Abstract
Class A β-lactamases are inactivated by the suicide inactivators sulbactam, clavulanic acid, and tazobactam. An examination of multiple alignments indicated that amino acids 216 to 218 differed among class A enzymes. By random replacement mutagenesis of codons 216 to 218 in PSE-4, a complete library consisting of 40,864 mutants was created. The library of mutants with mutations at positions 216 to 218 in PSE-4 was screened on carbenicillin and ampicillin with the inactivator sulbactam; a collection of 14 mutants was selected, and their bla genes were completely sequenced. Purified wild-type and mutant PSE-4 β-lactamases were used to measure kinetic parameters. One enzyme, V216S:T217A:G218R, was examined for its peculiar pattern of inhibition. There was an increase in the Km from 68 μM for the wild type to 271 μM for the mutant for carbenicillin and 33 to 216 μM for ampicillin. Relative to the wild-type PSE-4 enzyme, 37- and 30-fold increases in Ki values were observed for the mutant enzyme for sulbactam and tazobactam, respectively. The results that were obtained suggested that positions 216 to 218 are important for interactions with penicillanic acid sulfone inhibitors. In contrast, V216 and A217 in the TEM-1 class A β-lactamase do not tolerate amino acid residue substitutions. However, for the PSE-4 β-lactamase, 11 of 14 mutants from the library of mutants with mutations at positions 216 to 218 whose sequences were determined had substitutions at position 216 (G, R, A, S) and position 217 (A, S). The data showed the importance of residues 216 to 218 in their atomic interactions with inactivators in the PSE-4 β-lactamase structure.
The production of β-lactamases is one of several means by which bacteria can become resistant to β-lactam antibiotics. These enzymes hydrolyze the amide bond in the β-lactam ring of antibiotics, leading to a product that has lost its antibacterial properties (22). A way to counter this resistance is to use compounds that incapacitate the β-lactamase and that act in synergy with an antibiotic (19). These agents are known as suicide inactivators and include clavulanic acid and the penicillanic acid sulfones tazobactam and sulbactam (7).
Clavulanic acid inactivates group 2a, 2b, and 2be β-lactamases, rendering the combination of clavulanic acid and ticarcillin effective in vitro and in animal models of infections (2, 6, 11). Tazobactam has been shown to be an inactivator of many group 2 β-lactamases (6). This suicide inactivator acts irreversibly against both serine-based β-lactamases and metallo-β-lactamases (7). Studies have demonstrated that the combination of tazobactam and piperacillin has a wide spectrum of activity that includes gram-positive organisms such as staphylococci, as well as many gram-negative aerobic and anaerobic bacteria (9).
Wise et al. (32) have shown that the sulfone sulbactam enhances the activities of penicillin G, ampicillin, and carbenicillin against certain β-lactamase-producing bacteria like Bacteroides fragilis, Staphylococcus aureus, and Escherichia coli in vitro.
All three inactivators are used clinically in combination with antibiotics to treat intra-abdominal infections, skin and soft tissue infections, and upper and lower respiratory tract infections (9, 12, 20). Different combinations of antibiotics and inactivators are used: ticarcillin with clavulanate, amoxicillin with clavulanate, piperacillin with tazobactam, and ampicillin with sulbactam. These combinations are used to treat infections caused by bacteria producing enzymes in group 2, including Pseudomonas aeruginosa, Serratia marcescens, E. coli, and others (5, 6, 10, 12, 20, 30).
The model enzyme used in the study described here is PSE-4, a plasmid-derived β-lactamase from gram-negative bacteria. It was first found in P. aeruginosa (25). It is a class A β-lactamase of 271 amino acids, with the mature protein having a molecular mass of 29,810 Da. The PSE-4 β-lactamase has a very high rate of hydrolysis of carbenicillin and is genetically related to the PSE-1, CARB-3, and CARB-4 carbenicillinases (3). Analysis of the PSE-4 flanking DNA region revealed an integration site common to antibiotic resistance genes inserted into transposons of the Tn21 family (3).
In this report we describe the structural and functional features of a mutant PSE-4 β-lactamase, V216S:T217A:G218R, with different properties related to inhibition by penicillanic acid sulfones such as sulbactam and tazobactam as they relate to amino acids 216 to 218 (by the standard numbering system for class A enzymes of Ambler et al. [1]) in the PSE-4 enzyme. We suggest that residues 216 to 218 could be crucial amino acids that have atomic interactions with suicide inactivators. This was established by computer-assisted modeling and structural comparisons from a three-dimensional structure model of PSE-4 constructed for TEM-1 (18), S. aureus PC1 (13, 14), and Bacillus licheniformis 749/C (23) enzymes.
MATERIALS AND METHODS
Enzymes and antibiotics.
Polynucleotide kinase was purchased from Pharmacia Biotech (Baie d’Urfé, Québec, Canada), and restriction endonuclease BamHI was purchased from New England Biolabs Ltd. (Mississauga, Ontario, Canada). The Muta-Gene phagemid in vitro mutagenesis kit was purchased from Bio-Rad Laboratories Ltd. (Mississauga, Ontario, Canada). All antibiotics except nitrocefin were purchased from Sigma Diagnostic Canada (Mississauga, Ontario, Canada); nitrocefin was purchased from Oxoid (Basingstoke, England). Sulbactam and clavulanic acid were kindly provided by Pfizer Inc. (Groton, Conn.) and SmithKline Beecham Laboratories (Bristol, Tenn.), respectively. Tazobactam was a generous gift from Synphar Laboratories Inc. (Edmonton, Alberta, Canada).
Bacterial strains.
Four strains of E. coli were used: CJ236 (dut ung thi relA; pCJ105 (Cmr) [Bio-Rad]), JM101 [F′ traD36 lacqD(lacZ)M15 proA+B+/supE thiD(lac-proAB], DH5αF′ [F′ endA1 hsdR17 (rk− mk−) supE44 thi-1 recA1 gyrA (Nal′) relA1 D(lacIZYA-argF) U169 deoR (f80dlacD(lacZ) M15)], and BL21(DE3) [F− ompT hsdSB (rB− mB−) gal dcm (DE3; purchased from Novagen, Madison, Wis.)]. Plasmid pMON711 is a recombinant plasmid constructed in the cloning vector pBGS19+ by inserting a 1.1-kb DNA fragment from pMON707 containing the blaPSE-4 gene (3).
Plasmid isolation and DNA sequencing.
Single-stranded plasmid DNA was prepared for mutagenesis and DNA sequencing as described previously (29). Sequencing reactions were done with the Taq sequencing dye terminator from ABI, Perkin-Elmer Corp. (Foster City, Calif.) loaded on an Applied Biosystems 373A system. Plasmid DNA was prepared from E. coli by the alkaline lysis procedure (29).
Oligonucleotides and random replacement mutagenesis.
Mutagenesis and phosphorylation of oligonucleotides were carried out as described by Bio-Rad by using the Muta-Gene kit and a mutagenesis method (21). The oligonucleotide primers used for mutagenesis were synthesized with the Oligo1000 DNA synthesizer (Beckman). Construction of the random library was done as described by Petrosino and Palzkill (28). The first mutagenesis step used a 40-mer oligonucleotide for the insertion of a unique BamHI restriction site within the targeted nucleotides and changing of the blaPSE-4 open reading frame.
DNA was electroporated into E. coli CJ236, screening was done on Trypticase soy agar (TSA) plates, and selection was done with kanamycin (50 μg/ml) and chloramphenicol (30 μg/ml). Proper insertion and the fidelity of the DNA sequence were confirmed by nucleotide sequencing. The second mutagenesis restored the open reading frame, oligonucleotides were designed with 14-base arms for recognition in the bla gene, and 9-base randomized DNA sequences for library construction as follows: 5′-TGGTGAACAATCAANNC/G (position 216) NNC/G (position 217) NNC/G (position 218) AATTTACTACGTTC-3′ (where N is any of the four nucleotides).
Mutants were screened on separate TSA plates for resistance or susceptibility to inhibitors via ampicillin with sulbactam (78 and 8 μg/ml, respectively) and carbenicillin with sulbactam (1,250 and 8 μg/ml, respectively). Proper insertion of the linker and random replacement of the three-codon sequence as well as the complete sequence of the blaPSE-4 gene were confirmed by DNA sequencing.
The probability that the least probable (W, W, W = 1/32 × 1/32 × 1/32) or the most probable (L, L, L = 3/32 × 3/32 × 3/32) replacement mutants were present in each experimental pool was calculated by using the Poisson distribution P, where P = λxe−λ/x!. For these calculations λ is equal to np (where n is the pool size and p is the probability of the least or most common replacement), and x was the number of times that the sequence occurs in the pool size. For these calculations, x is fixed equal to zero, giving the equation P = 1 − e−np, which is the probability that the sequence occurs one or more times in the pool (26). To construct a library of size n, libraries were plated as single colonies and pooled for DNA preparation, the mutation efficiency was evaluated by digestion with BamHI, and 10 randomly picked colonies for the library were tested for the presence of the BamHI linker. A complete library of mutants was created from five small libraries pooled together to generate the complete library consisting of 40,864 mutants.
Antibiotic susceptibility.
MICs were obtained by a microdilution method with Mueller-Hinton broth in 96-well microtiter plates. Selected β-lactamase-producing E. coli DH5α cells were grown to 109 CFU/ml, diluted to 105 CFU/ml, and inoculated with 100 μl of broth; and serial twofold dilutions were tested with each antibiotic. When carbenicillin MICs were higher than 10,000 μg/ml, we used concentrations of carbenicillin that varied from 15,000 to 30,000 μg/ml, with an increment of 1,000 μg/ml for each concentration tested. Plates were examined after 20 h at 37°C, and the lowest concentration of antibiotic which inhibited visual growth was estimated to be the MIC. Quality controls included the standard strain E. coli ATCC 29522, values for which were compared with the values determined by the National Committee for Clinical Laboratory Standards.
β-Lactamase purification.
Strains were grown for 3 h in Terrific Broth (29) containing 50 μg of kanamycin and ampicillin per ml, and 1 mM isopropyl-1-thio-β-d-galactopyranoside was added. The cultures were incubated overnight at 37°C. Periplasmic proteins (including β-lactamase protein) were isolated by osmotic shock (24). The crude extract obtained was filtered on PD-10 Sephadex 25 columns (Pharmacia Biotech) and was directly loaded onto an anion-exchange chromatography Econo-Pac Q cartridge (Bio-Rad). Fractions containing β-lactamases were identified with nitrocefin and by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Selected fractions were concentrated and buffers were exchanged by ultrafiltration with Centriplus 10 and Centricon 10 filters (Amicon Canada Ltd. Oakville, Ontario, Canada). Molecular sieve chromatography was done with HiPrep 26/60 Sephacryl S-100 (Pharmacia Biotech) and a filtration step with 50 mM sodium phosphate buffer–0.15 M NaCl (pH 7.0) at a flow rate of 1.3 ml/min. Chromatography was done on a ConSep LC100 apparatus from Millipore (Nepean, Ontario, Canada). Fractions were selected as described above, and enzyme purity was estimated by SDS-PAGE with Coomassie blue and Sypro orange staining (Bio-Rad); the relative densities of the protein bands were estimated with NIH Image software (version 1.60). Enzymes were stabilized in 50% glycerol and 300 μg of ultrapure bovine serum albumin per ml and were kept in aliquots at −20°C.
Enzyme kinetics.
Kinetic analyses were done at 30°C in 50 mM sodium phosphate buffer (pH 7.0) for 30 s in a CARY 1 spectrophotometer (Varian). Kinetic parameters were determined for substrates with the corresponding change in molar extinction coefficients: for nitrocefin, 485 nm and 14,060 M−1 cm−1; for carbenicillin, 232 nm and 1,190 M−1 cm−1; and for ampicillin, 232 nm and 912 M−1 cm−1. The kinetic parameters Vmax and Km were determined from the rates of hydrolysis calculated from the initial linear portion of the curve, by using a least-squares calculation. The concentrations of the substrates varied from 15 to 1,000 μM for both wild-type and mutant enzymes. In a 1-ml reaction volume in a quartz cuvette, the concentration of the wild type enzyme was 3.20 nM and those for the mutants enzymes varied from 7.80 to 10.60 nM. All experiments were done in triplicate.
Kinetic parameters were also examined for tazobactam, sulbactam, and clavulanic acid by using carbenicillin as the substrate. The kinetic parameter Ki was determined from the rate of hydrolysis calculated in triplicate from the initial linear portion of the curve, by using a least-squares calculation. For the wild-type enzyme, the tazobactam was used at concentrations of 1.25 and 2.50 μM, sulbactam was used at concentrations of 25, 50, and 75 μM, and clavulanic acid was used at concentrations of 10 and 20 μM, with various concentrations of substrate (100 to 900 μM) used with all three compounds. The enzyme concentration varied between 3.20 and 8.40 nM. For the mutant, tazobactam was used at concentrations of 10 and 20 μM, sulbactam was used at concentrations of 100 and 200 μM, and clavulanic acid was used at concentrations of 20 and 40 μM, while the substrate concentration was varied (500 to 1,000 μM) for all three compounds. Enzyme was used at a concentration of 7.80 nM for all three assays of inhibitors. Analysis of enzyme kinetic data was done with Leonora, a program for robust regression of enzyme data, and a biweighting regression system (8). The Ki was computed from a double-reciprocal plot of 1/Vmax versus 1/[S] with varying [I], where [S] is the slope, [I] is the ordinate intercept, the slope is Km/Vmax, and the ordinate intercept is 1/Vmax.
Progressive inhibition determinations.
A concentration of 2 μM enzyme (PSE-4 and V216S:T217A:G218R) and various concentrations of inactivators (sulbactam and clavulanate) were incubated together at 25°C in a volume of 50 μl. A control containing enzyme with no inactivator was prepared in 50 mM sodium phosphate buffer (pH 7.0). Samples of 2 μl were withdrawn at regular intervals and were immediately diluted in 1,000 μl containing 500 or 700 μM carbenicillin. Reaction rates were monitored for 5 min in a CARY 1 spectrophotometer (Varian). A value of the percentage of the control activity was obtained by dividing an experimental hydrolysis rate from the linear portion of the curve by the value of the control activity obtained at identical time points. This analysis generated a partition ratio defined as the number of inhibitor molecules required to inactivate one enzyme molecule, which is based upon the inhibitor-enzyme ratio that resulted in >90% enzyme inactivation after 18 h of incubation (7).
Values of kcat and kinact were determined for both clavulanate and tazobactam for both the wild-type PSE-4 and the V216S:T217A:G218R variant enzymes. A kinact value was determined by using different inhibitor-to-enzyme ratios in a reaction volume of 100 to 150 μl at 4°C. Enzyme activity was assayed at different time intervals at between 0 and 6 min by taking 10 μl for the wild-type PSE-4 and 20 μl for the variant diluted in 1,000 μl containing 700 μM carbenicillin. Reaction rates were monitored for 30 s, and hydrolysis rates (V0) from the linear portion of the curve were obtained. A plot of V0 versus time was plotted, and the slope (kobs) of the linear portion was calculated. When plotting a double reciprocal of kobs versus the concentration of the inhibitor, the slope is equal to 1/kinact. By using the partition ratio (kcat/kinact) and kinact, a value of kcat is generated.
For tazobactam, inhibitor-to-enzyme ratios were between 0.1 and 2.0 with 258 nM wild-type PSE-4. For the variant enzyme, inhibitor-to-enzyme ratios were between 0.05 and 2.0 with 516 nM enzyme. For clavulanate, inhibitor-to-enzyme ratios of 0.1 to 5.0 with 258 nM enzyme were used for both the wild-type PSE-4 and the V216S:T217A:G218R variant enzymes.
RESULTS
Selection of mutants from the library of mutants with mutations at position 216 to 218.
Since the library was constructed to study interactions with suicide inactivators, mutants were selected on separate TSA plates containing ampicillin and sulbactam (78 and 8 μg/ml, respectively) and carbenicillin and sulbactam (1,250 and 8 μg/ml, respectively). These concentrations allowed the growth of E. coli producing wild-type PSE-4. To facilitate isolation of enzyme variants expressed in E. coli, selection was done with two- and fourfold the concentrations of substrates while maintaining a constant inhibitor concentration. No mutants were obtained at higher concentrations. However, only 16% of the library was recovered as bacterial colonies on plates with carbenicillin and sulbactam, and only 57% of the library was recovered on plates with ampicillin and sulbactam at concentrations similar to those that allowed the recovery of the wild type. The rest of the library could have comprised unstable or inactive mutants, and mutants were not selected for on this kind of selection medium.
Palzkill and Botstein (27) suggested that the ability to introduce amino acid substitutions in a target protein is central to protein structure and function studies. The tolerance of specific positions to amino acid substitutions defines the importance of the position for the structure and function of the protein. Positions that cannot tolerate substitutions are inferred to be essential for structure and/or function (27). DNA sequencing of the blaPSE-4 gene from the mutants showed gene integrity except at the mutagenesis position generating functional mutants with mutations at positions 216 to 218. As shown in Table 1, a variety of mutants had single, double, or triple amino acid changes. Of 14 variants sequenced, 11 had substitutions at position 216 (G, S, A, R), 11 had substitutions at position 217 (A, S), and 8 had substitutions at position 218 (R, S, A, Q, H). Multiple substitutions were seen in 10 variants. Two had substitutions at positions 216 and 217, one had substitutions at positions 216 and 218, one had substitutions at positions 217 and 218, and six had substitutions at all three positions (Table 1). Although there is variability in the mutants, for some variants such as pMON77020, pMON77022, pMON77023, pMON77024, pMON77025, pMON770228, and pMON77031, interesting patterns in the MICs of substrates or inactivators due to the mentioned substitutions were found, and these could have affected the structure and/or function of the enzyme (Tables 1 and 2).
TABLE 1.
Susceptibilities of E. coli DH5α strains to different antibiotics
| E. coli strain | Amino acid at the following codon positions:
|
MIC (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|---|
| 216 | 217 | 218 | Amp | Carb | Ticar | Pip | |
| DH5α | <20 | <20 | <20 | <20 | |||
| ATCC 25922 | <20 | <20 | <20 | <20 | |||
| DH5α(pBGS19+)b | <20 | <20 | <20 | <20 | |||
| DH5α(pMON711) (wild type) | V | T | G | 5,000 | 20,000c | 20,000 | 625 |
| DH5α mutants | |||||||
| pMON77020 | G | A | G | 625 | 2,500 | 1,250 | 313 |
| pMON77021 | V | A | G | 2,500 | 18,000c | 10,000 | 313 |
| pMON77022 | G | S | G | 1,250 | 5,000 | 2,500 | 313 |
| pMON77023 | G | S | R | 1,250 | 5,000 | 2,500 | 313 |
| pMON77024 | G | T | S | 1,250 | 5,000 | 2,500 | 313 |
| pMON77025 | G | S | A | 1,250 | 5,000 | 2,500 | 313 |
| pMON77026 | V | S | A | 5,000 | 18,000c | 10,000 | 625 |
| pMON77028 | R | T | G | 2,500 | 18,000c | 10,000 | 313 |
| pMON77029 | A | S | S | 2,500 | 10,000 | 10,000 | 313 |
| pMON77030 | A | S | Q | 2,500 | 10,000 | 10,000 | 313 |
| pMON77031 | S | A | R | 1,250 | 10,000 | 5,000 | 625 |
| pMON77033 | A | S | H | 2,500 | 18,000c | 10,000 | 313 |
Abbreviations: Amp, ampicillin; Carb, carbenicillin; Ticar, ticarcillin; Pip, piperacillin.
E. coli DH5α strain containing the pBGS19+ vector only.
MICs of between 15,000 and 30,000 μg/ml were determined.
TABLE 2.
Susceptibilities of E. coli DH5α strains to different combinations of antibiotics and inhibitors
| E. coli strain | Amino acid at the following codon positions:
|
MIC (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 216 | 217 | 218 | Carb + SLB | Amp + SLB | Carb + CLV | Amp + CLV | Carb + TZB | Amp + TZB | |
| DH5α | <10 | <10 | 2 | 2 | 8 | 8 | |||
| ATCC 25922 | <10 | <10 | 16 | 4 | 32 | 16 | |||
| DH5α(pBGS19+)b | <10 | <10 | 2 | 2 | 16 | 16 | |||
| DH5α(pMON711) (wild type) | V | T | G | 2,500 | 156 | 500 | 16 | 1,000 | 500 |
| DH5α mutants | |||||||||
| pMON77020 | G | A | G | 625 | 156 | 63 | 8 | 500 | 250 |
| pMON77021 | V | A | G | 1,250 | 156 | 250 | 31 | 500 | 250 |
| pMON77022 | G | S | G | 1,250 | 156 | 63 | 8 | 500 | 250 |
| pMON77023 | G | S | R | 625 | 156 | 63 | 8 | 500 | 125 |
| pMON77024 | G | T | S | 1,250 | 156 | ND | ND | 250 | 125 |
| pMON77025 | G | S | A | 1,250 | 156 | 63 | 8 | 500 | 250 |
| pMON77026 | V | S | A | 1,250 | 156 | 500 | 31 | 500 | 250 |
| pMON77028 | R | T | G | 2,500 | 156 | 63 | 4 | 1,000 | 250 |
| pMON77029 | A | S | S | 2,500 | 313 | 250 | 31 | 500 | 250 |
| pMON77030 | A | S | Q | 2,500 | 313 | 500 | 31 | 500 | 250 |
| pMON77031 | S | A | R | 2,500 | 625 | 63 | 8 | 500 | 250 |
| pMON77033 | A | S | H | 1,250 | 156 | 250 | 31 | 250 | 125 |
Abbreviations: Carb, carbenicillin; Amp, ampicillin; SLB, sulbactam; TZB, tazobactam; CLV, clavulanic acid. ND, not determined. Sulbactam, clavulanic acid, and tazobactam were each used at 8 μg/ml.
E. coli DH5α strain containing the pBGS19+ vector only.
Antibiotic and inhibitor susceptibilities.
The MICs of ampicillin and carbenicillin were different for the PSE-4 variants that were recovered. Some variants were more susceptible to ampicillin, as seen for variants with V216G:T217A and V216S:T217A:G218R substitutions, in which there were eight- and fourfold increases in susceptibility, respectively (Table 1). When carbenicillin was used, the V216G:T217A variant also had an eightfold increase in susceptibility, some variants had fourfold increases in susceptibility, while for others no significant changes in the MICs were found (Table 1). We noted that all no significant changes in the MICs of piperacillin were found for mutants with mutant enzymes (Table 1). Some variants showed eightfold increases in susceptibility to ticarcillin compared to that of the wild type, but the greatest difference was observed for the V216G:T217A variant, for which the MIC decreased 16-fold (Table 1).
The MICs of three suicide inactivators, sulbactam, tazobactam, and clavulanic acid in combination with ampicillin and carbenicillin as substrates, were determined. For tazobactam with ampicillin or carbenicillin, some mutants showed increased susceptibility compared to that of the wild type. While the MICs of carbenicillin and ampicillin combined with tazobactam for strains carrying the wild-type PSE-4 were 1,000 and 500 μg/ml, respectively, the MICs of both substrates with tazobactam for the mutants decreased two- or fourfold (Table 2).
The susceptibilities of the E. coli DH5α variants to clavulanic acid with ampicillin were not significantly different; some mutants had a twofold difference, which represents experimental error. Only for the V216R variant was there a fourfold decrease in the MIC. For clavulanic acid with carbenicillin, an eightfold increase in susceptibility occurred in variants pMON77020, pMON77022, pMON77023, pMON77025, pMON77028, and pMON77031 (Table 2).
For most of the mutants no more than a twofold decrease in the MICs of sulbactam with carbenicillin compared to the MICs for the wild type (2,500 μg/ml) were found. For two variants, V216G:T217A and V216G:T217S:G218R, however, the MICs were 625 μg/ml (Table 2). The only significant changes in the MICs of sulbactam with ampicillin was a fourfold decrease in susceptibility from 156 μg/ml for the wild-type to 625 μg/ml for the V216S:T217A:G218R variant (Table 2).
Kinetic analysis.
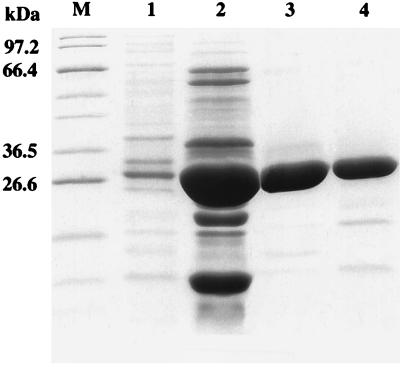
From among the 14 variant enzymes whose sequences were determined, 1 enzyme was purified to study the effects of these mutations on catalytic activity. This mutant was chosen because of the observed pattern in the MICs of suicide inactivators. The mutant showed a slight decrease in susceptibility to sulbactam in combination with ampicillin but increased susceptibility to clavulanic acid in combination with carbenicillin. Therefore, kinetic parameters for all three suicide inactivators were determined by using carbenicillin as the substrate because PSE-4 is classified as a carbenicillin-hydrolyzing enzyme (6). The V216S:T217A:G218R β-lactamase from pMON77031 was purified to a level of purity evaluated by SDS-PAGE to be ≥99%, as shown in Fig. 1, lane 3. Kinetic parameters for ampicillin and carbenicillin hydrolysis were also determined.
FIG. 1.
SDS-PAGE of the purified β-lactamase mutant. Lanes are as follows: M, broad-host-range protein marker (New England Biolabs); 1, osmotic shock fraction; 2, anion-exchange fraction; 3 and 4, molecular sieve fractions. A total of 2.45 μg of enzyme was loaded into lane 1, 60 μg was loaded into lane 2, 1.59 μg was loaded into lane 3, and 7.87 μg was loaded into lane 4.
As depicted in Table 3, there were similar values for the catalytic efficiencies (kcat/Km) of each enzyme for ampicillin and carbenicillin. The enzyme variant had a 10-fold decrease in its kcat/Km for both substrates. For carbenicillin, there was a fourfold increase in Km and a threefold decrease in kcat, explaining the 10-fold reduction in kcat/Km. For ampicillin, a different and more pronounced effect was seen on the Km value, which was sixfold the value for the wild-type enzyme. There was no significant change in the kcat of the V216S:T217A:G218R variant for ampicillin (Table 3).
TABLE 3.
Kinetic parameters for wild-type and mutant β-lactamases
| β-Lactamase | Carbenicillin
|
Ampicillin
|
||||||
|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1 [106]) | Relative valuea | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1 [106]) | Relative value | |
| PSE-4 (wild type) | 68 ± 4 | 1,190 ± 124 | 17 ± 0.2 | 1 | 33 ± 3 | 1,170 ± 134 | 35 ± 0.5 | 1 |
| PSE-4 mutant (216VTG→SAR) | 271 ± 30 | 422 ± 46 | 1.6 ± 0.2 | 0.094 | 216 ± 13 | 849 ± 87 | 4 ± 0.5 | 0.11 |
Ratio of kcat/Km for the mutant relative to that for the wild type.
As seen in Table 4, the mutations at positions 216 to 218 had a drastic effect on the kinetic parameters for tazobactam and sulbactam. There was a 30-fold increase in the Ki value for tazobactam relative to that of the wild-type enzyme, an indication of a decreased affinity of the enzyme for the inactivator (Table 4). A similar effect was reported for sulbactam, for which a 37-fold increase in the Ki value relative to that of the wild-type enzyme was noted. There was no significant change in the Ki value for clavulanic acid (Table 4).
TABLE 4.
Kinetic parameters for wild-type and mutant β-lactamases for inhibitors assayed with carbenicillina
| β-Lactamase | TZB
|
CLV
|
SLB
|
|||
|---|---|---|---|---|---|---|
| Ki (μM) | Relative valueb | Ki (μM) | Relative value | Ki (μM) | Relative value | |
| PSE-4 (wild type) | 0.33 ± 0.06 | 1 | 22 ± 3 | 1 | 6 ± 0.9 | 1 |
| PSE-4 mutant (216VTG→SAR) | 10 ± 1.2 | 30 | 32 ± 4 | 1.45 | 222 ± 4 | 37 |
Abbreviations: TZB, tazobactam; SLB, sulbactam; CLV, clavulanic acid.
Ratio of Ki for the mutant relative to that for the wild type.
Progressive inhibition determinations.
The activities of the suicide inactivators were estimated by determining the progressive inhibition of the enzyme. For clavulanic acid the ratio for turnover number before inactivation gave an inhibitor-to-enzyme ratio of 80 for both wild-type PSE-4 and the V216S:T217A:G218R variant. For tazobactam, a partition ratio of 1,000 was found for the wild-type enzyme, while that for the variant was 250, a fourfold difference (Table 5). This decrease in the partition ratio was due to a fivefold reduction in the kcat value of the V216S:T217A:G218R variant (Table 5). However, no significant change was seen in clavulanate, for which the kcat of the wild-type PSE-4 was 72 s−1 while the kcat of the variant was 58 s−1 (Table 5). We observed no significant difference in the kinact values of the wild-type and variant enzymes for both inactivators (Table 5).
TABLE 5.
Kinetic characteristics for the inactivation chemistry of the PSE-4 β-lactamase
| β-Lactamase | Clavulanate
|
Tazobactam
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | kinact (s−1) | kcat/kinact | kcat (s−1) | kinact (s−1) | kcat/kinact | |
| PSE-4 (wild type) | 72 | 0.90 | 80 | 1,030 | 1.03 | 1,000 |
| PSE-4 mutant (216VTG→SAR) | 58 | 0.73 | 80 | 221 | 0.88 | 250 |
DISCUSSION
From the random replacement mutagenesis done at residues 216 to 218 in the PSE-4 β-lactamase, we report on a mutant with a decreased affinity for the suicide inactivators sulbactam and tazobactam. This mutant exhibited substitutions at all three positions, V216S, T217A, and G218R.
Recent studies have shown that for the TEM-1 class A β-lactamase, V216 and A217 do not tolerate amino acid residue substitutions (15). For the PSE-4 β-lactamase, this is not the case because of the 14 mutants from the library of mutants with mutations at positions 216 to 218 whose sequences were determined, 11 had substitutions at positions 216 (G, R, A, S) and 217 (A, S). An examination of these residues in the three-dimensional structures of the TEM-1 (18), B. licheniformis 749/C (23), and S. aureus PC1 (13, 14) enzymes indicated that the environment around several of these residues differed among the class A enzymes (15).
In the structure of the TEM-1 enzyme, the V216 side chain is 31% solvent exposed and points into the active site toward the catalytic S70 domain (18). It is suggested that because amino acid residue substitutions are not tolerated at this position, there could be an important role for V216 in ampicillin hydrolysis (15). This leads to an interesting point because for PSE-4 mutant V216S:T217A:G218:R, there was a sixfold increase in the Km for ampicillin and a 10-fold decrease in catalytic efficiency. This suggested that although V216 in the PSE-4 mutant tolerated amino acid substitutions, it could have a role similar to that of TEM-1 in the hydrolysis of ampicillin.
Susceptibility studies confirmed that mutant V216S:T217A:G218:R exhibited a slight decrease in susceptibility to sulbactam with ampicillin and an increase in susceptibility to clavulanic acid with carbenicillin, while no significant changes in susceptibility to tazobactam with both substrates were observed. From the kinetic studies, 37- and 30-fold increases in Ki values relative to those for the wild-type PSE-4 were shown for sulbactam and tazobactam, respectively. No significant difference was found in the Ki value for clavulanic acid. It is interesting that the mutant exhibited a slightly lower level of susceptibility to sulbactam than to tazobactam. This could be explained by the fact that this mutant was selected on medium containing sulbactam.
We compared the MICs and Ki values of the three inactivators. We observed that clavulanate is more potent against E. coli containing PSE-4 β-lactamase. Although tazobactam showed the highest affinity to the active site of the enzyme compared to the affinities of the other inactivators, clavulanate had the largest effect on MICs. The MIC of sulbactam with carbenicillin for the strain producing wild-type PSE-4 decreased 8-fold, and the MIC of tazobactam for the same strain decreased 20-fold. However, the MIC of clavulanate decreased 40-fold compared to the MICs without any inactivators. For the strain producing the parent PSE-4, the MICs of ampicillin with sulbactam, tazobactam, and clavulanate decreased 32-, 10-, and 312-fold, respectively. Clavulanate had the largest effect on the MIC for the strain with V216S:T217A:G218R, with 320- and 625-fold decreases with carbenicillin and ampicillin, respectively (Tables 1 and 2). This suggested that although clavulanate does not have the highest affinity to the enzyme, the inhibitory effect of clavulanate toward the PSE-4 wild-type and V216S:T217A:G218R enzymes is more efficient than those of the other inactivators.
The interaction of clavulanic acid with residues 216 to 218 is very different from those of sulbactam and tazobactam, or, rather, clavulanic acid has no interactions with these residues in the PSE-4 enzyme. These results were consistent with those reported by Imtiaz et al. (17), whose proposed mechanism of inhibition by the penicillanic acid sulfones was different from that for clavulanic acid. It was demonstrated that protonation by the conserved crystallographic water for sulbactam is not thought to be required for the formation of the inactivating species as it is for clavulanic acid (17). In addition, the nature of the leaving group at C-5 is very different between sulbactam and clavulanic acid, explaining the differences in the mechanisms of inactivation (4). It has also been shown that R244 plays an important role during the inactivation reaction by both clavulanate and sulbactam (16, 17). However, the mechanisms of action and the interactions of these compounds with the TEM-1 β-lactamase were shown to be different (16, 17).
From the three-dimensional model constructed for PSE-4 by using atomic coordinates from crystallized TEM-1 (18), S. aureus PC1 (13, 14), and B. licheniformis 749/C (23) enzymes, we observed that amino acids 216 to 218 were present on a random coil between the all-α domain and the α/β domain. Although the PSE-4 enzyme has not yet been crystallized, Strynadka et al. (31) state that the active-site regions of the crystallized TEM-1, PC1, and B. licheniformis enzymes have very similar conformations, so that mechanistic deductions based on the structure of TEM-1 are applicable to other class A enzymes. Therefore, we refer to the mechanistic results obtained for TEM-1 and apply them to the PSE-4 enzyme. It should be noted that TEM-1 and PSE-4 have 41% amino acid sequence identity.
The purified PSE-4 mutant enzyme from pMON77031 had an important change at position 218, where a G residue, a small polar uncharged amino acid, has been substituted for an R residue, a large positively charged amino acid. Residues 216 to 218 are found at the interface of the two domains of the enzyme, and the active site lies between these two domains. A substitution at position 218 from a G to an R could increase the stability of the enzyme by making a favorable electrostatic interaction with E273 found at the beginning of the α-11 helix of the α/β domain of the enzyme.
The Ki values obtained indicated increases in Ki values for sulbactam and tazobactam of 37- and 30-fold, respectively, but no changes in the Ki value for clavulanate. This difference in the dissociation constant suggests that the enzyme has less affinity for the sulfone inactivators sulbactam and tazobactam, while the mutations at positions 216 to 218 did not affect clavulanate binding. However, while no significant changes in the kcat and kinact values for clavulanate were observed between wild-type PSE-4 and variant enzymes, a significant decrease in the kcat for tazobactam was observed for the variant V216S:T217A:G218S enzyme compared to that for the wild-type PSE-4 enzyme. This suggests that other than having a role in tazobactam binding, amino acids 216 to 218 are also implicated in the inactivation chemistry. On the other hand positions 216 to 218 are not implicated in either the binding of clavulanate or inactivation chemistry in the PSE-4 β-lactamase, as seen from the kinetic characteristics for this inactivator.
For the TEM-1 β-lactamase, in the preacylation complex the hydroxyl group of S235 forms a strong hydrogen bond with the carboxylate of clavulanate. This carboxylate also forms hydrogen bonds with invariant residues S130, R244, and crystallographic water W673, so that a total of four hydrogen bonds exist around the carboxyl anion (16). In the preacylation complex of TEM-1 with sulbactam, Imtiaz et al. (17) have demonstrated that there is a strong electrostatic attraction for the carboxylate of sulbactam with K234, S235, and R244. Therefore, there are only three hydrogen bonds in the preacylation complex with sulbactam, but four are suggested for clavulanate. This means that the latter has a stronger interaction in the active site than the former.
Therefore, the interactions between residues in the active site and the suicide inactivators are different for sulbactam, tazobactam, and clavulanic acid. In the V216S:T217A:G218R variant, the mutations in this case affected only residues implicated with sulbactam and tazobactam but not with clavulanate, as seen from the kinetic study results. Imtiaz et al. (16) have demonstrated that in TEM-1 the C-2 substituent of clavulanate lies near the guanidinium moiety of R244. A water molecule, W673, links R244, the clavulanate carboxyl group, and the main-chain carbonyl group of residue 216. In our case a change from V to S at position 216 did not appear to affect the hydrogen bonding between W673 and V216 with the clavulanate carboxyl group. However, other mutations might hinder the accessibility of the main-chain carbonyl group of residue 216 and affect inactivation by clavulanate.
One possible role of residues 216 to 218 is to maintain a kinetically favorable environment around the active site. A substitution at these positions could affect electrostatic interactions with substrates and with suicide inactivators, inducing changes to the enzyme’s properties as demonstrated in these studies. Further structural studies based on the crystal structure of PSE-4 β-lactamase are eventually required to confirm these results and will help us understand the mechanisms of β-lactamase inactivation.
ACKNOWLEDGMENTS
We express our gratitude to Lindsay Eltis for suggestions and comments in the kinetics analysis and in using the Leonora software.
R.C.L. is a Scholar of Exceptional Merit from the Fonds de la Recherche en Santé du Québec. Work in R.C.L.’s laboratory is funded by the Medical Research Council of Canada and by the Centers of Excellence via the Canadian Bacterial Diseases Network.
REFERENCES
- 1.Ambler R P, Coulson F W, Frère J-M, Ghuysen J-M, Joris B, Forsman M, Levesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–272. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry A L, Ayers L W, Gaven T L, Gerlach E N, Jones R N. In vitro activity of ticarcillin plus clavulanic acid against bacteria isolated in three centers. Eur J Clin Microbiol. 1984;3:203–206. doi: 10.1007/BF02014879. [DOI] [PubMed] [Google Scholar]
- 3.Boissinot M, Levesque R C. Nucleotide sequence of the PSE-4 carbenicillinase gene and correlations with the Staphylococcus aureus PC1 β-lactamase crystal structure. J Biol Chem. 1990;265:1225–1230. [PubMed] [Google Scholar]
- 4.Bonomo R A, Dawes C G, Knox J R, Shlaes D M. β-Lactamase mutations far from the active site influence inhibitor binding. Biochem Biophys Acta. 1995;1247:121–125. doi: 10.1016/0167-4838(94)00188-m. [DOI] [PubMed] [Google Scholar]
- 5.Brismar B, Malmborg A S, Tunevall G, Wretlind B, Bergman L, Mentzing L O, Nystrom P O, Kihlstrom E, Backstrand B, Skau T, Kasholm-Tengve B, Sjöberg L, Olsson-Liljequist B, Tally F P, Gatenbeck L, Eklund A E, Nord C E. Piperacillin-tazobactam versus imipenem-cilastatin for treatment of intra-abdominal infections. Antimicrob Agents Chemother. 1992;36:2766–2773. doi: 10.1128/aac.36.12.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamase and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush K, Macalintal C, Rasmussen B A, Lee V J, Yang Y. Kinetic interactions of tazobactam with β-lactamases from all major structural classes. Antimicrob Agents Chemother. 1993;37:851–858. doi: 10.1128/aac.37.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornish-Bowden A. Analysis of kinetic data. New York, N.Y: Oxford Science Publications; 1995. [Google Scholar]
- 9.Daniel K P, Krop L C. Piperacillin-tazobactam: a new β-lactam–β-lactamase inhibitor combination. Pharmacotherapy. 1996;16:149–162. [PubMed] [Google Scholar]
- 10.Fink M P, Helsmoortel C M, Arous E J, Doem G V, Moriarty K P, Fairchild P G, Townsed P L. Comparison of the safety and efficacy of parenteral ticarcillin/clavulanate and clindamycin/gentamicin in serious intra-abdominal infections. J Antimicrob Agents. 1989;24:147–156. doi: 10.1093/jac/24.suppl_b.147. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs P C, Barry A L, Thornsberry C, Jones R N. In vitro activity of ticarcillin plus clavulanic acid against 622 clinical isolates. Antimicrob Agents Chemother. 1984;25:392–394. doi: 10.1128/aac.25.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart S M, Bailey E M. A practical look at the clinical usefulness of the beta-lactam/beta-lactamase inhibitor combinations. Ann Pharmacother. 1996;30:1130–1140. doi: 10.1177/106002809603001013. [DOI] [PubMed] [Google Scholar]
- 13.Herzberg O. Refined crystal structure of β-lactamase from Staphylococcus aureus PC1 at 2.0 Å resolution. J Mol Biol. 1991;217:701–720. doi: 10.1016/0022-2836(91)90527-d. [DOI] [PubMed] [Google Scholar]
- 14.Herzberg O, Moult J. Bacterial resistance of β-lactam antibiotics: crystal structure of β-lactamase from Staphylococcus aureus PC1 at 2.5 Å resolution. Science. 1987;236:694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- 15.Huang W, Petrosino J, Hirsch M, Shenkin P S, Palzkill T. Amino acid sequence determinants of β-lactamase structure and activity. J Mol Biol. 1996;258:688–703. doi: 10.1006/jmbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- 16.Imtiaz U, Billings E, Knox J R, Manavathu E K, Lerner S A, Mobashery S. Inactivation of class A β-lactamase by clavulanic acid: the role of arginine-244 in a proposed nonconcerted sequence of events. J Am Chem Soc. 1993;115:4435–4441. [Google Scholar]
- 17.Imtiaz U, Billings E M, Knox J R, Mobashery S. A structure-based analysis of the inhibition of class A β-lactamases by sulbactam. Biochemistry. 1994;33:5728–5738. doi: 10.1021/bi00185a009. [DOI] [PubMed] [Google Scholar]
- 18.Jelsch C, Mourey L, Masson J-M, Samama J-P. Crystal structure of Escherichia coli TEM-1 β-lactamase at 1.8 Å resolution. Proteins Struct Funct Genet. 1993;16:364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]
- 19.Knowles J R. Penicillin resistance: the chemistry of β-lactamase inhibition. Acc Chem Res. 1985;18:97–104. [Google Scholar]
- 20.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 21.Lode H M. Clinical indications for β-lactamase inhibitors in comparison to other antibiotics. Int J Antimicrob Agents. 1996;7:S3–S7. doi: 10.1016/0924-8579(96)00312-3. [DOI] [PubMed] [Google Scholar]
- 22.Medeiros A A. β-Lactamases. Br Med Bull. 1984;40:18–27. doi: 10.1093/oxfordjournals.bmb.a071942. [DOI] [PubMed] [Google Scholar]
- 23.Moews P C, Knox J R, Dideberg O, Charlier P, Frère J-M. β-Lactamase of Bacillus licheniformis 749/C at 2 Å resolution. Proteins. 1990;7:156–171. doi: 10.1002/prot.340070205. [DOI] [PubMed] [Google Scholar]
- 24.Neu H C, Heppel L A. The release of enzymes from Escherichia coli by osmotic shock during the formation of spheroplasts. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 25.Newsom S W B. Carbenicillin-resistant Pseudomonas. Lancet. 1969;ii:1140. doi: 10.1016/s0140-6736(69)90742-9. [DOI] [PubMed] [Google Scholar]
- 26.Palzkill T, Botstein D. Probing β-lactamase structure and function using random replacement mutagenesis. Proteins. 1992;14:29–44. doi: 10.1002/prot.340140106. [DOI] [PubMed] [Google Scholar]
- 27.Palzkill T, Botstein D. Extracting information from protein sequences using random replacement mutagenesis. Methods. 1991;3:155–164. [Google Scholar]
- 28.Petrosino J F, Palzkill T. Systematic mutagenesis of the active-site omega-loop of TEM-1 β-lactamase. J Bacteriol. 1996;178:1821–1828. doi: 10.1128/jb.178.7.1821-1828.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Shlaes M D, Baughman R, Boylen C T, Chan J C, Charan N B, Norden C. Piperacillin/tazobactam compared with ticarcillin/clavulanate in community-acquired bacterial lower respiratory tract infection. J Antimicrob Chemother. 1994;34:565–577. doi: 10.1093/jac/34.4.565. [DOI] [PubMed] [Google Scholar]
- 31.Strynadka N C J, Adachi H, Jensen S E, Johns K, Sielecki A, Betzel C, Sutoh K, James M N G. Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 Å resolution. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 32.Wise R, Andrews J M, Bedford K A. Clavulanic acid and CP-45,899: a comparison of their in vitro activity in combination with penicillins. J Antimicrob Chemother. 1980;6:197–206. doi: 10.1093/jac/6.2.197. [DOI] [PubMed] [Google Scholar]