Abstract
Introduction
Neonatal mortality remains a critical concern, particularly in developing countries. The advent of machine learning offers a promising avenue for predicting the survival of at-risk neonates. Further research is required to effectively deploy this approach within distinct clinical contexts. Objective: This study aimed to assess the applicability of machine learning models in predicting neonatal mortality, drawing from maternal and clinical characteristics of pregnant women within an intensive care unit (ICU).
Methods
Conducted as an observational cross-sectional study, the research enrolled pregnant women receiving care in a level III national hospital's ICU in Peru. Detailed data encompassing maternal diagnosis, maternal characteristics, obstetric characteristics, and newborn outcomes (survival or demise) were meticulously collected. Employing machine learning, predictive models were developed for neonatal mortality. Estimations of beta coefficients in the training dataset informed the model application to the validation dataset.
Results
A cohort of 280 pregnant women in the ICU were included in this study. The Gradient Boosting approach was selected following rigorous experimentation with diverse model types due to its superior F1-score, ROC curve performance, computational efficiency, and learning rate. The final model incorporated variables deemed pertinent to its efficacy, including gestational age, eclampsia, kidney infection, maternal age, previous placenta complications accompanied by hemorrhage, severe preeclampsia, number of prenatal checkups, and history of miscarriages. By incorporating optimized hyperparameter values, the model exhibited an impressive area under the curve (AUC) of 0.98 (95 % CI: 0.95–1), along with a sensitivity of 0.98 (95 % CI: 0.94–1) and specificity of 0.98 (95 % CI: 0.93–1).
Conclusion
The findings underscore the utility of machine learning models, specifically Gradient Boosting, in foreseeing neonatal mortality among pregnant women admitted to the ICU, even when confronted with maternal morbidities. This insight can enhance clinical decision-making and ultimately reduce neonatal mortality rates.
Keywords: Artificial intelligence, Machine learning, Pregnancy complication, Intensive care units, Neonate, Mortality
1. Introduction
Neonatal mortality remains a critical health indicator worldwide [1], particularly in developing countries, constituting 60 % of infant deaths [2]. These disparities are evident in figures such as the estimated neonatal mortality rate of approximately 27 per thousand births in low-income countries [3,4], compared to Peru's rate of 12 per thousand births [5,6]. In contrast, developed nations boast a neonatal mortality rate as low as 3 per thousand live births [3,4].
Although neonatal mortality in Peru has remained stable over a decade [7], its proportional increase in infant mortality is a concerning trend [8,9]. The causes of neonatal deaths span socioeconomic and biological factors, with complications like preterm birth, birth asphyxia/trauma, congenital anomalies, and neonatal sepsis prevailing [10,11].
Although scales and predictive models have been used for neonatal mortality prediction [12,13], artificial intelligence concepts, specifically machine learning, have recently enhanced healthcare practices [14]. Machine learning models can discern patterns in data without human intervention, imitation, or explicit programming [15]. Predictive models provide specific predictions through machine learning and data collection, thus enhancing decision-making performance [16]. These models find utility across various scenarios, including diagnostic assessments, dosage standardization, clinical-laboratory value control, and case-based reasoning to inform appropriate decision-making [15,17].
The application of machine learning extends across various healthcare domains. Notably, in cardiology, it has demonstrated remarkable utility in predicting conditions like cardiac arrhythmias and heart failure [18]. Similarly, machine learning has propelled advancements in prognosticating metabolic, endocrine, and cancer-related disorders [14]. Within the field of communicable diseases, it has revolutionized tuberculosis diagnosis, while other conditions have benefited from convolutional neural networks [19].In neonatal mortality prediction, the application of models varies depending on the specific timing concerning pregnant women and newborns [[20], [21], [22]].
A prior investigation introduced a predictive machine learning model for neonatal death, utilizing secondary population data. This model incorporated XGBoost and ADASYN as learning systems and identified variables like maternal age, race, gestational age, support during childbirth, and newborn condition as key risk factors [14]. Similarly, another study identified predictors of neonatal mortality, including marital status, number of live-born children, birth order, and wealth index [23].
The ability to forecast neonatal mortality early can enhance survival rates and reduce morbidity [12]. Consequently, there is a demand for models with improved detection capabilities, prompting the application of machine-learning techniques in medical contexts [18,19,24]. However, it is noteworthy that the performance may vary depending on the specific clinical scenario in which they are deployed [21,22].
This study aimed to evaluate the effectiveness of machine learning models in predicting neonatal death by leveraging maternal and clinical attributes of pregnant women within an intensive care unit.
2. Materials and methods
2.1. Study design
A cross-sectional and retrospective observational study design was employed.
2.2. Sample
The study included pregnant women admitted to an intensive care unit and their respective newborns who attended the National Maternal Perinatal Institute between 2017 and 2019. Inclusion criteria comprised pregnant women admitted to the intensive care unit (ICU), those receiving childbirth care at the same institution, and pregnant women with live newborns at delivery. Exclusion criteria encompassed pregnant women who passed away before delivery, those with fetal malformations, those with a gestational age at delivery under 21 weeks or a neonatal weight of 500 g or less, and those lacking pertinent variables in their medical history records.
2.3. Sample size and sampling
As per Sen and Cohen [25], when dealing with machine-learning models, sample sizes exceeding 120 observations might result in marginal changes in the effect size estimation, ranging from 85 % to 99 %. Consequently, the current research incorporated a sample of 280 observations. Determining this sample size involved estimating proportions for a population with no defined endpoint, factoring in a neonatal mortality rate of 35 % [26]. This was done with a 95 % confidence level and an error of 0.055. Employing a simple random sampling method, these 280 observations were culled from the tangible records of medical histories pertaining to pregnant women and their infants from January 2017 to December 2019.
2.4. Data collection
Data extracted from clinical records encompassed various parameters, including maternal age, gestational age upon admission to the maternal intensive care unit, count of prior pregnancies, history of miscarriages, number of prenatal check-ups, maternal comorbidities (pre-existing health conditions predating pregnancy, like hypertension, cancer, stroke, epilepsy, or ailments affecting other organs), obstetric morbidity (disorders diagnosed in pregnant individuals upon ICU admission, such as severe preeclampsia, eclampsia, HELLP syndrome, chorioamnionitis, maternal sepsis, hyperemesis gravidarum, pneumonia, previous placental issues, placental accreta, placental abruption, along with other documented pathologies), and the well-being of the newborn (survival status until hospital discharge or within a follow-up duration of up to 28 days, whichever came first).
2.5. Procedures for information collection
Identification of pregnant women in the ICU via the electronic hospitalization database from 2017 to 2019. Medical records of admitted pregnant women were then reviewed, employing simple random sampling through Stata version 16. From maternal medical records, data on their newborns were extracted to ascertain newborn survival or mortality. The process continued until 280 pregnant women admitted to the ICU were enrolled.
The variables indicated in this study were collected using a data form specifically designed for this purpose. This form meticulously documented the presence, absence, or instances of missing data for each of the variables encompassed in the study. Through this instrument, comprehensive information about each expectant mother was amassed, alongside the outcomes of survival or mortality of their newborns. In total, 23 variables were meticulously recorded, encompassing maternal age, count of previous pregnancies, instances of prior miscarriages, number of living children, frequency of prenatal check-ups, gestational age upon admission to the ICU, presence of arterial hypertension, chronic hypertension compounded by preeclampsia, occurrences of cancer, cirrhosis, history of stroke, severe preeclampsia, eclampsia, HELLP syndrome, kidney infection, chorioamnionitis, hyperemesis gravidarum in conjunction with metabolic disorders, pneumonia, placenta accreta, placenta previa accompanied by bleeding, and untimely placental abruption associated with coagulation abnormalities. Notably, neonatal mortality served as the dependent variable for analysis.
2.6. Preparation of the database and development of machine learning models
This study used the feature-importance method to estimate and evaluate multiple machine learning models coupled with variable selection. The schematic depiction of the employed methodology is presented in Fig. 1.
Fig. 1.
Description of techniques applied in the machine learning modeling process.
SVC: support-vector machines.
Source: elaborated by the authors
The dataset was collected and organized in Excel format (xlsx), serving as the basis for subsequent processing on the accessible Google Colab platform. This platform harnessed the capabilities of the Python 3.7 programming language through Jupyter Notebooks.
Addressing the inherent imbalance in the target variable (with a distribution of 7.86 % within the deceased class), a common practice involved the application of oversampling techniques to enhance the reliability of outcomes. To this end, a synthetic minority oversampling technique (SMOTE) was implemented using Python 3.7. This approach entailed maintaining the class with a larger data representation while augmenting the class with fewer instances, thereby attaining a harmonized portrayal of both data categories. The generated data was produced with a random seed of 17 and 5 nearest neighbors, culminating in a balanced dataset where both categories were equally represented at approximately 50 % (approximately 236 records associated with deceased cases).
Two distinct datasets were formulated: the training set and the validation set. An 80 % random stratified sampling strategy was adopted for the training set, grounded in the dependent variable (neonatal mortality). The remaining 20 % was channeled into the validation set. In the pursuit of constructing machine learning models, predictor variables were enlisted as inputs to predict a range of models.
In machine learning algorithms, this study engaged logistic regression, decision trees, support vector machines, and gradient boosting. Each of these models was computed using both the original and oversampled datasets. Furthermore, all models underwent execution through a 10-fold cross-validation methodology to ensure robustness and unbiased assessment.
2.7. Data analysis plan
Categorical variables were described using frequencies and relative proportions, while numerical variables were characterized using mean, median, standard deviation, and range measures. Comparative analyses employed the Student's t-test and Fisher's exact test, with significance set at 0.05. Machine learning techniques were utilized to build neonatal death prediction models for the training dataset. Machine learning algorithms were developed using logistic regression, decision trees, gradient boosting, and support vector machine options based on the training dataset.
Given the nature of the classification task, the limited occurrence of events (neonatal death) alongside a higher frequency of non-events prompted the adoption of the F1-Score metric for evaluating the diverse machine-learning models. Ranging between 0 and 1, the F1-Score was leveraged to determine the algorithm that exhibited the value closest to 1, reflecting optimal performance.
Upon assessing the F1-Score values, the Gradient Boosting model emerged as the preferred choice among algorithms. Its selection was primarily attributed to including a learning rate parameter, facilitating iterative error correction during model estimation, thus mitigating errors.
After model selection, the process of hyperparameter tuning ensued. Hyperparameters, characterized by their arbitrary nature, are pivotal in enhancing predictive capabilities. Different values were trialed for the learning rate (0.1, 0.3, 0.5, and 0.7) and the maximum depth of the model (2, 3, 4 and 5). The optimal configuration materialized as a model featuring a learning rate 0.3 and a maximum depth of 3.
In pursuit of extracting pertinent predictor variables and identifying the minimal count of predictors within the model, the significance of each predictor was computed through the interaction computation weights inherent in the employed machine learning algorithm.
The constructed models underwent comprehensive evaluation, emphasizing discerning the model boasting the highest area under the curve, neonatal death prediction accuracy, sensitivity, specificity, and predictive values. The model achieving the highest neonatal death prediction accuracy was subsequently deployed on the validation set to gauge its performance and juxtapose the indicators against the training dataset outcomes.
To ascertain the robustness of the machine learning algorithm, a thousand iterations of new model estimations were executed through randomization. This iterative process assessed the consistency of the F1-Score and the accuracy of neonatal death prediction across the machine learning algorithm's iterations.
2.8. Ethical considerations
The study received approval from the Institutional Ethics Committee of the National Maternal Perinatal Institute of Peru. The study adhered to CIOMS ethical guidelines to safeguard participant data. Confidentiality was ensured through alphanumeric coding during data storage and processing, with the collected information used exclusively for the study.
3. Results
A total of 280 pregnant women admitted to the ICU and their neonates were included in this study. The mean maternal age was 28.2 ± 7.2 years, and the mean gestational age was 33.5 ± 4.2 weeks. The most prevalent maternal morbidities were severe preeclampsia (68.2 %), kidney infection (12.1 %), HELLP syndrome (10.7 %), and pneumonia (10.4 %). The neonatal mortality rate was 7.9 % (Table 1).
Table 1.
Maternal characteristics of pregnant women admitted to the ICU.
| Maternal Characteristics (N = 280) | n (%) |
|---|---|
| Maternal age (mean, SD) | 28.2±7.2 |
| Previous pregnancies (median, range) | 1 (0–8) |
| Previous miscarriages (median, range) | 0 (0–4) |
| Number of living children (median, range) | 1 (0–5) |
| Number of prenatal check-ups (mean, SD) | 4.2±3.2 |
| Gestational age (mean, SD) | 33.5±4.2 |
| Arterial hypertension | 18 (6.43 %) |
| Cancer | 1 (0.36 %) |
| Cirrhosis | 2 (0.71 %) |
| Epilepsy | 1 (0.36 %) |
| Preeclampsia with severity criteria | 191 (68.21 %) |
| HELLP | 30 (10.71 %) |
| Kidney infection | 34 (12.14 %) |
| Eclampsia | 14 (5.00 %) |
| Chorioamnionitis | 13 (4.64 %) |
| Hyperemesis gravidarum | 1 (0.36 %) |
| Pneumonia | 29 (10.36 %) |
| Chronic hypertension + PE | 10 (3.57 %) |
| Placental accreta | 15 (5.36 %) |
| Placenta previa | 10 (3.57 %) |
| Placental abruption | 4 (1.43 %) |
| Deceased newborn | 22 (7.86 %) |
HELLP syndrome: syndrome of hemolysis, elevated liver enzymes, and low platelets.
PE: Preeclampsia.
Source: elaborated by the authors based on the data of the study.
The sample was split into training (80 %) and validation (20 %) sets using simple random sampling. Predictive models were analyzed using the training set to obtain coefficients for subsequent comparison with the validation set.
Due to class imbalance, the SMOTE technique was applied to the validation set. Various models were evaluated based on F1-Score and receiver operating characteristic (ROC) curve values. Predictive models based on original data exhibited higher specificity, sensitivity, and F1-Score values but had a lower ROC curve area. Conversely, predictive models using data with SMOTE showed improved ROC curve performance owing to the handling of imbalanced data (Table 2).
Table 2.
Predictive modeling results using validation data.
| Model | Data | Accuracy | Precision | Specificity | Sensitivity | F1-score | AUC |
|---|---|---|---|---|---|---|---|
| Logistic Regression | Original | 0.95 | 1 | 1 | 0.25 | 0.93 | 0.62 |
| SMOTE | 0.9 | 0.86 | 0.85 | 0.96 | 0.9 | 0.9 | |
| Decision Tree Classifier | Original | 0.88 | 0.29 | 0.9 | 0.5 | 0.89 | 0.7 |
| SMOTE | 0.93 | 0.89 | 0.88 | 0.98 | 0.93 | 0.93 | |
| SVC | Original | 0.93 | 0 | 1 | 0 | 0.89 | 0.5 |
| SMOTE | 0.88 | 0.82 | 0.79 | 0.96 | 0.87 | 0.88 | |
| Gradient Boosting Classifier | Original | 0.93 | 0.5 | 0.96 | 0.5 | 0.93 | 0.73 |
| SMOTE | 0.93 | 0.91 | 0.9 | 0.96 | 0.93 | 0.93 | |
| Ensemble | Original | 0.95 | 0.67 | 0.98 | 0.5 | 0.94 | 0.74 |
| SMOTE | 0.92 | 0.88 | 0.87 | 0.98 | 0.92 | 0.92 |
AUC: Area Under the Curve of the Receiver Operating Characteristic curve.
SVC: support-vector machines.
Source: elaborated by the authors based on the data of the study
Gradient Boosting and Ensemble demonstrated the best F1-Score and ROC curve performance among various models in the validation set. Given minimal differences and computational efficiency, Gradient Boosting was selected. This model incorporates a learning rate parameter for error correction between iterations when estimating successive models to reduce errors in the next iteration (Table 2).
Comparing ROC curves among models highlighted the effectiveness of SMOTE in enhancing the area under the ROC curve for all models. The ROC curve areas were as follows: Logistic Regression 0.9 (95 % CI: 0.84–0.96), Decision Tree 0.93 (95 % CI: 0.88–0.98), Support Vector Machine (SVM) 0.88 (95 % CI: 0.81–0.95), Gradient Boosting 0.93 (95 % CI: 0.88–0.98), and Ensemble 0.92 (95 % CI: 0.86–0.98) (Fig. 2).
Fig. 2.
Comparison of the area under the curve of predictive models using the SMOTE data technique
AUC: Area Under the Curve of the Receiver Operating Characteristic curve.
Source: elaborated by the authors based on the data of the study
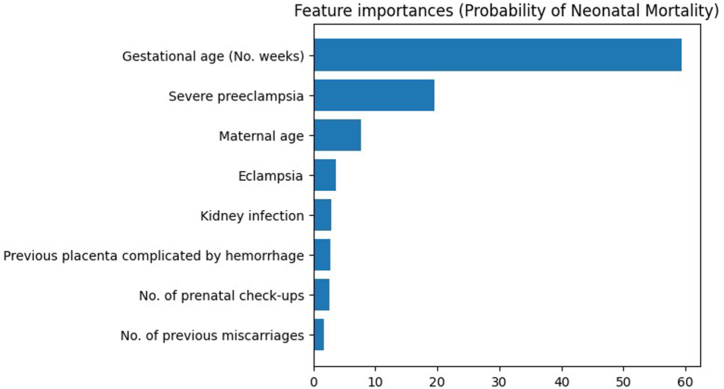
In order to streamline the prediction process, minimize effort, and expedite outcome estimation, the focus was placed on extracting the most relevant variables while identifying the minimal essential predictors. This strategic approach aimed to optimize efficiency. To achieve this, the Gradient Boosting model's feature importance was computed. The established criterion stipulated that a variable would be considered significant if its contribution to prediction surpassed the 1 % threshold. By applying this criterion, 14 variables were excluded due to their limited impact, resulting in a refined set of eight predictor variables for neonatal mortality. These variables were ranked based on importance, arranged in descending order: gestational age, eclampsia, kidney infection, maternal age, previous placenta complications involving hemorrhage, severe preeclampsia, number of prenatal check-ups, and number of previous miscarriages.
Consequent to variable selection, the next step involved hyperparameter tuning to further elevate predictive capabilities by identifying the model exhibiting the highest F1-score. Diverse values were explored during this process, encompassing the learning rate (0.1, 0.3, 0.5, and 0.7), maximum depth of the model (2, 3, 4 and 5), and maximum number of regressors (square root or logarithm of 2). Multiple methodologies demonstrated an impressive 98 % accuracy across various metrics, though the F1-score posed challenges in terms of singling out a definitive victor (Table 3).
Table 3.
Predictive model results with hyperparameter calibration.
| LR | MD | MF | A | E | S | F1-score | AUC |
|---|---|---|---|---|---|---|---|
| 0.1 | 2 | sqrt | 0.94 | 0.94 | 0.94 | 0.94 | 0.94 |
| 0.1 | 2 | log2 | 0.94 | 0.94 | 0.94 | 0.94 | 0.94 |
| 0.1 | 3 | sqrt | 0.94 | 0.92 | 0.96 | 0.94 | 0.94 |
| 0.1 | 3 | log2 | 0.95 | 0.94 | 0.96 | 0.95 | 0.95 |
| 0.1 | 4 | sqrt | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 |
| 0.1 | 4 | log2 | 0.95 | 0.94 | 0.96 | 0.95 | 0.95 |
| 0.1 | 5 | sqrt | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 |
| 0.1 | 5 | log2 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 |
| 0.3 | 2 | sqrt | 0.95 | 0.94 | 0.96 | 0.95 | 0.95 |
| 0.3 | 2 | log2 | 0.95 | 0.92 | 0.98 | 0.95 | 0.95 |
| 0.3 | 3 | sqrt | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| 0.3 | 3 | log2 | 0.97 | 0.96 | 0.98 | 0.97 | 0.97 |
| 0.3 | 4 | sqrt | 0.96 | 0.94 | 0.98 | 0.96 | 0.96 |
| 0.3 | 4 | log2 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| 0.3 | 5 | sqrt | 0.97 | 0.98 | 0.96 | 0.97 | 0.97 |
| 0.3 | 5 | log2 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| 0.5 | 2 | sqrt | 0.95 | 0.96 | 0.94 | 0.95 | 0.95 |
| 0.5 | 2 | log2 | 0.97 | 0.96 | 0.98 | 0.97 | 0.97 |
| 0.5 | 3 | sqrt | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 |
| 0.5 | 3 | log2 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| 0.5 | 4 | sqrt | 0.97 | 0.98 | 0.96 | 0.97 | 0.97 |
| 0.5 | 4 | log2 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| 0.5 | 5 | sqrt | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| 0.5 | 5 | log2 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| 0.7 | 2 | sqrt | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 |
| 0.7 | 2 | log2 | 0.95 | 0.92 | 0.98 | 0.95 | 0.95 |
| 0.7 | 3 | sqrt | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| 0.7 | 3 | log2 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| 0.7 | 4 | sqrt | 0.97 | 0.96 | 0.98 | 0.97 | 0.97 |
| 0.7 | 4 | log2 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| 0.7 | 5 | sqrt | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 |
| 0.7 | 5 | log2 | 0.97 | 0.96 | 0.98 | 0.97 | 0.97 |
LR: learning rate, MD: maximum depth, MF: maximum number of regressors, A: accuracy, E: specificity, S: sensitivity, AUC: area under the curve.
Source: elaborated by the authors based on the data of the study
Notwithstanding the challenge, a pragmatic approach was employed to select the most suitable configuration. This encompassed avoiding excessively low or high learning rates, opting for a balanced maximum depth, and employing the square root for calculating the maximum number of variables. As a result of this decision-making process, the optimal model configuration featured a learning rate of 0.3, a maximum depth of 3, and the calculation of the maximum number of variables using the square root (sqrt) (Table 3).
Incorporating hyperparameters, specifically a learning rate of 0.3, a maximum depth of 3 for the model, and a maximum number of regressors based on the square root led to notable enhancements across performance metrics for the gradient-boosting predictive model. This optimization manifested as augmented accuracy, an expanded area under the ROC curve, heightened sensitivity, specificity, and positive and negative predictive values (Table 4).
Table 4.
Results of estimated metrics in the machine learning model with hyperparameter calibration.
| Metric | Values CI (95 %) |
|---|---|
| Accuracy | 0.98 (0.95–1) |
| Specificity | 0.98 (0.93–1) |
| Sensitivity | 0.98 (0.94–1) |
| F1-score | 0.98 (0.95–1) |
| Area Under the Curve | 0.98 (0.95–1) |
Source: elaborated by the authors based on the data of the study
With these refined hyperparameters seamlessly integrated into the model, a recalibration of variable importance was warranted due to their impact on the calculations. This analysis yielded a discernible hierarchy of weight for predictive variables influencing neonatal mortality. Notably, gestational age surfaced as the paramount determinant, exercising the most profound influence. Subsequently, variables such as severe preeclampsia, maternal age, eclampsia, kidney infection, previous placenta complications involving hemorrhage, number of prenatal check-ups, and number of previous miscarriages followed in descending order of significance (Fig. 3).
Fig. 3.
Feature importance of final variables in the predictive model selected.
Source: elaborated by the authors based on the data of the study
To gain insights into the general directional impact of each variable on neonatal mortality prediction, the SHAP values method (Shapley Additive ExPlanations) was employed. This approach allowed us to quantify the contribution of each variable in augmenting or diminishing the likelihood of neonatal mortality. Interpretation involved two aspects: firstly, discerning the color of the points, and secondly, observing their positioning relative to the vertical axis.
Using the SHAP values method, variable impact directions were assessed. Higher gestational age decreased the probability of neonatal mortality, while lower values increased it. The absence of a previous placenta complication indicated reduced neonatal mortality probability, whereas its presence raised the possibility. Severe preeclampsia displayed a nonlinear effect on neonatal mortality probability. Kidney infection was absent in neonatal deaths, and identified this pattern. Conversely, neonatal mortality cases often lacked a history of miscarriage, indicating this pattern. Notably, these predictive variables contribute collectively to improving prediction accuracy rather than indicating causality (Fig. 4).
Fig. 4.
SHAP values of the predictive model selected.
Source: elaborated by the authors based on the data of the study
Illustrated in Fig. 4, discernible patterns emerged regarding the influence of higher gestational age values, which were associated with a reduction in the probability of neonatal mortality. Conversely, lower gestational age values were correlated with an increase in the likelihood of the outcome. Conversely, the absence of prior placenta complications involving hemorrhage contributed to a decrease in the probability of neonatal mortality, while the presence of such complications escalated the likelihood of the outcome.
Among the variables, the one denoting severe preeclampsia exhibited a nonlinear effect, evident by the dispersed distribution of colored points on both sides of the vertical line. This nonlinear behavior led to intricate fluctuations in the probability of neonatal mortality.
The revelation related to kidney infection was noteworthy. Notably, none of the 22 neonates born to mothers with kidney infections succumbed, thus showcasing a distinct pattern that the SHAP method effectively highlighted. On the contrary, within cases of neonatal mortality, 80 % were associated with mothers lacking a history of miscarriages. The model similarly identified this distinct pattern. It is essential to underline that the predictive variables within these machine learning models aren't construed as explanatory or causal factors. Instead, their collective contribution is fortifying prediction accuracy without necessarily offering descriptive insights.
Model consistency was demonstrated through repeated estimation (1000 times) using randomized training and validation data with SMOTE. F1-Score and Accuracy values consistently exceeded 0.85, with a mean F1-Score of 0.944 and a standard deviation of 0.02 in the validation set (Fig. 5).
Fig. 5.
Graph of F1-score re-estimation of the selected model, according to training data and validation with SMOTE for prediction of neonatal mortality.
Source: elaborated by the authors based on the data of the study
4. Discussion
In the context of predicting neonatal mortality through machine learning, several relevant maternal variables were identified, including gestational age, maternal age, number of previous miscarriages, kidney infection, number of previous pregnancies, number of prenatal checkups, previous placenta complications, placenta accreta, hypertension, HELLP syndrome, pneumonia, hyperemesis gravidarum, and premature placental detachment. Among various machine learning algorithms, the Gradient Boosting algorithm exhibited the highest performance, achieving an area under the curve (AUC), sensitivity, and specificity of 98 %.
In a previous study encompassing machine learning techniques such as artificial neural networks, decision trees, support vector machines (SVM), and Bayesian networks to predict neonatal mortality in an ICU, SVM and Ensemble models attained the highest AUC values of 0.98. Notably, the Random Forest (RF) model showcased the best accuracy (0.98) and specificity (0.94), while the SVM model demonstrated the highest accuracy (0.94) and sensitivity (0.95) [27].
In our cross-sectional study focusing on pregnant women admitted to an ICU, we encountered a neonatal mortality rate of 7.8 %. In this scenario, the Gradient Boosting method displayed superior predictive performance (AUC: 0.98; sensitivity and specificity of 0.98) compared to Logistic Regression, Decision Tree, and Support Vector Machine models in the context of predicting neonatal mortality.
One key advantage of machine learning models is their versatility across diverse clinical contexts. Specifically, the Gradient Boosting method has showcased its effectiveness in scenarios with infrequent event outcomes [28], which resonates with our study findings. Additionally, we opted for this method due to its ability to learn and rectify errors iteratively during model accuracy estimation. This mechanism relies on backpropagation, wherein estimated parameters are adjusted to minimize error in subsequent computational iterations [[27], [28], [29], [30]].
The machine learning model employed in our study demonstrated exceptional performance, boasting a 98 % AUC, sensitivity, and specificity. Conversely, a distinct predictive model using machine learning on secondary population databases showed an AUC ranging from 70 % during pregnancy to 80 % at delivery [31]. Such variations can be attributed to the temporal application and clinical context of predictive models [22].
In a study focusing on high-risk neonates, an AUC range of 75 %–95 % was reported, alongside a sensitivity of 65 % and specificity of 98 % [12]. Similarly, in preterm infants within intensive care units, neural networks outperformed logistic regression models in predicting neonatal mortality [20]. Similarly, random forest techniques in very low birth weight newborns showcased superior accuracy in neonatal death prediction compared to other mortality predictor classifiers [32]. These disparities underscore the contextual variability of machine learning model applications.
Differences between our study and previous research can be attributed to factors such as study design, machine learning model selection, predictor variability, and target population. Unlike previous studies, our investigation exclusively concentrated on maternal characteristic predictors within an ICU setting. Despite contextual variations, a prior systematic review underscored the accuracy of machine learning models in predicting neonatal mortality, emphasizing the significance of practical and accessible applications for healthcare professionals to enhance neonatal care and outcomes [33].
This study underscores the adaptability of machine learning models to specific clinical contexts, indicating their potential as precise and accessible tools for healthcare providers in neonatal death prediction. This holds substantial promise for ameliorating neonatal healthcare and curbing mortality within specific clinical environments. Scientifically, this study's contribution lies in its application of machine learning and artificial intelligence algorithms to the field of neonatal death prediction.
Nevertheless, inherent limitations stem from the study's retrospective observational design and limited sample size. Further research in this field is required. Nevertheless, machine learning models evolve with each interaction, allowing for tailored adjustments to their specific applications. Despite its retrospective nature, this study sourced information from medical records, diagnoses, and clinical data documented by specialized medical professionals. Data collection personnel were duly trained for these purposes.
5. Conclusion
Machine learning models, particularly Gradient Boosting, offer valuable utility in predicting neonatal mortality among pregnant women admitted to intensive care units, particularly when considering maternal morbidities. Among the maternal characteristics studied; the Gradient Boosting machine learning model emerged with the highest predictive prowess for neonatal death. The selected predictive model, enriched with hyperparameter values, showcased an impressive sensitivity and specificity of 98 %.
We strongly advocate for an extended exploration of integrating machine learning techniques within hospital settings to forecast neonatal mortality. This pursuit should encompass larger and more diverse sample sizes, spanning various timeframes. The potential of artificial intelligence-driven models lie in their capacity to assimilate past outcomes and dynamically adjust to evolving contexts.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/tables/figures. Material/referenced in article. Any other information will be made available on request.
CRediT authorship contribution statement
Marcos Espinola-Sánchez: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Silvia Sanca-Valeriano: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. Andres Campaña-Acuña: Conceptualization, Formal analysis, Methodology, Validation, Writing – original draft, Writing – review & editing. José Caballero-Alvarado: Conceptualization, Formal analysis, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.United Nations Children’s Fund (UNICEF) World Bank; 2020. Levels and Trends in Child Mortality: Report.https://www.unicef.org/reports/levels-and-trends-child-mortality-report-2020 Assessable at: [Google Scholar]
- 2.Tura G., Fantahun M., Worku A. The effect of health facility delivery on neonatal mortality: systematic review and meta-analysis. BMC Pregnancy Childbirth. 2013;13:18. doi: 10.1186/1471-2393-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.United Nations Children's Fund (UNICEF) Peru Newborns: Press releases. United nations children's fund. 2018. https://www.unicef.org/peru/comunicados-prensa/el-mundo-no-esta-cumpliendo-con-los-recien-nacidos-dice-unicef# Available in:
- 4.World Health Organization (WHO) 2022. Newborn Mortality.https://www.who.int/news-room/fact-sheets/detail/levels-and-trends-in-child-mortality-report-2021 Assessable at: [Google Scholar]
- 5.National Institute of Statistics and Informatics (INEI) 2018. Peru. Infant and Childhood Mortality.https://www.inei.gob.pe/media/MenuRecursivo/publicaciones_digitales/Est/Lib1656/pdf/cap007.pdf Lima, Peru. [Accessed February 25, 2022]. Available in: [Google Scholar]
- 6.Torres-Cantero A.M., Álvarez León E.E., Morán-Sánchez I., et al. Health impact of COVID pandemic. SESPAS Report 2022. Gac. Sanit. 2022;36(Suppl 1):S4–S12. doi: 10.1016/j.gaceta.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cárdenas M., Franco G., Riega-López P. Neonatal mortality: a challenge for the country and the university. An. Fac. Med. 2019;80(3):281–282. S12. [Google Scholar]
- 8.Choton M., Gonzales L. Evolution of neonatal mortality rate in the amazonas region, Peru. UNTRM Scientific Journal: Social Sciences and Humanities. 2020;3(1):66–71. doi: 10.25127/rcsh.20203.575. 2005 - 2018. [DOI] [Google Scholar]
- 9.Daemi A., Ravaghi H., Jafari M. Risk factors of neonatal mortality in Iran: a systematic review. Med. J. Islam. Repub. Iran. 2019;33:87. doi: 10.34171/mjiri.33.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendoza L.A., Gómez D., Gómez D., Osorio M.A., Villamarín E.A., Arias M.D. Biological determinants of neonatal mortality in a population of adolescent and adult women at a hospital in Colombia. Rev. Chil. Obstet. Ginecolog. 2017;82(4):424–437. doi: 10.4067/s0717-75262017000400424. [DOI] [Google Scholar]
- 11.Chowdhury H.R., Thompson S., Ali M., Alam N., Yunus M., Streatfield P.K. Causes of neonatal deaths in a rural subdistrict of Bangladesh: implications for intervention. J. Health Popul. Nutr. 2010;28(4):375–382. doi: 10.3329/jhpn.v28i4.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Márquez-González H., Jiménez-Báez M.V., Muñoz-Ramírez C.M., et al. Development and validation of the Neonatal Mortality Score-9 Mexico to predict mortality in critically ill neonates. Arch. Argent. Pediatr. 2015;113(3):213–220. doi: 10.5546/aap.2015.eng.213. [DOI] [PubMed] [Google Scholar]
- 13.Noboa Salgado M., González-Andrade F., Villagómez Aroca D.A. Proposal of a novel predictive model for mortality in high-risk neonates and evaluation of its performance. Rev. Ecuat. Pediatr. 2021;22(1):1–13. doi: 10.52011/0095. [DOI] [Google Scholar]
- 14.Teji J.S., Jain S., Gupta S.K., Suri J.S. NeoAI 1.0: machine learning-based paradigm for prediction of neonatal and infant risk of death. Comput. Biol. Med. 2022;147 doi: 10.1016/j.compbiomed.2022.105639. [DOI] [PubMed] [Google Scholar]
- 15.Pedrero V., Reynaldos-Grandón K., Ureta-Achurra J., Cortez-Pinto E. Overview of machine learning and its application in the management of emergency services. Rev. Med. Chile. 2021;149(2):248–254. doi: 10.4067/s0034-98872021000200248. [DOI] [PubMed] [Google Scholar]
- 16.Del Río R., Thió M., Bosio M., Figueras J., Iriondo M. Prediction of mortality in premature neonates. An updated systematic review. An. Pediatr. 2020;93(1):24–33. doi: 10.1016/j.anpedi.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Lugo-Reyes S.O., Maldonado-Colín G., Murata C. Artificial intelligence to assist clinical diagnosis in medicine. Rev. Alerg. Mex. 2014;61(2):110–120. [PubMed] [Google Scholar]
- 18.Dorado-Díaz P.I., Sampedro-Gómez J., Vicente-Palacios V., Sánchez P.L. Applications of artificial intelligence in cardiology. The future is already here. Rev. Esp. Cardiol. 2019;72(12):1065–1075. doi: 10.1016/j.rec.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Curioso W.H., Brunette M.J. Artificial intelligence and innovation to optimize the tuberculosis diagnostic process. Rev. Peru. Med. Exp. Salud Pública. 2020;37(3):554–558. doi: 10.17843/rpmesp.2020.373.5585. [DOI] [PubMed] [Google Scholar]
- 20.Rezaeian A., Rezaeian M., Khatami S.F., Khorashadizadeh F., Moghaddam F.P. Prediction of mortality of premature neonates using neural network and logistic regression. J. Ambient Intell. Hum. Comput. 2020;13:1269–1277. doi: 10.1007/s12652-020-02562-2. [DOI] [Google Scholar]
- 21.Houweling T.A.J., van Klaveren D., Das S., et al. A prediction model for neonatal mortality in low- and middle-income countries: an analysis of data from population surveillance sites in India, Nepal and Bangladesh. Int. J. Epidemiol. 2019;48(1):186–198. doi: 10.1093/ije/dyy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mboya I.B., Mahande M.J., Mohammed M., Obure J., Mwambi H.G. Prediction of perinatal death using machine learning models: a birth registry-based cohort study in northern Tanzania. BMJ Open. 2020;10(10) doi: 10.1136/bmjopen-2020-040132. Published 2020 Oct 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mfateneza E., Rutayisire P.C., Biracyaza E., Musafiri S., Mpabuka W.G. Application of machine learning methods for predicting infant mortality in Rwanda: analysis of Rwanda demographic health survey 2014-15 dataset. BMC Pregnancy Childbirth. 2022;22(1):388. doi: 10.1186/s12884-022-04699-8. Published 2022 May 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pesapane F., Codari M., Sardanelli F. Artificial intelligence in medical imaging: threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur Radiol Exp. 2018;2(1):35. doi: 10.1186/s41747-018-0061-6. 2018 Oct 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sen S., Cohen A.S. Sample size requirements for applying diagnostic classification models. Front. Psychol. 2021;11 doi: 10.3389/fpsyg.2020.621251. Published 2021 Jan 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubio A., González A., González E., Gonzalez G. Morbidity and maternal and fetal mortality in patients with severe preeclampsia. Progresos Obstet. Ginecol. 2011;54(1):4–8. doi: 10.1016/j.pog.2010.11.002. [DOI] [Google Scholar]
- 27.Sheikhtaheri A., Zarkesh M.R., Moradi R., Kermani F. Prediction of neonatal deaths in NICUs: development and validation of machine learning models. BMC Med. Inf. Decis. Making. 2021;21(1):131. doi: 10.1186/s12911-021-01497-8. Published 2021 Apr 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helm J.M., Swiergosz A.M., Haeberle H.S., et al. Machine learning and artificial intelligence: definitions, applications, and future directions. Curr Rev Musculoskelet Med. 2020;13(1):69–76. doi: 10.1007/s12178-020-09600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z., Jung C. GBDT-MO: gradient-boosted decision trees for multiple outputs. IEEE Transact. Neural Networks Learn. Syst. 2021;32(7):3156–3167. doi: 10.1109/TNNLS.2020.3009776. [DOI] [PubMed] [Google Scholar]
- 30.Konstantinov A., Utkin L. Interpretable machine learning with an Ensemble of gradient boosting machines. Knowl. Base Syst. 2021;222 doi: 10.1016/j.knosys.2021.106993. [DOI] [Google Scholar]
- 31.Shukla V.V., Eggleston B., Ambalavanan N., et al. Predictive modeling for perinatal mortality in resource-limited settings. JAMA Netw. Open. 2020;3(11) doi: 10.1001/jamanetworkopen.2020.26750. Published 2020 Nov 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaskari J., Myllärinen J., Leskinen M., Rad A.B., Hollmén J., Andersson S., Särkkä S. Machine learning methods for neonatal mortality and morbidity classification. IEEE Access. 2020;8:123347–123358. doi: 10.1109/ACCESS.2020.3006710. [DOI] [Google Scholar]
- 33.Mangold C., Zoretic S., Thallapureddy K., Moreira A., Chorath K., Moreira A. Machine learning models for predicting neonatal mortality: a systematic review. Neonatology. 2021;118(4):394–405. doi: 10.1159/000516891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/tables/figures. Material/referenced in article. Any other information will be made available on request.