Abstract
Regorafenib improves the survival of patients with metastatic colorectal cancer (mCRC); however, it is also characterized by detrimental dermal side effects that may require treatment cessation or modified dosing. In our previous prospective pharmacokinetic, pharmacodynamic, and pharmacogenetic studies, 17.5% (7/40) of the patients with mCRC had grade 3 erythema multiforme (EM) that caused treatment discontinuation. Haplotypes in genes encoding human leukocyte antigen (HLA) are associated with EM following the administration of drugs, such as allopurinol. This study examined the association between HLA haplotypes and regorafenib‐induced EM. Regorafenib was administered orally at 160 mg/body once daily for weeks 1–3 of each 4‐week cycle. To determine the HLA haplotypes, we used the WAKFlow HLA Typing Kit HLA‐A, ‐B, or ‐C. The carrier frequency of HLA‐C*01:02 in patients with EM (6/7) was higher than that in tolerant controls (8/33; odds ratio [OR] = 18.8, 95% confidence interval [CI] = 1.95–180, p = 0.00437). HLA‐B*46:01 was also associated with EM (OR = 11.6, 95% CI = 1.47–92.1, p = 0.0299). These associations were no longer significant after Bonferroni correction for multiple testing. Therefore, regorafenib‐induced EM in Japanese patients appears to be associated with specific HLA haplotypes but further validation is needed.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Regorafenib‐induced severe erythema multiforme (EM) has been observed in clinical trials and clinical practice, resulting in treatment discontinuation. However, the scientific basis of regorafenib‐related symptoms remains unclear.
WHAT QUESTION DID THIS STUDY ADDRESS?
As EM is an immune‐mediated mucocutaneous disease, a specific human leukocyte antigen (HLA) may be associated with the pathogenesis of its symptoms.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
We found that regorafenib‐induced EM in Japanese patients with metastatic colorectal cancer was associated with specific HLA haplotypes, HLA‐C*01:02 and HLA‐B*46:01.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
These HLA haplotypes may have clinical value in identifying patients susceptible to EM before regorafenib treatment, which may enable us to treat patients who develop severe EM appropriately.
INTRODUCTION
Regorafenib is an orally administered, small‐molecule, multi‐kinase inhibitor that blocks the activity of various protein kinases involved in the regulation of tumor angiogenesis, oncogenesis, and the tumor microenvironment. 1 Regorafenib is sequentially metabolized primarily in the liver by CYP3A4 to form two major pharmacologically active metabolites, regorafenib (pyridine)‐N‐oxide (M‐2), and N‐desmethyl regorafenib (pyridine)‐N‐oxide (M‐5). 2 , 3 Because a phase III CORRECT trial demonstrated significant survival benefit of regorafenib in patients with metastatic colorectal cancer (mCRC) who had progressed on all available standard therapies or could not tolerate standard therapies, 4 treatment with regorafenib can benefit these patients, and it has been approved for the treatment of mCRC in many countries.
Despite the significant survival advantages of regorafenib treatment, it induces characteristic dermal adverse events. 4 , 5 Erythema multiforme (EM) is a cutaneous and mucosal hypersensitivity reaction with characteristic target lesions triggered by certain antigenic stimuli, including therapeutic drugs. 6 Regorafenib is known to induce EM in clinical trials, 4 and also in clinical practice, 7 , 8 which frequently results in treatment cessation. Accordingly, the Japanese regorafenib package insert describes caution regarding these adverse events. In our prospective pharmacokinetic, pharmacodynamic, and pharmacogenetic studies, 17.5% (7/40) of patients with mCRC had grade 3 EM that caused treatment discontinuation. 9 Because EM is an immune‐mediated mucocutaneous disease, 10 the pathogenesis of the symptoms could be associated with a specific human leukocyte antigen (HLA). 11 Evidence has revealed an association between cutaneous adverse events induced by therapeutic drugs and specific HLA haplotypes. 11 Associations between carbamazepine‐induced EM and specific HLA haplotypes 12 suggest the potential of HLA haplotypes to be associated with regorafenib‐induced EM.
Based on this background, we retrospectively examined the association between HLA haplotypes and regorafenib‐induced EM by applying patients’ clinical data and genome samples collected in our previous prospective study on regorafenib. 9
METHODS
Study design
This was a retrospective study with data and samples obtained from our previous prospective pharmacokinetic, pharmacodynamic, and pharmacogenetic studies of patients with mCRC who were candidates for the administration of regorafenib at Showa University Hospital (Tokyo, Japan). 9 Our main objective was to examine the relationship between regorafenib‐induced EM and HLA haplotypes. The Institutional Review Board of Showa University approved the study protocol. The study was registered in Japan's University Hospital Medical Information Network‐Clinical Trials Registry (UMIN000013939). The eligibility of patients, treatment with regorafenib, and toxicity and efficacy evaluations have been described previously. 9
HLA typing
Genomic DNA was extracted from peripheral blood, which had been stored at −80°C until analysis, using a QIAamp Blood Kit (QIAGEN). Because many cases of drug‐induced cutaneous adverse reactions are known to be associated with HLA‐class I, 11 , 13 , 14 haplotypes in HLA‐A, ‐B, and ‐C were analyzed. To determine the HLA type, we used the WAKFlow HLA Typing Kit HLA‐A, ‐B, or ‐C (Wakunaga). The results were analyzed using the Bio‐Plex 200 system (Bio‐Rad Laboratories).
Pharmacokinetic analysis
The details of the pharmacokinetic analysis of regorafenib and its two active metabolites have been described in our previous study. 9 The total plasma concentration base and unbound plasma concentration base area under the plasma concentration‐time curve (AUC t and AUC u ) of regorafenib, M‐2, and M‐5 from time zero to the last sampling time were calculated.
Statistical analysis
Associations between two groups of categorical and continuous variables, and between two groups of categorical variables were analyzed using the Wilcoxon and Fisher's exact tests, respectively. Bonferroni correction for multiple comparisons of HLA haplotypes (12 for HLA‐A, 9 for ‐B, and 6 for ‐C) was performed to obtain corrected p values. Progression‐free survival (PFS) was calculated by the Kaplan–Meier method. Correlations of the PFS with categorical variables were assessed by the log‐rank test. All analyses were performed using the JMP software (version 16.0; SAS Institute). Associations were considered statistically significant when the two‐tailed p value was less than 0.05.
RESULTS
Patient characteristics
Forty patients were enrolled in the study between December 2013 and October 2018. The patient characteristics are shown in Table S1. All patients had liver and kidney function test results that met the eligibility criteria. Thirty‐eight patients received three or more lines of treatment. Among the 40 patients, seven had grade 3 EM, which resulted in the discontinuation of regorafenib treatment. The median duration of EM onset was 13 days (14–16 days). The total bilirubin, serum creatinine, and eGFR observed in patients with and without EM were not significantly different (p > 0.05), indicating almost equal liver and kidney functions between the two groups.
Pharmacokinetics of regorafenib, M‐2, and M‐5, and EM induced by regorafenib
First, we examined the association between the AUC t or AUC u of regorafenib, M‐2, and M‐5 and regorafenib‐induced EM. No significant differences were observed between systemic exposure to regorafenib and its active metabolites and regorafenib‐induced grade 3 EM (Figure 1).
FIGURE 1.
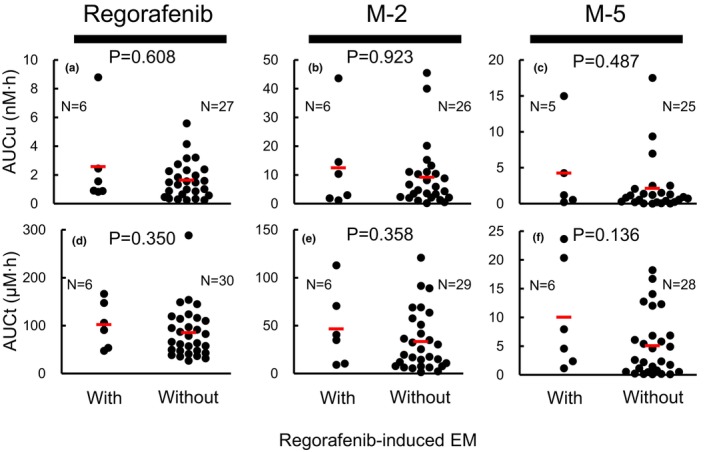
Associations of AUC u or AUC t values of regorafenib (a or d), M‐2 (b or e), and M‐5 (c or f) with regorafenib‐induced EM. No significant differences between AUC u or AUC t values of regorafenib, M‐2, and M‐5 with regorafenib‐induced EM were observed. AUC u and AUC t were analyzed for 36 patients, which included 6 patients with grade 3 EM. We failed to determine AUC u or AUC t values in some patients because of the plasma concentrations below the limit of quantitation. Wilcoxon test. Bars represent the means. AUC u , unbound plasma concentration base area under the plasma concentration‐time curve; AUC t , total plasma concentration base area under the plasma concentration‐time curve; EM, erythema multiforme.
Associations of HLA haplotypes and regorafenib‐induced EM
Next, we investigated the association between HLA haplotypes and regorafenib‐induced EM (Table 1). The carrier frequency of HLA‐C*01:02 in patients with EM (6/7) was higher than that in the tolerant controls (8/33; odds ratio [OR] = 18.8, 95% confidence interval = 1.95–180, p = 0.00437, Fisher's exact test). HLA‐B*46:01 was also associated with EM (OR = 11.6, 95% confidence interval = 1.47–92.1, p = 0.0299, Fisher's exact test). These associations were no longer significant after Bonferroni correction for multiple testing (Table 1). Diplotype configurations of HLA haplotypes seen in all 40 patients are shown in Table S2. There were no patients homozygous for HLA‐C*01:02 or HLA‐B*46:01.
TABLE 1.
Association between HLA haplotypes and regorafenib‐induced grade 3 EM.
| HLA haplotype | EM | OR (95% CI) | p | Corrected‐p | |
|---|---|---|---|---|---|
| Yes (N = 7) | No (N = 33) | ||||
| HLA‐A | |||||
| A*24:02 | 1 (14.3) a | 12 (36.4) | 0.292 (0.03–2.72) | 0.393 | 1.00 |
| A*02:01 | 1 (14.3) | 9 (27.3) | 0.444 (0.05–4.22) | 0.656 | 1.00 |
| A*02:06 | 1 (14.3) | 6 (18.2) | 0.750 (0.08–7.44) | 1.00 | 1.00 |
| A*11:01 | 2 (28.6) | 4 (12.1) | 2.90 (0.41–20.28) | 0.279 | 1.00 |
| A*31:01 | 1 (14.3) | 5 (15.2) | 0.933 (0.09–9.51) | 1.00 | 1.00 |
| A*33:03 | 1 (14.3) | 4 (12.1) | 1.21 (0.11–12.81) | 1.00 | 1.00 |
| A*02:07 | 2 (28.6) | 3 (9.09) | 4.00 (0.53–30.28) | 0.204 | 1.00 |
| A*26:03 | 1 (14.3) | 4 (12.1) | 1.21 (0.11–12.81) | 1.00 | 1.00 |
| A*11:02 | 1 (14.3) | 1 (3.03) | 5.33 (0.29–97.49) | 0.323 | 1.00 |
| A*02:03 | 1 (14.3) | 1 (3.03) | 5.33 (0.29–97.49) | 0.323 | 1.00 |
| A*26:05 | 1 (14.3) | 0 (0.00) | 15.5 (0.57–423.02) | 0.175 | 1.00 |
| A*24:07 | 1 (14.3) | 0 (0.00) | 15.5 (0.57–423.02) | 0.175 | 1.00 |
| HLA‐B | |||||
| B*51:01 | 2 (28.6) | 6 (18.2) | 1.80 (0.28–11.60) | 0.611 | 1.00 |
| B*15:01 | 1 (14.3) | 5 (15.2) | 0.933 (0.09–9.51) | 1.00 | 1.00 |
| B*54:01 | 3 (42.9) | 4 (12.1) | 5.44 (0.88–33.76) | 0.0877 | 1.00 |
| B*44:03 | 1 (14.3) | 5 (15.2) | 0.933 (0.09–9.51) | 1.00 | 1.00 |
| B*46:01 | 3 (42.9) | 2 (6.06) | 11.6 (1.47–92.14) | 0.0299 | 0.807 |
| B*48:01 | 1 (14.3) | 4 (12.1) | 1.21 (0.11–12.81) | 1.00 | 1.00 |
| B*59:01 | 1 (14.3) | 0 (0.00) | 15.5 (0.57–423.02) | 0.175 | 1.00 |
| B*15:18 | 1 (14.3) | 2 (6.06) | 2.58 (0.2–33.24) | 0.448 | 1.00 |
| B*08:01 | 1 (14.3) | 0 (0.00) | 15.5 (0.57–423.02) | 0.175 | 1.00 |
| HLA‐C | |||||
| C*01:02 | 6 (85.7) | 8 (24.2) | 18.8 (1.95–180.01) | 0.00437 | 0.118 |
| C*07:02 | 1 (14.3) | 8 (24.2) | 0.933 (0.09–9.51) | 1.00 | 1.00 |
| C*08:01 | 2 (28.6) | 6 (18.2) | 1.80 (0.28–11.60) | 0.611 | 1.00 |
| C*14:02 | 3 (42.9) | 7 (21.2) | 2.79 (0.5–15.46) | 0.338 | 1.00 |
| C*14:03 | 1 (14.3) | 5 (15.2) | 0.933 (0.09–9.51) | 1.00 | 1.00 |
| C*04:01 | 1 (14.3) | 1 (3.03) | 5.33 (0.29–97.49) | 0.323 | 1.00 |
Note: HLA haplotypes observed in patients who suffer grade 3 EM are listed.
Abbreviations: CI, confidence interval; EM, erythema multiforme; OR, odds ratio.
Number of patients (%).
Associations of HLA haplotypes and efficacy
We examined whether PFS was associated with HLA haplotypes. PFS was not associated with both HLA‐C*01:02 (p = 0.403) and HLA‐B*46:01 haplotypes (p = 0.875).
DISCUSSION
Cutaneous adverse events induced by therapeutic drugs have been revealed to be associated with specific HLA haplotypes. Carbamazepine‐induced Stevens‐Johnson syndrome or toxic epidermal necrosis which is associated with HLA‐B*15:02, and allopurinol‐induced severe cutaneous adverse drug reactions which occur in patients carrying HLA‐B*58:01 are observed in East Asian population. 11 Associations with both HLA‐C*01:02 and HLA‐B*46:01 have also been reported in East Asian populations in relation to cutaneous adverse reactions to other drugs, 15 , 16 , 17 but the current report is the first for regorafenib‐induced EM. According to the prescription information of regorafenib released by the US Food and Drug Administration in 2013, the incidence of serious adverse reactions such as EM in phase III CORRECT study 4 was 0.2%, 18 much lower than that observed in our study (17.5%). In the CORRECT study, a relatively small number of Japanese patients with mCRC (n = 67) were treated with regorafenib compared to non‐Japanese patients (n = 438). The frequency of HLA‐C*01:02 observed in East Asians, including Japanese (~20%), was higher than in Western populations (~3.0%). 19 In addition, although HLA‐B*46:01 is observed in East Asians, such as Japanese (5%–20%), the frequency of the haplotype in Western populations is almost zero. 19 The higher incidence of regorafenib‐induced EM observed in our Japanese patients appears to be attributed to the higher frequencies of these HLAs in Japanese patients. However, the lower incidence of regorafenib‐induced severe EM observed in the CORRECT study might reflect the small proportion of Japanese patients participating in the international phase III trial compared to Western patients. Cases of regorafenib‐induced severe EM observed in clinical practice have been reported in Japan, 7 , 8 but not in Western countries. Considering these results, Japanese patients will may be more likely to develop regorafenib‐related EM, although the grade and frequency have not been comprehensively elucidated. HLA‐C*01:02 and HLA‐B*46:01 might have clinical values to identify patients susceptible to EM before initiating regorafenib treatment, which enables us to treat patients who tend to develop severe EM appropriately. Further clinical studies are required to confirm the reproducibility of these results.
Because Ikeda et al. 20 demonstrated that the haplotype frequencies of HLA‐C*01:02 and HLA‐B*46:01 in the Japanese population were 17.6% and 4.77%, respectively, ~32% and 9.3% of Japanese are estimated to possess these haplotypes, respectively. The frequencies of HLA‐C*01:02 and HLA‐B*46:01 carriers observed in our 33 control patients were 24.2% and 6.06%, roughly consistent with the calculated values for the Japanese population. These results suggested that our control patients were adequate for estimating the risk of these haplotypes in regorafenib‐induced EM.
Our patients were enrolled between 2013 and 2018, shortly after the introduction of regorafenib into clinical practice. During this early period, the emergence of toxicities, such as hand‐foot syndrome, limited the use of regorafenib despite the observed survival benefits. According to the results obtained from clinical trials, 21 , 22 physicians in clinical practice have adopted a lower initial dose of regorafenib (<160 mg/day) to safely start the therapy, and then increase the dose depending on the toxicities. However, because EM is an immune‐mediated mucocutaneous disease, the dose of regorafenib does not appear to be associated with the incidence and the grade of EM. Our results showed no association between EM, and systemic exposure to regorafenib and its active metabolites. Therefore, dose reduction might not effectively manage EM. We recommend medical oncologists to discontinue regorafenib treatment when patients develop EM. The identification of patients predisposed to EM by HLA typing before regorafenib treatment may warn the physicians in terms of careful management of such patients. HLA typing is already available widely in Japanese hospitals. Therefore, this typing kit could be used for HLA typing in patients with cancer who are candidates for regorafenib administration.
Although drug‐induced severe cutaneous adverse reactions are generally known to be associated with a specific HLA haplotype, 11 , 13 , 14 phenytoin‐related severe cutaneous adverse reactions were reported to be associated with the plasma concentration profile of the therapeutic drug. 23 Conversely, we could not observe associations between regorafenib‐induced EM and AUC of regorafenib and active metabolites, suggesting the substrate‐dependent relations between drug‐induced severe cutaneous adverse reactions and pharmacokinetics.
This study has several limitations. First, the sample size was small and insufficient to obtain significant results after the Bonferroni correction. Although 160 mg/day was the initial dose of regorafenib in this study, many patients in recent clinical practice are initiated with a reduced dose of regorafenib to manage regorafenib‐induced toxicities. Therefore, it might be difficult to initiate regorafenib with a dose of 160 mg/day, even though we can enroll additional patients. Second, we did not validate the present findings. Therefore, further studies involving more patients are necessary to validate their application in clinical practice.
In conclusion, we revealed for the first time that regorafenib‐induced grade 3 EM observed in Japanese patients with mCRC was associated with specific HLA‐C*01:02 and HLA‐B*46:01 haplotypes.
AUTHOR CONTRIBUTIONS
K.F., N.M., and R.M. wrote the manuscript; K.F., H.I., and Y.K. designed the study; K.F., N.M., R.M., K.T., H.I., and Y.K. performed the research; K.F., N.M., R.M., and K.T., analyzed the data.
FUNDING INFORMATION
This study was supported in part by Grant‐in‐Aid for Scientific Research (C) (19K07204 to K.F.) from the Japan Society for the Promotion of Science (JSPS).
CONFLICT OF INTEREST STATEMENT
The authors declared no competing interests for this work.
Supporting information
Table S1
Table S2
ACKNOWLEDGMENTS
The authors thank Dr Taisei Mushiroda for his valuable advice on the haplotype analysis of human leucocyte antigen genes.
Fujita K‐i, Matsumoto N, Murase R, Takeshima K, Ishida H, Kubota Y. Associations of HLA‐C*01:02 and HLA‐B*46:01 with regorafenib‐induced erythema multiforme in Japanese patients with metastatic colorectal cancer. Clin Transl Sci. 2023;16:1741‐1747. doi: 10.1111/cts.13589
REFERENCES
- 1. Roed Skarderud M, Polk A, Kjeldgaard Vistisen K, Larsen FO, Nielsen DL. Efficacy and safety of regorafenib in the treatment of metastatic colorectal cancer: a systematic review. Cancer Treat Rev. 2018;62:61‐73. [DOI] [PubMed] [Google Scholar]
- 2. US Food and Drug Administration (FDA) . Clinical pharmacology and biopharmaceutics review(s) . https://www.accessdata.fda.gov/drugsatfda_docs/nda/2012/203085Orig1s000ClinPharmR.pdf. Accessed 29 January, 2020
- 3. European Medicines Agency (EMA) . Summary of product characteristics . https://www.ema.europa.eu/en/documents/product‐information/stivarga‐epar‐product‐information_en.pdf Accessed 29 January, 2020.
- 4. Grothey A, Van Cutsem E, Sobrero A, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo‐controlled, phase 3 trial. Lancet. 2013;381:303‐312. [DOI] [PubMed] [Google Scholar]
- 5. Yoshino T, Komatsu Y, Yamada Y, et al. Randomized phase III trial of regorafenib in metastatic colorectal cancer: analysis of the CORRECT Japanese and non‐Japanese subpopulations. Invest New Drugs. 2015;33:740‐750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hafsi W, Badri T. Erythema multiforme. StatPearls. StatPearls Publishing; 2023. [PubMed] [Google Scholar]
- 7. Matsunaga M, Ushijima T, Fukahori M, Tanikawa K, Miwa K. Erythema multiforme induced by regorafenib. J Gen Fam Med. 2017;18:90‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tashiro K, Shinto E, Kajiwara Y, et al. Systemic steroid treatment can desensitize the skin reaction due to regorafenib in a recurrence colorectal cancer patient. Int Cancer Conf J. 2019;8:164‐169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kubota Y, Fujita KI, Takahashi T, et al. Higher systemic exposure to unbound active metabolites of Regorafenib is associated with short progression‐free survival in colorectal cancer patients. Clin Pharmacol Ther. 2020;108:586‐595. [DOI] [PubMed] [Google Scholar]
- 10. Volkers SM, Meisel C, Terhorst‐Molawi D, et al. Clonal expansion of CD4(+)CD8(+) T cells in an adult patient with mycoplasma pneumoniae‐associated erythema multiforme majus. Allergy Asthma Clin Immunol. 2021;17:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pan RY, Dao RL, Hung SI, Chung WH. Pharmacogenomic advances in the prediction and prevention of cutaneous idiosyncratic drug reactions. Clin Pharmacol Ther. 2017;102:86‐97. [DOI] [PubMed] [Google Scholar]
- 12. Sousa‐Pinto B, Correia C, Gomes L, et al. HLA and delayed drug‐induced hypersensitivity. Int Arch Allergy Immunol. 2016;170:163‐179. [DOI] [PubMed] [Google Scholar]
- 13. Kloypan C, Koomdee N, Satapornpong P, Tempark T, Biswas M, Sukasem C. A comprehensive review of HLA and severe cutaneous adverse drug reactions: implication for clinical pharmacogenomics and precision medicine. Pharmaceuticals (Basel). 2021;14:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mushiroda T. Avoidance of cutaneous adverse drug reactions induced by antiepileptic drugs based on pharmacogenomics. J Hum Genet. 2023;68:227‐230. [DOI] [PubMed] [Google Scholar]
- 15. Jiang ML, Wang LT, Chen SA, et al. Association between HLA‐B*46:01 and cutaneous adverse drug reactions in Han Chinese. J Bio‐X Res. 2018;1:73‐78. [Google Scholar]
- 16. Kim SH, Kim M, Lee KW, et al. HLA‐B*5901 is strongly associated with methazolamide‐induced Stevens‐Johnson syndrome/toxic epidermal necrolysis. Pharmacogenomics. 2010;11:879‐884. [DOI] [PubMed] [Google Scholar]
- 17. Yang F, Xuan J, Chen J, et al. HLA‐B*59:01: a marker for Stevens‐Johnson syndrome/toxic epidermal necrolysis caused by methazolamide in Han Chinese. Pharmacogenomics J. 2016;16:83‐87. [DOI] [PubMed] [Google Scholar]
- 18. FDA . Highlights of prescription information (Regorafenib) . https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/203085s001lbl.pdf. Accessed March 13, 2023.
- 19. AFND . Allele frequency net database . http://www.allelefrequencies.net/hla.asp. Accessed March 14, 2023.
- 20. Ikeda N, Kojima H, Nishikawa M, et al. Determination of HLA‐A, ‐C, ‐B, ‐DRB1 allele and haplotype frequency in Japanese population based on family study. Tissue Antigens. 2015;85:252‐259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Argiles G, Mulet N, Valladares‐Ayerbes M, et al. A randomised phase 2 study comparing different dose approaches of induction treatment of regorafenib in previously treated metastatic colorectal cancer patients (REARRANGE trial). Eur J Cancer. 2022;177:154‐163. [DOI] [PubMed] [Google Scholar]
- 22. Bekaii‐Saab TS, Ou FS, Ahn DH, et al. Regorafenib dose‐optimisation in patients with refractory metastatic colorectal cancer (ReDOS): a randomised, multicentre, open‐label, phase 2 study. Lancet Oncol. 2019;20:1070‐1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chung WH, Chang WC, Lee YS, et al. Genetic variants associated with phenytoin‐related severe cutaneous adverse reactions. JAMA. 2014;312:525‐534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2


