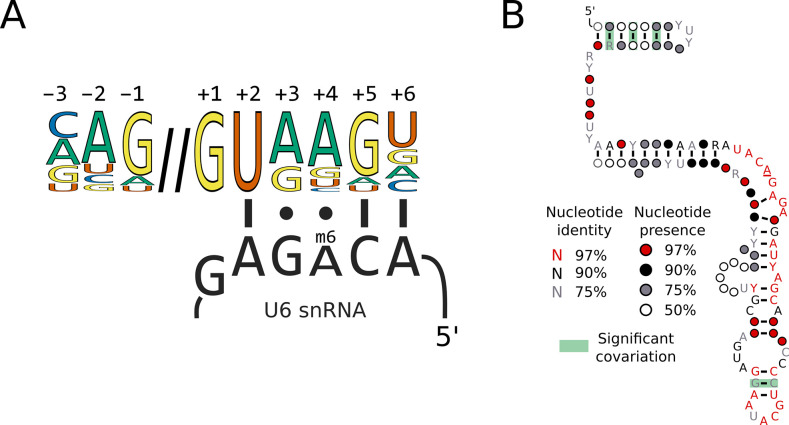
Figure 3. Switches in 5’SS sequence preference are not associated with compensatory changes in U6 snRNA sequence.
(A) Diagram showing the interaction of the U6 snRNA ACAGA box with the approximate human 5'SS consensus sequence. The methylated position of the ACAGA box is located opposite the 5'SS +4 position. (B) Conservation of U6 snRNA positions calculated from best U6 snRNA sequences predicted from genomes for 240 species (including 230 Saccharomycotina and 10 outgroups) using Infernal and plotted onto the predicted structure using R-scape and R2R. The predicted structure is derived from the predicted human U6 snRNA structure (Aoyama et al., 2020). All the positions in the ACAGA box were 100% conserved across all 240 species.