Abstract
Objective
Autoimmune hepatitis (AIH) is a chronic immune-mediated inflammatory liver disease. Intestinal flora disturbance in AIH is closely related to TFH/TFR cell imbalances. As a new method of microbial therapy, the role of fecal microbiota transplantation (FMT) in AIH remains elusive. Here, we attempted to verify the functional role and molecular mechanism of FMT in AIH.
Methods
An experimental autoimmune hepatitis (EAH) mouse model was established to mimic the characteristics of AIH. H&E staining was used to detect histological features in mouse liver tissues. Serological tests were employed to identify several liver function biomarkers. Flow cytometry was utilized to examine the status of TFH/TFR cell subsets. Western blotting was used to evaluate TLR pathway-associated protein abundance. RT‒qPCR was applied to evaluate Treg cell markers and inflammation marker levels in mouse liver tissues.
Results
There was significant liver inflammation and dysregulated TFR/TFH cells with elevated levels of liver inflammation-associated biomarkers in EAH mice. Interestingly, transferring therapeutic FMT into EAH mice dramatically reduced liver injury and improved the imbalance between splenic TFR and TFH cells. FMT treatment also reduced elevated contents of serum alanine transaminase (ALT), aspartate aminotransferase (AST), and total bilirubin (TBIL) in EAH mice. Furthermore, therapeutic FMT reversed the increased levels of IL-21 while promoting IL-10 and TGF-β cytokines. Mechanistically, FMT regulated TFH cell response in EAH mice in a TLR4/11/MyD88 pathway-dependent manner.
Conclusion
Our findings demonstrated that liver injury and dysregulation between TFR and TFH cells in EAH might be reversed by therapeutic FMT via the TLR4/11-MyD88 signaling pathway.
Keywords: Experimental autoimmune hepatitis, Fecal microbiota, Transplantation, TFR/TFH cell imbalance, TLR pathway
1. Introduction
Autoimmune hepatitis (AIH) is a common chronic, immune-mediated inflammatory disease of the liver characterized by the presence of circulating autoantibodies, elevated serum levels of IgG and transaminase, and damaged hepatocytes [1,2]. The incidence of AIH occurs in all age groups with a female dominance, and its incidence is predicted to be 4/100,00–24.5/100,00 per year [3,4]. If AIH is left unaddressed, it can progress to some detrimental conditions, such as cirrhosis and liver failure. Several hypotheses, including genetic susceptibility together with environmental factors leading to the breakdown of hepatic immune homeostasis, have been proposed [5,6]; however, its exact etiology and pathogenic mechanisms remain elusive.
Increasing evidence suggests that T follicular helper (TFH) cells play a vital role in immune homeostasis and modulate the clinical course of the disease, treatment outcome, and overall survival. It is commonly accepted that TFH cells are essential for germinal center (GC) B cells to produce autoantibodies. TFH cells provide a helping signal to B cells to proliferate, survive, and promote the Ig class switch. Dysregulated TFH cells, however, can lead to aberrant GC B responses that result in the production of autoantibodies and promote autoimmune disorders. Previously, it was reported that excessive activation of TFH cells and elevated levels of IL-21 resulted in hypergammaglobulinemia, which augmented the immunopathological events of AIH [5]. Interestingly, TFH cells can be inhibited by follicular T regulatory (TFR) cells, which can inhibit the production of autoantibodies. TFR cells originate from peripheral T regulatory (Treg) cells that constitutively express Foxp3 and CXCR5 and Toll-like receptors (TLRs), such as TLR2 and TLR4 receptors [6]. TFR cells have a phenotype similar to both Treg and TFH cells [7]. Previous research has found that the imbalance between TFH/TFR cells could induce dysregulated immune homeostasis and that the overproduction of autoantibodies could contribute to the development of AIH [8], suggesting that the TFH/TFR cell balance is the maintenance and aggravating factor of AIH pathogenesis. Therefore, maintaining the balance between TFH/TFR cells is essential for preventing AIH.
The gut microbiota has drawn much attention in the search for potential novel and efficient treatment alternatives, largely as a result of human observational studies and animal research. The human gut microbiota is a distinctive system that subtly influences host metabolism and immunity [9]. The term “gut-liver axis” is frequently used to describe the intimate relationship between the gut and liver via the portal vein. Alterations in gut microbiota characterized by perturbations in microbial composition and function have been observed in AIH [10]. A previous report confirmed that there is an alteration in the composition and function of the gut microbiota, which is engaged in AIH pathogenesis [11]. Accordingly, intestinal microbiomes were shown to be involved in the pathogenesis of AIH in mouse model-based experiments [12,13].
Research has been performed on the therapeutic modulation of the gut-liver axis and dysbiosis of the gut microbiota. Numerous promising preliminary studies on the efficacy and clinical applicability of novel therapeutic strategies for microbiota regulation, including multibiotic and fecal microbiota transplantation (FMT), have been proposed [14,15]. FMT has recently received renewed attention and is regarded as a promising course of therapy to regulate gut microbiota directly [16]. FMT dates back to 1700 years ago in the Elbow Backup Emergency Recipe written by Ge Hong during the Eastern Jin Dynasty in China, the related records of using feces to treat food poisoning and digestive diseases [17]. FMT refers to the transfer of beneficial bacteria from a healthy donor to the digestive tract of patients via different routes of administration to rebalance a healthy microbial environment in the gut [[18], [19], [20]]. Various therapeutic applications of FMT have been reported in the literature. FMT is effective in treating persistent and refractory Clostridium difficile infections (CDIs) [21]. Similarly, FMT has also been demonstrated to be useful in treating obesity, type 2 diabetes, inflammatory bowel diseases (IBDs), intractable functional constipation, and other conditions [[22], [23], [24]]. Moreover, supplementation with sodium butyrate, a metabolite of the intestinal flora, has been shown to relieve liver injury and block the migration of intestinal pathobionts into AIH mouse livers [25]. In parallel with these important data, several studies have examined the association between gut microbiota and TFH cells. There is evidence that the gut microbiota regulates the autoimmune disease arthritis through TFH cells but not Th17 cells [26]. More recently, Ma et al. demonstrated that FMT is capable of controlling AIH by regulating the TFH/TFR cell imbalance by restoring intestinal microbiota dysbiosis [27]. However, it was not shown through which molecular mechanism the TFH/TFR cell imbalance was regulated. Therefore, this study attempted to validate whether FMT could regulate the TFH/TFR cell imbalance in AIH mice and explored the involved signaling pathway, which may provide novel insight for seeking therapeutic plans to treat AIH.
2. Materials and methods
2.1. Animals, reagents, and antibodies
Specific pathogen-free (SPF)-class female C57BL/6 mice (6–8 weeks, 18–20 g) from Shanghai Experimental Animal Center of the Chinese Academy of Sciences (Shanghai, China); biochemistry automatic analyzer from Roche Diagnostics (Branchburg, USA); Ficoll-Paque Plus from Amersham Biosciences (Little Chalfont, UK); flow cytometry machine LSR II Instrument from BD Biosciences (California, USA); primary antibodies including anti-TRL1/2/5/6/11, anti-MyD88 and anti-GAPDH and horseradish peroxidase-labeled secondary antibody from Abcam (Shanghai, China); anti-TRL4 antibody from Santa Cruz Biotechnology (USA); RNApure Kits, Super M-MLV and Power Taq PCR Master Mix from Bioteke (Beijing, China); SYBR Green from Solarbio LIFE SCIENCES; RIPA lysis buffer and Enhanced BCA Protein Assay Kit from Beyotime (Shanghai, China).
2.2. Induction of experimental autoimmune hepatitis (EAH)
All animal experiments were performed in accordance with the guidelines of the Care and Use of Laboratory Animals and approved by the Laboratory Animal Ethics Committee of the First People's Hospital of Changzhou, The Third Affiliated Hospital of Soochow University. Mice were randomly divided into the control and EAH groups with 20 mice in each group. An experimental autoimmune hepatitis (EAH) mouse model was established according to an earlier autoimmune hepatitis (EAH) mouse model, as described previously [28]. Liver antigen (S-100) was freshly prepared from female C57BL/6 mice after liver perfusion with PBS. After homogenizing the livers on ice, nuclei and remaining intact cells were subjected to centrifugation at 100,000×g for 60 min. Then, the supernatant was collected and applied for immunization. SPF class C57BL/6 female mice were induced into the EAH model by intravenously injecting freshly prepared S-100 antigen at a dose of 0.5–2 mg/ml in 0.5 ml PBS that was emulsified in an equal volume of complete Freund's adjuvant (CFA) on day 0. These mice were also injected with a booster dose at day 7. Mice in the control group received equal amounts of normal saline and CFA.
2.3. Fecal microbiota transplantation
The experimental mice in the control and EAH groups were subdivided into the control + PBS, control + FMT, EAH + PBS and EAH + FMT groups according to a random number table, with 10 mice in each group. For FMT, fresh murine fecal specimens (50 g) were harvested from 10 age- and sex-matched SPF-class C57BL/6 mice, pooled and dissolved in 250 ml sterile PBS. The supernatant was used for murine donor suspension under anaerobic conditions. Anesthetized mice in the EAH + FMT and Control + FMT groups received treatment with 0.3 ml of murine donor suspension via the anus, while those in the Control + PBS and EAH + PBS groups received PBS at the same amount (0.3 ml) via the anus. Mice were subjected to euthanasia and histological analysis on day 28 when the peak of disease activity was observed.
2.4. Histological analysis
Liver tissues from sacrificed animals were harvested for histological analysis. The samples were fixed with 4 % formaldehyde overnight, dehydrated, paraffin-embedded and sectioned into 4-μm sections using a microtome (Leica HistoCore BIOCUT, China). The sections were dewaxed with xylene and treated with gradient ethanol. Then, the sections were stained with hematoxylin (Beyotime) for 5 min and eosin (Beyotime) for 2 min to reveal and estimate the degree of inflammatory cell infiltration. The pathological changes in liver tissues were observed through a light microscope (200 × ) (Olympus, Japan).
2.5. Serological test
Peripheral blood specimens were collected from sacrificed animals through eyeball extraction. Serum alanine transaminase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL) and albumin levels were assessed using a biochemistry automatic analyzer (Hitachi 7600 automatic analyzer, Tokyo, Japan).
2.6. Enzyme-linked immunosorbent assay (ELISA)
The cytokine levels in the serum of mice were measured by ELISA using commercially available interleukin-21 (IL-21), (IL-10), and transforming growth factor beta (TGF-β) (Boster, Wuhan, China) following the manufacturer's guidelines. A multifunctional enzyme analyzer was applied to detect the optical density (OD) at 450 nm.
2.7. Flow cytometry
Spleen mononuclear cells (SMNCs) were isolated using density-gradient centrifugation via Ficoll-Paque Plus. To detect TFH/TFR cell subsets, 1 × 106 SMNCs were stained with BV510-anti-CD4 (cat. no. 563106, BD Biosciences), PerCP-Cy5.5-anti-CXCR5 (cat. no. 560528, BD Biosciences), and FITC-anti-GITR (cat. no. 126308, BioLegend) for 30 min, and the cells were kept at 4 °C in the dark. After washing the cells with PBS, the frequencies of CD4+CXCR5+GITR+TFR and CD4+CXCR5+GITR− TFH cells in EAH mice were measured by flow cytometry analysis. The cells were gated on living lymphocytes and then gated on CD4+CXCR5+ T cells using FlowJo software (v5.7.2).
2.8. Protein isolation and western blotting
Total protein was isolated from the mouse liver tissues by RIPA lysis buffer (Thermo Fisher, USA). Protein quantification was carried out using a BCA kit following the manufacturer's guidelines (Beyotime). Total protein (20 μg) from each sample was separated by 10 % SDS‒PAGE and transferred to PVDF membranes (Millipore, Billerica, MA, USA). After blocking the membranes with 5 % skimmed milk powder at room temperature for 2 h, membranes were incubated with the indicated primary antibodies overnight at 4 °C. Then, secondary antibodies (1:2000, cat. no. ab6721, Abcam; 1:2000, cat. no. ab6728, Abcam, UK) were added, incubated for another 2 h at room temperature, and washed with TBS. Finally, the membranes were revealed by enhanced chemiluminescence reagent (Beyotime). The relative densities of the protein bands were analyzed by ImageJ (v1.8.0; National Institutes of Health). The primary antibodies used were as follows: TLR1 (1:1000, cat. no. ab189337, Abcam), TLR2 (1:1000, cat. no. ab209216, Abcam), TLR4 (1:1000, cat. no. sc-293072, Santa Cruz Biotechnology, USA), TLR5 (1:1000, cat. no. ab13876, Abcam), TLR6 (1:1000, cat. no. ab228424, Abcam), TLR11 (1:1000, cat. no. ab21274, Abcam), MyD88 (1:1000, cat. no. ab219413, Abcam), and GAPDH (1:2500, cat. no. ab9485, Abcam) served as the loading control.
2.9. RNA extraction and RT‒qPCR
Total RNA was extracted from mouse liver tissues using an RNApure Kit following the manufacturer's instructions (Qiagen). cDNA was reverse-transcribed via Super M-MLV (R&D Systems) and a SYBR PrimeScript RT‒qPCR Kit (Qiagen). Quantitative real-time reactions were carried out under the following cycling conditions: 50 °C for 30 min, 95 °C for 15 min, and 40 cycles of 95 °C for 20 s, followed by 56 °C for 30 s and 72 °C for 30 s before a final primer sequence extension incubation at 72 °C for 5 min. GAPDH was used as an internal control reference. The relative gene expression was obtained with the 2–ΔΔCt method. The primer sequences are presented in Table 1.
Table 1.
Primer sequences used in this study.
| Name | Sequences |
|---|---|
| Foxp3 | forward: 5′-TCCTTCCCAGAGTTCTTCC-3′ |
| reverse: 5′-GATAAGGGTGGCATAGGTG-3′ | |
| IL-21 | forward: 5′- GGACCCTTGTCTGTCTGGTAG-3′ |
| reverse: 5′-TGTGGAGCTGATAGAAGTTCAGG-3′ | |
| IL-10 | forward: 5′-CTTACTGACTGGCATGAGGATCA-3′ |
| reverse: 5′-GCAGCTCTAGGAGCATGTGG-3′ | |
| TGF-β | forward: 5′-TGACGTCACTGGAGTTGTACGG-3′ |
| reverse: 5′-GGTTCATGTCATGGATGGTGC-3′ | |
| GAPDH | forward: 5′-ACTCTTCCACCTTCGATGC-3′ |
| reverse: 5′-CCGTATTCATTGTCATACCAGG-3′ | |
| E. coli | forward: 5′-CGGACTTTCTGCGTGCTAAGA-3′ |
| reverse: 5′-CAATTGGATTTTTGACTTCTG-3′ | |
| Lactobacillus | forward: 5′-AGCAGTAGGGAATCTTCCA-3′ |
| reverse: 5′-CACCGCTACACATGGAG-3′ | |
| Bifidobacterium | forward: 5′-TTACGTCCAGGGCTTCACG-3′ |
| reverse: 5′-ATTACTAGCGACTCCGCCTTCA-3′ | |
| Clostridium leptum | Forward: 5′-GCACAAGCAGTGGAGT-3′ |
| Reverse: 5′-CTTCCTCCGTTTTGTCAA-3′ |
2.10. Fecal microbiota analysis
The ZR Fecal DNA Kit (Zymo Research, CA, USA) was used to extract DNA from mouse fecal samples (100 mg wet weight for each sample) according to the manufacturer's protocol, followed by quantification with the Nanodrop ND-1000 Spectrophotometer at 260 nm. Then, RT‒qPCR was conducted to assess the abundance of selected bacteria, such as E. coli and Lactobacillus, using specific primers. The primer sequences used are shown in Table 1.
2.11. Statistical analysis
SPSS 20.0 software was used for statistical analysis. The data are expressed as the mean ± standard deviation (±SD). Student's t-test was used to analyze significant differences between two groups, and one-way analysis of variance followed by Tukey's post hoc test was used to compare significant differences among multiple experimental groups. A P value of <0.05 was considered significant.
3. Results
3.1. TLR/MyD88 pathway is activated in EAH mice
Our first objective was to induce a successful EAH mouse model to investigate the involved signaling pathway. We first attempted to establish an EAH mouse model to mimic the characteristics of AIH. H&E staining demonstrated that the structure of hepatic lobules was complete, hepatic cords were neatly arranged, and hepatocyte morphology in the portal vein and vicinity of the central vein was clear and complete without necrotic cells in the control group (Fig. 1A). In contrast, the EAH group had severely disordered hepatic lobules and hepatocyte cords, damaged hepatocyte morphology in the hepatic portal area and vicinity of the central vein and scattered and infiltrated inflammatory cells (Fig. 1A). Serological test results demonstrated that ALT, AST and TBIL concentrations were significantly higher while albumin content showed no difference in EAH mice relative to those in the control group (Fig. 1B–E). These results indicate the successful induction of the EAH mouse model. Subsequently, we collected splenocytes from these mice and detected TFR and TFH cell subsets through FACS. As shown in Fig. 1F, there was an increased frequency of CD4+CXCR5+GITR+ TFH cells and a reduced frequency of CD4+CXCR5+GITR− TFR cells in the EAH group compared with the control group.
Fig. 1.
Activation of the TLR/MyD88 pathway in EAH mice. (A) Hematoxylin and eosin (HE) staining displaying histological features of liver tissues after standard induction of EAH and control on day 28 (magnification, 200×, scale bar = 100 μm). (B–E) Serological levels of ALT, AST, TBIL, and albumin in the EAH and control groups. (F) Flow cytometry analysis of the TFR/TFH cell subset in spleen mononuclear cells from EAH and control mice. (G) Western blot analysis of TLR/MyD88 pathway-associated protein abundance in liver tissues from EAH and control mice. Data are representative of four independent experiments. **P < 0.01, ***P < 0.001.
Given that the TLR4/MyD88 pathway is involved in regulating gut microbiota [29], we explored whether the TFR/TFH response in EAH mice was mediated by this pathway. We measured several TLR proteins in mouse liver tissues using western blotting. As shown in Fig. 1G, there was no difference in the protein expression levels of TLR1/2/5/6 in EAH and control group mice. In contrast, TLR4/11 and MyD88 protein expression levels were significantly elevated in EAH mice relative to the control group mice (Fig. 1G), suggesting that the TLR4/11/MyD88 signaling pathway was activated in EAH mice. Collectively, these data indicate that EAH triggers liver injury by upregulating the frequencies of TFH cells while downregulating TFR cells by activating the TLR/MyD88 signaling pathway.
3.2. FMT ameliorates liver injury in EAH mice
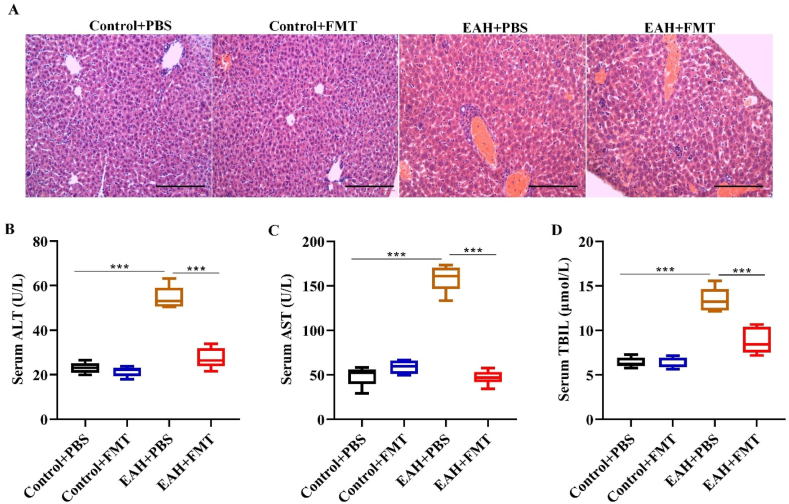
In the next step, we intended to assess whether FMT could exert a therapeutic effect on liver injury in AIH, EAH and control group mice subjected to murine fecal microbiota transplantation (FMT). As shown by H&E staining, FMT rescued the disordered hepatic lobule structure and damaged hepatocyte morphology as well as inflammatory cell infiltration in the EAH + FMT mouse group (Fig. 2A). In contrast, EAH mice that were administered PBS (EAH + PBS) had aberrant hepatic lobule structure with damaged hepatocyte morphology, while the control group mice had normal hepatic morphology. In addition, the elevated serum ALT, AST and TBIL contents in EAH mice were significantly reversed by FMT treatment (Fig. 2B–D). These results imply that therapeutic FMT could ameliorate liver injury in an EAH mouse model.
Fig. 2.
FMT alleviated liver injury in EAH mice. (A) Representative histological picture of liver tissues in different groups of mice after treatment with PBS or therapeutic FMT (scale bar = 100 μm). (B–D) Serum levels of liver functional biomarkers (ALT, AST, and TBIL) in mice under the indicated treatment. Data are representative of four independent experiments. ***P < 0.001.
3.3. FMT regulates TFR/TFH cell imbalances in EAH mice
We next explored whether FMT could modulate the EAH-triggered TFR/TFH cell imbalances in the splenic tissues of control and EAH group mice. FACS data demonstrated that FMT rescued the depleted CD4+CXCR5+GITR+TFR cells and elevated CD4+CXCR5+GITR−TFH cells in EAH mice to the same level as that of the control groups (Fig. 3A and B). Foxp3 is one of the key transcription factors controlling Treg cell development and function [30]. IL-21, a vital inflammation marker, exerts an inflammatory function in B cells [31], while IL-10 and TGF-β are regarded as anti-inflammatory and immunosuppressive cytokines [32]. We detected the gene expression levels of these important cytokines in mouse liver tissues. RT‒qPCR demonstrated that Foxp3, IL-10, and TGF-β gene expression levels were significantly reduced, while IL-21 gene expression levels were higher in the EAH group mice than in the control group mice (Fig. 3C–E). Interestingly, administrating FMT into EAH mice significantly reversed these changes by upregulating Foxp3, IL-10, and TGF-β gene expression while downregulating IL-21 gene expression. To further support these data, we also measured the serum levels of IL-21, IL-10, and TGF-β cytokines in these mice and found that FMT therapy prominently downregulated the production of inflammatory IL-21 cytokines while promoting the secretion of anti-inflammatory IL-10 and TGF-β cytokines (Fig. 3G–I). Collectively, these results suggest that FMT could regulate TFR/TFH cell imbalances by controlling inflammatory/anti-inflammatory cytokines in EAH mice.
Fig. 3.
FMT suppressed TFR/TFH cell imbalance in EAH mice. (A–B) Representative flow cytometry analysis of the TFR/TFH cell subsets in spleen mononuclear cells of the indicated groups. RT‒qPCR analysis of (C) IL-21, (D) Foxp3, (E) IL-10, and (F) TGF-β gene expression levels in mouse liver tissues in the indicated groups. Serum levels of (G) IL-21, (H) IL-10, and (I) TGF-β cytokines in the indicated groups. Data are representative of four independent experiments. **P < 0.01, ***P < 0.001.
3.4. FMT affects gut microbiota in EAH mice
Studies have demonstrated that disturbances in gut microbiota are critically involved in autoimmune disorders, including EAH [33,34]. Therefore, the effects of FMT on the gut microbiota in EAH mice were further investigated. The results of real-time PCR revealed that the reduction in the quantities of Lactobacillus, Bifidobacterium, and Clostridium leptum and the increase in the quantity of E. coli in the EAH group were significantly restored by FMT to normal levels, suggesting that FMT could regulate the gut microbiota in EAH progression (Fig. 4A–D).
Fig. 4.
FMT restored the gut microbiota to normal levels in EAH mice. Real-time PCR was conducted to examine the amount of (A) E. coli, (B) Lactobacillus, (C) Bifidobacterium and (D) Clostridium leptum in the fecal samples of mice in each group. *P < 0.05, **P < 0.01.
3.5. FMT inhibits TLR/MyD88 pathway activation in EAH mice
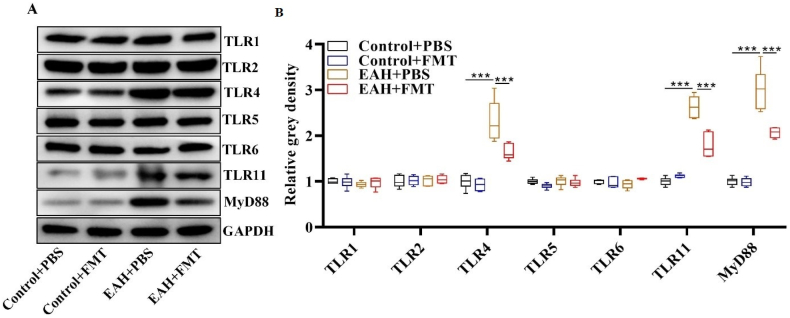
Finally, we explored whether FMT could inhibit the activation of TLR/MyD88 signaling, which is involved in AIH progression in an EAH mouse model. Western blot analysis demonstrated that the elevation in TLR4, TLR11 and MyD88 protein abundances in EAH mice was rescued after FMT administration (Fig. 5A and B). These data suggest that FMT could facilitate TLR/MyD88 signaling inactivation in EAH mice.
Fig. 5.
FMT inhibited TLR/MyD88 pathway activation in EAH mice. (A)Western blot measurement of TLR/MyD88 pathway-associated protein abundance in mouse liver tissues of the indicated groups. (B) Representative quantification of Western blot data. Data are representative of four independent experiments. ***P < 0.001.
4. Discussion
The human intestinal flora is a complex and enormous microecosystem, and the intestinal mucosa is a natural barrier that maintains intestinal flora homeostasis [35]. Once the intestinal mucosa is damaged, the intestinal flora is unbalanced, and a large amount of toxins enter the liver through the portal vein, thus exceeding the ability of the liver to detoxify and metabolize [36]. Intestinal microbiota dysbiosis has been reported to be associated with numerous autoimmune disorders, including inflammatory bowel diseases (IBDs), hepatic encephalitis, and diabetes mellitus [22,37,38]. In addition, numerous studies have demonstrated that the liver-gut axis plays a significant role in the etiology of chronic hepatitis, including NAFLD and primary biliary cirrhosis (PBC) [39,40]. In AIH patients, the unique microbial composition is correlated with disease progression. Cai et al. demonstrated a close association between AIH and the microbiota using transgenic AIH mice carrying HLA-DR3 [41]. In another study, it was shown that the intestinal flora composition was significantly different in AIH patients when compared to healthy individuals [42]. Moreover, intestinal flora disturbance has a close association with TFR/TFH cell imbalances [27]. Previous studies have reported that TFR/TFH plays an important role in various autoimmune diseases [43,44]. In a recent study, it was demonstrated that the TFR/TFH cell ratio was dysregulated and promoted the pathogenesis of EAH [27]. In the present study, we noticed that there were severely disordered hepatic lobules, damaged hepatocytes and infiltrated inflammatory cells in EAH mice. We found that the gut microbiota, such as E. coli, was altered in EAH mice and restored to normal levels by FMT. Our results also demonstrated a dysregulation between TFR/TFH cells, which might contribute to the pathogenesis of AIH. We noted greater numbers of TFH cells and a reduced number of TFR cells together with elevated serum levels of AST, ALT and TBIL in the EAH mice, which are consistent with previous findings [43,44]. Moreover, we also noticed that the onset of AIH was associated with IM alterations, as treatment of EAH mice with FMT ameliorated liver injury, rebalanced the number of TFR/TFH cells and restored the serum levels of AST, ALT and TBIL in these mice, supporting the concept that FMT might be a therapeutic option for treating AIH [21,24].
It is widely accepted that the gut microbiota controls T-cell activation and differentiation in a DC-dependent manner [45]. Intestinal DCs recognize gut microbiota through TLRs, leading to the activation of the MyD88 signaling axis that in turn results in T-cell activation and differentiation. A previous report demonstrated that functionally specialized DCs activate the TLR/MyD88 pathway to promote inflammatory Th17 cells. Several other studies have shown the involvement of the TLR/MyD88 pathway in different liver diseases. For example, chronic intermittent hypoxia induces liver fibrosis in mice via the TLR4/MyD88/MAPK/NF-kB pathway [46]. Taraxasterol suppresses liver injury by the TLR-4/MyD88/NF-κB pathway [47]. Therefore, we investigated whether the TLR/MyD88 pathway is involved in EAH and whether FMT regulates the imbalance between TFR/TFH cells via this pathway. Our data demonstrated no difference in the protein levels of TLR1/2/5/6 but showed prominently upregulated TLR4/11 and MyD88 protein levels in EAH mice. Interestingly, FMT treatment downregulated the levels of TLR4/11 and MyD88 in EAH mice. Notably, we also found that FMT significantly upregulated the gene expression levels of Foxp3 and anti-inflammatory cytokine IL-10 while downregulating pro-inflammatory IL-21. These data suggest that the TLR4/11/MyD88 signaling pathway is activated in EAH and that FMT treatment inhibits this signaling axis, thus impeding the TFH response in AIH.
5. Limitations
This study also had some limitations. First, the gut microbiota signature influenced by fecal microbiota transplantation in the EAH mouse model still requires further investigation. Second, the sample size is relatively small, and the duration is short, which may limit the precise outcomes of our study. Third, the correlation between the fecal microbiota and immune system regulatory cells was not deeply investigated. Future studies are required to investigate the effects of fecal microbiota transplantation on gut microbiota composition in the EAH mouse model as well as in AIH patients and the correlation with immune-system regulatory cells in AIH progression in a large sample size.
6. Conclusion
In summary, our data support the notion that FMT could control hepatitis progression by regulating TFR/TFH cell imbalance by inhibiting the TLR4/11-MyD88 signaling pathway. Thus, FMT might be an effective practice for treating AIH.
Funding statement
We have updated this section again.“This study was Sponsored by by grants from the Top Talent of Changzhou “The 14th Five-Year Plan” High-Level Health Talents Training Project (2022260), the Changzhou Sci&Tech Program (Grant No. CZQM2020011; CZQM2020079), Natural Science Foundation of Xinjiang Uygur Autonomous Region (2021D01F64), the Applied Basic Research Programs of Science, Technology Department of Changzhou city (CJ20210109; CJ20190095; CJ20160031), Natural Science Foundation of Xinjiang Uygur Autonomous Region (2022D01A307), Xinjiang Kirgiz Autonomous Prefecture Medical and Health Technology Project (Grant No. 43; No.44); the National Natural Science Foundation of China (No. 81700500) .”
Data availability statement
Original data generated in this study can be acquired from corresponding authors under reasonable requests.
Author contribution statement
Liang Ma; Jianguo Song; Xueping Chen: Performed the experiments; Analyzed and interpreted the data.
Duan Dai: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Jianping Chen; Liwen Zhang: Conceived and designed the experiments; Wrote the paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e20591.
Contributor Information
Jianping Chen, Email: 1517442002@stu.suda.edu.cn.
Liwen Zhang, Email: zhangliwenlove827@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
figs7.
figs8.
figs9.
figs10.
figs11.
figs12.
figs13.
figs14.
References
- 1.Sebode M., et al. Autoimmune hepatitis: from current knowledge and clinical practice to future research agenda. Liver Int. 2018;38(1):15–22. doi: 10.1111/liv.13458. [DOI] [PubMed] [Google Scholar]
- 2.Vuerich M., et al. Dysfunctional immune regulation in autoimmune hepatitis: from pathogenesis to novel therapies. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.746436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EASL clinical practice guidelines: autoimmune hepatitis. J. Hepatol. 2015;63(4):971–1004. doi: 10.1016/j.jhep.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Ngu J.H., et al. Population-based epidemiology study of autoimmune hepatitis: a disease of older women? J. Gastroenterol. Hepatol. 2010;25(10):1681–1686. doi: 10.1111/j.1440-1746.2010.06384.x. [DOI] [PubMed] [Google Scholar]
- 5.Ma L., et al. Tfh and plasma cells are correlated with hypergammaglobulinaemia in patients with autoimmune hepatitis. Liver Int. 2014;34(3):405–415. doi: 10.1111/liv.12245. [DOI] [PubMed] [Google Scholar]
- 6.Levy M., et al. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017;17(4):219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 7.Chung Y., et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 2011;17(8):983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang M., et al. Dysregulated TFR and TFH cells correlate with B-cell differentiation and antibody production in autoimmune hepatitis. J. Cell Mol. Med. 2020;24(7):3948–3957. doi: 10.1111/jcmm.14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommer F., et al. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017;15(10):630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 10.Wei Y., et al. Alterations of gut microbiome in autoimmune hepatitis. Gut. 2020;69(3):569–577. doi: 10.1136/gutjnl-2018-317836. [DOI] [PubMed] [Google Scholar]
- 11.Luo Y., et al. Gut microbiota regulates mouse behaviors through glucocorticoid receptor pathway genes in the hippocampus. Transl. Psychiatry. 2018;8(1):1–10. doi: 10.1038/s41398-018-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., et al. Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases. Autoimmun. Rev. 2017;16(9):885–896. doi: 10.1016/j.autrev.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Yuksel M., et al. A novel “humanized mouse” model for autoimmune hepatitis and the association of gut microbiota with liver inflammation. Hepatology. 2015;62(5):1536–1550. doi: 10.1002/hep.27998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pai N., et al. Results of the first pilot randomized controlled trial of fecal icrobiota transplant in pediatric ulcerative colitis: lessons, limitations, and future prospects. Gastroenterology. 2021;161(2):388–393.e3. doi: 10.1053/j.gastro.2021.04.067. [DOI] [PubMed] [Google Scholar]
- 15.Davar D., et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371(6529):595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Groot P.F., et al. Fecal microbiota transplantation in metabolic syndrome: history, present and future. Gut Microb. 2017;8(3):253–267. doi: 10.1080/19490976.2017.1293224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasbarrini G., Bonvicini F., Gramenzi A. vol. 50. 2016. Probiotics history. J clin gastroenterol; pp. S116–s119. (Proceedings from the 8th Probiotics, Prebiotics & New Foods for Microbiota and Human Health Meeting Held in Rome, Italy on September 13-15). Suppl 2. 2015. [DOI] [PubMed] [Google Scholar]
- 18.Wang J.W., et al. Fecal microbiota transplantation: review and update. J. Formos. Med. Assoc. 2019;118(Suppl 1):S23–s31. doi: 10.1016/j.jfma.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Vindigni S.M., Surawicz C.M. Fecal microbiota transplantation. Gastroenterol. Clin. N. Am. 2017;46(1):171–185. doi: 10.1016/j.gtc.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Antushevich H. Fecal microbiota transplantation in disease therapy. Clin. Chim. Acta. 2020;503:90–98. doi: 10.1016/j.cca.2019.12.010. [DOI] [PubMed] [Google Scholar]
- 21.Bakken J.S., et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011;9(12):1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palacios T., et al. Targeting the intestinal microbiota to prevent type 2 diabetes and enhance the effect of metformin on glycaemia: a randomised controlled pilot study. Nutrients. 2020;12(7) doi: 10.3390/nu12072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Wietmarschen H.A., et al. Probiotics use for antibiotic-associated diarrhea: a pragmatic participatory evaluation in nursing homes. BMC Gastroenterol. 2020;20(1):151. doi: 10.1186/s12876-020-01297-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F., et al. Microbiota transplantation: concept, methodology and strategy for its modernization. Protein Cell. 2018;9(5):462–473. doi: 10.1007/s13238-018-0541-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J.L., et al. Sodium butyrate ameliorates S100/FCA-induced autoimmune hepatitis through regulation of intestinal tight junction and toll-like receptor 4 signaling pathway. Immunol. Lett. 2017;190:169–176. doi: 10.1016/j.imlet.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Block K.E., et al. Gut microbiota regulates K/BxN autoimmune arthritis through follicular helper T but not Th17 cells. J. Immunol. 2016;196(4):1550–1557. doi: 10.4049/jimmunol.1501904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang M., et al. Fecal microbiota transplantation controls progression of experimental autoimmune hepatitis in mice by modulating the TFR/TFH immune imbalance and intestinal microbiota composition. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.728723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma L., et al. Exploration the significance of Tfh and related molecules on C57BL/6 mice model of experimental autoimmune hepatitis. J. Microbiol. Immunol. Infect. 2021;54(2):221–227. doi: 10.1016/j.jmii.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Bo R., et al. Mechanism of Lycium barbarum polysaccharides liposomes on activating murine dendritic cells. Carbohydr. Polym. 2019;205:540–549. doi: 10.1016/j.carbpol.2018.10.057. [DOI] [PubMed] [Google Scholar]
- 30.Wang J., et al. Foxp3(+) T regulatory cells as a potential target for immunotherapy against primary infection with echinococcus multilocularis eggs. Infect. Immun. 2018;86(10) doi: 10.1128/IAI.00542-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshizaki A., Tedder T.F. [IL-21 induces regulatory B cell differentiation and immunosuppressive effect through cognate interaction with T cells] Nihon Rinsho Meneki Gakkai Kaishi. 2015;38(1):57–64. doi: 10.2177/jsci.38.57. [DOI] [PubMed] [Google Scholar]
- 32.Li L., et al. Effects of immune cells and cytokines on inflammation and immunosuppression in the tumor microenvironment. Int Immunopharmacol. 2020;88 doi: 10.1016/j.intimp.2020.106939. [DOI] [PubMed] [Google Scholar]
- 33.Wei Y., et al. Alterations of gut microbiome in autoimmune hepatitis. Gut. 2020;69(3):569–577. doi: 10.1136/gutjnl-2018-317836. [DOI] [PubMed] [Google Scholar]
- 34.Liu Q., et al. Probiotics alleviate autoimmune hepatitis in mice through modulation of gut microbiota and intestinal permeability. J. Nutr. Biochem. 2021;98 doi: 10.1016/j.jnutbio.2021.108863. [DOI] [PubMed] [Google Scholar]
- 35.Zhou B.H., et al. Effects of Eimeria tenella infection on the barrier damage and microbiota diversity of chicken cecum. Poultry Sci. 2020;99(3):1297–1305. doi: 10.1016/j.psj.2019.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amornphimoltham P., et al. Gut leakage of fungal-derived inflammatory mediators: Part of a gut-liver-kidney Axis in bacterial sepsis. Dig. Dis. Sci. 2019;64(9):2416–2428. doi: 10.1007/s10620-019-05581-y. [DOI] [PubMed] [Google Scholar]
- 37.Levy M., et al. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017;17(4):219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 38.Hevia A., et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio. 2014;5(5) doi: 10.1128/mBio.01548-14. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saltzman E.T., et al. Intestinal microbiome shifts, dysbiosis, inflammation, and non-alcoholic fatty liver disease. Front. Microbiol. 2018;9:61. doi: 10.3389/fmicb.2018.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnabl B., Brenner D.A. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146(6):1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai W., et al. Intestinal microbiome and permeability in patients with autoimmune hepatitis. Best Pract. Res. Clin. Gastroenterol. 2017;31(6):669–673. doi: 10.1016/j.bpg.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 42.Lou J., et al. Fecal microbiomes distinguish patients with autoimmune hepatitis from healthy individuals. Front. Cell. Infect. Microbiol. 2020;10:342. doi: 10.3389/fcimb.2020.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sage P.T., et al. Suppression by T(FR) cells leads to durable and selective inhibition of B cell effector function. Nat. Immunol. 2016;17(12):1436–1446. doi: 10.1038/ni.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng J., et al. Dysregulation of circulating tfr/tfh ratio in primary biliary cholangitis. Scand. J. Immunol. 2017;86(6):452–461. doi: 10.1111/sji.12616. [DOI] [PubMed] [Google Scholar]
- 45.Stagg A.J. Intestinal dendritic cells in health and gut inflammation. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang H.H., et al. Chronic intermittent hypoxia induces liver fibrosis in mice with diet-induced obesity via TLR4/MyD88/MAPK/NF-kB signaling pathways. Biochem. Biophys. Res. Commun. 2017;490(2):349–355. doi: 10.1016/j.bbrc.2017.06.047. [DOI] [PubMed] [Google Scholar]
- 47.Li Z., et al. Effects of taraxasterol against ethanol and high-fat diet-induced liver injury by regulating TLR4/MyD88/NF-κB and Nrf2/HO-1 signaling pathways. Life Sci. 2020;262 doi: 10.1016/j.lfs.2020.118546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data generated in this study can be acquired from corresponding authors under reasonable requests.