Some years ago the Staphylococcus aureus DNA topoiso-merase IV genes were cloned and named grlA and grlB (2). Alterations in the GrlA subunit have been associated with fluoroquinolone resistance in clinical isolates (2). Further work with laboratory mutants supported data which indicated that DNA topoisomerase IV was a primary target of fluoroquinolones in S. aureus (1).
In this work, a total of 55 strains of S. aureus isolated from patients from two hospitals in Spain together with the type strain ATCC 25923 were analyzed by single-stranded chain polymorphism (SSCP) analysis and DNA sequencing in order to find point mutations associated with fluoroquinolone resistance in grlA.
MICs of nalidixic acid, norfloxacin, and ciprofloxacin were determined by dilution methods. DNA amplification by PCR was performed by using standard protocols. The primers used were G1 (5′-ATTCAAGTGGTAATACACAC-3′, positions 149 to 168) and G2 (5′-TGACTTAAACGGACCATTGC-3′, positions 253 to 272), and they gave rise to a 124-bp DNA fragment. For SSCP analysis, the PCR products were then loaded on a 10% polyacrylamide gel (29:1 acrylamide/bis-acrylamide ratio) containing 8% glycerol. Electrophoresis was performed at 200 V in 0.5× TBE buffer at 12°C for 3 h. Bands were visualized by silver staining procedures.
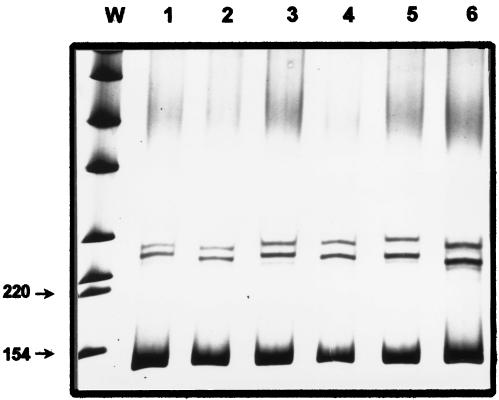
In the 55 clinical isolates, six SSCP patterns were found (Fig. 1). A wild-type control strain, in addition to some clinical strains demonstrated pattern 1. The nucleotide sequences of several strains representing each pattern were determined. All nucleotide sequences from strains with the same SSCP pattern were identical. Except for those in the wild-type sequence, three mutations were detected: Ser-80 (TCC) to Phe (TTC) (patterns 2 and 6), Ser-80 (TCC) to Tyr (TAC) (pattern 3), and Glu-84 (GAA) to Lys (AAA) (pattern 5). A silent mutation (CAT to CAC) in codon 77 (patterns 4, 5, and 6) was also detected.
FIG. 1.
SSCP analysis of the grlA gene. W, molecular weight markers for double-stranded DNA. Lane 1, wild-type DNA fragment; lane 2, TCC-to-TTC mutant (Ser-80 to Phe); lane 3, TCC-to-TAC mutant (Ser-80 to Tyr); lane 4, CAT-to-CAC mutant in codon 77 (silent); lane 5, GAA-to-AAA mutant (Glu-84 to Lys) and CAT-to-CAC mutant in codon 77 (silent); lane 6, TCC-to-TCC mutant (Ser-80 to Phe) and CAT-to-CAC mutant in codon 77 (silent). In all cases the lower band is the double-stranded 124-bp amplified fragment. Numbers to the left indicate molecular weight (in thousands).
The 18 strains, including the control strain, for which MICs of ciprofloxacin were ≤0.125 mg/liter, which were also the most susceptible to norfloxacin and nalidixic acid, contained the wild-type DNA fragment. Strains with low-level resistance (MICs of ciprofloxacin, 0.5 to 1 mg/liter) contained the wild-type sequence (seven strains) or a mutation: Glu-84 to Lys (two strains) or Ser-80 to Phe (two strains). The most highly resistant isolates, for which MICs of ciprofloxacin were ≥2 mg/liter and MICs of norfloxacin were ≥8 mg/liter, harbored a mutation in codon 80: Ser to Phe (n = 25) or Ser to Tyr (n = 2).
It can be concluded from these results that mutations in grlA are associated with quinolone resistance, as were changes in gyrA gene (3). However, alterations in grlA alone are not enough to obtain high levels of quinolone resistance. Since the quinolone MICs for strains harboring the same mutation in grlA were different, other factors, such as mutations in as gyrA, gyrB, or norA (2, 4–6), are implicated in quinolone resistance. However, it can be seen that mutations in the grlA gene are necessary to reach high-level quinolone resistance.
Finally, it should be noted that SSCP analysis of grlA is a sensitive method for the detection of mutations, because the six different patterns were associated with different sequences and vice versa.
Part of this work was supported by an FIS grant of the Spanish government (96/0018).
REFERENCES
- 1.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 3.Goswitz J J, Willard K E, Fasching C E, Peterson L R. Detection of gyrA gene mutations associated with ciprofloxacin resistance in methicillin-resistant Staphylococcus aureus: analysis by polymerase chain reaction and automated direct DNA sequencing. Antimicrob Agents Chemother. 1992;36:1166–1169. doi: 10.1128/aac.36.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito H, Yoshida H, Bogaki Shonai M, Niga T, Hattori H, Nakamura S. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2014–2023. doi: 10.1128/aac.38.9.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob Agents Chemother. 1994;38:1345–1355. doi: 10.1128/aac.38.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]