Abstract
9-cis-retinoic acid (9cRA) binds retinoic acid receptors (RAR) and retinoid X receptors (RXR) with nanomolar affinities, in contrast to all-trans-retinoic acid (atRA), which binds only RAR with nanomolar affinities. RXR heterodimerize with type II nuclear receptors, including RAR, to regulate a vast gene array. Despite much effort, 9cRA has not been identified as an endogenous retinoid, other than in pancreas. By revising tissue analysis methods, 9cRA quantification by liquid chromatography-tandem mass spectrometry becomes possible in all mouse tissues analyzed. 9cRA occurs in concentrations similar to or greater than atRA. Fasting increases 9cRA in white and brown adipose, brain and pancreas, while increasing atRA in white adipose, liver and pancreas. 9cRA supports FoxO1 actions in pancreas β-cells and counteracts glucose actions that lead to glucotoxicity; in part by inducing Atg7 mRNA, which encodes the key enzyme essential for autophagy. Glucose suppresses 9cRA biosynthesis in the β-cell lines 832/13 and MIN6. Glucose reduces 9cRA biosynthesis in 832/13 cells by inhibiting Rdh5 transcription, unconnected to insulin, through cAMP and Akt, and inhibiting FoxO1. Through adapting tissue specifically to fasting, 9cRA would act independent of atRA. Widespread occurrence of 9cRA in vivo, and its self-sufficient adaptation to energy status, provides new perspectives into regulation of energy balance, attenuation of insulin and glucose actions, regulation of type II nuclear receptors, and retinoid biology.
Keywords: autophagy, FoxO1, insulin, liquid chromatography, mass spectrometry, nuclear receptors, pancreas, RAR, Rdh5, retinoic acid, RXR, transcription
The parent retinoid, retinol, produces multiple metabolites, with all-trans-retinoic acid (atRA) functioning as a major autacoid (1, 2). atRA activates retinoic acid (RA) receptors (RAR), but also acts through noncanonical mechanisms (3, 4). An atRA isomer, 9-cis-retinoic acid (9cRA) binds with high affinity to both RAR and retinoid X receptors (RXR) in vitro (5). 9cRA occurring in tissue concentrations similar to atRA would verify presence of an autacoid with potentially more extensive regulatory impact than atRA. Since its identification as a high-affinity RAR and RXR ligand, however, the in vivo occurrence of 9cRA has been controversial (5, 6, 7). Multiple searches failed to demonstrate 9cRA as an endogenous tissue retinoid (8). These included liquid chromatography-tandem mass spectrometry (LC/MS/MS) assays validated for tissue analyses of RA isomers (9, 10, 11). Inability to quantify 9cRA in vivo initiated searches for an endogenous RXR ligand. Phytanic acid, docosahexaenoic acid, and several polyunsaturated fatty acids were proposed as RXR ligands, but each requires 4 to >10 μM to activate RXR, that is, several orders of magnitude less potent than 9cRA (12, 13, 14, 15). None attains in vivo concentrations as free acids capable of activating RXR, and none activate RAR. A more recent conclusion that 9-cis-13,14-dihydroretinoic acid serves as a physiological RXR ligand faces several drawbacks (16). It does not bind RAR at all, and its more active R-enantiomer has no affinity for RXR at 100 nM; at 10 μM it exerts only ∼40% of 9cRA’s binding affinity. Its affinity was estimated by a reporter assay with an RXR ligand-binding domain transfected into COS cells, which did not achieve saturation. These limitations pose substantial concern about the relevance of 9-cis-13,14-dihydroretinoic acid as an RXR ligand, especially if 9cRA were present in tissues.
The question of 9cRA as an endogenous retinoid was partially resolved by its quantitation in pancreas in concentrations similar to or higher than atRA (17, 18). Pancreas 9cRA relates inversely to serum glucose and insulin in mice, and induces glucose intolerance during glucose-tolerance tests. 9cRA in nanomolar concentrations represses glucose-stimulated insulin secretion (GSIS) in pancreatic β-cells and in mouse islets through inhibiting glucose uptake by Glut2 and glucose phosphorylation by glucokinase (Gck), posttranscriptionally. 9cRA also suppresses Pdx-1 and HNF4α mRNA transcriptionally. Pdx-1 induces Gck, whereas HNF4α induces Glut2, and both stimulate insulin release through enhancing mitochondrial metabolism (19, 20, 21). Defects in GK, Pdx-1, and HNF4 underlie maturity onset diabetes of the young (MODY1, 2, and 4). 9cRA decreases in mouse models of reduced β-cell content, such as heterozygous Akita mice and streptozotocin-treated mice. In contrast, 9cRA rises to abnormally high levels in glucose-intolerant mice with β-cell hypertrophy, including mice with diet-induced obesity, as well as ob/ob and db/db mice. These data established 9cRA as a pancreatic autacoid that modifies GSIS, and beg the question whether 9cRA occurs endogenously in other tissues.
Widespread occurrence of 9cRA in vivo would inform its therapeutic applications. As the drug alitretinoin, 9cRA exerts ameliorating effects on skin cancers, such as cutaneous T-cell lymphoma and Kaposi’s sarcoma, therapy-resistant lichen planus, and chronic hand eczema (22, 23, 24). 9cRA also has been evaluated for chemoprevention and chemotherapy, because it promotes neuroblastoma cell differentiation, inhibits growth of the human myeloid leukemia cell line HL-60 with greater potency than atRA, induces differentiation of isolated human leukemia cells, and inhibits growth of the estrogen receptor-positive breast cancer cell line MCF7 (25, 26).
Potential for new therapeutic agents has spawned synthesis and evaluation of 9cRA analogs that bind specifically with RXR, known as rexinoids (27, 28). The Food and Drug Administration has approved one of these, Targretin (bexarotene), for treating cutaneous T cell lymphoma (29, 30). Rexinoids also decrease hyperglycemia and hyperinsulinemia by improving skeletal muscle insulin sensitivity in mouse models of obesity and noninsulin–dependent diabetes (31, 32, 33, 34, 35). Although rexinoids may prove useful as insulin sensitizers and to reduce obesity, they also present with deleterious side effects. These include suppressing thyroid hormone action, increasing serum triglycerides and inducing hepatomegaly (34). The diverse actions of rexinoids likely result from RXR heterodimerization with type II nuclear receptors, including FXR, LXR, PPAR, PXR, RAR, THR, and VDR (36). Understanding locations and regulation of 9cRA in tissues should provide data useful to evaluating clinical application of rexinoids.
The present work aimed to determine whether 9cRA occurs in tissues other than pancreas, and if so, to provide insight into its presence vis-à-vis atRA, and regulation of its biosynthesis. We report widespread tissue occurrence of 9cRA in concentrations similar to atRA. 9cRA and atRA vary tissue specifically and independently in tissues with fasting versus refeeding. In β-cells, glucose reduces Rdh5-catalyzed 9cRA biosynthesis, accounting for decreased 9cRA during the fed state. Variation of 9cRA with energy status complements actions of type II nuclear receptors, which regulate genes that modify energy metabolism, among other functions.
Results
Prevention of RA isomerization
Skepticism about 9cRA as an autacoid has been fostered by failure to identify it in tissues using analyses optimized for quantifying atRA. To address this, we revised recovery and liquid chromatography (LC) protocols for quantifying RA isomers in serum and tissues by returning to tissue homogenization with methanol, rather than with saline/ethanol, and modifying LC elution to improve resolution (37). The original LC mobile phase resolved four RA isomers (Fig. 1). The revised LC protocol improved resolution (Fig. 1, B and C). The original saline/ethanol homogenization resulted in atRA isomerizing partially into 13cRA (Fig. 1D) and 9cRA isomerizing partially into atRA (9,13-di-cis-RA (9,13dcRA) and 13-cis-RA (13cRA) (Fig. 1F). In contrast, methanol-based homogenization maintained the integrity of extracted atRA and 9cRA (Fig. 1, E and G). To verify reliability of the methanol-based procedure, we added 9cRA to a pancreas homogenate, extracted the sample, and compared it to a sample without added 9cRA (Fig. 2). The methanol-based procedure recovered 9cRA without isomerization.
Figure 1.
Comparison of RA resolution and isomerization during extraction.A, structures of RA isomers. Standards of RA isomers (100 fmol each) were resolved by either (B), original LC conditions or (C), revised LC conditions. D, atRA and (F), 9cRA standards in saline/ethanol were extracted and resolved by the original LC conditions. E, atRA and (G), 9cRA standards in methanol were extracted and resolved by the revised LC conditions. Analyses were done by LC/MS/MS using multiple reaction monitoring with a triple quadrupole mass spectrometer, which first isolated the molecular ion at m/z 301 [M + H]+ in Q1 and then quantified its product ion at m/z 205 in Q3. 9cRA, 9-cis-retinoic acid; atRA, all-trans-retinoic acid; LC, liquid chromatography; LC/MS/MS, LC-tandem mass spectrometry; RA, retinoic acid.
Figure 2.
Recovery of 9cRA from pancreas without isomerization.A, one microliter of a 100 nM solution of 9cRA in methanol (100 fmol) was injected into LC/MS/MS. B, a pancreas from a male mouse was homogenized in 1.5 ml methanol. Half of the homogenate was analyzed using the revised protocol. C, ten microliters of a 100 nM solution of 9cRA was added to the remaining half of the pancreas homogenate and analyzed using the revised protocol. Recovery was 96%. 9cRA, 9-cis-retinoic acid; LC/MS/MS, LC-tandem mass spectrometry.
Full-scan mass spectrum of endogenous 9cRA
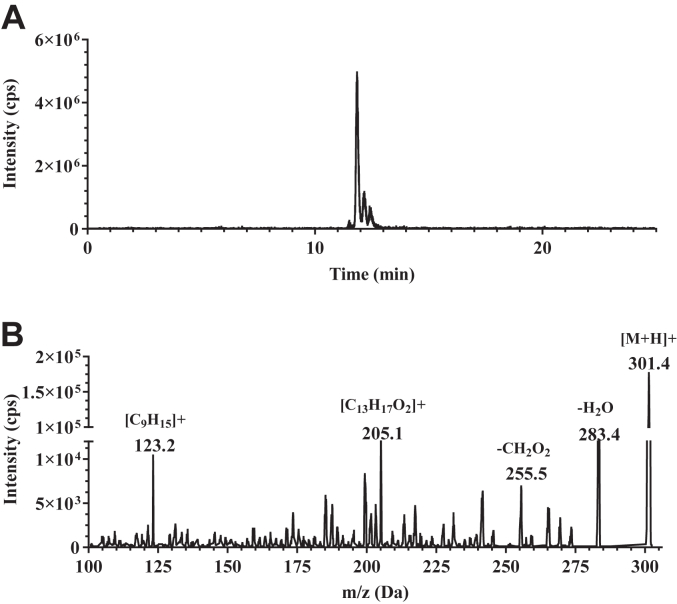
To further confirm 9cRA authenticity, 46 pooled mouse pancreata were analyzed after methanol homogenization, followed by extraction with hexane. Endogenous 9cRA was resolved from other RA isomers by high-performance LC. Reinjection of the collected 9cRA fraction for a Q3 mass scan generated a mass spectrum consistent with that of a 9cRA standard (Fig. 3) (38). The protonated molecular ion occurred at m/z 301. Characteristic product ions were present at m/z 283, 255, 205, and 123. These ions identified the molecular weight as an RA, with loss of water, loss of the carbonyl group, presence of the side-chain plus a portion of the β-ionone ring and the β-ionone ring, respectively.
Figure 3.
Mass spectrum of endogenous 9cRA. Pancreata from 46 mice were pooled, homogenized in methanol, and applied to LC. The 11.5 to 12.5 min fractions were collected, concentrated, and reinjected for a Q3 product ion scan. A, total ion chromatogram of the reinjected sample. B, Q3 mass scan of endogenous 9cRA. 9cRA, 9-cis-retinoic acid; LC, liquid chromatography.
Verification of 9cRA by derivatization with 3-NPH
Derivatization with 3-nitropenylhydrazine (3-NPH) provided another independent confirmation of 9cRA authenticity (Fig. 4A). First, we tested derivatizing atRA with 3-NPH using N-(3-dimethylaminopropryl)-N′-carbodiimide as catalyst. Mass transitions of atRA-3-NPH were determined by Q1 and Q3 scans (Fig. S1). Having validated the structure of the atRA 3-NPH derivative, six liver samples were homogenized individually with methanol and extracted with hexane. One of 40 μl was injected into the LC-atmospheric pressure chemical ionization (APCI)-MS/MS to confirm presence of 9cRA and atRA. The remaining 39 μl from each sample were pooled, concentrated, derivatized, extracted, and compared to RA 3-NPH standards. The endogenous 9cRA-3-NPH derivative comigrated with a synthetic 9cRA-3-NPH derivative and had identical mass characteristics through Q1 and Q3 (Fig. 4, B and C).
Figure 4.

Derivatization of endogenous 9cRA with 3-NPH.A, reaction of 9cRA with 3-NPH. B, chromatogram of 9,13dcRA-3-NPH, 9cRA-3-NPH and atRA-3-NPH standards (300 fmol each). C, livers from six mice were homogenized with methanol and analyzed by LC/MS/MS for comparison to authentic 3-NPH derivatives. The Y-axes shows intensity of the product ion in Q3 (m/z 283.3). 3-NPH, 3-nitropenylhydrazine; 9cRA, 9-cis-retinoic acid; LatRA, all-trans-retinoic acid; C/MS/MS, LC-tandem mass spectrometry.
Assay of tissue RA isomers
Both protocols were applied to mouse tissues to determine impact on the composition of RA isomers. Identification of RA isomers consisted of LC comigration with authentic standards, selection of the molecular ion at m/z 301 by Q1, and quantification of the characteristic product ion at m/z 205 in Q3. The ethanol protocol provided results equivalent to the original reports authenticating 9cRA in pancreas and presence of 9,13dcRA, but not 13cRA (Fig. 5A), and failing to observe 9cRA above the limits of quantification outside pancreas (17, 18). Serum and liver included atRA, relatively high concentrations of 9,13dcRA, and undetectable 13cRA, as reported originally. The brain showed a high level of 9,13dcRA, with atRA and 9cRA below limits of quantification. A screen of kidney, testis, epididymal white adipose tissue (eWAT), inguinal white adipose tissue (iWAT) and brown adipose tissue (BAT) revealed similar results. Comparatively, the methanol-based protocol revealed presence of 9cRA and atRA in the pancreas, but did not include 9,13dcRA or 13cRA (Fig. 5B). The methanol-based protocol showed 9,13dcRA only in serum, confirming a previous report (39). The revised protocol detected 9cRA in all other tissues assayed: the liver, the brain, the kidney, testes, eWAT, BAT, and iWAT, and did not show 9,13dcRA.
Figure 5.
Assay of tissue RA.A, mouse tissues were homogenized in saline/ethanol and analyzed by the original LC protocol. B, mouse tissues were homogenized in methanol and analyzed by the revised LC protocol. In both cases, detection was done by LC/MS/MS including comigration with authentic standards, selection of the molecular ion at m/z 301 in Q1, and quantification of the characteristic product ion at m/z 205 in Q3. Y-axes indicate intensity of the Q3 signal. LC, liquid chromatography; LC/MS/MS, LC-tandem mass spectrometry; RA, retinoic acid.
Fasting affects 9cRA and atRA tissue concentrations independently
9cRA concentrations exceeded atRA in the brain, the kidney, eWAT and fasted pancreas (Table 1). atRA concentrations exceeded 9cRA in the liver and testis. 9cRA and atRA occurred at comparable levels in BAT and iWAT. A 16-h fast increased 9cRA 3-fold and atRA 2-fold in pancreas and >4-fold in eWAT. 9cRA levels also were increased by fasting in the BAT (1.7-fold) and brain (2.3-fold), whereas atRA levels were not affected. Fasting decreased 9cRA ∼50% in the kidney. Fasting increased atRA ∼1.9-fold in the liver, consistent with previous reports (40, 41, 42). Fasting/refeeding did not affect atRA levels in serum, BAT, iWAT, kidney, brain, and testis.
Table 1.
Variations of 9cRA and atRA with fasting versus refeeding
| Tissue (ng/mg protein) | 9cRA |
atRA |
||
|---|---|---|---|---|
| Fasted | refed | Fasted | refed | |
| Serum (nM) | -- | -- | 2.9 ± 0.4 | 2.9 ± 0.6 |
| Pancreas | 9.7 ± 0.6 | 3.1 ± 0.4 (p < 0.02) | 6.4 ± 0.6 | 3.1 ± 0.4 (p < 0.002) |
| Liver | 4.8 ± 0.4 | 4.1 ± 0.4 | 12.3 ± 1.3 | 6.6 ± 0.6 (p < 0.003) |
| Brain | 34 ± 2.2 | 14.7 ± 1.3 (p < 0.0001) | 4.0 ± 0.5 | 4.3 ± 0.7 |
| Kidney | 6.5 ± 1.1 | 13.5 ± 1.3 (p < 0.003) | 2.2 ± 0.4 | 3.1 ± 0.4 |
| Testes | 2.8 ± 0.4 | 3.5 ± 0.5 | 7.3 ± 0.2 | 8.0 ± 0.7 |
| eWAT | 18.8 ± 1.4 | 4.3 ± 0.7 (p < 0.0001) | 11.2 ± 1.2 | 2.5 ± 0.5 (p < 0.0001) |
| BAT | 3.6 ± 0.3 | 2.1 ± 0.3 (p < 0.002) | 2.8 ± 0.5 | 3.1 ± 0.5 |
| iWAT | 7.0 ± 0.9 | 7.0 ± 1.1 | 7.5 ± 0.9 | 7.8 ± 1.5 |
Mice were fasted 16 h or fasted 16 h and refed 6 h (n = 6, means ± SE). p values refer to the differences between fasted and refed values. Serum 9cRA samples were lost.
Glucose reduces 9cRA biosynthesis
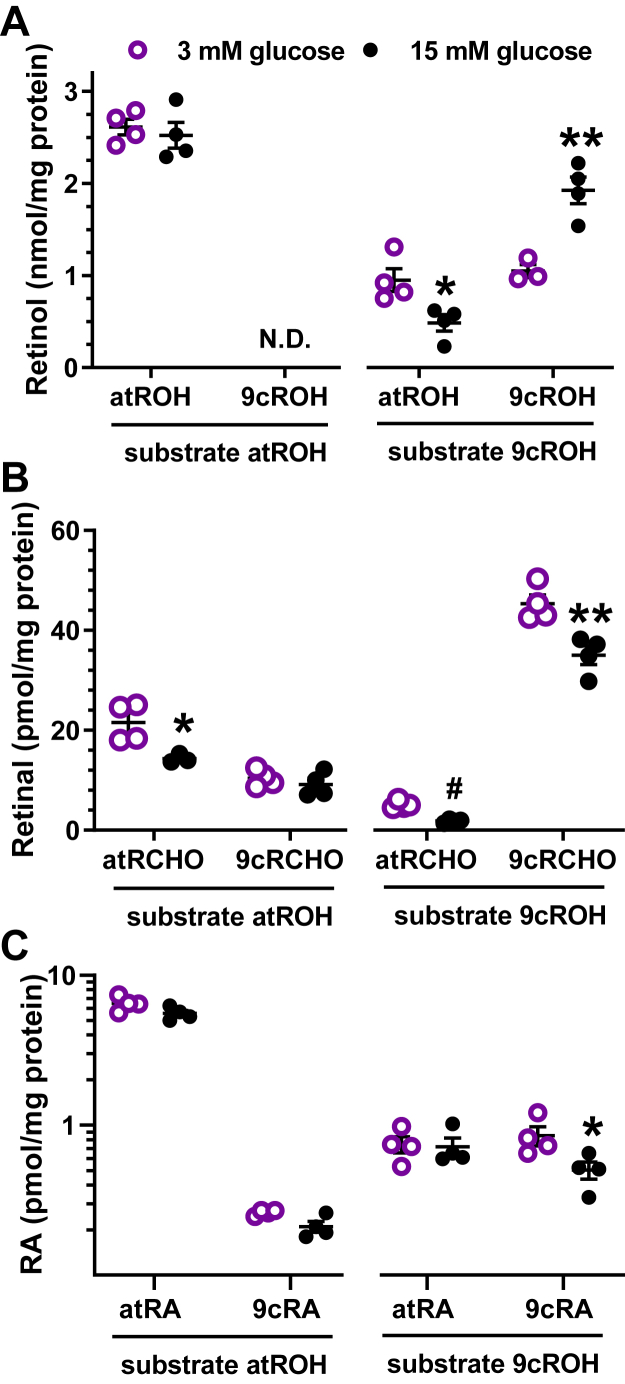
Mouse models with depleted β-cell populations indicate that β-cells account for ∼70% of pancreas 9cRA. Moreover the β-cell model 832/13 produces 9cRA from 9-cis-retinol (17). GSIS in β-cells has been investigated extensively in 832/13 cells (43, 44, 45). We used 832/13 cells to determine whether glucose regulates 9cRA biosynthesis. We quantified retinol uptake and production of retinal and RA isomers in 832/13 cells treated with 250 nM all-trans-retinol or 9-cis-retinol (Fig. 6). Exposure to all-trans-retinol increased intracellular all-trans-retinol, but did not result in the presence of 9-cis-retinol (Fig. 6A). The medium glucose concentration did not affect the amount of intracellular all-trans-retinol. Cells treated with 9-cis-retinol had both all-trans-retinol and 9-cis-retinol present. Fifteen millimolar glucose decreased the amount of all-trans-retinol by 49%, whereas it increased 9-cis-retinol by 1.8-fold compared to the 3 mM glucose-treated group. All-trans-retinol generated mostly all-trans-retinal and about half as much 9-cis-retinal (Fig. 6B). In contrast, 9-cis-retinol generated predominantly 9-cis-retinal, with minor amounts of all-trans-retinal. Retinal concentrations with 3 mM glucose exceeded those of 15 mM glucose, with exception of 9-cis-retinal during all-trans-retinol treatment. Both atRA and 9cRA were detected with all-trans-retinol substrate, with much higher atRA than 9cRA (Fig. 6C). 9-cis-retinol generated similar levels of atRA and 9cRA. Fifteen millimolar glucose decreased 9cRA biosynthesis from 9-cis-retinol ∼40%.
Figure 6.
9cRA biosynthesis in a β-cell line. 832/13 cells were FBS starved 16 h in medium with 11.1 mM glucose and then incubated 4 h with 250 nM all-trans-retinol or 9-cis-retinol and 3 mM or 15 mM glucose with continuing FBS starvation. Retinoids were quantified by LC/MS/MS. A, retinol isomers recovered. B, retinal isomers generated. C, atRA and 9cRA generated: ∗p < 0.05, ∗∗p < 0.02, #p < 0.001 versus 3 mM glucose. 9cRA, 9-cis-retinoic acid; atRA, all-trans-retinoic acid; FBS, fetal bovine serum; LC/MS/MS, LC-tandem mass spectrometry.
Glucose suppresses Rdh5 transcription
Tissues express Rdh5 widely and recombinant Rdh5 catalyzes conversion of 9-cis-retinol into 9cRA efficiently (46). Decreases in 9-cis-retinal and 9cRA with 15 mM glucose suggest that refeeding versus fasting affects Rdh5 mRNA or Rdh5 activity, as it does for Rdh1 and Rdh10 in the liver (40, 41, 42). Refeeding after a fast decreased Rdh5 mRNA in pancreas, with reductions of 40 and 67% in 4 and 6 h, respectively (Fig. 7A). Reductions after refeeding were not limited to pancreas (Fig. 7B). Six hours after refeeding Rdh5 mRNA decreased in the liver, kidney, and brown adipose tissue by 68, 93, and 92%, respectively, but increased 15% in eWAT and 80% in retinal pigment epithelium. Regulation of Rdh5 mRNA by glucose was tested in 832/13 cells (Fig. 7C). Glucose (15 mM) reduced Rdh5 mRNA ∼60% relative to 3 mM glucose after 6 h, without affecting Rdh10. We also evaluated the mouse pancreas cell line MIN6, another model of β-cell function (Fig. 7D) (47). Rdh5 mRNA in the presence of 3 mM glucose exceeded that of 15 mM glucose by 1.8-fold. Actinomycin D (ActD) prevented the increase in Rdh5 mRNA prompted by 3 mM glucose, indicating that low glucose permits activation of Rdh5 transcription (Fig. 7E). ActD did not decrease Rdh5 mRNA further than 15 mM glucose, indicating 15 mM glucose prevents Rdh5 transcription. To confirm that Rdh5 catalyzes 9cRA biosynthesis, 832/13 cells were transfected with Rdh5 (Fig. 7F). Rdh5 overexpressing cells increased Rdh5 mRNA and net conversion of 9-cis-retinol into 9cRA.
Figure 7.
Regulation of Rdh5 mRNA.A, mouse pancreas Rdh5 mRNA after 16 h fasting or after 6 h refeeding following a 16 h fast (3–5 mice per condition). B, Rdh5 expression in mouse tissues after a 16 h fast, or after 6 h refeeding following a 16 h fast. Open symbols, 16 h fast. Filled symbols, 6 h refeeding following a 16 h fast. Data show a representative experiment of 1 to 4 (5–10 mice per condition; ∗∗∗p < 0.005, ##p < 0.0001 versus fasted. BAT, brown adipose tissue; eWAT, epididymal white adipose tissue; RPE, retinal pigment epithelium. C, glucose represses Rdh5 mRNA in the 832/13 β-cell model. Cells were maintained 6 h in serum-free medium with 3 mM glucose (open symbols) or 15 mM glucose (filled symbols): ##p < 0.02 versus 3 mM glucose. D, effect of glucose on Rdh5 mRNA in MIN6 cells. Cells were incubated 16 h in serum-free medium with 3 mM or 25 mM glucose: ∗∗∗p < 0.005 versus 3 mM glucose. E, 832/13 cells were incubated overnight in serum-free medium containing 15 mM glucose to decrease Rdh5 mRNA (reference condition at 0 h); then incubated 6 h in serum-free medium containing 3 mM (open symbols) or 15 mM (filled symbols) glucose ±0.1 μg/ml actinomycin D. Data show a representative experiment of four with 4 to 5 replicates per condition: ∗p < 0.05 from 0 h group. F, 832/13 cells were transfected with 10 ng pSG5 Rdh5 or control vector and treated 24 h in serum-free medium containing 3 mM glucose, then 4 h in serum-free medium containing 3 mM glucose with 250 nM 9-cis-retinol. Rdh5 mRNA and net 9-cis-retinal were normalized to cell protein: ∗∗∗p < 0.005, ##p < 0.0001 versus control vector.
Glucose reduces 9cRA biosynthesis through Akt independent of insulin
We tested insulin to determine the possibility of negative feedback on 9cRA biosynthesis (Fig. 8A). Insulin had no effect on Rdh5 mRNA, regardless of the medium glucose concentration. We then applied pharmacological approaches to major aspects of glucose signaling to identify pathways linking glucose to Rdh5 expression. Glucose increases cAMP in β-cells and 3-isobutyl-l-methylxanthine (IBMX) increases cAMP and cGMP (48). IBMX decreased Rdh5 expression by 52 to 56% in 3 mM and 15 mM glucose, respectively (Fig. 8B). Glucagon-like peptide-1 (GLP-1) targets β-cells, among others, and increases β-cell cAMP (49). The GLP-1 analog exendin-4, however, had no effect on Rdh5 mRNA, regardless of the glucose concentration. Increased glucose via cAMP activates Crem (cAMP response element modulator), which encodes transcription factors that regulate gene expression in β-cells (50). IBMX increased Crem mRNA by 46-fold and 5-fold with 3 mM or 15 mM glucose, respectively. This result does not support a contribution of Crem-encoded proteins to Rdh5 regulation, because unlike CREM, IBMX affected Rdh5 mRNA to the same extent despite the glucose concentration. Exendin-4 had no effect on Crem mRNA regardless of the glucose concentration, excluding GLP-1 effects. Because IBMX increases both cAMP and cGMP, we tested the impact of riociguat, a guanylate cyclase activator (Fig. 8D). Riociguat suppressed Rdh5 transcription only in the presence of low glucose conditions, indicating that cGMP suppresses Rdh5 transcription only during low glucose and confirming cAMP suppresses Rdh5 transcription in the presence of high glucose. Next, we investigated signaling downstream of glucose-ATP-Ca2+. During low glucose, the FoxO1 inhibitor AS1842856 reduced Rdh5 expression by ∼50%, that is, to the same extent as high glucose (Fig. 8E). With high glucose, the FoxO1 inhibitor suppressed Rdh5 expression another 50%. These data indicate that FoxO1 serves as the proximate inducer of Rdh5. During low glucose, neither the CaMK inhibitor nor the Akt inhibitor affected Rdh5 expression. In 15 mM glucose, CaMK and Akt inhibitors prevented Rdh5 repression, indicating involvement of both in Rdh5 transcription. This again indicates a difference in mechanism between low and high glucose. Inhibitors of PKA, GSK3β, or rapamycin had no effect on high glucose-mediated Rdh5 repression (Fig. 8G). These data suggest that CaMK transmits glucose signaling through Akt and Foxo1 to Rdh5.
Figure 8.
Mechanism of glucose action. β-cells (832/13) were incubated 6 h with serum-free medium containing 3 mM or 15 mM glucose plus the additions noted. A, insulin effects on Rdh5 mRNA. Cells were incubated in the presence or absence of insulin (100 ng, n = 6 plates/group, ∗∗∗p < 0.005 versus 3 mM glucose. B, IBMX (10 μM) and exendin-4 (10 nM) effects on Rdh5 mRNA (n = 6, ##p < 0.0001 versus 3 mM glucose; xp < 0.0001 versus 3 mM glucose and IBMX. C, IBMX (10 μM) and exendin-4 (10 nM) effects on Crem mRNA (##p < 0.0001 versus 3 mM glucose; ∗∗∗p < 0.005 versus 3 mM glucose with IBMX; xp <0.0001 versus 15 mM glucose. D, riociguat (Rio) effects on Rdh5 mRNA (∗p < 0.05 versus 3 mM glucose). E, effects of FoxO1 (100 nM AS1842856), CaMK (1 μM CK59), or Akt (1 μM Triciribine hydrate) inhibitors (##p < 0.0001 versus 3 mM glucose, ∗∗∗p < 0.005 versus 15 mM glucose. F, effect of 9cRA on Atg7 mRNA (##p < 0.0001 versus 3 mM glucose). 9cRA (1 nM) increases Atg7 mRNA in 832/13 cells in the presence of 15 mM glucose (##p < 0.0001 versus 3 mM glucose). G, effects of inhibitors of mTOR (10 nM rapamycin), PKA (400 nM 14–22 amide) or GSK3β (100 nM XXV) (#p< 0.001 versus 3 mM glucose). 9cRA, 9-cis-retinoic acid; IBMX, 3-isobutyl-l-methylxanthine.
9cRA induces β-cell autophagy
Autophagy maintains β-cell mass and function (51). RAR induces autophagy by binding to a retinoic acid receptor response element in the Atg7 promoter (52, 53). High glucose decreased Atg7 mRNA relative to lower glucose (Fig. 8F). 9cRA restored Atg7 mRNA during high glucose to that of low glucose levels.
Discussion
This report establishes existence of 9cRA in multiple tissues at concentrations similar to or greater than atRA. Identification rested on multiple criteria including the following: (1) selective ion monitoring of multiple tissues relying on comigration of endogenous 9cRA with synthetic 9cRA during high-resolution LC, accompanied by selection of the molecular ion in Q1 at m/z 301 [M + H+], followed by verification of a characteristic product ion in Q3 at m/z 205; (2) a full-scan mass spectrum of endogenous 9cRA revealing the typical molecular ion and product ions after isolation by high-resolution LC; (3) derivatization and comigration of the endogenous 9cRA derivative with a synthetic 9cRA derivative during high-resolution LC. Each of these procedures alone would have authenticated the analyte as 9cRA. The three together should resolve unmistakably the presence of 9cRA in tissues. This work also resolved why 9cRA has not been observed previously in tissues other than pancreas. Apparently, a combination of ethanol/saline-based homogenization and tissue-specific matrix effects causes 9cRA to isomerize and/or to extract poorly, reducing it below limits of quantification. Methanol precipitates proteins more efficiently than the saline/ethanol mixture. Homogenization in methanol, with removal of precipitated proteins before hexane extraction, diminishes matrix effects and prevents isomerization.
atRA has been accepted as an autacoid with diverse actions, that is, an endogenous retinoid effector of diverse biological processes (54). Debate over the biological significance of atRA was resolved by demonstrating its occurrence in tissues (55) and the discovery of RAR (56, 57). 9cRA satisfies the criteria that established the significance of atRA, and therefore should experience acceptance as a retinoid-derived autacoid. The many therapeutic actions of 9cRA and rexinoids contribute to this realization (58). Dissimilar tissue-specific distributions of 9cRA and atRA, and their individual responses to fasting/refeeding cycles, indicates that they contribute independently to retinoid action, with potentially distinct activities.
Because of its catalytic activity with 9-cis-retinol, and its relatively high mRNA expression in pancreas, this work examined the possibility that Rdh5 participates in pancreas 9cRA biosynthesis. Rdh5 is better known as one of the 11-cis-retinol dehydrogenases important to the visual cycle (59, 60). Mutations in Rdh5 cause the disease fundus albipunctatus which retards dark adaptation and results in night blindness (61, 62). Rdh5, however, has equivalent catalytic efficiency (kcat/Km) with 9-cis-retinol and has widespread mRNA expression (46). The current data connects Rdh5 with the first and rate-limiting step of 9cRA biosynthesis. This does not imply that Rdh5 represents the only Rdh that participates in 9cRA biosynthesis. Rdh1 and Rdh10, for example, have activity with cis-retinols (63) Moreover, 9-cis-carotenoids occur in nature and undergo metabolism into 9-cis-retinal, thereby providing a source of cis-retinoids (64). This report also confirms the reports that a glucose challenge reduces pancreas 9cRA. In addition to the nongenomic mechanism of decreasing 9cRA, glucose reduces Rdh5 mRNA by inhibiting its transcription. Glucose, therefore, counteracts the biological functions of 9cRA by reducing its concentrations through both transcriptional and posttranscriptional mechanisms.
Chronic hyperglycemia creates glucotoxicity through raising ATP and cAMP levels, which impair GSIS and trigger β-cell dysfunction (65). cAMP induces Crem, which encodes inducible cAMP early repressor (ICER), a protein that reduces the insulin content of islets and contributes to impairing GSIS (50, 66, 67, 68). Ultimately, these and many other gene expression abnormalities set up β-cell failure and apoptosis. 9cRA counteracts glucotoxicity by posttranscriptionally reducing Glut2 and Gck activities, and transcriptionally repressing Pdx1 and HNF4α, two transcription factors that induce Glut2 and Gck expression (19, 20, 69, 70). Glucotoxicity also impairs β-cell function by preventing autophagy, a process essential for maintaining healthy β-cells (71, 72). 9cRA counteracts glucotoxicity by limiting uptake and metabolism of glucose and by inducing Atg7, the key enzyme essential for autophagy (Fig. 9).
Figure 9.
Glucose and 9cRA exert opposing actions on β-cells. The glucose transporter Glut2 enables glucose uptake by β-cells. Glucokinase (Gck) catalyzes phosphorylation of intracellular glucose, which undergoes glycolysis to generate pyruvate. Acetyl-CoA formed from pyruvate enters the mitochondria to produce ATP. The increase in ATP induces depolarization of the K+ channel, which prompts insulin release ((91)). Increased ATP also stimulates cAMP biosynthesis via adenylate cyclase (Adcy), which induces calmodulin-dependent protein kinase (CaMK). CamK activates Akt, which phosphorylates the transcription factor FoxO1, resulting in its expulsion from the nucleus. Glucose metabolism also results in the repression of the vital autophagy regulator Atg7. FoxO1 induces expression of Rdh5, which catalyzes the first and rate-limiting reaction in 9cRA biosynthesis. The current data and previous reports show that 9cRA exerts diverse actions on β-cells (17, 18). 9cRA rapidly reduces activities of Glut2 and Gck through nongenomic mechanisms. It represses transcription of Pdx1 and HNF4α. The latter two transcription factors induce transcription of Glut2 and Gck, decreasing glucose metabolism and thereby tempering GSIS. 9cRA also induces Atg7, maintaining β-cell autophagy and vigor. Even though Adcy, CaMK, and Akt affect the activities of PKA, mTOR, and GSK3β, the latter do not affect Rdh5 mRNA. 9cRA, 9-cis-retinoic acid; GSIS, glucose-stimulated insulin secretion.
The substantial concentration of 9cRA in brain relative to atRA, and its adjustment to fasting suggests a critical contribution to regulating feeding behavior and energy use. 9cRA at low nM concentrations affects expressions of multiple genes in human neural stem cells (73). Much less is known about 9cRA neural effects compared to atRA. But atRA actions could inform those of 9cRA. atRA exerts both posttranscriptional and transcriptional effects on neurite outgrowth, brain development, memory formation, and various neurological diseases, including fetal alcohol spectrum disorders, Alzheimer’s disease, Parkinson’s disease, and glaucoma (74, 75, 76, 77, 78, 79). Aberrant RAR and RXR signaling results in neuropathology, whereas atRA and rexinoids alleviate neuronal diseases (80, 81, 82). For both RAR and RXR, either defective signaling or overstimulation by pharmacological ligand concentrations cause pathology (83, 84). Therefore, it seems likely that 9cRA neurological effects would overlap with and exceed those of atRA. Future research should address these possibilities.
FoxO1 serves as the nexus between the counter effects of glucose versus 9cRA in β-cells. Insulin in liver phosphorylates Akt/Pkb, which phosphorylates FoxO1. Phosphorylated FoxO1 translocates from the nucleus to undergo cytosolic proteolysis. The absence of nuclear FoxO1 deactivates the RA biosynthetic pathway by reducing transcription of Rdh1, Rdh10, Dhrs9, Raldh1, Raldh3, and Rbp1 (85). The current work shows that insulin does not affect β-cell 9cRA biosynthesis and Rdh5 mRNA. Instead, glucose metabolism activates Akt independently of insulin to suppress FoxO1. FoxO1 has multiple beneficial effects on β-cells, such as reducing oxidative stress, regulating mitochondria-mediated metabolism, promoting autophagy, and reducing DNA damage, all key processes related to glucotoxicity (86). Activation of Rdh5 transcription by FoxO1 indicates that supporting 9cRA biosynthesis involves another mechanism whereby FoxO1 promotes β-cell fitness.
Calléja et al. (87) excluded an RA serving as a universal ligand for RXRα partnered with PPARβ/δ or RARγ in epidermal suprabasal keratinocytes, in which RARγ transcriptionally represses unknown genes. Formation of lamellar granules in these skin cells requires transcriptional activation by liganded PPARβ/δ/RXRα and transcriptional repression by unliganded RARγ/RXRα, with RARγ dominating RXRα. They concluded that an RA binding to both RARγ and RXRα would produce defective lamellar granules by activating RARγ, Clearly, both RARγ and RXRα cannot undergo activation simultaneously in this context, but neither does RXRα activation seem obligatory. Therefore, explanations other than the one proposed emerge as plausible. For example, the authors noted that retinoids do not occur in mouse keratinocytes. This reveals a physiological mechanism to promote lamellar granule formation, Cyp26s have a well-known function to catabolize RAs to protect retinoid-sensitive locations (83). This established mechanism eclipses the nonphysiological interventions that fostered the conclusion. Suppression of RAR by alternative mechanism(s) might also occur. For example, the glucocorticoid receptor represses RA-induced transcription by interacting with the RAR/RXR transcription complex (41). Finally, with respect to a role for 9cRA as a signaling molecule, it has at least 5-fold higher affinity for RAR than RXR (88). This disparity introduces the possibility that 9cRA may prioritize RAR activation, depending on the tissue concentrations of each RA. This report establishes the presence of 9cRA as an endogenous retinoid in multiple tissues, in concentrations similar to or greater than atRA. Tissue concentrations of 9cRA vary with fasting and refeeding independently of atRA. Glucose decreases 9cRA concentrations and biosynthesis in pancreatic β-cells, whereas 9cRA counterbalances glucotoxicity. These data establish 9cRA as a retinoid-derived autacoid that moderates GSIS in pancreas, promotes β-cell vigor, and likely affects energy use in general.
Experimental procedures
All aspects of these experiments were performed under yellow lights to prevent retinoid isomerization.
Materials
Optima LC/MS grade methanol, acetonitrile, water, and formic acid were purchased from Thermo Fisher Scientific. Standards of atRA, 9cRA, 13cRA, and 3-nitrophenylhydrazine hydrochloride (3-NPH·HCl) and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC·HCl) were purchased from Sigma-Aldrich. Standard 9,13-di-cis-RA was purchased from Santa Cruz Biotechnology.
Animal experiments
C57BL/6J (strain# 000664) mice were purchased from The Jackson Laboratory. Upon arrival, mice were fed a purified AIN93G diet (Research Diets, D10012G) containing 4 IU retinyl acetate/g and 7% fat and bred in-house through >3 generations. Blood, liver, pancreas, kidney, testis, epididymal and inguinal white adipose tissues (eWAT and iWAT), brown adipose tissue (BAT), and brain samples were collected from male 8∼9 weeks-old-mice, fasted 16 h and compared to those refed 6 h after a 16 h fast. Mice were euthanized in an isoflurane chamber, followed by cervical dislocation. Blood samples, drawn from the vena cava, were allowed to clot on ice and centrifuged 30 min at 12,000g and 4 °C. Sera and tissues were snap-frozen in liquid nitrogen and stored at −80 °C until assayed. Animal experimental protocols were approved by the University of California Berkeley Animal Care and Use Committee.
Saline/ethanol homogenization
Tissues (50∼150 mg) or sera (100 μl) were homogenized in ice-cold 0.9% saline using a glass pestle tissue grinder (PYREX, 77245) with a motorized homogenizer (Heidolph, RZR1) at 280 rpm to produce a 10% homogenate. The homogenate (0.5 ml) was transferred to a 23 ml round-bottom glass tube and mixed with 10 μl internal standard (100 nM 3,3-dimethyl-RA in acetonitrile) and 3 ml of 25 mM KOH, prepared in ethanol, by vortexing for 10 s. Ten milliliters hexane was added. The mixture was vortexed 20 s and centrifuged 2 min at 2000g. The upper hexane phase was discarded. One hundred eighty microliters of 4 M HCl were added to the remaining aqueous phase. RAs were extracted with 10 ml hexane by vortexing 20 s. The upper hexane phase was transferred to a new round bottom glass tube and evaporated under a gentle stream of nitrogen in a water bath (Organomation Associates, N-EVAP 112) at 25 to 30 °C. The residue was reconstituted in 40 μl acetonitrile with 20 s vortexing, spun for 10 s at 1200g, and transferred to a glass vial insert. One microliter was injected for LC-APCI-MS/MS analysis.
Methanol homogenization
Tissue samples (50∼150 mg) or sera (100 μl) were placed into a glass pestle tissue grinder with 1.5 ml of ice-cold methanol and 10 μl of the 100 nM internal standard, prepared in acetonitrile. Samples were homogenized using the motorized homogenizer at 280 rpm. The homogenate was transferred to a 15 ml glass conical tube (PYREX, 8060) and centrifuged 2 min at 1200g at room temperature. The supernatant was transferred to a clean glass round-bottom tube and mixed with 100 μl of 750 mM KOH, prepared in methanol, and vortexed 10 s. To remove hydrophobic impurities, 10 ml of hexane was added, vortexed 10 s, and spun 10 s at 1200g. The upper hexane layer was discarded. The bottom methanol phase was mixed with 180 μl 4 M HCl by vortexing 10 s and extracted with 10 ml hexane. The mixture was centrifuged 2 min at 1200g. The upper hexane layer was transferred to a new round-bottom glass tube and evaporated under a gentle stream of nitrogen with heating at 25 to 30 °C in a water bath. The residue was reconstituted in 40 μl of acetonitrile with 20 s vortexing, spun 10 s at 1200g, and transferred to a glass vial insert. One microliter was injected for LC-APCI-MS/MS analysis.
Original LC conditions
Chromatography was done with an Agilent 1290 series liquid chromatograph consisting of a binary pump, temperature-controlled column compartment, and temperature-controlled autosampler. The column compartment and autosampler were maintained at 40 °C and 10 °C, respectively. The mobile phase included two components: component A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile). The following gradient was used: 0 to 3 min, hold at 70% B; 3 to 15 min, 70% B to 95% B; 15 to 20 min, hold at 95% B; 20 to 21 min, 95% B to 70% B; 21 to 25 min, re-equilibrate at 70% B. An Ascentis RP-Amide column resolved analytes (150 × 2.1 mm, 3 μm, Supelco 565302-U) fitted with a preceding guard column (Supelguard ABZ + PLUS, 20 × 2.1 mm, 5 μm, Supelco 59605), at a flow rate of 400 μl/min.
Revised LC conditions
Resolution of RA isomers was achieved with an analytical Ascentis Express RP-Amide column (100 × 2.1 mm, 2.7 μm, Sigma-Aldrich, 53913-U) fitted with a preceding guard column at a flow rate of 500 μl/min. The following were the mobile phases used: (A) 0.1% formic acid in water/methanol (6:4); (B) 0.1% formic acid in acetonitrile/methanol (6:4). A gradient was applied over 25 min: 0 to 2 min, 40% B; 2 to 10 min, 40 to 55% B; 10 to 18 min, 55 to 95%; 18 to 21 min, holding at 95% B; 21 to 23 min, 95 to 40% B; 23 to 25 min, holding at 40% B.
Tandem mass spectrometry (MS/MS)
Quantitative data were obtained using a Sciex API-4000 triple-quadrupole mass spectrometer, equipped with an APCI source, operated in positive ion mode. The instrument was controlled by Analyst v1.4 software (https://sciex.com/products/software/analyst-software) and operated in multiple reaction–monitoring mode. RAs were monitored using a m/z 301.1 [M + H]+ to m/z 205.0 transition. The internal standard 3,3-dimethyl-RA was monitored using a m/z 329.4 [M + H]+ to m/z 151.3 transition. The dwell time for both RA and 3,3-dimethyl-RA was 150 ms. Optimum positive APCI conditions included the following: collision gas, 7 psig; curtain gas, 10 psig; gas1, 70 psig; nebulizer current, 3 μA; source temperature, 350 °C; declustering potential, 55 V; entrance potential, 10 V; collision energy, 17 eV; and collision exit potential, 5 V.
RA derivatization with 3-NPH
Standards of RA isomers (30 pmol each) were prepared in 50 μl acetonitrile. Remaining samples after the injection for RA extraction/quantification were pooled, evaporated under a gentle stream of nitrogen, and reconstituted in 50 μl acetonitrile. Twenty millimolar of 3-NPH was prepared in 40 mM HCl and ethanol (v/v 3:1). One hundred millimolar of EDC was prepared in pyridine and acetonitrile (v/v 1:1). In a 1.8 ml amber glass vial, 50 μl of sample was mixed with 100 μl 3-NPH and 75 μl EDC by vortexing 10 s, then incubated 1 h at room temperature. The reaction mixture was transferred to a 23 ml round-bottom glass tube and extracted with 10 ml hexane as described in the revised extraction section.
LC conditions for RA-3-NPH
Resolution of RA-3-NPH isomers was achieved with the analytical Ascentis Express RP-Amide column with the preceding guard column at a flow rate of 400 μl/min. The following were the mobile phases used: (A) 0.1% formic acid in water; (B) 0.1% formic acid in methanol. The following gradient was applied over 25 min: 0 to 21 min, 30 to 85% B; 21 to 22.5 min, holding at 85% B; 22.5 to 23.5 min, 85 to 40% B; 23.5 to 25 min, holding at 30% B.
MS/MS conditions for RA-3-NPH
RA-3-NPH was monitored using an m/z 436.7 [M + H]+ to m/z 283.3 transition. Positive APCI conditions included the following: collision gas, 7 psig; curtain gas, 10 psig; gas1, 70 psig; nebulizer current, 1 μA; source temperature, 350 °C; declustering potential, 55 V; entrance potential, 10 V; collision energy, 20 eV; collision exit potential, 5 V.
Quantification of retinol and retinal
Retinol isomers were measured by HPLC/ultraviolet absorbance detection (89). Retinal was quantified using a published method (90) with LC modifications for improved resolution of retinal isomers: retinal-O-ethyloxime isomers were resolved via an Acclaim C30 HPLC Column (100 × 2.1 mm, 3 μm, Thermo Fisher Scientific, 078665) with a flow rate of 400 μl/min (mobile phase A: 0.1% formic acid in water, B: 0.1% formic acid in methanol) at a gradient: 0 to 2 min, hold at 80% B; 2 to 2.5 min, 80% B to 85% B; 2.5 to 17.5 min, 85% B to 95% B; 17.5 to 20 min, hold at 95% B; 20 to 20.5 min, 95% B to 80% B; 20.5 to 25 min, re-equilibrate at 80% B at 40 °C.
Cell culture
The rat insulinoma cell line 832/13 was a gift from the Newgard lab (43). Cells were maintained in growth medium (RPMI 1640 containing 11.1 mM glucose, 10% heat-inactivated fetal bovine serum (FBS) (Gibco, 10082), 2 mM L-glutamine, 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol, 100 U/ml penicillin, 100 μg/ml streptomycin and 10 mM Hepes) under 5% CO2 in T75 flasks or 6-well plates. To minimize effects of unknown factors, cells were cultured in FBS-free growth medium containing 11.1 mM glucose 18 h prior to experiments. MIN6 cells were maintained in growth medium (Dulbecco's modified Eagle's medium containing 4.5 g/l glucose) under 5% CO2 in T75 flasks or 6-well plates.
RNA isolation and qPCR
RNA was isolated from tissues and cells with TRI reagent (Sigma-Aldrich, T9424) following manufacturer’s instructions. Complementary DNA (cDNA) was reverse-transcribed using iScript cDNA Synthesis Kit (Bio-Rad, 1708891). Real-time quantitative PCR (qPCR) was prepared with PrimeTime Gene Expression Master Mix (IDT, 1055772) and qPCR probes (mouse Actb, Mm00607939_s1; mouse Rdh5, Mm00506111_m1; rat Rdh5, Rn.PT.56a.11212766.g; rat Gusb, Rn.PT.39a.22214822.g, rat Rdh10, Rn00710727_m1; rat Crem, Rn01538528_m1) and run on CFX Connect Real Time System (Bio-Rad). Gene expression was analyzed by the ΔΔ-Ct method, normalized to Actb (mouse) or Gusb (rat), and expressed as fold-change relative to expression in reference condition.
Inhibitor assays
Inhibitors were purchased from: FoxO1, AS1842856 (EMD Millipore, 344355); GSk3β, XXV (EMD Millipore, 361568); PKA, 14 to 22 Amide (Sigma-Aldrich, 476485); Akt, triciribine hydrate (Sigma-Aldrich, T3830); mTOR, rapamycin (Sigma-Aldrich, R8781); CaMK, CK59 (Sigma-Aldrich, 208922). 832/13 cells were seeded on 6-well plates and maintained in growth medium. FBS-starved cells were treated with serum-free growth medium containing 3 mM or 15 mM glucose ± inhibitors for 6 h, followed by RNA isolation, cDNA synthesis, and qPCR.
Data presentation and statistics
Data are expressed as mean ± SEM. Statistical analyses were made by two-tailed unpaired t tests using GraphPad Prism ver. 9.5.1.
Data availability
All data are present in this manuscript.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This research was supported by grants from USA National Institutes of Health (DK112754 and DK107773). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We are grateful to Dr Yinghua Deng for developing RA derivatization with 3-NPH.
Author contributions
H. S. Y., K. O. M., and J. L. N. conceptualization; H. S. Y., K. O. M., and J. L. N. writing–original draft; H. S. Y., K. O. M., M. A. C., and W. W. investigation.
Reviewed by members of the JBC Editorial Board. Edited by Qi-Qun Tang
Footnotes
Present addresses for: Kristin Obrochta Moss, BioMarin Pharmaceutical Inc, San Rafael, California, USA, 94901; Michael A. Cockrum, 2301 Market Street, San Francisco, California 94114, USA; Wonsik Woo, 900 University Ave., 2460B Geology Building, Riverside, California 92521, USA.
Supporting information
References
- 1.Gudas L.J. Retinoid metabolism: new insights. J. Mol. Endocrinol. 2022;69:T37–T49. doi: 10.1530/JME-22-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Connor C., Varshosaz P., Moise A.R. Mechanisms of feedback regulation of vitamin A metabolism. Nutrients. 2022;14:1312. doi: 10.3390/nu14061312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petkovich M., Chambon P. Retinoic acid receptors at 35 years. J. Mol. Endocrinol. 2022;69:T13–T24. doi: 10.1530/JME-22-0097. [DOI] [PubMed] [Google Scholar]
- 4.Nhieu J., Lin Y.-L., Wei L.-N. Noncanonical retinoic acid signaling. Methods Enzymol. 2020;637:261–281. doi: 10.1016/bs.mie.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Kane M.A. Analysis, occurrence, and function of 9-cis-retinoic acid. Biochim. Biophys. Acta. 2012;1821:10–20. doi: 10.1016/j.bbalip.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Heyman R.A., Mangelsdorf D.J., Dyck J.A., Stein R.B., Eichele G., Evans R.M., et al. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 7.Levin A.A., Sturzenbecker L.J., Kazmer S., Bosakowski T., Huselton C., Allenby G., et al. 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXR alpha. Nature. 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 8.Wolf G. Is 9-cis-retinoic acid the endogenous ligand for the retinoic acid-X receptor? Nutr. Rev. 2006;64:532–538. doi: 10.1111/j.1753-4887.2006.tb00186.x. [DOI] [PubMed] [Google Scholar]
- 9.Kane M.A., Chen N., Sparks S., Napoli J.L. Quantification of endogenous retinoic acid in limited biological samples by LC/MS/MS. Biochem. J. 2005;388:363–369. doi: 10.1042/BJ20041867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kane M.A., Folias A.E., Wang C., Napoli J.L. Quantitative profiling of endogenous retinoic acid in vivo and in vitro by tandem mass spectrometry. Anal. Chem. 2008;80:1702–1708. doi: 10.1021/ac702030f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold S.L.M., Amory J.K., Walsh T.J., Isoherranen N. A sensitive and specific method for measurement of multiple retinoids in human serum with UHPLC-MS/MS. J. Lipid Res. 2012;53:587–598. doi: 10.1194/jlr.D019745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitareewan S., Burka L.T., Tomer K.B., Parker C.E., Deterding L.J., Stevens R.D., et al. Phytol metabolites are circulating dietary factors that activate the nuclear receptor RXR. Mol. Biol. Cell. 1996;7:1153–1166. doi: 10.1091/mbc.7.8.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Urquiza A.M., Liu S., Sjöberg M., Zetterström R.H., Griffiths W., Sjövall J., et al. Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science. 2000;290:2140–2144. doi: 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]
- 14.Lengqvist J., Mata De Urquiza A., Bergman A.-C., Willson T.M., Sjövall J., Perlmann T., et al. Polyunsaturated fatty acids including docosahexaenoic and arachidonic acid bind to the retinoid X receptor alpha ligand-binding domain. Mol. Cell. Proteomics. 2004;3:692–703. doi: 10.1074/mcp.M400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Calderon F., Kim H.-Y. Role of RXR in neurite outgrowth induced by docosahexaenoic acid. Prostaglandins Leukot. Essent. Fatty Acids. 2007;77:227–232. doi: 10.1016/j.plefa.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rühl R., Krzyżosiak A., Niewiadomska-Cimicka A., Rochel N., Szeles L., Vaz B., et al. 9-cis-13,14-Dihydroretinoic acid is an endogenous retinoid acting as RXR ligand in mice. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane M.A., Folias A.E., Pingitore A., Perri M., Obrochta K.M., Krois C.R., et al. Identification of 9-cis-retinoic acid as a pancreas-specific autacoid that attenuates glucose-stimulated insulin secretion. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21884–21889. doi: 10.1073/pnas.1008859107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kane M.A., Folias A.E., Pingitore A., Perri M., Krois C.R., Ryu J.Y., et al. CrbpI modulates glucose homeostasis and pancreas 9-cis-retinoic acid concentrations. Mol. Cell. Biol. 2011;31:3277–3285. doi: 10.1128/MCB.05516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H., Maechler P., Antinozzi P.A., Hagenfeldt K.A., Wollheim C.B. Hepatocyte nuclear factor 4alpha regulates the expression of pancreatic beta -cell genes implicated in glucose metabolism and nutrient-induced insulin secretion. J. Biol. Chem. 2000;275:35953–35959. doi: 10.1074/jbc.M006612200. [DOI] [PubMed] [Google Scholar]
- 20.Bartoov-Shifman R., Hertz R., Wang H., Wollheim C.B., Bar-Tana J., Walker M.D. Activation of the insulin gene promoter through a direct effect of hepatocyte nuclear factor 4 alpha. J. Biol. Chem. 2002;277:25914–25919. doi: 10.1074/jbc.M201582200. [DOI] [PubMed] [Google Scholar]
- 21.Kaneto H., Miyatsuka T., Kawamori D., Yamamoto K., Kato K., Shiraiwa T., et al. PDX-1 and MafA play a crucial role in pancreatic beta-cell differentiation and maintenance of mature beta-cell function. Endocr. J. 2008;55:235–252. doi: 10.1507/endocrj.k07e-041. [DOI] [PubMed] [Google Scholar]
- 22.Ramchatesingh B., Martínez Villarreal A., Arcuri D., Lagacé F., Setah S.A., Touma F., et al. The use of retinoids for the prevention and treatment of skin cancers: an updated review. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232012622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolios A.G.A., Marques Maggio E., Gubler C., Cozzio A., Dummer R., French L.E., et al. Oral, esophageal and cutaneous lichen ruber planus controlled with alitretinoin: case report and review of the literature. Dermatology. 2013;226:302–310. doi: 10.1159/000349980. [DOI] [PubMed] [Google Scholar]
- 24.King T., McKenna J., Alexandroff A.B. Alitretinoin for the treatment of severe chronic hand eczema. Patient Prefer. Adherence. 2014;8:1629–1634. doi: 10.2147/PPA.S38830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redfern C.P., Lovat P.E., Malcolm A.J., Pearson A.D. Gene expression and neuroblastoma cell differentiation in response to retinoic acid: differential effects of 9-cis and all-trans retinoic acid. Eur. J. Cancer. 1995;31A:486–494. doi: 10.1016/0959-8049(95)00066-r. [DOI] [PubMed] [Google Scholar]
- 26.de Almeida N.R., Conda-Sheridan M. A review of the molecular design and biological activities of RXR agonists. Med. Res. Rev. 2019;39:1372–1397. doi: 10.1002/med.21578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dawson M.I., Xia Z. The retinoid X receptors and their ligands. Biochim. Biophys. Acta. 2012;1821:21–56. doi: 10.1016/j.bbalip.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atigadda V.R., Xia G., Deshpande A., Wu L., Kedishvili N., Smith C.D., et al. Conformationally defined rexinoids and their efficacy in the prevention of mammary cancers. J. Med. Chem. 2015;58:7763–7774. doi: 10.1021/acs.jmedchem.5b00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominguez M., Alvarez S., de Lera A.R. Natural and structure-based RXR ligand Scaffolds and their functions. Curr. Top. Med. Chem. 2017;17:631–662. doi: 10.2174/1568026616666160617072521. [DOI] [PubMed] [Google Scholar]
- 30.Leal A.S., Reich L.A., Moerland J.A., Zhang D., Liby K.T. Potential therapeutic uses of rexinoids. Adv. Pharmacol. 2021;91:141–183. doi: 10.1016/bs.apha.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee R., Davies P.J., Crombie D.L., Bischoff E.D., Cesario R.M., Jow L., et al. Sensitization of diabetic and obese mice to insulin by retinoid X receptor agonists. Nature. 1997;386:407–410. doi: 10.1038/386407a0. [DOI] [PubMed] [Google Scholar]
- 32.Shen Q., Cline G.W., Shulman G.I., Leibowitz M.D., Davies P.J.A. Effects of rexinoids on glucose transport and insulin-mediated signaling in skeletal muscles of diabetic (db/db) mice. J. Biol. Chem. 2004;279:19721–19731. doi: 10.1074/jbc.M311729200. [DOI] [PubMed] [Google Scholar]
- 33.Villarroya F., Iglesias R., Giralt M. Retinoids and retinoid receptors in the control of energy balance: novel pharmacological strategies in obesity and diabetes. Curr. Med. Chem. 2004;11:795–805. doi: 10.2174/0929867043455747. [DOI] [PubMed] [Google Scholar]
- 34.Pérez E., Bourguet W., Gronemeyer H., de Lera A.R. Modulation of RXR function through ligand design. Biochim. Biophys. Acta. 2012;1821:57–69. doi: 10.1016/j.bbalip.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Ren G., Kim T., Kim H.-S., Young M.E., Muccio D.D., Atigadda V.R., et al. A small molecule, UAB126, reverses diet-induced obesity and its associated metabolic disorders. Diabetes. 2020;69:2003–2016. doi: 10.2337/db19-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefebvre P., Benomar Y., Staels B. Retinoid X receptors: common heterodimerization partners with distinct functions. Trends Endocrinol. Metab. 2010;21:676–683. doi: 10.1016/j.tem.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Napoli J.L., Pramanik B.C., Williams J.B., Dawson M.I., Hobbs P.D. Quantification of retinoic acid by gas-liquid chromatography-mass spectrometry: total versus all-trans-retinoic acid in human plasma. J. Lipid Res. 1985;26:387–392. [PubMed] [Google Scholar]
- 38.Jones J.W., Pierzchalski K., Yu J., Kane M.A. Use of fast HPLC multiple reaction monitoring cubed for endogenous retinoic acid quantification in complex matrices. Anal. Chem. 2015;87:3222–3230. doi: 10.1021/ac504597q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horst R.L., Reinhardt T.A., Goff J.P., Nonnecke B.J., Gambhir V.K., Fiorella P.D., et al. Identification of 9-cis,13-cis-retinoic acid as a major circulating retinoid in plasma. Biochemistry. 1995;34:1203–1209. doi: 10.1021/bi00004a013. [DOI] [PubMed] [Google Scholar]
- 40.Obrochta K.M., Krois C.R., Campos B., Napoli J.L. Insulin regulates retinol dehydrogenase expression and all-trans-retinoic acid biosynthesis through FoxO1. J. Biol. Chem. 2015;290:7259–7268. doi: 10.1074/jbc.M114.609313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoo H.S., Rodriguez A., You D., Lee R.A., Cockrum M.A., Grimes J.A., et al. The glucocorticoid receptor represses, whereas C/EBPβ can enhance or repress CYP26A1 transcription. iScience. 2022;25 doi: 10.1016/j.isci.2022.104564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoo H.S., Cockrum M.A., Napoli J.L. Cyp26a1 supports postnatal retinoic acid homeostasis and glucoregulatory control. J. Biol. Chem. 2023;299 doi: 10.1016/j.jbc.2023.104669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hohmeier H.E., Mulder H., Chen G., Henkel-Rieger R., Prentki M., Newgard C.B. Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes. 2000;49:424–430. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- 44.Schuit F.C., Huypens P., Heimberg H., Pipeleers D.G. Glucose sensing in pancreatic beta-cells: a model for the study of other glucose-regulated cells in gut, pancreas, and hypothalamus. Diabetes. 2001;50:1–11. doi: 10.2337/diabetes.50.1.1. [DOI] [PubMed] [Google Scholar]
- 45.Prentki M., Matschinsky F.M., Madiraju S.R.M. Metabolic signaling in fuel-induced insulin secretion. Cell Metab. 2013;18:162–185. doi: 10.1016/j.cmet.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 46.Wang J., Chai X., Eriksson U., Napoli J.L. Activity of human 11-cis-retinol dehydrogenase (Rdh5) with steroids and retinoids and expression of its mRNA in extra-ocular human tissue. Biochem. J. 1999;338:23–27. [PMC free article] [PubMed] [Google Scholar]
- 47.Ishihara H., Asano T., Tsukuda K., Katagiri H., Inukai K., Anai M., et al. Pancreatic beta cell line MIN6 exhibits characteristics of glucose metabolism and glucose-stimulated insulin secretion similar to those of normal islets. Diabetologia. 1993;36:1139–1145. doi: 10.1007/BF00401058. [DOI] [PubMed] [Google Scholar]
- 48.Holz G.G., Habener J.F. Signal transduction crosstalk in the endocrine system: pancreatic beta-cells and the glucose competence concept. Trends Biochem. Sci. 1992;17:388–393. doi: 10.1016/0968-0004(92)90006-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doyle M.E., Egan J.M. Mechanisms of action of glucagon-like peptide 1 in the pancreas. Pharmacol. Ther. 2007;113:546–593. doi: 10.1016/j.pharmthera.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y.-P., Marlen K., Palma J.F., Schweitzer A., Reilly L., Gregoire F.M., et al. Overexpression of repressive cAMP response element modulators in high glucose and fatty acid-treated rat islets. A common mechanism for glucose toxicity and lipotoxicity? J. Biol. Chem. 2003;278:51316–51323. doi: 10.1074/jbc.M307972200. [DOI] [PubMed] [Google Scholar]
- 51.Jung H.S., Chung K.W., Won Kim J., Kim J., Komatsu M., Tanaka K., et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 52.Wang S., Feng R., Shi Y., Chen D., Weng H., Ding H., et al. Intracellular alpha-fetoprotein interferes with all-trans retinoic acid induced ATG7 expression and autophagy in hepatocellular carcinoma cells. Sci. Rep. 2021;11:2146. doi: 10.1038/s41598-021-81678-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brigger D., Schläfli A.M., Garattini E., Tschan M.P. Activation of RARα induces autophagy in SKBR3 breast cancer cells and depletion of key autophagy genes enhances ATRA toxicity. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iskakova M., Karbyshev M., Piskunov A., Rochette-Egly C. Nuclear and extranuclear effects of vitamin A. Can. J. Physiol. Pharmacol. 2015;93:1065–1075. doi: 10.1139/cjpp-2014-0522. [DOI] [PubMed] [Google Scholar]
- 55.McCormick A.M., Napoli J.L. Identification of 5,6-epoxyretinoic acid as an endogenous retinol metabolite. J. Biol. Chem. 1982;257:1730–1735. [PubMed] [Google Scholar]
- 56.Giguere V., Ong E.S., Segui P., Evans R.M. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 57.Petkovich M., Brand N.J., Krust A., Chambon P. A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature. 1987;330:444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- 58.Ahuja H.S., Szanto A., Nagy L., Davies P.J.A. The retinoid X receptor and its ligands: versatile regulators of metabolic function, cell differentiation and cell death. J. Biol. Regul. Homeost. Agents. 2003;17:29–45. [PubMed] [Google Scholar]
- 59.Maeda A., Maeda T., Sun W., Zhang H., Baehr W., Palczewski K. Redundant and unique roles of retinol dehydrogenases in the mouse retina. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19565–19570. doi: 10.1073/pnas.0707477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farjo K.M., Moiseyev G., Takahashi Y., Crouch R.K., Ma J. The 11-cis-retinol dehydrogenase activity of RDH10 and its interaction with visual cycle proteins. Invest. Ophthalmol. Vis. Sci. 2009;50:5089–5097. doi: 10.1167/iovs.09-3797. [DOI] [PubMed] [Google Scholar]
- 61.Sergouniotis P.I., Sohn E.H., Li Z., McBain V.A., Wright G.A., Moore A.T., et al. Phenotypic variability in RDH5 retinopathy (fundus albipunctatus) Ophthalmology. 2011;118:1661–1670. doi: 10.1016/j.ophtha.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 62.Makiyama Y., Ooto S., Hangai M., Ogino K., Gotoh N., Oishi A., et al. Cone abnormalities in fundus albipunctatus associated with RDH5 mutations assessed using adaptive optics scanning laser ophthalmoscopy. Am. J. Ophthalmol. 2014;157:558–570.e1–4. doi: 10.1016/j.ajo.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 63.Napoli J.L. Post-natal all-trans-retinoic acid biosynthesis. Methods Enzymol. 2020;637:27–54. doi: 10.1016/bs.mie.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maeda T., Perusek L., Amengual J., Babino D., Palczewski K., von Lintig J. Dietary 9-cis-β,β-carotene fails to rescue vision in mouse models of leber congenital amaurosis. Mol. Pharmacol. 2011;80:943–952. doi: 10.1124/mol.111.074732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mukherjee N., Lin L., Contreras C.J., Templin A.T. β-Cell death in diabetes: past discoveries, present understanding, and potential future advances. Metabolites. 2021;11:796. doi: 10.3390/metabo11110796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abderrahmani A., Cheviet S., Ferdaoussi M., Coppola T., Waeber G., Regazzi R. ICER induced by hyperglycemia represses the expression of genes essential for insulin exocytosis. EMBO J. 2006;25:977–986. doi: 10.1038/sj.emboj.7601008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Favre D., Niederhauser G., Fahmi D., Plaisance V., Brajkovic S., Beeler N., et al. Role for inducible cAMP early repressor in promoting pancreatic beta cell dysfunction evoked by oxidative stress in human and rat islets. Diabetologia. 2011;54:2337–2346. doi: 10.1007/s00125-011-2165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salvi R., Abderrahmani A. Decompensation of β-cells in diabetes: when pancreatic β-cells are on ICE(R) J. Diabetes Res. 2014;2014 doi: 10.1155/2014/768024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ashizawa S., Brunicardi F.C., Wang X.-P. PDX-1 and the pancreas. Pancreas. 2004;28:109–120. doi: 10.1097/00006676-200403000-00001. [DOI] [PubMed] [Google Scholar]
- 70.Babu D.A., Deering T.G., Mirmira R.G. A feat of metabolic proportions: Pdx1 orchestrates islet development and function in the maintenance of glucose homeostasis. Mol. Genet. Metab. 2007;92:43–55. doi: 10.1016/j.ymgme.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sheng Q., Xiao X., Prasadan K., Chen C., Ming Y., Fusco J., et al. Autophagy protects pancreatic beta cell mass and function in the setting of a high-fat and high-glucose diet. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-16485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mainz L., Sarhan M.A.F.E., Roth S., Sauer U., Kalogirou C., Eckstein M., et al. Acute systemic knockdown of Atg7 is lethal and causes pancreatic destruction in shRNA transgenic mice. Autophagy. 2022;18:2880–2893. doi: 10.1080/15548627.2022.2052588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kubickova B., Martinkova S., Bohaciakova D., Nezvedova M., Liu R., Brozman O., et al. Effects of all-trans and 9-cis retinoic acid on differentiating human neural stem cells in vitro. Toxicology. 2023;487 doi: 10.1016/j.tox.2023.153461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chiang M.Y., Misner D., Kempermann G., Schikorski T., Giguère V., Sucov H.M., et al. An essential role for retinoid receptors RARbeta and RXRgamma in long-term potentiation and depression. Neuron. 1998;21:1353–1361. doi: 10.1016/s0896-6273(00)80654-6. [DOI] [PubMed] [Google Scholar]
- 75.McCaffery P., Koul O., Smith D., Napoli J.L., Chen N., Ullman M.D. Ethanol increases retinoic acid production in cerebellar astrocytes and in cerebellum. Brain Res. Dev. Brain Res. 2004;153:233–241. doi: 10.1016/j.devbrainres.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 76.Chen N., Napoli J.L. All-trans-retinoic acid stimulates translation and induces spine formation in hippocampal neurons through a membrane-associated RARalpha. FASEB J. 2008;22:236–245. doi: 10.1096/fj.07-8739com. [DOI] [PubMed] [Google Scholar]
- 77.Khillan J.S. Vitamin A/retinol and maintenance of pluripotency of stem cells. Nutrients. 2014;6:1209–1222. doi: 10.3390/nu6031209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Serio R.N., Gudas L.J. Modification of stem cell states by alcohol and acetaldehyde. Chem. Biol. Interact. 2020;316 doi: 10.1016/j.cbi.2019.108919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pouso M.R., Cairrao E. Effect of retinoic acid on the neurovascular unit: a review. Brain Res. Bull. 2022;184:34–45. doi: 10.1016/j.brainresbull.2022.03.011. [DOI] [PubMed] [Google Scholar]
- 80.Wietrzych M., Meziane H., Sutter A., Ghyselinck N., Chapman P.F., Chambon P., et al. Working memory deficits in retinoid X receptor gamma-deficient mice. Learn. Mem. 2005;12:318–326. doi: 10.1101/lm.89805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szanto A., Narkar V., Shen Q., Uray I.P., Davies P.J.A., Nagy L. Retinoid X receptors: X-ploring their (patho)physiological functions. Cell Death Differ. 2004;11:S126–S143. doi: 10.1038/sj.cdd.4401533. [DOI] [PubMed] [Google Scholar]
- 82.Kawahara K., Suenobu M., Ohtsuka H., Kuniyasu A., Sugimoto Y., Nakagomi M., et al. Cooperative therapeutic action of retinoic acid receptor and retinoid x receptor agonists in a mouse model of Alzheimer’s disease. J. Alzheimers Dis. 2014;42:587–605. doi: 10.3233/JAD-132720. [DOI] [PubMed] [Google Scholar]
- 83.McCaffery P.J., Adams J., Maden M., Rosa-Molinar E. Too much of a good thing: retinoic acid as an endogenous regulator of neural differentiation and exogenous teratogen. Eur. J. Neurosci. 2003;18:457–472. doi: 10.1046/j.1460-9568.2003.02765.x. [DOI] [PubMed] [Google Scholar]
- 84.Napoli J.L. Effects of ethanol on physiological retinoic acid levels. IUBMB Life. 2011;63:701–706. doi: 10.1002/iub.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Napoli J.L. Retinoic acid: sexually dimorphic, anti-insulin and concentration-dependent effects on energy. Nutrients. 2022;14:1553. doi: 10.3390/nu14081553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gui T., Burgering B.M.T. FOXOs: masters of the equilibrium. FEBS J. 2022;289:7918–7939. doi: 10.1111/febs.16221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Calléja C., Messaddeq N., Chapellier B., Yang H., Krezel W., Li M., et al. Genetic and pharmacological evidence that a retinoic acid cannot be the RXR-activating ligand in mouse epidermis keratinocytes. Genes Dev. 2006;20:1525–1538. doi: 10.1101/gad.368706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Allegretto E.A., McClurg M.R., Lazarchik S.B., Clemm D.L., Kerner S.A., Elgort M.G., et al. Transactivation properties of retinoic acid and retinoid X receptors in mammalian cells and yeast. Correlation with hormone binding and effects of metabolism. J. Biol. Chem. 1993;268:26625–26633. [PubMed] [Google Scholar]
- 89.Kane M.A., Folias A.E., Napoli J.L. HPLC/UV quantitation of retinal, retinol, and retinyl esters in serum and tissues. Anal. Biochem. 2008;378:71–79. doi: 10.1016/j.ab.2008.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang J., Yoo H.S., Obrochta K.M., Huang P., Napoli J.L. Quantitation of retinaldehyde in small biological samples using ultrahigh-performance liquid chromatography tandem mass spectrometry. Anal. Biochem. 2015;484:162–168. doi: 10.1016/j.ab.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Komatsu M., Takei M., Ishii H., Sato Y. Glucose-stimulated insulin secretion: a newer perspective. J. Diabetes Investig. 2013;4:511–516. doi: 10.1111/jdi.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are present in this manuscript.