Abstract
Number of factors, including newly emerging infectious diseases and an increase in multi-drug resistant microbial pathogens with particular relevance for Gram-positive bacteria, make the treatment of infectious diseases in hospital-based healthcare a major challenge in the medical community. 4-Aminobenzoic acid (PABA), has demonstrated a variety of biological actions particularly, antimicrobial activity. In our study we coupled this vitamin-like molecule with different isatin derivatives. We investigated the antibacterial activity of the synthesized Schiff's bases. The compounds showed high selective activity against Gram-positive bacteria and showed weak or no activity against both Gram-negative bacteria and fungi. Compound 2a showed highest activity against S. aureus and B. subtilis (MIC 0.09 mmol/L). Additionally, these substances exhibit strong anti-B. Subtilis biofilm formation. We were able to shed insight on the binding mode of these new inhibitors using in silico docking of the compounds in the binding sites of a 3D structure of B. subtilis histidine kinase/Walk. The binding free energy of the compound 2a to the catalytic domain walk, of histidine kinase enzyme of B. subtilis bacteria, was calculated using molecular mechanics/generalized born surface area scoring. The key residues for macromolecule–ligand binding were postulated. The optimized 3D protein–ligand binding modes shed light on the B. subtilis HK/Walk–ligand interactions that afford a means to assess binding affinity to design new HK/Walk inhibitor as antibacterial agents.
Keywords: Aminobenzoic acid, Antibacterial activity, Schiff’s bases, Synthesis, Docking study
1. Introduction
The treatment of bacterial illnesses, especially those brought on by Gram-positive bacteria, requires the use of antibiotics. Gram-positive bacteria are capable of causing variety of infections, including those of the skin, soft tissues, respiratory system, and even the life-threatening bloodstream. It can be difficult to design and create antibiotics that work well against Gram-positive bacteria. These bacteria's cell walls have a thick layer of peptidoglycan that serves as a defence against antibiotics. Furthermore, the development of resistance mechanisms against commonly used antibiotics in Gram-positive bacteria makes the hunt for novel and enhanced antibacterial drugs even more crucial. Understanding the precise methods by which these bacteria cause illnesses and discovering potential targets for inhibition are necessary for the discovery of anti-Gram-positive medicines. Enzymes that are involved in the production of cell walls, machinery used in protein synthesis, and crucial metabolic pathways are some of the targets for Gram-positive bacteria. The main objective is to create medications that may selectively target these bacterial components without having an adverse effect on the host.
In addition, bacterial cells adhere to surfaces in an irreversible manner, leading to the formation of quorum-sensing QS molecules, which in turn causes the formation of biofilm. Pseudomonas aeruginosa produces virulence factors as exotoxin A, lection, pyocyanin, and elastase through QS, whereas Staphylococcus aureus produce other virulence factors as protein A, enterotoxin, lipases, hemolysins, and fibronectin through QS (Yarwood et al., 2004; Carnes et al., 2010). The rapid rise in antibiotic resistance brought on by unchecked antibiotic use has become a serious health concern (Zhong and Zhao, 2018; Ma et al., 2019; Sarkar et al., 2020). Despite the fact that using antibiotics is the first line of defense against bacterial infections. Most antibiotics are designed to target the machinery involved in protein synthesis, which in turn causes the pathogenic cells to be destroyed (Nikaido, 2009). Sessile cells that produce biofilms are frequently unaffected, which help the infection survive. To battle the biofilm, this problem justifies the investigation of novel medications or substances that act as biofilm inhibitor antibacterial.
One of the main objectives of medicinal chemistry is the creation of novel therapeutic agents. In medical chemistry, 4-aminobenzoic acid (also known as p-aminobenzoic acid; PABA) is a well-known amino acid molecule with numerous uses in chemical reactions. PABA is an essential component of this metabolic process and is a substrate for the in vivo production of folic acid in numerous microbes and plants. PABA has been demonstrated to be an essential component in both the folate metabolism and microbial pathogenicity, as well as a precursor to coenzyme Q. However, neither PABA's biosynthesis nor necessity for mammals are known (Akberova, 2002, Crisan et al., 2014, Kadhum et al., 2016, Patel et al., 2019, Sowinska et al., 2019). When paired with other antibiotics, PABA offers a synergistic antibacterial activity for many microbial strains, such as Staphylococcus aureus or Pseudomonas aeruginosa, in contrast to its necessity for some germs. Listeria monocytogenes, Salmonella enteritidis, and E. coli were all directly susceptible to PABA's direct antibacterial activity. Low pH levels enhance this effect. Similar findings on the adjunctive/synergistic activity of antivirotics were made (Akberova, 2002, Kadhum et al., 2016, Patel et al., 2019). Numerous antioxidant and antiviral advantages have also been demonstrated for it. PABA and its derivatives are used as an active ingredient in sunscreens to filter UVB rays since they are ROS scavengers (Akberova, 2002, Kadhum et al., 2016, Patel et al., 2019). Phase II biotransformation takes place after the acid is passively absorbed. PABA can either be N-acetylated or bound to glycine in the liver to produce 4-aminohippuric acid. The complete urine excretion of PABA and its metabolites ensure their non-toxic and well-tolerated characters (Chang and Hu, 1996, Nortje et al., 2015). Sulfonamides, which are used to treat a variety of pathogenic-microbe illnesses, serve as structural analogs of PABA and competitively inhibit dihydropteroate synthase (Yun et al., 2012). Dihydropteroate synthase can also be affected by 4-acetamidobenzoic acid (acedoben), a naturally occurring metabolite of PABA (Kluczyk et al., 2002). Dihydropteroate synthase uses the antitubercular drug 4-aminosalicylic acid in place of PABA and incorporates it into the folate pathway to produce a hydroxyl dihydrofolate antimetabolite that inhibits the activity of dihydrofolate reductase, disrupting the purines and thymidylate biosynthesis (Yun et al., 2012, Zheng et al., 2013). A PABA is extensively employed in medicinal chemistry to develop medications, and many different pharmaceuticals with different therapeutic effects contain this scaffold. We can differentiate between more complicated PABA derivatives that additionally have their benzene rings substituted and “simple” PABA-derived medicines that primarily have their carboxyl or/and amino groups substituted. The carboxyl can exist in three different states: free, salt, or modified by the formation of an ester or amide bond. Typically, the amino group might be unalkylated, mono- or di-alkylated, or it can be part of an amide (Kluczyk et al., 2002). Isatin has been known for almost 150 years to have biological effects on mammals that are comparable to those of oxindole and endogenous polyfunctional heterocyclic chemicals (Da Silva et al., 2001). Isatin was first identified in 1841 as orange monoclinic crystals made by oxidising indigo dye with nitric and chromic acids (Varun, et al., 2019). Prior to its isolation from the Isatis genus of plants (Borad et al., 2014), Calanthe discolor LINDL (Yoshikawa et al., 1998), the cannonball tree fruits, as a the parotid gland’s secretions of Bufo frogs (Khan and Maalik, 2015), and as adrenalin metabolite in humans (Chiyanzu et al., 2003), this isatin was thought to be a synthetic moiety. Due to its advantageous electronic nature, the design and development of analogues with medicinal activity now include isatin. Isatin analogues such jp-8 g, MDM2-p53 inhibitors, nintedanib, orantinib, semaxanib, SPX-F, SU-9561, and sunitinib are among the medications that are currently on the market (Abdelrahman et al., 2022). Researchers are actively investigating chemical compounds having isatin scaffold for their antibacterial properties. These compounds exhibit therapeutic potential against a variety of harmful microorganisms. There several classes of isatin derivatives include benzylidines I (Süreyya and Semiha, 2009), Mannich bases II (Daisley and Shah, 1984), imines III, hydrazones IV (Radwan et al., 2022), hydrazide-hydrazones V (Lian et al., 2016), spiro derivatives VI-IX (Singh et al., 2018, Ibrahim et al., 2010; (Kenchappa et al., 2017), chalcones X (Ibrahim et al., 2010) (Fig. 1). Isatin scaffold have several biological activities such as isatine-5-sulphonamide anticancer agents (Eldeeb et al., 2022), indolinone incorporated thiosemicarbazone, thiazole and piperidinosulfonyl moieties anticonvulsant agents (Fayed et al., 2021), 3-[(1H-pyrazol-3-yl)imino]indolin-2-one derivatives as DNA gyrase inhibitor antimicrobial agents (Alzahrani et al., 2022), 2-one derivative incorporating thiazole moiety as DHFR and quorum sensing inhibitors (Alzahrani A.Y. et al., 2022), isatin-metal coordinates antimicrobial agents (Noreen and Sumrra, 2022, Sumrra et al., 2022) and spiro-oxindoles based on uracil derivatives as SARS-CoV-2 inhibitors targeting RNA polymerase and spike glycoprotein (El-Kalyoubi et al., 2022).
Fig. 1.
Chemical structure of classes of isatin derivatives.
Our current research focuses on the preparation of Schiff bases of substituted isatin with analogues of aminobenzoic acids, as well as their assessment as possible antibacterial agents. Through the imine bond, these condensed molecules combine two pharmacophores. Following that, target compounds were verified by mass spectrometry, 1H and 13C NMR, and NMR. Then, their ability to suppress biofilm formation and their efficacy against Gram-positive bacteria were assessed. Docking experiments validated the results from biological screening by predicting likely binding interactions of the target compounds with histidine kinase/Walk active sites using the crystal structure of the histidine kinase/Walk protein retrieved from the Protein Data Bank (3sl2) and Autodock software. The stability of the histidine kinase/Walk active site was assessed using molecular dynamic simulations to support its inhibitory activity.
2. Experiment
2.1. Chemistry
Alkyl bromides, substituted-aminobenzoic acids, and starting isatin were acquired from Sigma-Aldrich (St. Louis, MO, USA). An electrothermal melting point device (Stuart Scientific, Stone, Staffordshire, UK) was used to measure the melting points. Thin layer chromatography (TLC) was performed using precoated silica gel plates (Kieselgel 0.25 mm, 60G F254, Merck, Darmstadt, Germany) with a chloroform/methanol (8:2) developing system. The King Saud University, Saudi Arabia, research center conducted the spectral analysis of the newly synthesized substances. Nuclear magnetic resonance (NMR) spectra were collected using an NMR spectrophotometer (Bruker, Flawil, Switzerland) running at 500 MHz for 1H and 125.76 MHz for 13C, while infrared (IR) spectra were done on KBr discs using an FTIR spectrophotometer (Perkin Elmer, Shelton, CT, USA). A model 320 MS spectrometer from Bruker, Bremen, Germany, was used to capture mass spectra, and a model 2400 Perkin Elmer from Waltham, Massachusetts, USA, was used to conduct elemental analysis.
2.2. Synthesis of N-(substituted)isatin (1a,b)
A round bottom flask was equipped with a solution of potassium carbonate (13 mmol) ind DMF (100 mL), which was then agitated for five minutes at room temperature. Isatin (10 mmol) was added to the reaction mixture and stirring was continued for 30 min. The reaction mixture received 11 mmol of methyl/isobutyl bromide and continued stirring for 48 h at room temperature. The reaction mixture was poured into 200 mL of ice-cold water, extracted with ethyl acetate, dried over anhydrous sodium sulfate, and concentrated under vacuum. By utilizing a mixture of 1:9 ethyl acetate and hexane to purify the resulting residue, analytically pure N-substituted isatin was produced in quantifiable amounts.
1-(2-methylallyl)indoline-2,3-dione (1a). Yield 1.9 g, 94 %, mp 94–93 °C; IR (KBr, cm−1) 3265 (NH), 3059–2914 (aromtic C–H str), 1724 (C = O), 1601 (C = O), 956, 816 (arom C–H out of plane bending); 1H NMR (CDCl3, δ ppm): 1.78 (s, 3H, CH3), 4.31 (s, 2H, CH2), 4.98, 5.01 (2H, two s, non-equiv. CH2 = ), 6.90–7.63 (m, 4H, arom CH). 13C NMR (CDCl3, δ ppm): 19.79 (CH3), 46.04 (>CH2), 111.02 (=CH2), 113.42 (ArC), 117.25 (ArC), 123.77 (ArC), 125.26 (ArC), 138.0 (ArC), 138.32 (>C = ), 151.03 (ArC), 158.05 (C = O), 183.18 (C = O). MS (m/z) [M+] 201.53. Anal. calcd. for C12H11NO2: C, 71.63; H, 5.51; N, 6.96. Found: C, 71.48; H, 5.39; N, 6.82.
1-isobutylindoline-2,3-dione (1b). Yield 1.82 g, 90 %, mp 90–91 °C; IR (KBr, cm−1) 3230 (NH), 1722 (C = O), 1603 (C = O), 915, 746 (arom C–H out of plane bending),; 1H NMR (CDCl3, δ ppm): 0.99 (s, 6H, two CH3), 2.11–2.19 (m, 1H, CH), 3.54 (d, 2H, CH2), 6.90–7.60 (m, 4H, arom CH). 13C NMR (CDCl3, δ ppm): 20.19 (CH3), 27.00 (CH), 47.72 (CH2), 110.47 (ArCH), 117.49 (ArCH), 123.59 (ArCH), 125.36 (ArCH), 138.33 (ArCH), 151.48 (ArC), 158.42 (C = O), 183.61 (C = O). MS (m/z) [M+] 203.38. Anal. calcd. for C12H13NO2: C, 70.92; H, 6.45; N, 6.89. Found: C, 70.75; H, 6.32; N, 6.77.
2.3. Synthesis of (E)-4-(1-(2-methylallyl)-2-oxoindolin-3-ylideneamino)benzoic acid derivatives (2a,b-7a,b)
A mixture of ethanol (40 mL), glacial acetic acid (1 mL),1.5 g of isatin (10 mmol) and a suitable aminobenzoic acid compound (10 mmol) was refluxed overnight. After cooling the reaction mixture, the solvent was drawn out under reduced pressure. The residue was then mixed with water and ethylacetate (50/50), and the organic layer was separated and washed twice in diluted sulfuric acid and twice in brine. Under reduced pressure, the solvent was evaporated, and the precipitate was then crystallized from ethanol.
(E)-4-(1-(2-methylallyl)-2-oxoindolin-3-ylideneamino)benzoic acid (2a). Yield 2.8 g, 87 %, mp 208–207 °C; IR (KBr, cm−1), 2710–3100 (carboxylic O–H str), 1705 (C = O), 1645 (C = N), 1592 (C = O), 904, 800 (ArCH out of plane bending); 1H NMR (CDCl3, δ ppm): 1.82 (3H, s, CH3), 4.4 (2H, s, CH2), 5.01 (2H, s, =CH2), 6.58–9.19 (8H, m, ArCH), 10.35 (1H, s, COOH). 13C NMR (CDCl3, δ ppm): 19.71 (CH3), 45.71 (CH2), 113.02 (=CH2), 117.09 (ArC), 123.57 (ArC), 124.77 (ArC), 125.94 (ArC), 127.59 (ArC), 131.1 (ArC), 137.7 ((>C = ), 147.34 (ArC), 150.74 (ArC), 154.16 (ArC), 157.84 (ArC), 162.44 (C = N), 167.58 (C = O), 183.03 (C = O). MS (m/z) [M+] 320.89. Anal. calcd. for C19H16N2O3: C, 71.24; H, 5.03; N, 8.74. Found: C, 71.11; H, 5.08; N, 8.66.
(E)-2-(1-(2-methylallyl)-2-oxoindolin-3-ylideneamino)benzoic acid (3a). Yield 2.5 g, 78 %, mp 131–132 °C IR (KBr, cm−1) 752–913 (arom C–H out of plane bending), 3085–2650 (carboxylic O–H str)1582 (C = N), 1607 (C = O), 1727 (C = O),; 1H NMR (CDCl3, δ ppm): 1.64 (3H, s, CH3), 4.16 (2H, s, CH2), 4.87 (2H, s, =CH2), 6.73–8.02 (8H, m, ArCH), 10.2 (1H, s, COOH. 13C NMR (CDCl3, δ ppm): 19.81 (CH3), (CH2), 45.91, 111.07 (=CH2), 113.34 (ArC), 117.36 (ArC), 123.78 (ArC), 125.16 (ArC), 129.56 (ArC), 131.86 (ArC), 134.1 (ArC), 135.07 (ArC), 136.84 (ArC), 137.91 (>C = ), 138.4 (ArC), 150.97 (ArC), 158.04 (C = O), 169.59 (C = N), 183.21 (C = O). MS (m/z) [M+] 320.38. Anal. calcd. for C19H16N2O3: C, 71.24; H, 5.03; N, 8.74. Found: C, 71.11; H, 5.08; N, 8.66.
(E)-2-(1-(2-methylallyl)-2-oxoindolin-3-ylideneamino)-5-chlorobenzoic acid (4a). Yield 2.9 g, 82 %, mp 113–114 °C; IR (KBr, cm−1) 3125–2519 (carboxylic O–H str), 1728 (C = O), 1672 (C = O), 1578 (C = N), 757–907 (arom C–H out of plane bending); 1H NMR (CDCl3, δ ppm): 1.69 (3H, s, CH3), 4.2 (2H, s, CH2), 4.89 (2H, s, =CH2), 6.77–7.99 (8H, m, ArCH), 10.74 (1H, s, COOH). 13C NMR (CDCl3, δ ppm): 19.71 (CH3), 45.73 (CH2), 111.0 (=CH2), 113.08 (ArC), 117.18 (ArC), 119.26 (ArC), 121.55 (ArC), 123.63 (ArC), 124.89 (ArC), 130.67 (ArC), 133.42 (ArC), 137.75 (ArC), 138.32 (ArC), 146.75 (>C = ), 150.79 (ArC), 157.90 (C = N), 168.68 (C = O), 183.08 (C = O). MS (m/z) [M+] 354.64. Anal. calcd. for C19H15ClN2O3: C, 64.32; H, 4.26; N, 7.90. Found: C, 64.12; H, 4.14; N, 7.83.
(E)-2-(1-(2-methylallyl)-2-oxoindolin-3-ylideneamino)-4-chlorobenzoic acid (5a). Yield 2.65 g, 75 %, mp 128–129 °C; IR (KBr, cm−1) 3115–2528 (carboxylic O–H str), 1726 (C = O), 1609 (C = O), 1578 (C = N), 904, 755 (arom C–H out of plane bending); 1H NMR (CDCl3, δ ppm): 1.65 (3H, s, CH3), 4.17 (2H, s, CH2), 4.82 (2H, s, =CH2), 6.50–8.64 (m, 8H, arom CH), 11.30 (1H, s, COOH). 13C NMR (CDCl3, δ ppm): 19.76 (CH3), 45.85 (CH2), 111.06 (=CH2), 113.22 (ArC), 117.26 (ArC), 123.71 (ArC), 123.75 (ArC), 125.02 (ArC), 125.11 (ArC), 133.06 (ArC), 137.82 (>C = ), 138.39 (ArC), 139.51 (ArC), 150.88 (ArC), 158.02 (C = N),169.27 (C = O), 183.19 (C = O). MS (m/z) [M+] 354.48. Anal. calcd. for C19H15ClN2O3: C, 64.32; H, 4.26; N, 7.90. Found: C, 64.66; H, 4.02; N, 7.81.
(E)-4-(1-(2-methylallyl)-2-oxoindolin-3-ylideneamino)-2-hydroxybenzoic acid (6a). Yield 2.5 g, 74 %, mp > 300 °C; IR (KBr, cm−1) 3307 (phenolic OH), 2673–3085 (carboxylic O–H str), 1715 (C = O), 1608 (C = O), 1569 (C = N), 901, 749 (arom C–H out of plane bending); 1H NMR (CDCl3, δ ppm): 2.24 (s, 3H, CH3), 3.94 (s, 2H, CH2), 4.56, 4.62 (two s, 2H, non-equiv. CH2 = ), 6.46–7.31 (m, 7H, arom CH). 13C NMR (CDCl3, δ ppm): 19.68, 45.62, 102.32, 110.97, 117.3, 122.88, 123.56, 124.79, 125.75, 126.87, 129.30, 130.15, 131.68, 137.71, 138.27, 150.72, 157.84, 171.93, 176.95. MS (m/z) [M+] 336.47. Anal. calcd. for C19H16N2O4: C, 67.85; H, 4.79; N, 8.33. Found: C, 67.73; H, 4.72; N, 8.48.
(E)- 4-((1-(2-methylallyl)-2-oxoindolin-3-ylideneamino)methyl)benzoic acid (7a). Yield 2.65 g, 79 %, mp 111–112 °C; IR (KBr, cm−1) 3120–2553 (carboxylic O–H str), 1721 (C = O), 1638 (C = N), 1604 (C = O), 905, 752 (arom C–H out of plane bending); 1H NMR (CDCl3, δ ppm): 1.79 (3H, s, CH3), 4.32 (2H, s, CH2), 5.02 (s, 2H, =CH2), 6.9–8.19 (8H, m, ArH), 10.3 (1H, s, COOH). 13C NMR (CDCl3, δ ppm): 19.85 (CH3), 46.11 (CH2), 69.34 (CH2), 111.05 (=CH2), 113.48, 117.51, 123.82 (ArC), 125.34 (ArC), 129.56 (ArC), 130.76 (ArC), 134.36 (ArC), 138.06 (>C = ), 138.34 (ArC), 151.09 (ArC), 158.11 (C = N), 183.23 (C = O), 191.61 (C = O). MS (m/z) [M+] 334.68. Anal. calcd. for C20H18N2O3: C, 71.84; H, 5.43; N, 8.38. Found: C, 71.71; H, 5.48; N, 8.25.
(E)-4-(1-isobutyl-2-oxoindolin-3-ylideneamino)benzoic acid(2b). Yield 2.45 g, 76 %, mp 190–191 °C; IR (KBr, cm−1) 3088–2667 (carboxylic O–H str), 1726 (C = O), 1676 (C = N), 1598 (C = N), 931, 754 (arom C–H out of plane bending),; 1H NMR (CDCl3, δ ppm): 0.84 (d, 6H, two CH3), 2.00–2.43 (m, 1H, CH), 3.45 (d, 2H, CH2), 6.39–7.98 (m, 8H, arom CH). 13C NMR (CDCl3, δ ppm): 20.12, 26.88, 47.56, 113.60, 117.22, 122.52, 123.54, 125.12, 126.26, 131.57, 134.53, 138.38, 147.95, 151.20, 162.92, 167.9, 168.51. MS (m/z) [M+] 322.58. Anal. calcd. for C19H18N2O3: C, 70.79; H, 5.63; N, 8.69. Found: C, 70.88; H, 5.56; N, 8.53.
(E)-2-(1-isobutyl-2-oxoindolin-3-ylideneamino)benzoic acid (3b). Yield 2.6 g, 81 %, mp 177–178 °C; IR (KBr, cm−1) 3095–2528 (carboxylic O–H str), 1728 (C = O), 1664 (C = N), 1604 (C = O), 800, 745 (arom C–H out of plane bending); 1H NMR (CDCl3, δ ppm): 1.01 (6H, d, (two CH3), 2.12–2.19 (1H, m, CH), 3.54 (1H,1H, d, CH2), 6.66–7.93 (8H, m, ArCH), 10.11 (1H, s, COOH). 13C NMR (CDCl3, δ ppm): 20.20 (CH3), 27.02 (CH), 47.73 (CH2), 109.61 (ArC), 110.46 (ArC), 116.42 (ArC), 116.82 (ArC), 117.51 (ArC), 123.62 (ArC), 125.40 (ArC), 132.11 (ArC), 135.06 (ArC), 138.32 (ArC), 151.12 (ArC), 151.47 (ArC), 158.47 (C = N), 173.53 (C = O), 183.62 (C = O). MS (m/z) [M+] 322.72. Anal. calcd. for C19H18N2O3: C, 70.79; H, 5.63; N, 8.69. Found: C, 70.64; H, 5.58; N, 8.62.
(E)-2-(1-isobutyl-2-oxoindolin-3-ylideneamino)-5-chlorobenzoic acid (4b). Yield 2.7 g, 76 %, mp 185–186 °C; IR (KBr, cm−1) 3066–2515 (carboxylic O–H str), 1728 (C = O), 1672 (C = N), 1587 (C = N), 817, 761 (ArCH out of plane bending),; 1H NMR (CDCl3, δ ppm): 1.08 (6H, d, (two CH3), 2.22–2.28 (1H, m, CH), 3.57 (1H,1H, d, CH2), 6.75–7.96 (7H, m, 7H, ArCH), 11.36 (1H, s, COOH). 13C NMR (CDCl3, δ ppm): 24.99 (CH3), 31.70 (CH), 52.37 (CH2), 115.45 (ArC), 116.19 (ArC), 122.15 (ArC), 122.88 (ArC), 123.87 (ArC), 128.36 (ArC), 129.88 (ArC), 135.32 (ArC), 138.29 (ArC), 143.35 (ArC), 154.73 (ArC), 156.15 (ArC), 163.18 (C = N), 173.89 (C = O), 188.41 (C = O). MS (m/z) [M+] 356.25. Anal. calcd. for C19H17ClN2O3: C, 70.79; H, 5.63; N, 8.69. Found: C, 70.62; H, 5.54; N, 8.61.
(E)-2-(1-isobutyl-2-oxoindolin-3-ylideneamino)-4-chlorobenzoic acid (5b). Yield 2.6 g, 73 %, mp 240–241 °C; IR (KBr, cm−1) 3089–2527 (carboxylic O–H str), 1738 (C = O), 1655 (C = N), 1605 (C = O), 896, 754 (arom C–H out of plane bending); 1.05 (d, 6H, two CH3), 2.23–2.31 (m, 1H, CH), 3.54 (d, 2H, CH2), 6.95–7.82 (m, 7H, arom CH). 13C NMR (CDCl3, δ ppm): 23.67, 31.32, 52.05, 114.64, 115.72, 121.96, 122.55, 123.73, 128.01, 129.56, 134.22, 137.88, 142.62, 154.68, 155.95, 162.44, 173.31, 187.35. MS (m/z) [M+] 356.28. Anal. calcd. for C19H17ClN2O3: C, 63.96; H, 4.80; N, 7.85. Found: C, 63.95; H, 4.79; N, 7.93.
(E)-4-(1-isobutyl-2-oxoindolin-3-ylideneamino)-2-hydroxybenzoic acid (6b). Yield 2.3 g, 68 %, mp > 300 °C; IR (KBr, cm−1) 3363 (phenolic OH), 3055–2642 (carboxylic O–H str), 1715 (C = O), 1678 (C = N), 1606 (C = O), 846, 749 (arom C–H out of plane bending); 1H NMR (CDCl3, δ ppm): 1.03 (d, 6H, two CH3), 2.05–2.11 (m, 1H, CH), 3.41 (d, 2H, CH2), 6.72–7.86 (m, 7H, arom CH). 13C NMR (CDCl3, δ ppm): 20.38, 26.74, 47.88, 109.51, 110.32, 115.66, 116.18, 117.07, 122.95, 124.68, 132.03, 134.85, 137.77, 150.15, 151.22, 157.37, 172.58, 183.06. MS (m/z) [M+] 338.48. Anal. calcd. for C19H18N2O4: C, 67.44; H, 5.36; N, 8.28. Found: C, 67.21; H, 5.18; N, 8.34.
(E)-4-((1-isobutyl-2-oxoindolin-3-ylideneamino)methyl)benzoic acid (7b). Yield 2.25 g, 67 %, mp 136–137 °C; IR (KBr, cm−1) 3084–2624 (carboxylic O–H str), 1718 (C = O), 1690 (C = N) 1608 (C = O), 838, 747 (arom C–H out of plane bending); 1H NMR (CDCl3, δ ppm): 1.01 (6H, d, two CH3), 2.15–2.20 (1H, m, CH), 3.55 (2H, d, CH2), 5.65 (2H, s, CH2), 6.54–8.28 (8H, m, ArCH), 10.26 (s, 1H, COOH). 13C NMR (CDCl3, δ ppm): 20.31 (CH3), 27.03 (CH), 47.74 (CH2), 49.41 (CH2), 110.43, 117.54, 123.60 (ArC), 125.43 (ArC), 126.39 (ArC), 129.64 (ArC), 129.88 (ArC), 130.75 (ArC), 138.26 (ArC), 151.50 (ArC), 158.45 (C = N), 177.04 (C = O), 183.06 (C = O). MS (m/z) [M+] 336.32. Anal. calcd. for C20H20N2O3: C, 71.41; H, 5.99; N, 8.33. Found: C, 71.52; H, 5.94; N, 8.25.
2.4. Assessment of antimicrobial activity
Gram-positive bacteria S. aureus (ATCC29213), B. subtilis (ATCC10400), Gram negative E. coli (ATCC25922), P. aeruginosa (ATCC27855), and the fungus C. albicans (ATCC10231) were used to test the antimicrobial properties of isatin derivatives. The agar well diffusion method, recommended by the Clinical and Laboratory Standards Institute (CLSI, 2004), was used to measure the activity of the compounds under test. 107CFU (Colony Forming Unit) of each microorganism were seed inoculated on Muller Hinton agar (MHA) plates for bacteria or Sabouraud Dextrose Agar (SDA) plates for fungi after being adjusted spectrophotometrically. A cork borer was then used to punch wells (12 mm in diameter) in the agar media. The final concentration of each isatin derivative (2a,b-7a,b) was then diluted to 1 mg/mL in phosphate-buffer saline (pH 7). All isatin derivatives (2a,b-7a,b) were dissolved in DMSO to a concentration of 10 mg/mL. The solution was put into the wells in a volume of 100 µL. Nystatin (for fungus) and chloramphenicol (for bacteria) were employed as positive controls, and DMSO (1 % in phosphate-buffered saline) was used as a negative control. MHA plates were incubated at 37 °C overnight whereas SDA plates were incubated at 25 °C for 48 h prior to determining the widths of the inhibition zones.
2.5. Determination of MIC
The broth microdilution technique outlined in the CLSI recommendations (CLSI, 2012) was used to calculate the antibacterial minimum inhibitory concentrations (MIC) of isatin compounds against the same isolates (Wayne PA, 2015). In microtiter plates, isatin derivative solutions were serially diluted using Sabouraud dextrose broth for fungi and Mueller Hinton Broth for bacteria. The solution's ultimate isatin derivative concentrations ranged from 2048 to 0.5 µg/mL (2048, 1024, 512, 256, 128 to 64 to 32 to 16 to 8 to 4 to 2 to 0.5 µg/mL). The indicated microorganisms were inoculated and isatin compounds were serially diluted, resulting in an inoculum density per well of 106 CFU/mL. Negative controls included uninoculated wells, whereas positive controls included fluconazole (for fungi) and imipenem (for bacteria) at the same concentrations. The lowest concentrations that could stop detectable microbial growth after 24 h at 37 °C for bacteria and 48 h at 25 °C for fungi were described as MICs.
2.6. Effect on biofilm formation
The biofilm-forming microorganisms on SA ATCC29213 and MRSA ATCC35501 was tested to see if isatin derivatives with promising antibacterial activity could prevent biofilm formation at half their MICs (Plyuta et al., 2013). After being incubated at 37 °C overnight, tubes containing double-strength nutritional broth, were inoculated with freshly cultured slants. Bacterial counts were adjusted to 1 × 108 CFU/mL, and suspensions were centrifuged for five minutes at 10,000 rpm. In 96-well flat-bottomed plates from NuncTM Thermo ScientificTM, this inoculum was distributed as 100 µL after being diluted 1:20 in fresh media. Plates were incubated at 37 °C for 24 h after the addition of isatin derivatives 2a −5a (100 µL; 0.5 MIC). Triplicates of each experiment were carried out. Each plate contained wells that had not been infected as a negative control, wells that had just bacterial suspensions as a positive control, and wells that had been treated with Chloramphenicol (0.5 MIC) as a standard. The biofilms were then properly rinsed with 100 µL of phosphate-buffered saline (pH 7.2; NaCl: 137 mM; Na2HPO4: 10 mM; KH2PO4: 1.8 mM) to get rid of any free bacteria after the incubation time. The biofilms were dried after being stained for two minutes at room temperature with 100 µL of 0.1 % crystal violet solution. Wells were properly cleaned with PBS after excess dye was aspirated. The dye from the stained biofilms was released into the solution using glacial acetic acid (100 µL, 33 %), which was then used to destain the biofilms before the absorbance of the stained biofilms was evaluated spectrophotometrically at 630 nm using microplate reader (Bio-Tek, Elx-808, Winooski, VT, USA). The following equation, which was reported (Lemos et al., 2018) and was used in calculating the percentage of biofilm inhibition:
2.7. Docking study
The Autodock4.2 program was used to conduct a molecular docking investigation (Morris et al., 2009). Under physiological circumstances, the chemical structures of compounds 2a,b-7a,b were produced in the protonated state. B. subtilis' histidine kinase subunit A's crystal structure (3SL2.pdb) was identified (Celikel et al., 2012). The co-crystallized ligand was docked in the original protein structure using the program's default settings, and a 10 Å diameter sphere was defined around the binding pocket's center. Based on the heavy atom RMSD values (1 Å), the resulting binding poses were selected. All of the compound's torsion angles were set to free rotation during docking, and 10 binding conformers for each ligand were produced. For interactions between ligands and receptors, the binding pose with the highest overall score was taken into account.
2.8. Molecular dynamics simulations
The HK-ligand complex that the docking method produced was subjected to MD simulations. 100 ns of MD simulations for the complex of the macromolecule and chemical 2a were run. The simulations required the protein–ligand complex system to store the atom coordinates at 1-ps intervals. By using the initial structures from the MD simulation as the reference structure, the CPPTRAJ module of AmberTools22 (Case et al., 2022) was utilised to calculate RMSDs in order to confirm the convergence of the MD simulation processes. By computing root-mean-square fluctuations (RMSFs), structural flexibilities of the 3sl2-peptide structure was computed using the CPPTRAJ module in Amber Tools 22 (Daniel et al 2013). The trajectories from the last 10 ns of the equilibration MD simulations were used to calculate the RMSF values, and the average structures from those trajectories served as the reference structures. The CPPTRAJ module was used to look at the hydrogen bonds that exist between the ligand and the nearby amino acid residues. High occupancy hydrogen bonding for the complex system was observed over the simulation period. Molecular Mechanics Poisson-Boltzmann Surface Area (MM/PBSA) pairwise decomposition was used to calculate the interaction energy of the residues involved in high-occupancy hydrogen bonding to the ligand during the last 10 ns of the simulation (Miller III et al., 2012). The MM/GBSA approach was used to determine the binding free energy (Gbinding), Eq. (1) for receptor-binder complex systems (Kollman et al., 2000). A total of 1000 snapshot were taken along the MD simulation process at 10-ns intervals in order to determine the MM/GBSA free energy difference, Eq. (1).
| (1) |
where ΔGgas is molecular mechanics energy of the gas phase, including electrostatic (Eelectrostatic) and van der Waals (Evdw); The solvation energy, ΔGsolv comprises the nonpolar (ΔGSA) and polar (ΔGGB) roles; Gprotein, Gligand and Gcomplex, refer to the free energies of the protein, ligand and complex respectively. Using w = 80 and p = 1.0, Onufriev et al.'s (Onufriev et al.. 2004) modified GB model was used to estimate the polar contribution ΔGGB and the probe sphere radius was set to 1.4 Å in order to calculate the solvent-accessible surface area (SASA) (Weiser et al., 1999). The nonpolar solvation free energy for a point solute (b) was set to 0.00 kcal mol-1, and the surface tension proportionality constant (γ) was set to 0.0072 kcal mol–1Å−2.
3. Results
3.1. Chemistry
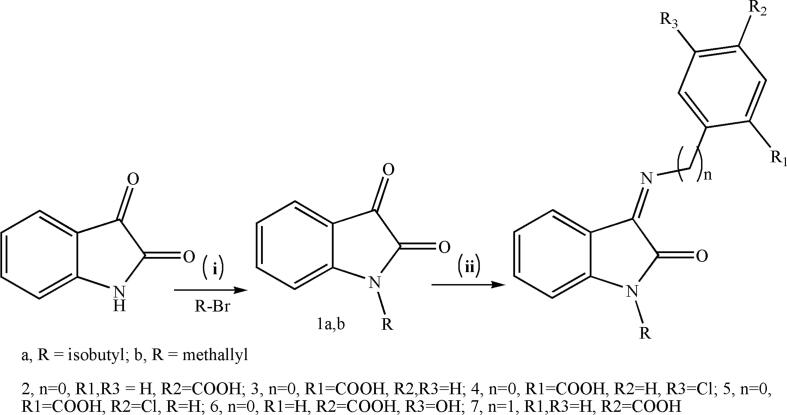
Compounds 1a, b–7a, b's synthesis route is depicted in Scheme 1. In the presence of anhydrous potassium carbonate, isatin was condensed with the appropriate alkylhalide to produce the compounds 1- (2-methylallyl)indoline-2,3-dione (1a) or isobutylindoline-2,3-dione (1b). Compounds 1a,b underwent condensation with the appropriate substituted aminobenzoic acid resulting in (E)-(1-(2-methylallyl)-2-oxoindolin-3-ylideneamino)benzoic acid derivatives (2a-7a) and (E)-(1-isobutyl-2-oxoindolin-3-ylideneamino)benzoic acid derivatives (2b-7b).
Scheme 1.
General synthesis of compounds 1a,b-7a,b. Reagents and conditions: (i) K2CO3, DMF, r.t. overnight; (ii) Ethanol, AcOH, reflux overnight.
3.2. Antimicrobial screening
The findings of the antimicrobial screening of compounds 2a,b-7a,b are displayed in Table 1. The compounds had little to no antifungal and anti-Gram negative activity, but they demonstrated strong activity against S. aureus and B. subtilis, two Gram-positive bacteria.
Table 1.
Results of minimum inhibitory concentrations (MIC mmol/L) of compounds 2a,b-7a,b.
| Compd |
Gram-positive bacteria |
|
|---|---|---|
|
B. subtilis |
S. aureus |
|
| MIC (mmol/L) | MIC (mmol/L) | |
| 2a | 0.090 | 0.090 |
| 3a | 0.2 | 0.4 |
| 4a | 0.362 | 0.181 |
| 5a | 0.181 | 0.181 |
| 6a | 0.190 | 0.190 |
| 7a | 0.191 | 0.191 |
| 2b | 0.198 | 0.198 |
| 3b | 0.198 | 0.397 |
| 4b | 0.180 | 0.360 |
| 5b | 0.360 | 0.360 |
| 6b | 0.190 | 0.190 |
| 7b | 0.192 | 0.192 |
| Imipenem | 0.036 | 0.036 |
| Fluconazole | ND | ND |
3.3. Effect on biofilm formation
The sub-inhibitor concentrations was used to evaluate the inhibitory effect of compounds 2a, 5a on the biofilm formation of S. aureus and MRSA strains (Fig. 2).
Fig. 2.
Percent Biofilm Inhibition against S. aureus ATCC29213 and MRSA ATCC35501 Biofilms.
3.4. Docking study
The correctness of the docking strategy was supported by the RMSD value of the posture resulting from the redocking of the ATP structure itself (0.635 Å) (Fig. 3). The binding site delineated by the side chains of Asn50, Ser53, Asn54, Ala55, Lys57, Tyr58, Ser59, Gly86, Ile87, Gly88, Ile89, Pro90, Val104, Asp105, Leu119. Fig. 4 and Table 2 show the docking results of the synthesized compounds 2a,b-7a,b.
Fig. 3.
The co-crystallized ATP (from 3sl2.pdb, colored grey) and the redocked ATP structure (colored cyan).
Fig. 4.
The newly synthesized compounds docked inside the binding site, hydrogen bonds are displayed in yellow color.
Table 2.
Results of docking study of compounds 2a,b-7a,b.
| Compd | Binding energy. (Kcal Mol−1) | Estimated inhibition value Ki (nM) | H bond Amino acid residue |
|---|---|---|---|
| 2a | −10.36 | 25.64 | Asn54 |
| 3a | −9.28 | 157.82 | Lys57, Arg103 |
| 4a | −9.81 | 64.71 | Lys57 |
| 5a | −9.81 | 64.52 | Lys57, Arg103 |
| 6a | −9.67 | 81.94 | Tyr58, Arg103 |
| 7a | −9.4 | 129.01 | Arg103 |
| 2b | −9.77 | 69.44 | Asn54 |
| 3b | −9.0 | 253.19 | Lys57, Val104 |
| 4b | −9.59 | 92.89 | Lys57 |
| 5b | −9.56 | 98.43 | Lys57, Arg103 |
| 6b | −8.75 | 383.04 | Tyr58, Arg103 |
| 7b | −9.37 | 134.81 | Tyr58, Arg103 |
3.5. Molecular dynamics simulations
The simulation strategy made it possible to observe how the conformations of chemical 2a changed within the binding-site of the macromolecule. In Fig. 5, the dynamic stabilities of the 3sl2-compound 2a complex system are displayed using the root mean-square deviation (RMSD) values.
Fig. 5.
Root-mean-square deviation of 3sl2-enzyme-compound 2a complex. 3sl2 peptide backbones (green lines), proteins with all atoms (red lines), and compound 2a (black lines).
To identify the portions of a system that are more flexible and to evaluate each component's flexibility in respect to other system components, a root mean square fluctuation (RMSF) analysis can be used. The RMSF was displayed in Fig. 6 for the past 10 ns.
Fig. 6.
The RMSF chart of amino acid residues of 3sl2 protein during simulation process.
The carboxylate group of Asp105 formed a hydrogen bond with compound 2a, according to the CPPTRAJ hydrogen bond analysis of the final 10 ns frame period. The binding free energies of compound 2a to the 3sl2 peptide were calculated using MM/GBSA techniques utilising the MD simulation trajectories for the final 4 ns (Table 3).
Table 3.
The binding free energy of the 3sl2 protein-compound 2a complex as estimated by MM/GBSA. (kcal mol−1).
| ΔGvdw | ΔGelec | ΔGpolara | ΔGSurfb | ΔGMM/GBSA |
|---|---|---|---|---|
| −36.3498 | −43.7817 | 53.4821 | −4.5686 | −31.2180 |
a ΔGelectrostatic + ΔGpolar are the whole electrostatic contribution.
B ΔGnp = ΔGvdw + ΔGsurf are the whole non polar contribution.
The results of the MM/PBSA-pairwise-residue decomposition analysis are shown in Table 4. Additionally, Fig. 7 displays the average structure for the 3sl2-compound 2a structure generated from the last 10 ns trajectory data of the MD simulations process. The carboxylic groups of the benzoic acid fragment and Asp105 form a hydrogen bond, which anchors compound 2a inside the binding site of the 3sl2 structure. The Van der Waal interactions with the hydrophobic side chains of the amino acid residues Asn54, Tyr58, Ile89, Pro90, Val97, Val104, Lys106, Ala107, Leu119, and Thr143 support the bound conformation of compound 2a. While the benzene ring of the benzoic acid fragment is positioned horizontally among the hydrophobic side chains of Val97, Val104, Lys106, Ala107, Leu119, and Thr143, the isatin moiety is sandwiched between the hydrophobic side chains of amino acid residues Asn54, Tyr58, Ile89, and Pro90.
Table 4.
The results of the MM/PBSA-pairwise-residue decomposition analysis of 3sl2-compoud 2a complex.
| Asn54 | Tyr58 | Asn72 | Asp84 | Ile89 | Pro90 | Val104 | Asp105 | Lys106 |
|---|---|---|---|---|---|---|---|---|
| −1.729 | −3.625 | −5.999 | −1.372 | −2.258 | −1.3 | −3.430 | −8.074 | −1.416 |
Fig. 7.
The representative complex structure of 3sl2-compound 2a as determined by the averaged structure from the last 10 ns MD simulation. The hydrogen bonds are shown in yellow solid lines. The interacting residues are beige-colored sticks and the ligand is a cyan-colored stick.
4. Discussions
4.1. Chemistry
On the basis of data from elemental analysis, mass spectrum analysis, 1H NMR, IR, and 13C NMR, the compounds' structures were determined. The compounds 1a,b's IR spectra showed two distinguishing bands at 1724–1722 cm−1 and 1603–1601 cm−1 (two C = O groups), as well as one band at 3230–3265 cm−1 (NH). The latter band vanished when the compounds 1a,b were transformed into compounds 2a,b-7a,b, which displayed four distinguishing bands. One characteristic band appeared at 3125(3055)-2673(2515) cm−1 due to stretching band of the carboxylic OH group. The bands appeared at ranges 1738–1705 cm−1 due to carboxylic carbonyl group, 1690–1635 cm−1 due to the Schiff’s base imino groups and 1611–1592 cm−1 due to the carbonyl group of 2-oxoindoline moiety. 1H NMR spectra of compounds 1a-7a were characterized by the appearance of the characteristic signals of methallyl substituent at δ 2.11–1.75 ppm of methyl group, δ 4.4–3.94 ppm of aliphatic methylene group and at δ 5.05–4.65 ppm of the olefinic methylene signals. The characteristic signal of carboxyl group at δ 10.38–10.01 ppm appeared with compounds 2a-7a. 1H NMR spectra of compounds 1b-7b were characterized by the appearance of the characteristic signals of isobutyl substituent at δ 0.86–1.08 ppm of the methyl-protons doublet signals, at δ 2.0–2.42 ppm of the methine-protons multiplet signals and δ 3.41–3.61 ppm of the methylene-protons doublet signals. The characteristic signal of carboxyl group at δ 10.35–10.08 ppm appeared with compounds 2b-7b. 13C NMR spectra of compounds 1a-7a revealed characteristic signals of methallyl substituents at δ 24.87–19.1 ppm (CH3 group), δ 60.35–44.62 ppm (CH2 group), δ 111.19–110.15 ppm (=CH2 group) and δ 141.33–138.21 ppm (>C = group). Moreover, compound 1a showed characteristic signals at δ 158.02 ppm (C = O) and at δ 183.15 (C = O) while the latter signal disappeared in compounds 2a-7a and was replaced by a signal at δ 163.24–157.88 ppm (>C = N group) and another two peaks signals appeared at δ 183.06–163.02 (2-oxoindoline-C2) and at δ 191.68–187.84 ppm (-(C = O)OH). 13C NMR spectra of compounds 1b-7b revealed characteristic signals of isobutyl substituents at δ 24.99–20.12 ppm (CH3 group), δ 31.7–26.74 ppm (>CH– group) and δ 52.37–47.56 ppm (–CH2- group). In addition, compound 1b showed characteristic signals at δ 158.42 (C = O) and at δ 183.61 (C = O) while the latter signal disappeared in compounds 2a-7a and was replaced by a signal at δ 163.24–157.88 ppm (>C = N group) and another two peaks signals appeared at δ 177.04–162.92 (2-oxoindoline-C2) and at δ 188.41–168.51 ppm (-(C = O)OH). The weight percentage values for carbon, hydrogen, and nitrogen were in agreement with the calculated percentage values, according to data from the synthetic compounds 1a,b-7a,b's elemental analysis.
4.2. Antimicrobial screening
In comparison with reference antibacterial drug (Table 1), imipenem (MIC 0.040 mmol/L) compounds 2ab-7a,b showed promising activity against Gram-positive bacteria with high selectivity against S. aureus and B. subtilis. Compounds 2a, 5a are the most active against both B. subtilis and S. aureus (MIC 0.09 mmol/L and 0.181 mmol/L respectively). The N-Methallylindoline-2,3-dione derivatives 2a-7a showed higher activity (MIC 0.09––0.181 mmol/L) than 1-isobutylindoline-2,3-dione derivatives 2b-7b (MIC 0.19–––0.36 mmol/L) compared with imipenem reference drug (MIC 0.040 mmol/L). The methallyl group is less hydrophobic than the isobutyl group, thus the methallyl group is supposed to result in total hydrophobicity optimum enough for the compound to penetrate the microbial cell and the double bond of methallyl group is supposed to increase interaction with bacterial target protein through π…H bond.
4.3. Effect on biofilm formation
As Fig. 2 indicated, biofilm formation of S. aureus and MRSA (intense biofilm producing strains) was significantly reduced after treatment with compounds 2a or 2b for 24 h (OD630 ∼ 55 %), compared to the positive control (∼20 %). The biofilm formation-inhibition effects obtained with compounds 2a and 5a suggested that the antibacterial effects of compounds are associated with the inhibition of the histidine kinase-mediated pathway (Hirakawa et al., 2020). Since, bacterial two-component systems (TCSs), which include histidine kinase/Walk and response regulators (RR), are crucial for the development of bacterial biofilms (Prüß BM, 2017), the tested compounds are suggested as HK/Walk inhibitors. The results prompted us toward in silico study of binding mechanism of compound 2a with HK/walk enzyme.
4.4. Docking study
Bacterial two-component systems (TCSs), which include histidine kinase/Walk and response regulators (RR), are crucial for the development of bacterial biofilms (Prüß BM, 2017). TCSs have gained increased interest as possible targets for the development of antibacterial and anti-biofilm medicines since they lack a comparable protein or in humans (Bem et al., 2015). In S. aureus and B. subtilis, the histidine kinase/Walk YycG is one of the TCSs that regulates cell wall metabolism, drug resistance, virulence factors, and biofilm formation (Martin et al., 1999). The crystal structure of a monomer histidine kinase domain complex with ATP (3sl2.A) of B. subtilis was retrieved from the protein database in order to study the potential interactions between compounds 2a,b-7a,b and the amino acids that make up the binding site of S. aureus histidine kinase (Celikel et al., 2012) (Fig. 3). Compounds 2a,b-7a,b's docking poses revealed hydrogen bonds between the C-3 hydroxyl group of the isatin moiety, the C-2 carbonyl oxygen of the isatin moiety, the carbonyl oxygen of the acetophenone moiety, and the NH group of the binding site residues Asn54, Lys57, Tyr58, Arg103 and Val104 Fig. 4 and Table 2. However, we used the in vivo microbial model and molecular modeling to derive qualitative structure–activity relationship of compounds 2a,b-7a,b, there are relative agreements between the estimated docking scores for compounds 2a,b-7a,b and the determined MICs against either S. aureus or B. Subtilis.
4.5. Molecular dynamics simulations
During the simulation period, the conformational changes of compound 2a were detected. Root Mean Square Deviation (RMSD) was used to calculate the difference between the initial structural conformation of the protein–ligand coordinates and its final coordinates. The deviation from the initial conformation that occurred during the simulation served as a gauge of the system's stability. The more stable the structure, the fewer the variances Fig. 5. Identification of flexible areas and comparison of the relative flexibility of different protein–ligand complex system components were done using a root mean square fluctuation (RMSF) analysis. In order to identify some important trends for the protein-binder complex system, we looked at how flexibility changed during MD simulations. The RMSF at the C-α of each amino acid residue was taken into consideration for the average flexibility of each residue in the 3sl2 peptide for the 3sl2-compound 2a complex. During the final 10 ns of simulation process, the RMSF have been depicted in Fig. 6. Peaks show the regions that fluctuate the most during the simulation. The residues of the amino acids Asn54, Tyr58, Asn72, Asp84, Ile89, Pro90, Val104, Asp105, and Lys106 showed the least fluctuation. The side chains of these amino acids have hydrophobic characteristics, which suggests that hydrophobic interactions have a significant impact on the free energy of binding of the models. The diagram demonstrates how these amino acid residues define the s3sl2-active site and play a significant role in how the enzyme interacts with the binder. The CPPTRAJ hydrogen bond analysis of the MD simulation data revealed the hydrogen bonds of the active site residue Asp105 to compound 2a using the final 10 ns frame period. The binding free energies of the compound 2a to the 3sl2 structure were calculated using MD simulation trajectories during the previous 4 ns (Table 3). The interactions between the binding site of 3sl2 and compound 2a were revealed by the MM/PBSA-pairwise decomposition analysis. Absolute decomposed energy in Table 4 shows that the amino acid residue Asp105 is essential for efficient binding interactions with chemical 2a.
5. Conclusions
In conclusion, a number of isatin compounds carrying analogues of p-aminobenzoic acids were created and biologically tested for their antibacterial properties. The produced chemicals had no effect on Gram-negative bacteria or fungi but were most effective against B. subtilis and S. aureus, two gram-positive species. The most effective compounds against B. subtilis and S. aureus were (E)-4-(1-(2-methylallyl)-2-oxoindolin-3-ylideneamino)-2-hydroxybenzoic acid 6a and (E)-4-(1-isobutyl-2-oxoindolin-3-ylideneamino)-2-hydroxybenzoic acid 6b (MIC 0.095 mmol/L and 0.190 mmol/L respectively). The 3D structure of B. subtilis histidine kinase was studied to determine the binding modes of compounds 2a,b-7a,b. It was found that these compounds had similar orientations in the binding site and similar hydrogen bonds to that with the same Xray ligand ATP. The results of molecular dynamcis simulations showed that the hydrogen bonding between Asp105 residue to compound 2a suggested Asp105 has a key role for the activity. Compound2a is suggested as a lead for further optimization to design and synthesize more potent and selective anti-Gram-positive bacteria, particularly S. aureus and B. subtilis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research. (IFKSURC-1-0814).
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2023.101781.
Contributor Information
Awwad A. Radwan, Email: aradwan@ksu.edu.sa.
Fares K. Al-Anazi, Email: afars@ksu.edu.sa.
Mohammed Al-Agamy, Email: malagamy@ksu.edu.sa.
Adel F. Alghaith, Email: afalghaith@ksu.edu.sa.
Gamal M. Mahrous, Email: gmmarous@ksu.edu.sa.
Mohammad R. Alhuzani, Email: malhuzani@ksu.edu.sa.
Abdulrhman S.A. Alghamdi, Email: Absalghamdi@ksu.edu.sa.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Abdelrahman, M.A., Almahli, H., Al-Warhi, T., Majrashi, T.A., Abdel-Aziz, M.M., Eldehna, W.M., Said, M.A. 2022. Development of novel isatin-tethered quinolines as anti-Tubercular agents against multi and extensively drug-resistant Mycobacterium tuberculosis. Molecules 27, 8807-8818. https://doi.org/ 10.3390/molecules27248807. [DOI] [PMC free article] [PubMed]
- Akberova S.I. New biological properties of p-aminobenzoic acid. Biol. Bull. 2002;29:390–393. doi: 10.1023/A:1016871219882. [DOI] [PubMed] [Google Scholar]
- Alzahrani A.Y., Ammar Y.A., Salem M.A., Abu-Elghait M., Ragab A. Design, synthesis, molecular modeling, and antimicrobial potential of novel 3-[(1H-pyrazol-3-yl)imino]indolin-2-one derivatives as DNA gyrase inhibitors. Arch. Pharm. 2022;355:e2100266. doi: 10.1002/ardp.202100266. [DOI] [PubMed] [Google Scholar]
- Alzahrani A.Y., Ammar Y.A., Abu-Elghait M., Abu-Elghait M., Salem M.A., Assiri M.A., Ali T.E., Ragab A. Development of novel indolin-2-one derivative incorporating thiazole moiety as DHFR and quorum sensing inhibitors: Synthesis, antimicrobial, and antibiofilm activities with molecular modelling study. Bioorg. Chem. 2022;119 doi: 10.1016/j.bioorg.2021.105571. [DOI] [PubMed] [Google Scholar]
- Bem, A. E., Velikova, N., Pellicer, M. T., Baarlen, P.v, Marina, A., Wells, J. M., 2015. Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem Biol. 10, 213–224. https://doi.org/10.1021/cb5007135. [DOI] [PubMed]
- Borad, M.A., Bhoi, M.N., Prajapati, N.P., Patel, H.D., Francis, 2014. Review of synthesis of spiro heterocyclic compounds from isatin. Synth. Commun. 44, 897–922. http://dx.doi.org/10.1080/00397911.2013.843196.
- Case, D.A., Aktulga, H.M., Belfon,K., Ben-Shalom, I.Y., Berryman, J.T., Brozell, S.R., Cerutti, D.S., T.E. et al 2022. Amber 2022, University of California, San Francisco.
- Celikel R., Veldore V.H., Mathews I., Devine K.M., Varughese K.I. ATP forms a stable complex with the essential histidine kinase Walk (YycG) domain. Acta Crystallogr. D Biol. Crystallogr. 2012;68:839–845. doi: 10.1107/S090744491201373X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.-Y., Hu M.-L. Concentrations and lipid peroxidation in tissues and toxicity of para-aminobenzoic acid fed to rats in drinking water. J. Nutr. Biochem. 1996;7:408–413. doi: 10.1016/S0955-2863(96)00065-4. [DOI] [Google Scholar]
- Chiyanzu I., Hansell E., Gut J., Rosenthal P.J., McKerrow J.H., Chibale K. Synthesis and evaluation of isatins and thiosemicarbazone derivatives against cruzain, falcipain-2 and rhodesain. Bioorg. Med. Chem. Lett. 2003;13:3527–3530. doi: 10.1016/s0960-894x(03)00756-x. [DOI] [PubMed] [Google Scholar]
- CLSI 2004, Method for Antifungal Disk Diffusion Susceptibility Testing of Yeasts, Approved Guideline. CLSI document M44-A. CLSI, 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898, USA.
- CLSI 2012, Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard, 7th ed., CLSI document M02-A11. Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087, USA.
- Crisan M.E., Bourosh P., Maffei M.E., Forni A., Pieraccini S., Sironi M., Chumakov Y.M. Synthesis, crystal structure and biological activity of 2-hydroxyethylammonium salt of p-aminobenzoic acid. PLoS One. 2014;9:e101892. doi: 10.1371/journal.pone.0101892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva J.F.M., Garden S.J., Pinto A.C. The chemistry of isatins: A review from 1975 to 1999. J. Braz. Chem. Soc. 2001;12:273–324. doi: 10.1590/S0103-50532001000300002. [DOI] [Google Scholar]
- Daisley R.W., Shah V.K. Synthesis and antibacterial activity of some 5-nitro-3-phenyliminoindol-2(3H)-ones and their N-Mannich bases. J. Pharm. Sci. 1984;73:407–408. doi: 10.1002/jps.2600730333. [DOI] [PubMed] [Google Scholar]
- Daniel R.R., Thomas E., Cheatham I.I.I. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- Eldeeb M., Sanad E.F., Ragab A., Ammar Y.A., Mahmoud K., Ali M.M., Hamdy N.M. Anticancer effects with molecular docking confirmation of newly synthesized isatin sulfonamide molecular hybrid derivatives against hepatic cancer cell lines. Biomedicines. 2022;10(722):1–15. doi: 10.3390/biomedicines10030722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kalyoubi S.A., Ragab A., Abu Ali O.A., Ammar Y.A., Seadawy M.G., Ahmed A., Fayed E.A. One-Pot synthesis and molecular modeling studies of new bioactive spiro-oxindoles based on uracil derivatives as SARS-CoV-2 inhibitors targeting RNA polymerase and spike glycoprotein. Pharmaceuticals. 2022;15:376. doi: 10.3390/ph15030376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayed E.A., Ragab A.E.E., Rogy R.B., Ashraf H., Ammar Y.A. n vivo screening and toxicity studies of indolinone incorporated thiosemicarbazone, thiazole and piperidinosulfonyl moieties as anticonvulsant agent. Bioorg. Chem. 2021;116 doi: 10.1016/j.bioorg.2021.105300. [DOI] [PubMed] [Google Scholar]
- Hirakawa H., Kurushima J., Hashimoto Y., Tomita H. Progress overview of bacterial two-component regulatory systems as potential targets for antimicrobial chemotherapy. Antibiotics (basel) 2020;9(635):1–15. doi: 10.3390/antibiotics9100635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, M. A., EL-Messmary, M. F., Elarfi, M.G.A., 2010. Synthesis of spiro heterocyclic compounds. J. Chem. 7, 55-58. https://doi.org/10.1155/2010/604549.
- Kadhum W.R., Oshizaka T., Ichiro H., Todo H., Sugibayashi K. Usefulness of liquid-crystal oral formulations to enhance the bioavailability and skin tissue targeting of p-amino benzoic acid as a model compound. Eur. J. Pharm. Sci. 2016;88:282–290. doi: 10.1016/j.ejps.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Kenchappa R., Bodke Y.D., Telkar S., Nagaraja O. Synthesis and antimicrobial activity of fused isatin and diazepine derivatives derived from 2-acetyl benzofuran. Russ. J. Gen. Chem. 2017;87:2027–2038. doi: 10.1134/S1070363217090195. [DOI] [Google Scholar]
- Khan F., Maalik A. Advances in pharmacology of isatin and its derivatives: A review. Trop. J. Pharm. Res. 2015;14:1937. doi: 10.4314/tjpr.v14i10.28. [DOI] [Google Scholar]
- Kluczyk A., Popek T., Kiyota T., de Macedo P., Stefanowicz P., Lazar C., Konishi Y. Drug evolution: p-aminobenzoic acid as a building block. Curr. Med. Chem. 2002;9:1871–1892. doi: 10.2174/0929867023368872. [DOI] [PubMed] [Google Scholar]
- Kollman, P.A. Massova, I.R., Carolina, K.,B., Huo, S.C., Lillian, L., M., Lee, T.D., Yong, W., Wei, D.,Oreola, C.,P., Srinivasan, J., Case, David A., Cheatham, T.E. 2000. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 33, 889–897. [DOI] [PubMed]
- Lemos A.S.O., Campos L.M., Melo L., Guedes M.C.M.R., Oliveira L.G., Silva T.P., Melo R.C.N., Rocha V.N., Aguiar J.A.K., Apolônio A.C.M., Scio E., Fabri R.L. Antibacterial and antibiofilm activities of psychorubrin, a pyranonaphthoquinone isolated from Mitracarpus frigidus (Rubiaceae) Front. Microbiol. 2018;9(724):1–10. doi: 10.3389/fmicb.2018.00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Z.-M., Sun J., Zhu H.-L. Design, synthesis and antibacterial activity of isatin derivatives as FtsZ inhibitors. J. Mol. Struct. 2016;1117:8–16. doi: 10.1016/j.molstruc.2016.03.036. [DOI] [Google Scholar]
- Martin P.K., Li T., Sun D., Biek D.P., Schmid M.B. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 1999;181:3666–3673. doi: 10.1128/JB.181.12.3666-3673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B.R., III, McGee T.D., Jr., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Comput. Chem. 2009;16:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noreen S., Sumrra S.H. Correlating the charge transfer efficiency of metallic sulfa-isatins to design efficient NLO materials with better drug designs. Biometals. 2022;35:519–548. doi: 10.1007/s10534-022-00385-6. [DOI] [PubMed] [Google Scholar]
- Nortje C., Jansen van Rensburg P., Cooke C., Erasmus E. The simultaneous detection and quantification of p-aminobenzoic acid and its phase 2 biotransformation metabolites in human urine using LC–MS/MS. Bioanalysis. 2015;7:1211–1224. doi: 10.4155/bio.15.54. [DOI] [PubMed] [Google Scholar]
- Onufriev, A., Bashford., D., Case, D.A. 2004. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55,383–394. https://doi.org/10.1002/ prot.20033 PMID: 15048829. [DOI] [PubMed]
- Patel H.M., Bhardwaj V., Sharma P., Noolvi M.N., Lohan S., Bansal S., Sharma A. Quinoxaline-PABA bipartite hybrid derivatization approach: Design and search for antimicrobial agents. J. Mol. Struct. 2019;1184:562–568. doi: 10.1016/j.molstruc.2019.02.074. [DOI] [Google Scholar]
- Plyuta V., Zaitseva J., Lobakova E., Zagoskina N., Kuznetsov A., Khmel I. Effect of plant phenolic compounds on biofilm formation by Pseudomonas aeruginosa. APMIS. 2013;121:1073–1081. doi: 10.1111/apm.12083. [DOI] [PubMed] [Google Scholar]
- Prüß B.M. Involvement of two-component signaling on bacterial motility and biofilm development. J. Bacteriol. 2017;199:e00259–e00317. doi: 10.1128/JB.00259-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan A.A., Aanazi F.K., Al-Agamy M., Mahrous G.M. Design, synthesis and molecular modeling study of substituted indoline-2-ones and spiro[indole-heterocycles] with potential activity against Gram-positive bacteria. Acta Pharm. 2022;72:79–95. doi: 10.2478/acph-2022-0004. [DOI] [PubMed] [Google Scholar]
- Singh G., Arora A., Singh A., Kalra P., Rani S., Singh K., Maurya I.K., Mandal R.S. Molecular design, synthesis, computational screening, antimicrobial evaluation and molecular docking study of acetylinic isatin hybrids. ChemistrySelect. 2018;3:1942–1952. doi: 10.1002/slct.201703051. [DOI] [Google Scholar]
- Sowinska, M., Morawiak, M., Bochy ´nska-Czẏz, M., Lipkowski, A.W., Ziemi ´nska, E., Zabłocka, B., Urbanczyk-Lipkowska, Z., 2019. Molecular antioxidant properties and in vitro cell toxicity of the p-aminobenzoic acid (PABA) functionalized peptide dendrimers. Biomolecules 9, 89. https://doi.org/10.3390/biom9030089. [DOI] [PMC free article] [PubMed]
- Sumrra S.H., Mushtaq F., Ahmad F., Hussain R., Zafar W., Imran M., Zafar M.N. Coordination behavior, structural, statistical and theoretical investigation of biologically active metal-based isatin compounds. Chem. Pap. 2022;76:3705–3727. doi: 10.1007/s11696-022-02123-1. [DOI] [Google Scholar]
- Varun S., Kakkar R. Isatin and its derivatives: A survey of recent syntheses, reactions, and applications. Med. Chem. Commun. 2019;10:351–368. doi: 10.1039/C8MD00585K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser J., Shenkin P.S., Still W.C. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO) J. Comput. Chem. 1999;20:217–230. doi: 10.1002/(SICI)1096-987X(19990130)20:2<217::AID-JCC4>3.0.CO;2-A. [DOI] [Google Scholar]
- Yoshikawa M., Murakami T., Kishi A., Sakurama T., Matsuda H., Nomura M., Matsuda H., Kubo M. Novel indole S, O-bisdesmoside, calanthoside, the Precursor glycoside of tryptanthrin, indirubin, and isatin, with increasing skin blood flow promoting effects, from two calanthe species (Orchidaceae) Chem. Pharm. Bull. 1998;46:886–888. doi: 10.1248/cpb.46.886. [DOI] [PubMed] [Google Scholar]
- Yun M.-K., Wu Y., Li Z., Zhao Y., Waddell M.B., Ferreira A.M., Lee R.E., Bashford D., White S.W. Catalysis and sulfa drug resistance in dihydropteroate synthase: crystal structures reveal the catalytic mechanism of DHPS and the structural basis of sulfa drug action and resistance. Science. 2012;335:1110–1114. doi: 10.1126/science.1214641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Rubin E.J., Bifani P., Mathys V., Lim V., Au M., Jang J., Nam J., Dick T., Walker J.R., Pethe K., Camacho L.R. para-Aminosalicylic acid is a prodrug targeting dihydrofolate reductase in Mycobacterium tuberculosis. J. Biol. Chem. 2013;288:23447–23456. doi: 10.1074/jbc.M113.475798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.