Abstract
Researches indicate miR-3200 is closely related to tumorigenesis, However, the role of miR-3200 in human hepatocarcinogenesis is still unclear. In this study, we clearly demonstrate that miR-3200 accelerates the growth of liver cancer cells in vivo and in vitro. Obviously, these findings are noteworthy that miR-3200 affects the transcriptional regulation for several genes, including DSP,BABAM2, Rab7A,SQSTM1,PRKAG2,CDK1,ABCE1,BECN1,PTEN,UPRT. And miR-3200 affects the transcriptional ability of several genes, such as, upregulating CADPS, DSP,FBXO32, PPCA,SGK1, PATXN7L1, PLK2,ITGB5,FZD3,HOXC8,HSPA1A,C-Myc,CyclnD1,CyclinE,PCNA and down –regulating SUV39H1, MYO1G, OLFML3, CBX5, PPDE2A, HOXA7, RAD54L, CDC45,SHMT7,MAD2L1,P27,IQGAP3,PTEN,P57,SCAMP3,etc...On the other hand, it is obvious that miR-3200 affects the translational ability of several genes, such as, upregulating GNS,UPRT,EIFAD,YOS1,SGK1,K-Ras,PKM2,C-myc,Pim1,CyclinD1,mTOR,erbB-2,CyclinE,PCNA,RRAS,ARAF,RAPH1,etc.. and down-regulating KDM2A, AATF, TMM17B, RAB8B, MYO1G,P21WAF1/Cip1,GADD45,PTEN,P27,P18,P57,SERBP1,RPL34,UFD1,Bax,ANXA6,GSK3β. Strikingly, miR-3200 affects some signaling pathway in liver cancer, including carbon metabolism signaling pathway, DNA replication pathway, FoxO signaling pathway, Hippo signaling pathway, serine and threonine metabolism signaling pathway, mTOR signaling pathway, Fatty acid biosynthesis signaling pathway, carcinogenesis-receptor activation signaling pathway, autophagy signaling pathway. Furthermore, our results suggest that miR-3200 enhances expression of RAB7A, and then Rab7A regulates the carcinogenic function of miR-3200 by increasing telomere remodeling in human liver cancer. These results are of great significance for the prevention and treatment of human liver cancer.
Keywords: miR-3200, Liver cancer cells, RAB7A, Telomere
1. Introduction
Researches indicate miR-3200 is closely related to tumorigenesis. For examples, miR-3200 promotes ferroptosis by targeting ATF4, increasing cell proliferation and metastasis [1].Moreover, miR-3200 exerts tumor-suppressive effects [2]and miR-3200 promotes cell invasion via BRMS1 suppression[3]. Also,miR-3200 promotes the development of gastric carcinoma [4]and miR-3200 regulates the proliferation and metastasis by suppressing the expression of CAMK2A [5]. In particular, knockdown of long non-coding RNA PEG10 inhibits growth, migration and invasion of gastric carcinoma cells via up-regulating miR-3200 [6], and silencing of LINC00324 inhibited the proliferation, migration, and invasion of GC cells through regulating the miR-3200-5p/BCAT1 axis [7]. However, the role of miR-3200 in human hepatocarcinogenesis are still unclear.
The research suggests RAB7A shuttles between late endosomes and mitochondria to enable mitophagy [8] and RAB7A promotes the migration of MSCs [9]. Up to now, the roles of RAB7a has not been fully elucidated in human hepatocarcinogenesis.
In this study, we clearly demonstrate that miR-3200 affects transcriptome, proteome,and some signaling pathway in liver cancer and accelerate the growth of liver cancer cells through RAB7A. These results are of great significance for the prevention and treatment of human liver cancer, so it is worth further study.
2. Materials and methods
Cell Lines, Lentivirus Human liver cancer cell line (Hep3B) was maintained in Dulbecco's modified Eagle medium(Gibco) supplemented with 10 % fetal bovine serum (Gibco) in a humidified atmosphere of 5 % CO2 incubator at 37 °C. rLV, rLV-miR-3200 were purchased from Wu Han viraltherapy Technologies Co. Ltd. pGFP-V-RS was purchased from Origene (Rockville, MD,USA).
Cell infection Pre-miR-3200 (RefSeq: MI0014249) precursor sequence (GGUGGUCGAGGGAAUCUGAGAAGGCGCACAAGGUUUGUGUCCAAUACAGUCCACACCUUGCGCUACUCAGGUCUGCUCGUGCCCU) was cloned into the lentiviral vector pLVX-ZsGreen-miRNA-Puro (pLVX-miR-3200) and prepared rLV-miR-3200 lentivirus. Cells were infected with Lentivirus according to manufacturer's instructions. Five microliters of 108 titers of lentivirus was added to a 6-well plate containing cells and the culture medium was changed 48 h later. Next, the purinomycin (0.5 μg/mL) was added in 96 h later to screen stable cell lines. the expression of green fluorescent protein was observed under a fluorescence microscope and extract total RNA to identify the expression of miR-3200.
microRNA Detection Total RNA was extracted from cultured cells with a mirVana miRNA Isolation Kit (Ambion) according to manufacturer's instructions. The real-time RT-PCR-based detection of mature miR-3200 and U6 snRNA was achieved with a mirVana miRNA Detection Kit..Each sample was run in triplicate. Ct values for mature miR-3200 were calculated and normalized to Ct values for U6snRNA.
RT-PCR Total RNA was purified using Trizol (Invitrogen) according to manufacturer's instructions followed by treatment with RNase-free DNase (QIAGEN). cDNA was prepared by using oligonucleotide (dT15-18), random primers, and a SuperScript First-Strand Synthesis System (Invitrogen). General PCR was performed in accordance with the manufacturers' protocols. β-actin was used as an internal control.
Protein extraction The logarithmically growing cells were washed twice with ice-cold phosphate-buffered saline (Hyclone) and lysed in RIPA protein lysis buffer on ice for 30 min.Next, the cells lysates were centrifuged at 14,000g for 25 min at 4 °C, and then the supernatants were separated. Protein concentration was measured using the BCA protein assay assay kit(Beyotime).
Western blotting After being boiled for 10 min in the 2 × protein denaturation solution, the samples containing proteins were separated on a 10 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose membranes (Beyotime). The blots were incubated with appropriate dilution antibody overnight at 4 °C. Following three washes, membranes were then incubated with secondary antibody at 4 °C overnight in TBST. Signals were visualized by enhanced chemiluminescence plus kit(Beyotime).
Co-immunoprecipitation(IP) Five-hundred-microliter cell lysates was used in immunoprecipitation with antibody. Western blot was performed with a another related antibody indicated in Western blotting according to the manufacturer's instructions.
Super-EMSA Cells were washed and scraped in ice-cold PBS to prepare nuclei for electrophoretic gel mobility shift assay with the use of the gel shift assay system (Beyotime) modified according to the manufacturer's instructions.
Chromatin immunoprecipitation (CHIP) assay Cells were cross-linked with 1 % (v/v) formaldehyde (Sigma) for 10 min at room temperature and stopped with 125 mM glycine for 5 min. Crossed-linked cells were washed with phosphate-buffered saline, resuspended in lysis buffer, and sonicated for 10 min. Chromatin extracts were pre-cleared with Protein-A/G-Sepharose beads, and immunoprecipitated with specific antibody on Protein-A/G-Sepharose beads. After washing, elution and de-cross-linking, the ChIP DNA was detected by PCR.
ChIP-3C assays Antibody-specific immunoprecipitated chromatin was obtained as described above for ChIP assays. Chromatin still bound to the antibody-Protein-A-Sepharose beads was digested with restriction enzyme, ligated with T4 DNA ligase, eluted, and de-cross-linked. After purification, the ChIP-3C material was detected using PCR.
Cells proliferation CCK8 Assay The cell proliferation reagent CCK8 is purchased from Beyotime and the operation according to the manufacturer instruction.
Colony-Formation Efficiency Assay Cells were plated on the six wells and DMEM containing 10%FBS was added in the three replicate. Then these cells were incubated at 37 °C in humidified incubator for 7 days. Cell colonies on the dishes were stained with 1 ml of 0. 5 % Crystal Violet for more than 1 h and the colonies were counted by MacBiophotonics Image J.
Xenograft transplantation in vivo Four-weeks male athymic Balb/C mice per group were injected with liver cancer cells at the armpit area subcutaneously. The mice were observed over 4 weeks, and then sacrificed to recover the tumors. The use of mice for this work was reviewed and approved by the institutional animal care and use committee in accordance with China national institutes of health guidelines.
Chip-Seq CHIP sequencing analysis was performed according to according to the manufacturer operation manual (Novogene Co., Ltd., Beijing Nuohe Zhiyuan Technology Co., Ltd.)
RNA sequencing analysis Total RNAs were extracted and RNA sequencing analysis was performed according to according to the manufacturer operation manual (Shanghai Majorbio Bio-pharm Technology Co.,Ltd)
Mass spectrometric analysis Total proteins were extracted and mass spectrometric analysis of enzyme hydrolyzed peptides of protein without label free was performed according to according to the manufacturer operation manual (Shanghai Majorbio Bio-pharm Technology Co.,Ltd)
Statistical analysis The significant differences between mean values obtained from at least three independent experiments. The Student's t-test was used for comparisons, with P < 0.05 considered significant.
3. Results
3.1. miR-3200 promotes the proliferation of liver cancer cells in vitro and in vivo
To study the effect of miR-3200 on liver cancer cells, rLV and rLV-miR-3200 were infected liver cancer cell Hep3B(Fig. 1A). miR-3200 was overexpressed in the rLV-miR-3200 group compared to the rLV group(Fig. 1B and C). The proliferation ability was significantly increased in the rLV-miR-3200 group compared to rLV group (24th hour: P<0.01; 48th hour: P<0.01) (Fig. 1D). The cellular colony formation ability was significantly increased in the rLV-miR-3200 group compared to rLV group (12 ± 2.78 % versus 56.27 ± 10.56 %, P = 0.000599; 20.27 ± 3.3 % vs 56.67 ± 4.3 %, P = 0.00012) (Fig. 1Ea &b,Fig. 1A&B). The weight of transplanted tumors was significantly increased in the rLV-miR-3200 group compared to rLV group (0.352 ± 0.038g vs 0.923 ± 0.047 g, P = 0.00000019) and the appearance of transplanted tumors(xenograft) was significantly decreased in the rLV-miR-3200 group compared to rLV group (13.5 ± 3.02 days vs 8.17 ± 0.753 days, P = 0.0052) (Fig. 1Fa-c). The well differentiated cells were significantly decreased in the rLV-miR-3200 group compared to rLV group and the poorly differentiated cells were significantly increased in the rLV-miR-3200 group compared to rLV group. (Fig. S1C).
Fig. 1.
miR-3200 promotes the growth of liver cancer cells in vitro. A. Hep3B cells were infected with rLV-miR-3200 and the pictures were taken under fluorescence microscope. B. The precursor of miR-3200 was detected by reverse transcription polymerase chain reaction (RT-PCR). β-actin was used as internal reference gene.C. Quantitative RT-PCR was used to detect the mature miR-3200.U6 was used as internal reference gene. The values of each group were expressed as mean ± SD (n = 3), * *, P < 0.01, and *, P < 0.05. D. CCK8 method was used to determine the cell proliferation ability. The values of each group were expressed as mean ± SD (n = 3), * *, P < 0.01, and *, P < 0.05. E.a-b. The colony forming ability of cells was measured. (right)photos of plate colonies. (left) analysis of colony forming ability of cells. The values of each group were expressed as mean ± standard deviation (bar ± SD, n = 3), * *, P < 0.01, *, P < 0.05. F. a.the xenograft tumor was dissected. b. Comparison of tumor size (g). The values of each group were expressed as mean ± SD (n = 6), * *, P < 0.01, and *, P < 0.05, respectively. c. Comparison of tumor appearance time(days). The values of each group were expressed as mean ± SD (n = 6), * *, P < 0.01, and *, P < 0.05, respectively.
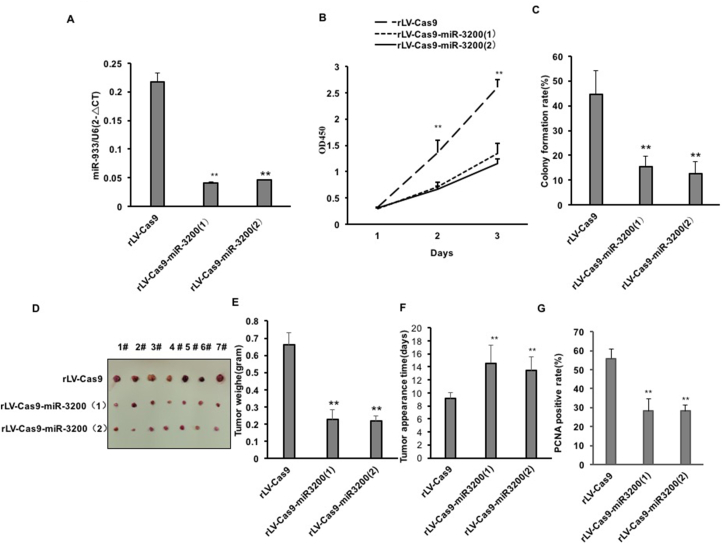
Furthermore, mature miR-3200 was decreased in the rLV-Cas9-miR-3200(1) group and rLV-Cas9-miR-3200(2) group compared to rLV-Cas9 group (Fig. 2A). The proliferation ability was significantly decreased in the rLV-Cas9-miR-3200(1)group and rLV-Cas9-miR-3200(2) group compared to rLV-Cas9 group (24th hour: P = 0.0085,0.024; 48th hour: P = 0.0016,0.0013) (Fig. 2B). The cellular colony formation ability was decreased in the rLV-Cas9-miR-3200(1)group(44.66 ± 9.66 % versus 15.32 ± 4.39 %, P = 0.00036) and rLV-Cas9-miR-3200(2) group compared to rLV-Cas9 group (44.66 ± 9.66 % versus 12.78 ± 4.86 %, P = 0.000039) (Fig. 2C,Fig. 2A). The weight of transplanted tumors(xenograft) was significantly decreased in the rLV-Cas9-miR-3200(1)group (0.66 ± 0.07g vs 0.226 ± 0.057 g, P = 0.000019) and rLV-Cas9-miR-3200(2) group compared to rLV-Cas9 group (0.66 ± 0.07g vs 0.217 ± 0.032 g, P = 0.0000006) (Fig. 2D&E). The appearance time of transplanted tumors(xenograft) was significantly increased in the rLV-Cas9-miR-3200(1)group (9.14 ± 0.89 days vs 14.57 ± 2.76 days, P = 0.00178) and rLV-Cas9-miR-3200(2) group compared to rLV-Cas9 group (09.14 ± 0.89 days vs 13.43 ± 2.15 days, P = 0.00172) (Fig. 2F). As shown in Fig. 2G&Figs. 2B–C, the well differentiated cells were significantly increased in the rLV-Cas9-miR-3200 group compared to rLV-Cas9 group and the poorly differentiated cells were significantly decreased in the rLV-Cas9-miR-3200 group compared to rLV-Cas9 group. The expression of proliferating cell nuclear antigen (PCNA) were significantly decreased in the rLV-Cas9-miR-3200(1)group(55.81 ± 4.81 % versus 28.07 ± 6.72 %, P = 0.00016) and rLV-Cas9-miR-3200(2) group compared to rLV-Cas9 group (55.81 ± 4.81 % versus 28.05 ± 3.31 %, P = 0.000023) (Fig. 2H). Collectively, these results suggest that miR-3200 accelerates the growth ability of liver cancer cells to grow in vivo and in vitro.
Fig. 2.
miR-3200 knockdown inhibits the growth of liver cancer cells in vitro and in vivo. A. Hep3B cells were infected with rLV-Cas9-miR-3200 and miR-3200 was detected by real-time reverse transcription polymerase chain reaction (RT-PCR). U6 was used as internal reference gene. B. CCK8 method was used to determine the cell proliferation ability. The values of each group were expressed as mean ± SD (n = 3), * *, P < 0.01, and *, P < 0.05. C. The colony forming ability of cells was measured. a. photos of plate colonies. b. analysis of colony forming ability of cells. The values of each group were expressed as mean ± standard deviation (bar ± SD, n = 6), * *, P < 0.01, *, P < 0.05. D. the xenograft tumor was dissected. E. Comparison of tumor size (g). F. Comparison of tumor appearance time(days). The values of each group were expressed as mean ± SD (n = 6), * *, P < 0.01, and *, P < 0.05, respectively. G. The transplanted tumor tissue sections (4 μ m) fixed in 4 % formaldehyde and embedded in paraffin were stained with anti-PCNA immunohistochemical staining.PCNA positive rate(%).
3.2. miR-3200 affects epigenetic regulation in human liver cancer cells
To explore how miR-3200 affect the epigenetic regulation of human live cancer cells Hep3B, chromatin immunoprecipitation sequencing (Chip-Seq) with anti-H3K27me3 was performed. The agarose gel electrophoresis showed that the majority of DNA fragments were 500-1000bp after ultrasonic DNA fragmentation (Fig. 3). Visualization of the reads comparison results showed the modification distribution of H3K27me3 on 23 pairs of chromosomes in rLV group and rLV-miR-3200 group (Fig. 3A). Hierarchical clustering analysis showed peak differences between the two groups (Fig. 3B). Visualization of the reads comparison results showed there is significantly difference of the modification distribution of H3K27me3 on some genes between rLV group and rLV-miR-3200 group,for example, Rab7A,SQSTM1,CDK1,CLASP2, KDM2A,CADPS, BCAR3,DSP,PDE2A,PCCA,PRKAG2,ATXN7L1,BABAM2,UPRT,CNOT9,etc..(Fig. 3C-E,Fig. 4A&B). Compared to rLV group, the laoding of H3K27me3 on DSP,KDM2A,BABAM2 promoter region were significantly increased and the laoding of H3K27me3 on ASQSTM1,PRKAG2,UPRTpromoter region were significantly decreased in rLV-miR-3200 group(Fig. 4C-H). There are differences in peak binding motif in the genomic region between rLV group and rLV-miR-3200 group,e.g. AGAGAGAG,GAGAGAGA,ACACACAC,GTCTGTGT,CACACGCA,ATATATAT,ATTCCATT,GGAGAGAG,GTGTGTGT(Fig. 5).Collectively,these results suggest that miR-3200 effects on epigenetic regulation in human liver cancer cells.
Fig. 3.
Chromatin immunoprecipitation sequencing (Chip-Seq) with anti-H3K27me3 high-throughput analysis was performed in human liver cancer cells. A. IGV browser interface (Demo): visualization of the reads of the modification distribution of H3K27m3 on 23 pairs of chromosomes in rLV group and rLV-miR-3200 group. B.Hierarchical clustering analysis. C-E.Chromatin immunoprecipitation sequencing (Chip-Seq) with anti-H3K27me3 high-throughput analysis was performed in human liver cancer cells. IGV browser interface (Demo): visualization of the reads of modification distribution of H3K27me3 on gene region.
3.3. miR-3200 effect on transcriptome in liver cancer
To study the effect of miR-3200 on the transcriptome of human liver cancer cells, the RNA sequencing was performed. First, The RNA was detected by electrophoresis in rLV group and rLV-miR-3200 group (Fig. 4A). Volcanic Map analysis (Fig. 6) and Hierarchical clustering (Heat Map) analysis (Fig. 4B) showed that 47 genes were up-regulated expression and 102 genes were down regulated expression. As shown in Fig. 4C-D&Fig. 8A, there are several up-regulated genes, including GEN,TMTC4,VANGL2,CADPS,DSP,TSC22D1,RIN2,WDR59,COL7A1, CCN1, ADAM22, SQSTM1, LAMB1,CABP1,CCN2,FBXO32,PCCA,DUSP5,EPAS1,HBP1,TXNRD1,MXI1,SGK1,RPL17-C18orf32, HOXC8, RAB7A, NDST3, RNF157, SH3PXD2A, ATXN7L1,PLK2,SLC35A1,PDCD4,THBS1,SINHCAF,ITGB5,CACHD1,TES,CNBP,HSPA1A,ASB3,FZD3,GCLM et al.. As shown in Fig. 4E-F&&Fig. 8B, there are several down-regulated genes, including TUBG2, AAAS,NUP85,GPC1,SNX1,SUV39H1,MYO1G,OLFML3,LSM5,TPM2,SEZ6,PRKAG2,CYCSP1,PLCD1,CBX5,PSMF1,BMP5,INPP5J,PIM2,SEPTIN3,CHD5,GAR1,MYBL2,WNT11,H2AZ1,HTRA2,PDE2A,POLE,HOXA7,OSTC,ESPL1,ADPGK,RACGAP1,RAD54L,ODF2,PCK2,TUBA1B,PRADC1,RRM2,BCAR3,DTL,PDE1B,MCM5,DUT,ABCF2-H2BE1, PP1H, CDC45, MX1, UBE2C, LACTB, MCM3,TUBA3C,ZNF282,STMN1,CELF2,RAB13,H2AX,NR5A2,SHMT2,RPS27L,CACNA1G,HAT1,HR,TUBG1,ADGRL1,ZHX2,PSAT1,HOXA6,CDK5,CCNA2,SLC25A11,PLXNA2,LOXL4,MAD2L1,IQGAP3,ABCF2,CDC20,GRIA4,FEN1,RPIA,PGP,AUP1,NCAPD2,PAPSS1,MOGS,HOXA9,CCT7,LMNB1,SNPH,etc.Compared to rLV group, the transcriptional ability of CADPS, DSP, FBXO32,PPCA,SGK1,PATXN7L1,PLK2,ITGB5,FZD3,HOXC8,HSPA1A,C-Myc, CyclnD1,CyclinE,PCNA were significantly increased (Fig. 8A) and the transcriptional ability of SUV39H1, MYO1G, OLFML3, CBX5, PPDE2A, HOXA7,RAD54L,CDC45,SHMT7,MAD2L1,P27,IQGAP3,PTEN,P57,SCAMP3 were significantly decreased in rLV-miR-3200 group(Fig. 8B).The differential regulated KEGG (Kyoto Encyclopedia of genes and genes) includes cell cycle signaling pathway, carbon metabolism signaling pathway, DNA replication pathway,FoxO signaling pathway,cellular senescence,Hippo signaling pathway,serine and threonine metabolism signaling pathway,mTOR signaling pathway,amino acid metabolism signaling pathway, carbohydrate metabolism signaling pathway, cell growth and death signaling pathway, etc..(Fig. 9A&B).Collectively, these results suggest that miR-3200 affects on the transcriptome of human liver cancer cells.
Fig. 4.
miR-3200 affects on the transcriptome of human liver cancer cells. A. Total RNA was extracted and detected by 1 % agarose gel electrophoresis.B. Heat map analysis (cluster) of all gene expression in the two groups. C-D. Up-regulated genes. E-F. Down-regulated genes.
3.4. miR-3200 affects proteome in liver cancer
To study the effect of miR-3200 on the proteomics of human liver cancer cells, the proteolytic peptides label free mass spectrometry anasis was performed. First, the proteins were analyzed in rLV group and rLV-miR-3200 group (Fig. 10).Volcanic Map analysis (Fig. 11) and Hierarchical clustering (Heat Map) analysis (Fig. 5A) showed that 189 genes were up-regulated expression and 326 genes were down regulated expression. As shown in Fig. 5B-C&Fig. 12A, these are several up-regulated genes, including ACTG1,PFN2,PRKAR2B,RANBP2,RALA,GNS,CLASP2,YOS1,AARSD1,FBXL20,UPRT,DPM1,EIFAD,RDH11,MYOF,ZNF326,MRPL48,ZCCHC8,DDHD,TMEM91,etc... As shown in Fig. 5D-E&Fig. 12B, these are several down-regulated genes, including HSPA2, KDM2A,ANAPC4, OSGEP,RPE, RAB8B,AATF, HINT2,CNOT9, BABAM2,NAA11,DHX16,OGFOD3,WDR3,TIMM17B,STK38,CCDC88A,OCIAD1,SCAMP3,SPTLC2,etc.. Compared to rLV group, the expression of RRAS,ARAF,CARM1,UPF3B,METTL7A,K-Ras,PKM2,C-myc,Pim1,CyclinD1,mTOR,erbB-2,CyclinE,PCNA were significantly increased (Fig. 12C) and the expression of KDM2A,SERBP1, TMM17B,RPL34,UFD1, P21WAF1/Cip1, GADD45, PTEN, P27, P18,P57,Bax,ANXA6,GSK3β,SCAMP3 were significantly decreased in rLV-miR-3200 group(Fig. 12D).The KEGG mainly involves in amino acid metabolism signaling pathway, engery metabolism signaling pathway, cell growth and death signaling pathway, cancer signaling pathway etc.. The up-regulated KEGG includes hepatocellular carcinoma signaling pathway,Fatty acid biosynthesis signaling pathway,ECM-receptor interaction signaling pathway,Tyrosine metabolism signaling pathway. The up-regulated KEGG includes Biosynthesis of amino acids signaling pathway,Glycine, serine and threonine metabolism signaling pathway,Metabolic pathways signaling pathway, fatty acid degradation signaling pathway, phagosome signaling pathway.(Fig. 13A&B). Collectively, these results suggest that miR-3200 affects the proteomics in liver cancer.
Fig. 5.
miR-3200 affects proteome in liver cancer A. Differential protein cluster Heatmap. The vertical is the clustering of samples and the horizontal is the clustering of proteins. B–C. Histogram of up-regulated proteins. D-E. Histogram of down-regulated proteins.
3.5. miR-3200 enhances the expression of Rab7A in liver cancer
Given that CHIP-seq indicates miR-3200 affects the loading of H3K27me3 on Rab7A promoter region, we will further explore whether miR-3200 affects the expression of Rab7A in liver cancer cells. The binding ability of H3K27me3 to the promoter region of Rab7A was significantly decreased in rLV-miR-3200 group compared with rLV group (Fig. 6A). The binding ability of RNAPolII to the promoter-enhancer loop of Rab7A was significantly increased in rLV-miR-3200 group compared with rLV group (Fig. 6B). The binding ability of RNAPolII to the Rab7A promoter probe was significantly increased in rLV-miR-3200 group compared with rLV group (Fig. 6C). Compared to rLV, the Rab7A promoter luciferase activity was significantly increased in rLV-miR-3200 group (11002.93 ± 1221.46 vs 87388.96 ± 9550.96, P = 0.0032) (Fig. 6D). The transcription level of Rab7A was significantly increased in rLV-miR-3200 group compared with rLV group (Fig. 6E). Therefore, the expression of Rab7A was significantly increased in rLV-miR-3200 group compared with rLV group (Fig. 6F). Taken together, these observations suggest that miR-3200 enhances the expression of Rab7A in liver cancer.
Fig. 6.
miR-3200 enhances the expression of Rab7A in liver cancer. A. Chromatin immunocoprecipitation (CHIP) analysis was performed by anti-H3K27me3. The PCR amplification was carried out by using primers designed according to the DNA of Rab7A promoter region. IgG CHIP was used as the negative control. B. 3C-Chromatin immunoprecipitation (CHIP) analysis was performed by anti-RNAPolII. C.anti-RNAPolII super-EMSA analysis. D.Rab7A promoter luciferase activity analysis. E. The transcriptional ability of Rab7A was detected by RT-PCR. β-actin was used as the internal reference gene. F. The translational ability of Rab7A was detected by Western blot. β-actin was used as the internal reference gene.
3.6. Rab7A regulates the carcinogenic function of miR-3200 by increasing telomere activity in liver cancer
Given that miR-3200 enhances the expression of Rab7A in liver cancer, we consider whether miR-3200 results in the carcinogenic function dependent on Rab7A. Rab7A was significantly increased in rLV-miR-3200 group and decreased in rLV-miR-3200+rLV-shRNA-Rab7A group compared with rLV group, respectively (Fig. 7A). miR-3200 was significantly increased in rLV-miR-3200 group and rLV-miR-3200+rLV-shRNA-Rab7A group compared with rLV group (Fig. 7B). Although the proliferation ability was significantly increased in rLV-miR-3200 group compared with rLV group (24h: P = 0.00083; 48h: P = 0.00067), it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group, respectively (24h: P = 0.1269; 48h:P = 0.13376) (Fig. 7C). Although the colony formation ability was significantly increased in rLV-miR-3200 group compared with rLV group (23.73 ± 6.07 % vs 64.77 ± 7.49 %, P = 0.00016), it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group (23.73 ± 6.07 % vs 22.29 ± 4.59 %, P = 0.7316) (Fig. 7D&Fig. 14A). Although the weight of transplanted tumors was significantly increased in the rLV-miR-3200 group compared to rLV group (0.216 ± 0.043 g vs 0.703 ± 0.072 g, P = 0.0000093), it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group (0.216 ± 0.043g vs 0.235 ± 0.077g, P = 0.316) (Fig. 7E&Fig. 14B). Although the appearance time of transplanted tumors was significantly decreased in the rLV-miR-3200 group compared to rLV group (9.83 ± 1.33 days vs 7.33 ± 0.816 days, P = 0.0088), it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group (9.83 ± 1.33 days vs 9.5 ± 2.43 days, P = 0.3476) (Fig. 7F).Although the expression of proliferating cell nuclear antigen (PCNA) were significantly increased in the rLV-miR-3200 group compared to rLV group (33.91 ± 4.64 % vs 77.73 ± 11.004 %, P = 0.000283), it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group (33.91 ± 4.64 % vs 37.19 ± 8.17 %, P = 0.272) (Figs. 14C–E). Although the expression of Rab7A,K-Ras,PKM2,C-myc, CyclinD1 were significantly increased and the expression of GADD45,PTEN,P57,P27,P18,GSK3β,SCAMP3 were significantly decreased in the rLV-miR-3200 group compared to rLV group, it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group (Fig. 15). Although the interaction between CST/AAF and POT1 were significantly decreased and the interaction between TPP1 and POT1, TPP1 and EXOI, TTP1 and TRNF2,TRF2 and SNM1B,TRF2 and RETL,TRF2 and TIN2 were significantly increased in the rLV-miR-3200 group compared to rLV group, it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group (Fig. 16). Although the interaction between TERT and Cbf5,TCAB1,Reptin, Pontin were significantly increased in the rLV-miR-3200 group compared to rLV group, it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group (Fig. 7G). Although the interaction between TERT and TERT was significantly increased and the interaction between TERT and TERRA was significantly decreased in the rLV-miR-3200 group compared to rLV group, it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group (Fig. 17).Although the telomerase activity was significantly increased in the rLV-miR-3200 group compared to rLV group(0.0159 ± 0.0025 vs 0.0795 ± 0.0077, P = 0.00000035), it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group(0.0159 ± 0.0025 vs 0.0166 ± 0.0048, P = 0.3056) (Fig. 7H). Although the telomere length was significantly increased in the rLV-miR-3200 group compared to rLV group(3.69 ± 0.55 vs 9.35 ± 0.551, P = 0.00279), it was not significantly changed in rLV-miR-3200+rLV-ShRNA-Rab7A group compared with rLV group(3.69 ± 0.55 vs 4.247 ± 0.296, P = 0.102) (Fig. 7I). Collectively, these observations suggest that Rab7A regulates the carcinogenic function of miR-3200 by increasing telomere activity in liver cancer.
Fig. 7.
Rab7A regulates the carcinogenic function of miR-3200 by increasing telomere activity in liver cancer A. Rab7A was detected with Western blot in rLV group,rLV-miR-3200 group,rLV-miR-3200+rLV-ShRNA-Rab7A group.β-actin was used as internal reference gene. B. Quantitative RT-PCR was used to detect the mature miR-3200.U6 was used as internal reference gene. The values of each group were expressed as mean ± SD (n = 3), * *, P < 0.01, and *, P < 0.05. C. CCK8 method was used to determine the cell proliferation ability. The values of each group were expressed as mean ± SD (n = 3), * *, P < 0.01, and *, P < 0.05. D. The colony forming ability of cells was measured. The analysis of colony forming ability of cells. The values of each group were expressed as mean ± standard deviation (bar ± SD, n = 6), * *, P < 0.01, *, P < 0.05. E. the xenograft tumor was dissected. b. Comparison of tumor size (g). F. Comparison of tumor appearance time(days). The values of each group were expressed as mean ± SD (n = 6), * *, P < 0.01, and *, P < 0.05, respectively. G. Co-immunoprecipitation(IP) analysis. H. telomerase activity assay. I. telomere length assay. J. The schematic diagram that miR-3200 accelerates the growth of liver cancer cells.
4. Discussion
To date, we clearly demonstrate that miR-3200 affects transcriptome, proteome,and some signaling pathways in liver cancer and accelerates the growth of liver cancer cells through RAB7A and telomere remodeling(Fig. 7J). These results are of great significance for the prevention and treatment of human liver cancer.
Obviously, these findings are noteworthy that miR-3200 affects the transcriptional regulation for several genes, including RAB7A, SQSTM1,CDK1,ABCE1,BECN1,PTEN. The study indicates p62/SQSTM1 is a stress-inducible cellular protein, which interacts with various signaling proteins to regulate a variety of cellular functions [10] and SQSTM1/p62 activates NFE2L2/NRF2 and protects mouse liver from lipotoxicity [11].Moreover, cyclin-dependent kinase 1 (CDK1) promotes phosphorylation of RAPTOR, causing dissociation of mTORC1 [12], and CDK1 was identified as a positive regulator of global translation [13].Also, ABCE1mediated the progression of N-MYC–driven and c-MYC–driven cancers [14]. Furthermore,miR-153 inhibits the resistance of lung cancer to gefitinib dependent on ABCE1 [15]. In particular, the downregulation of PTEN promotes proliferative of bladder cancer cells [16]. We speculate that these genes play an important role in the carcinogenic function of miR-3200, but further research is needed.
It is worth mentioning that miR-3200 affects the transcriptional ability of several genes, such as, GEN1,TMTC4, TSC22D1, SGK1, ITGB5,SNX1, SUV39H1, CBX5,HOXA7, RAD54L.The study indicates that the immunotherapy based on GEN1 is good for ovarian cancer [17]. Moreover, the deletion of TMTC4 activates the unfolded protein [18]. Also, RNA-binding protein MEX3D promotes cervical carcinoma tumorigenesis by destabilizing TSC22D1 (TSC22 domain family protein 1) Mrna [19]. In particular, serum and glucocorticoid-induced protein kinase 1 (SGK1), a member of the ‘AGC’ subfamily of protein kinases, inhibits autophagy-dependent apoptosis via the mTOR-Foxo3a pathway [20]. Strikingly, SNHG16 promotes neuroblastoma progression [21]. In addition, RAD54L promotes progression of bladder cancer [22]. Thus, we infer that these genes play an important role in the carcinogenic function of miR-3200, but further research is needed.
On the other hand, it is obvious that miR-3200 affects the translational ability of several genes, such as,ACTG1,PRKAR2B, EIFAD,MYOF, TMEM91,HSPA2,KDM2A, AATF, OCIAD1, SPTLC2. It has been reported that the overexpression of Actin gamma 1 (ACTG1) could promote hepatocellular carcinoma proliferation by regulating the cell cycle in HCC cells [23].Moreover, RRAD suppresses the Warburg effect by downregulating ACTG1 in hepatocellularcarcinoma [24]. Also, PRKAR2B promotes aerobic glycolysis and tumour growth in prostate cancer [25]. And myoferlin affects tumor aggressiveness [26]. In addition, HSPA2 promotes proliferation via ERK1/2 pathway in lung adenocarcinoma [27].Interestingly, RNF144A functions as a tumor suppressor in breast cancer [28] and KDM2A promotes proliferation [29]. So, we speculate that these genes play an important role in the carcinogenic function of miR-3200, but further research is needed.
In particular, our results suggest that miR-3200 enhances Rab7A in liver cancer. The study suggests that Rab7A influences AKT and PAK activity, which favors cell migration [30]. Also, Rab7A functioned as an oncogene in breast cancer [31].In particular, RAB7A phosphorylation promotes mitophagy via the PINK-PARKIN pathway [32].
Strikingly, our findings indicate miR-3200 some signaling pathway in liver cancer, including cell cycle signaling pathway, carbon metabolism signaling pathway, DNA replication pathway, FoxO signaling pathway, Hippo signaling pathway, serine and threonine metabolism signaling pathway, mTOR signaling pathway, amino acid metabolism signaling pathway, RNA degradation signaling pathway, Fatty acid biosynthesis signaling pathway, Tyrosine metabolism signaling pathway, Biosynthesis of amino acids signaling pathway, Fatty acid degradation signaling pathway, carcinogenesis-receptor activation signaling pathway, autophagy signaling pathway. The activity of cell-cycle proteins is tightly controlled by their cell-cycle-specific transcription, protein degradation, as well as by several CDK inhibitor proteins [33]. Also, arginine methylation of several proteins is prevalent in breast, lung and colon cancers and leukaemia [34].Moreover, the synthesis and consumption of pyruvate is tightly controlled and is often differentially regulated in cancer cells [35]. In particular, the increased glucose consumption can be observed with clinical tumour imaging [36]. Interestingly, a conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span [37] and the Hippo-signaling pathway is an important regulator of cellular proliferation and organ size. Moreover, Hippo pathway activity is essential for the maintenance of the differentiated hepatocyte state([38,39]. In addition, DNA mismatch repair promotes APOBEC3-mediated diffuse hypermutation in human cancers [40]. Also, PI3K-Akt-mTOR signaling pathway promotes neuronal differentiation [41]. Thus, we speculate that these signaling pathways play an important role in the carcinogenic functions of miR-3200.
In summary, miR-3200 affects transcriptome, proteome,and some signaling pathway in human liver cancer and accelerate the growth of liver cancer cells through enhancing RAB7A and telomere remodeling. These results are of great significance for the prevention and treatment of human liver cancer, so it is worth further study.
Ethics approval and consent to participate
All methods were carried out in “accordance” with the approved guidelines. All experimental protocols “were approved by” a Tongji university institutional committee. Informed consent was obtained from all subjects. The study was reviewed and approved by the China national institutional animal care and use committee.
Consent for publication
Not applicable.
Funding
This study was supported by grants from National Natural Science Fundation of China (NCSF No.82073130) and by grants from Science and Technology Commission of Shanghai Municipality Shanghai Science and Technology Plan Basic Research Field Project(20JC1411400).
Authors' contributions
Dongdong Lu conceived the study and participated in the study design, performance, coordination and manuscript writing. Shuting Song, Sijie Xie, Xinlei Liu, Shujie Li, Liyan Wang, Xiaoxue Jiang, performed the research. All authors have read and approved the final manuscripts.
CRediT authorship contribution statement
Shuting Song: Investigation. Sijie Xie: Investigation. Xinlei Liu: Investigation. Shujie Li: Investigation. Liyan Wang: Investigation. Xiaoxue Jiang: Investigation. Dongdong Lu: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgements
This study was supported by grants from National Natural Science Fundation of China (NCSF No.82073130) and by grants from Science and Technology Commission of Shanghai Municipality Shanghai Science and Technology Plan Basic Research Field Project(20JC1411400).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2023.10.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Guan L., Wang F., Wang M., Han S., Cui Z., Xi S., Xu H., Li S. Downregulation of HULC induces ferroptosis in hepatocellular carcinoma via targeting of the miR-3200-5p/ATF4 Axis. Oxid. Med. Cell. Longev. 2022 May 16;2022 doi: 10.1155/2022/9613095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y., Wei G., Zhang L., Zhou H., Wang W., Guo P., Cheng C., Ji L., Cai Q., Feng Y., Tu H. LncRNA DARS-AS1 aggravates the growth and metastasis of hepatocellular carcinoma via regulating the miR-3200-5p-Cytoskeleton associated protein 2 (CKAP2) axis. Bioengineered. 2021 Dec;12(1):8217–8232. doi: 10.1080/21655979.2021.1982272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li G., Li L., Sun Q., Wu J., Ge W., Lu G., Cai M. MicroRNA-3200-5p promotes osteosarcoma cell invasion via suppression of BRMS1. Mol. Cell. 2018 Jun;41(6):523–531. doi: 10.14348/molcells.2018.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu K., Qin X., Shao Y., Zhou Y., Ye G., Xu S. Circular RNA MTO1 suppresses tumorigenesis of gastric carcinoma by sponging miR-3200-5p and targeting PEBP1. Mol. Cell. Probes. 2020 Aug;52 doi: 10.1016/j.mcp.2020.101562. [DOI] [PubMed] [Google Scholar]
- 5.Wang H., Zeng Z., Yi R., Luo J., Chen J., Lou J. MicroRNA-3200-3p targeting CAMK2A modulates the proliferation and metastasis of glioma in vitro. Bioengineered. 2022 Mar;13(3):7785–7797. doi: 10.1080/21655979.2022.2048995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J., Chu X.Q., Zhang D., Kong D.F. Knockdown of long non-coding RNA PEG10 inhibits growth, migration and invasion of gastric carcinoma cells via up-regulating miR-3200. Neoplasma. 2018 Sep 19;65(5):769–778. doi: 10.4149/neo_2018_171204N794. [DOI] [PubMed] [Google Scholar]
- 7.Wang S., Cheng Y., Yang P., Qin G. Silencing of long noncoding RNA LINC00324 interacts with MicroRNA-3200-5p to attenuate the tumorigenesis of gastric cancer via regulating BCAT1. Gastroenterol. Res. Pract. 2020 doi: 10.1155/2020/4159298. 2020 Aug 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan B.R., Li T., Coyaud E., Laurent E.M.N., St-Germain J., Zhou Y., Kim P.K., Raught B., Brumell J.H. C5orf51 is a component of the Mon1-CCZ1 complex and controls RAB7A localization and stability during mitophagy. Autophagy. 2022 Apr;18(4):829–840. doi: 10.1080/15548627.2021.1960116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K., Du B., Zhang Y., Wu C., Wang X., Zhang X., Wang L. Vimentin-Rab7a pathway mediates the migration of MSCs and lead to therapeutic effects on ARDS. Stem Cell. Int. 2021 doi: 10.1155/2021/9992381. 2021 Jul 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao M., Liu T., You Q., Jiang Z. p62 as a therapeutic target for tumor. Eur. J. Med. Chem. 2020 May 1;193 doi: 10.1016/j.ejmech.2020.112231. [DOI] [PubMed] [Google Scholar]
- 11.Lee D.H., Park J.S., Lee Y.S., Han J., Lee D.K., Kwon S.W., Han D.H., Lee Y.H., Bae S.H. SQSTM1/p62 activates NFE2L2/NRF2 via ULK1-mediated autophagic KEAP1 degradation and protects mouse liver from lipotoxicity. Autophagy. 2020 Nov;16(11):1949–1973. doi: 10.1080/15548627.2020.1712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odle R.I., Florey O., Ktistakis N.T., Cook S.J. CDK1, the other 'master regulator' of autophagy. Trends Cell Biol. 2021 Feb;31(2):95–107. doi: 10.1016/j.tcb.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Haneke K., Schott J., Lindner D., Hollensen A.K., Damgaard C.K., Mongis C., Knop M., Palm W., Ruggieri A., Stoecklin G. CDK1 couples proliferation with protein synthesis. J. Cell Biol. 2020 Mar 2;219(3) doi: 10.1083/jcb.201906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J., Jung M., Mayoh C., Venkat P., Hannan K.M., Fletcher J.I., Kamili A., Gifford A.J., Kusnadi E.P., Pearson R.B., Hannan R.D., Haber M., Norris M.D., Somers K., Henderson M.J. Suppression of ABCE1-mediated mRNA translation limits N-MYC-Driven cancer progression. Cancer Res. 2020 Sep 1;80(17):3706–3718. doi: 10.1158/0008-5472.CAN-19-3914. [DOI] [PubMed] [Google Scholar]
- 15.Wang L., Lv X., Fu X., Su L., Yang T., Xu P. MiR-153 inhibits the resistance of lung cancer to gefitinib via modulating expression of ABCE1. Cancer Biomarkers. 2019;25(4):361–369. doi: 10.3233/CBM-190094. [DOI] [PubMed] [Google Scholar]
- 16.Man X., Piao C., Lin X., Kong C., Cui X., Jiang Y. USP13 functions as a tumor suppressor by blocking the NF-kB-mediated PTEN downregulation in human bladder cancer. J. Exp. Clin. Cancer Res. 2019 Jun 14;38(1):259. doi: 10.1186/s13046-019-1262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thaker P.H., Borys N., Fewell J., Anwer K. GEN-1 immunotherapy for the treatment of ovarian cancer. Future Oncol. 2019 Feb;15(4):421–438. doi: 10.2217/fon-2018-0423. [DOI] [PubMed] [Google Scholar]
- 18.Li J., Akil O., Rouse S.L., McLaughlin C.W., Matthews I.R., Lustig L.R., Chan D.K., Sherr E.H. Deletion of Tmtc4 activates the unfolded protein response and causes postnatal hearing loss. J. Clin. Invest. 2018 Nov 1;128(11):5150–5162. doi: 10.1172/JCI97498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Z., Chen X., Cai X., Lin H., Xu J., Cheng X. RNA-binding protein MEX3D promotes cervical carcinoma tumorigenesis by destabilizing TSC22D1 mRNA. Cell Death Dis. 2022;8(1):250. doi: 10.1038/s41420-022-01049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W., Wang X., Liu Z., Wang Y., Yin B., Yu P., Duan X., Liao Z., Chen Y., Liu C., Li X., Dai Y., Tao Z. SGK1 inhibition induces autophagy-dependent apoptosis via the mTOR-Foxo3a pathway. Br. J. Cancer. 2017 Oct 10;117(8):1139–1153. doi: 10.1038/bjc.2017.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang Y., Yu J., Zhao S., Cao X., Wang Q. HOXA7 promotes the metastasis of KRAS mutant colorectal cancer by regulating myeloid-derived suppressor cells. Cancer Cell Int. 2022 Feb 19;22(1):88. doi: 10.1186/s12935-022-02519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mun J.Y., Baek S.W., Park W.Y., Kim W.T., Kim S.K., Roh Y.G., Jeong M.S., Yang G.E., Lee J.H., Chung J.W., Choi Y.H., Chu I.S., Leem S.H. E2F1 promotes progression of bladder cancer by modulating RAD54L involved in homologous recombination repair. Int. J. Mol. Sci. 2020 Nov 27;21(23):9025. doi: 10.3390/ijms21239025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Y., Xu H., Zhang L., Zhou X., Qian X., Zhou J., Huang Y., Ge W., Wang W. RRAD suppresses the Warburg effect by downregulating ACTG1 in hepatocellular carcinoma. OncoTargets Ther. 2019;12:1691–1703. doi: 10.2147/OTT.S197844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Y., Xu H., Zhang L., Zhou X., Qian X., Zhou J., Huang Y., Ge W., Wang W. RRAD suppresses the Warburg effect by downregulating ACTG1 in hepatocellular carcinoma. OncoTargets Ther. 2019 Feb 28;12:1691–1703. doi: 10.2147/OTT.S197844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia L., Sun J., Xie S., Chi C., Zhu Y., Pan J., Dong B., Huang Y., Xia W., Sha J., Xue W. PRKAR2B-HIF-1α loop promotes aerobic glycolysis and tumour growth in prostate cancer. Cell Prolif. 2020 Nov;53(11) doi: 10.1111/cpr.12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rademaker G., Hennequière V., Brohée L., Nokin M.J., Lovinfosse P., Durieux F., Gofflot S., Bellier J., Costanza B., Herfs M., Peiffer R., Bettendorff L., Deroanne C., Thiry M., Delvenne P., Hustinx R., Bellahcène A., Castronovo V., Peulen O. Myoferlin controls mitochondrial structure and activity in pancreatic ductal adenocarcinoma, and affects tumor aggressiveness. Oncogene. 2018 Aug;37(32):4398–4412. doi: 10.1038/s41388-018-0287-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao L., Yuan X., Bao F., Lv W., He Z., Tang J., Han J., Hu J. Downregulation of HSPA2 inhibits proliferation via ERK1/2 pathway and endoplasmic reticular stress in lung adenocarcinoma. Ann. Transl. Med. 2019 Oct;7(20):540. doi: 10.21037/atm.2019.10.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y.L., Zhang Y., Li D.D., Zhang F.L., Liu H.Y., Liao X.H., Xie H.Y., Lu Q., Zhang L., Hong Q., Dong W.J., Li D.Q., Shao Z.M. RNF144A functions as a tumor suppressor in breast cancer through ubiquitin ligase activity-dependent regulation of stability and oncogenic functions of HSPA2. Cell Death Differ. 2020 Mar;27(3):1105–1118. doi: 10.1038/s41418-019-0400-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu W.H., Liang D.Y., Wang Q., Shen J., Liu Q.H., Peng Y.B. Knockdown of KDM2A inhibits proliferation associated with TGF-β expression in HEK293T cell. Mol. Cell. Biochem. 2019;456(1–2):95–104. doi: 10.1007/s11010-018-03493-5. [DOI] [PubMed] [Google Scholar]
- 30.Romano R., Calcagnile M., Margiotta A., Franci L., Chiariello M., Alifano P., Bucci C. RAB7A regulates vimentin phosphorylation through AKT and PAK. Cancers. 2021;13(9):2220. doi: 10.3390/cancers13092220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie J., Yan Y., Liu F., Kang H., Xu F., Xiao W., Wang H., Wang Y. Knockdown of Rab7a suppresses the proliferation, migration, and xenograft tumor growth of breast cancer cells. Biosci. Rep. 2019;39(2) doi: 10.1042/BSR20180480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heo J.M., Ordureau A., Swarup S., Paulo J.A., Shen K., Sabatini D.M., Harper J.W. RAB7A phosphorylation by TBK1 promotes mitophagy via the PINK-PARKIN pathway. Sci. Adv. 2018;4(11) doi: 10.1126/sciadv.aav0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherr C.J., Roberts J.M. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18(22):2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 34.Wu Q., Schapira M., Arrowsmith C.H., Barsyte-Lovejoy D. Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 2021;20(7):509–530. doi: 10.1038/s41573-021-00159-8. [DOI] [PubMed] [Google Scholar]
- 35.Olson K.A., Schell J.C., Rutter J. Pyruvate and metabolic flexibility: illuminating a path toward selective cancer therapies. Trends Biochem. Sci. 2016;41(3):219–230. doi: 10.1016/j.tibs.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 37.Lehtinen M.K., Yuan Z., Boag P.R., Yang Y., Villén J., Becker E.B., DiBacco S., de la Iglesia N., Gygi S., Blackwell T.K., Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125(5):987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 38.Yimlamai D., Christodoulou C., Galli G.G., Yanger K., Pepe-Mooney B., Gurung B., Shrestha K., Cahan P., Stanger B.Z., Camargo F.D. Hippo pathway activity influences liver cell fate. Cell. 2014;157(6):1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulkkinen H.H., Kiema M., Lappalainen J.P., Toropainen A., Beter M., Tirronen A., Holappa L., Niskanen H., Kaikkonen M.U., Ylä-Herttuala S., Laakkonen J.P. BMP6/TAZ-Hippo signaling modulates angiogenesis and endothelial cell response to VEGF. Angiogenesis. 2021;24(1):129–144. doi: 10.1007/s10456-020-09748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mas-Ponte D., Supek F. DNA mismatch repair promotes APOBEC3-mediated diffuse hypermutation in human cancers. Nat. Genet. 2020;52(9):958–968. doi: 10.1038/s41588-020-0674-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He J., Zhang N., Zhu Y., Jin R., Wu F. MSC spheroids-loaded collagen hydrogels simultaneously promote neuronal differentiation and suppress inflammatory reaction through PI3K-Akt signaling pathway. Biomaterials. 2021;265 doi: 10.1016/j.biomaterials.2020.120448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.