Summary
The constant emergence of mutated pathogens poses great challenges to the existing vaccine system. A screening system is needed to screen for antigen designs and vaccination strategies capable of inducing cross-protective immunity. Herein, we report a screening system based on DNA vaccines and a micro-electroporation/electrophoresis system (MEES), which greatly improved the efficacy of DNA vaccines, elevating humoral and cellular immune responses by over 400- and 35-fold respectively. Eighteen vaccination strategies were screened simultaneously by sequential immunization with vaccines derived from wildtype (WT) SARS-CoV-2, Delta, or Omicron BA.1 variant. Sequential vaccination of BA.1-WT-Delta vaccines with MEES induced potent neutralizing antibodies against all three viral strains and BA.5 variant, demonstrating that cross-protective immunity against future mutants can be successfully induced by existing strain-derived vaccines when a proper combination and order of sequential vaccination are used. Our screening system could be used for fast-seeking vaccination strategies for emerging pathogens in the future.
Subject areas: Computational molecular modelling, Immune response, Immunology
Graphical abstract

Highlights
-
•
A screening system is developed to rapidly identify effective vaccination strategies
-
•
This system employs DNA vaccines and micro-electroporation/electrophoresis (MEES)
-
•
MEES greatly strengthens the induction of both humoral and cellular immune responses
-
•
Cross-protective immunity is induced by rationally combining DNA vaccines and MEES
Computational molecular modelling; Immune response; Immunology
Introduction
Vaccines are the most cost-effective means of combating infectious diseases. Respiratory viruses such as influenza and SARS-CoV-2 frequently break through the immune defenses established by traditional vaccines because of their high infectivity and variability. Over the past 20 years, antigenically drifted influenza viruses emerged several times, resulting in a precipitous decline in vaccine effectiveness in those flu seasons.1,2,3 Recently, SARS-CoV-2 has produced several iterations of mutated strains, also called variants of concern (VOCs), in just two years, making the existing SARS-CoV-2 vaccine less effective in preventing infection with the latest mutated strains.4,5,6 Unfortunately, it is very likely that highly pathogenic mutated strains will continue to emerge in the near future, just as there have been four influenza pandemics in the past century. Besides influenza and SARS-CoV-2 vaccines, the development of vaccines against many viruses, such as HIV-1, is also hindered by the high heterogeneity and frequent mutations in the viral genome.7 Therefore, the development of universal vaccines capable of inducing cross-protective immunity is a common but unmet need for a large number of viral vaccines. A screening system that can rapidly prepare a large variety of antigens and screen their combinations is essential to achieve this goal.
In order to establish an efficient screening system, the choice of vaccine type is the first factor to be considered. Taking SARS-CoV-2 as an example, several types of vaccines have been developed, including clinical approved inactivated vaccines, recombinant protein vaccines, viral vector vaccines, mRNA vaccines,8 and DNA vaccines that are still in phase I/II/III clinical trials.9,10,11 Compared to several other vaccine types, DNA vaccines can be designed and prepared rapidly, making them more suitable for the rapid screening of large numbers of antigen designs. In contrast, the preparation process for inactivated vaccines and recombinant vaccines has to be developed individually for each new antigen, which is very time-consuming and costly. Viral vectors may not be suitable for screening vaccination schedules that require multiple administrations in weeks, since preexisting immunity would be a concern for at least some, if not all, vectors. RNA vaccines do not have these problems, but DNA vaccines are easier and faster to prepare and less costly in comparison. More importantly, the results of screening on DNA vaccines can be readily applied to RNA vaccines.
Immunogenicity is another crucial factor for vaccines used in a screening system, since those vaccines require two or more doses to elicit a sufficient level of immune responses would apparently delay the screening process and complicate the results. DNA, as a negatively charged macromolecule, is unable to cross cell membranes autonomously to enable protein expression.12 Therefore, the delivery system is particularly important. Over the past two decades, electroporation-based delivery systems have stepped up to become one of the leading delivery methods for DNA vaccines, successfully delivering DNA vaccines or therapeutic DNA into small and large animals.13,14 Further, electroporation-facilitated DNA vaccines entered phase II/III clinical trials for cervical cancer treatment15 and SARS-CoV-2 DNA vaccine.11 However, conventional electrodes for in vivo electroporation, require the application of high voltage to achieve successful DNA delivery due to the bulk separation between electrodes,16,17 which may cause physical and psychological discomfort to the recipients. On the other hand, DNA vaccines used for screening systems need to have an extremely high ability to induce immune responses, preferably with only one shot to induce a high titer of neutralizing antibodies, while existing vaccines, even when augmented by a delivery system, often require two more doses.18,19,20 Therefore, how to further reduce the voltage while further increasing the delivery efficiency is the direction of delivery system optimization, which is important for both rapid screening and clinical application of DNA vaccines.
Intradermal delivery is a promising strategy to realize the potential of vaccines.21,22 As the first line of immune defense, the skin contains a large number of immune cells that are absent in muscles, especially antigen-presenting cells (APC), including Langerhans cells (LC) and various types of dermal dendritic cells (DC).23 DCs are key cells bridging innate and adaptive immune responses and are particularly important for the induction of CD4+ and CD8+ T responses.24 Theoretically, compared to subcutaneous and intramuscular injections, intradermal delivery of nucleic acid vaccines not only promotes the uptake of exogenously expressed antigens by DCs, but also enables the direct transfection of DCs, resulting in endogenous expression of antigens in DCs. This process facilitates the induction of CD8+ T cell responses via the MHC I pathway.25 Nevertheless, enhancing the efficiency of nucleic acid entry into the cytoplasm remains a key issue to be addressed for the intradermal delivery of DNA vaccines.
Needle electrodes developed for skin application scenarios could provoke cellular electroporation, thereby improving the effectiveness of intradermally injected DNA vaccines.26 To further reduce pain and skin damage, shorter microneedle-electrode arrays have been developed and were also effective in improving the efficiency of siRNA and DNA delivery by inducing the electroporation of skin cells.27,28,29 By adjusting the size of the microneedles, the targeted genes can be precisely expressed in the epidermis, a superficial skin layer that contains abundant keratinocytes and APCs.30 Moreover, the use of microneedle-electrode arrays with close electrode spacing could further reduce the voltage and current required for electroporation, which would further improve compliance.29
Besides electroporation which causes the perforation of the cell membrane by high-voltage narrow pulses, electrophoresis of DNA driven by a series of low-voltage long pulses has been demonstrated to be crucial for an effective transfection of cultured cells.31 Electrophoresis promotes the migration of DNA and its contact with the cell membrane, allowing it to eventually pass through the pores generated by electroporation. Electrophoresis is particularly important for in vivo electrotransfection in the skin, which contains dense connective tissue that can greatly impede the migration of DNA. A recent study reported that longer pulses increased the electromobility of plasmid DNA and resulted in higher transfection rates for cells embedded in collagen gels.32 Therefore, the combination of electroporation plus electrophoresis was found to be more effective in the needle electrode-based transfection system, but electroporation voltages above 1000 V/cm were often required for the skin.26,33 It is not clear whether this strategy works for microneedle arrays, and whether microneedle arrays are able to further reduce the electric field required remains unknown. Moreover, the effect of the electric field on the skin’s immune microenvironment has been rarely studied.
In this study, we developed a microneedle array-based micro-electroporation/electrophoresis system (MEES), in which a relatively low electric field (600 V/cm for electroporation, 300 V/cm for electrophoresis) was highly effective in boosting the effectiveness of DNA vaccines (Figure 1A). The MEES not only enhanced the expression of antigens, but also promoted antigen uptake by recruiting DCs into the vaccination site. Further, a screening system based on DNA vaccines and MEES was subsequently established for surveying vaccination strategies with the capability to induce cross-protective immune responses. Results indicated that cross-protective immune responses against mutant viruses could be successfully induced by existing strain-derived vaccines given that a proper combination and order of sequential vaccination were used. The MEES developed in this work presented an effective and versatile platform for the delivery and screening of DNA vaccines, which possesses promising potential for fast-seeking vaccination strategies for emerging pathogens in the future.
Figure 1.
The development of the MEES-assisted screening system
(A) Illustration of the MEES-assisted fast screening process for SARS-CoV-2 vaccination strategies. A large variety of DNA vaccines could be quickly prepared according to nucleic acid sequences of various strains, such as wildtype (WT), Delta or Omicron BA.1 variant. The MEES was developed and incorporated into the screening system to improve the efficacy of DNA vaccines, elevating humoral and cellular immune responses. The system could be used for fast-seeking vaccination strategies for emerging, mutated, and highly pathogenic SARS-CoV-2 variants. As an example, cross-protective immune responses against future mutant viruses can be successfully induced by existing strain-derived vaccines when a proper combination and order of sequential vaccinations are used.
(B) 304H stainless steel was processed using laser etching technology to fabricate microneedles.
(C) Diagram of a single microneedle with grooves.
(D) Images of a row of microneedles (left) and a single microneedle (right) by a stereomicroscope. scale bar = 1 mm (left) or 0.2 mm (right).
(E) Diagram of MEES with circuits.
(F) An assembled microneedle array was photoed using a stereomicroscope. scale bar = 1 mm.
Results
The development of micro-electroporation/electrophoresis system
The microneedle array was fabricated by laser cutting on 200 μm thick 304H stainless steel (Figure 1B). With the assistance of high-precision laser etching technology, tips were fabricated with a shape feature, while an additional groove structure was carved on each needle extending to the tip as a way to increase the sharpness of the needle and reduce the damage to the skin when piercing and pulling it out (Figure 1C). Each needle was 1 mm in height. The bottom of each needle was 220 μm wide, gradually narrowing to 150 μm and finally forming the tip. The width of the groove in the middle was 110 μm, while the depth was 50 μm (Figure 1D). The final microneedle array consisted of a row of 9 needles, each spaced 600 μm apart (Figures 1D and 1E). Four rows of microneedles were assembled onto a 3D-printed basement, with a 1 mm separation between every two rows (Figures 1E and 1F), while the microneedle patches alternatively served as cathode or anode. Finally, the two rows of cathode and anode pins were subsequently connected to an external power supply, forming the MEES (Figure 1E).
Computational simulation on electroporation settings
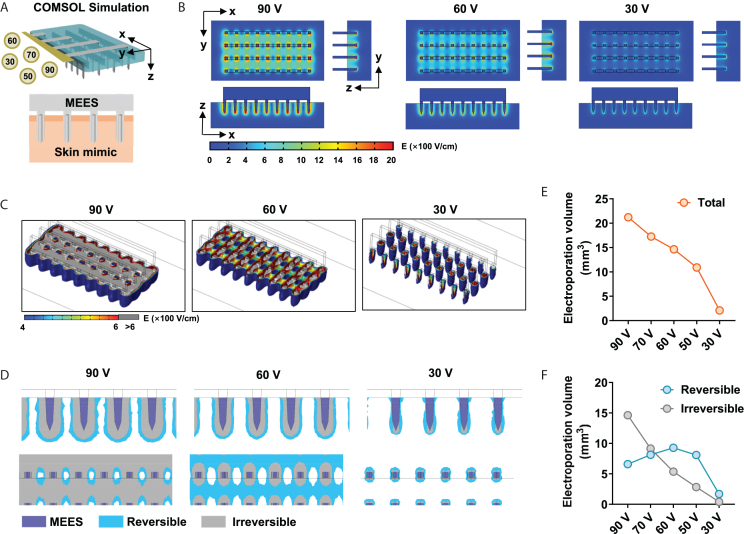
To reveal the electric field distribution in the skin during the microneedles-induced electroporation process, the finite element analysis (FEA) was computed by COMSOL Multiphysics 5.4. Based on the physical setup or microneedles and skin, a 3D model was built, while the physical properties (e.g., conductivity) were referenced to the actual parameters. The conductivity was set at 1 × 107 S/m or 0.2 S/m for microneedles or skin, respectively, while an insulating layer was set between the base of the needle and the skin to mimic the stratum corneum.34 Five electroporation conditions at various voltages ranging from 30 V to 90 V were applied to the microneedle, and the steady-state electric field distribution was analyzed (Figure 2A). The electric field intensity was found to be highest at the tip of the microneedle, and gradually decreased along the direction from the tip to the base until it disappeared completely (Figure 2B). As expected, higher pulse voltage resulted in a higher electric field intensity distributed in the skin (Figure 2B). We further calculated the 3D spatial distribution of the electric field intensity to more intuitively represent the total volume of electroporation regions (Figure 2C). Although any electric field density greater than 400 V/cm was considered to be the critical threshold for causing cell membrane perforation, excessive electric field densities might cause irreversible electroporation inducing excessive cell damage and cell death. Here, 400–600 V/cm was considered to be the suitable condition, also called the reversible electroporation interval.35 As seen from the electric field distribution, although the overall electroporation volume was larger under the 90 V condition, the corresponding irreversible electroporation volume was also larger, compared to the 60 V condition (Figure 2C, 1st and 2nd panels).
Figure 2.
FEA simulation for the reversible electroporation volume
(A) Schematic diagram of FEA simulation on microneedle electroporation in the skin.
(B) The steady electric field strength distributions under 90 V (left), 60 V (middle) or 30 V (right) pulses were shown.
(C) 3D electric field strength distributions were reconstructed and shown.
(D) The electroporation area was shown as a side view (upper) and a top view (lower), in which the irreversible electroporation area (E > 600 V/cm) was marked as gray and the reversible electroporation area (400 < E < 600 V/cm) was marked as blue.
(E) The volume of the total electroporation area (E > 400 V/cm) was calculated.
(F) The irreversible electroporation area and the reversible electroporation area were calculated.
We then quantified the electroporation volume for Figure 2D, in which reversible and irreversible regions were marked in blue and gray respectively. As expected, the total electroporation volume was largest at 90 V and decreased sequentially as the applied voltage decreased (Figure 2E). The irreversible electroporation volume also showed a similar decreasing trend (Figure 2F). While the total electroporation volume was maximum at 90 V, most of the region was suggested to irreversible electroporation that was unbeneficial for DNA delivery. The reversible electroporation volume was maximum at 60 V, while a less irreversible electroporation region was observed, indicating that 60 V might achieve the best electroporation effect for in vivo DNA transfection (Figure 2F). In terms of electrophoresis conditions, since skin tissue is very dense, it is preferable to choose the maximum voltage that does not cause electroporation. Under 30 V, there was only a very small amount of electroporation volume in very close proximity to the needle, indicating that 30 V might not cause electroporation in the skin by MEES and would be more suitable for electrophoresis (Figures 2B–2F).
Micro-electroporation/electrophoresis system was highly effective in enhancing in vivo DNA transfection
Based on the above simulation results, we speculated that a combination of electroporation at 60 V and electrophoresis at 30 V might be an appropriate strategy. The in vivo transfection experiments were performed, in which mice were first intradermally (i.d.) injected with plasmids coding luciferase (pLuc) followed by the MEES treatment (Figure 3A). We first compared three settings of pulse conditions, including 60 V/50 μs for 2 pulses (EP1), 60 V/50 μs for 10 pulses (EP2) and 60 V/50 μs for 2 pulses +30 V/10 ms for 8 pulses (EP3), since a previous study has demonstrated that 8 × 10 ms treatment was able to enhance the mobility of DNA (Figure 3B).32 The efficiency of in vivo DNA transfection was evaluated by bioluminescence imaging on the expression level of luciferase, where the results showed that the combination of electroporation and electrophoresis (EP3) could significantly increase the expression level of luciferase compared to electroporation alone (EP1) (Figures 3B and 3C). Notably, even when increasing the electroporation alone from 2 to 10 times (EP2), the effect was still significantly inferior to that of electroporation plus electrophoresis (EP3), indicating that the electrophoresis would enhance the intradermal delivery of DNA (Figures 3B and 3C).
Figure 3.
Optimizing electrical settings of MEES for in vivo DNA transfection
(A) Schematic diagram of MEES-based DNA transfection in mouse skin. Plasmids coding luciferase (pLuc) or EGFP (pEGFP) were i.d. injected into the skin, followed by the MEES treatment.
(B) C57BL/6J mice were injected with 10 μg plasmid, then the different electrical pulses were applied. EP1, 60 V/50 μs for 2 pulses; EP2, 60 V/50 μs for 10 pulses; EP3, 60 V/50 μs for 2 pulses +30 V/10 ms for 8 pulses. Live bioluminescence imaging was performed to visualize luciferase expression 48 h after pLuc delivery.
(C) Luciferase expression under different settings was summarized. n = 3–5.
(D) Schematic view of electrical pulse protocols.
(E) The detailed parameters of electrical pulses.
(F) Representative bioluminescence images showing luciferase expression 48 h after plasmid delivery in mice receiving 10 μg (upper) or 1 μg (lower) pLuc, followed by MEES with indicated electrical pulse (EE) protocol. The mock received plasmids only.
(G and H) Luciferase expression under different settings in (F) was summarized. The ROI (total flux, photons per second) was calculated from the mice treated with 10 μg (G) or 1 μg (H) pLuc, respectively. n = 3.
(I) Linear regression was performed on the ROI of the 10 μg group and the predicted reversible electroporation volume.
(J) Linear regression was performed on the ROI of the 1 μg group and the predicted reversible electroporation volume.
(K) Mice received i.d. injection of 10 μg pEGFP, followed by MEES treatment under the setting of 60 V/50 μs for 2 pulses +30 V/10 ms for 8 pulses (MEES + pEGFP), or by inserting the microneedle array without electroporation and electrophoresis (MN + pEGFP). Representative intravital microscopy images of EGFP expression in living mice 24 h after the treatment were shown. scale bar = 1000 μm (left) or 500 μm (right).
(L) Quantitative analysis of the mean fluorescence intensity (MFI) of pEGFP. n = 4–5. Data were shown as mean ± SEM. p values were calculated by one-way ANOVA with Tukey’s multiple comparisons test (C), Pearson correlation coefficients (I, J), or two-tailed Student’s t test (L). ∗p < 0.05, ∗∗p < 0.01 and ns, no significance.
Next, we explored whether 60 V was indeed the best electroporation condition as predicted by computational simulation (Figure 2F). Five different settings of electroporation pulses, including the use of electric voltage at 30, 50, 60, 70, and 90 V, were employed, following a same electrophoresis treatment at 30 V (Figures 3D and 3E). When mice received 10 μg plasmid, protocol 1 (60 V) was found to mediate the highest luciferase expression level, compared to other protocols with other voltage conditions (Figures 3F and 3G). The expression of luciferase level under different conditions correlated with the simulation results that an intermediate voltage pulse was optimized for electroporation (Figure 3I, r = 0.8752, p = 0.0224). Meanwhile, the expression level of luciferase was low when all pulses were at 30 V, suggesting that electrophoresis alone was insufficient for an effective in vivo transfection (Figure 3G). Based on these results, the pulse condition of 60 V/50 μs for 2 pulses +30 V/10 ms for 8 pulses was determined as the optimized setting for MEES.
The dosage of plasmids might also have a nonnegligible impact on the outcome. When mice received a low dosage of plasmids (1 μg), all electroporation conditions elicited only weak reporter gene expression, and there was little difference between the outcomes of these conditions (Figures 3F and 3H). Moreover, the correlation between the in vivo transfection efficiency and the simulation results was not significant when 1 μg plasmids were used (Figure 3J). To unveil this impact of the dosage, we explored the effect of various dosages (1, 5, 10, 20 μg/injection) on the transfection result, revealing that luciferase expression reached the plateau at a dosage of 10 μg (Figure S1). Thus, 10 μg plasmids were used for the following experiments unless otherwise stated.
In addition to the pLuc, we also used a plasmid expressing enhanced green fluorescent protein (EGFP) to verify the capability of MEES. Mice were i.d. injected with pEGFP followed by MEES or by inserting the microneedle array with neither electroporation nor electrophoresis as the control group (MN). The expression of EGFP was imaged by confocal microscopy and quantified by ImageJ (Figure 3K). Again, MEES was highly effective in enhancing the in vivo transfection of pEGFP in mouse skin by ∼3-fold compared to the control group (Figure 3L).
Examination on the safety of micro-electroporation/electrophoresis system
Next, we investigated whether the application of MEES induced excessive inflammation in the skin. The histological morphologies of mice skin were monitored for one week after MEES or MN treatment. Skin samples were collected on the indicated days followed by H&E staining. As shown in Figure 4A, microneedles penetrated the skin effectively leaving tiny wounds, which were closed within one day, restoring the barrier function of the skin. Both MEES and MN caused the infiltration of inflammatory cells (Day 1), but the inflammation was confined to the area around the pinhole and was transient, resolving rapidly by the next day (Day 2) (Figures 4A and 4B). The skin was fully recovered within 7 days. In terms of histology and cell infiltration, MN and MEES groups showed little significant difference after treatments (Figures 4A and 4B). Since excessive electroporation often causes irreversible cell death, we also labeled dead cells in the skin while observing EGFP expression (Figure 3K).22 A small number of dead cells surrounded the needle holes in both treatments, indicating that these deaths were caused by acupuncture rather than electroporation (Figure 4C). Compared to MN, MEES did not cause an increased amount of cell death, either at 2 or 24 h later (Figure 4C). Taken together, these results suggested that MEES had a similar safety profile to that of MN treatment. The electroporation and electrophoresis settings of MEES were sufficiently mild to avoid excessive cell death in vivo.
Figure 4.
Safety assessment of MEES
(A) C57BL/6J mice received MEES treatment under the setting of 60 V/50 μs for 2 pulses +30 V/10 ms for 8 pulses (MEES), or by inserting the microneedle array without electroporation and electrophoresis (MN). The mock did not receive any treatment. Representative images of H&E staining of the skin at various time points after the treatment were shown. Scale bar = 200 μm.
(B) Infiltrated cells in the skin were quantified from H&E staining by ImageJ. n = 4–12.
(C) Mice were i.d. injected with 10 μg pEGFP, followed by MN or MEES treatment. The dead cells were stained by DRAQ7 (red). Scale bar = 500 μm (left) or 250 μm (right).
Micro-electroporation/electrophoresis system efficiently boosted the effectiveness of DNA vaccines
The potency of MEES was further investigated in a DNA vaccine coding a model antigen ovalbumin (OVA). DNA vaccine alone was insufficient to elicit humoral immune responses in most animals, regardless of one or two doses (Figures 5A–5H). In contrast, with the assistance of MEES, a single dose of vaccine successfully induced immune responses in 75% of the animals (Figure 5A). In the prime vaccination, the induced immune response was slightly T helper 2 (Th2)-biased, indicated by a preferable induction of IgG1 (Th2) rather than IgG2c (Th1) (Figures 5B–5D). After booster immunization, vaccine + MEES induced high titers of OVA-specific IgG in all mice (Figure 5E). The geometric mean of the titers (GMT) in the MEES group was 400-fold higher than that in the vaccine-alone (pOVA) group. MEES further boosted the induction of IgG1 and IgG2c simultaneously, resulting in a more balanced IgG2c/IgG1 ratio (Figures 5F–5H).
Figure 5.
Immune responses after DNA vaccination with MEES
Mice were i.d. immunized with 10 μg DNA vaccine encoding ovalbumin (pOVA), with or without MEES treatment.
(A–D) Sera were collected 14 days after the prime vaccination. Total IgG (A), IgG2c (B), or IgG1 (C) titers were measured by ELISA. IgG2c/IgG1 ratios were shown in (D). n = 6–7.
(E–H) Sera were collected 7 days after the boost vaccination. IgG (E), IgG2c (F), or IgG1 (G) titers were measured. IgG2c/IgG1 ratios were shown in (H). n = 6–7.
(I) Splenocytes were isolated 7 days after the boost vaccination. CD8+ T cells producing IFNγ were measured by flow cytometry after the peptide (OVA257-264) stimulation. n = 6–7.
(J–L) Mice were i.d. immunized with 10 μg pOVA with or without MEES. Some mice were immunized with 100 μg OVA protein alone. IgG1 (J) and IgG2c (K) titers were measured by ELISA. IgG2c/IgG1 ratios were shown in (L). n = 6–8.
(M) Schematic diagram of intravital microscope imaging on CD11c-cre; tdTomatofl/wt transgenic mice, in which DCs expressed a fluorescent protein, tdTomato.
(N) Transgenic mice were i.d. injected with 10 μg pEGFP, followed by MN or MEES treatment. The recruitment DCs into the skin was visualized 24 or 48 h after injection. Different layers of skin were scanned and the number in each panel indicated the depth from the skin surface. Images represented similar results from 3 mice in each group. Scale bar = 500 μm (left) or 250 μm (right).
(O–R) Mice were i.d. immunized with 10 μg pOVA with MN or MEES treatment. Flow cytometry was performed 48 h after treatment. Absolute numbers of total cDC (O), cDC1 (P), cDC2 (Q), and LCs (R) in the skin of untreated or treated mice were quantified. n = 3 mice. The results were shown as geometric mean ±95% Cl (A-H, J-L), or mean ± SEM (I, O-R). Statistical analysis, two-tailed Mann-Whitney U-test (A-H), Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test (J-L), two-tailed Student’s t test (I), or one-way ANOVA with Dunnett’s multiple comparisons test (O-R). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ns, no significance.
MEES not only improved humoral immune responses but also effectively improved the cellular immune response, especially for antigen-specific CD8+ T cells which were elevated by 35-fold (Figure 5I). The effect of MEES on elevating CD4+ T cells was moderate (Figure S2). Subsequently, we also compared the differences between the injection of pOVA alone, vaccination of pOVA with MEES (pOVA + MEES) and the injection with protein vaccines (OVA protein) only. Immunization with 100 μg OVA protein alone or 10 μg pOVA in the presence of MEES induced comparable titers of IgG1, whereas only pOVA + MEES were able to induce high titers of IgG2c (Figures 5J and 5K). As a result, the OVA protein vaccine profoundly elicited Th2-skewed immune responses. In sharp contrast, the immune response induced by pOVA + MEES was very balanced (Figure 5L). This is particularly important for viral vaccines for which Th1 immune responses are indispensable.
Micro-electroporation/electrophoresis system recruited dendritic cells into the skin
Next, the mechanism of the capability of MEES in inducing high levels of Th1 immune responses, especially CD8+ T cells, was investigated. To track the dynamic changes of local DCs distribution profile after MEES treatment, we established a CD11c-cre; tdTomatofl/wt mouse strain (Figure 5M). DCs and LCs from these mice expressed the fluorescent protein tdTomato and thus could be tracked in real-time. CD11c-cre; tdTomatofl/wt mice were first i.d. injected with pEGFP in the presence of MEES or MN, and were subsequently subject to intravital microscopy (Figure 5M). Intriguingly, MEES dramatically recruited DCs into the local skin area, while MN had a very weak recruitment effect (Figure 5N). This recruitment began to show up at 24 h and culminated at 48 h. As a result, a large number of DCs gathered around the pinhole in a circular shape, but kept some distance from the center (Figure 5N). Accumulation of DCs at 48 h coincided with the histology data showing more infiltrated cells were found in MEES at this time point (Figure 4B). The flow cytometry was used to further confirm the recruitment of DCs. WT C57BL/6 mice received pOVA with MN or MEES treatment. Skin cells were isolated 48 h later and analyzed for their expression of CD45, CD64, CD11c, MHC II, CD11b, CD207 and XCR1 (Figure S3).36 The number of total conventional DC (cDC, CD45+ CD64− CD11c+ MHC II+) was significantly higher in the skin treated with MEES than that treated with MN (Figure 5O). In detail, MEES profoundly recruited cDC2 (CD45+ CD64− CD11c+ MHC II+ CD11b+ CD207-) into the skin (Figure 5Q), whereas no difference was found for the numbers of cDC1 (CD45+ CD64− CD11c+ MHC II+ CD11b− CD207+ XCR1+) and LCs (CD45+ CD64− CD11c+ MHC II+ CD11b+ CD207+) between MN and MEES (Figures 5P and 5R). These results suggested that the recruitment of DCs might be responsible, at least in part, for the superior capability of MEES to promote the induction of CD8+ T cell responses.
Micro-electroporation/electrophoresis system greatly enhanced the immunogenicity of SARS-CoV-2 DNA vaccines
We then set out to test what kind of DNA vaccine would be the best partner for MEES. There were two main classes of antigens encoded by existing SARS-CoV-2 nucleic acid vaccines, Receptor-binding domain (RBD) or full-length spike protein (S).37,38,39 S protein is the major surface antigen of SARS-CoV-2, while RBD is the key subunit responsible for binding to the angiotensin-converting enzyme II (ACE2) of target cells. Different delivery systems mediate different biological processes, such as the kinetics of DC recruitment, and therefore need to be coupled with the right vaccine to maximize its efficacy. Thus, we designed two types of DNA vaccines. The RBD vaccine contained a DNA sequence coding RBD and a human IgE signal peptide (GenBank: AH005278.2) to facilitate the secretion of RBD protein (Figures S4A–S4C).40 On the other hand, the S6P vaccine coded the full-length S protein comprising a transmembrane domain, resulting in a membrane-bound antigen (Figures S4F–S4H). Compared to natural S proteins, S6P has proline substitutions at six sites (F817P, A892P, A899P, A942P, K986P, V987P) on the S2 region to improve structural stability.41
MEES effectively enhanced the immunogenicity of the S6P vaccine, but was not effective for the RBD vaccine in inducing humoral immune responses (Figures S4D, S4E, S4I, and S4J). For cellular immune responses, elevated CD8+ T cell immune responses against the S510 epitope were indeed detected in mice receiving 10 μg of RBD vaccine and MEES (Figures S5A and S5D). S510 is the dominant epitope of the RBD region in C57BL/6 mice.42 Although the S6P vaccine did not induce significant levels of CD8+ T cell responses against the S510 epitope even with the assistance of MEES (Figures S5B and S5E), it did induce a high level of CD8+ T cell responses against another dominant epitope, S538, provided that MEES was used (Figures S5C and S5F). Concretely, MEES had augmented the S6P-induced CD8+ T cell response by 19.5-fold (Figures S5C and S5F). Since only 10 μg of S6P vaccine was immunogenic, this dosage was used in the following experiments (Figures S4 and S5). The fact that the S6P vaccine, but not the RBD, elicited a high level of immune response with the assistance of MEES was in line with our observation that the peak of DC recruitment by MEES was at 48 h, as the membrane-bound S protein might be able to stay at the vaccination site for a longer period of time, offering the late arriving DCs the opportunity to take them up.
We then designed the S6P vaccine encoding the Omicron BA.1 variant (pBA.1) and compared it with the vaccine encoding the WT strain (pWT). The immunogenicity of pBA.1 was slightly higher than that of pWT. Vaccination with pBA.1 vaccine alone could induce antigen-specific IgGs in some animals albeit the titer was low (1:59), whereas pWT did not induce any humoral immune response (<1:25), regardless of whether one or two injections were given (Figures 6A and 6B). In the presence of MEES, only one dose of pWT or pBA.1 vaccine was sufficient to elevate GMT of antigen-specific IgGs to 1:3456 and 1:25600, respectively, within 14 days of vaccination (Figures 6A and 6B). In other words, MEES increased the capability of pBA.1 in eliciting humoral immune responses by 433-fold. A booster shot of pWT + MEES further increased the IgG titer by 9-fold to 1:29863 (Figure 6A). In contrast, a second shot of pBA.1 + MEES could not increase the IgG titer further, probably because one shot was potent enough to plateau the immune response (Figure 6A). We also explored whether there was cross-protection between the immune responses induced by the two vaccines using a pseudovirus neutralization assay. Unsurprisingly, pWT-induced antibodies did not effectively neutralize the BA.1 strain, and vice versa (Figures S6A and S6B).
Figure 6.
Cross-neutralizing antibodies induced by sequential vaccination of multiple SARS-CoV-2 DNA vaccines
Mice were i.d. immunized with 10 μg SARS-CoV-2 S6P DNA vaccine coding the S protein of WT virus (pWT) or Omicron BA.1 variant (pBA.1) with or without MEES on day 0 and boosted on day 14. Sera were collected on day 14 (prime) or day 21 (boost).
(A) IgG titers against WT S protein were measured by ELISA. n = 8–9.
(B) IgG titers against BA.1 S protein were measured by ELISA. n = 8–9.
(C–I) Mice were i.d. immunized with 10 μg SARS-CoV-2 S6P DNA vaccine encoding S protein of WT strain, Delta variant, or BA.1 variant on days 0, 21, and 42 with or without MEES. Sera were collected on day 56. Neutralization assays were performed to measure NAb titers against pseudoviruses whose S protein was from WT (C), Delta (D), BA.1 (F) or BA.5 (H) viral strains. n = 6–9 mice. (E) p values of the pairwise comparison of (D). (G) p values of the pairwise comparison of (F).
(I) p values of the pairwise comparison of (H). Vaccines, immunized with DNA vaccine alone. +MEES, immunized with DNA vaccine followed by MEES treatment. The results were presented as geometric mean ±95% Cl (A-D, F, H). Statistical analysis, two-tailed Mann-Whitney U-test. ∗p < 0.05, ∗∗∗∗p < 0.0001, ns, no significance.
These results suggested the need for a screening system to identify appropriate vaccine strategies that were effective in inducing cross-protective immune responses. Because of its high immunogenicity, the combination of the S6P DNA vaccine and MEES would be a promising candidate for this screening system.
The screening system revealed the impact of vaccination orders on inducing cross-protective immune responses
To test the capability of this MEES-based screening system, we tested a scenario of inducing cross-neutralizing antibodies against emerging strains (BA.5) by combining existing vaccines (WT, Delta, BA.1). Mice were immunized with three doses of vaccines, including homologous vaccines and combinations of three in various orders, with or without MEES on day 0, 21, and 42. Sera were collected on day 56. Thus, 9 combinations and 18 strategies were screened simultaneously (Figure S7). Serum neutralizing antibodies (NAbs) against SARS-CoV-2 WT strain, Delta variant, Omicron BA.1, or Omicron BA.5 variants were measured using a pseudovirus neutralization assay and quantified as half pseudovirus neutralization titers (PVNT50). First of all, MEES was demonstrated to be essential for the screening system, as it was difficult to effectively induce neutralizing antibodies against all of these strains in the absence of MEES, regardless of the vaccination strategy (Figures 6C, 6D, 6F, and 6H).
In the presence of MEES, mice immunized with three doses of WT or Delta DNA vaccines elicited potent NAbs against WT and Delta (Figures 6C and 6D), but NAbs against Omicron BA.1 were not induced (Figure 6F). On the other hand, mice immunized with three doses of BA.1 vaccine were equipped with robust NAbs against BA.1 and WT (Figures 6C and 6F), but not against Delta (Figure 6D). None of these homologous vaccination strategies was effective for inducing NAbs against BA.5. Only 28.6% of participants had NAbs against BA.5 after receiving three doses of BA.1 vaccine (Figure 6H).
Overall, all combinations of the three vaccines were able to induce cross-neutralizing antibodies against WT, Delta and BA.1, irrespective of the vaccination order (Figures 6C, 6D, and 6F). However, we found that the order BA.1-Delta-WT induced lower NAbs against Delta (Figures 6D and 6E), while the ability of Delta-WT-BA.1 to induce NAbs against BA.1 was weaker (Figures 6F and 6G). The Geometric Mean (GM) of PVNT50 of each strategy was summarized in Figure 7A. For the emerging strain BA.5, most strategies only induce cross-neutralizing antibodies in a small proportion of animals (Figures 6H and 7B). The ability of Delta-BA.1-WT to induce NAbs against BA.5 (BA.5-NAbs) was the weakest among all the heterologous immunizations (Figure 6H). BA.1-WT-Delta and WT-Delta-BA.1 were the only two strategies that significantly induced BA.5-NAbs after the addition of MEES (Figure 6I). Whilst BA.1-WT-Delta was the only strategy capable of inducing BA.5-NAbs in >50% (66%) of the mice (Figure 7B). Taken together, we found that the order of immunization had an important impact on the induction of cross-neutralizing antibodies. Some strategies, such as BA.1-Delta-WT, Delta-BA.1-WT, Delta-WT-BA.1, were less effective for one of the four tested strains, while the strategy BA.1-WT-Delta was among the best strategies for each strain (Figures 7A and 7B).
Figure 7.
Cellular immune responses induced by sequential vaccinations and analysis of epitopes
(A) The geometric means (GM) of PVNT50 against multiple viral stains after sequential vaccinations were summarized as a heatmap.
(B) The proportions of mice with PVNT50 above the median calculated from all mice were summarized as a heatmap.
(C) Mutations of SARS-CoV-2 variants were labeled on the structure of WT S protein (PDB ID: 6VYB) using Pymol 2.6. (D) Mutation sites and CD8+ T cell epitopes in the S1 subunit of Delta, BA.1, and BA.5 were summarized and compared.
(E) Mutation sites and CD8+ T cell epitopes in the S2 subunit were summarized and compared.
(F) Mice were sequentially immunized with 10 μg of WT-BA.1-Delta or BA.1-WT-Delta DNA vaccines on day 0, day 21, and day 42.
(G and H) Splenocytes were isolated on day 56. CD8+ T cells producing IFNγ after the S538 epitope peptide (G) or peptide pool (H) stimulation were measured by flow cytometry. The Peptide pool was a mixture of S263, S471, S510, S538, and S820 epitopes. n = 3–4. The results were shown as mean ± SEM. Statistical analysis, two-tailed Student’s t test. ∗p < 0.05 and ∗∗p < 0.01.
To understand how mutations affect the induction of NAbs, we further analyzed mutations of different variants based on the structure of the S protein.43,44,45 Mutations on S proteins are mainly concentrated in the N-terminal domain (NTD) and RBD (Figure 7C). BA.1 has more mutations than Delta, and BA.5 in turn has more mutations than BA.1 (Figures 7D and 7E). Although BA.1 shared mutations with BA.5 in the S2 region (Figure 7E), BA.5 had evolved multiple de novel mutations in the S1 region that were not present in BA.1, which might explain the inability of the BA.1 vaccine to effectively induce cross-neutralizing antibodies against BA.5 (Figure 7D). Notably, the mutation L452R occurred only in Delta and BA.5, suggesting that the addition of Delta vaccine would be beneficial for the induction of BA.5-NAbs (Figure 7D). These results suggested that heterologous boosting strategies could cover more mutations and were more likely to facilitate the induction of cross-neutralizing antibodies.
In addition to B-cell epitopes, we also analyzed the major CD8+ T cell epitopes, which are important for the clearance of infected cells.46 As shown in Figures 7D and 7E, these epitopes of SARS-CoV-2 were mainly concentrated in conserved regions, suggesting that CD8+ T cell responses might be able to provide good cross-protection, as we have observed in the influenza vaccine.3 We selected the two combinations, BA.1-WT-Delta which induced the strongest BA.5-NAbs, and WT-BA.1-Delta which induced mild BA.5-NAbs, to test their ability to induce CD8+ T cell responses. Intriguingly, both strategies could successfully induce comparable cellular immune responses against the S538 epitope or the epitope pool, provided that MEES was available (Figures 7F–7H). This data indicated that, unlike humoral immune responses, the induction of CD8+ T cell responses against conserved epitopes was always effective by DNA vaccine + MEES, regardless of vaccination orders.
Discussion
In this work, we have demonstrated that sequential immunization with different monovalent DNA vaccines based on existing viral stains could elicit a broader spectrum of NAbs capable of neutralizing emerging strains and CD8+ T cell responses against conserved epitopes. Sequential immunization with vaccines from multiple strains was more effective in inducing cross-protective immune responses than repeated immunization with vaccines from the same strain. It is also important to note that not every immunization order is competent. Thus, a screening system such as DNA vaccine + MEES is favorable to exclude poor candidates and identify the best immunization order.
Currently, new mutations of SARS-CoV-2 continue to emerge at an annual pace. Emerging variants often easily escape existing immune defenses and cause breakthrough infections, resulting in recurrent peaks in morbidity and mortality worldwide.4,47,48 A similar situation occurs every few years for influenza viruses as well.49 Vaccines based on ancestral viral strains do not provide sufficient protection against emerging variants. Many individuals who had received two or even three doses of SARS-CoV-2 vaccines still suffered from breakthrough infections in the Omicron predominant period.4,47 On the other hand, the development of vaccines often lags behind the evolution of viruses. By the time a new vaccine is developed and launched, the virus may have accumulated enough new mutations to make the vaccine much less effective. The ultimate solution is to develop a universal vaccine de novo to combat a large number of mutated strains, including those that may emerge in the future. However, many of the key technologies and theories involved in the development of a universal vaccine are being explored, and it may be difficult to have a universal vaccine on the market in a short period of time. Alternatively, the possibility of inducing cross-protection against future strains by combining vaccines against existing strains is another direction of interest. If feasible, it would be a faster, though not the best, solution.
The latter option was proved to be feasible in this study. Considering the high lethality of WT strain, as well as the high transmissibility of Delta and Omicron variants, we generated three kinds of DNA vaccines encoding spike protein of WT, Delta, or Omicron viruses, respectively. The results showed that three doses of the WT monovalent DNA vaccines (WT × 3) failed to elicit NAbs against BA.1 or BA.5 variants, supported by real-world evidence.4,47 Unlike two doses of the BA.1 vaccine, which could not induce NAbs against WT strain, three doses of the BA.1 vaccine did induce NAbs against WT. One possible explanation is that the third dose causes the emergence of new B cell clones and the increment of antibody breadth.50,51 However, three doses of BA.1 were still ineffective in inducing neutralizing antibodies against BA.5, which is consistent with our previous clinical findings that repeated administration of the same vaccine could not increase cross-neutralizing antibodies against the mutant strain in an unlimited manner.6
Compared with repeated injections of the same monovalent vaccine, sequential immunization with three kinds of monovalent DNA vaccines could elicit potent NAbs against WT, Delta, BA.1, and BA.5. Notably, the order of sequential immunization discernibly affects BA.5-NAbs. By comparing the efficiency of different vaccination strategies and the mutation profiles of variants, we speculated the reason why BA.1-WT-Delta was more effective is that the sequential immunization of WT-Delta might induce NAbs against L452R mutation shared by Delta and BA.5, as well as the existence of immune imprinting, which were recently reported in SARS-CoV-2 booster vaccination.52,53,54 As reported, repeated vaccination with inactivated vaccine back-boosts ancestral memory but dampens immune response to Omicron variants in patients suffering from Omicron breakthrough infection.52 In triple-vaccinated individuals, Omicron infection after previous ancestral infection provided weaker boosting for Omicron-specific response than Omicron infection with prior naive infection.53 The phenomenon of immune imprinting also existed in some other infections. For example, in the influenza pandemic of 1918, the peak in mortality among adults occurred in those born in 1890 (28-year-olds) when H3Nx virus was in a pandemic. Because of childhood exposure to H3Nx virus, H1N1 infection boosted immune response to the previous virus, but responses to H1N1 itself were muted.55 Consequently, these results suggested that sequential immunization with vaccines derived from various strains could cover more mutation sites, and subsequently induce broader neutralizing antibodies. On the other hand, due to the immune imprinting by previous antigenic exposure, we should pay special attention to the order of sequential immunization to induce robust immunity against emerging variants. Again, a screening system such as DNA vaccine + MEES would facilitate the identification of the best immunization order.
The MEES developed in this work remarkably potentiated the capacity of DNA vaccine to induce both humoral and cellular immunity. For example, MEES elevated the humoral and cellular immune responses induced by pOVA by over 400- and 35-fold respectively. There are two possible reasons for the effect of electric fields to promote the effectiveness of a nucleic acid vaccine. One is to facilitate the cellular entry of nucleic acids by generating temporary and reversible pores on cell membranes (electroporation effect) and helping negatively charged nucleic acids to migrate through the dense tissue and counter the negative charge repulsion on the cell surface (electrophoresis effect).56 By integrating electroporation and electrophoresis, MEES could very effectively enhance the expression of pLuc and pGFP in the skin, better than either the electroporation setting or the electrophoretic setting alone.
In addition to promoting the entry of nucleic acids into cells, MEES may also confer adjuvant effects by increasing the recruitment of DCs into the vaccination site. To the best of our knowledge, immunological effects of electric fields in the skin, especially of weak electric fields like those used in the present study, have rarely been reported. To study these immunological effects, we utilized our previously developed method for tracking cell death in live mice and newly constructed transgenic mice (CD11c-cre; tdTomatofl/wt) that can be used to observe DC changes in vivo. Compared to widely used MHC II-EGFP mice, CD11c-tdTomato mice enable imaging DCs in deeper tissues.21 MEES recruited a large number of DCs into the skin around the microneedles. Previous studies including ours have demonstrated that dead cells caused by needle piercing or lasers could recruit dendritic cells and confer adjuvant effects.21,22,57 However, our live imaging results showed that the addition of a weak electric field did not lead to increased cell death. Both MEES- and MN-treated skin exhibited only a small amount of cell death due to microneedle insertion, so dead cells were not the leading cause of DC recruitment. Nevertheless, weak electric stimulation can activate the innate immune pathway and thus cause DC recruitment in some manner, but the exact mechanism needs to be further investigated. In addition, since the massive recruitment of DCs takes 24–48 h, how to match the time of antigen expression and retention with the kinetics of DC recruitment will likely be an important direction to maximize the effectiveness of MEES.
In summary, a DNA vaccine and MEES-based screening system was developed in this work, which enabled fast screening enabled fast preparing a large variety of vaccines according to nucleic acid sequences of various existing viral strains and screening for the optimal vaccination strategy to induce cross-protective immunity against emerging viral variants. The gentle electrical stimulation of MEES could recruit DCs into the vaccination sites and was highly effective in boosting both humoral and cellular immune responses of S6P DNA vaccines for SARS-CoV-2. In the presence of MEES, cross-protective immune responses against future mutant viruses could be successfully induced by existing strain-derived vaccines when a proper combination and order of sequential vaccination were used, emphasizing the importance of a screening system. The as-developed MEES possessed high potential as an effective and versatile platform for intradermal delivery of DNA vaccine for fast vaccine screening. This strategy is promising for fast-seeking vaccination strategies to combat emerging mutated pathogens such as SARS-CoV-2 and Influenza viruses in the future.
Limitations of the study
Our study has several limitations. First, MEES, despite its excellent ability to enhance DNA vaccines, relies on sophisticated and expensive Arbitrary Waveform Generator and Amplifiers, so it is still an R&D device for vaccine strategy screening rather than a medical device for mass vaccination. Encouragingly, a recent elegant study reported the possibility of using a piezoelectric pulser to generate electrical pulses, thus making the electrotransfection device much smaller and cheaper.27 This will be the direction of future improvements of MEES for clinical applications. Second, the injection of DNA vaccine and MEES treatment were performed separately in this study, requiring an additional procedure of intradermal injection of the vaccine using a syringe. The development of an integrated microneedle system that incorporates injection and MEES will be another important direction in the future. Lastly, the mechanisms by which electrical stimulation interacts with the innate immune system in the skin are unclear. Elucidating these mechanisms will likely bring new advances in the delivery systems and adjuvants for nucleic acid vaccines.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HRP-conjugated sheep anti-mouse IgG | Cytiva | Cat #NA931 |
| HRP-conjugated goat anti-mouse IgG1 | Southern Biotech | Cat #1073-05 |
| HRP-conjugated goat anti-mouse IgG2c | Southern Biotech | Cat #1079-05 |
| anti-mouse CD28 | Biolegend | Cat #102102; RRID: AB_312867 |
| anti-mouse CD16/32 | Biolegend | Cat #101302; RRID: AB_312801 |
| FITC anti-mouse CD4 | Biolegend | Cat #100406; RRID: AB_312691 |
| PE anti-mouse CD8b.2 | Biolegend | Cat #140408; RRID: AB_10644002 |
| APC anti-mouse IFNγ | Biolegend | Cat #505810; RRID: AB_315404 |
| FITC anti-mouse CD45 | Biolegend | Cat #103107; RRID: AB_312972 |
| PE/Cyanine7 anti-mouse CD64 | Biolegend | Cat #139314; RRID: AB_2563904 |
| Alexa Fluor(R) 488 anti-mouse CD11c | Biolegend | Cat #117311; RRID: AB_389306 |
| Brilliant Violet 711(TM) anti-mouse I-A/I-E | Biolegend | Cat #107643; RRID: AB_2565976 |
| Pacific Blue (TM) anti-mouse/human CD11b | Biolegend | Cat #101224; RRID: AB_755986 |
| PE anti-mouse/human CD207 (Langerin) | Biolegend | Cat #144203; RRID: AB_2561498 |
| APC anti-mouse/rat XCR1 | Biolegend | Cat #148205; RRID: AB_2563931 |
| APC/Cyanine7 anti-mouse/human CD45R/B220 | Biolegend | Cat #103223; RRID: AB_313006 |
| SARS-CoV-2 Spike RBD antibody | Sino Biological | Cat #40592-T62 |
| HA-Tag antibody | CST | Cat #3724S |
| Tubulin beta antibody | Affinity Biosciences | Cat #AF7011; RRID: AB_2827688 |
| anti-rabbit IgG HRP-linked Antibody | CST | Cat #7074S |
| Bacterial and virus strains | ||
| pcDNA3-OVA | Addgene | Cat #64599 |
| pcDNA3.1(+)-SARS-CoV-2 Spike (WT) | Wang et al.6 | N/A |
| pcDNA3.1(+)-SARS-CoV-2 Spike (Omicron BA.1) | Wang et al.6 | N/A |
| psPAX2 | Wang et al.6 | N/A |
| pLenti-CMV-Puro-Luc (168w-1) | Wang et al.6 | N/A |
| pCMV-SARS-CoV-2 Spike (Delta) | Sino Biological | Cat #VG40804-UT |
| pCMV-SARS-CoV-2 Spike (Omicron BA.5) | Sino Biological | Cat #VG40930-UT |
| Chemicals, peptides, and recombinant proteins | ||
| Isoflurane | RWD | Cat #R510 |
| Hair removal cream | Nair | N/A |
| D-luciferin | Beyotime | Cat #ST198 |
| DRAQ7 | Biolegend | Cat #424001 |
| Ovalbumin | Sigma-Aldrich | Cat #A5503 |
| WT SARS-CoV-2 spike | Sino Biological | Cat #40589-V08B1 |
| WT SARS-CoV-2 RBD | Sino Biological | Cat #40592-V08H |
| Omicron BA.1 SARS-CoV-2 spike | Sino Biological | Cat #40589-V08H26 |
| ELISA coating buffer | Solarbio | Cat #C1055 |
| Tween-20 | Solarbio | Cat #T8220 |
| BSA | Genview | Cat #FA016-100G |
| TMB | Beyotime | Cat #P0209 |
| ELISA stopping solution | Beyotime | Cat #P0215 |
| Lipo8000™ | Beyotime | Cat #C0533 |
| Polybrene | Beyotime | Cat #C0351 |
| Firefly luciferase lysis buffer | Beyotime | Cat #RG126M |
| Luciferase substrate | Beyotime | Cat #RG058M |
| Erythrocyte lysate | Biosharp | Cat #BL503B |
| OVA257-264 (SIINFEKL) | Synpeptide | N/A |
| OVA323-339 (ISQAVHAAHAEINEAGR) | Synpeptide | N/A |
| S263 (AAYYVGYL) | Synpeptide | N/A |
| S471 (EIYQAGST) | Synpeptide | N/A |
| S510 (VVVLSFEL) | Synpeptide | N/A |
| S538 (CVNFNFNGL) | Synpeptide | N/A |
| S820 (DLLFNKVTL) | Synpeptide | N/A |
| Brefeldin A solution | Biolegend | Cat #420601 |
| Zombie Violet™ Fixable Viability Kit | Biolegend | Cat #423113 |
| 4% PFA | leagene | Cat #DF0135 |
| Saponin | Beyotime | Cat #P0095 |
| Collagenase type IV | Sigma | Cat #C5138 |
| Dispase II | Roche | Cat #4942078001-1 |
| DNase I | Roche | Cat #11284932001 |
| Hyaluronidase | Solarbio | Cat #8030 |
| Fixable Viability Stain 440UV | BD | Cat #566332 |
| RIPA buffer | Beyotime | Cat #P0013B |
| PMSF | Beyotime | Cat #ST506 |
| 5 × SDS-PAGA Loading Buffer | Beyotime | Cat #P0015 |
| 10% polyacrylamide gel | ACE | Cat #ET12010LGel |
| PVDF membrane | Merck Millipore | Cat #ISEQ00010 |
| TBST | Biosharp | Cat #JXF003 |
| ECL Western blot substrate reagents | Fude | Cat #FD8020 |
| Deposited data | ||
| Original Western blot images | This paper; Mendeley Data | https://doi.org/10.17632/zmtf5ykwkf.1 |
| Experimental models: Cell lines | ||
| Human: HEK293T cells | Wang et al.6 | N/A |
| Human: hACE2-293T cells | Wang et al.6 | N/A |
| Experimental models: Organisms/strains | ||
| WT C57BL/6J mice | GemPharmatech Co. Ltd | N/A |
| Mouse: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (or tdTomatofl/fl) | The Jackson Laboratory | JAX: 007909 |
| Mouse: B6.Cg-Tg(Itgax-cre)1-1Reiz/J (or CD11c-cre) | The Jackson Laboratory | JAX: 008068 |
| Recombinant DNA | ||
| pcDNA3.1-Luc | This paper | N/A |
| pcDNA3.1-EGFP | This paper | N/A |
| pcDNA3.1-SARA-CoV-2 RBD (WT) | This paper | N/A |
| pcDNA3.1-SARA-CoV-2 S6P (WT) | This paper | N/A |
| pcDNA3.1-SARA-CoV-2 S6P (Delta) | This paper | N/A |
| pcDNA3.1-SARA-CoV-2 S6P (Omicron BA.1) | This paper | N/A |
| Software and algorithms | ||
| COMSOL Multiphysics 5.4 | COMSOL | N/A |
| IVIS Lumina III | PerkinElmer | N/A |
| Living Image Software v 4.4 | PerkinElmer | N/A |
| ImageJ software (version 1.51) | National Institutes of Health | N/A |
| KFbio Slide Viewer (KF-PRO-020) | KFBIO | N/A |
| Varioskan Lux Microplate Reader | Thermo Fisher | N/A |
| GraphPad Prism (version 9.51) | GraphPad Software, Inc. | N/A |
| FACSymphony A3 | BD | N/A |
| Pymol 2.6 | Schrodinger | N/A |
| FlowJo (version 10.0) | BD | N/A |
| Other | ||
| Arbitrary Waveform Function Generator | RIGOL | DG1022Z |
| High Voltage Amplifier | Aigtek | ATA-214 |
| Microneedle array | This paper | N/A |
| The structure of WT SARA-CoV-2 Spike protein | PDB | ID: 6VYB |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Ji Wang (wangj683@mail.sysu.edu.cn).
Materials availability
pcDNA3.1-Luc, pcDNA3.1-EGFP, pcDNA3.1-SARS-CoV-2-RBD (WT), pcDNA3.1-SARS-CoV-2-S6P (WT), pcDNA3.1-SARS-CoV-2-S6P (Delta), and pcDNA3.1-SARS-CoV-2-S6P (Omicron BA.1) generated in this study. These plasmids will be made available upon request.
Experimental model and study participant details
Animals
Specific pathogen-free (SPF) 6-8 weeks-old female C57BL/6J mice were purchased from GemPharmatech Co. Ltd (Nanjing, China). In order to generate CD11c-cre;tdTomatofl/wt mice, CD11c-cre transgenic mice were mated with homozygous tdTomatofl/fl mice. Both mouse lines were from Jackson Laboratory. All animal experiments were approved by the Institutional Animal Care and Use Committee, Sun Yat-Sen University (Approval No. SYSU-IACUC-2021-000203, SYSU-IACUC-2022-000614), and IEC for Clinical Research and Animal Trials of the First Affiliated Hospital of Sun Yat-sen University (Approval No. [2021]869).
Cell lines
Human ACE2 overexpression stable HEK293T (hACE2-293T, PackGene Biotech) cells were cultured under an atmosphere of 5% CO2 at 37°C in Dulbecco’s modified Eagle’s medium (DMEM, #10-013-CVRC, Corining) supplemented with 10% inactivated FEB (#FSP500, ExCellBio), non-essential amino acids (NEAA, #11140-050, Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin (#SV30010, HyClone). Specially, 1 μg/ml Puromycin was applied for hACE2-293T culture. hACE2-293T cell lines were passaged less than 15 generations and examined the mycoplasma by PCR kit (#G1900, Servicebio).
Plasmids
The DNA encoding luciferase or EGFP was cloned into pcDNA3.1(+) vector to construct pcDNA3.1-Luc or pcDNA3.1-EGFP plasmid, respectively. The pcDNA3-OVA plasmid was obtained from Addgene (#64599). The pcDNA3.1(+) plasmids encoding the spike protein from ancestral WT strain, and Omicron BA.1 variant, as well as psPAX2 and pLenti-CMV-Puro-Luc (168w-1) plasmids, were described in our previous study.6 The pCMV plasmids expressing the optimized spike protein from Delta (VG40804-UT) or Omicron BA.5 (VG40930-UT) variants were purchased from Sino Biological (Beijing, China).
Method details
Fabrication of MEES
The MEES was fabricated by assembling a microneedle array with the circuit to receive electrical pulses. The electrical pulses generation device contains an Arbitrary Waveform Function Generator (DG1022Z, RIGOL, China) and a High Voltage Amplifier (ATA-214, Aigtek, China). In detail, a microneedle array was fabricated by assembling four rows of stainless steel microneedles in a 3D-printed insulative basement. Each row had nine microneedles, each spaced 600 μm apart measured from bottom to bottom. The adjacent microneedle rows were connected to opposite electrical polarity, and the distance between the adjacent anode and cathode was 1 mm. The high-precision laser etching technology was used to prepare the microneedles with grooves. Each needle was 1 mm in height. The bottom of each needle is 220 μm wide, gradually narrowing to 150 μm and finally forming the tip. A groove was carved into each needle from the lower middle to the tip. The groove was 110 μm in wide and 50 μm in depth.
COMSOL modeling
To reveal the electric field distribution in the skin during the electroporation process, the FEA was computed by the Electric Currents (ec) interface of COMSOL Multiphysics 5.4, which the main equation is as follows:
Where V is potential, E refers to the intensity of the electric field, J refers to current density, σ is the material conductivity, and ∇ refers to Hamiltonian. Simultaneous above equations resulted in a Laplace equation for the calculation of electric potential and electric field:
Considering the extra boundary of the model was insulated and the tips of the microneedle were totally inserted into the skin, the frame of the device and the stratum corneum of the skin were omitted. The conductivity of the microneedle was set to 1 × 107 S/m, and the conductivity of the skin was set to 0.2 S/m. The back of the four microneedles was set as ground and potential boundary alternately. In the study of steady electric field distribution in different potentials, the potential was set from 30 V to 90 V.
Bioluminescence imaging
Different doses of pLuc (1, 3, 5, 10, 15, and 20 μg) were dissolved in 20 μL PBS. Mice were anesthetized by the inhalation anesthetic isoflurane (#R510, RWD, China), at 5% for anesthetic induction and 2% for maintenance. The hair on the lower back of the mice was removed using the hair removal cream (Nair, Australia) for visualization of the skin and proper microneedle insertion. Plasmids were intradermally injected into one or both flanks of the mice with a 29G insulin needle (#320312, BD, USA). Immediately after injection, MEES treatment was performed. In brief, the microneedle array was inserted into the injected spot, and electric pulses described in Figures 3B and 3D were applied. After 48 hours, mice were intraperitoneally injected with 250 μL D-luciferin (15 mg/mL, #ST198, Beyotime, China), and in vivo bioluminescent imaging was performed 15 minutes later with an IVIS Lumina III (PerkinElmer, Waltham, USA). The total flux in the region of interest (ROI) was measured using the Living Image Software v 4.4.
Intravital confocal imaging
For EGFP expression imaging, 10 μg pEGFP plasmid dissolved in 20 μL PBS was intradermally injected into the lower back of CD11c-cre;tdTomatofl/wt transgenic mice. Subsequently, the injected spots were applied with MEES or MN treatment. EGFP expression and accumulation of DCs were then imaged by confocal microscopy (Olympus FV-3000). For dead cell imaging, 100 μL of DRAQ7 (10 μM) (#424001, Biolegend, USA) was subcutaneously injected the pcDNA3.1-EGFP plasmid-treated site of wild-type C57BL/6J mice. Then EGFP expression and dead cells at the treated site were imaged by confocal microscopy. In this experiment, 60 V/50 μs for 2 pulses + 30 V/10 ms for 8 pulses was set for MEES. The mean fluorescence intensity of EGFP was quantified by the ImageJ software (version 1.51).
Histological examination
The lower dorsal skin of mice was treated with 20 μL of PBS followed by MN or MEES application (60 V/50 μs for 2 pulses + 30 V/10 ms for 8 pulses). Then the mice were sacrificed at indicated times, and the treated skin tissue was dissected, fixed and stained using a standard H&E procedure. The slides were photoed and analyzed by KFbio Slide Viewer (KF-PRO-020, China). Cell numbers in the skin were quantified by the ImageJ software (version 1.51).
Immunization study in mice
For mice immunization with DNA vaccine, female C57BL/6J mice were injected intradermally with DNA vaccines with or without MEES, which was given immediately after the injection. Electroporation/electrophoresis conditions of 60 V/50 μs for 2 pulses + 30 V/10 ms for 8 pulses was set for MEES. For OVA-encoding DNA plasmid vaccination, the mice were immunized with two vaccinations on days 0 (prime) and 14 (boost) and sera were collected on days 14 and 21. To screen out a more efficient DNA vaccine for SARS-CoV-2 WT strain, the mice were immunized with different doses (2.5 or 10 μg per mouse) of DNA vaccines encoding RBD or S6P protein of SARS-CoV-2 WT strain. Two vaccinations on days 0 and 14 were applied and sera were collected on days 14 and 21. For OVA protein vaccination, mice were immunized with 100 μg OVA protein alone on days 0 and 14, and sera were collected on days 14 and 21. For three doses of various DNA vaccines (expressing S6P protein of WT, Delta, or BA.1 strains) immunization, the mice received 10 μg per dose of DNA vaccines on days 0, 21, and 42, and sera were collected on day 56. Pre-immunization sera were collected before starting the primary immunization. Splenocytes were isolated immediately after the last sera collection. The sera were kept at −20°C before use.
ELISA
The proteins OVA (#A5503, Sigma-Aldrich, USA), WT SARS-CoV-2 spike (#40589-V08B1, Sino Biological, China), RBD (#40592-V08H, Sino Biological, China), or Omicron BA.1 SARS-CoV-2 spike (#40589-V08H26, Sino Biological, China) were used to coat the 96-well ELISA plates (#655061, Greiner Bio-one, Germany) at a final concentration of 1 μg/mL in coating buffer (#C1055, Solarbio, China) at 4°C overnight, respectively. Plates were washed three times with PBS containing 0.5% Tween-20 (#T8220, Solarbio, China) (PBST), followed by blocking using 5% BSA (#FA016-100G, Genview, China) in PBST (blocking buffer) for 1 h at room temperature. Plates were washed 3 times with PBST. Then, 100 μL serially diluted sera were added and incubated for an additional 1 h at room temperature. Detailedly, sera were first diluted 25-fold, followed by 4-fold serial dilution. Plates were washed 4 times with PBST, and incubated with 50 μL/well HRP-conjugated sheep anti-mouse IgG (1:4000, #NA931, Cytiva, USA), HRP-conjugated goat anti-mouse IgG1 (1:3000, #1073-05, Southern Biotech, USA), or HRP-conjugated goat anti-mouse IgG2c (1:3000, #1079-05, Southern Biotech, USA) antibodies in blocking buffer for 30 min at room temperature. Subsequently, the plates were washed five times using PBST and developed with 100 μL/well TMB (#P0209, Beyotime, China) for 15 min at room temperature. The reactions were stopped by 50 μL/well of the stopping solution (#P0215, Beyotime, China). OD450 was measured using Varioskan Lux Microplate Reader (Thermo Fisher, USA).
Pseudovirus neutralization assay
The pseudovirus neutralization assay was performed by measuring the infection of hACE2-293T.6 To generate pseudoviruses with S protein of WT, Delta, BA.1, or BA.5 strains, pcDNA3.1-spike (WT, Delta, BA.1, or BA.5), psPAX2 and pLenti-CMV-Puro-Luc (168w-1) plasmids were co-transfected to HEK293T using lipo8000™ (#C0533, Beyotime, China) according to the manufacturer’s instruction. The culture supernatants containing Luc-expressing pseudoviruses were harvested after 72 h and stored at −80°C until use. The hACE2-293T cells (2 × 104 cells/well) were seeded in the black flat-bottom 96-well plates and grown overnight. Inactivated sera were first diluted 10-fold then 4-fold serially diluted in DMEM, and subsequently co-incubated with the same volume of pseudovirus for 1 h at 37°C. The mixtures were subjected with 10 μg/mL polybrene (#C0351, Beyotime, China) to hACE2-293T cells for 6-h absorption. The mixtures were discarded, and fresh culture medium was added and incubated for an additional 42 h at the cell incubator. After incubation, the infected cells were lysed using firefly luciferase lysis buffer (#RG126M, Beyotime, China), and the luciferase substrate (#RG058M, Beyotime, China) was applied for the luciferase assay. The relative light unit (RLU) was measured using Varioskan Lux Microplate Reader (Thermo Fisher, USA). The half pseudovirus neutralization titers (PVNT50) were determined with a four-parameter nonlinear regression curve (GraphPad Prism, version 9.51).
Intracellular cytokine staining
Splenocytes were isolated from mice 7 d after the second vaccination or 14 d after the third vaccination, by passing the spleen through a 40 μm cell strainer (#93040, SPL life sciences, Korea), followed by lysis of red blood cells using 5 mL of erythrocyte lysate (#BL503B, Biosharp, China) for 5 min on ice. Cells at 1 × 107 cells/mL were stimulated for 2 h at 37°C with OVA or SARS-CoV-2 spike epitope peptides (5 μg/mL) in the presence of anti-mouse CD28 (1:125, #102102, Biolegend, USA). The peptides used include OVA257-264 (SIINFEKL) for OVA CD8+ T cell epitope, OVA323-339 (ISQAVHAAHAEINEAGR) for OVA CD4+ T cell epitope, as well as SARS-CoV-2 spike CD8+ T cell epitope containing S263 (AAYYVGYL), S471 (EIYQAGST), S510 (VVVLSFEL), S538 (CVNFNFNGL), and S820 (DLLFNKVTL). Brefeldin A solution (1:1000, #420601, Biolegend, USA) was applied for the culture and incubated for an additional 4 h. The stimulated cells were collected and blocked with anti-mouse CD16/32 (1:500, #101302, Biolegend, USA). Then the cells were stained with Zombie Violet™ Fixable Viability Kit (1:400, #423113, Biolegend, USA) and anti-mouse CD4 (FITC, 1:100, #100406, Biolegend, USA) and anti-mouse CD8b.2 (PE, 1:100, #140408, Biolegend, USA). Cells were subsequently fixed using 4% PFA (#DF0135, leagene, China) and permeabilized with Saponin (#P0095, Beyotime, China) and stained with anti-mouse IFNγ (APC, 1:100, #505810, Biolegend, USA). The stained cells were acquired on a FACSymphony A3 (BD) and analyzed using FlowJo (version 10.0).
Flow cytometry
Skin samples (1 × 0.6 cm full-thickness pieces) were cut into small fragments and incubated in a digestion solution (RPMI1640 containing 10% FBS, HEPES, 0.2 mg/ml Collagenase type IV (#C5138, Sigma, USA), 0.2 mg/ml Dispase II (#4942078001-1, Roche, Switzerland), 0.67 mg/ml DNase I (#11284932001, Roche, Switzerland), and 0.2 mg/ml Hyaluronidase (#8030, Solarbio, China) for 1 hour at 37°C. Then skin cells were filtered on a cell strainer with 40-μm pores (#93040, SPL life sciences, Korea) to prepare single-cell suspensions. The cells were washed with PBS containing 2% FBS, and incubated with anti-mouse CD16/32 antibody (1:100, #101302, Biolegend, USA) for 20 min on ice. Subsequently, surface molecules were stained by incubation for 1 hour on ice with antibodies against CD45 (FITC, 1:100, #103107, Biolegend), CD64 (PE/Cyanine 7, 1:100, #139314, Biolegend), CD11c (Alexa Fluor 488, 1:100, #117311, Biolegend), MHC II (Brilliant Violet 711, 1:100, #107643, Biolegend), CD11b (Pacific Blue, 1:100, #101224, Biolegend), CD207 (PE, 1:100, #144203, Biolegend), XCR1 (APC, 1:100, #148205, Biolegend), and B220 (APC/Cyanine 7, 1:100, #103223, Biolegend), followed by staining with Fixable Viability Stain 440UV (1:200, #566332, BD, USA) for 10 min on ice. Stained samples were analyzed in a FACSymphony A3 (BD) and data was analyzed using FlowJo (version 10.0).
Western blot
Plasmids pcDNA3.1-SARA-CoV-2 RBD (WT) or pcDNA3.1-SARA-CoV-2 S6P (WT) were transfected into HEK293T cells using lipo8000™ (#C0533, Beyotime, China) following standard manufacturer’s instructions. Cell culture supernatants were collected 72 hours later, and cells were lysed in RIPA buffer (#P0013B, Beyotime, China) containing PMSF (#ST506, Beyotime, China). Samples were mixed with 5 × SDS-PAGA Loading Buffer (#P0015, Beyotime, China), inactivated, and separated on a 10% polyacrylamide gel (#ET12010LGel, ACE, China). After electrophoresis, the separated proteins were transferred to PVDF membrane (#ISEQ00010, Merck Millipore, USA), which was blocked using 5% fat-free milk in TBST (#JXF003, Biosharp, China) for 1 hour at room temperature, and subsequently incubated with specific primary antibodies at 4 °C overnight. Anti-RBD antibody (1:2000, #40592-T62, SinoBiological, Cina) was used to detect RBD protein expression. Spike protein expression was analyzed using antibodies (1:2000, #3724S, CST, USA) against HA tag which was fused with the C-terminal of Spike protein. The expression of the internal reference gene was detected using anti-β-Tubulin antibody (1:2000, #AF7011, Affinity Biosciences). After incubation, HRP-conjugated anti-rabbit IgG Antibody (1:3000, #7074S, CST, USA) was used as the secondary antibody, incubating for 1 hour at room temperature. The signals were detected using ECL Western blot substrate reagents (#FD8020, Fude, China).
Quantification and statistical analysis
All statistical analyses were performed using GraphPad Prism (version 9.51). For the data fitting a normal distribution, the results were shown as mean ± SEM, and p values were calculated by one-way ANOVA with Tukey’s multiple comparisons test, one-way ANOVA with Dunnett’s multiple comparisons test, two-tailed Student’s t test, or Pearson correlation coefficients. For the data not fitting a normal distribution, the results were shown as geometric mean with 95% Cl, and p values were calculated by two-tailed Mann-Whitney U-test or Kruskal-Wallis ANOVA with Dunn’s multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns, no significance. Details were shown in the figure legends.
Acknowledgments
We thank Dr. Lu Lu at Fudan University for the kind help in the pseudovirus neutralization assay. Schematic diagrams were created with BioRender.com. The work is supported by the National Key Research and Development Program of China (2022YFC2305800, 2021YFA0911100), the National Natural Science Foundation of China (82341042, 31970870, 32270993, 22007105, 62105380, 32171399, T2225010), China Postdoctoral Science Foundation (2021M693686, 2022T150763), Key Area Research and Development Program of Guangdong Province (2022A1111070001), the Science and Technology Program of Guangzhou (202103000076), Guangzhou Health Science and Technology Project (20201A011040), the Fundamental Research Funds for the Central Universities, Sun Yat-sen University (22qntd3201, 22dfx02), the Independent Fund of the State Key Laboratory of Optoelectronic Materials and Technologies, Sun Yat-sen University (OEMT-2022-ZRC-04), and Open Funds of State Key Laboratory of Oncology in South China (HN2022-01).
Author contributions
J.W. and X.X. conceived and supervised the study. YY.L., J.M., J.L., C.D., M.L., Z.H., J.J., X.W., L.S., and X.L. performed the experiments and collected data. YY.L., J.M., J.L., and Y.L. performed the statistical analysis, simulation assay, and diagram drawing. YY.L., J.M., J.L., and Y.L. drafted the article. J.W. and X.X. made a critical revision. All authors approved the final version before submission.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: September 28, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108086.
Contributor Information
Xi Xie, Email: xiexi27@mail.sysu.edu.cn.
Ji Wang, Email: wangj683@mail.sysu.edu.cn.
Supplemental information
Data and code availability
-
•
Original western blot images have been deposited at Mendeley data and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
References
- 1.Wei C.J., Crank M.C., Shiver J., Graham B.S., Mascola J.R., Nabel G.J. Next-generation influenza vaccines: opportunities and challenges. Nat. Rev. Drug Discov. 2020;19:239–252. doi: 10.1038/s41573-019-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tenforde M.W., Kondor R.J.G., Chung J.R., Zimmerman R.K., Nowalk M.P., Jackson M.L., Jackson L.A., Monto A.S., Martin E.T., Belongia E.A., et al. Effect of antigenic drift on influenza vaccine effectiveness in the united states-2019-2020. Clin. Infect. Dis. 2021;73:e4244–e4250. doi: 10.1093/cid/ciaa1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Li P., Yu Y., Fu Y., Jiang H., Lu M., Sun Z., Jiang S., Lu L., Wu M.X. Pulmonary surfactant-biomimetic nanoparticles potentiate heterosubtypic influenza immunity. Science. 2020;367 doi: 10.1126/science.aau0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhlmann C., Mayer C.K., Claassen M., Maponga T., Burgers W.A., Keeton R., Riou C., Sutherland A.D., Suliman T., Shaw M.L., Preiser W. Breakthrough infections with SARS-cov-2 omicron despite mrna vaccine booster dose. Lancet. 2022;399:625–626. doi: 10.1016/S0140-6736(22)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J., Peng P., Cao X., Wu K., Chen J., Wang K., Tang N., Huang A.L. Increased immune escape of the new SARS-cov-2 variant of concern omicron. Cell. Mol. Immunol. 2022;19:293–295. doi: 10.1038/s41423-021-00836-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J., Deng C., Liu M., Liu Y., Li L., Huang Z., Shang L., Jiang J., Li Y., Mo R., et al. A fourth dose of the inactivated SARS-cov-2 vaccine redistributes humoral immunity to the n-terminal domain. Nat. Commun. 2022;13:6866. doi: 10.1038/s41467-022-34633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nomaguchi M., Doi N., Koma T., Adachi A. Hiv-1 mutates to adapt in fluxing environments. Microb. Infect. 2018;20:610–614. doi: 10.1016/j.micinf.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Creech C.B., Walker S.C., Samuels R.J. SARS-cov-2 vaccines. J. Am. Med. Assoc. 2021;325:1318–1320. doi: 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- 9.Momin T., Kansagra K., Patel H., Sharma S., Sharma B., Patel J., Mittal R., Sanmukhani J., Maithal K., Dey A., et al. Safety and immunogenicity of a dna SARS-cov-2 vaccine (zycov-d): results of an open-label, non-randomized phase i part of phase i/ii clinical study by intradermal route in healthy subjects in india. Eclinicalmedicine. 2021;38 doi: 10.1016/j.eclinm.2021.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinicaltrials.gov. phase i_ii study of covid-19 dna vaccine (ag0302-covid19 high-dose) https://www.clinicaltrials.gov/ct2/show nct04993586
- 11.Inovio pharmaceuticals. inovio reports fda partial clinical hold for planned phase 2/3 trial of covid-19 vaccine candidate ino-4800. https://ir.inovio.com/news-releases/news-releases-details/2020/inovio-reports-fda-partial-clinical-hold-for-planned-phase-2--3-trial-of-covid-19-vaccine-candidate-ino-4800/default.aspx
- 12.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 13.Muthumani K., Falzarano D., Reuschel E.L., Tingey C., Flingai S., Villarreal D.O., Wise M., Patel A., Izmirly A., Aljuaid A., et al. A synthetic consensus anti–spike protein dna vaccine induces protective immunity against middle east respiratory syndrome coronavirus in nonhuman primates. Sci. Transl. Med. 2015;7:301ra132. doi: 10.1126/scitranslmed.aac7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esquivel R.N., Patel A., Kudchodkar S.B., Park D.H., Stettler K., Beltramello M., Allen J.W., Mendoza J., Ramos S., Choi H., et al. In vivo delivery of a dna-encoded monoclonal antibody protects non-human primates against zika virus. Mol. Ther. 2019;27:974–985. doi: 10.1016/j.ymthe.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinicaltrials.gov. reveal 2 trial (evaluation of vgx-3100 and electroporation for the treatment of cervical hsil) https://www.clinicaltrials.gov/ct2/show/nct03721978
- 16.Gilbert R.A., Jaroszeski M.J., Heller R. Novel electrode designs for electrochemotherapy. Biochim. Biophys. Acta. 1997;1334:9–14. doi: 10.1016/s0304-4165(96)00119-5. [DOI] [PubMed] [Google Scholar]
- 17.Sersa G., Stabuc B., Cemazar M., Jancar B., Miklavcic D., Rudolf Z. Electrochemotherapy with cisplatin: potentiation of local cisplatin antitumour effectiveness by application of electric pulses in cancer patients. Eur. J. Cancer. 1998;34:1213–1218. doi: 10.1016/s0959-8049(98)00025-2. [DOI] [PubMed] [Google Scholar]
- 18.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the chadox1 ncov-19 vaccine (azd1222) against SARS-cov-2: an interim analysis of four randomised controlled trials in brazil, south africa, and the uk. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the bnt162b2 mrna covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., et al. Phase 1-2 trial of a SARS-cov-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Shah D., Chen X., Anderson R.R., Wu M.X. A micro-sterile inflammation array as an adjuvant for influenza vaccines. Nat. Commun. 2014;5:4447. doi: 10.1038/ncomms5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., Li B., Wu M.X. Effective and lesion-free cutaneous influenza vaccination. Proc. Natl. Acad. Sci. USA. 2015;112:5005–5010. doi: 10.1073/pnas.1500408112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabashima K., Honda T., Ginhoux F., Egawa G. The immunological anatomy of the skin. Nat. Rev. Immunol. 2019;19:19–30. doi: 10.1038/s41577-018-0084-5. [DOI] [PubMed] [Google Scholar]
- 24.Del Prete A., Salvi V., Soriani A., Laffranchi M., Sozio F., Bosisio D., Sozzani S. Dendritic cell subsets in cancer immunity and tumor antigen sensing. Cell. Mol. Immunol. 2023;20:432–447. doi: 10.1038/s41423-023-00990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., et al. Systemic rna delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 26.Roos A.K., Moreno S., Leder C., Pavlenko M., King A., Pisa P. Enhancement of cellular immune response to a prostate cancer dna vaccine by intradermal electroporation. Mol. Ther. 2006;13:320–327. doi: 10.1016/j.ymthe.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Xia D., Jin R., Byagathvalli G., Yu H., Ye L., Lu C.Y., Bhamla M.S., Yang C., Prausnitz M.R. An ultra-low-cost electroporator with microneedle electrodes (epatch) for SARS-cov-2 vaccination. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2110817118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang T., Huang D., Li C., Zhao D., Li J., Zhang M., Chen Y., Wang Q., Liang Z., Liang X.J., et al. Rolling microneedle electrode array (romea) empowered nucleic acid delivery and cancer immunotherapy. Nano Today. 2021;36 doi: 10.1016/j.nantod.2020.101017. [DOI] [Google Scholar]
- 29.Wei Z., Zheng S., Wang R., Bu X., Ma H., Wu Y., Zhu L., Hu Z., Liang Z., Li Z. A flexible microneedle array as low-voltage electroporation electrodes for in vivo dna and sirna delivery. Lab Chip. 2014;14:4093–4102. doi: 10.1039/c4lc00800f. [DOI] [PubMed] [Google Scholar]
- 30.Chang H., Chew S.W.T., Zheng M., Lio D.C.S., Wiraja C., Mei Y., Ning X., Cui M., Than A., Shi P., et al. Cryomicroneedles for transdermal cell delivery. Nat. Biomed. Eng. 2021;5:1008–1018. doi: 10.1038/s41551-021-00720-1. [DOI] [PubMed] [Google Scholar]
- 31.Pavlin M., Kandušer M. New insights into the mechanisms of gene electrotransfer--experimental and theoretical analysis. Sci. Rep. 2015;5:9132. doi: 10.1038/srep09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meglič S.H., Pavlin M. The impact of impaired dna mobility on gene electrotransfer efficiency: analysis in 3d model. Biomed. Eng. Online. 2021;20:85. doi: 10.1186/s12938-021-00922-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.André F.M., Gehl J., Sersa G., Préat V., Hojman P., Eriksen J., Golzio M., Cemazar M., Pavselj N., Rols M.P., et al. Efficiency of high- and low-voltage pulse combinations for gene electrotransfer in muscle, liver, tumor, and skin. Hum. Gene Ther. 2008;19:1261–1271. doi: 10.1089/hum.2008.060. [DOI] [PubMed] [Google Scholar]
- 34.Pavselj N., Miklavcic D. A numerical model of permeabilized skin with local transport regions. IEEE Trans. Biomed. Eng. 2008;55:1927–1930. doi: 10.1109/TBME.2008.919730. [DOI] [PubMed] [Google Scholar]
- 35.Huang D., Zhao D., Wang X., Li C., Yang T., Du L., Wei Z., Cheng Q., Cao H., Liang Z., et al. Efficient delivery of nucleic acid molecules into skin by combined use of microneedle roller and flexible interdigitated electroporation array. Theranostics. 2018;8:2361–2376. doi: 10.7150/thno.23438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashem S.W., Haniffa M., Kaplan D.H. Antigen-presenting cells in the skin. Annu. Rev. Immunol. 2017;35:469–499. doi: 10.1146/annurev-immunol-051116-052215. [DOI] [PubMed] [Google Scholar]
- 37.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., et al. A vaccine targeting the rbd of the s protein of SARS-cov-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 38.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mrna-1273 SARS-cov-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R., et al. A thermostable mrna vaccine against covid-19. Cell. 2020;182:1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., et al. Modified mrna vaccines protect against zika virus infection. Cell. 2017;168:1114–1125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh C.L., Goldsmith J.A., Schaub J.M., Divenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-cov-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davenport B.J., Morrison T.E., Kedl R.M., Klarquist J. Conserved and novel mouse cd8 t cell epitopes within SARS-cov-2 spike receptor binding domain protein identified following subunit vaccination. J. Immunol. 2021;206:2503–2507. doi: 10.4049/jimmunol.2100195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walls A.C., Park Y.J., Tortorici M.A., Wall A., Mcguire A.T., Veesler D. Structure, function, and antigenicity of the SARS-cov-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu P., Faraone J., Evans J.P., Zou X., Zheng Y.M., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., et al. Neutralization of the SARS-cov-2 omicron ba.4/5 and ba.2.12.1 subvariants. N. Engl. J. Med. 2022;386:2526–2528. doi: 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ou J., Lan W., Wu X., Zhao T., Duan B., Yang P., Ren Y., Quan L., Zhao W., Seto D., et al. Tracking SARS-cov-2 omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Signal Transduct. Targeted Ther. 2022;7:138. doi: 10.1038/s41392-022-00992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhuang Z., Lai X., Sun J., Chen Z., Zhang Z., Dai J., Liu D., Li Y., Li F., Wang Y., et al. Mapping and role of t cell response in SARS-cov-2-infected mice. J. Exp. Med. 2021;218 doi: 10.1084/jem.20202187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leung K., Lau E.H.Y., Wong C.K.H., Leung G.M., Wu J.T. Estimating the transmission dynamics of SARS-cov-2 omicron bf.7 in beijing after adjustment of the zero-covid policy in november–december 2022. Nat. Med. 2023;29:579–582. doi: 10.1038/s41591-023-02212-y. [DOI] [PubMed] [Google Scholar]
- 48.Tan S.T., Kwan A.T., Rodríguez-Barraquer I., Singer B.J., Park H.J., Lewnard J.A., Sears D., Lo N.C. Infectiousness of SARS-cov-2 breakthrough infections and reinfections during the omicron wave. Nat. Med. 2023;29:358–365. doi: 10.1038/s41591-022-02138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrova V.N., Russell C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018;16:47–60. doi: 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- 50.Muecksch F., Wang Z., Cho A., Gaebler C., Ben Tanfous T., Dasilva J., Bednarski E., Ramos V., Zong S., Johnson B., et al. Increased memory b cell potency and breadth after a SARS-cov-2 mrna boost. Nature. 2022;607:128–134. doi: 10.1038/s41586-022-04778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y., Zeng Q., Deng C., Li M., Li L., Liu D., Liu M., Ruan X., Mei J., Mo R., et al. Robust induction of b cell and t cell responses by a third dose of inactivated SARS-cov-2 vaccine. Cell Discov. 2022;8:10. doi: 10.1038/s41421-022-00373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao B., He L., Bao Y., Chen Y., Lu G., Zhang Y., Xu Y., Su B., Xu J., Wang Y., Yeap L.S. Repeated vaccination of inactivated SARS-cov-2 vaccine dampens neutralizing antibodies against omicron variants in breakthrough infection. Cell Res. 2023;33:258–261. doi: 10.1038/s41422-023-00781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reynolds C.J., Pade C., Gibbons J.M., Otter A.D., Lin K.M., Muñoz Sandoval D., Pieper F.P., Butler D.K., Liu S., Joy G., et al. Immune boosting by b.1.1.529 (omicron) depends on previous SARS-cov-2 exposure. Science. 2022;377:q1841. doi: 10.1126/science.abq1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q., Bowen A., Valdez R., Gherasim C., Gordon A., Liu L., Ho D.D. Antibody response to omicron ba.4-ba.5 bivalent booster. N. Engl. J. Med. 2023;388:567–569. doi: 10.1056/NEJMc2213907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worobey M., Han G.Z., Rambaut A. Genesis and pathogenesis of the 1918 pandemic h1n1 influenza a virus. Proc. Natl. Acad. Sci. USA. 2014;111:8107–8112. doi: 10.1073/pnas.1324197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bart Geboers M.H.J.S., Sanne Nieuwenhuizen M.R.S.P., Rafael V., Davalos M.P.B.R., Damijan Miklavčič M.P.M.R. High-voltage electrical pulses in oncology: irreversible electroporation, electrochemotherapy, gene electrotransfer, electrofusion, and electroimmunotherapy. Radiology. 2020:254–272. doi: 10.1148/radiol.2020192190. [DOI] [PubMed] [Google Scholar]
- 57.Depelsenaire A.C.I., Meliga S.C., Mcneilly C.L., Pearson F.E., Coffey J.W., Haigh O.L., Flaim C.J., Frazer I.H., Kendall M.A.F. Colocalization of cell death with antigen deposition in skin enhances vaccine immunogenicity. J. Invest. Dermatol. 2014;134:2361–2370. doi: 10.1038/jid.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Original western blot images have been deposited at Mendeley data and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.