Abstract
Introduction
Translating narrative clinical guidelines to computable knowledge is a long‐standing challenge that has seen a diverse range of approaches. The UK National Institute for Health and Care Excellence (NICE) Content Advisory Board (CAB) aims ultimately to (1) guide clinical decision support and other software developers to increase traceability, fidelity and consistency in supporting clinical use of NICE recommendations, (2) guide local practice audit and intervention to reduce unwarranted variation, (3) provide feedback to NICE on how future recommendations should be developed.
Objectives
The first phase of work was to explore a range of technical approaches to transition NICE toward the production of natively digital content.
Methods
Following an initial ‘collaborathon’ in November 2022, the NICE Computable Implementation Guidance project (NCIG) was established. We held a series of workstream calls approximately fortnightly, focusing on (1) user stories and trigger events, (2) information model and definitions, (3) horizon‐scanning and output format. A second collaborathon was held in March 2023 to consolidate progress across the workstreams and agree residual actions to complete.
Results
While we initially focussed on technical implementation standards, we decided that an intermediate logical model was a more achievable first step in the journey from narrative to fully computable representation. NCIG adopted the WHO Digital Adaptation Kit (DAK) as a technology‐agnostic method to model user scenarios, personae, processes and workflow, core data elements and decision‐support logic. Further work will address indicators, such as prescribing compliance, and implementation in document templates for primary care patient record systems.
Conclusions
The project has shown that the WHO DAK, with some modification, is a promising approach to build technology‐neutral logical specifications of NICE recommendations. Implementation of concurrent computable modelling by multidisciplinary teams during guideline development poses methodological and cultural questions that are complex but tractable given suitable will and leadership.
Keywords: clinical decision support systems, computable knowledge, decision modelling, practice guideline
1. INTRODUCTION
A long‐standing challenge for clinical decision support is the knowledge engineering of traditional narrative clinical guidelines and recommendations into computable knowledge objects such as logical models or executable code. 1 Numerous methods and technical formalisms have been used over several decades, 2 , 3 , 4 as described in overviews and histories of clinical decision support. 5 , 6 , 7 We refer readers to these papers for descriptions of key concepts in the field.
In the existing landscape of guideline‐based clinical decision support (CDS) software, multiple product suppliers independently repeat this knowledge engineering exercise with varying degrees of fidelity, traceability and consistency. 8 In this context: ‘fidelity’ means how faithful the product is to the guideline narrative—is it a correct representation or has it missed something or introduced extra material without explanation; ‘traceability’ means the ability to show explicitly how each element of the guideline narrative is modelled in the product and how each element of the product is derived from the narrative; ‘consistency’ means that the supplier takes a standardised approach to guideline translation across a variety of topics and that the resulting logic is safely consistent with how other products render the narrative in computable form. The current extensive duplication of effort increases the amount of expert input that is required and means that some products have suboptimal content, unacceptable variation or even preventable errors. 9 , 10
Sittig et al proposed that one of ten grand challenges for CDS was ‘to create a set of standards‐based interfaces to externally maintained clinical decision support services that any [Electronic Health Record] could ‘subscribe to’, in such a way that healthcare organizations and practices can implement new state of the art clinical decision support interventions with little or no extra effort on their part’. 11 Based on a similar aspiration, the Mobilizing Computable Biomedical Knowledge (MCBK) movement began in 2017 with an inaugural meeting at the University of Michigan and is now spreading globally. 12 MCBK defines its meaning as ‘enabling the curation, dissemination and application of medical knowledge at a global scale’. 13 The 2018 MCBK Manifesto describes a vision that includes disseminating biomedical knowledge in ‘computable formats that can be shared and integrated into health information systems and applications’ and the ‘evolution of an open computable biomedical knowledge ecosystem dedicated to achieving the FAIR principles: making [Computable Biomedical Knowledge] easily findable, universally accessible, highly interoperable and readily reusable’. 14
Important work has been achieved on the theoretical foundations of CDS, drawing upon decision theory and formal representation of knowledge, processes, organizations and agents. 15 The theoretical framework embodied in the CREDO knowledge ladder (Figure 1) provides an abstract model of the various conceptual ‘layers’ involved in decision support, 16 , 17 which we believe also offers a structure to assess the completeness and coherence of any particular approach.
FIGURE 1.

CREDO Knowledge Ladder (reproduced with permission from Fox 16 ).
The National Institute for Health and Care Excellence (NICE) was established by the UK Government to provide authoritative guidance on clinical practice and health technology. Formally, it now operates as a non‐departmental public body of the Department of Health and Social Care in England. NICE's mission is to help practitioners and commissioners get the best care to people while ensuring value to the taxpayer. The NICE strategy 2021 to 2026 and subsequent business plans articulate that NICE will produce advice that is useful and useable, and will be part of a system that continually learns from data and implementation. 18 The strategic objectives include new approaches to guideline development, such as ‘a more modular approach’ and ‘guideline recommendations produced in an interactive, digitalised format’. In November 2021, NICE formed a Content Advisory Board (CAB), a panel of external experts to help inform and shape their content strategy to achieve these goals. In addition, NICE wants to track the extent to which their advice is being implemented, for example by monitoring use of guideline recommendations that are embedded in CDS systems.
In this paper, we report on the initial phase of MCBK‐UK work with NICE to model clinical narrative as computable knowledge.
2. RESEARCH QUESTIONS
The ultimate aims of the NICE CAB are:
To guide CDS/software developers to increase traceability, fidelity and consistency in supporting clinical use of NICE recommendations.
To guide local practice audit and intervention to reduce unwarranted variation.
To feed back to NICE on how future recommendations should be developed.
The first phase of work had the objective to explore a range of technical approaches to transition NICE toward the production of natively digital content and sought to answer the following questions:
What are the user personae, user stories and events we need to model the recommendations?
What information model (data items and relationships) is required and what definitions must be supplied to avoid ambiguity?
What format should be used to model these entities? What else is out there that we can re‐use?
3. METHODS
3.1. Proposal
In May 2022, a proposal was made by the co‐chair of MCBK‐UK (first author of this paper) and agreed by the NICE CAB, to organise two ‘collaborathons’ to explore ways of representing guidelines in a computable format. The term ‘collaborathon’ rather than ‘hack day’ or similar expressions was used on the basis that it ‘aims to be inclusive and appeal to all equally, maximising benefit from diverse representation’. 19 The work was unfunded and delivered by a volunteer team of industry, academics and clinicians between May 2022 and March 2023, with further work planned (Figure 2). Participants were invited from the CAB and informal professional networks, with no restriction placed on who could take part. The group included clinicians, medical publishers, standards developers (HL7 UK 20 and the UK Professional Record Standards Body [PRSB] 21 ) and software developers. Most were from industry, with a few academics.
FIGURE 2.

Timeline.
The aim of the collaborathons was to explore a range of technical approaches to transition NICE toward the production of natively digital content for education and standards‐based computable decision support. The specific objectives were: (1) to bring together relevant stakeholders who are needed to design the ecosystem for producing and utilising natively digital NICE content, (2) to demonstrate a selection of use cases, knowledge types, usage patterns, integration options, decision types and technical standards, and (3) to educate the stakeholder community (as represented by participants), generate new ideas and assess the relative merits of the variety of methods undertaken.
3.2. Preparation
Several clinical use cases were suggested in CAB meetings: GP urgent cancer referral guidelines, medication in type 2 diabetes, antimicrobial prescribing, rare disease diagnosis or ‘something’ in mental health (not further defined, but based on a feeling that mental health is often the last sector to be considered in technology projects). After discussion, NICE made the decision and selected the recommendations for adult type 2 diabetes (NICE Guideline [NG] 28) given its importance in population health and its moderate complexity, to maximise learning from the analysis. 22 To maintain a manageable scope in the first collaborathon, we focused on glucose management (NG28 section 1.6) and medication management (NG28 section 1.7).
We used a modified version of the knowledge levels defined by Boxwala et al, 23 as shown in Table 1. The purpose of these knowledge levels is to categorise the spectrum of knowledge types from entirely human‐readable narrative with no computability (L1), through intermediate levels of structured or semi‐structured content that goes some way toward computability (L2‐3), all the way to actual executable software (L4). The project adopted Health Level 7 (HL7) Clinical Quality Language (CQL), CDS Hooks and Business Process Modelling Plus for Health (BPM+) as the initial set of technical standards to consider, based on the prior knowledge of the participants, the anticipated utility of knowledge fragments in a CDS Hooks implementation, the de facto dominance of Business Process Modelling Notation (BPMN) and associated modelling tools in business process analysis, and the natural fit of this stack with HL7 Fast Healthcare Interoperability Resources (FHIR) as the anticipated predominant data interchange standard. 24 , 25 , 26 , 27 We added the L2A ‘Tagged fragment’ level to our modified Boxwala diagram to cover the use case of a knowledge component (eg, a paragraph or sentence from a NICE recommendation) that might be called by technology patterns like CDS Hooks.
TABLE 1.
Computable knowledge levels (modified from Boxwala et al 23 ).
| Knowledge level | Example |
|---|---|
| L1: Narrative | NICE recommendation |
| L2A: Tagged fragment | Deep link to section/paragraph of NICE recommendation (like HL7 Info button or BMJ Best Practice) |
| L2B: Semi‐structured | Structured natural language or diagrammatic logic based on NICE recommendation |
| L3: Structured | Computable CQL expressions or BPM+ model representing the logic specification |
| L4: Executable |
Same as L3 if the app/EPR directly supports CQL/BPM+ or Internal EPR/CDS software code to implement the CQL/BPM+ logic |
3.3. First collaborathon
The first collaborathon was held in November 2022, and the participants were organised into two sets of teams that interacted and consulted each other as the discussions proceeded. The primarily clinical team had the task to decompose the selected NICE content (L1) into useful tagged sub‐sections (L2A) and note any common principles for content decomposition across topics. They were asked to identify the kind of clinical questions that would lead a user to search the glucose management section of the recommendations, specifically:
what range of practitioners might be asking the question (or prompted to ask)
whether it would be practitioner‐initiated search or data‐triggered suggestion
what data items would be required to know if the question is relevant
what data items would be required to apply the recommendation.
They were asked to decompose the section into useful sub‐sections or fragments to answer specific questions, starting on blank flipchart sheets then consolidating into tables with the suggested column headings (modified as they wished): clinical question, fragment description, practitioner type, search/trigger, data for question, data for application.
The primarily technical team had the tasks to represent a selected NICE recommendation (L1) or pathway (L2B) in a fully specified logical model (L3) using CQL or BPM+ and note any common principles for how to translate L1/L2B into L3. Their scope was the medication management section of the recommendations, in particular the visual summary. 28 They were asked to identify:
any ambiguities or inconsistencies in the visual summary diagram or its notation style
any obvious inconsistencies between the narrative in section 1.7 and the visual summary
content which looks like an algorithm or rule
content which looks like a pathway or process model
content which is hard to categorise
logical sub‐sections of the visual summary, to decide which elements to use in the modelling task.
The results are summarised below
3.4. NCIG workstream sprints
The subsequent work was constituted as a mini‐project named NICE Computable Implementation Guidance (NCIG) to iteratively progress the work in preparation for the second collaborathon in March 2023. We formed three workstreams, which worked on (1) user stories and trigger events, (2) information model and definitions, (3) horizon‐scanning and output format. We held approximately fortnightly calls online for the whole team, with separate workstream calls in between. We initially shared our project work on github.com, but were later granted access to use the community Confluence platform managed by HL7 International. 29
As noted above, in the first collaborathon the technical teams had a focus on developing technical implementation models as direct representations of the clinical guideline logic. In NCIG, we stepped back from technical implementation standards to start with a user‐centred modelling approach to the clinical guideline content, drawing upon principles of domain‐driven modelling and requirements engineering practices such as user stories, personae and event modelling. 30 , 31 , 32
We started with a fairly informal approach but actively sought an existing methodology we could apply. During our project iterations, we considered the technology‐agnostic logical models and processes from the Common KADS knowledge‐acquisition methodology. 33 Some of Common KADS approach seems outdated, but its basic principles of modelling remain valid. Its main elements are logical models of the organization, task, agent, knowledge, communication and design. We were aware of related work in the FHIR community, particularly the FHIR Clinical Guidelines Implementation Guide, 34 however we did not pursue this further for this phase given our conscious decision not to focus on technical implementation standards. Our active horizon‐scanning later discovered the recent World Health Organization (WHO) Digital Adaptation Kit (DAK). 35 The WHO DAK was designed to support multiple health domains, consistent with the WHO SMART guidelines. 36 , 37 While the DAK is neither a formal ‘standard’ nor a mature methodology with a strong evidence base, its development has involved formal standards‐development bodies, and it comes with the imprimatur of the WHO so we believed there was a prima facie case to consider it. There is also a ‘worked example’ available, for antenatal care. 38 We therefore adopted the WHO DAK as our modelling approach in this phase. The principal components of the DAK are:
data dictionary (which we have also called ‘lexicon’)
decision logic
outcome indicators
operational requirements (functional and non‐functional)
personae
user scenarios
process models.
3.4.1. Second collaborathon
In the second collaborathon, we consolidated all the work of the NCIG workstream sprints as reported here and discussed this with a wider group of participants who had not been involved in the project work. This enabled us to identify the further work needed, as described below.
4. RESULTS
4.1. First collabarathon
Eighteen participants attended the first collaborathon (less than planned due to a UK railway strike), which produced:
Some terms that could be immediately concept‐coded using SNOMED CT and a list of terms that needed further definition
An extensive and inclusive list of all the relevant user roles
A list of user scenarios and trigger events
Fragmentary CQL and BPM+ models.
We also identified some confusion in the presentation and interpretation of NG28 section 1.7 visual summary, 28 partly because it used a non‐standard bespoke notation, and concluded that it was more of a psychological model than a strictly logical model, as it seems to be depicting clinical thinking and problem‐solving rather than a fully specified operational process flow.
Although it was noted as an objective, time did not permit addressing the question of common principles for content decomposition across topics.
4.2. NCIG workstream sprints
The subsequent NCIG iterations refined the initial products in the three multi‐disciplinary workstreams.
4.2.1. User scenarios and personae
The extensive list of scenarios and events was critically reviewed, and it was concluded that the two events when the guidance was likely to be most rigorously applied were the time at which a prescription was being initiated and when a review of the patient's diabetes was being undertaken. For the purposes of building a single illustrative end‐to‐end computable process it was collectively decided that the event to use for the implementation guide would be the initiation of the first diabetes‐related prescription. Box 1 illustrates a narrative user scenario.
BOX 1. Illustrative user scenario.
Jane booked an appointment with the practice because she was falling asleep after meals. The doctor confirmed that, despite her extensive efforts to follow the lifestyle and diet advice that had been given, her HbA1c (taken the previous week) was still not within an adequate range. The computer system confirmed that, based on her encoded information in her clinical record, she was eligible to be prescribed a standard dose of metformin. The prescriber confirmed that no new conditions had been diagnosed, or test results had been taken that may not be visible to the practice and that she was not taking any medications (for diabetes or otherwise) that the practice was not aware of. An up‐to‐date weight and BMI was recorded, the prescriber confirmed that Jane has no immediate plans to become pregnant and that she was not at risk of ketoacidosis.
This tightening of the use case sparked a refinement of the list of personae and it made sense to anchor on the term ‘Prescriber’ to encompass all legally approved prescribing decision makers. The model allowed for a supporting role and, reinforcing the particular importance of shared decision‐making in diabetes management, the secondary role of ‘Patient’ was documented.
The DAK user scenario table was used to document the data that would need to be used by the prescriber in conjunction with the NICE guidance. This was found to be a mix of coded data that could realistically be expected to be present in the electronic health record (EHR) as well as that which would need to be gathered in conversation with the patient at the time of the event. Diabetes management includes a significant amount of unstructured knowledge unlikely to be coded and some subjective judgement, for example, NG28 1.6.5 says ‘Discuss and agree an individual HbA1c target with adults with type 2 diabetes. Encourage them to reach their target and maintain it, unless any resulting adverse effects’. This kind of knowledge was also documented but the focus of the hand over to the decision logic team was the computable coded knowledge. Table 2 shows an extract of the related DAK table indicating the cross‐mapping between data items and decision logic.
TABLE 2.
Subset of a user scenario table for diabetes review.
| Data in record | Additional data elements to be collected (if any) | Decision support logic to be embedded |
|---|---|---|
| Adult (over 18) | [To identify patient record in scope] | |
| Type 2 diabetes | ||
| HbA1c >48 mmol/mol (6.5%) | Update HbA1c | Clinical review |
| Medications—absence of diabetes treatment | Update medications list | Is not on diabetes treatment—medication required |
| Pregnancy status—not pregnant | Confirm not planning to become pregnant | Is not pregnant—medication indicated |
| Previous target set (likely free text?) | Previous target set | Target not met—medication is required |
| Hyperglycaemia recorded | Update hyperglycaemia status | Symptoms indicate hyperglycaemia is present—consider insulin or sulfonylurea at any time in therapy/pathway |
4.2.2. Data dictionary
We adopted a hybrid approach to identifying and defining key terms in the narrative. Part of the exercise involved clinical participants manually reviewing content within the guidelines and providing manual definitions (with cited sources) and automated concept tagging, mapping, and annotation using natural language processing (NLP) by one of our industry participants.
Firstly, a subject matter expert (SME) analysed key features and semantic patterns within the guideline. Three classification schemas were formed (Automate, Both, Clinical) and then applied as viewed in the table below:
| Classification schema | Definition for schema | Example | Rationale for categorization |
|---|---|---|---|
| A. Automate | Must‐have, viable, safe to guide users explicitly by defined guidelines. | ‘Stable/unstable DM’ | There are known parameters defined within stable and unstable DM which can be codified (eg, Hb1AC has known values) |
| B. Both | Needs clinician input for definition. | ‘Abnormal HB disturbed erythrocyte turnover or abnormal haemoglobin type’ | Requires external parameters plus clinician domain knowledge to answer or interpret accordingly |
| C. Clinical | Needs clinician input for guidance/determination; not enough information in guideline to make informed and recommended decision to produce reproducible guidance. | ‘Investigate unexplained discrepancies between HbA1c and other glucose measurements. Seek advice from a team with specialist expertise in diabetes or clinical biochemistry’ | Here, a source of ground truth is not defined but left to clinical domain expertise |
Figure 3 provides a breakdown of the classification schema with respective categories showing an actual count and percent distribution for each of the categories.
FIGURE 3.

Comparing classification schemas for input, output, and combined. A: Automate, B: Both and C: Clinical.
In the NLP analysis (Figure 4), terms were parsed and normalized using a query parsing engine using Named Entity Recognition (NER) feature selection. Then a gap analysis identified unique concepts which were mapped to international medical standards: UMLS meta thesaurus, SNOMED CT, and ICD10 value sets. All concepts included unique identifiers, a hierarchical taxonomy, and ontological relationships which were also captured. Additionally, semantic categories for each of the concepts were included, providing visibility of the types of terms that are constructing the lexicon. There was a 98% match to terms within the existing taxonomy, the 2% of missing terms were added.
FIGURE 4.
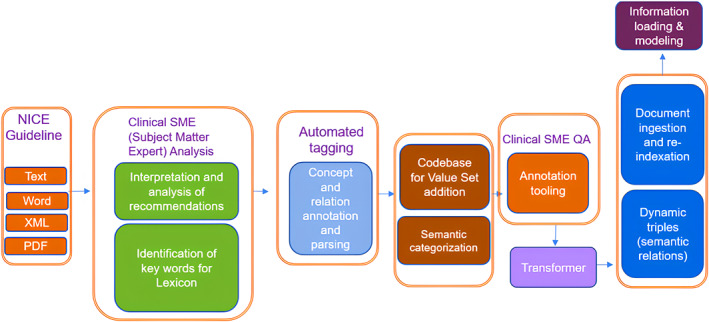
NLP concept identification and mapping.
4.2.3. Decision logic and process models
A simple high‐level process model was produced and the prescribing decision logic was defined in the relevant DAK table (Table 3).
TABLE 3.
Extract of decision logic table.
| Inputs | Output | Action | Annotations | |
|---|---|---|---|---|
| Latest HbA1c >6.5% | Previous HbA1c >6.5% | Discuss with patient. Enter consultation note. |
Prescribe metformin unless contraindicated. Specify prescription review date. |
This rule can be modified by adding a rule defining contraindication |
| Latest HbA1c >11.0 mmol/L | Discuss with patient. Enter consultation note. | Prescribe metformin unless contraindicated. Request follow‐up HbA1c test. Specify the prescription review date. | This rule can be modified by adding a rule defining contraindication | |
4.3. Second collaborathon
The NCIG workstream sprints produced a partial DAK model of NG28, and the second collaborathon was an opportunity for each workstream to consolidate its output and update NICE stakeholders and other CAB members. Our review discussion highlighted two remaining pieces of work to demonstrate real‐world feasibility: (1) implementation in a widely‐used electronic patient record system and/or in a clinical decision support system, and (2) using the decision logic for the prescribing recommendation to produce a query definition for outcome indicators (eg, defining a query on aggregate data of actual diabetes prescribing decisions to assess guideline conformance). Further work beyond that would need to address usability and clinician satisfaction as important elements of the DAK requirements specification.
For task (1) we propose to test ease of implementation and efficacy of approach using templates in general practice computer systems. For task (2) we propose to work with one of the UK national primary care data sources to build appropriate query sets.
5. DISCUSSION
The various components of the DAK seem to offer a sufficiently comprehensive and intuitive structure to address all the conceptual layers identified in the CREDO knowledge ladder (Figure 1 and Table 4). The user‐story‐driven approach is consistent with modern software engineering practice. We conclude that the WHO DAK, with some modification, offers a promising approach for representing complex narrative clinical guidelines in a technology‐agnostic logical model. The linkage of decision logic with outcome indicators also offers the opportunity both to assess guideline adherence to identify unwarranted variation, and also to detect legitimate positive deviance—a form of real‐world evidence that could indicate the need to update a guideline. 39 , 40
TABLE 4.
Representation of CREDO knowledge layers in WHO DAK components.
| CREDO layer | Relevant DAK component(s) |
|---|---|
| Agents | Personae, user scenarios, requirements |
| Plans | Process models |
| Decisions | Decision logic |
| Rules | Decision logic, indicators |
| Descriptions | Data dictionary |
| Concepts | Data dictionary |
| Symbols | Data dictionary (implicitly) |
This work has demonstrated the feasibility of post facto completion of a DAK logical model from a typical NICE narrative guideline section. The bigger challenge is to change the process of guideline development so that the necessary precision and definition of concepts, use cases, personae, decision logic, information model and indicators are built‐in to concurrent, multidisciplinary (not just clinical) work on developing a guideline and its equivalent computable form. 41 , 42
We suggest that using a technology‐agnostic and commercially neutral logical model such as the WHO DAK is (a) a more achievable first step when starting from scratch in an unfunded project and (b) more stable than a technical implementation model for a particular stack, and (c) has the advantage of not being subject to regulation as it is not directly executable. 43 Having said that, we see the FHIR Clinical Guidelines implementation guide as an important area for future work. We also believe there are improvements that can be made to the DAK, which we shall report elsewhere.
The example work that we report here is relatively straightforward and we accept there is a challenge of combinatorial explosion once the approach is tested with more complex guidelines or patients with multimorbidities; however, we argue that starting from the outset with user stories, definable logic and conceptual precision will help to manage the complexity. That said, it cannot be assumed that all narrative clinical guidance is susceptible to high‐fidelity, high‐precision computable modelling. Also, we reflect that our pragmatic focus on offering realistic actionable recommendations to consumers of guidance rests on having a behaviour change goal in focus when publishing recommendations.
Furthermore, in the reality of clinical practice, there is often a lack of ‘ground truth’ regarding which guideline recommendations to follow, which may differ between individual guidelines on the same disease. The guideline eco‐system is now a constantly evolving, moving target of intertwined recommendations from multiple sources—for example, NICE and international guideline producers, such as the European Society of Cardiology (ESC). Even guidelines from the same source but covering different topics may also conflict, although this is generally actively avoided. There are many potential explanations for such conflicts including differing interpretations of clinical evidence, prioritisation of different clinical outcomes, different intended timescales of benefit or different weightings given to cost‐effectiveness.
Guidelines may differ in choices of treatments (eg, in cardiology, the ESC recommended routine use of SGLT2 inhibitors in patients with high cardiovascular risk in 2019, whereas NICE recommended these from 2022 onwards). They may also differ on treatment thresholds: ESC recommend a low‐density lipoprotein (LDL) cholesterol target of <1.4 mmol/L in patients with prior myocardial infarction, whereas NICE recommend a target reduction of non‐high density lipoprotein cholesterol by 40%. In addition, patients now frequently have multiple comorbidities and the treatment recommendations for each individual comorbidity may also conflict with each other. Exceptional cases and special circumstances, such as comorbidities (particularly advanced age, frailty, and poor renal function), are now more frequently covered in guidelines, but may be included in a separate guideline that is not referenced by the first.
Clinicians must ultimately resolve all these differing and complex recommendations to produce a final management decision. However, there is no universally accepted method of ratifying conflicting recommendations. In general, most clinicians are likely to follow recommendations in order of locality. For example, in the UK, secondary care clinicians are likely to follow local NHS hospital guidelines, followed by NICE, followed by international guidelines. Many additional factors also influence these decisions including personal opinions of clinicians regarding utility and cost‐effectiveness, perceived validity of the source and local availability of management options. Modern electronic health record systems can call upon multiple CDS systems and bring recommendations back into workflows, leaving the clinician to make an overall judgement. Whilst an initial minimum viable product may focus on a single guideline, it is important that these issues are taken into account during the design of the process for translating narrative into computable guidance and the technology products that implement it so that both can flexibly scale and adapt.
The work reported here was loosely coupled to the evolving guideline development methodology within NICE. More progress was possible as they progressed at the same time, with work on computable guidance happening alongside work on a content management system for the NICE product lifecycle.
An important step toward moving from narrative guidelines to digital decision support is to define what is to be looked for in the record when following the guideline, and what is to be recorded in the record. This is a minimal viable product that NICE would need to provide to underpin any digitally enabled recommendations, providing significant value in its own right.
The project explored a standards‐based approach to digital guidelines. A critical aspect of standards and their development is community building, supporting collaboration within the community of intended users. The project suggested benefits for NICE in working with standards organisations such as HL7 UK and the PRSB.
While the focus of this article is computable knowledge as utilised in CDS, some of the general principles that we discuss are potentially also applicable in educational, research and public health use cases 14 and operational matters like fraud detection and economic predictions. An obvious parallel is translating a textbook into an e‐learning resource, where many analogous steps (and difficulties) are involved. 44
6. CONCLUSION
Well‐designed guideline‐based clinical decision support can enhance accessibility, streamline usage, and minimise errors stemming from interpretation and translation of narrative guidance as long as it is based upon consistent, traceable and faithful representation in a computable form. We suggest that the WHO DAK is an important step toward this goal, as it offers a sufficiently comprehensive and intuitive structure that is consistent with modern software engineering practice. The indicators to identify unwarranted variation in care delivery and to detect legitimate positive deviance provide additional value.
CONFLICT OF INTEREST STATEMENT
Several authors work in companies that provide computable knowledge products, related data science services, patient record systems and healthcare technology consultancy: Elia Lima‐Walton and Klara Brunnhuber (Elsevier Ltd), Polly Shepperdson (First Data Bank UK Ltd), Ben McAlister (Oracle Health), Charlie McCay (Ramsey Systems), Mark Thomas (Medicaite Ltd) and Justin Whatling (Palantir Technologies Inc.). Several authors work in NICE: Susan Faulding, Felix Greaves, Michaela Heigl, Shaun Rowark and Justin Whatling. Two authors are involved in standards‐development bodies: Charlie McCay (PRSB) and Ben McAlister (HL7 UK). Philip Scott is co‐chair of MCBK‐UK. No author received funding for this project.
ACKNOWLEDGEMENTS
We thank all the participants who gave their time and intellectual effort free of charge.
We thank Elsevier for use of their taxonomy and NLP expertise in the lexicon development.
Scott P, Heigl M, McCay C, et al. Modelling clinical narrative as computable knowledge: The NICE computable implementation guidance project. Learn Health Sys. 2023;7(4):e10394. doi: 10.1002/lrh2.10394
REFERENCES
- 1. National Academy of Medicine . Optimizing Strategies for Clinical Decision Support. NAM, Washington, DC; 2017. [PubMed] [Google Scholar]
- 2. Peleg M. Computer‐interpretable clinical guidelines: a methodological review. J Biomed Inform. 2013;46(4):744‐763. doi: 10.1016/j.jbi.2013.06.009 [DOI] [PubMed] [Google Scholar]
- 3. Iglesias N, Juarez JM, Campos M. Comprehensive analysis of rule formalisms to represent clinical guidelines: selection criteria and case study on antibiotic clinical guidelines. Artif Intell Med. 2020;103:101741. doi: 10.1016/j.artmed.2019.101741 [DOI] [PubMed] [Google Scholar]
- 4. Zhou L, Karipineni N, Lewis J, et al. A study of diverse clinical decision support rule authoring environments and requirements for integration. BMC Med Inform Decis Mak. 2012;12(1):128. doi: 10.1186/1472-6947-12-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Middleton B, Sittig DF, Wright A. Clinical decision support: a 25 year retrospective and a 25 year vision. Yearb Med Inform. 2016;25(S01):S103‐S116. doi: 10.15265/IYS-2016-s034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Greenes RA, Bates DW, Kawamoto K, Middleton B, Osheroff J, Shahar Y. Clinical decision support models and frameworks: seeking to address research issues underlying implementation successes and failures. J Biomed Inform. 2018;78:134‐143. doi: 10.1016/j.jbi.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 7. Sutton RT, Pincock D, Baumgart DC, Sadowski DC, Fedorak RN, Kroeker KI. An overview of clinical decision support systems: benefits, risks, and strategies for success. npj Dig Med. 2020;3:17. doi: 10.1038/s41746-020-0221-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O'Sullivan J, Wyatt J, Scott P. Building a ‘Clinical Satnav’ for Practitioners and Patients; 2022. https://www.bcs.org/policy-and-influence/health-and-care/building-a-clinical-satnav-for-practitioners-and-patients/
- 9. Riches N, Panagioti M, Alam R, et al. The effectiveness of electronic differential diagnoses (DDX) generators: a systematic review and meta‐analysis. PloS One. 2016;11(3):e0148991. doi: 10.1371/journal.pone.0148991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iacobucci G. Computer error may have led to incorrect prescribing of statins to thousands of patients. BMJ. 2016;353:i2742. doi: 10.1136/bmj.i2742 [DOI] [PubMed] [Google Scholar]
- 11. Sittig DF, Wright A, Osheroff JA, et al. Grand challenges in clinical decision support. J Biomed Inform. 2008;41(2):387‐392. doi: 10.1016/j.jbi.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khan N, Rubin J, Williams M. Summary of fifth annual public MCBK meeting: mobilizing computable biomedical knowledge (CBK) around the world. Learn Health Syst. 2023;7(1):e10357. doi: 10.1002/lrh2.10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. University of Michigan . Welcome to the MCBK Community; 2021. https://mobilizecbk.med.umich.edu/
- 14. Wyatt J, Scott P. Computable knowledge is the enemy of disease. BMJ Health Care Inform. 2020;27(2):e100200. doi: 10.1136/bmjhci-2020-100200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fox J, Glasspool D, Patkar V, et al. Delivering clinical decision support services: there is nothing as practical as a good theory. J Biomed Inform. 2010;43(5):831‐843. doi: 10.1016/j.jbi.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 16. Fox J. Cognitive systems at the point of care: the CREDO program. J Biomed Inform. 2017;68:83‐95. doi: 10.1016/j.jbi.2017.02.008 [DOI] [PubMed] [Google Scholar]
- 17. Fox J, Khan O, Curtis H, et al. Rapid translation of clinical guidelines into executable knowledge: a case study of COVID‐19 and online demonstration. Learn Health Syst. 2021;5(1):e10236. doi: 10.1002/lrh2.10236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. NICE . The NICE Strategy 2021 to 2026; 2021. https://www.nice.org.uk/about/who-we-are/corporate-publications/the-nice-strategy-2021-to-2026
- 19. Conway A. OneHealthTech Collaborathon at eHealth Week 2017; 2017. https://fabnhsstuff.net/fab-stuff/onehealthtech-collaborathon-ehealth-week-2017-angela-conway
- 20. HL7 UK . HL7 Delivers Healthcare Interoperability Standards; 2023. https://www.hl7.org.uk/
- 21. PRSB . Professional Record Standards Body; 2023. https://theprsb.org/
- 22. NICE . Type 2 Diabetes in Adults: Management; 2022. https://www.nice.org.uk/guidance/ng28/chapter/Recommendations
- 23. Boxwala AA, Rocha BH, Maviglia S, et al. A multi‐layered framework for disseminating knowledge for computer‐based decision support. J Am Med Inform Assoc. 2011;18(Suppl 1):i132‐i139. doi: 10.1136/amiajnl-2011-000334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Object Management Group . BPM+ Health; 2023. https://www.bpm-plus.org/
- 25. HL7 International . HL7 CDS Hooks; 2023. https://cds-hooks.hl7.org/
- 26. HL7 International . HL7 FHIR; 2023. https://hl7.org/fhir/
- 27. Andrikopoulou E, Scott P. Experiences of Creating Computable Knowledge Tutorials Using HL7 Clinical Quality Language; 2022. doi: 10.3233/SHTI220914 [DOI] [PubMed]
- 28. NICE . Type 2 Diabetes in Adults: Choosing Medicines; 2022. https://www.nice.org.uk/guidance/ng28/resources/visual-summary-full-version-choosing-medicines-for-firstline-and-further-treatment-pdf-10956472093
- 29. HL7 International . HL7 International Confluence pages; 2023. https://confluence.hl7.org/
- 30. Evans E. Domain‐Driven Design Reference; 2015. https://www.domainlanguage.com/wp-content/uploads/2016/05/DDD_Reference_2015-03.pdf
- 31. Pruitt J, Grudin J. Personas. Proceedings of the 2003 Conference on Designing for User Experiences, 1–15; 2003. doi: 10.1145/997078.997089 [DOI]
- 32. Lucassen G, Dalpiaz F, van der Werf JMEM, Brinkkemper S. Forging high‐quality User Stories: Towards a discipline for Agile Requirements. In: 2015 IEEE 23rd International Requirements Engineering Conference (RE); 2015:126–135.
- 33. Schreiber G. Knowledge Engineering. Amsterdam: Elsevier B.V; 2008:929‐946. [Google Scholar]
- 34. HL7 International . FHIR Clinical Guidelines. Retrieved August 16, 2023, from FHIR Clinical Guidelines Implementation Guide.
- 35. Tamrat T, Ratanaprayul N, Barreix M, et al. Transitioning to digital systems: the role of World Health Organization's digital adaptation kits in operationalizing recommendations and interoperability standards. Glob Health: Sci Pract. 2022;10(1):e2100320. doi: 10.9745/GHSP-D-21-00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. WHO . SMART Guidelines; 2023. https://www.who.int/teams/digital-health-and-innovation/smart-guidelines
- 37. Pretty F, Tamrat T, Ratanaprayul N, et al. Experiences in aligning WHO SMART guidelines to classification and terminology standards. BMJ Health Care Inform. 2023;30(1):e100691. doi: 10.1136/bmjhci-2022-100691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. WHO . Digital Adaptation Kit for Antenatal Care: Operational Requirements for Implementing WHO Recommendations in Digital Systems; 2021. https://www.who.int/publications/i/item/9789240020306
- 39. NICE . Real‐World Evidence Framework; 2023. https://www.nice.org.uk/about/what-we-do/real-world-evidence-framework
- 40. Lawton R, Taylor N, Clay‐Williams R, Braithwaite J. Positive deviance: a different approach to achieving patient safety. BMJ Qual Saf. 2014;23(11):880‐883. doi: 10.1136/bmjqs-2014-003115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scott PJ, Heitmann KU. Team competencies and educational threshold concepts for clinical information modelling. Stud Health Technol Inform. 2018;255:252‐256. doi: 10.3233/978-1-61499-921-8-252 [DOI] [PubMed] [Google Scholar]
- 42. Goud R, Hasman A, Strijbis A‐M, Peek N. A parallel guideline development and formalization strategy to improve the quality of clinical practice guidelines. Int J Med Inform. 2009;78(8):513‐520. doi: 10.1016/j.ijmedinf.2009.02.010 [DOI] [PubMed] [Google Scholar]
- 43. Wyatt JC, Scott P, Ordish J, et al. Which computable biomedical knowledge objects will be regulated? Results of a UK workshop discussing the regulation of knowledge libraries and software as a medical device. Learn Health Syst. 2023;e10386. doi: 10.1002/lrh2.10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bennett K, McGee P. Transformative power of the learning object debate. Open Learn. 2005;20(1):15‐30. doi: 10.1080/0268051042000322078 [DOI] [Google Scholar]


