Summary
Persistent HIV-1 reservoirs of infected CD4 T cells are a major barrier to HIV-1 cure, although the mechanisms by which they are established and maintained in vivo remain poorly characterized. To elucidate host cell gene expression patterns that govern virus gene expression, we analyzed viral RNA+ (vRNA) CD4 T cells of untreated simian immunodeficiency virus (SIV)-infected macaques by single-cell RNA sequencing. A subset of vRNA+ cells distinguished by spliced and high total vRNA (7–10% of reads) expressed diminished FOS, a component of the Activator protein 1 (AP-1) transcription factor, relative to vRNA-low and -negative cells. Conversely, FOS and JUN, another AP-1 component, were upregulated in HIV DNA+ infected cells compared to uninfected cells from people with HIV-1 on suppressive therapy. Inhibiting c-Fos in latently infected primary cells augmented reactivatable HIV-1 infection. These findings implicate AP-1 in latency establishment and maintenance and as a potential therapeutic target to limit HIV-1 reservoirs.
Subject areas: Molecular biology, Immunology, Microbiology
Graphical abstract

Highlights
-
•
SIV RNA+ T cells comprise low and high viral RNA subsets during acute infection
-
•
SIV RNAhigh T cells express decreased FOS, an AP-1 transcription factor subunit
-
•
AP-1 mRNA and activity are elevated in HIV DNA+ T cells from ART-suppressed PWH
-
•
Inhibiting c-Fos during HIV infection increases susceptibility to latency reversal
Molecular biology; Immunology; Microbiology
Introduction
Effective HIV-1 cure strategies have proven elusive due to a persistent population of immune cells harboring integrated proviral DNA able to withstand decades of antiretroviral therapy and support virus replication when therapy is interrupted.1 These infected cells, or reservoirs, are established within days of infection and are difficult to target due to limited viral transcript and protein expression, rendering them invisible to immune surveillance.2,3 To induce viral transcription and increase susceptibility of reservoirs to viral cytopathicity or immune-mediated cytotoxicity, therapeutic approaches have heavily focused on the development of latency-reversal agents. However, clinical trials evaluating these agents have largely failed to demonstrate biologically meaningful reductions in reservoirs,4,5,6,7,8 highlighting the need to better understand the basic mechanisms regulating latent and active infection in vivo.
The process by which latent infection is established and maintained is complex and incompletely understood. Multiple conditions are known to contribute to HIV-1 entry into transcriptional latency,9,10,11 herein defined as a cell state in which transcription of an integrated provirus is minimal or absent. These include: (1) low levels of activating host transcription factors that promote proviral expression; (2) insufficient viral transactivator of transcription factor protein (Tat); (3) repressive epigenetic modifications of the HIV-1 promoter; and (4) other chromatin regulatory factors such as histone modifiers.12,13,14,15,16,17,18,19 Several host transcription factors (e.g., NF-κB, NFAT, STAT5, Sp1, and TATA binding protein) bind the HIV-1 genome and are particularly critical for efficient HIV-1 mRNA expression.20,21 However, many of the over 50 confirmed or predicted transcription factor binding sites in the HIV-1 5′ long terminal repeat (LTR) promoter were identified in cell line models of latency,22 and their function in primary cells of people with HIV-1 (PWH) is not well established.
Efforts to elucidate host factors involved in HIV-1 reservoir establishment and maintenance have increasingly turned to single-cell transcriptomics and high-content phenotyping to identify signatures of infected cells.23,24,25,26,27,28,29 Isolation of infected cells directly from PWH has historically been challenging due to their rarity and the lack of definitive markers displayed without manipulation.1,30,31,32,33 As a result, many analyses rely on in vitro stimulation to induce viral protein or RNA expression from patient-derived cells,34,35,36 which may not faithfully recapitulate host cell properties in vivo. However, recent technological advances have enabled direct detection of cells harboring HIV-1 DNA, RNA, or protein without stimulation. These analyses have culminated in novel revelations about the properties of infected cells in PWH on and off suppressive ART, such as preferential infection of Th1-polarized, cytotoxic effector memory cells during early infection37; enrichment of specific transcription factor motifs38; upregulation of pro-survival and immune checkpoint markers39; increased cell proliferation and HIV-1 silencing signatures40; phenotypically distinct T cell subsets targeted in blood and lymph nodes41; and differential expression of host restriction factors.42 Despite these gains, a knowledge gap remains regarding the cellular environment conducive to latency establishment in vivo, which requires detailed viral transcriptional activity information to distinguish productive versus non-productive infection combined with the host transcriptome. This is supported by findings from in vitro models in which latently infected cells exhibit distinct transcriptomic profiles from virus-expressing cells.43,44
Here, we report single-cell, full-length mRNA sequencing of memory CD4 T cells directly ex vivo to define the viral mRNA (vRNA) content of infected cells and identify host factors associated with distinct stages of the viral life cycle in SIV-infected macaques, a model which recapitulates most aspects of HIV-1 infection in people. Surface marker phenotyping and TCR clonotyping was used to further interrogate immunologic properties of infected cells. To corroborate findings from SIV-infected cells, relevant host factors were evaluated for differential expression in HIV-1 DNA+ human CD4 T cells from PWH on suppressive ART as well as for functional involvement in regulating latent infection in a primary cell model of HIV-1 latency.40,45 Our results reveal a dueling role for the Activator protein (AP-1) transcription factor in both the restriction and reactivation of HIV-1 replication that is context-dependent. AP-1 may represent a target for therapeutic strategies to limit HIV-1 reservoirs.
Results
Single-cell viral RNA profile of in vivo SIV-infected CD4 T cells
To identify and characterize CD4 T cells harboring SIV in vivo during the earliest stages of infection, we isolated CD4 T cells from two rhesus macaques at 10 and 21 days post SIVmac251 and SIVmac239 infection, respectively (Table 1), when latent reservoirs are first seeded and infected cells are abundant,2,3 including transcriptionally active populations.46 Memory CD4 T cells (CD95+) from a mesenteric lymph node (animal AY69, n = 657 cells) and PBMC (animal T034, n = 941 cells; Figure S1A; Table 1) were single-cell index sorted by flow cytometry. Single-cell RNA sequencing (scRNA-seq) was performed on sorted cells using cDNA generated by SMART-Seq technology to produce full-length cDNA and maximize sensitivity for alternatively spliced viral transcripts.47 To limit alterations to their transcriptomic profile and best capture their in vivo state, cells were analyzed without stimulation or any ex vivo manipulations. We focused our analysis on memory CD4 T cells due to negligible vRNA detected in naive CD4 T cells during acute SIV infection.46 Unsupervised clustering was used to identify and filter potential contaminating cells exhibiting hallmarks of non-T cell lineages (Figures S1B and S1C). Reads from scRNA-seq for each cell were mapped to the SIVmac251 reference genome and revealed broad coverage across the viral genome (Figure 1A), indicating intact provirus. Well-characterized splice junctions present in transcripts encoding singly-spliced env and multi-spliced tat/rev mRNA (SD1-SA5 and SD4-SA7, respectively) were readily observed, allowing identification of cells expressing spliced viral mRNA (svRNA) species as putative markers of replication-active infection.46,48
Table 1.
Characteristics of SIV-infected macaque specimens interrogated by combined CD4 T cell flow cytometry and single-cell gene expression
| Animal | Virus | Infection Route |
Days PI | pVL (x106) | Tissue | Cells sorted | QC Cells analyzed | vRNA- cells | vRNA+, svRNA- cells | vRNA+, svRNA+ cells | Gene expression method | Estimated svRNA+ cells (%)a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AY69 | SIVmac251 | IV | 10 | 24 | LNMC | 657 | 593 | 364 | 132 | 95 | scRNA-seq | 15 |
| T034 | SIVmac239X | IR | 21 | 170 | PBMC | 941 | 672 | 543 | 64 | 55 | scRNA-seq | 7 |
| 12C174 | SIVmac239X | IV | 14 | 44 | PBMC | 768 | 695 | 282 | 374 | 39 | scRT-qPCR | 4 |
| TOTAL | 1960 | 1189 | 570 | 189 | ||||||||
IV, intravenous; IR, intrarectal; PI, post-infection.
Estimated by Poisson distribution probability applied to SIV gene RT-qPCR analysis of cells sorted at serial limiting dilution (3–1000 cells/well, replicates of 6–30). Poisson distribution experiments were performed separately from single-cell experiments.
Figure 1.
Single-cell RNA sequencing to reveal SIV mRNA profile of infected CD4 T cells
(A) RNA-seq reads mapping to SIV genome from a single vRNA+ CD4 T cell isolated from a mesenteric lymph node of SIVmac251-infected rhesus macaque (AY69, day 10 p.i.). Histogram (top) depicts read counts aligning to corresponding position of the genome. Arch graph depicts spliced vRNA spanning discontinuous regions of the genome. Vertical dotted lines represent relevant splice donor and acceptor sites.
(B) Distribution of SIV read content among CD4 T cells from two SIV-infected macaques (AY69) and (T034), displayed as percentage of total reads. Individual cells are colored by vRNA content (top) as follows: vRNA-, gray; vRNA+ lacking svRNA, blue; vRNA+ and svRNA+, red; vRNA+ not encompassed by above categories, pink (see also Figure S2); open symbols, positive for one of two vRNA splice junctions and SIV reads <1% of total. Bar chart depicts frequency of cells with the indicated vRNA content.
(C) mRNA counts for single CD4 T cells (dots) classified by vRNA status (colored bars) for the indicated genes.
Bars depict mean ± SD with significant differences indicated (∗∗∗∗, p < 0.0001; Mann-Whitney test).
(D‒G) Relationship between unspliced (gag) vRNA reads and percentage of mRNA corresponding to SIV (D) or total vRNA reads (E); unspliced (gag) and multiply spliced (ms; tat/rev) reads (F); and multiply spliced and singly spliced (ss; env) reads (G). Spearman rank (rho) and p value are indicated. Neg., negative.
To define the vRNA content of SIV-infected CD4 T cells, we determined the proportion of transcripts mapping to SIV within each cell. Of the 657 and 941 memory CD4 T cells sorted from the two animals, 593 and 672 cells passed quality control filter, respectively, of which 38% and 15% were vRNA+ (Figure 1B, Table 1). The vRNA content of these cells varied from 0.00001% to 41% of all reads. While there is the potential for PCR amplification bias when generating cDNA libraries and subsequent inflation of vRNA abundance, similar values have been reported for in vitro infected cells using other technologies.49,50 Notably, the vRNA content displayed a bimodal distribution, with the majority of cells containing <1% vRNA and a smaller subset with >1% vRNA, suggesting two distinct populations of vRNA+ infected cells.
The infection status of vRNA+ cells was further assessed by mapping SIV reads to unspliced (gag) and spliced (tat/rev, env) vRNA, using the splice junction regions described above. Strikingly, cells with <1% vRNA were largely positive for gag, but negative for svRNA, while vRNA-high cells were positive for both spliced and unspliced vRNA. In the latter group, vRNA comprised a median of 10% and 8% of the transcriptome in the LN and PBMC specimens, respectively (IQR 6–15%, maximum 41%; IQR 4–11%, maximum 32%), indicating a substantial fraction of transcription devoted to vRNA production. svRNA+ infected cells expressed not only the most total SIV RNA, but also abundant unspliced gag (Figure 1C). gag vRNA represented ∼10% of the total SIV RNA while the spliced tat/rev and env vRNA each comprised ∼1%. Similar results were observed in both animals. In contrast, svRNA- cells contained approximately 100-fold less total SIV and gag vRNA than the svRNA+ cells and median vRNA content was 0.05% and 0.001%, respectively. Based on these results we considered cells as resembling three distinct stages of the viral life cycle: 1) uninfected or transcriptionally silent infected cells lacking measurable vRNA; 2) infected cells harboring low levels of vRNA without evidence of substantial viral transcription and therefore unlikely to be supporting viral replication; and 3) infected cells expressing svRNA coincident with large quantities of vRNA (Figure S2). Of note, the frequency of cells positive for env and tat/rev transcripts, 14.9% in LN and 6.3% in PBMC, was comparable to estimates determined by orthogonal, qualified RT-qPCR methodology with single-cell sensitivity (Table 1; Figure S3A),46,48,51 supporting the accuracy and sensitivity of vRNA detection by our scRNA-seq method.
We explored the relationship between viral transcript levels for each vRNA class within individual infected CD4 T cells. Unspliced gag transcripts were highly correlated with total vRNA burden, as assessed by both the fraction of mRNA corresponding to SIV and the total SIV read count (Figures 1D and 1E). Unspliced gag also correlated with multiply spliced tat/rev mRNA levels, though many gag+ cells lacked these svRNAs (Figure 1F). And lastly, singly- (env) and multiply-spliced vRNA expression were highly correlated among svRNA+ cells (Figure 1G). Again, both animals exhibited similar vRNA co-expression patterns.
To confirm the results obtained by scRNA-seq, memory CD4 T cells from peripheral blood of a third SIV-infected animal, 12C174 (SIVmac239, day 14 p.i.; Table 1), were analyzed by single-cell RT-qPCR (scRT-qPCR) for the unspliced gag and spliced tat/rev transcripts using a targeted proteo-transcriptional evaluation method.48 Of 768 memory CD4 T cells sorted, 49% were positive for unspliced gag RNA and 5% were positive for both unspliced and multiply spliced tat/rev RNA. Again, gag and tat/rev transcripts were positively correlated (Figure S3B). Overall, these results from three independent animals analyzed by either scRNA-seq or scRT-qPCR show that SIV RNA+ infected CD4 memory T cells comprise two subpopulations distinguished by markedly different vRNA content: either low levels of unspliced vRNA in the absence of spliced vRNA or abundant levels of both spliced and unspliced vRNA. The latter phenotype is consistent with cells supporting active viral replication.
Down-regulation of CD3 and CD4 protein on svRNA+ CD4 T cell surface
Both HIV-1 and SIV proteins are well-characterized for their ability to down-modulate specific host proteins from the surface of in vitro infected CD4 T cells, properties which are believed to promote immune evasion and viral release, for example.52,53,54 To assess the vRNA+ T cells for surface protein down-modulation as evidence of viral protein expression and independently confirm viral transcription, we analyzed CD3 and CD4 protein expression on the cell surface measured by flow cytometry when cells were single-cell index sorted for sequencing. Both CD3 and CD4 surface expression varied with the proportion of the cellular transcriptome devoted to SIV. Cells with higher vRNA content (>∼1%) displayed diminished CD3 and CD4 staining (Figures 2A and 2B), while cells with < ∼1% vRNA maintained high expression of both proteins. Further, these proteins were downmodulated in a coordinated fashion, whereby loss of expression of both markers was most apparent in cells with the greatest vRNA content (Figure 2C). Using the cell grouping strategy described above to classify cells by vRNA status, we quantified the average staining fluorescence intensity for cells in each group and observed significant reductions in both CD3 and CD4 expression among the svRNA+ cells relative to both the vRNA- and svRNA- cells (Figure 2D). These markers were also downregulated in animal 12C174. MHC class I and CD28, additional host proteins downmodulated from the cell surface by both SIV and HIV-1 Nef, were also diminished on svRNA+ cells in animal T034. These data provide strong evidence of viral protein expression in the svRNA+ cells, further supporting our classification scheme for cellular infection status and confirming that these post-translational host protein modulations occur in vivo in macaques.
Figure 2.
Cell surface CD3 and CD4 protein expression varies with vRNA burden
(A and B) CD3 and CD4 (B) protein expression on the surface of T cells isolated from SIV-infected macaque mesenteric lymph node (top, AY69) and PBMC (bottom, T034) is plotted against SIV reads (percent of total reads) for each cell, represented as individual dots. Protein expression measured by surface staining and flow cytometric detection is reported as the fluorescence intensity (FI).
(C) Co-expression of surface CD3 and CD4 protein. (A-C) Fluorescence values within the autofluorescence range (<100, “negative”) are distributed randomly between 25 and 100 for visualization. Neg., negative.
(D) Single-cell surface CD3, CD4, MHC class I, and CD28 protein expression on cells grouped by vRNA content for each animal. vRNA- cells lacking surface CD4 expression (CD4 fluorescence intensity <500) were excluded to limit analyses to cells most likely to represent CD4 T cells. Lines connect medians; significant nonparametric Wilcoxon comparison differences between groups are indicated: ∗∗∗∗, p < 0.0001; ∗∗∗, p = 0.001–0.0001.
Host surface protein expression profile of vRNA+ CD4 T cells
Surface expression of protein markers on memory T cells is useful for defining the activation, memory, and effector profile of infected cells to elucidate preferred host cell targets and properties. To study the detailed phenotype of the vRNA+ cells, we analyzed a panel of 10 surface markers by cell staining and flow cytometric detection during cell sorting (Figure S1A). The cellular activation marker, HLA-DR, was expressed at higher levels on svRNA+ cells compared to vRNA- and vRNA+sVRNA- cells in all animals (Figures 3A and S4A). Increased CD69 and CD38, additional activation markers, were also apparent in AY69. Of the homing receptors assessed, SIV replication was biased toward cells expressing higher levels of CCR6 in AY69 LNMC and decreased CCR6 in T034 PBMC, the former consistent with a viral preference for Th17 cells.55 In general, neither CXCR3 nor CXCR5 differed by cellular infection status, with the exception of increased CXCR5, a LN homing and T follicular helper marker, among vRNA+svRNA- cells relative to vRNA- cells. svRNA+ cells expressed higher levels of CD95, a marker of T cell memory and receptor for programmed cell death signaling. Expression of PD-1 and TIGIT, activation and/or exhaustion markers, were also upregulated in svRNA+ cells from animals AY69 and T034. IFITM1, an interferon-induced HIV-1 restriction factor that inhibits viral infectivity and protein synthesis,56 was elevated on svRNA+ infected cells compared to vRNA- cells in these animals. Finally, neither ICOS nor integrin β7 preferentially marked vRNA+ cells in a consistent way; rather, β7 expression was diminished on svRNA+ cells relative to vRNA- cells in T034 PBMC and a similar trend in AY69 LNMC (p = 0.09). These findings indicate infection of heterogeneous T cell subsets, with increased activation among a subset of svRNA+ cells.
Figure 3.
Activation, homing, and exhaustion surface marker profiling of SIV RNA+ cells CD4 T cells
(A) Surface protein expression measured by flow cytometry for SIV-infected macaque mesenteric lymph node (AY69) and PBMC (T034, 12C174). Single-cell fluorescence intensity is displayed; lines connect median values. ∗∗∗∗, p < 0.0001; ∗∗∗, p = 0.001–0.0001; ∗∗, p = 0.01–0.001; nonparametric Wilcoxon comparisons between groups.
(B) Multivariate surface marker expression clustering displayed for each vRNA cell group. vRNA+ cells are overlaid upon the vRNA- cells (gray).
(C) Relative protein expression among the eight cell clusters identified by FlowSOM (see Figure S4).
(D) Proportion of each vRNA cell grouping comprised of the eight FlowSOM clusters.
To examine the combined protein profile of infected cells incorporating multiple markers, we performed multivariate clustering analysis by unsupervised hierarchical clustering using a Self-Organizing Map visualization technique, FlowSOM.57 Eight clusters were identified among the memory CD4 T cells pooled from the three animals, with evidence of specific subset enrichment among svRNA+ cells in animals AY69 and T034 (Figures 3B, 3C, and S4B). In AY69 LNMC, clusters 6 and 7 together comprised nearly 50% of all svRNA+ cells, compared to ∼10% and 20% of vRNA- and vRNA+svRNA- cells, respectively (Figure 3D). These clusters were defined by high expression of CD69 alone (cluster 7) or in combination with elevated CD38, TIGIT, PD-1, CXCR5, CXCR3, IFITM1, ICOS, HLA-DR, CD95, MHC class I and CD28 (cluster 6), indicating preferential viral transcription in recently activated cells, some of which co-express additional activation markers as well as hallmarks of both Th1 and T follicular helper cells. In contrast, T034 PBMC svRNA+ cells were enriched for cluster 2, characterized by diminished expression of most of these proteins, including activation markers. Increased expression of IFITM1, HLA-DR, PD-1, TIGIT, and IFITM1 by T034 svRNA+ cells relative to vRNA- cells at the univariate level (Figure 3A) suggests that these proteins are not elevated on a discrete subset of cells. vRNA+svRNA- cells comprise similar clusters as vRNA- cells for all three animals, as were 12C174 svRNA+ cells. UMAP clusters were distinct between PBMC and LNMC specimens but did not differ between the PBMC samples interrogated by different gene expression methods (Figure S4C).
Host cell genes differentially expressed in SIV-replicating T cells
To determine host factors involved in regulating viral transcription and infected cell persistence, we compared host gene expression among SIV-infected cells and vRNA- cells by analyzing the transcriptomes obtained by scRNA-seq. Exploring host gene profiles that distinguish infected cells, we performed multivariate clustering analysis of sorted single cells. We did not observe specific sub-clustering of memory CD4 T cells by infection status (replication-active or -inactive cells) in either AY69 or T034 (Figure 4A), indicating that these vRNA+ cell subsets fail to exhibit major substantial shifts in gene expression patterns that readily differentiate them from their vRNA- counterparts.
Figure 4.
Single-cell RNA-seq analysis of SIV-infected macaques identifies host genes associated with productive infection
(A) UMAP clustering of CD4 T cells from macaque AY69 and T034 displaying the distribution of vRNA-, vRNA+svRNA- and svRNA+ cells. Cells (dots) are colored by vRNA status.
(B) Differentially expressed genes (Bonferroni p < 0.05) between vRNA- and svRNA+ cells in AY69 and their corresponding values in T034. Genes with differential expression in the same direction in both animals. Natural log fold-change is indicated (FC; vRNA-/svRNA+).
(C) FOS expression in cells grouped by vRNA status. Significant (p < 0.05) nonparametric Wilcoxon comparisons are indicated for 12C174 (scRT-qPCR).
Using a univariate approach to identify individual differentially expressed genes (DEG) in the context of active viral transcription, we compared the transcriptome of svRNA+ (n = 95) and vRNA- (n = 364) cells in animal AY69. The large number of svRNA+ cells in this animal enabled a well-powered analysis. Nine genes were differentially expressed in the svRNA+ cells (Figure 4B, Bonferroni p < 0.05), including elevated expression of the viral coreceptor, CCR5. As validation, expression of these 9 genes was assessed in animal T034 (svRNA+, n = 55; vRNA-, n = 543). One gene, FOS, was differentially expressed in the same direction in both animals. FOS was downregulated in svRNA+ cells compared to vRNA- cells 2.1- and 4.4-fold in the two animals, respectively (Bonferroni p < 0.05, after multiple testing correction). FOS encodes c-Fos, a component of the AP-1 transcription factor. We further validated this finding using a more stringent cell exclusion criteria to limit any additional potential non-CD4 T cell contaminants by filtering clusters enriched for surface CD4-negative cells for animal AY69 (Figure S1D). FOS remained the top differentially expressed gene (Bonferroni p = 1.4E-06).
Of the differentially expressed genes in svRNA+ cells, FOS was the most pronounced in animal AY69 (p = 2.39E-06). In addition, the reduction in FOS expression relative to vRNA+svRNA- cells was nominally significant in both AY69 and T034 (Figure 4C; unadjusted p < 0.05). To verify these findings using an orthogonal approach, we assessed differential host gene expression by high-throughput scRT-qPCR on 695 memory CD4 T cells from animal 12C174 (PBMC). Again, FOS expression was lower in svRNA+ cells compared to both vRNA- and vRNA+svRNA- cells by 2.2-fold and 2.3-fold, respectively (p = 0.01 and 0.003; Figure 4C). These data indicate that high levels of FOS within a cell coincide with reduced likelihood of viral transcription, a potentially surprising result given extensive literature on AP-1 as an activator of HIV-1 transcription.
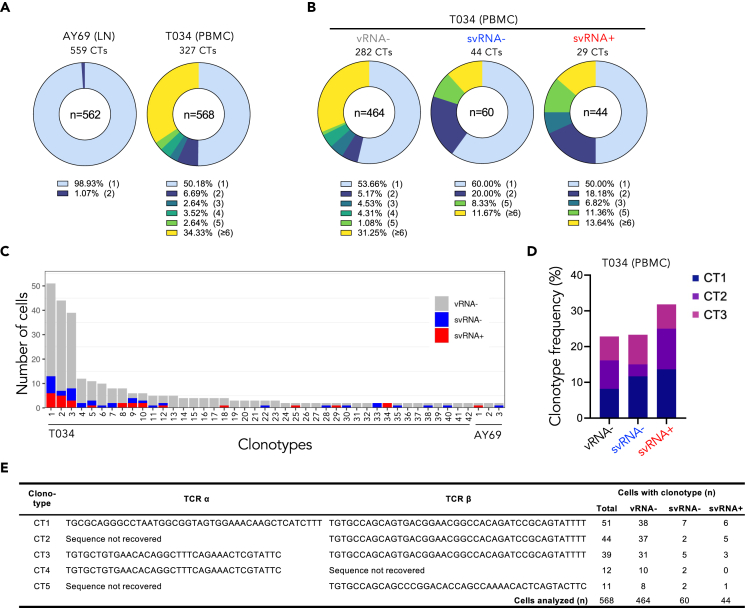
SIV-infected CD4 T cell clonal diversity
Reservoirs of HIV-1-infected memory CD4 T cells are maintained in PWH on antiretroviral therapy by multiple mechanisms, including antigen-driven proliferation, general immune activation, homeostatic proliferation, and provirus-driven expansion.58,59,60,61,62 Early provirus clonal expansion of SIV-infected cells has been reported in untreated acute infection in macaques by integration site analysis, with evidence of one early clone persisting for at least one year on ART.63 To build on these findings, we investigated the extent to which expanded T cell clones contribute to the pool of infected cells during early acute infection by T cell receptor (TCR) repertoire analysis. TCR clonotyping was performed by paired TCRα and TCRβ CDR3 nucleotide analysis of the scRNAseq memory CD4 T cell data. TCR sequence information was obtained for at least TCRα or TCRβ for a total of 562 and 568 cells in animals AY69 (LN) and T034 (PBMC), respectively; the TCR clonotypes comprised 559 and 327 unique sequences (Figure 5A). In both samples, unique clonotypes (present in one cell only) predominated: representing 99% and 50% of cells in the LN and PBMC specimens. Clonal expansion was more apparent in T034 (21 days p.i.), with 34% of cells among highly frequent clonotypes (≥6 cells) and the remaining 15% comprised smaller expanded clones (2–5 cells). These data indicate extensive T cell expansion in one of two animals during acute SIV infection, with a substantial proportion of memory CD4 T cells among highly expanded clones.
Figure 5.
Expansion of T cell receptor clonotypes in memory CD4 T cells during acute SIV infection
(A) TCR clonotype analysis of two macaques infected with SIV is shown for lymph node (animal AY69) and PBMC (T034) specimens. Pie charts depict the proportion of cells with unique (light blue) or shared (other colors) clonotypes (CTs). Other colors represent cells with clonotypes shared by 2, 3, 4, or 5 cells or by 6 or more cells. The total number of cells analyzed is indicated in center of pie chart and percentage at bottom represents the proportion of cells in each slice.
(B) Shared clonotype distribution among T cells separated by vRNA status for animal T034 PBMC.
(C) Bar graph depicts clonotypes occurring in >1 cell in T034 or AY69 and clonotype overlap among cells in the different vRNA cell groupings. Unique clonotypes are indicated along the x axis for each animal (1–42 for T034; 1–3 for AY69).
(D) Frequency of cells with the three most abundant clonotypes (CT) observed within each vRNA-defined group of animal T034.
(E) TCR nucleotide sequence and prevalence of most common clonotypes in animal T034. NA, not available due to lack of sequence obtained.
To determine whether expanded clones harbor vRNA and are enriched for viral transcription, we assessed TCR clonality among vRNA+ and vRNA- cells in animal T034, where clonal expansion was prevalent. Unique clones were the most frequent in each T cell subset regardless of vRNA status, ranging from 50 to 60% of cells (Figure 5B). We examined TCR sharing by comparing the predominant TCR sequences among the vRNA+ and vRNA- cells. Consistent with the limited clonal expansion observed in AY69, clonotype sharing was minimal among cells across the vRNA groupings (Figure 5C). In T034, by contrast, TCR clones were shared among cells in each of the three vRNA groups. Analysis of the three most abundant clonotypes revealed identical clonotypes present at similar cell frequencies among vRNA-, vRNA+svRNA- and vRNA+svRNA+ cells (Figures 5D and 5E). Of note, these three clonotypes combined comprised approximately 20–30% of each cell group, and shared the same TCRβ sequence, indicating a widely used TCRβ sequence in this animal’s TCR repertoire. Furthermore, the size of the two largest clonotypes may be an underestimate, as the TCRα chain sequence was undetermined for a substantial number of cells with this common TCRβ. Predominance of the same shared clonotypes across all three cell groups distinguished by vRNA status indicates that the same large clones were present among cells independent of their vRNA status, suggesting limited if any infection bias toward specific clonotypes.
Differential FOS expression in HIV-1 latently infected T cells from PWH
To further explore the relationship between FOS expression and virus transcription in vivo, we reanalyzed public transcriptome data from HIV-1 DNA+ memory CD4 T cells obtained from people receiving long-term antiretroviral therapy (ART) initiated during chronic infection. In a previous study,40 HIV DNA+ cells from these individuals showed transcriptomic signatures of HIV silencing, indicating quiescence of infection enforced by distinctive host gene expression patterns in some of these cells. These cells were found to harbor minimal HIV-1 RNA (<0.05%), consistent with latent infection or defective HIV-1 provirus. In a DEG analysis, we found that FOS and JUN, which encodes c-Jun, another major component of AP-1, were elevated in HIV DNA+ memory infected T cells compared to uninfected cells (2- and 3-fold, respectively; n = 72–300 HIV DNA+ cells per person; Figure 6A). In addition, genes with known activating or inhibitory relationships with FOS were differentially expressed in a manner consistent with increased AP-1 activity (p = 1.12E-02, Z score 2.164; Figures 6B and S5), providing further evidence of elevated FOS activity in HIV DNA+ cells. These findings suggest that increased expression and transcriptional activity of FOS and JUN in infected cells from PWH might be consistent with virus latency, rather than active virus expression.
Figure 6.
AP-1 activity in latently HIV-1 infected T cells and c-Fos inhibition in vitro
(A) RNA sequencing of HIV DNA+ and DNA-memory CD4 T cells from PBMC of PWH on suppressive ART (n = 3). FOS and JUN mRNA expression levels are depicted as counts per million (CPM). Each dot represents a 100-cell aliquot. Red bars depict median values. FDR p value for Wald’s test between two groups is reported.
(B) c-Fos target genes with known relationships to c-Fos activity and differential expression in HIV DNA+ compared to HIV DNA-cells. Genes predicted to be up- and down-regulated by c-Fos are indicated at top (orange bar) and bottom (green bar), respectively.
(C) c-Fos inhibition during infection increases HIV-1 reactivation. Human primary CD4 T cells (n = 9 donors) were cultured with or without T-5224 for 3 days during NL4-3 infection (left; d10-13, culture days 10–13), followed by 4 days of ART and then anti-CD3/CD28 stimulation (right, culture day 17). HIV-1 expression is plotted as p24+ (CD4−) cell frequency. Bars depict mean values. Wilcoxon matched-pairs signed rank test p value is indicated.
(D) c-Fos inhibition during latency reactivation in J-Lat cells stimulated with either PMA or TNFα following pre-treatment with either DMSO (vehicle) or T-5224 at the indicated concentration. HIV-1 reactivation was measured by flow cytometry as the frequency of HIV-expressing cells (GFP+, left) or the median GFP fluorescence (MFI, right). Data are represented as mean ± SD.
(E) c-Fos inhibition during latency reactivation in primary memory CD4 T cells stimulated with anti-CD3/CD28 (n = 7 donors; d17, culture day 17).
c-Fos inhibition during HIV-1 infection increases susceptibility to reactivation
Since FOS expression was diminished in productive T cell infection (SIV) and upregulated in latent infection (HIV-1), we sought to directly test the hypothesis that c-Fos participates in latency establishment and maintenance. T-5224 is a c-Fos-specific small molecule AP-1 inhibitor that targets DNA-binding activity.64 Human CD4 T cells were cultured with T-5224 during in vitro HIV-1NL4-3 infection in a primary cell model of latency reactivation using replication-competent virus.65 After inoculation and infection spreading, the frequency of infected cells did not differ between T-5224-treated and untreated cultures, indicating that c-Fos inhibition does not alter productive HIV-1 infection efficiency in activated CD4 T cells (Figure 6C). However, upon HIV-1 reactivation by anti-CD3/CD28 stimulation following four days of latency establishment, T-5224 pre-treatment increased reactivation by an average of 23% relative to untreated cells (p = 0.03). The small sample size limited statistical power to identify potential sex effects on T-5224 treatment susceptibility, though stratification by sex revealed stronger trends in females (p = 0.12; n = 4) than males (p = 0.44, n = 5). No drug toxicity was observed, with comparable cell viability across conditions. These data suggest that inhibiting c-Fos at the time of HIV-1 infection increases cell susceptibility to viral reactivation, supporting a role for c-Fos/AP-1 in establishing and maintaining latency.
AP-1 has been extensively characterized for its ability to activate HIV-1 transcription,66,67 a property that seems at odds with promoting latency. We therefore sought to determine whether c-Fos inhibition at the time of latency reversal would have the opposite effect. Previously, AP-1 was implicated in HIV-1 latency reversal by synergizing with NF-κB upon TCR stimulation in Jurkat cells using a MAPK inhibitor that blocked c-Fos nuclear localization.68 To determine whether c-Fos/AP-1 exerts an activating function in the setting of HIV-1 reactivation, we evaluated latency reversal in the presence of T-5224 using the J-Lat latent HIV-1 cell line model.69 T-5224 was added to cells just prior to stimulation with either PMA or TNFα. c-Fos inhibition decreased HIV-1 reactivation, as assessed by the frequency of GFP+ cells, in a dose-dependent manner with both reactivating agents (Figure 6D). HIV-1 transcription per cell (GFP fluorescence intensity) was also reduced by T-5224. Again, no toxicity was observed (Figure S6). Similar experiments in the primary cell model described above did not demonstrate HIV-1 reactivation inhibition by T-5224 (Figure 6E), which may be due to the crucial role of NFAT rather than NF-κB in HIV-1 reactivation in primary memory CD4 T cells.70,71 Taken together, these data provide direct evidence that c-Fos/AP-1 participates in HIV-1 reactivation from latency in some contexts and indicate a dueling function in the early stages of the viral life cycle by which c-Fos promotes latency (Figure S7).
Gene co-expression networks differentially expressed in SIV-infected cells
Since few genes were identified by DEG analysis of SIV RNA+ using our statistical significance criteria, we sought exploratory analyses to identify larger groups of genes that might differ between cell types but for which individual genes lacked independent statistical support. We performed weighted gene co-expression network analysis (WGCNA),72 which identifies networks of co-expressed genes. Using cells from the two RNA sequenced animals, five modules were generated after network construction (Figure 7A). Modules 4 and 5 were elevated in vRNA- cells relative to svRNA+ cells in both animals (Figure 7B). Enrichment analysis showed similar gene pathways represented in these two modules, including processes involved in viral infection, RNA metabolism, peptide metabolism, and ribonucleoprotein complex biogenesis (Figure 7C). Of note, FOS was the second highest ranked gene within module 5 for intramodular gene connectivity (Table S1), indicating a prominent role for FOS as a module 5 hub gene highly co-expressed with other module members. Further, greater expression of module 5 among vRNA- cells relative to svRNA+ cells is consistent with diminished FOS in svRNA+ cells (Figures 4B and 4C).
Figure 7.
Gene co-expression network analyses identifies that FOS belongs to a module strongly related to metabolism and viral infection
(A) Five modules (M) consisting of co-expressed genes identified from combined scRNA-seq data from animals AY69 and T034.
(B) Differential expression of network modules between vRNA- and svRNA+ cells in animal AY69 (top) and T034 (bottom) is depicted as log2 fold-change. Differences >0 indicate increased expression in vRNA- cells; <0 are elevated in svRNA+ cells. Significant differences in module expression are indicated (Padjusted <0.05); X, nonsignificant. Circle size reflects the number of genes in each module.
(C) Gene ontology of pathways enriched in each module.
Expression of two modules, 1 and 2, were increased in svRNA+ cells relative to vRNA- cells in AY69 and T034, respectively. Numerous infection and antiviral pathways were represented in module 1, including genes involved in HIV-1 infection, interferon responses, DNA damage and chromatin remodeling, as well as the module 4 and 5 enriched pathways. Among the top hub genes in module 1 were interferon-induced protein family members, IFITM1, IFIT1, IFIT2, IFIT3, MX1 and RSAD2, and ribosomal proteins, such as RPS18, RPLP1, RPL31, RPL18A, and RPL21. While module 1 was not elevated in svRNA+ cells of animal T034, lower cell numbers limited statistical power. Module 2 comprised a subset of the pathways found in module 1. These data indicate svRNA+ cells exhibit increased expression of multiple pathways associated with pathogen infection, interferon and cytokine antiviral responses, nucleic acid metabolism, and proteolytic processes, while networks enriched among vRNA- cells were limited to a discrete subset of these pathways. These profiles may reflect distinct processes triggered by active viral replication within svRNA+ cells versus more generalized systemic pro-inflammatory innate responses among vRNA- cells caused by acute SIV infection. Taken together our data show differential expression of FOS among infected cells and implicate AP-1/c-Fos in the regulation of active and latent infection in vivo, highlighting the importance of cellular factors as potential targets for therapeutic intervention.
Discussion
Defining the events that contribute to the establishment and persistence of infected CD4 T cells able to support HIV-1 replication remain key targets in HIV-1 prevention and cure strategies. We phenotyped over 2,000 memory CD4 T cells from three acutely SIV-infected macaques directly ex vivo, i.e., without any in vitro stimulation, by single-cell transcriptomic and cell surface protein expression, identifying ∼800 vRNA+ cells. Well-powered analyses of viral and host factors distinguished vRNA+ cell subsets from one another and from their vRNA- counterparts, leading to several key findings. First, vRNA+ cells exhibited a striking bimodal vRNA content distribution, which coincided with svRNA expression, supporting vRNA+ cell classification into cells either actively expressing vRNA and proteins or cells lacking evidence of viral replication. Second, CD4 T cell clonal expansion was observed in one of two animals and across vRNA+ and -negative cell groupings, supporting early T cell expansion during acute infection as a mechanism of infected cell oligoclonality. Third, host factors discriminated svRNA+ cells, including decreased expression of FOS, a component of the AP-1 transcription factor. Further investigation of AP-1 activity in the setting of HIV-1 revealed upregulation of FOS and JUN in HIV-1 DNA+ CD4 T cells from PWH on ART, while inhibiting c-Fos activity during HIV-1 infection increased TCR-induced viral reactivation from latency. These results reveal a new mechanism that may determine cell fate with respect to establishment of latent versus productive infection and identify c-Fos/AP-1 as an important modulator of viral transcription (Figure S7).
The identification of two distinct subsets of cells harboring vRNA in primary cells likely reflects different stages of the viral life cycle. The svRNA- group harbored ∼100-fold lower total and unspliced vRNA without evidence of major spliced transcripts. This population is therefore consistent with early, abortive, or latent stages of infection. Future analyses incorporating measurement of integrated provirus are warranted. Given the dynamic nature of acute infection with abundant free virions and frequent cell-virion interactions, unspliced vRNA in these cells may represent virion-derived genomic mRNA rather than de novo transcription. The low vRNA content may also reflect early transcription events with inefficient elongation or multiple splicing, as observed in latently infected cells.73 Of note, we cannot exclude the possibility that cells lacking vRNA are also infected, harboring integrated proviral DNA that is not transcribed.
The transcriptionally active subset was characterized by the presence of svRNA, high levels of unspliced vRNA, >1% of the cellular transcriptome devoted to SIV (median 8–10%), and surface CD4 and CD3 downregulation, indicating robust viral transcription and viral protein synthesis.30,46,74,75 In cells with the largest viral burden, SIV reads constituted a remarkable 30–40% of mRNA while total mRNA per cell was unchanged, implicating substantial replacement of host-cell mRNA with viral mRNA. Previous studies of synchronized, bulk HIV-infected cell lines obtained similar values, reporting 18%, 30% and 38% viral transcripts with increasing time post-infection.49,50 The heterogeneity in vRNA content observed in the svRNA+ subset may therefore reflect cells spanning early to later stages of productive infection. The similar extent to which viral transcription usurps host-cell transcription in these in vitro model systems indicates that SIV infection in rhesus cells recapitulates aspects of HIV-1 infection in human cells. vRNA content heterogeneity may also stem from variation in cellular activation status, as activated CD4 T cells produce more virus than resting cells.76 While a subset of cells displaying an activated phenotype (CD69+) was enriched among svRNA+ in the AY69 LN specimen, many svRNA- cells expressed similar levels of these canonical markers and only ∼50% of svRNA+ cells exhibited this phenotype. Thus, we deem activation status as unlikely to be the primary determinant of cell infection fate with respect to progressing to active infection. Discordant infected-cell phenotypes between the AY69 and T034 specimens may be due to either differences in the duration of infection or the compartment, both of which influence infected cell dynamics during acute HIV-1 infection in people.41
Homeostatic proliferation of infected T cells is one mechanism believed to contribute to maintenance of HIV-1 reservoirs in PWH on ART.77 Our TCR repertoire analysis showed T cell expansion in one of two animals studied. Again, this may reflect duration of infection, with longer infection (21 versus 10 days) allowing more time for clonal expansion. Alternatively, peripheral blood and lymph node may exhibit varying T cell clonal diversity, with blood characterized by a recirculating representation of cells stemming from all secondary lymphoid tissue and thus more oligoclonal. However, greater infected cell clonal expansion in blood versus lymph node was not observed in people with acute HIV-1.41 The trend of less TCR clonal diversity among infected cells may be due to preferential infection of proliferating T cell clones responding to either viral antigen or innate inflammatory cytokines. Shared TCRs among vRNA+ and vRNA- T cells suggests that clonal expansion is independent of cellular infection status during acute infection. Analysis of the specificity of the expanded clones among vRNA+ cells will elucidate potential mechanisms driving enrichment of T cell clones among the infected pool.
The AP-1 heterodimer of c-Fos and c-Jun regulates chromatin remodeling induced by co-stimulation during T cell activation and has been implicated primarily in HIV-1 transcriptional activation.78 SIV and HIV-1 share many promoter elements, including one AP-1 binding site in the enhancer region of the LTR U3 (−105 to −78)79 and three AP-1 binding sites downstream of the transcriptional start site in the R/U5 region. Several lines of evidence support an activating role for the latter AP-1 sites.80 This includes substantially decreased HIV-1 expression upon deletion of these sites and increased LTR transactivation upon c-Fos and c-Jun overexpression.66,67,81,82,83 c-Fos nuclear localization is also temporally associated with HIV-1 reactivation by TCR stimulation in latently infected cells.68
Our finding of diminished FOS among svRNA+ CD4 T cells and increased FOS and JUN among latently infected cells in PWH suggests that AP-1 can also repress viral transcription. These data are consistent with less appreciated evidence that AP-1 negatively regulates HIV-1 transcription in some contexts. There is precedent for dueling regulatory activities by AP-1, as AP-1 is well-documented to exert contrasting functions in cancer settings, acting as either an activator or a repressor of transcription depending on the components of the AP-1 dimer or other context-dependent variables.84,85 In prior HIV-1 studies, while c-Jun overexpression enhanced Tat-mediated HIV-1 transcription, it inhibited basal HIV-1 LTR promoter activity in the absence of Tat.83 Further, the AP-1 motif in the U3 enhancer element was required for latency establishment in a cell line model.86 Of note, among the AP-1 factors expressed in T cells, c-Fos was the most active as well as the predominant factor bound to the U3 AP-1 site in primary human CD4 T cells, followed by c-Jun. Reduced FOS expression in svRNA+ infected cells reported here strongly suggests c-Fos participates in viral transcription suppression upon infection. Conversely, increased FOS in transcriptionally quiescent HIV-1 DNA+ cells indicates that persistent elevated AP-1 may actively maintain transcriptional repression. In vitro experiments inhibiting c-Fos/AP-1 activity also indicated opposing roles of c-Fos dependent on the stage of infection: blockade during HIV-1 infection increased T cell susceptibility to subsequent reactivation, while blockade at the time of latency reversal inhibited HIV-1 reactivation. Taken together, these data support a model of HIV-1/SIV latency through LTR suppression determined by AP-1 levels (or composition) within a cell at the time of infection, whereby c-Fos/AP-1 restricts productive infection during initial viral exposure through promoting latency establishment and its maintenance (Figure S7). This function appears to be distinct from AP-1-mediated HIV-1 transcription activation upon subsequent TCR stimulation reactivation in cells with established latent infection. Other mechanisms of latency, including epigenetic transcriptional silencing by a restrictive chromatin environment,87,88,89,90,91 are not excluded by this model.
Several prior studies interrogating the transcriptome of HIV-1-infected cells identified FOS and AP-1-related genes among differentially expressed host factors. First, FOS or AP-1 activity was elevated in in vitro infected CD4 T cells supporting active viral replication relative to uninfected cells or relative to latently infected cells.43,44,92 Of note, these in vitro model systems employed TCR stimulation to activate T cells, which is known to induce AP-1 and therefore generates a transcription factor profile distinct from the unstimulated, direct ex vivo analysis performed here. In this context, AP-1 would most likely exert its HIV-1 activating function.68 Second, blood cells from PWH on suppressive ART identified FOS downregulation among a subset of memory CD4 T cells with reduced susceptibility to HIV-1 reactivation,93 though this cell population was not enriched for HIV-1 DNA or RNA. And third, ATACseq epigenetic analysis of infected memory CD4 T cells from PWH indicated increased accessibility of AP-1-related motifs, including FOS and JUN family members, in HIV-1 DNA+ cells compared to their DNA- counterparts.38 Here, we provide direct evidence of elevated FOS and JUN transcription in HIV-1 DNA+ cells from ART-suppressed PWH.
Future studies evaluating AP-1 expression levels in HIV-1-infected cells from PWH in untreated acute infection will extend our understanding of the role AP-1 plays in establishing HIV-1 latent infection in CD4 T cells in vivo. Non-memory CD4 T cell reservoir populations governed by different mechanisms of latency also warrant investigation.94,95,96 Similarly, transcriptomic analyses of HIV-1 mRNA quantity and splicing with single-cell resolution will be important to corroborate and define HIV-1-infected cell life cycle stages that occur in vivo, including identification of additional host factors that modulate HIV-1 transcription. Leveraging host regulatory factors such as AP-1 to prevent HIV-1 latency establishment and reservoir maintenance represents a potential therapeutic approach to limit the size and persistence of long-lived infected CD4 T cells.
Limitations of the study
The sample size of our study included single-cell gene expression analysis of three rhesus macaques (n = 1960 CD4 T cells) and bulk sequencing analysis for three PWH (n = 72–300 cells per person). Future studies confirming our findings in larger datasets will be valuable. An additional caveat to our study is that the HIV-1 DNA+ cells interrogated from PWH cannot be definitively classified as latently infected. Though these cells harbored HIV-1 read percentages 100-fold less than that reported for HIV-1-expressing cells stimulated in vitro,36,40 limited HIV-1 RNA content may be due to proviral defects. We are unable to comment on the influence of sex on the gene expression analyses as all infected macaques and PWH were male. While in vitro c-Fos inhibition experiments included PBMC from both sexes, larger studies are required to determine potential sex effects on these results.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD4-BV786 | BD Biosciences | Cat#563914, clone L200, lot 8099675; RRID: AB_2738485 |
| CD8-BUV496 | BD Biosciences | Cat#564804, clone RPA-T8, lot 7193592; RRID: AB_2744460 |
| CD3-BV650 | BD Biosciences | Cat#563918, clone SP34-2, lot 7018763; RRID: AB_2738487 |
| CD95-BUV737 | BD Biosciences | Cat#564710, clone DX2, lot 8072740; RRID: AB_2738907 |
| CD28-BV711 | BioLegend | Cat#302948, clone CD28.2, lot 236771; RRID: AB_2616857 |
| CD14-BV510 | BioLegend | Cat#301842, clone M5E2, lot 237626; RRID: AB_2561379 |
| CD16-BV510 | BioLegend | Cat# 302048, clone 3G8, lot 238638; RRID: AB_2561380 |
| CD20-BV510 | BioLegend | Cat#302340, clone 2H7, lot 244953; RRID: AB_2561941 |
| CXCR3-PE-Cy5 | BD Biosciences | Cat#551128, clone 1C6, lot 7040790; RRID: AB_394061 |
| CD38-PE | NHP Reagent Resource | clone OKT10, lot 060816CK |
| CD69-BUV395 | BD Biosciences | Cat#564364, clone FN50, lot 7108931; RRID: AB_2738770 |
| ICOS-Alex Fluor 700 | BioLegend | Cat#313531, clone C398.4A, lot 227702; RRID: AB_2566130 |
| HLA-DR-APC-H7 | BD Biosciences | Cat#641393, clone L243, lot 9051562; RRID: AB_1645739 |
| HLA-ABC | BioLegend | Cat#311431, clone W6/32, lot 256257; RRID: AB_2566150 |
| TIGIT-PerCP-eFluor 710 | eBioscience | Cat#S469500-41, clone MBSA43, lot 4290851 |
| CXCR5-FITC | ThermoFisher | Cat#11-9185-42, clone MU5UBEE, lot 4325074; RRID: AB_2572526 |
| PD-1-PE-eFluor 610 | eBioscience | Cat#61279942, clone eBioJ105, lot 4323887; RRID: AB_2574598 |
| Integrin β7-BV421 | BD Biosciences | Cat#564283, clone FIB 504, lot 6270954; RRID: AB_2738728 |
| CCR6-PE-Cy7 | BD Biosciences | Cat#564659, clone 11A9, lot 8152573; RRID: AB_2738881 |
| IFITM1 | AB Biosciences | Cat#224063, rabbit polyclonal, lot GR110366-1 |
| Goat-anti-rabbit-APC | Jackson Immuno Research | Cat# 111-136-144; RRID:AB_233798, lot 126640 |
| Aqua Live/Dead stain | ThermoFisher | Cat#L34957, lot 1596087 |
| Bacterial and virus strains | ||
| SIVmac251 | NIH HIV Reagent Program | Cat#ARP-253 |
| SIVmac239X | Del Prete et al.97 | GenBank M33262.1 |
| Biological samples | ||
| Peripheral blood of people without HIV-1 | Homo sapiens | |
| Lymph Node Mononuclear Cell | Macaca mulatta | |
| Chemicals, peptides, and recombinant proteins | ||
| Raltegravir | NIH HIV Reagent Program | Cat#HRP-11680 |
| Nelfinavir | NIH HIV Reagent Program | Cat#ARP-4621 |
| T-5224 | APExBIO Technology | Cat#B4664 |
| CD3/CD28 Dynabeads™ | ThermoFisher Scientific | Cat#11141D |
| TNF-α | Peprotech | Cat# 300-01A |
| DMSO | Sigma | D5879 |
| Phorbol-myristate-acetate (PMA) | Invitrogen | J63916.MCR |
| Critical commercial assays | ||
| SMART-Seq Ultra Low Input RNA kit | Takara | Cat# 634891 |
| CellsDirect One-Step qRT-PCR kit | ThermoFisher Scientific | Cat#11753100 |
| EasySep™ Human Naive CD4+ T cell Isolation Kit | Stem Cell Tech | Cat#19555RF 19555 |
| Deposited data | ||
| Raw scRNA-seq data from SIV-infected m. mulatta | This paper | GEO: GSE232998 |
| RNA sequencing data from people with HIV-1 on suppressive ART | Clark et al.40 | dbGaP; phs003095.v1.p1 |
| Experimental models: Cell lines | ||
| J-Lat 10.6 cells | NIH HIV Reagent Program | Cat#ARP-9849 |
| Experimental models: Organisms/strains | ||
| Macaca mulatta | ||
| Homo sapiens | ||
| Oligonucleotides | ||
| FOS TaqMan assay | ThermoFisher | Hs01119267_g1 |
| SIV gag primers and probe | Bolton et al.46 | N/A |
| SIV D1-A5 splice junction primers and probe | Bolton et al.46 | N/A |
| SIV D4-A7 splice junction primers and probe | Bolton et al.46 | N/A |
| Software and algorithms | ||
| GraphPad Prism (v.8) and (v.9.3.1) | N/A | https://www.graphpad.com/features |
| JMP (v.12) | N/A | https://www.jmp.com/en_us/home.html |
| FlowJo (v10.8.1) | N/A | https://www.flowjo.com/ |
| Python | N/A | https://www.python.org/ |
| R Bioconductor package (v3.6) | McCarthy et al.98 | https://smorabit.github.io/hdWGCNA/ |
| Seurat packages (v3.1.5) | N/A | https://satijalab.org/seurat/ |
| HISAT2 (v2.1.0) | Kim et al.99 | Nat Biotechnol 37, 907–915. https://doi.org/10.1038/s41587-019-0201-4 |
| htseq-count (v0.9.1) | Anders et al.100 | Bioinformatics 31, 166–169. https://doi.org/10.1093/bioinformatics/btu638 |
| BLAT (v36 × 1) | Kent et al.101 | Genome Res 12, 656664. https://doi.org/10.1101/gr.229202 |
| Harmony | Korsunsky et al.102 | https://portals.broadinstitute.org/harmony/articles/quickstart.html |
| Metascape | Zhou et al.103 | Nat Commun 10, 1523. https://doi.org/10.1038/s41467-019-09234-6 |
| Trimmomatic (v0.36) | Bolger et al.104 | Bioinformatics 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 |
| scater/scran (v1.12.2) | McCarthy et al.98 | https://bioconductor.org/packages/release/bioc/html/scater.html |
| MixCR (v2.1.5) | Bolotin et al.105 | Nat Methods 12, 380–381. https://doi.org/10.1038/nmeth.3364 |
| scRepertoire (v1.3.2) | Borcherding et al.106 | F1000Res9, 47. https://doi.org/10.12688/f1000research.22139.2 |
| circlize (v0.4.13) | Gu et al.107 | Bioinformatics 30, 2811–2812. https://doi.org/10.1093/bioinformatics/btu393 |
| FIND-seq | Clark et al.108 | https://www.protocols.io/view/find-seq-protocol-v1-0-cez3tf8n.html |
| CLC Genomics Workbench (v. 21) | Qiagen | Cat#832021 |
| Ingenuity Pathway Analysis | Qiagen | summer release 2021 |
| R hdWGCNA package | Morabito et al.109 | https://cran.r-project.org/web/packages/WGCNA/index.html#:∼:text=WGCNA%3A%20Weighted%20Correlation%20Network%20Analysis&text=Includes%20functions%20for%20rudimentary%20data,for%20data%20manipulation%20and%20visualization. |
| Other | ||
| Macaca mulatta genome | reference assembly 8.0.1 | N/A |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Diane Bolton (DBolton@hivresearch.org).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Animals and SIV infection
All work involving research animals was conducted under NIH Institute & Center Animal Care & Use Committee (ACUC)-approved animal use protocols of the NIH National Cancer Institute or the NIH Vaccine Research Center in AAALAC International-accredited facilities with a Public Health Services Animal Welfare Assurance and in compliance with the Animal Welfare Act and other federal statutes and regulations relating to laboratory animals (Animal Welfare Assurance number D16-00602). Macaques were housed at the NIH-Bethesda or NIH-Poolesville facilities and cared for in accordance with the NIH “Guide for the Care and Use of Animals” (National Research Council; 2011; National Academies Press; Washington, D.C.). Prior to study initiation, all animals were free of cercopithecine herpesvirus 1, SIV, simian type-D retrovirus, and simian T lymphotropic virus type 1.
Three colony-bred Indian-origin male rhesus macaques (age 3-10 years old) infected with SIV were analyzed in this study (Table 1). AY69 was inoculated intravenously with 100 MID50 of SIVmac251 (VRC animal protocol 356) with a virus preparation of 1.0 ml via the saphenous vein. T034 was inoculated intrarectally with 300 IU of SIVmac239X97 (ACUC protocol AVP-051); 12C174 was intravenously inoculated daily for 5 consecutive days with single SIVmac239X clonotypes, 20 IU each (ACUC protocol AVP-059). Infection duration ranged from 10-21days. LNMC were prepared by mincing with scissors followed by grinding through a 70 μm cell strainer. Whole blood was collected by venipuncture into EDTA Vacutainer tubes (BD) and PBMC were isolated by Ficoll-Paque density gradient separation. Blood and lymph biopsies were collected from sedated animals. People with HIV-1.
Human transcriptome data was generated from a previously reported study examining latently infected cells from three male PWH aged 57-60 years old under long-term suppressive ART.40 Participants were White/European American (n=2) and Black/African American (n=1). The San Francisco General Hospital IRB approved study participant recruitment (SCOPE protocol, NCT00187512).
Cell lines and primary cell cultures
The latent HIV-1 infected human T cell line J-Lat 10.669 and primary human CD4 T cells were cultured in RPMI 1640 with 10% FBS, 1% penicillin-streptomycin and 1% L-glutamine (Invitrogen) and maintained at 37°C under 5% CO2. J-Lat cells were obtained from the NIH HIV Reagent Program and a parallel vial tested negative for mycobacterium negative. Cells were not authenticated. PBMC were from nine people without HIV-1 (Gulf Coast Regional Blood Center); n=4 females and n=5 males, aged 36-67.
Method details
Flow cytometry and cell sorting
Cells were thawed and stained with fluorescent labeled monoclonal antibodies specified in the key resources table. Cells were first incubated with Aqua Live/Dead stain and anti-IFITM1 antibody, then washed and stained with a cocktail of all the remaining antibodies including secondary APC-labeled antibody for IFITM1 detection. Immediately after staining, memory CD4 T cells were isolated by single-cell sorting on a BD FACSAriaTM III SORP, such that one cell was deposited per well in 96-well LowBind plates (Framestar clear 96 semi-skirted PCR plate, 4titute®Ltd) for lysis and cDNA library prep. To minimize impact on cellular RNA content and protein expression prior to analysis, cell samples were maintained on ice at all times, with the exception of the surface stain (15 min) and elapsed sorting time (∼30 min). All samples were cryopreserved, which does not impact viral gene expression.46 Surface staining was recorded for each cell by well position. vRNA- cells lacking surface CD4 expression (CD4 fluorescence intensity <500) were excluded from downstream analyses of differential host protein expression to limit comparisons to cell populations most likely to represent CD4 T cells; this cut-off is consistent with undetected CD4 and CD40L mRNA by scRT-qPCR among vRNA- but not vRNA+ cells.46 For display purposes, fluorescence intensity values were transformed for each marker by adding the minimum value to achieve positive values. Multivariate flow cytometry data were analyzed using FlowJo (v10.8.1) and visualized using uniform manifold approximation and projection (UMAP)-dimensionality reduction on concatenated CD4 T cells combined from all animals. Concatenated cells were then further grouped using FlowSOM to create a self-organizing map for cluster based on surface protein marker expression.57
Single-cell RNA-seq and data analysis
Single cells were lysed in SMART-Seq lysis buffer supplemented with RNAse inhibitor for 2 minutes at room temperature, vortexed, centrifuged, and frozen at -80°C until cDNA library preparation. Whole cDNA library was prepared using the SMART-Seq Ultra Low Input RNA kit and sequenced on the Illumina NovaSeq Platform as described previously.110,111,112 Sequencing reads were trimmed with Trimmomatic (v0.36)104 and aligned to the Macaca mulatta genome (reference assembly 8.0.1) combined with either the SIVmac251 (GenBank KU892415.1) or SIVmac239 (GenBank M33262.1) genome for AY69 or T034, respectively, using HISAT2 (v2.1.0).99 Global gene expression was quantified using htseq-count (v0.9.1).100 Spliced and unspliced viral mRNA were examined using an in-house pipeline that included BLAT (v36 × 1) and custom Python scripts.101 Classification of cells was carried out based on presence of vRNA and svRNA (Figure S2). Cells lacking SIV reads were classified vRNA-. vRNA+ cells were considering transcriptionally active if they met at least one of the following two criteria: 1) positive for svRNA encoding both tat/rev (reads mapping to D4-A7 splice junction) and env (read mapping to D1-A5 splice junction); or 2) vRNA exceeding 1% of total mRNA and positive for at least one of the above svRNA species. Cells with SIV reads <1% lacking svRNA reads were classified as replication-inactive. Cells with SIV reads mapping to only one svRNA species and with <1% total SIV mRNA were excluded due to ambiguous infection status, as were cells with SIV reads higher than 1% but no svRNA. Quality control and normalization was performed using the scater/scran (v1.12.2) and Seurat packages (v3.1.5) in the R Bioconductor package (v3.6).98,113 Cells were excluded from analysis if they met one or more of the following exclusion criteria: high ERCC percentage, low library size, or low total features. To exclude non-CD4 T cells, principal component analysis was performed on the entirety of sorted single cells and revealed distinct clusters. A putative B cell population, characterized by absence of CD3D and high expression of CD19, JCHAIN, MEF2C, FCRL, MAMU-DRB1 (Figure S1B), was excluded from further analysis in animal AY69, with 593 cells remaining. For animal T034, small clusters of myeloid, NK, and B cells were identified and excluded, leaving 672 cells after filtering. An average of 2159 (st. dev. 499) genes were detected per cell in AY69, and 1520 (st. dev. 316) in T034. An average of 31,133 and 45,676 reads mapped to the SIV genome in AY69 and T034, respectively. Among detected genes, AY69 had an average of 801 reads per gene and T034 had 4130 reads per gene. DEG analysis between cell classifications was performed using a Wilcoxon Rank Sum test in Seurat using the Bonferroni adjustment for multiple testing. For animal T034, DEG and multiple testing correction was performed on only the subset of genes that were significant in animal AY69. RPS and RPL genes were filtered out after differential analyses. MixCR (v2.1.5), scRepertoire (v1.3.2), and circlize (v0.4.13) were used to generate, analyze, and visualize TCR sequences.105,106,107
qPCR analysis
Differential expression of selected genes was confirmed by qPCR, using cells that were single-cell sorted into 5.75 μl of the SMART-Seq lysis buffer, as described above. 3.8 μl of lysis buffer containing one sorted single cell was combined with 16.2 μl of the CellsDirect One-Step qRT-PCR kit (ThermoFirsher Scientific), containing TaqMan assays for host and SIV genes.46 cDNA was synthesized by 18 cycles of preamplification and served as template for high-throughput qPCR using the Gene expression 96.96 Dynamic array platform on the Fluidigm Biomark microfluidic chip platform as described previously.46 Absolute RNA copies were calculated as 2(Et-13), where Et=40-Ct. Cells with undetected FOS gene expression by RT-qPCR were assigned Et=13, which approximates a single RNA molecule using this protocol.51 Estimations of vRNA+ cell frequency by RT-qPCR were performed by flow cytometric sorting of limiting-cell dilutions and Poisson distribution.46 Results were analyzed using JMP (v.12) and GraphPad Prism (v.8).
Transcriptome analysis of HIV DNA+ cells
HIV DNA+ and HIV DNA- memory CD4 T cells were isolated using FIND-seq technology and their transcriptomes sequenced using Illumina’s HiSeq 4000 platform.40,108 Resulting data were analyzed using the CLC Genomics Workbench (v. 21). Sequencing read counts per million values for FOS and JUN were extracted and plotted using GraphPad Prism (v.9.3.1) and the fold change and FDR was obtained from differential gene expression between HIV DNA+ and HIV DNA- samples. Upstream regulator analysis was performed in Ingenuity Pathway Analysis (Qiagen, summer release 2021) using differentially expressed genes with an absolute fold change of ≥1.5 and FDR ≤ 0.05.
Latent HIV-1 in vitro infection and reactivation
Naïve CD4+ T cells were isolated via negative selection (EasySep™ Human Naïve CD4+ T Cell Isolation Kit) from PBMC of people without HIV-1 (n=9; n=4 female, n=5 male; 36-67 y.o., median 40 y.o.) and cultured in a latent HIV-1 in vitro infection model.65 Briefly, cells were activated for 72h, expanded for 96h, inoculated with HIV-1NL4-3 for 72h, then cultured with or without 10 μM c-Fos inhibitor T-5224 (APExBIO Technology) for 72h during the crowding phase (culture days 10-13). T-5224 was washed out and cells were removed from crowding conditions and cultured in the presence of ART (1 μM Raltegravir, 0.5 μM Nelfinavir; NIH AIDS Reagent Program) for 96h. Cells were then reactivated with CD3/CD28 Dynabeads™ (ThermoFisher Scientific; culture day 17) for 48h at a concentration of 1 bead per cell. Alternatively, cells were treated with T-5224 for 90 min just prior to reactivation CD3/CD28 Dynabeads™ (n=7; n=3 female, n=4 male; subset of people without HIV mentioned above). 3x105 J-Lat 10.6 cells69 were co-cultured with DMSO or T-5224 at 20 μM, 40 μM or 80 μM for 90min and then stimulated with 5 ng/mL phorbol-myristate-acetate or 10 ng/mL TNF-α for 12h. All cells were cultured in RPMI 1640 with 10% FBS, 1% penicillin-streptomycin and 1% L-glutamine (Invitrogen) and maintained at 37°C under 5% CO2.
Gene co-expression network analysis
Cells from animals AY69 and T034 were combined into one Seurat object for WGCNA.72 Gene expression was normalized and scaled to make a principal component analysis. Harmony was used to account for batch correction based on the source animal origin of the cells (AY69 versus T032).102 The R package hdWGCNA was used to generate metacells with original gene expression clusters from each animal on the Harmony reduction with nondefault parameters (k = 20, max_shared = 10, and min_cells = 40).109 A network was constructed using a soft power of 9. Module eigengenes generated for the resulting modules were also corrected for source animal using Harmony. Gene connectivity (or “module membership”) was quantified by correlating gene expression and module eigengenes (kME, or eigengene connectivity). Genes with large correlation coefficients (high kME) were considered to be highly connected “hub genes” within their respective modules. Wilcoxon rank sum test was used to compare the distribution of module eigengenes between the vRNA- and svRNA+ cell categories within each animal. The average difference between module eigengenes in the two groups is the difference between the means. The lists of genes from all modules were used as input for gene ontology enrichment analysis using Metascape.103
Quantification and statistical analysis
Differential expression of surface proteins was determined by nonparametric Wilcoxon comparisons between cell groups. DEG analysis of scRNA-seq data was assessed by Wilcoxon Rank Sum test in Seurat using the Bonferroni adjustment for multiple testing. scRT-qPCR FOS gene expression differences were analyzed by nonparametric Wilcoxon comparison. Differential FOS and JUN expression for HIV DNA+ cells was assessed by Wald’s test between two groups with FDR adjustment. Wilcoxon matched-pairs signed rank test was applied to in vitro culture experiments with and without c-Fos inhibitor. Additional statistical details of experiments can be found in the figure legends.
Additional resources
Human gene expression data analyzed in this study was previously generated by analysis of samples from participants in the SCOPE study (NCT00187512).
Acknowledgments
We thank Matthew Creegan (MHRP, HJF) for flow cytometry cell sorting; Michael Eller, and Hannah King (MHRP, HJF) for insightful scientific discussion; and Kathryn Foulds and Mario Roederer (Vaccine Research Center, NIAID, NIH) for generous contribution of macaque specimens. We appreciate data analysis input from Biju Isaac (formerly MHRP, HJF), Greg Finak (formerly Fred Hutchinson Cancer Research Center), and Brandon Keele (AIDS and Cancer Virus Program, Frederick National Laboratory). We also thank the Nonhuman Primate Research Support Core (AIDS and Cancer Virus Program) for specimen processing and inventory management. The views expressed are those of the authors and should not be construed to represent the positions of the US Army or the Department of Defense (DoD). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This work was supported by a cooperative agreement (W81XWH-18-2-0040) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the DoD and in part by federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. 75N91019D00024/HHSN261201500003I.
Author contributions
Conceptualization, A.T., R.T., and D.L.B.; Methodology, V.C.J., A.G., R.T., and D.L.B.; Formal Analysis, A.G., K.M., D.P.P., C.S., P.M., R.T., and D.L.B.; Investigation, V.C.J., A.T., P.E., S.D., N.H., and A.Z.; Resources, G.D.Q., R.T., and D.L.B.; Writing – Original Draft, V.C.J., A.T., and D.L.B; Writing – Reviewing and Editing, G.Q.D., E.A.B., A.B., R.T., and D.L.B.; Supervision, E.A.B., A.B., R.T., and D.L.B.
Declaration of interests
D.L.B. is an inventor on a related HIV-1 therapeutic patent (US Provisional Application 63/500,996 filed 9 May 2023). The other authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. We support inclusive, diverse, and equitable conduct of research.
Published: September 22, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108015.
Contributor Information
Rasmi Thomas, Email: rthomas@hivresearch.org.
Diane L. Bolton, Email: dbolton@hivresearch.org.
Supplemental information
Genes are listed in rank order of degree of connectivity with other genes within the module.
Data and code availability
-
•
Single-cell RNA-seq data have been deposited at GEO. Bulk human RNA-seq data were previously deposited at dbGaP. All data are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
References
- 1.Finzi D., Hermankova M., Pierson T., Carruth L.M., Buck C., Chaisson R.E., Quinn T.C., Chadwick K., Margolick J., Brookmeyer R., et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Colby D.J., Trautmann L., Pinyakorn S., Leyre L., Pagliuzza A., Kroon E., Rolland M., Takata H., Buranapraditkun S., Intasan J., et al. Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat. Med. 2018;24:923–926. doi: 10.1038/s41591-018-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitney J.B., Hill A.L., Sanisetty S., Penaloza-MacMaster P., Liu J., Shetty M., Parenteau L., Cabral C., Shields J., Blackmore S., et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014;512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archin N.M., Kirchherr J.L., Sung J.A., Clutton G., Sholtis K., Xu Y., Allard B., Stuelke E., Kashuba A.D., Kuruc J.D., et al. Interval dosing with the HDAC inhibitor vorinostat effectively reverses HIV latency. J. Clin. Invest. 2017;127:3126–3135. doi: 10.1172/JCI92684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archin N.M., Liberty A.L., Kashuba A.D., Choudhary S.K., Kuruc J.D., Crooks A.M., Parker D.C., Anderson E.M., Kearney M.F., Strain M.C., et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott J.H., McMahon J.H., Chang C.C., Lee S.A., Hartogensis W., Bumpus N., Savic R., Roney J., Hoh R., Solomon A., et al. Short-term administration of disulfiram for reversal of latent HIV infection: a phase 2 dose-escalation study. Lancet. HIV. 2015;2:e520–e529. doi: 10.1016/S2352-3018(15)00226-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen T.A., Tolstrup M., Brinkmann C.R., Olesen R., Erikstrup C., Solomon A., Winckelmann A., Palmer S., Dinarello C., Buzon M., et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet. HIV. 2014;1:e13–e21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- 8.Søgaard O.S., Graversen M.E., Leth S., Olesen R., Brinkmann C.R., Nissen S.K., Kjaer A.S., Schleimann M.H., Denton P.W., Hey-Cunningham W.J., et al. The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori L., Valente S.T. Key Players in HIV-1 Transcriptional Regulation: Targets for a Functional Cure. Viruses. 2020;12:529–564. doi: 10.3390/v12050529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruelas D.S., Greene W.C. An integrated overview of HIV-1 latency. Cell. 2013;155:519–529. doi: 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coiras M., López-Huertas M.R., Pérez-Olmeda M., Alcamí J. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat. Rev. Microbiol. 2009;7:798–812. doi: 10.1038/nrmicro2223. [DOI] [PubMed] [Google Scholar]
- 12.Emiliani S., Fischle W., Ott M., Van Lint C., Amella C.A., Verdin E. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 1998;72:1666–1670. doi: 10.1128/JVI.72.2.1666-1670.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emiliani S., Van Lint C., Fischle W., Paras P., Jr., Ott M., Brady J., Verdin E. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. USA. 1996;93:6377–6381. doi: 10.1073/pnas.93.13.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yukl S., Pillai S., Li P., Chang K., Pasutti W., Ahlgren C., Havlir D., Strain M., Günthard H., Richman D., et al. Latently-infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity. Virology. 2009;387:98–108. doi: 10.1016/j.virol.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaczmarek K., Morales A., Henderson A.J. T Cell Transcription Factors and Their Impact on HIV Expression. Virology. 2013;2013:41–47. doi: 10.4137/VRT.S12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez G., Zaikos T.D., Khan S.Z., Jacobi A.M., Behlke M.A., Zeichner S.L. Targeting IkappaB proteins for HIV latency activation: the role of individual IkappaB and NF-kappaB proteins. J. Virol. 2013;87:3966–3978. doi: 10.1128/JVI.03251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakre S., Chavez L., Shirakawa K., Verdin E. Epigenetic regulation of HIV latency. Curr. Opin. HIV AIDS. 2011;6:19–24. doi: 10.1097/COH.0b013e3283412384. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen K., Das B., Dobrowolski C., Karn J. Multiple Histone Lysine Methyltransferases Are Required for the Establishment and Maintenance of HIV-1 Latency. mBio. 2017;8 doi: 10.1128/mBio.00133-17. e00133–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Lint C., Bouchat S., Marcello A. HIV-1 transcription and latency: an update. Retrovirology. 2013;10:67. doi: 10.1186/1742-4690-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ne E., Palstra R.J., Mahmoudi T. Transcription: Insights From the HIV-1 Promoter. Int. Rev. Cell Mol. Biol. 2018;335:191–243. doi: 10.1016/bs.ircmb.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Bosque A., Nilson K.A., Macedo A.B., Spivak A.M., Archin N.M., Van Wagoner R.M., Martins L.J., Novis C.L., Szaniawski M.A., Ireland C.M., et al. Benzotriazoles Reactivate Latent HIV-1 through Inactivation of STAT5 SUMOylation. Cell Rep. 2017;18:1324–1334. doi: 10.1016/j.celrep.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira L.A., Bentley K., Peeters A., Churchill M.J., Deacon N.J. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giri M.S., Nebozhyn M., Showe L., Montaner L.J. Microarray data on gene modulation by HIV-1 in immune cells: 2000-2006. J. Leukoc. Biol. 2006;80:1031–1043. doi: 10.1189/jlb.0306157. [DOI] [PubMed] [Google Scholar]
- 24.Mehla R., Ayyavoo V. Gene array studies in HIV-1 infection. Curr. HIV AIDS Rep. 2012;9:34–43. doi: 10.1007/s11904-011-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rato S., Golumbeanu M., Telenti A., Ciuffi A. Exploring viral infection using single-cell sequencing. Virus Res. 2017;239:55–68. doi: 10.1016/j.virusres.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Ciuffi A., Rato S., Telenti A. Single-Cell Genomics for Virology. Viruses. 2016;8:123–133. doi: 10.3390/v8050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradley T., Ferrari G., Haynes B.F., Margolis D.M., Browne E.P. Single-Cell Analysis of Quiescent HIV Infection Reveals Host Transcriptional Profiles that Regulate Proviral Latency. Cell Rep. 2018;25:107–117.e3. doi: 10.1016/j.celrep.2018.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golumbeanu M., Desfarges S., Hernandez C., Quadroni M., Rato S., Mohammadi P., Telenti A., Beerenwinkel N., Ciuffi A. Proteo-Transcriptomic Dynamics of Cellular Response to HIV-1 Infection. Sci. Rep. 2019;9:213. doi: 10.1038/s41598-018-36135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darcis G., Van Driessche B., Van Lint C. HIV Latency: Should We Shock or Lock? Trends Immunol. 2017;38:217–228. doi: 10.1016/j.it.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds M.R., Piaskowski S.M., Weisgrau K.L., Weiler A.M., Friedrich T.C., Rakasz E.G. Ex vivo analysis of SIV-infected cells by flow cytometry. Cytometry A. 2010;77:1059–1066. doi: 10.1002/cyto.a.20960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grau-Expósito J., Serra-Peinado C., Miguel L., Navarro J., Curran A., Burgos J., Ocaña I., Ribera E., Torrella A., Planas B., et al. A Novel Single-Cell FISH-Flow Assay Identifies Effector Memory CD4(+) T cells as a Major Niche for HIV-1 Transcription in HIV-Infected Patients. mBio. 2017;8 doi: 10.1128/mBio.00876-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eriksson S., Graf E.H., Dahl V., Strain M.C., Yukl S.A., Lysenko E.S., Bosch R.J., Lai J., Chioma S., Emad F., et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasternak A.O., Lukashov V.V., Berkhout B. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology. 2013;10:41. doi: 10.1186/1742-4690-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fromentin R., Bakeman W., Lawani M.B., Khoury G., Hartogensis W., DaFonseca S., Killian M., Epling L., Hoh R., Sinclair E., et al. CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baxter A.E., Niessl J., Fromentin R., Richard J., Porichis F., Charlebois R., Massanella M., Brassard N., Alsahafi N., Delgado G.G., et al. Single-Cell Characterization of Viral Translation-Competent Reservoirs in HIV-Infected Individuals. Cell Host Microbe. 2016;20:368–380. doi: 10.1016/j.chom.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohn L.B., da Silva I.T., Valieris R., Huang A.S., Lorenzi J.C.C., Cohen Y.Z., Pai J.A., Butler A.L., Caskey M., Jankovic M., Nussenzweig M.C. Clonal CD4(+) T cells in the HIV-1 latent reservoir display a distinct gene profile upon reactivation. Nat. Med. 2018;24:604–609. doi: 10.1038/s41591-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collora J.A., Liu R., Pinto-Santini D., Ravindra N., Ganoza C., Lama J.R., Alfaro R., Chiarella J., Spudich S., Mounzer K., et al. Single-cell multiomics reveals persistence of HIV-1 in expanded cytotoxic T cell clones. Immunity. 2022;55:1013–1031.e7. doi: 10.1016/j.immuni.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu V.H., Nordin J.M.L., Nguyen S., Joy J., Mampe F., Del Rio Estrada P.M., Torres-Ruiz F., González-Navarro M., Luna-Villalobos Y.A., Ávila-Ríos S., et al. Profound phenotypic and epigenetic heterogeneity of the HIV-1-infected CD4(+) T cell reservoir. Nat. Immunol. 2023;24:359–370. doi: 10.1038/s41590-022-01371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun W., Gao C., Hartana C.A., Osborn M.R., Einkauf K.B., Lian X., Bone B., Bonheur N., Chun T.W., Rosenberg E.S., et al. Phenotypic signatures of immune selection in HIV-1 reservoir cells. Nature. 2023;614:309–317. doi: 10.1038/s41586-022-05538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark I.C., Mudvari P., Thaploo S., Smith S., Abu-Laban M., Hamouda M., Theberge M., Shah S., Ko S.H., Pérez L., et al. HIV silencing and cell survival signatures in infected T cell reservoirs. Nature. 2023;614:318–325. doi: 10.1038/s41586-022-05556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gantner P., Buranapraditkun S., Pagliuzza A., Dufour C., Pardons M., Mitchell J.L., Kroon E., Sacdalan C., Tulmethakaan N., Pinyakorn S., et al. HIV rapidly targets a diverse pool of CD4(+) T cells to establish productive and latent infections. Immunity. 2023;56:653–668.e5. doi: 10.1016/j.immuni.2023.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geretz A., Ehrenberg P.K., Clifford R.J., Laliberté A., Prelli Bozzo C., Eiser D., Kundu G., Yum L.K., Apps R., Creegan M., et al. Single-cell transcriptomics identifies prothymosin alpha restriction of HIV-1 in vivo. Sci. Transl. Med. 2023;15 doi: 10.1126/scitranslmed.adg0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jefferys S.R., Burgos S.D., Peterson J.J., Selitsky S.R., Turner A.M.W., James L.I., Tsai Y.H., Coffey A.R., Margolis D.M., Parker J., Browne E.P. Epigenomic characterization of latent HIV infection identifies latency regulating transcription factors. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ratnapriya S., Harris M., Chov A., Herbert Z.T., Vrbanac V., Deruaz M., Achuthan V., Engelman A.N., Sodroski J., Herschhorn A. Intra- and extra-cellular environments contribute to the fate of HIV-1 infection. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosque A., Planelles V. Studies of HIV-1 latency in an ex vivo model that uses primary central memory T cells. Methods. 2011;53:54–61. doi: 10.1016/j.ymeth.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolton D.L., McGinnis K., Finak G., Chattopadhyay P., Gottardo R., Roederer M. Combined single-cell quantitation of host and SIV genes and proteins ex vivo reveals host-pathogen interactions in individual cells. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X., He Y., Zhang Q., Ren X., Zhang Z. Direct Comparative Analyses of 10X Genomics Chromium and Smart-seq2. Dev. Reprod. Biol. 2021;19:253–266. doi: 10.1016/j.gpb.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tokarev A., Creegan M., Eller M.A., Roederer M., Bolton D.L. Single-cell Quantitation of mRNA and Surface Protein Expression in Simian Immunodeficiency Virus-infected CD4+ T Cells Isolated from Rhesus macaques. J. Vis. Exp. 2018 doi: 10.3791/57776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang S.T., Sova P., Peng X., Weiss J., Law G.L., Palermo R.E., Katze M.G. Next-generation sequencing reveals HIV-1-mediated suppression of T cell activation and RNA processing and regulation of noncoding RNA expression in a CD4+ T cell line. mBio. 2011;2 doi: 10.1128/mBio.00134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corbeil J., Sheeter D., Genini D., Rought S., Leoni L., Du P., Ferguson M., Masys D.R., Welsh J.B., Fink J.L., et al. Temporal gene regulation during HIV-1 infection of human CD4+ T cells. Genome Res. 2001;11:1198–1204. doi: 10.1101/gr.gr-1802r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dominguez M.H., Chattopadhyay P.K., Ma S., Lamoreaux L., McDavid A., Finak G., Gottardo R., Koup R.A., Roederer M. Highly multiplexed quantitation of gene expression on single cells. J. Immunol. Methods. 2013;391:133–145. doi: 10.1016/j.jim.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bell I., Ashman C., Maughan J., Hooker E., Cook F., Reinhart T.A. Association of simian immunodeficiency virus Nef with the T-cell receptor (TCR) zeta chain leads to TCR down-modulation. J. Gen. Virol. 1998;79(Pt 11):2717–2727. doi: 10.1099/0022-1317-79-11-2717. [DOI] [PubMed] [Google Scholar]
- 53.Swigut T., Iafrate A.J., Muench J., Kirchhoff F., Skowronski J. Simian and human immunodeficiency virus Nef proteins use different surfaces to downregulate class I major histocompatibility complex antigen expression. J. Virol. 2000;74:5691–5701. doi: 10.1128/jvi.74.12.5691-5701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benson R.E., Sanfridson A., Ottinger J.S., Doyle C., Cullen B.R. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J. Exp. Med. 1993;177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stieh D.J., Matias E., Xu H., Fought A.J., Blanchard J.L., Marx P.A., Veazey R.S., Hope T.J. Th17 Cells Are Preferentially Infected Very Early after Vaginal Transmission of SIV in Macaques. Cell Host Microbe. 2016;19:529–540. doi: 10.1016/j.chom.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee W.Y.J., Fu R.M., Liang C., Sloan R.D. IFITM proteins inhibit HIV-1 protein synthesis. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-32785-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Gassen S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., Saeys Y. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 58.Lau C.Y., Adan M.A., Maldarelli F. Why the HIV Reservoir Never Runs Dry: Clonal Expansion and the Characteristics of HIV-Infected Cells Challenge Strategies to Cure and Control HIV Infection. Viruses. 2021;13 doi: 10.3390/v13122512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simonetti F.R., Zhang H., Soroosh G.P., Duan J., Rhodehouse K., Hill A.L., Beg S.A., McCormick K., Raymond H.E., Nobles C.L., et al. Antigen-driven clonal selection shapes the persistence of HIV-1-infected CD4+ T cells in vivo. J. Clin. Invest. 2021;131 doi: 10.1172/JCI145254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chomont N., El-Far M., Ancuta P., Trautmann L., Procopio F.A., Yassine-Diab B., Boucher G., Boulassel M.R., Ghattas G., Brenchley J.M., et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maldarelli F., Wu X., Su L., Simonetti F.R., Shao W., Hill S., Spindler J., Ferris A.L., Mellors J.W., Kearney M.F., et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagner T.A., McLaughlin S., Garg K., Cheung C.Y.K., Larsen B.B., Styrchak S., Huang H.C., Edlefsen P.T., Mullins J.I., Frenkel L.M. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferris A.L., Wells D.W., Guo S., Del Prete G.Q., Swanstrom A.E., Coffin J.M., Wu X., Lifson J.D., Hughes S.H. Clonal expansion of SIV-infected cells in macaques on antiretroviral therapy is similar to that of HIV-infected cells in humans. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aikawa Y., Morimoto K., Yamamoto T., Chaki H., Hashiramoto A., Narita H., Hirono S., Shiozawa S. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat. Biotechnol. 2008;26:817–823. doi: 10.1038/nbt1412. [DOI] [PubMed] [Google Scholar]
- 65.Martins L.J., Bonczkowski P., Spivak A.M., De Spiegelaere W., Novis C.L., DePaula-Silva A.B., Malatinkova E., Trypsteen W., Bosque A., Vanderkerckhove L., Planelles V. Modeling HIV-1 Latency in Primary T Cells Using a Replication-Competent Virus. AIDS Res. Hum. Retroviruses. 2016;32:187–193. doi: 10.1089/aid.2015.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roebuck K.A., Rabbi M.F., Kagnoff M.F. HIV-1 Tat protein can transactivate a heterologous TATAA element independent of viral promoter sequences and the trans-activation response element. AIDS. 1997;11:139–146. doi: 10.1097/00002030-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 67.Roebuck K.A., Gu D.S., Kagnoff M.F. Activating protein-1 cooperates with phorbol ester activation signals to increase HIV-1 expression. AIDS. 1996;10:819–826. doi: 10.1097/00002030-199607000-00004. [DOI] [PubMed] [Google Scholar]
- 68.Hokello J., Lakhikumar Sharma A., Tyagi M. AP-1 and NF-kappaB synergize to transcriptionally activate latent HIV upon T-cell receptor activation. FEBS Lett. 2021;595:577–594. doi: 10.1002/1873-3468.14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jordan A., Bisgrove D., Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bosque A., Planelles V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood. 2009;113:58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cron R.Q., Bartz S.R., Clausell A., Bort S.J., Klebanoff S.J., Lewis D.B. NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clin. Immunol. 2000;94:179–191. doi: 10.1006/clim.1999.4831. [DOI] [PubMed] [Google Scholar]
- 72.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yukl S.A., Kaiser P., Kim P., Telwatte S., Joshi S.K., Vu M., Lampiris H., Wong J.K. HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rose J.J., Janvier K., Chandrasekhar S., Sekaly R.P., Bonifacino J.S., Venkatesan S. CD4 down-regulation by HIV-1 and simian immunodeficiency virus (SIV) Nef proteins involves both internalization and intracellular retention mechanisms. J. Biol. Chem. 2005;280:7413–7426. doi: 10.1074/jbc.M409420200. [DOI] [PubMed] [Google Scholar]
- 75.Wei B.L., Arora V.K., Foster J.L., Sodora D.L., Garcia J.V. In vivo analysis of Nef function. Curr. HIV Res. 2003;1:41–50. doi: 10.2174/1570162033352057. [DOI] [PubMed] [Google Scholar]
- 76.Reilly C., Wietgrefe S., Sedgewick G., Haase A. Determination of simian immunodeficiency virus production by infected activated and resting cells. AIDS. 2007;21:163–168. doi: 10.1097/QAD.0b013e328012565b. [DOI] [PubMed] [Google Scholar]
- 77.Gantner P., Pagliuzza A., Pardons M., Ramgopal M., Routy J.P., Fromentin R., Chomont N. Single-cell TCR sequencing reveals phenotypically diverse clonally expanded cells harboring inducible HIV proviruses during ART. Nat. Commun. 2020;11:4089. doi: 10.1038/s41467-020-17898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yukawa M., Jagannathan S., Vallabh S., Kartashov A.V., Chen X., Weirauch M.T., Barski A. AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J. Exp. Med. 2020;217 doi: 10.1084/jem.20182009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murakami K., Li Y. AP1-Related Factors Interact with Simian Immunodeficiency Virus Long Terminal Repeats. J. Biomed. Sci. 1996;3:170–177. doi: 10.1007/BF02253097. [DOI] [PubMed] [Google Scholar]
- 80.Yang X., Chen Y., Gabuzda D. ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-kappaB. J. Biol. Chem. 1999;274:27981–27988. doi: 10.1074/jbc.274.39.27981. [DOI] [PubMed] [Google Scholar]
- 81.Van Lint C., Amella C.A., Emiliani S., John M., Jie T., Verdin E. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J. Virol. 1997;71:6113–6127. doi: 10.1128/JVI.71.8.6113-6127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Franza B.R., Jr., Rauscher F.J., 3rd, Josephs S.F., Curran T. The Fos complex and Fos-related antigens recognize sequence elements that contain AP-1 binding sites. Science. 1988;239:1150–1153. doi: 10.1126/science.2964084. [DOI] [PubMed] [Google Scholar]
- 83.van der Sluis R.M., Derking R., Breidel S., Speijer D., Berkhout B., Jeeninga R.E. Interplay between viral Tat protein and c-Jun transcription factor in controlling LTR promoter activity in different human immunodeficiency virus type I subtypes. J. Gen. Virol. 2014;95:968–979. doi: 10.1099/vir.0.059642-0. [DOI] [PubMed] [Google Scholar]
- 84.Eferl R., Wagner E.F. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 85.Hess J., Angel P., Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 86.Duverger A., Wolschendorf F., Zhang M., Wagner F., Hatcher B., Jones J., Cron R.Q., van der Sluis R.M., Jeeninga R.E., Berkhout B., Kutsch O. An AP-1 binding site in the enhancer/core element of the HIV-1 promoter controls the ability of HIV-1 to establish latent infection. J. Virol. 2013;87:2264–2277. doi: 10.1128/JVI.01594-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tyagi M., Pearson R.J., Karn J. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 2010;84:6425–6437. doi: 10.1128/JVI.01519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pearson R., Kim Y.K., Hokello J., Lassen K., Friedman J., Tyagi M., Karn J. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 2008;82:12291–12303. doi: 10.1128/JVI.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van Lint C., Emiliani S., Ott M., Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 90.He G., Ylisastigui L., Margolis D.M. The regulation of HIV-1 gene expression: the emerging role of chromatin. DNA Cell Biol. 2002;21:697–705. doi: 10.1089/104454902760599672. [DOI] [PubMed] [Google Scholar]
- 91.Kauder S.E., Bosque A., Lindqvist A., Planelles V., Verdin E. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Imbeault M., Giguère K., Ouellet M., Tremblay M.J. Exon level transcriptomic profiling of HIV-1-infected CD4(+) T cells reveals virus-induced genes and host environment favorable for viral replication. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rasmussen T.A., Zerbato J.M., Rhodes A., Tumpach C., Dantanarayana A., McMahon J.H., Lau J.S.Y., Chang J.J., Gubser C., Brown W., et al. Memory CD4(+) T cells that co-express PD1 and CTLA4 have reduced response to activating stimuli facilitating HIV latency. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cochrane C.R., Angelovich T.A., Byrnes S.J., Waring E., Guanizo A.C., Trollope G.S., Zhou J., Vue J., Senior L., Wanicek E., et al. Intact HIV Proviruses Persist in the Brain Despite Viral Suppression with ART. Ann. Neurol. 2022;92:532–544. doi: 10.1002/ana.26456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Heesters B.A., Lindqvist M., Vagefi P.A., Scully E.P., Schildberg F.A., Altfeld M., Walker B.D., Kaufmann D.E., Carroll M.C. Follicular Dendritic Cells Retain Infectious HIV in Cycling Endosomes. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pinzone M.R., Weissman S., Pasternak A.O., Zurakowski R., Migueles S., O'Doherty U. Naive infection predicts reservoir diversity and is a formidable hurdle to HIV eradication. JCI Insight. 2021;6 doi: 10.1172/jci.insight.150794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Del Prete G.Q., Park H., Fennessey C.M., Reid C., Lipkey L., Newman L., Oswald K., Kahl C., Piatak M., Jr., Quiñones O.A., et al. Molecularly tagged simian immunodeficiency virus SIVmac239 synthetic swarm for tracking independent infection events. J. Virol. 2014;88:8077–8090. doi: 10.1128/JVI.01026-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McCarthy D.J., Campbell K.R., Lun A.T.L., Wills Q.F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kent W.J. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 106.Borcherding N., Bormann N.L., Kraus G. scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Res. 2020;9:47. doi: 10.12688/f1000research.22139.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gu Z., Gu L., Eils R., Schlesner M., Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 108.Clark I.C., Wheeler M.A., Lee H.G., Li Z., Sanmarco L.M., Thaploo S., Polonio C.M., Shin S.W., Scalisi G., Henry A.R., et al. Identification of astrocyte regulators by nucleic acid cytometry. Nature. 2023;614:326–333. doi: 10.1038/s41586-022-05613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morabito S., Reese F., Rahimzadeh N., Miyoshi E., Swarup V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Rep. Methods. 2023;3 doi: 10.1016/j.crmeth.2023.100498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 111.Ehrenberg P.K., Shangguan S., Issac B., Alter G., Geretz A., Izumi T., Bryant C., Eller M.A., Wegmann F., Apps R., et al. A vaccine-induced gene expression signature correlates with protection against SIV and HIV in multiple trials. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aaw4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shangguan S., Ehrenberg P.K., Geretz A., Yum L., Kundu G., May K., Fourati S., Nganou-Makamdop K., Williams L.D., Sawant S., et al. Monocyte-derived transcriptome signature indicates antibody-dependent cellular phagocytosis as a potential mechanism of vaccine-induced protection against HIV-1. Elife. 2021;10 doi: 10.7554/eLife.69577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes are listed in rank order of degree of connectivity with other genes within the module.
Data Availability Statement
-
•
Single-cell RNA-seq data have been deposited at GEO. Bulk human RNA-seq data were previously deposited at dbGaP. All data are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request