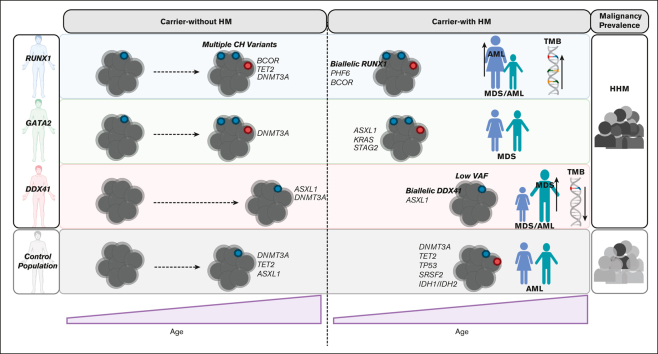
Key Points
-
•
Germline RUNX1, GATA2, and DDX41 HHMs are associated with driver somatic variants during leukemogenesis which are unique for each syndrome.
-
•
Ongoing molecular monitoring of germline carriers without HM is needed to assess the risk profile and clinical actionability of somatic markers.
Visual Abstract
Abstract
Individuals with germ line variants associated with hereditary hematopoietic malignancies (HHMs) have a highly variable risk for leukemogenesis. Gaps in our understanding of premalignant states in HHMs have hampered efforts to design effective clinical surveillance programs, provide personalized preemptive treatments, and inform appropriate counseling for patients. We used the largest known comparative international cohort of germline RUNX1, GATA2, or DDX41 variant carriers without and with hematopoietic malignancies (HMs) to identify patterns of genetic drivers that are unique to each HHM syndrome before and after leukemogenesis. These patterns included striking heterogeneity in rates of early-onset clonal hematopoiesis (CH), with a high prevalence of CH in RUNX1 and GATA2 variant carriers who did not have malignancies (carriers-without HM). We observed a paucity of CH in DDX41 carriers-without HM. In RUNX1 carriers-without HM with CH, we detected variants in TET2, PHF6, and, most frequently, BCOR. These genes were recurrently mutated in RUNX1-driven malignancies, suggesting CH is a direct precursor to malignancy in RUNX1-driven HHMs. Leukemogenesis in RUNX1 and DDX41 carriers was often driven by second hits in RUNX1 and DDX41, respectively. This study may inform the development of HHM-specific clinical trials and gene-specific approaches to clinical monitoring. For example, trials investigating the potential benefits of monitoring DDX41 carriers-without HM for low-frequency second hits in DDX41 may now be beneficial. Similarly, trials monitoring carriers-without HM with RUNX1 germ line variants for the acquisition of somatic variants in BCOR, PHF6, and TET2 and second hits in RUNX1 are warranted.
Introduction
Hereditary hematopoietic malignancies (HHMs) are hematologic syndromes characterized by Mendelian inheritance patterns and an increased lifetime risk for hematopoietic malignancies (HMs).1,2 Individuals with HHM-associated germ line variants have a highly variable risk for leukemogenesis, and many HHM-variant carriers do not develop malignancies (carriers-without HM).3 Very little is understood about the premalignant states in carriers-without HM, the molecular and genetic factors that affect leukemogenic risk, or the environmental factors that drive leukemogenesis in HHMs. This knowledge gap has hampered efforts to refine the clinical surveillance of carriers-without HM, identify individuals with the highest risk for HMs, and develop interventions that delay or prevent leukemogenesis in high-risk carriers-without HM. Moreover, treatments used for malignancies in HHM-variant carriers (carriers-with HM) are not tailored to these syndromes aside from DDX41 and GATA2 carriers, for which there is a limited role for lenalidomide therapy or prophylactic hematopoietic stem cell transplant, respectively.4, 5, 6 Instead, carriers-with HM are treated with standard-of-care therapies for sporadic HMs, which may carry an uncharacterized gene mutation–specific risk of additional treatment effects, such as engraftment failure or secondary therapy-related neoplasms. Given the paucity of HHM families at individual institutions, a coordinated, multi-institutional effort is required to understand the natural history of HHMs, leukemogenic mechanisms, and the unique biologic factors that may be present in individual HHM syndromes.
HHMs have been recognized phenotypically for over 100 years. Autosomal dominant (AD) predisposition to myeloid malignancies is the most well characterized, with more than 15 AD HHM-related genes identified to date.7 Pathogenic germ line variants in RUNX1, GATA2, and DDX41 collectively represent the most common causes of AD HHMs and are primarily associated with myeloid malignancies. These HHMs are more common than previously recognized and may have highly penetrant leukemogenic phenotypes. Germ line DDX41 carriers account for ∼2% to 4% of all patients with seemingly sporadic HMs, and GATA2 carriers have a 90% lifetime risk of developing HMs. RUNX1-driven HHMs were the first known HHM syndrome and have a high penetrance for HM (∼44%).3,4,8, 9, 10, 11 Identifying these syndromes can be challenging because of limited syndromic features, and recognition is often made based on a high-risk family history, an early-onset HM, or the identification of an HHM-associated variant on tumor-based molecular profiling.12 Individuals harboring germ line variants in these genes often present with cytopenias: RUNX1 most commonly with thrombocytopenia;13 GATA2 with monocytopenia, dendritic cell, B, and natural killer cell (NK) lymphoid deficiency;14 and DDX41 with variable cytopenias that can include leukopenia, neutropenia, and/or erythroid dysplasia.15,16 The age of myelodysplastic syndrome/acute myeloid leukemia (MDS/AML) diagnosis also differs between HHMs, with GATA2 carriers developing MDS/AML at a mean age of 19 years, RUNX1 carriers at 29 years, and DDX41 carriers at 67 years.16,17
The mechanisms driving leukemogenesis in these variant carriers are unclear. Most work to date has focused on germ line RUNX1 variant carriers.8 RUNX1 carriers have an increased risk for clonal hematopoiesis (CH) (67%-75% CH18,19). However, because of the rarity of RUNX1 HHM, single-center studies have limited numbers of patients available (919 and 318). Recent studies looking at CH in germ line GATA2 carriers have shown an association between CH and a hypocellular marrow while also linking specific somatic events with the likelihood of leukemic transformation.20,21 Similarly assessment of CH patterns in comparative cohorts of HHM-variant carriers may identify specific leukemogenic patterns for different HHMs and ultimately inform clinical trials to define guidelines for clinical surveillance of unaffected HHM-variant carriers.
To address this knowledge gap, we collected retrospective next-generation sequencing (NGS) data from hematopoietic tissue samples from an international cohort of patients with HHM driven by germ line RUNX1, GATA2, or DDX41 variants. Our cohort is the largest comparative HHM-focused cross-sectional collection of its kind, with 240 patient samples evenly distributed between carriers-without HM (n = 120) and carriers-with HM (n = 120). We used a uniform variant calling and curation approach to identify driver somatic variants in each sample. This unique distribution of samples from carriers-without HM and carriers-with HM and across multiple HHMs, in conjunction with a uniform bioinformatic approach, enabled us to determine driver somatic variants that develop within hematopoietic tissue in RUNX1, GATA2, and DDX41 variant carriers before and after diagnosis of a blood cancer in the HHM syndromes.
Patients and methods
Patient cohort
Clinical and genomics data from germ line RUNX1, GATA2, or DDX41 variant carriers were collected from the RUNX1 database (https://runx1db.runx1-fpd.org/),22 the Centre for Cancer Biology (Australia), the University of Chicago (USA), and the National Institutes of Health (USA). In total, data from 195 patients who had undergone genomics profiling (whole-exome sequencing or panel-based sequencing) were retrospectively collated to form the RUNX1, GATA2, and DDX41 cohorts. All procedures in this study involving human participants were performed in accordance with the Declaration of Helsinki. Studies were approved by institutional human research ethics committees and/or institutional research boards. All participants signed an informed consent form to share genomics and protected health information.
NGS reanalysis and variant calling pipeline
NGS data were collected and reanalyzed with the bioinformatics pipeline used for the RUNX1 database.22 Original FASTQ (textfile format for sequencing data) or Binary Alignment Map (BAM) files were obtained. Sequence reads were aligned to the GRCh37 (hs37d5) human reference genome with BWA-MEM (ver 0.7.12).23 Sambamba (ver 0.6.5)24 was used for marking polymerase chain reaction duplicates, and GATK (ver 3.8-1) was used to recalibrate base-quality scores. Freebayes (ver 1.2)25 was used to call single nucleotide variants (SNVs) and insertions/deletions (indels). Variant-, gene-, and protein-level annotations were performed using an in-house pipeline (https://github.com/SACGF/variantgrid). Somatic variant curation was performed as previously described22 (supplemental Methods). All data sets were independently curated by at least 3 variant curators.
Tumor mutation burden (TMB) analysis
SNVs and indels were identified with Seurat, Shimmer, Strelka, and SomaticSniper,26, 27, 28, 29 using paired germ line samples (cultured skin fibroblasts or hair) from the same patient to remove germ line variants. Somatic variants identified in 3 or more callers were included with high confidence. Variant calling thresholds were set at alternate allelic depth ≥3 and variant allele frequency (VAF) ≥ 5%. Somatic variants were filtered and annotated with the variant effect predictor package (hg19). The total number of somatic variants in the tumor exome was divided by the length of exome capture (38 Mb) to calculate the TMB.
Statistical analysis
GraphPad Prism 7.03 and RStudio Version 1.4.17 with tidyverse, ggplot2, ggrepel, caTools, and ROCR packages were used for statistical calculations and figures. ProteinPaint was used to create lollipop plots.30 Circos plots were created using ShinyCircos software.31 Unless otherwise stated, the P value was calculated using a one-way analysis of variance with Tukey multiple comparisons test using a single pooled variance. P value of sex differences were calculated using a two-sided Fisher exact test. The prop.test function/z-value was used as a 2-sample test for equality of proportions with continuity correction. Logistic regression modeling was used to determine the relationship between age and CH. The nonparametric Mann-Whitney U test was used to calculate the significance of the TMB between the RUNX1 and DDX41 cohorts.
Results
Genomic cohorts for germ line RUNX1, GATA2, or DDX41 HHMs
Through international data sharing, we created cohorts of carriers-without HM (no HM) and carriers-with HM (diagnosed with an HM) with germ line RUNX1, GATA2, or DDX41 variants (Figure 1; supplemental Methods). NGS data included samples from germ line controls, complete remission patients, carriers-without HM, and carriers-with HM. Multiple samples were collected from individuals when available, including longitudinal. The RUNX1 cohort included 66 carriers-without HM and 52 carriers-with HM individuals (including 80 and 66 independent NGS samples, respectively). The GATA2 cohort included 9 carriers-without HM and 13 carriers-with HM individuals (9 and 13 NGS samples, respectively). The DDX41 cohort included 22 carriers-without HM and 29 carriers-without HM individuals (including 31 and 41 independent NGS samples, respectively). Each cohort is summarized in supplemental Table 1 and supplemental Figures 1A and 2. We used a standardized bioinformatics and variant curation approach22 to identify clinically relevant and potentially clinically relevant somatic variants (driver somatic variants, detailed in the supplemental Methods).
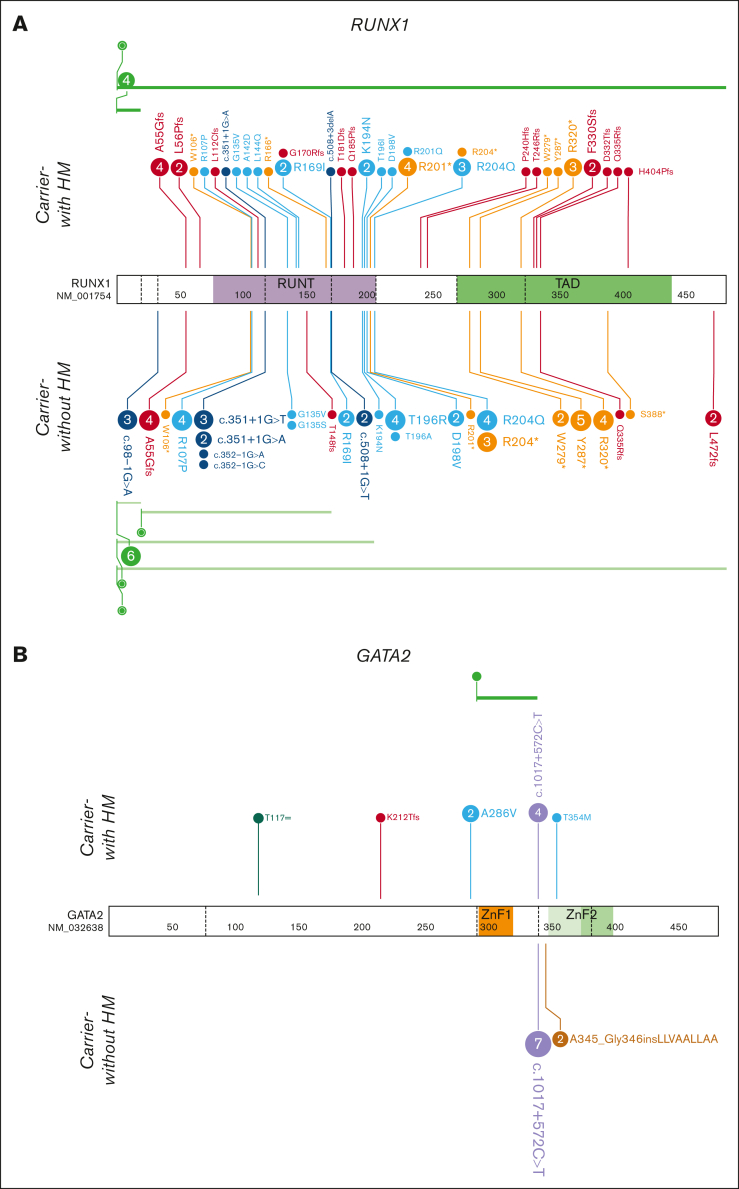
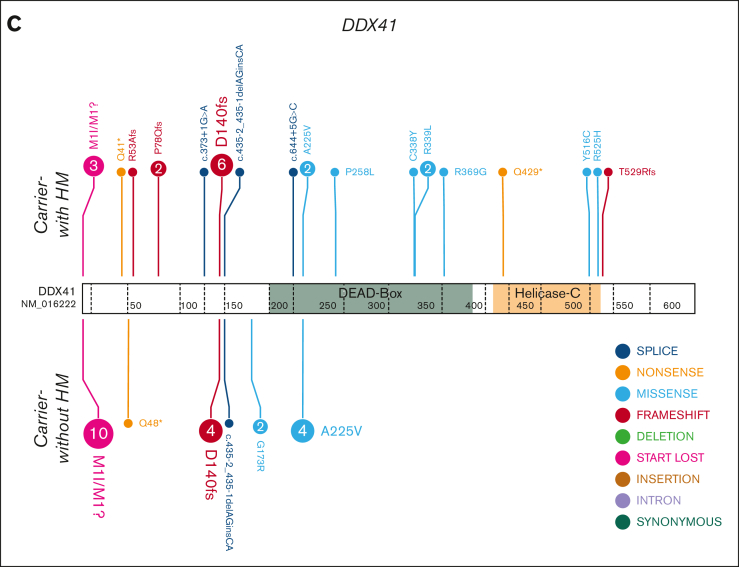
Figure 1.
HHM genomics cohorts. Germ line variants in the HHM cohorts were visualized using the ProteinPaint web application.30 Carriers-with HM cohorts (diagnosed with HM) are visualized above the protein. Carriers-without HM cohorts (no HM diagnosis) are below the protein. Variants (displayed as protein changes where possible) are color coded by variant type. The number of individuals with each variant is indicated within the circle when the number is greater than 1. (A) Germ line RUNX1 (66 carriers-without HM and 52 carriers-with HM individuals); (B) Germ line GATA2 (9 carriers-without HM and 13 carriers-with HM individuals); and (C) Germ line DDX41 (22 carriers-without HM and 29 carriers-with HM individuals) cohorts.
CH is prevalent in RUNX1 and GATA2 but not DDX41 HHM carriers-without HM
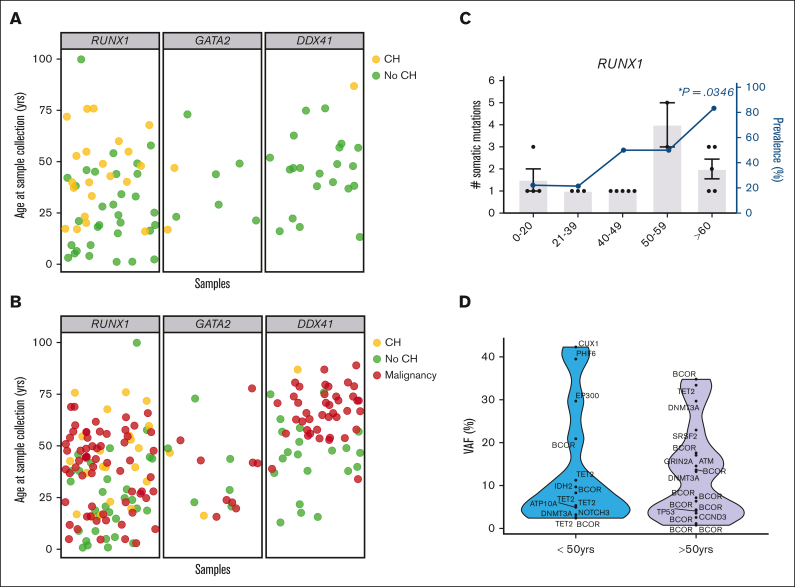
Age-related CH is frequently observed in healthy populations, with the prevalence of CH in HHMs an area of active investigation.18, 19, 20, 21,32, 33, 34 We evaluated our cross-sectional cohorts of RUNX1, GATA2, and DDX41 carriers-without HM for CH-related variants at the time of sample collection (Table 1; Figure 2A). We identified CH in 35% of RUNX1 carriers-without HM (23 of 66 individuals; Figure 2A,C) and 22% (2 of 9 individuals, Figure 2A; supplemental Figure 1B) of GATA2 carriers-without HM, respectively. The prevalence of CH was significantly lower (3%, 1 of 31 individuals, P = .002; Figure 2A; supplemental Figure 1C) in DDX41 carriers-without HM. The reduced prevalence of CH in the DDX41 cohort was independent of age, as the age distribution of samples was overlapping between HHM cohorts (supplemental Figure 1A). For germ line RUNX1, CH was identified in all age groups, and the prevalence of CH significantly increased with age (Figure 2C, P=.0267, logistic regression). In the RUNX1 cohort, 5 of 6 (83%) individuals 60 years of age and older had at least 1 CH variant (Figure 2C). The number of variants increased with age, as 92% of individuals under the age of 50 years with CH had only 1 CH variant, whereas 71% of patients over the age of 50 years had 2 or more CH variants (P = .001, Figure 2C,E). The median VAF of CH variants did not change significantly with age (Figure 2D). For all cohorts, no CH was identified in any individual younger than 16 years (n = 9).
Table 1.
Driver somatic variants detected in germ line RUNX1, GATA2, or DDX41 carriers-without HM
| Individual |
Phenotype |
Gene |
Somatic variant |
|||||
|---|---|---|---|---|---|---|---|---|
| Clinical presentation | Age | Sex | Genomic coordinates | c_HGVS | p_HGVS | VAF (%) | ||
| M09_1 | Thrombocytopenia | 16 | F | BCOR | X:39933270 CA>C | NM_001123385.2:c.1328del | p.Leu443CysfsTer8 | 8.3 |
| M01_3 | Thrombocytopenia | 17 | M | ATP10A | 15:25953381 G>A | NM_024490.3:c.2411C>T | p.Ala804Val | 5.0 |
| M03_2 | Thrombocytopenia | 17 | M | PHF6 | X:133527608 CTG>C | NM_032458.3:c.321_322del | p.Ala108IlefsTer3 | 39.4 |
| CUX1 | 7:101882720 G>A | NM_001202543.2:c.3776G>A | p.Arg1259Gln | 42.3 | ||||
| IDH2 | 15:90631934 C>T | NM_002168.3:c.419G>A | p.Arg140Gln | 9.7 | ||||
| G01_2 | Thrombocytopenia | 19 | F | NOTCH3 | 19:15276860 GC>G | NM_000435.3:c.5404del | p.Ala1802LeufsTer23 | 3.2 |
| V01_1 | Thrombocytopenia | 23 | M | EP300 | 22:41574723 TCCACACCACGTTTCC>T | NM_001429.3:c.7014_7028del15 | p.His2338_Pro2342del | 29.6 |
| G07_3 | Thrombocytopenia | 33 | M | BCOR | X:39932334 G>GT | NM_001123385.2:c.2264dup | p.Tyr755Ter | 2.4 |
| M06_2 | Thrombocytopenia | 37 | F | TET2 | 4:106164068 G>A | NM_001127208.3):c.3578G>A | p.Cys1193Tyr | 11.3 |
| G07_2 | Thrombocytopenia | 40 | M | TET2 | 4:106164787 C>T | NM_001127208.3:c.3655C>T | p.His1219Tyr | 5.3 |
| S_G_3 | Thrombocytopenia | 40 | F | TET2 | 4:106196461 T>G | NM_001127208.3:c.4794T>G | p.Tyr1598Ter | 5.0 |
| W01_3 | Thrombocytopenia | 40 | M | BCOR | X:39921617 CG>C | NM_001123385.2:c.4202del | p.Pro1401ArgfsTer83 | 20.8 |
| S_D_2 | Thrombocytopenia | 43 | M | DNMT3A | 2:25463242 AG>A | NM_022552.5:c.2250del | p.Phe751SerfsTer28 | 3.1 |
| 48 | M | DNMT3A | 2:25463242 AG>A | NM_022552.5:c.2250del | p.Phe751SerfsTer28 | 4.9 | ||
| U02_1 | Thrombocytopenia | 49 | M | TET2 | 4:106162587 G>A | NM_001127208.3:c.3500+1G>A | p.? | 2.6 |
| A01_1 | Thrombocytopenia | 53 | F | TET2 | 4:106158510 T>C | NM_001127208.3:c.3409+2T>C | p.? | 33.3 |
| DNMT3A | 2:25497943 CG>C | NM_022552.5:c.505del | p.Arg169GlyfsTer56 | 29.6 | ||||
| SRSF2 | 17:74732959 G>A | NM_003016.4:c.284C>T | p.Pro95Leu | 22.9 | ||||
| 56 | F | DNMT3A | 2:25497943 CG>C | NM_022552.5:c.505del | p.Arg169GlyfsTer56 | 42.8 | ||
| TET2 | 4:106158510 T>C | NM_001127208.3:c.3409+2T>C | p.? | 30.5 | ||||
| SRSF2 | 17:74732959 G>A | NM_003016.4:c.284C>T | p.Pro95Leu | 28.0 | ||||
| G02_2 | Thrombocytopenia | 55 | M | BCOR | X:39932898 T>TG | NM_001123385.2:c.1700dup | p.Ala568SerfsTer43 | 6.7 |
| BCOR | X:39923055 C>T | NM_001123385.2:c.3653G>A | p.Trp1218Ter | 4.3 | ||||
| BCOR | X:39911577 GA>G | NM_001123385.2:c.5052del | p.Pro1685GlnfsTer40 | 2.5 | ||||
| 54 | M | BCOR | X:39932898 T>TG | NM_001123385.2:c.1700dup | p.Ala568SerfsTer43 | 6.4 | ||
| BCOR | X:39923055 C>T | NM_001123385.2:c.3653G>A | p.Trp1218Ter | 2.2 | ||||
| BCOR | X:39933676 TG>T | NM_001123385.2:c.922del | p.Gln308ArgfsTer70 | 1.2 | ||||
| BCOR | X:39933416 TG>T | NM_001123385.2:c.1182del | p.Lys395ArgfsTer47 | 0.7 | ||||
| X01_1 | Asymptomatic | 60 | F | BCOR | X:39932109 ACT>A | NM_001123385.2:c.2488_2489del | p.Ser830CysfsTer6 | 17.6 |
| W01_2 | Thrombocytopenia | 68 | M | BCOR | X:39933593 A>AG | NM_001123385.2:c.1005dup | p.Ser336LeufsTer45 | 7.1 |
| I03_2 (BM) | Thrombocytopenia | 72 | F | DNMT3A | 2:25470029 T>C | NM_022552.5:c.1015-2A>G | p.? | 13.3 |
| BCOR | X:39933492 TG>T | NM_001123385.2:c.1106del | p.Ser369Ter | 4.0 | ||||
| F01_6 | Thrombocytopenia | 76 | M | BCOR | X:39916476 C>T | NM_001123385.2:c.4527G>A | p.Trp1509Ter | 13.6 |
| ATM | 11:108213949 G>A | NM_000051.3:c.8269G>A | p.Val2757Met | 17.0 | ||||
| GRIN2A | 16:9857831 G>C | NM_000833.4:c.3570C>G | p.His1190Gln | 14.5 | ||||
| F01_8 | Thrombocytopenia | 76 | M | BCOR | X:39921490 TG>T | NM_001123385.2:c.4329del | p.Thr1444ProfsTer40 | 34.7 |
| TP53 | 17:7578442 T>C | NM_000546.6:c.488A>G | p.Tyr163Cys | 4.1 | ||||
| CCND3 | 6:41903707 G>A | NM_001760.4:c.850C>T | p.Pro284Ser | 3.5 | ||||
| U02_3 | Asymptomatic | NA | M | TET2 | 4:106162587 G>A | NM_001127208.3:c.3500+1G>A | p.? | 1.2 |
| S_E_4 | Thrombocytopenia | NA | F | SRSF2 | 17:74732959 G>T | NM_003016.4:c.284C>A | p.Pro95His | 13.9 |
| ATR | 3:142281940 A>G | NM_001184.4:c.304T>C | p.Trp102Arg | 3.9 | ||||
| D01_2 | Thrombocytopenia | NA | M | DNMT3A | 2:25466797 C>T | NM_022552.5:c.1906G>A | p.Val636Met | 24.7 |
| BCOR | X:39923092 TA>T | NM_001123385.2:c.3615del | p.Lys1207AsnfsTer31 | 3.6 | ||||
| DNMT3A | 2:25464460 C>T | NM_022552.5:c.2053G>A | p.Gly685Arg | 3.2 | ||||
| BCOR | X:39933373 TGCCCGG>TT | NM_001123385.2:c.1220_1225delCCGGGCinsA | p.Pro407GlnfsTer31 | 3.1 | ||||
| D02_2 | Thrombocytopenia | NA | F | PTPN11 | 12:112926887 G>A | NM_002834.4:c.1507G>A | p.Gly503Arg | 25.2 |
| U02_4 | NA | NA | F | TET2 | 4:106157215 C>T | NM_001127208.3:c.2116C>T | p.Gln706Ter | 7.3 |
| Family_53_8 | Asymptomatic | 16.5 | F | KDM5A | 12:416952 C>CT | NM_001042603.3:c.3597dup | p.Gly1200ArgfsTer7 | 3.2 |
| Family_53_3 | Asymptomatic | 47 | M | DNMT3A | 2:25457171 T>A | NM_022552.5:c.2716A>T | p.Lys906Ter | 3.6 |
| Family_0127.041 | Asymptomatic | 87 | F | ASXL1 | 20:31022576 TAC>T | NM_015338.5:c.2062_2063del | p.Thr688fs29 | 4.2 |
| DNMT3A | 2:25467022 A>G | NM_022552.5:c.1851+2T>C | p.? | 4.1 | ||||
Figure 2.
Defining the spectrum of CH in germline GATA2, RUNX1, and DDX41 carriers-without HM. (A) Each individual in the carriers-without HM cohort was defined as having CH (yellow) or no identifiable CH (no-CH, green), based on the identification of somatic clinically relevant variants, driver somatic variants. The age of the individual at the time of sample collection is indicated. (B) Correlation of the ages of malignancy development (HM, red) observed in the germ line malignancy cohorts with the carriers-without HM cohort, with (yellow) and without CH (green). (C) Demographics of individuals with CH variants in the RUNX1 germ line carriers-without HM cohorts. Column graph shows the number of somatic variants identified in individuals with CH. Error bars show the standard error of the mean. Line graphs show the prevalence of CH in the carriers-without HM germ line cohort in different age groups. ∗P < .05, logistic regression model. (D) Violin plots showing the distribution of VAFs of driver somatic variants (shown in panel A) in the germ line RUNX1 carriers-without HM cohort in individuals under the age of 50 years or >50 years old. VAFs for X-chromosome genes were normalized in male individuals to compensate for ploidy, enabling comparison with autosomal genes. HM, hematologic malignancy.
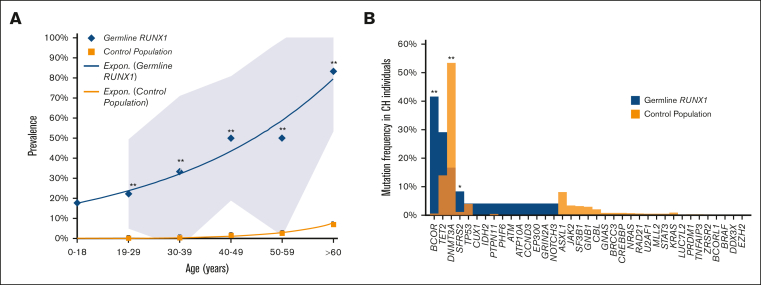
CH is increased in RUNX1 carriers-without HM relative to population controls
We then compared the prevalence of CH in our cohort of RUNX1 carriers-without HM to population controls from Jaiswal et al and Genovese et al (n = 27 783).32,33 The prevalence of CH was higher in RUNX1 carriers-without HM in every age group (Figure 3A, Z test of proportions, P < .0001). The prevalence of CH was 0.2% in controls between the ages of 19 and 29 years but was 22.2% in RUNX1 carriers-without HM in the same age group. In individuals aged 60 years or older, CH was detectable in 7% of controls and 83% of RUNX1 carriers-without HM, demonstrating that RUNX1 carriers-without HM have an increased prevalence of CH at all ages compared with population controls. We investigated the frequency of variants in prototypical CH genes. Variants in the epigenetic regulators DNMT3A (54%), TET2 (29%), and ASXL1 (8%) are the most frequent CH-related genes in the general population.32,35,36 Surprisingly, the most frequently mutated CH-related gene in RUNX1 carriers-without HM was BCOR (42%), which is mutated in only 0.6% of population controls with CH (Figure 3B, P < .0001).32,35,36 DNMT3A was mutated in 17% (P < .0001), TET2 in 14% (P = .2182), and ASXL1 was not mutated in RUNX1 carriers-without HM with CH (Figure 3B). These findings demonstrate that the mechanism of CH in RUNX1 carriers-without HM is distinct for this syndrome as compared with population controls.
Figure 3.
Spectrum of CH in age-related CH compared with the germ line RUNX1 carriers-without HM cohort. (A) Prevalence of CH in the control population compared with the germ line RUNX1 carriers-without HM cohort. The control population includes cohorts from Jaiswal et al32 and Genovese et al33 (n = 27 783). The 0 to 18 age group is only available for the RUNX1 cohort. (B) Mutational spectrum of CH in the control population compared with the germ line RUNX1 carriers-without HM cohort. Graph shows the frequency of variants in individuals with CH in each cohort. The control population includes cohorts from Desai et al,36 Abelson et al,35 and Jaiswal et al.32 ∗P < .05, ∗∗P < .001, 2-proportions Z test, approximate to normal distribution, ±95% confidence interval.
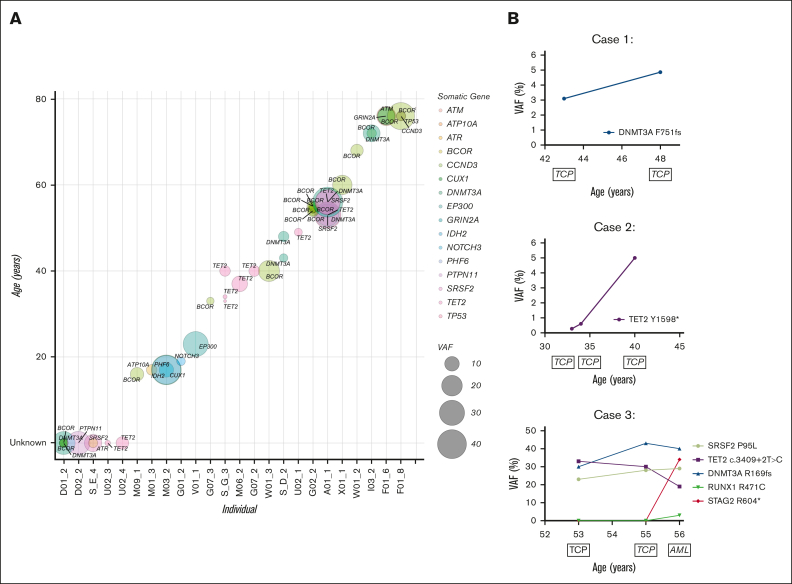
Clonal structure and evolution in RUNX1 carriers-without HM
Further examining the clonal composition of somatic variants in carriers, we postulated the order of mutation acquisition in samples with multiple variants by using relative VAFs (Figure 4A). We observed that BCOR, as well as being the most frequently mutated gene, was also present across the entire age spectrum, from carriers as young as 16 years to 76 years, found as a first hit (Figure 4A; Table 1). Consistent with the overall data, there was a general increase in BCOR VAF with age and additional mutations, which could be both additional BCOR mutations as well as mutations in other genes including TP53 and ATM, with 1 case where DNMT3A was antecedent to BCOR variants (Figure 4A; Table 1). Three RUNX1 carriers-without HM had longitudinal peripheral blood samples available, which allowed us to track the temporal evolution of CH (Figure 4B). Case 1: a male with thrombocytopenia and a germ line RUNX1 p.R169I variant had a somatic DNMT3A p.F751fs variant detected at a VAF of 3.1% at 43 years of age. The clone increased to a VAF of 5.0% over 5 years without any clinical-level changes, including leukemogenesis, or the development of additional clones. Case 2: a female with thrombocytopenia and a germ line RUNX1 p.R320∗ variant who developed a TET2 p.Y1598∗ somatic variant that persisted for more than 7 years, increasing from a VAF of <1% to 5%, without clinical changes. Case 3: a female with a germ line RUNX1 c.351+1G>A splicing variant and thrombocytopenia who developed AML 3 years later. We identified 3 somatic variants (DNMT3A, SRSF2, and TET2) in the patient’s initial sample, collected at age 53 years. These variants persisted for 2 years with persistent thrombocytopenia but no leukemogenesis. The patient then developed AML with additional somatic RUNX1 and STAG2 variants at 56 years of age. The initial DNMT3A and SRSF2 CH-related variants remained stable, whereas the TET2 variant was outcompeted during leukemogenesis.
Figure 4.
Molecular monitoring of germ line RUNX1 carriers with CH. (A) Driver somatic variants identified in RUNX1 carriers-without HM individuals across age. Circle size = increasing VAF, color = gene. Individuals with unknown age were given the value 0 (D01_2, D02_2, S_E_4, U02_3, U02_4). (B) Longitudinal case studies with the VAF of detected driver somatic variants plotted across years. Clinical diagnosis at the time of monitoring is indicated below the age of the individual at which the sample was collected. AML, acute myeloid leukemia; TCP, thrombocytopenia.
Somatic variants in germ line RUNX1, GATA2, and DDX41 malignancy samples
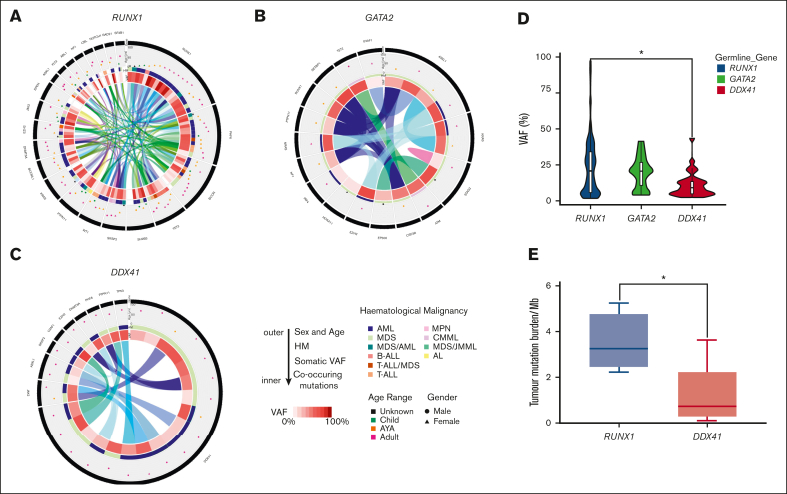
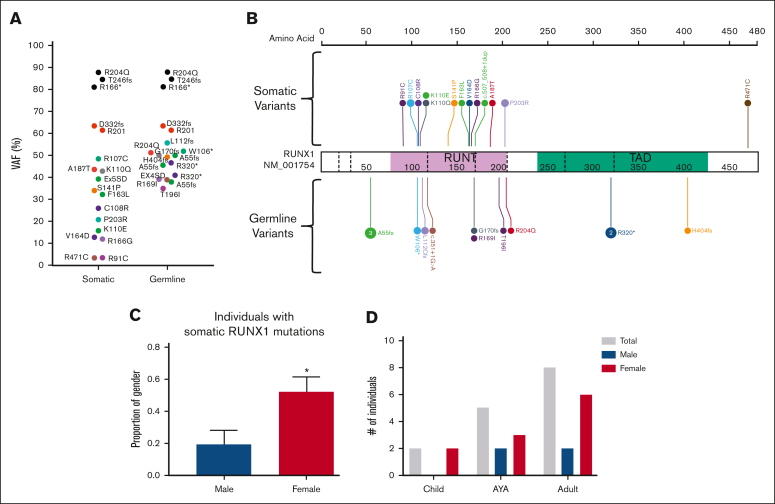
We next sought to define the landscape of driver somatic variants in our carriers-with HM cohorts who had developed malignancies. In the RUNX1 carriers-with HM cohort, at least 1 driver somatic variant was detected in 46 of 52 (88%) individuals diagnosed with an HM. No association between the number of driver somatic variants and the histologic subtype of malignancy was observed (supplemental Figure 2C). Driver somatic variants were identified in 64 unique genes, and 22 genes were mutated in more than 1 individual (Figure 5A; supplemental Figure 3A). Second hits in RUNX1 were the most frequent somatic mutations, with variants detected in 18 individuals (41% of patients with complete sequencing coverage of RUNX1 [supplemental Methods]). Three types of somatic RUNX1 variants were identified: small indels and SNVs (unique from the germ line variant, 72%), copy neutral loss of heterozygosity variants (17%), and trisomy 21 (somatic amplification of the germ line RUNX1 variant, 11%). Somatic second hits in RUNX1 included 12 missense variants in the exons coding for the RUNT domain as well as a splice-site variant (c.507_508+1dupAGG, Figure 6A-B). Cytogenetic analyses identified 2 individuals with +21 (VAF > 60%) and 3 individuals with a mutant VAF >80% (copy neutral loss of heterozygosity variants) (supplemental Figure 3A). We did not identify associations between individual germ line and driver somatic variant pairs. Most individuals (78%) with a somatic RUNX1 variant were female (P = .02, Figure 6C). Somatic RUNX1 variants were identified in all age groups, and no association was established between individual somatic RUNX1 variants and the age of HM diagnosis (Figure 6D). A female sex bias for HM was observed in all age groups (Figure 6D). Besides second hits in RUNX1, a series of established cancer genes were mutated in the HM cohort: PHF6 (21%), BCOR (20%), TET2 (13%), SH2B3 (11%), and SRSF2 (11%) (Figure 5A; supplemental Figure 3A). AML was the predominant malignancy in germ line RUNX1 variant carriers, with a sex bias for female AML diagnosis (23 of 29 females, 9 of 23 males, P = .004, supplemental Figure 4). Among individuals with somatic RUNX1 variants, 15 (83%) had AML, and 12 of 15 (80%) were females. These data from germ line RUNX1 variant carriers support a female sex bias for AML leukemogenesis driven by somatic RUNX1 variants.
Figure 5.
Clinically relevant somatic variants identified in the germ line carriers-with HM cohorts. Distribution of the clinically relevant somatic variants, driver somatic variants, identified in the carriers-with HM cohorts. From outside to inside: 1. Gene with the somatic variant. The length of the black bar indicates the frequency of variant within the germ line carriers-with HM cohort. 2. The age and sex of the individual with the somatic variant. Triangle = female, circle = male. Age groups are indicated by colors (green = child [≤14 years], orange = AYA [15-39 years], adult = pink [≥40 years], black = the age of the individual is unknown). 3. The type of HM is indicated by the color of the bar. 4. VAF of the somatic variant in the sample as represented on a sliding scale (darker = high VAF, lighter = low VAF). 5. The inner ring depicts the association of different somatic variants in the sample. The colored ribbon depicts a unique sample and the associated somatic variants observed in the sample. (A) Germ line RUNX1 carriers-with HM cohort. Only shown are the genes that are identified as somatically mutated in 2 or more individuals. (B) Germ line GATA2 carriers-with HM cohort, showing all driver somatic variants and (C) Germ line DDX41 carriers-with HM cohort showing all driver somatic variants. (D) Violin plot displaying the distribution of driver somatic variant VAFs observed in the germ line carriers-with HM cohorts. Boxes represent the 25th and 75th percentiles, with the horizontal line in the middle indicating the median, and the vertical lines representing the 95th percentile cohorts. ∗P < .05, 1-way analysis of variance of log-transformed values, with Tukey multiple comparison test. (E) TMB in germ line RUNX1 and DDX41 carriers-with HM cohorts. TMB is the number of SNV and INDELs divided by 38Mb coding region. Only malignancy samples where we had available a matched germ line control tissue were used for analysis. Boxes represent the 25th and 75th percentiles, with the horizontal line in the middle indicating the median, and the vertical lines representing the max and min values. ∗P < .05 nonparametric Mann-Whitney U test. AYA, adolescents and young adults; AML, acute myeloid leukemia; AL, acute leukemia; B-ALL, B-cell acute lymphoblastic leukemia; CML, chronic myeloid leukemia; CMML, chronic myelomonocytic leukemia; JMML, juvenile myelomonocytic leukemia; MPN, myeloproliferative neoplasms; T-ALL, T-cell acute lymphoblastic leukemia.
Figure 6.
Somatic variants in RUNX1 are the most common event in the germ line RUNX1 carriers-with HM cohort. (A) Plot of acquired somatic RUNX1 variants and associated germ line RUNX1 variants. Data points are colored according to the somatic and associated germ line variant observed in the patient. VAF of more than 60% indicates a copy neutral loss of heterozygosity (CNLOH) or Trisomy 21. (B) Somatic and germ line RUNX1 variants are visualized using the ProteinPaint web application.30 Variants are colored according to the somatic and associated germ line variant observed in the patient. The number of probands for each variant is indicated within the circle where the number is greater than 1. All variants are annotated to RUNX1c; NM_001754.4; LRG_ 482. (C) The proportion of male and females harboring a somatic RUNX1 variant is significantly different. ∗P < .05. (D) Sex and age distribution of individuals with a somatic RUNX1 variant; adult ≥ 40 years, AYA = 15 to 39 years, children ≤14 years. Data points are colored according to the somatic and associated germ line variant observed in the patient. AYA, adolescents and young adults.
No somatic second hits in GATA2 were detected in our cohort of 13 germ line GATA2 variant carriers (Figure 5B; supplemental Figure 3B). We detected at least 1 driver somatic variant in 69% (9 of 13) of germ line GATA2 variant carriers who had developed malignancies. Analysis of the GATA2 cohort was limited by low sample numbers (supplemental Figure 4A), but the lack of second hits in GATA2 suggests biallelic variants are not a common leukemogenic mechanism in germ line GATA2 variant carriers.37
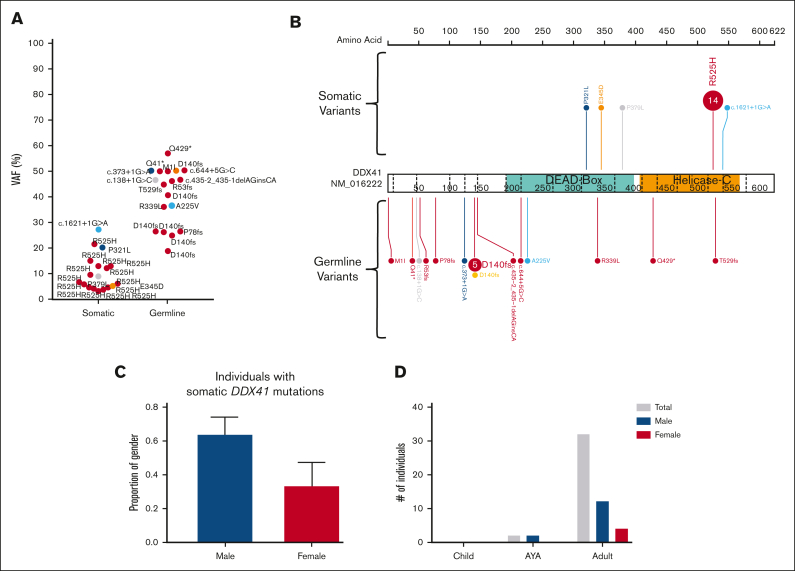
In the DDX41 carriers-with HM cohort, we identified at least 1 driver somatic variant in 10 unique genes in 18 individuals (62%, Figure 5C; supplemental Figure 3C). Only 3 genes were mutated in more than 1 individual (DDX41, ASXL1, and JAK2 p.Val617Phe). The most frequent somatic event was a second hit in DDX41, which was observed in 62% (n = 18) of individuals with HM. Apart from a single splice-site variant (c.1621+1G>A), all somatic DDX41 variants were missense variants in the DEAD-box domain (3 of 18) or the recurrent p.R525H variant in the helicase C domain (14 of 18 DDX41 somatic variants, 78%, Figure 7A-B). We observed a significant sex bias for DDX41 malignancies (3:1 male:female, P = .0002), which correlated with males presenting with a somatic DDX41 variant (14 of 18 males, Figure 7C-D). No association between specific somatic variants and germ line DDX41 variants, age of malignancy diagnosis, or histologic subtype of malignancy was observed.
Figure 7.
A somatic variant in DDX41 is the most common event in the germ line DDX41 carriers-with HM cohort. (A) Plot of acquired somatic DDX41 variants and associated germ line DDX41 variants. Data points are colored according to the somatic and associated germ line variant observed in the patient. (B) Somatic and germ line DDX41 variants are visualized using the ProteinPaint web application.30 Variants are colored according to the somatic variant and associated germ line variant observed in the patient. The number of probands for each variant is indicated within the circle where the number is greater than 1. All variants are annotated to DDX41; NM_016222.4; LRG_ 1386. (C) The proportion of male and females harboring a somatic DDX41 variant shows no significant difference. (D) Sex and age distribution of individuals with a somatic DDX41; adult ≥40 years, AYA = 15 to 39 years, children ≤14 years. AYA, adolescents and young adults.
Mutational burden in germ line RUNX1, GATA2, or DDX41 malignancy samples
To better understand the somatic mutational burden in each syndrome, we evaluated the VAF of all driver somatic variants. A large distribution of VAFs was observed in RUNX1 carriers-with HM (median VAF = 22.4%, mean = 27.0%, mode = 5.7, 34%) and GATA2 carriers-with HM (median = 21.0%, mean = 20.6%, mode = 8, 27.2%). VAFs among RUNX1 carriers-with HM showed the largest distribution (Figure 5D). The DDX41 cohort harbored low VAF driver somatic variants (median VAF = 8.9%, mean 13.4%, Figure 5D). No association between age at malignancy diagnosis and the VAF of driver somatic variants was observed in any cohort (supplemental Figure 5). TMB was calculated for DDX41 (n = 14) and RUNX1 (n = 4) carriers-with HM with matched germ line/tumor samples. DDX41 had a lower TMB (0.75 mutations/Mb) than RUNX1 malignancies (3.3 mutations/Mb; P = .01, Figure 5E).
Discussion
The prevalence of HHMs is estimated to range from 7% to 14% in cohorts of patients with myeloid malignancies.38,39 Although the clinical recognition of these syndromes has improved since RUNX1-driven HHMs were first described,8 questions remain regarding the optimal approach to monitoring carriers-without HM and how malignancy-directed treatments may be individualized for affected patients. Currently it is challenging for clinicians to provide tailored risk-assessment to patients as the natural history of carriers-without HM is not well understood and there has been no approach to identify HHM individuals at highest risk for leukemogenesis. To address this gap, we have leveraged our HHM international collaborative network and assembled and characterized the most extensive cross-sectional comparative cohort of carriers-without HM and carriers-with HM germ line RUNX1, GATA2, or DDX41 variants (n = 191, 102 probands, Figure 1). We demonstrate RUNX1, GATA2, and DDX41 germ line variant carriers experience highly variable risk for CH and unique somatic drivers during CH relative to population controls. Each HHM is remarkable for mutational profiles during frank leukemogenesis that are also unique to each HHM syndrome.
The most significant risk factor for CH in the general population is aging, with ∼10% of individuals over the age of 70 years having detectable CH.32,33 Several studies investigating CH in the background of inherited bone marrow failure have shown an increased risk for CH.34,40,41 Interestingly, individuals without HM with HHM germ line variants have been shown to have variable risk for CH in a series of small studies (ANKRD26, ETV6, RUNX1).18,19,42,43 We have now performed the largest collective analysis of CH in RUNX1, GATA2, and DDX41 carriers without HM to date. This analysis extends studies of CH in the HHMs to novel phenotypes (DDX41) and suggests that HHM predisposition in GATA2, and RUNX1 carriers-without HM, may be driven by early-onset CH (22.2% in RUNX1 and 25% in GATA2). Recently, larger cohorts of patients with germ line GATA2 without HM,20,21 have also shown CH is common in patients without HM, with CH associated with a hypocellular marrow. Further investigation is required to determine if CH also correlates with cytopenias in germ line RUNX1 cohorts. In contrast, DDX41 carriers-without HM have a very low risk for CH at any age. RUNX1 patients (without HM) with CH also had unique somatic drivers relative to CH population controls, most notably a high prevalence of BCOR variants.32,33 This has similarity to aplastic anemia, where BCOR and BCORL1 are frequently mutated.44 BCOR variants alone did not appear sufficient to cause leukemogenesis in our cohort. This suggests additional co-operating variants are required for malignancy progression (including somatic RUNX1, TET2, DNMT3A, and BCORL1 variants [supplemental Figure 3A]). Some of these interactions are validated in in vivo models with conditional Bcor knockout mouse models combined with variants in Dnmt3a, Kras, or Tet2 sufficient to drive malignancy transformation.45, 46, 47 Notably, BCOR variants are frequent in the RUNX1 carriers-with HM cohort, supporting the notion that CH in this setting, is a risk factor for leukemic transformation. However further models are required to determine the functional effects of co-occurring BCOR and RUNX1 variants on hematopoetic stem and progenitor cell (HSPC) fitness and leukemic transformation.
The most frequent leukemogenic event in our cohort of DDX41 and RUNX1 carriers-with HM was biallelic somatic variants in DDX41 and RUNX1, respectively (supplemental Figure 6). In contrast, second hits in GATA2 germ line variant carriers with malignancies were not detected. In case study #3, for example, a germ line RUNX1 carrier initially presented with thrombocytopenia before progressing to AML. In this patient, leukemogenesis was associated with the acquisition of a biallelic RUNX1 variant, but at a late stage after the acquisition of TET2, DNMT3A, and SRSF2 variants. Given that we never detected somatic RUNX1 variants in our RUNX1 carriers-without HM cohort, in stark contrast to the high frequency of second-hit RUNX1 variants in our HM cohort, we suggest that somatic RUNX1 variants likely represent a later step that may be key to leukemogenic transformation. Interestingly, for DDX41, the lack of CH gene mutations in carriers was mirrored by a lack of CH gene mutations in malignancy (Figure 5C). This indicates that the molecular natural history of this disorder is quite different from both RUNX1 and GATA2 HHMs. Further longitudinal, lineage tracing, and single-cell sequencing studies are required to determine if these are initiating events in malignancy development and the timeline to disease progression.
Interestingly, both germ line RUNX1 and DDX41 cohorts presented with a sex bias for HM development, but this did not correlate with differences in X-linked somatic variants. Sex bias was not observed in our GATA2 cohort, as we have also observed previously.14 RUNX1 genomic alterations have a high correlation with hormone-related cancers, especially cancers common in female patients, and with estrogen known to play a role in hematopoiesis,48,49 we hypothesize that disruption of specific estrogen signaling pathways in germ line RUNX1 carriers could predispose females to AML.50, 51, 52, 53 In the germ line RUNX1 malignancy cohort, recurrent somatic gene variants are involved in epigenetic regulation and epigenetic dysregulation and can occur in leukemogenesis, with sex-specific differences in methylation observed in hematopoietic tissue.50,54,55 The innate immune response is also known to be increased in females relative to males.56 For DDX41, given its role as an intracellular pattern recognition receptor that triggers the innate immune response,57 a dysregulated immune response could exaggerate existing differences in innate immunity between males and females, contributing to the observed sex bias in malignancy penetrance. Further investigation is warranted to understand the interplay of these mechanisms on tumorigenesis, which may ultimately inform the development of sex-specific therapies that optimize outcomes for patients with HHM.
Despite a lack of definitive guidelines, limitations, and ongoing debate, molecular monitoring in clinical practice is becoming more widespread.43 Findings from this study have implications for clinical surveillance and counseling for different patients with HHM. For example, in RUNX1 and GATA2 HHMs, regular targeted sequencing of CH genes, even in younger carriers-without HM, will provide a tool to monitor the evolution of the clonal burden associated with these variants. In contrast, given the low VAF and high frequency of somatic DDX41 variants in DDX41 HHMs, serial high-depth sequencing of DDX41 for the common R525H mutation may be a preferred approach in DDX41 carriers-without HM. Although in the aging population, CH is a risk factor for leukemic transformation, the presence of CH in inherited bone marrow failure is in some situations associated with somatic rescue or normalization of HSPC fitness. Therefore, it is important to discriminate CH events that are associated with risk for leukemic transformation from CH, which results in normalization of function.58 Given that CH variants feasibly confer a step toward HM,36 the high frequency of BCOR and TET2 variants in our cohort of RUNX1 HHM malignancies and their presence in RUNX1 carriers-without HM, at least in the research study setting, warrant monitoring of these genes as potential molecular biomarkers of leukemogenesis. Changes in CH trajectory may eventually inform clinical decision-making, such as the timing of repeat bone marrow biopsies. These decisions will be made in conjunction with more classic clinical tools, such as the monitoring of peripheral blood cell counts.43
This study highlights the immense benefit of international collaboration and data sharing within the HHM and rare disease communities. We have established the framework for the continued accumulation of patient data, including longitudinal molecular monitoring, which is required to define the different risk states associated with leukemogenesis across these disorders. With continued progress, this work may lead to the establishment of a defined molecular risk stratification for leukemia progression in carriers and, with it, the ability to design and test in trials interventions to halt progression to full-blown HM in vulnerable HHM-variant carriers. For instance, with regular clinical surveillance, it may be possible to detect individuals who develop second hits in DDX41 or RUNX1 before a clinical diagnosis of HM. These individuals may benefit from intensive clinical surveillance or low-toxicity prophylactic therapies. In contrast, defining TET2, BCOR, or other epigenetic regulators as emerging vulnerabilities opens an avenue for the development of prophylactic treatments for HHM carriers via TET inhibitors, histone deacetylases (HDAC) inhibitors, hypomethylating agents, and combinatorial therapies that do not carry the morbidity and mortality of stem cell transplant. This study provides the most comprehensive investigation of leukemogenic molecular mechanisms in HHMs to date, informing the next generation of studies into the clinical management and surveillance of these disorders as well as potential insights into personalized and preemptive therapies for carriers.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the NIH Intramural Sequencing Center Comparative Sequencing Program appears in “Appendix.”
Acknowledgments
The authors thank the patients and their family members for participating in this research program.
This work is supported by a grant from the RUNX1 Research Program. This project is also proudly supported by funding from the Leukemia Foundation of Australia and project grants APP1145278 and APP1164601 from the National Health and Medical Research Council of Australia. This work was produced with the financial and other support of Cancer Council SA's Beat Cancer Project on behalf of its donors and the State Government of South Australia through the Department of Health (PRF Fellowship to H.S.S.). This work was supported by a Damon Runyon Cancer Research Foundation Physician Scientist Training Award (M.W.D.), the Edward P. Evans Foundation Young Investigator Award (M.W.D.), the Cancer Research Foundation (M.W.D.), and a National Institutes of Health (NIH) K12 Paul Calabresi award (M.W.D.). P.A. was supported by a fellowship from The Hospital Research Foundation. Part of this project was undertaken while P.A. was holding a Royal Adelaide Hospital Mary Overton Early Career Fellowship. L.M is supported by the Associazione Italiana per la Ricerca sul Cancro (Accelerator Award Project 22796; 5x1000 Project 21267; Investigator Grant 2017 Project 20125). L.C.F and P.V. are supported by Maddie Riewoldt’s Vision. L.A.G was supported by the Cancer Research Foundation. K.Y. and P.L. are supported by the Division of Intramural Research, National Human Genome Research Institute, NIH. T.R. is supported by a grant of the European Hematology Association and Federal Ministry of Education and Research (BMBF) MyPred (01GM1911B). C.B. is supported by the European Union’s Horizon 2020 Research and Innovation Program under grant agreement number 739593 and by the Ministry of Innovation and Technology of Hungary from the National Research, Development and Innovation Fund, financed under the ED-18-1-2019 to 001, TKP2021-EGA-24 and TKP2021-NVA-15 funding schemes.
NIH Intramural Sequencing Center Comparative Sequencing Program was involved in the generation of sequencing data from the NIH.
Authorship
Contribution: C.C.H., M.W.D., and K.Y. were involved in all aspects of the project including designing the research, manuscript preparation, collecting next-generation sequencing (NGS) and clinical data, American College of Medical Genetics and Genomics (ACMG)-variant classification, somatic-variant analysis, and curation and analysis of the data; A.L.B., L.A.G., P.L., and H.S.S. designed the research, contributed NGS and clinical data, manuscript preparation, and ACMG-variant classification; J.F., L.A.-M., M.J.P., K.E.M., T.H, M.A., P.W., A.W.S., E.K., and R. Sood designed bioinformatic pipelines and analysis; D.M.L. designed VariantGrid software used for somatic and germ line variant curation (VariantGrid); P.V., P.A., S.L.K.-S., J.C., and C.N.H. curated somatic and germ line variant data; B.P. advised on statistical analysis; C.B., A.B.C., M.C., E.D., C.D.D., N.D., R.F., S.F., A.R.-M., B.P., J.M.K., A.K., M.K., J.L., N.V.M., G.N., C.O., K.P.P., C.P., H. Raslova., H. Rienhoff., T.R., R. Susman., K.T., E.V., E.K., R. Schulte., A.P.H., S.M.H., K.P., N.K.P., M.B., A.H.W., C.F., H.M.F., I.D.L., J.C., R. Sood, L.C.F., P.B., D.S., D.H., B.Y., L.M., A.L.B., and C.N.H. contributed NGS data, clinical patient information, and scientific insight; and all authors critically reviewed and approved the manuscript.
Footnotes
∗C.C.H., M.W.D., and K.Y. contributed equally to this manuscript.
Access to RUNX1 genomics data is available through the RUNX1 database (https://runx1db.runx1-fpd.org/). Original data may be obtained by email request to the corresponding author, Anna L. Brown (anna.brown@sa.gov.au). Access to additional deidentified genomics data is available on request.
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.Tawana K, Brown AL, Churpek JE. Integrating germline variant assessment into routine clinical practice for myelodysplastic syndrome and acute myeloid leukaemia: current strategies and challenges. Br J Haematol. 2022;196(6):1293–1310. doi: 10.1111/bjh.17855. [DOI] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 3.Feurstein S, Drazer MW, Godley LA. Genetic predisposition to leukemia and other hematologic malignancies. Semin Oncol. 2016;43(5):598–608. doi: 10.1053/j.seminoncol.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Polprasert C, Schulze I, Sekeres MA, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27(5):658–670. doi: 10.1016/j.ccell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abou Dalle I, Kantarjian H, Bannon SA, et al. Successful lenalidomide treatment in high risk myelodysplastic syndrome with germline DDX41 mutation. Am J Hematol. 2020;95(2):227–229. doi: 10.1002/ajh.25610. [DOI] [PubMed] [Google Scholar]
- 6.Parta M, Shah NN, Baird K, et al. Allogeneic hematopoietic stem cell transplantation for GATA2 deficiency using a busulfan-based regimen. Biol Blood Marrow Transplant. 2018;24(6):1250–1259. doi: 10.1016/j.bbmt.2018.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godley LA, Shimamura A. Genetic predisposition to hematologic malignancies: management and surveillance. Blood. 2017;130(4):424–432. doi: 10.1182/blood-2017-02-735290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song WJ, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23(2):166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 9.Lewinsohn M, Brown AL, Weinel LM, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127(8):1017–1023. doi: 10.1182/blood-2015-10-676098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collin M, Dickinson R, Bigley V. Haematopoietic and immune defects associated with GATA2 mutation. Br J Haematol. 2015;169(2):173–187. doi: 10.1111/bjh.13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan Z, Han B. Clinical features of DDX41 mutation-related diseases: a systematic review with individual patient data. Ther Adv Hematol. 2021;12 doi: 10.1177/20406207211032433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drazer MW, Kadri S, Sukhanova M, et al. Prognostic tumor sequencing panels frequently identify germ line variants associated with hereditary hematopoietic malignancies. Blood Adv. 2018;2(2):146–150. doi: 10.1182/bloodadvances.2017013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown AL, Arts P, Carmichael CL, et al. RUNX1-mutated families show phenotype heterogeneity and a somatic mutation profile unique to germline predisposed AML. Blood Adv. 2020;4(6):1131–1144. doi: 10.1182/bloodadvances.2019000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homan CC, Venugopal P, Arts P, et al. GATA2 deficiency syndrome: a decade of discovery. Hum Mutat. 2021;42(11):1399–1421. doi: 10.1002/humu.24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sébert M, Passet M, Raimbault A, et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood. 2019;134(17):1441–1444. doi: 10.1182/blood.2019000909. [DOI] [PubMed] [Google Scholar]
- 16.Cheah JJC, Hahn CN, Hiwase DK, Scott HS, Brown AL. Myeloid neoplasms with germline DDX41 mutation. Int J Hematol. 2017;106(2):163–174. doi: 10.1007/s12185-017-2260-y. [DOI] [PubMed] [Google Scholar]
- 17.Brown AL, Hahn CN, Scott HS. Secondary leukemia in patients with germline transcription factor mutations (RUNX1, GATA2, CEBPA) Blood. 2020;136(1):24–35. doi: 10.1182/blood.2019000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiFilippo EC, Coltro G, Carr RM, et al. Spectrum of abnormalities and clonal transformation in germline RUNX1 familial platelet disorder and a genomic comparative analysis with somatic RUNX1 mutations in MDS/MPN overlap neoplasms. Leukemia. 2020;34(9):2519–2524. doi: 10.1038/s41375-020-0752-x. [DOI] [PubMed] [Google Scholar]
- 19.Churpek JE, Pyrtel K, Kanchi KL, et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood. 2015;126(22):2484–2490. doi: 10.1182/blood-2015-04-641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West RR, Calvo KR, Embree LJ, et al. ASXL1 and STAG2 are common mutations in GATA2 deficiency patients with bone marrow disease and myelodysplastic syndrome. Blood Adv. 2022;6(3):793–807. doi: 10.1182/bloodadvances.2021005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Largeaud L, Collin M, Monselet N, et al. Somatic genetic alterations predict haematological progression in GATA2 deficiency. Haematologica. 2023;108(6):1515–1529. doi: 10.3324/haematol.2022.282250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homan CC, King-Smith SL, Lawrence DM, et al. The RUNX1 Database (RUNX1db): establishment of an expert curated RUNX1 registry and genomics database as a public resource for familial platelet disorder with myeloid malignancy. Haematologica. 2021;106(11):3004–3007. doi: 10.3324/haematol.2021.278762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 Preprint posted online 16 March. [Google Scholar]
- 24.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31(12):2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012 Preprint posted online 17 July. [Google Scholar]
- 26.Christoforides A, Carpten JD, Weiss GJ, Demeure MJ, Von Hoff DD, Craig DW. Identification of somatic mutations in cancer through Bayesian-based analysis of sequenced genome pairs. BMC Genom. 2013;14:302. doi: 10.1186/1471-2164-14-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen NF, Gartner JJ, Mei L, Samuels Y, Mullikin JC. Shimmer: detection of genetic alterations in tumors using next-generation sequence data. Bioinformatics. 2013;29(12):1498–1503. doi: 10.1093/bioinformatics/btt183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28(14):1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 29.Larson DE, Harris CC, Chen K, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28(3):311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X, Edmonson MN, Wilkinson MR, et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat Genet. 2016;48(1):4–6. doi: 10.1038/ng.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y, Ouyang Y, Yao W. shinyCircos: an R/Shiny application for interactive creation of Circos plot. Bioinformatics. 2018;34(7):1229–1231. doi: 10.1093/bioinformatics/btx763. [DOI] [PubMed] [Google Scholar]
- 32.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy AL, Myers KC, Bowman J, et al. Distinct genetic pathways define pre-malignant versus compensatory clonal hematopoiesis in Shwachman-Diamond syndrome. Nat Commun. 2021;12(1):1334. doi: 10.1038/s41467-021-21588-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400–404. doi: 10.1038/s41586-018-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desai P, Mencia-Trinchant N, Savenkov O, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med. 2018;24(7):1015–1023. doi: 10.1038/s41591-018-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armes H, Rio-Machin A, Krizsán S, et al. Acquired somatic variants in inherited myeloid malignancies. Leukemia. 2022;36(5):1377–1381. doi: 10.1038/s41375-022-01515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F, Long N, Anekpuritanang T, et al. Identification and prioritization of myeloid malignancy germline variants in a large cohort of adult patients with AML. Blood. 2022;139(8):1208–1221. doi: 10.1182/blood.2021011354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feurstein SK, Trottier AM, Estrada-Merly N, et al. Germline predisposition variants occur in myelodysplastic syndrome patients of all ages. Blood. 2022;140(24):2533–2548. doi: 10.1182/blood.2022015790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahoo SS, Pastor VB, Goodings C, et al. Clinical evolution, genetic landscape and trajectories of clonal hematopoiesis in SAMD9/SAMD9L syndromes. Nat Med. 2021;27(10):1806–1817. doi: 10.1038/s41591-021-01511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perdigones N, Perin JC, Schiano I, et al. Clonal hematopoiesis in patients with dyskeratosis congenita. Am J Hematol. 2016;91(12):1227–1233. doi: 10.1002/ajh.24552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drazer MW, Homan CC, Yu K, et al. Clonal hematopoiesis in patients with ANKRD26 or ETV6 germline mutations. Blood Adv. 2022;6(15):4357–4359. doi: 10.1182/bloodadvances.2022007211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Homan CC, Scott HS, Brown AL. Hereditary platelet disorders associated with germline variants in RUNX1, ETV6 and ANKRD26. Blood. 2023;141(13):1533–1543. doi: 10.1182/blood.2022017735. [DOI] [PubMed] [Google Scholar]
- 44.Yoshizato T, Dumitriu B, Hosokawa K, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 2015;373(1):35–47. doi: 10.1056/NEJMoa1414799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tara S, Isshiki Y, Nakajima-Takagi Y, et al. Bcor insufficiency promotes initiation and progression of myelodysplastic syndrome. Blood. 2018;132(23):2470–2483. doi: 10.1182/blood-2018-01-827964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly MJ, So J, Rogers AJ, et al. Bcor loss perturbs myeloid differentiation and promotes leukaemogenesis. Nat Commun. 2019;10(1):1347. doi: 10.1038/s41467-019-09250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sportoletti P, Sorcini D, Guzman AG, et al. Bcor deficiency perturbs erythro-megakaryopoiesis and cooperates with Dnmt3a loss in acute erythroid leukemia onset in mice. Leukemia. 2021;35(7):1949–1963. doi: 10.1038/s41375-020-01075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakada D, Oguro H, Levi BP, et al. Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. Nature. 2014;505(7484):555–558. doi: 10.1038/nature12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heo H-R, Chen L, An B, Kim KS, Ji J, Hong SH. Hormonal regulation of hematopoietic stem cells and their niche: a focus on estrogen. Int J Stem Cells. 2015;8(1):18–23. doi: 10.15283/ijsc.2015.8.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sánchez-Aguilera A, Arranz L, Martín-Pérez D, et al. Estrogen signaling selectively induces apoptosis of hematopoietic progenitors and myeloid neoplasms without harming steady-state hematopoiesis. Cell Stem Cell. 2014;15(6):791–804. doi: 10.1016/j.stem.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 51.Riggio AI, Blyth K. The enigmatic role of RUNX1 in female-related cancers - current knowledge & future perspectives. FEBS J. 2017;284(15):2345–2362. doi: 10.1111/febs.14059. [DOI] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chimge N-O, Ahmed-Alnassar S, Frenkel B. Relationship between RUNX1 and AXIN1 in ER-negative versus ER-positive breast cancer. Cell Cycle. 2017;16(4):312–318. doi: 10.1080/15384101.2016.1237325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu D, Shilatifard A. Epigenetics of hematopoiesis and hematological malignancies. Genes Dev. 2016;30(18):2021–2041. doi: 10.1101/gad.284109.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singmann P, Shem-Tov D, Wahl S, et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenet Chromatin. 2015;8:43. doi: 10.1186/s13072-015-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghosh S, Klein RS. Sex drives dimorphic immune responses to viral infections. J Immunol. 2017;198(5):1782–1790. doi: 10.4049/jimmunol.1601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Omura H, Oikawa D, Nakane T, et al. Structural and functional analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide. Sci Rep. 2016;6:34756. doi: 10.1038/srep34756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai FD, Lindsley RC. Clonal hematopoiesis in the inherited bone marrow failure syndromes. Blood. 2020;136(14):1615–1622. doi: 10.1182/blood.2019000990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.