology Brazil, Takeda Oncology Brazil, BMS Brazil, Janssen Oncology Brazil, Pfizer Oncology Brazil, MD Health
Consulting or Advisory Role: MSD, AstraZeneca, Janssen Oncology Brazil, Novartis, Takeda Brazil, Roche Oncology Brazil
Speakers' Bureau: Roche Oncology Brazil, BMS Brazil, Janssen Oncology Brazil, MSD, Instituto D'Or de Pesquisa e Ensino, Takeda Brazil, AstraZeneca
Research Funding: AstraZeneca, Roche/Genentech, Janssen Oncology
Travel, Accommodations, Expenses: Janssen Oncology, Takeda
Pedro De Marchi
Honoraria: Sanofi
Consulting or Advisory Role: Merck Sharp & Dohme, AstraZeneca, Bristol Myers Squibb, Roche, Bayer
Speakers' Bureau: Merck Sharp & Dohme, Bristol Myers Squibb, Roche, AstraZeneca, Novartis, Merck, Boehringer Ingelheim, Janssen, Takeda
Research Funding: AstraZeneca, Amgen, Merck Sharp & Dohme, Roche
Travel, Accommodations, Expenses: Merck Sharp & Dohme, AstraZeneca, Roche, Bristol Myers Squibb
Carlos Barrios
Stock and Other Ownership Interests: MedSIR, Tummi
Honoraria: Novartis, Roche/Genentech, Pfizer, GlaxoSmithKline, Sanofi, Boehringer Ingelheim, Eisai, MSD, Lilly, Bayer, AstraZeneca, Zodiac Pharma
Consulting or Advisory Role: Boehringer Ingelheim, Roche/Genentech, Novartis, GlaxoSmithKline, Eisai, Pfizer, AstraZeneca, Libbs, MSD Oncology, United Medical, Lilly
Research Funding: Pfizer (Inst), Novartis (Inst), Amgen (Inst), AstraZeneca (Inst), Boehringer Ingelheim (Inst), GlaxoSmithKline (Inst), Roche/Genentech (Inst), Lilly (Inst), Sanofi (Inst), Taiho Pharmaceutical (Inst), Mylan (Inst), Merrimack (Inst), Merck (Inst), AbbVie (Inst), Astellas Pharma (Inst), Biomarin (Inst), Bristol Myers Squibb (Inst), Daiichi Sankyo (Inst), Abraxis BioScience (Inst), AB Science (Inst), Asana Biosciences (Inst), Medivation (Inst), Exelixis (Inst), ImClone Systems (Inst), LEO Pharma (Inst), Millennium (Inst), Janssen (Inst), Clinica Atlantis (Inst), INC Research (Inst), Halozyme (Inst), Covance (Inst), Celgene (Inst), inVentiv Health (Inst), Merck KGaA (Inst), Shanghai Henlius Biotech (Inst), Polyphor (Inst), PharmaMar (Inst)
Travel, Accommodations, Expenses: Roche/Genentech, Novartis, Pfizer, BMS Brazil, AstraZeneca, MSD Oncology, Lilly
Carolina Kawamura
Consulting or Advisory Role: BMS Brazil, AstraZeneca, MSD, Sanofi/Regeneron, Janssen, Takeda, Pfizer
Speakers' Bureau: BMS Brazil, MSD, AstraZeneca, Pfizer, Takeda, Janssen, Sanofi/Regeneron, Novartis
Research Funding: Janssen, Novartis, Pfizer
Travel, Accommodations, Expenses: Daiichi Sankyo/Astra Zeneca, AstraZeneca
Aknar Calabrich
Employment: Clínica AMO
Stock and Other Ownership Interests: Clínica AMO
Honoraria: MSD Oncology, BMS Brazil, Roche, AstraZeneca, Sanofi/Aventis, Janssen Oncology, GlaxoSmithKline, Takeda
Consulting or Advisory Role: AstraZeneca, MSD Oncology, BMS Brazil, Pfizer, Daiichi Sankyo/UCB Japan, GlaxoSmithKline
Speakers' Bureau: Roche, MSD Oncology, BMS Brazil, AstraZeneca, Pfizer, GlaxoSmithKline, Janssen Oncology, Takeda
Research Funding: AstraZeneca (Inst), Janssen Oncology
Travel, Accommodations, Expenses: MSD Oncology, BMS Brazil, AstraZeneca
Luiz Henrique Araújo
Consulting or Advisory Role: Merck Sharp & Dohme, Bristol Myers Squibb, AstraZeneca, Pfizer, Boehringer Ingelheim, Merck, Roche
Speakers' Bureau: AstraZeneca, Pfizer, Merck, Roche, Merck Sharp & Dohme, Bristol Myers Squibb, Boehringer Ingelheim
Research Funding: Merck Sharp & Dohme (Inst), Bristol Myers Squibb (Inst), AstraZeneca (Inst), Pfizer (Inst), Merck (Inst), Roche (Inst), Boehringer Ingelheim (Inst)
Gilberto Castro
Honoraria: AstraZeneca, Pfizer, Merck Sharp & Dohme, Bristol Myers Squibb, Novartis, Roche, Amgen, Janssen, Merck Serono, Lilly, Takeda
Consulting or Advisory Role: Boehringer Ingelheim, Pfizer, Bayer, Roche, Merck Sharp & Dohme, Bristol Myers Squibb, AstraZeneca, Yuhan, Merck Serono, Janssen, Libbs, Sanofi, Novartis, Lilly, Takeda
Speakers' Bureau: AstraZeneca, Bayer, Novartis, Roche, Merck Serono, Bristol Myers Squibb, Merck Sharp & Dohme, Boehringer Ingelheim, Pfizer, Janssen, Amgen, Takeda
Travel, Accommodations, Expenses: Merck Sharp & Dohme, Novartis, Pfizer, Roche, AstraZeneca, Boehringer Ingelheim, Bayer, Bristol Myers Squibb, Merck Serono
Carolina Bustamante
Employment: OCPM
André Santa Maria
Employment: AstraZeneca, Zodiac Pharma (I)
Carlos Gil Ferreira
Employment: Oncoclinicas & Co
Stock and Other Ownership Interests: Oncoclinicas & Co
Consulting or Advisory Role: AstraZeneca
Speakers' Bureau: BeiGene
No other potential conflicts of interest were reported.
SUPPORT
Supported by AstraZeneca Brazil.
AUTHOR CONTRIBUTIONS
Conception and design: Tatiane Montella, Mauro Zukin, Clarissa Baldotto, Carlos Barrios, Carolina Kawamura, Aknar Calabrich, Luiz Henrique Araújo, André Santa Maria, Carlos Gil Ferreira
Administrative support: Carlos Gil Ferreira
Provision of study materials or patients: Mariano Zalis, Mauro Zukin, Vladmir Claudio Cordeiro de Lima, Clarissa Baldotto, Paulo Salles, Clarissa Mathias, Luiz Henrique Araújo, Carlos Gil Ferreira
Collection and assembly of data: Tatiane Montella, Mauro Zukin, Clarissa Baldotto, Paulo Salles, Carolina Kawamura, Aknar Calabrich, Luiz Henrique Araújo, Gilberto Castro, Carolina Bustamante, André Santa Maria, Marcelo Reis, Carlos Gil Ferreira
Data analysis and interpretation: Tatiane Montella, Mariano Zalis, Mauro Zukin, Vladmir Claudio Cordeiro de Lima, Clarissa Baldotto, Pedro De Marchi, Clarissa Mathias, Carlos Barrios, Carolina Kawamura, Aknar Calabrich, Gilberto Castro, André Santa Maria, Marcelo Reis, Carlos Gil Ferreira
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/go/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Tatiane Montella
Consulting or Advisory Role: MSD Oncology, Sanofi, BMS Brazil, Takeda, Janssen
Speakers' Bureau: MSD Oncology, AstraZeneca, Sanofi, Roche, Takeda, Janssen Oncology
Travel, Accommodations, Expenses: Janssen Oncology, MSD Oncology
Mauro Zukin
Honoraria: Janssen Oncology, AstraZeneca Brasil, Roche, Lilly, AstraZeneca, Wyeth Brazil, Novartis Brazil
Consulting or Advisory Role: Janssen Oncology Panama, MSD, AstraZeneca
Speakers' Bureau: Roche, AstraZeneca, Janssen Oncology
Travel, Accommodations, Expenses: Mundipharma, MSD Oncology, Roche, BMS Brazil, MSD Oncology
Vladmir Claudio Cordeiro de Lima
Consulting or Advisory Role: BMS Brazil, AstraZeneca, MSD, Lilly, Pfizer, Janssen, Daiichi Sankyo, Amgen
Speakers' Bureau: BMS Brazil, MSD, Lilly, Novartis, AstraZeneca/Daiichi Sankyo
Research Funding: BMS (Inst)
Travel, Accommodations, Expenses: MSD, AstraZeneca, Boehringer Ingelheim, BMS Brazil
Clarissa Baldotto
Honoraria: AstraZeneca, MSD Oncology Brazil, Roche Oncology Brazil, Takeda Oncology Brazil, BMS Brazil, Janssen Oncology Brazil, Pfizer Oncology Brazil, MD Health
Consulting or Advisory Role: MSD, AstraZeneca, Janssen Oncology Brazil, Novartis, Takeda Brazil, Roche Oncology Brazil
Speakers' Bureau: Roche Oncology Brazil, BMS Brazil, Janssen Oncology Brazil, MSD, Instituto D'Or de Pesquisa e Ensino, Takeda Brazil, AstraZeneca
Research Funding: AstraZeneca, Roche/Genentech, Janssen Oncology
Travel, Accommodations, Expenses: Janssen Oncology, Takeda
Pedro De Marchi
Honoraria: Sanofi
Consulting or Advisory Role: Merck Sharp & Dohme, AstraZeneca, Bristol Myers Squibb, Roche, Bayer
Speakers' Bureau: Merck Sharp & Dohme, Bristol Myers Squibb, Roche, AstraZeneca, Novartis, Merck, Boehringer Ingelheim, Janssen, Takeda
Research Funding: AstraZeneca, Amgen, Merck Sharp & Dohme, Roche
Travel, Accommodations, Expenses: Merck Sharp & Dohme, AstraZeneca, Roche, Bristol Myers Squibb
Carlos Barrios
Stock and Other Ownership Interests: MedSIR, Tummi
Honoraria: Novartis, Roche/Genentech, Pfizer, GlaxoSmithKline, Sanofi, Boehringer Ingelheim, Eisai, MSD, Lilly, Bayer, AstraZeneca, Zodiac Pharma
Consulting or Advisory Role: Boehringer Ingelheim, Roche/Genentech, Novartis, GlaxoSmithKline, Eisai, Pfizer, AstraZeneca, Libbs, MSD Oncology, United Medical, Lilly
Research Funding: Pfizer (Inst), Novartis (Inst), Amgen (Inst), AstraZeneca (Inst), Boehringer Ingelheim (Inst), GlaxoSmithKline (Inst), Roche/Genentech (Inst), Lilly (Inst), Sanofi (Inst), Taiho Pharmaceutical (Inst), Mylan (Inst), Merrimack (Inst), Merck (Inst), AbbVie (Inst), Astellas Pharma (Inst), Biomarin (Inst), Bristol Myers Squibb (Inst), Daiichi Sankyo (Inst), Abraxis BioScience (Inst), AB Science (Inst), Asana Biosciences (Inst), Medivation (Inst), Exelixis (Inst), ImClone Systems (Inst), LEO Pharma (Inst), Millennium (Inst), Janssen (Inst), Clinica Atlantis (Inst), INC Research (Inst), Halozyme (Inst), Covance (Inst), Celgene (Inst), inVentiv Health (Inst), Merck KGaA (Inst), Shanghai Henlius Biotech (Inst), Polyphor (Inst), PharmaMar (Inst)
Travel, Accommodations, Expenses: Roche/Genentech, Novartis, Pfizer, BMS Brazil, AstraZeneca, MSD Oncology, Lilly
Carolina Kawamura
Consulting or Advisory Role: BMS Brazil, AstraZeneca, MSD, Sanofi/Regeneron, Janssen, Takeda, Pfizer
Speakers' Bureau: BMS Brazil, MSD, AstraZeneca, Pfizer, Takeda, Janssen, Sanofi/Regeneron, Novartis
Research Funding: Janssen, Novartis, Pfizer
Travel, Accommodations, Expenses: Daiichi Sankyo/Astra Zeneca, AstraZeneca
Aknar Calabrich
Employment: Clínica AMO
Stock and Other Ownership Interests: Clínica AMO
Honoraria: MSD Oncology, BMS Brazil, Roche, AstraZeneca, Sanofi/Aventis, Janssen Oncology, GlaxoSmithKline, Takeda
Consulting or Advisory Role: AstraZeneca, MSD Oncology, BMS Brazil, Pfizer, Daiichi Sankyo/UCB Japan, GlaxoSmithKline
Speakers' Bureau: Roche, MSD Oncology, BMS Brazil, AstraZeneca, Pfizer, GlaxoSmithKline, Janssen Oncology, Takeda
Research Funding: AstraZeneca (Inst), Janssen Oncology
Travel, Accommodations, Expenses: MSD Oncology, BMS Brazil, AstraZeneca
Luiz Henrique Araújo
Consulting or Advisory Role: Merck Sharp & Dohme, Bristol Myers Squibb, AstraZeneca, Pfizer, Boehringer Ingelheim, Merck, Roche
Speakers' Bureau: AstraZeneca, Pfizer, Merck, Roche, Merck Sharp & Dohme, Bristol Myers Squibb, Boehringer Ingelheim
Research Funding: Merck Sharp & Dohme (Inst), Bristol Myers Squibb (Inst), AstraZeneca (Inst), Pfizer (Inst), Merck (Inst), Roche (Inst), Boehringer Ingelheim (Inst)
Gilberto Castro
Honoraria: AstraZeneca, Pfizer, Merck Sharp & Dohme, Bristol Myers Squibb, Novartis, Roche, Amgen, Janssen, Merck Serono, Lilly, Takeda
Consulting or Advisory Role: Boehringer Ingelheim, Pfizer, Bayer, Roche, Merck Sharp & Dohme, Bristol Myers Squibb, AstraZeneca, Yuhan, Merck Serono, Janssen, Libbs, Sanofi, Novartis, Lilly, Takeda
Speakers' Bureau: AstraZeneca, Bayer, Novartis, Roche, Merck Serono, Bristol Myers Squibb, Merck Sharp & Dohme, Boehringer Ingelheim, Pfizer, Janssen, Amgen, Takeda
Travel, Accommodations, Expenses: Merck Sharp & Dohme, Novartis, Pfizer, Roche, AstraZeneca, Boehringer Ingelheim, Bayer, Bristol Myers Squibb, Merck Serono
Carolina Bustamante
Employment: OCPM
André Santa Maria
Employment: AstraZeneca, Zodiac Pharma (I)
Carlos Gil Ferreira
Employment: Oncoclinicas & Co
Stock and Other Ownership Interests: Oncoclinicas & Co
Consulting or Advisory Role: AstraZeneca
Speakers' Bureau: BeiGene
No other potential conflicts of interest were reported.
REFERENCES
- 1.WHO Cancer, 2022. https://www.who.int/news-room/fact-sheets/detail/cancer [Google Scholar]
- 2.Instituto Nacional de Câncer José Alencar Gomes da Silva: Estimativa 2020: incidéncia de câncer no Brasil / Instituto Nacional de Câncer José Alencar Gomes da Silva. Rio de Janeiro, Brazil, INCA, 2019 [Google Scholar]
- 3. Araujo LH, Baldotto C, de Castro G, Jr, et al. Lung cancer in Brazil. J Bras Pneumol. 2018;44:55–64. doi: 10.1590/S1806-37562017000000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kumarakulasinghe NB, van Zanwijk N, Soo RA. Molecular targeted therapy in the treatment of advanced stage non-small cell lung cancer (NSCLC) Respirology. 2015;20:370–378. doi: 10.1111/resp.12490. [DOI] [PubMed] [Google Scholar]
- 5. Hinrichs JW, Marja van Blokland WT, Moons MJ, et al. Comparison of next-generation sequencing and mutation-specific platforms in clinical practice. Am J Clin Pathol. 2015;143:573–578. doi: 10.1309/AJCP40XETVYAMJPY. [DOI] [PubMed] [Google Scholar]
- 6. Riely GJ, Politi KA, Miller VA, et al. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res. 2006;12:7232–7241. doi: 10.1158/1078-0432.CCR-06-0658. [DOI] [PubMed] [Google Scholar]
- 7. Castellanos E, Feld E, Horn L. Driven by mutations: The predictive value of mutation subtype in EGFR-mutated non-small cell lung cancer. J Thorac Oncol. 2017;12:612–623. doi: 10.1016/j.jtho.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 8. Tsoulos N, Papadopoulou E, Metaxa-Mariatou V, et al. Tumor molecular profiling of NSCLC patients using next generation sequencing. Oncol Rep. 2017;38:3419–3429. doi: 10.3892/or.2017.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sousa AC, Silveira C, Janeiro A, et al. Detection of rare and novel EGFR mutations in NSCLC patients: Implications for treatment-decision. Lung Cancer. 2020;139:35–40. doi: 10.1016/j.lungcan.2019.10.030. [DOI] [PubMed] [Google Scholar]
- 10. Tu HY, Ke EE, Yang JJ, et al. A comprehensive review of uncommon EGFR mutations in patients with non-small cell lung cancer. Lung Cancer. 2017;114:96–102. doi: 10.1016/j.lungcan.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 11. De Pas T, Toffalorio F, Manzotti M, et al. Activity of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with non-small cell lung cancer harboring rare epidermal growth factor receptor mutations. J Thorac Oncol. 2011;6:1895–1901. doi: 10.1097/JTO.0b013e318227e8c6. [DOI] [PubMed] [Google Scholar]
- 12. Cheema PK, Kuruvilla S. Fulfilling the potential of real-world evidence for lung cancer in Canada. Curr Oncol. 2019;26:157–159. doi: 10.3747/co.26.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn. 2018;20:129–159. doi: 10.1016/j.jmoldx.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 14. Freitas HC, Torrezan GT, da Cunha IW, et al. Mutational portrait of lung adenocarcinoma in Brazilian patients: Past, present, and future of molecular profiling in the clinic. Front Oncol. 2020;10:1068. doi: 10.3389/fonc.2020.01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Williams HL, Walsh K, Diamond A, et al. Validation of the Oncomine™ focus panel for next-generation sequencing of clinical tumour samples. Virchows Arch. 2018;473:489–503. doi: 10.1007/s00428-018-2411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Palacio S, Pontes L, Prado E, et al. EGFR mutation testing: Changing patterns of molecular testing in Brazil. Oncologist. 2019;24:e137–e141. doi: 10.1634/theoncologist.2018-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Lima Silva-Fernandes IJdL, Andrade LMN, Lima AESeO, et al. Characterization of patients with lung adenocarcinoma treated at a reference oncology center in northeastern Brazil and submitted to EGFR gene mutation research. J Clin Oncol. 2022;40(16 suppl) abstr e20548. [Google Scholar]
- 18. Graham RP, Treece AL, Lindeman NI, et al. Worldwide frequency of commonly detected EGFR mutations. Arch Pathol Lab Med. 2018;142:163–167. doi: 10.5858/arpa.2016-0579-CP. [DOI] [PubMed] [Google Scholar]
- 19. Crino L, Weder W, van Meerbeeck J, et al. Early stage and locally advanced (non-metastatic) non-small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(suppl 5):v103–v115. doi: 10.1093/annonc/mdq207. [DOI] [PubMed] [Google Scholar]
- 20.National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology—Non-small Cell Lung Cancer. NCCN Guidelines Version 4 2016. https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf [Google Scholar]
- 21. Giardina T, Robinson C, Grieu-Iacopetta F, et al. Implementation of next generation sequencing technology for somatic mutation detection in routine laboratory practice. Pathology. 2018;50:389–401. doi: 10.1016/j.pathol.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 22. O'Kane GM, Bradbury PA, Feld R, et al. Uncommon EGFR mutations in advanced non-small cell lung cancer. Lung Cancer. 2017;109:137–144. doi: 10.1016/j.lungcan.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 23. Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: A systematic review and global map by ethnicity (mutMapII) Am J Cancer Res. 2015;5:2892–2911. [PMC free article] [PubMed] [Google Scholar]
- 24. Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 25. Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 26. Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:213–222. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 27. Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 28. Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 29. Mitsudomi T, Tada H. EGFR gene mutations: Is it prognostic or predictive in surgically resected lung cancer? J Thorac Oncol. 2012;7:1739–1741. doi: 10.1097/JTO.0b013e3182743a0c. [DOI] [PubMed] [Google Scholar]
- 30. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 31. Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J Clin Oncol. 2011;29:2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 32. Galli G, Corrao G, Imbimbo M, et al. Uncommon mutations in epidermal growth factor receptor and response to first and second generation tyrosine kinase inhibitors: A case series and literature review. Lung Cancer. 2018;115:135–142. doi: 10.1016/j.lungcan.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 33. Russo A, Franchina T, Ricciardi G, et al. Heterogeneous responses to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in patients with uncommon EGFR mutations: New insights and future perspectives in this complex clinical scenario. Int J Mol Sci. 2019;20:1431. doi: 10.3390/ijms20061431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Attili I, Passaro A, Pisapia P, et al. Uncommon EGFR Compound Mutations in Non-Small Cell Lung Cancer (NSCLC): A Systematic Review of Available Evidence. Current Oncology 29:255-266, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mao L, Zhao W, Li X, et al. Mutation spectrum of EGFR from 21,324 Chinese patients with non-small cell lung cancer (NSCLC) successfully tested by multiple methods in a CAP-accredited laboratory. Pathol Oncol Res. 2021;27:602726. doi: 10.3389/pore.2021.602726. [DOI] [PMC free article] [PubMed] [Google Scholar]