Abstract
Mutations and gene expression are the two most studied genomic features in cancer research. In the last decade, the combined advances in genomic technology and computational algorithms have broadened mutation research with the concept of mutation density and expanded the traditional scope of protein-coding RNA to noncoding RNAs. However, mutation density analysis had yet to be integrated with non-coding RNAs. In this study, we examined long non-coding RNA (lncRNA) mutation density patterns of 57 unique cancer types using 80 cancer cohorts. Our analysis revealed that lncRNAs exhibit mutation density patterns reminiscent to those of protein-coding mRNAs. These patterns include mutation peak and dip around transcription start sites of lncRNA. In many cohorts, these patterns justified statistically significant transcription strand bias, and the transcription strand bias was shared between lncRNAs and mRNAs. We further quantified transcription strand biases with a Log Odds Ratio metric and showed that some of these biases are associated with patient prognosis. The prognostic effect may be exerted due to strong Transcription-coupled repair mechanisms associated with the individual patient. For the first time, our study combined mutational density patterns with lncRNA mutations, and the results demonstrated remarkably comparable patterns between protein-coding mRNA and lncRNA, further illustrating lncRNA’s potential roles in cancer research.
Keywords: Long Noncoding RNA, Mutation Density, Transcription Start Site, Mutation Strand Bias
Graphical Abstract
1. Introduction
Cancers are a major group of diseases characterized by the uncontrolled growth and spread of abnormal cells. They remain a major cause of death globally, with 19.3 million new cases reported in 2020, leading to over 10.0 million deaths worldwide [1]. Generally, cancers are caused by alterations in the genetic material, principally oncogenes, tumor-suppressor genes, and microRNA genes, which generally take the form of gene expression dysregulation or somatic mutations to DNA sequences that occur during cell division [2]. Early detection and diagnosis are key to successful treatment and improved outcomes for cancer patients.
Mutations in cancer have been studied extensively in recent years to facilitate understanding their development and potential treatments. One area of research that has received less attention, however, is the role of mutational density patterns in evaluating the role of mutations and their significance. Mutational density patterns in cancer refer to a volume of mutations that occur in a given region of the genome, which can be used to identify oncogenes and understand the underlying mechanisms of carcinogenesis [3]. Such patterns can also be used to predict the prognosis of a cancer patient and to identify potential therapeutic targets. This research has implications for understanding the underlying causes of cancer, as well as for developing new diagnostic and treatment strategies.
In addition to somatic mutation, another major cause of cancer is gene expression dysregulation. The majority of previous cancer gene expression studies have focused on protein-coding genes. However, over the last decade, more evidence has shown the relevance of non-coding RNAs. Long non-coding RNAs (lncRNAs) are a class of RNA molecules that are transcribed from the genome but do not encode proteins. LncRNAs have become the focus of intense research due to their potential role in the development and progression of cancer. One of the key functions of lncRNAs in cancer is their ability to act as molecular scaffolds, binding to and regulating the activity of specific proteins and DNA sequences[4]. Studies have shown that lncRNAs are involved in a variety of biological processes, including regulation of gene expression [5], chromatin organization, and DNA methylation [6]. Through these regulatory functions, lncRNAs exert their oncogenic or tumor suppressor functions [7]. Moreover, studies have further shown that lncRNA expression levels are often altered in cancer cells compared to normal cells. In addition, lncRNAs have been shown to play a role in regulating the activity of the immune system, which can impact the ability of the body to respond to cancer cells [8].
Previous work has found particular mutational density biases between coding and template RNA strands around the DNA replication origin regions and transcription start sites (TSSs) [3]. These findings indicate that mutational density patterns are potentially indicative of tumorigenesis history. The discernment of distinct mutation density patterns typically implies the presence of either known or unknown underlying biological mechanisms. Consequently, this encourages researchers to delve deeper into elucidating the relationship between the mutation density pattern and the associated biological mechanism. Given recent evidence demonstrating the relevance of non-coding RNAs to cancer, we hypothesized that essential lncRNAs exhibit similar mutational density strand biases as observed in protein-coding RNA. Such mutational density strand biases further evidence the cancer relevance of lncRNA.
2. Methods
2.1. Data collection
The International Cancer Genome Consortium (ICGC) (https://dcc.icgc.org/repositories) assembled mutation data from 81 global cancer cohorts. Among the 81 cancer cohorts, the French cohort of Liver Hepatocellular Macronodules (LIHM-FR) contributed the scarcest data by recruiting only four cancer patients and calling only 103 mutations in total. One mutation category, C>G (G>C, equivalently), found zero count in LIHM-FR. Because such a low mutation volume did not permit quantification of mutation density in sufficiently sized genomic regions, we excluded LIHM-FR at the beginning of our analysis workflow.
Our workflow is applicable to only single base substitutions (SBSs). Hence, SBS mutation data for cancer patients of 80 ICGC cohorts covering 57 unique cancer types were taken as the raw data. By design, multiple ICGC cohorts for a same cancer type originate from different distinct nations or territories, which usually feature distinct race compositions. For instance, cohorts from countries like China, Japan, or Korea exclusively comprise Asian patients, while cohorts from Western nations typically exhibit racial diversity, with Caucasians being the predominant group. Nevertheless, ICGC does not release detailed race information. Even if such information were available, variations in culture and dietary factors within the same cancer type across different geographic regions would likely introduce biases. Hence, we did not attempt to combine distinct cohorts toward a same cancer type, but instead conducted parallel workflows for each cohort individually.
Genomic coordinates of lncRNA transcripts were taken from LNCipedia (v5.2) [9]. The lncRNA coordinate data file downloaded from LNCipedia (v5.2) comprised 127,432 raw lncRNAs, presenting an extremely biased gene body length distribution. The longest lncRNA entity, identified as “lnc-MAT2B-3:28″, had a length of 1787,073 bp, whereas the median length was only 4.6 kb. Exceptionally long gene bodies of lncRNAs raise suspicion, and the corresponding boundaries are subject to future calibration. To prevent from unnecessary dilution of potential transcription strand bias due to inflated gene bodies, we excluded the top 5% raw lncRNAs of the longest lengths before merging overlapping transcripts. These pre-processing steps led to a set of 53,248 technically defined, mutually exclusive lncRNA gene bodies. The TSS of each lncRNA gene was extracted as the terminal position on either the 5′-end (gene on the forward strand) or the 3′-end (gene on the reverse strand). Accordingly, the transcription end site of each lncRNA gene was taken as the other terminal position of the gene body region. All data curation and analysis steps were done using the statistical analysis software R.
2.2. Mutation density pattern and transcription strand bias
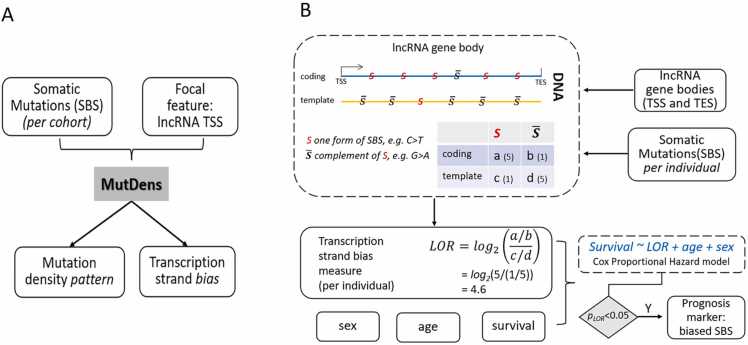
Due to the complementary property of DNA, the 12 possible single nucleotide mutations are classified into six mutational categories as C>A (C>A & G>T), C>G (C>G & G>C), C>T (C>T & G>A), T > A (T > A & A>T), T > C (T > C & A>G), and T > G (T > G & A>C). Of note, each mutational category consists of two mutually complementary forms, such as the pair of C>T and G>A. Mutation density analysis requires predefined focal genomic features. In a previous study, we demonstrated the transcription strand bias in the vicinity of the TSS of protein-coding RNAs [3]. In this study, our focal genomic feature is lncRNA TSS. To analyze the spatial patterns of mutation density in the vicinity of focal genomic features, we counted mutations in the immediate flanking regions which include 2000 nucleotides one either upstream or downstream flank. The bidirectional flanks were evenly divided into a total of 40 bins with a length of 100 nucleotides each. The mutations (distinct genomic positions) called from a cancer cohort were counted within each sequential 100-bp bin relative to all instances of lncRNA TSS. The mutations within each bin were further normalized by considering the G/C relative to A/T ratio in GRCh38, giving rise to the mutation density assessed in “Mutations Per Kilo total mutations per Megabase” (MPKM). The mutation densities of the two complementary strands were plotted using R for visualization. This section of analyses leveraged our previously developed R application MutDens [3].
Two primary statistical analyses were offered by MutDens and were conducted in the present study in regards to lncRNA TSS (Fig. 1A). First, we tried to detect whether a mutation peak or dip exists in the vicinity of lncRNA TSS. A peak denotes an visible sharp mutation density increase which can be observed centering the genomic feature or residing left (PeakL) or right (PeakR) to the genomic feature. A dip denotes a visible sharp mutation density decrease. To statistically detect peak or dip, a background mutation density in Poisson distribution was established using mutations far away from the focal genomic features. The mutation density of lncRNA for each ICGC cohort was compared to the background distribution to detect peaks or dips in mutation density. Nominal p < 1 × 10−5 out of the Poisson test was adopted to affirm a mutation density peak or dip. Next, we tested whether a strand bias exists using Wilcoxon Signed-Rank test. While MutDens outputted Wilcoxon Signed-Rank test results for three different sections (TSS upstream flank, TSS downstream flank, and bidirectional flanks), we only considered the TSS downstream test result to seek potential transcription strand bias with False Discovery Rate controlled at 0.2 (Benjamini-Horchberg adjustment).
Fig. 1.
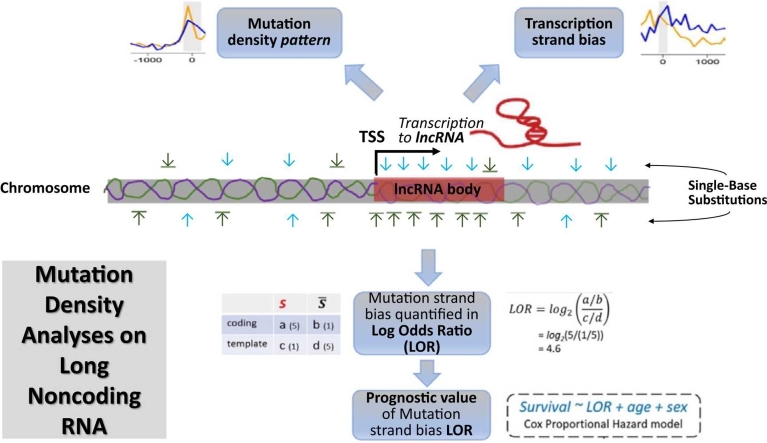
Workflow illustration. A) at the cohort level, somatic single base substitutions (SBSs) of one same mutational category were supplied to the published R application MutDens to enable analyses of mutation density patterns around lncRNA Transcription Start Sites (TSSs). B) at the individual subject level, somatic single base substitutions (SBSs) of one same mutational category were integrated with lncRNA gene body coordinates to calculate Log Odds Ratio (LOR) statistics as a per-individual transcription strand bias quantity; next, at the cohort level, a Cox model was built on LOR, age, and sex to fit overall survival of all patients. Of note, notation in the illustration corresponds to SBS′ in Eq. 1.
2.3. Prognostic bias strength of single base substitution
Beyond application of MutDens functionalities, we also employed a Cox Proportional Hazard model to assess prognostic value of single base substitution transcription bias (Fig. 1B). The transcriptional strand bias is visible at cohort level. However, in order to utilize it for survival analysis, quantification of transcriptional strand bias at individual level is needed. At individual level, the quantity of transcriptional strand bias will show a great variation due to tumor heterogeneity, genetic background differences etc. To quantify the prominence of transcription strand bias in each individual, we devised a metric that was inspired by Log Odds Ratio (LOR). In light of the coordinates of TSSs and transcription end sites of lncRNAs (details above), the genome-wide SBSs were reduced to those occurring in lncRNA gene bodies. Based on the strandedness of each lncRNA gene, a mutation reported at a particular genomic position was counted towards one of four groups: the defining SBS on the coding strand (SBScoding), the complementary SBS (i.e., SBS′) on the coding strand (SBS′coding), the defining SBS on the template strand (SBS′template), and the complementary SBS on the template strand (SBS′template). In the literature, researchers typically calculate the ratio of two paired mutation forms (e.g. C>A & G>T) for coding strands and template strands separately and further display a marked disparity between the two ratios. Hence, we captured the prominence of transcription strand bias with a LOR metric (Eq. 1). Given the proposed mechanistic explanations for transcription strand biases in lung and liver cancers [10], strong biases can be reflected as extreme LOR values in either the positive or the negative direction. A systemic bias of LOR to large positive values implies the coding strand favors one form of mutation (SBS), whereas a systemic bias of LOR to the negative direction implies the coding strand favors the complementary form of mutation (SBS′). Of note, a LOR should be calculated only when the mutations of an SBS category exceeds a minimum number, which was set at 50 in this work.
| (1) |
| (2) |
In cases where a given cancer cohort had 100 or more patients with information for both the LOR measure and overall survival, the LOR measure was built into a multivariate Cox proportional hazard survival model, along with the age and the sex of each individual (Eq. 2). The Cox models were built and resolved for all combinations of cancer types and mutation categories. In the case of Uterine Corpus Endometrial Carcinoma (UCEC), the sex variable was dropped because there were only female patients. Since up to six SBS types were analyzed separately for each cancer cohort, we performed Benjamini Hochberg multiple test adjustment within each cancer cohort. Within each cancer cohort, if nominal p < 0.05 and False Discovery Rate was less than 0.2, we reported the variable LOR for the specific SBS as a statistically significant prognostic marker. To visualize the prognostic value of LOR, we calculated a fitted hazard rate for each individual as the linear combination of the three variables (LOR, age, and sex) using the corresponding coefficients resolved from the Cox model (Eq. 2). Patients were evenly partitioned to a high risk group and a low risk group based on the linearly combined hazard rate, and Kaplan-Meier survival curves were plotted for the two groups respectively.
3. Results
3.1. Overall study design
Our workflow was divided into two components, purported for detecting mutation density patterns and prognostic markers respectively (Fig. 1). The detailed cancer cohort, type, and sample size information can be seen in Supplementary Table 1.
The same mutation density analysis workflow (Fig. 1A, via MutDens) was repeatedly applied to investigate 80 cancer cohorts of 57 cancer types. This component is considered a cohort-level analysis due to three facts. First, SBS mutations of all individuals were merged and distinct SBS sites were compiled into a wholesome mutation dataset for the cohort. Second, the MPKM values and mutation density values in vicinity of lncRNA TSSs were based on the cohort-specific mutation dataset, so their interpretation must be made with respect to the complete cohort, not a cancer patient. Third, if a significant trend of transcription strand bias is ascertained, it suggests that a cancer type tends to show transcription strand bias for a SBS category, but it can happen that a specific cancer patient does not demonstrate transcription strand bias. Previous studies have accrued transcription strand bias findings at the cohort level, affirming C>T bias in melanoma, G>T bias in lung cancer, etc. Our exhaustive screening in 57 cancer types for all six SBS categories provides an opportunity to detect potential trends of transcription strand bias in diverse cancer types, which may shed light on the tumorigenesis mechanisms in under-investigated cancer types and catalyze the concomitant diagnosis and therapeutic strategies.
As for the Cox survival analyses (Fig. 1B) that required a minimum of 100 patients with concurrent mutation and survival data, 20 cancer cohorts met the sample size requirement and they were analyzed in altogether 86 cancer-SBS scenarios. As we expounded in Section 2.3, this component is considered an individual-level analysis, because the quantitative bias metric, LOR, was assessed for each cancer patient separately. The Cox prognostic model was built and solved at the cohort level, though. If a Cox model sees statistically significant contribution from the bias variable (LOR), the ultimate prognostic prediction can be made for a specific cancer patient based on his or her LOR value.
3.2. Mutation density peaks and dips
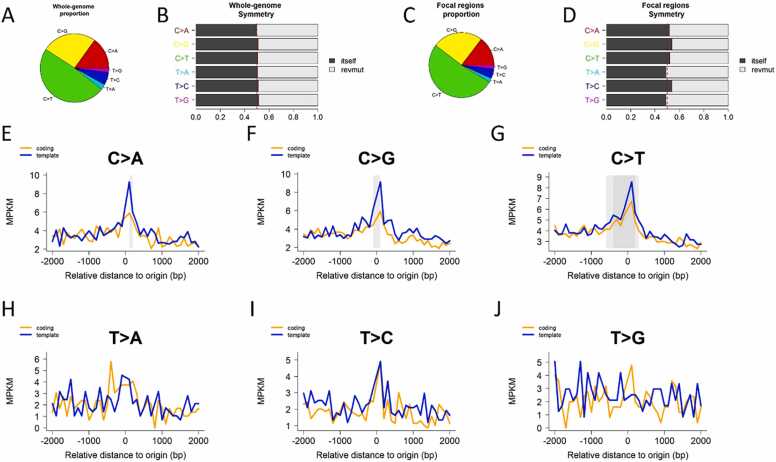
We conducted mutation density analysis using ICGC mutation data with lncRNA positions. For each ICGC cancer cohort, mutation density was summarized for each of three major genomic territories (protein exons, protein introns, intergenic regions) in human reference genome GRCh38 (Supplementary Table 2), with genomic territory segmentation inherited from our previous work AnnoGen [11]. Comparing among the three series of MPKM values across all cancer cohorts using Wilcoxon Signed-Rank test, we did not detect a significant cross-territory mutation density difference (p > 0.05). MutDens was employed to analyze each of 80 ICGC cohorts separately. For a representative set of results, the graphical output for bladder urothelial carcinoma US cohort is displayed in Fig. 2.
Fig. 2.
Mutational pattern analysis graphical output for BLCA-US cohort. A. Pie chart that depicts the genome-wide mutation type’s distribution. B. Barplot that depicts the genome-wide mutation type symmetry. The expected symmetry is at 0.5. C. Pie chart that depicts mutation type’s distribution around the lncRNA TSS regions. D. Barplot that depicts the mutation type symmetry around the lncRNA TSS regions. (E-J). Density plot of six categories of mutations around TSS regions of lncRNA: C>A (E), C>G (F), C>T (G), T > A (H), T > C (I), and T > G (J). Solid grey boxes indicate existence of significant mutation density patterns, in this case central peaks for C>A, C>G, and C>T.
First, we tested whether a mutation density peak or dip exists in three regions (upstream, downstream, and center) with respect to lncRNA TSS position using Poisson distribution. The tests were conducted for each of the six mutation types as defined in Methods, and potential central peaks (Peak), upstream-flank peaks (PeakL), right-flank peaks (PeakR), and central dips (Dip) were identified if raw Wilcoxon test p < 1 × 10−5. Considering that this round of analyses consist of 1920 tests in total (arising from 6 SBSs, 4 patterns (Peak, Dip, PeakL, PeakR), and 80 cancer cohorts), the adjusted p-value for any mutation density pattern would be less than 0.02 post Bonferroni adjustment. As a result, totally 178 significant results were identified, most of which (156) were central peaks (Supplementary Table 3). For C>A mutations, TSS-coincident and downstream peaks were observed for 29 and 2 cancer cohorts respectively, and one dip in one cancer cohort; for C>G mutations, TSS-coincident, TSS-upstream, and TSS-downstream peaks were observed for 31, 1, and 2 cancer cohorts, respectively; For C>T mutations, peaks were observed around lncRNA TSSs for 46 cancer cohorts and downstream for 1 cancer cohort; for T > A mutations, peaks were observed for 13 cancer cohorts (12 at TSS, 1 at downstream) and dip was identified for two cancer cohort; for T > C mutations, peaks were observed for 28 cancer cohorts around lncRNA TSS, three peaks downstream, and two dips around lncRNA TSS; for T > G mutations, 10 central peaks, 3 upstream peaks, and 1 downstream peak were observed, and a dip was also observed.
3.3. Cohort-level transcription strand bias
We employed False Discovery Rate less than 0.2 to affirm the existence of transcription strand bias at the cohort level. Overall, 65 significant results were identified, including 26 for C>A, 6 for C>G, 13 for C >T, 6 for T > A, 12 for T > C, and 2 for T > G (Supplementary Table 4). A total of 38 cancer cohorts demonstrated transcription strand bias, and 16 cancer cohorts showed bias in more than one mutation type. Two liver cancer cohorts (LICA-CN and LICA-FR) and one lung cancer (LUAD-US) each showed transcription strand bias in four mutation types. It is a noteworthy fact that transcription strand bias trends were shown across mutation categories for many cancer types. This concurs with a phenomenon that DNA damages in some cancer types are pervasive, not restricting to one specific type of mutation category. Like in bladder urothelial carcinoma US cohort, three mutation categories presented in substantial proportion (Fig. 2A). Similarly, prevalent occurrence of both C>A and C>T mutations were reported recently for liver cancer [12].
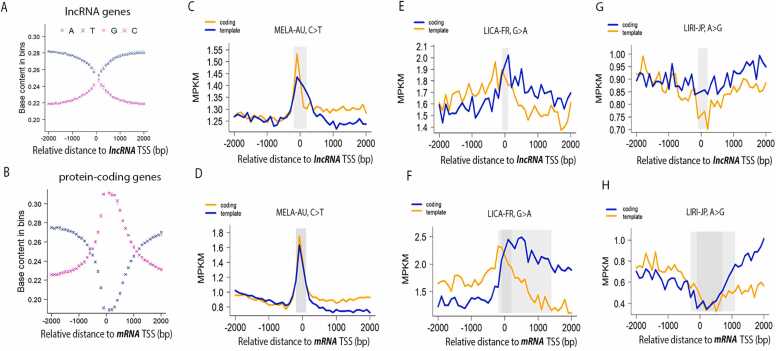
As we reasoned in the previous work, a SBS-wise mutation density metric must be normalized against the base content of the specific original bases, namely G/C or A/T, and the normalization can be locally dynamic or globally static [3]. Like mRNA TSS, lncRNA TSS has elevated G/C content in its bidirectional close proximity, but the highest G/C content in lncRNA reached only 48%, as compared to 62% for mRNA genes (Fig. 3, A and B). Given a much milder increase of GC content near lncRNA TSS and for the sake of minimal computational complexity, we applied a global normalization strategy by taking a constant G/C proportion of 40%. Under the global normalization strategy, the mutable bases for categories C>A, C>G, and C>T were quantified as 40% of all bases in the considered genomic regions, while the mutable bases for categories T > A, T > C, T > G were multiplied with a coefficient of 60%. The eventual mutation density value for a specific mutation category was calculated as the ratio of actual mutation counts over the percentage-adjusted mutable bases in the considered genomic regions.
Fig. 3.
Comparable mutation density patterns between mRNA TSS vicinity and lncRNA TSS vicinity. A) G/C content dynamics within upstream and downstream 2 kb of lncRNA TSS. B) G/C content dynamics within upstream and downstream 2 kb of mRNA TSS. C) mutation density curves for C>T SBS on coding strands and template strands of lncRNA genes, in MELA-AU. D) mutation density curves for C>T SBS on coding strands and template strands of protein-coding genes, in MELA-AU. E) mutation density curves for G>A SBS on coding strands and template strands of lncRNA genes, in LICA-FR. F) mutation density curves for G>A SBS on coding strands and template strands of protein-coding genes, in LICA-FR. G) mutation density curves for G>A SBS on coding strands and template strands of lncRNA genes, in LIRI-JP. H) mutation density curves for G>A SBS on coding strands and template strands of protein-coding genes, in LIRI-JP. Solid grey boxes indicate existence of significant mutation density patterns: central peaks (C, D, E, F) and central dips (G and H).
Generally, TSS-centered mutation density curves show comparable trends between lncRNA genes and protein-coding genes. Three representative cancer cohorts were selected to illustrate such striking parallelism between lncRNAs and mRNAs (Fig. 3, C-H). Looking at C>T mutations in MELA-AU, both lncRNA genes and mRNA genes clearly display a TSS-coincident peak and higher mutation density on the coding strand than the template strand in the TSS downstream (Fig. 3, C, and D). Still obvious divergence is seen in the coding/template strands for G>A (i.e., the C>T category) mutations in LICA-FR, with more mutations on the coding strand (Fig. 3, E, and F). The G>A (equivalently, C>T) mutations in LICA-FR do not form a peak as sharp as in the case of C>T in MELA-AU. Lastly, the A>G (i.e., the T > C category) mutation density curves display dips rather than peaks around lncRNA/mRNA TSS, and an obvious divergence between the coding strand and the template strand is clearly revealed for both lncRNA and mRNA (Fig. 3, G and H).
3.4. Individual-level transcription strand bias and prognosis
The DNA transcriptional-coupled repair mechanism predominantly engages in the repair of the template strand, commencing from the Transcription Start Site (TSS) to release the DNA-lesion-caused blockage of RNA polymerase. Consequently, lower mutation density is discernible in the template strand when compared to the coding strand. This observed transcriptional strand bias aligns with the conventional understanding of the transcription-coupled repair mechanism. It is plausible to conjecture that the extent of transcriptional strand bias could serve as an indicator of transcription-coupled repair proficiency. In the context of chemotherapy, which often targets specific genes by inducing DNA damage, individuals with robust Transcription-coupled repair capabilities may potentially mitigate the effects of drug-induced damage, thereby potentially diminishing the efficacy of chemotherapy and resulting in poorer survival outcomes.
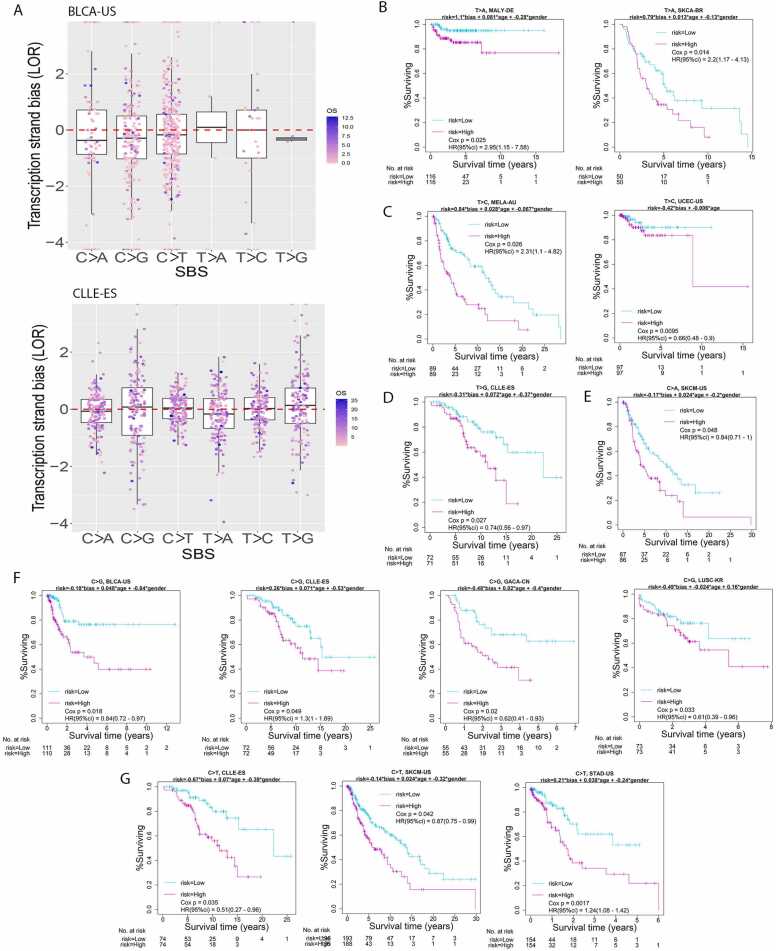
Patients of the same cancer cohort displayed a spectrum of transcription strand bias. For example, the bladder urothelial carcinoma US cohort carried C>G or C>T mutations most frequently (Fig. 2, A and C), and these two mutation categories showed higher variation in LOR than the least frequent category T > G, with standard deviations 1.3 and 1.2 for C>G and C>T, and 0.14 for T > G. Less prevalent mutation categories may also display considerable variation in the transcription strand bias measure, as exemplified in T > A and T > C with sd= 1.6 and 1.5 respectively (Fig. 4A, top). Another cancer cohort, CLLE-ES, carried the six mutation categories more evenly, and these six categories all showed moderate variation in LOR among the patients (Fig. 4A, bottom). Nevertheless, the two categories that had the highest LOR variation (sd=1.3 for C>G and sd=1.1 for T > G) had fewer mutation sites than the other mutation categories.
Fig. 4.
Quantification of individual-level transcription strand bias and its association with cancer survival. A. Patients in one were each assessed for their prominence of transcription strand bias with respect to a mutation category. Shown were two example cohorts: BLCA-US (top) and CLLE-ES (bottom). B. MALY-DE and SKCA-BR showed prognosis significance for T > A transcription strand bias. C. MELA-AU and UCEC-US showed prognosis significance for T > C transcription strand bias. D. CLLE-DE showed prognosis significance for T > G transcription strand bias. E. SKCM-US showed prognosis significance for C>A transcription strand bias. F. BLCA-US, CLLE-ES, GACA-CN, and LUSC-KR showed prognosis significance for C>G transcription strand bias. G. CLLE-ES, SKCM-US, and STAD-US showed prognosis significance for C>T transcription strand bias.
Given transcription strand bias quantified as LOR for each patient, we were able to assess the prognostic potential of biased SBS through a Cox survival model adjusted for age and sex. For all 86 cancer-SBS combinations that had ample sample size, 13 cancer-SBS combinations displayed potential prognostic value at nominal p < 0.05 and False Discovery Rate lower than 0.2, involving 10 cancer cohorts (Supplementary Table 5; Fig. 4, B-G). All six mutation categories found prognosis significance for transcription strand bias in certain cancer types; C>G and C>T were most noteworthy, reaching prognostic significance in four (Fig. 4F) and three (Fig. 4G) cancer cohorts, respectively. For the total 13 scenarios, C>A in SKCM-US, C>G in BLCA-US, C>T in SKCM-US, and T > C in UCEC-US also demonstrated cohort-level significant transcription strand bias with Wilcoxon p < 0.05.
Considering both the cohort-level LOR bias direction and the coefficient sign of LOR in the Cox risk model, we could investigate if a more extreme LOR value indeed predicts poorer survival. To ascertain the initiative mutation form in context of strand bias, one must examine the relative predominance of the two mutation density curves at TSS downstream. For example, C>T is the initiative mutation form in melanoma (MELA-AU), and accordingly the “coding” mutation density is higher than the “template” mutation density (Figs. 3C and 3D). In such a straightforward case, a large positive LOR value indicates a strong transcription bias. In a flipped scenario, as exemplified in the liver cancer cohort LICA-FR (Figs. 3E and 3F), the counter-intuitive predominance of “template” over “coding” simply indicates the complementary G>A mutations are the genuine initiative mutation form, rather than the form of C>T. In such a mutation-form-flipped case, a large negative LOR value indicates a strong transcription bias.
Combining the bias direction at the cohort level and the coefficient sign for LOR in the Cox model, the data showedthat the strength of transcription strand bias in SKCM-US (C>A), BLCA-US (C>G), or UCEC-US (T > C) was a risk factor for overall survival. Only SKCM-US (C>T) was an exception, with a stronger bias associated with a higher survival chance. This reversal, counter-intuitive trend may caution the existence of false positive observation at either the cohort-level analyses or at the individual-level analyses, or may signify involvement of additional, complicating biological mechanisms other than the sole Transcription-coupled repair effect. The other nine scenarios did not reveal cohort-level significant transcription strand bias, involving CLLE-ES in C>T, C>G, and T > G mutations, GACA-CN and LUSC-KR in C>G mutation, STAD-US in C>T mutation, MALY-DE and SKCA-BR in T > A mutation, and MELA-AU in T > C mutation.
4. Discussion
By September 2023, there have been 86 clinical trials based on lncRNA, of which 30 were related to cancer. Even though the results of non-coding RNA based clinical trials have been ambivalent [13], the importance and necessity of lncRNA in cancer research have been recognized throughout the research community. Over the last decade, even though the majority of the lncRNA study have been focused on the expression of lncRNA potentially due to the abundant availaibity of RNAseq data, some studies also highlighted the importance of mutations in lncRNAs. For example, mutations in lncRNA MALAT1 was shown to cause alternative splicing in cancers [14]; mutations in lncRNA RMRP impair mouse and human T cell activation [15]; mutations in lncRNA TCL6 is predicted to affect binding afficacy of RNA-binding proteins [16]. As a natural follow-up to our previous mutational pattern study on protein-coding RNA, a.k.a. mRNA [3], we designed a follow-up study with the focus shifted to lncRNA. This lncRNA study utilized the same mutation datasets used in the preceeding mRNA study, thus enabling proper comparison between lncRNA and mRNA.
While lncRNAs do not serve as templates for protein synthesis, lncRNAs do exhibit many similar characteristics to protein-coding RNAs. For example, both lncRNA and protein-coding RNA are transcribed from DNA and can be processed post-transcriptionally. Like mRNAs, most annotated lncRNAs are RNA polymerase II transcribed and the lncRNA transcripts may share some structural similarity with mRNA [17]. After transcription, both lncRNA and mRNA undergo processing to produce mature functional RNA molecules. In the case of mRNA, this includes splicing, capping, and polyadenylation. In the case of lncRNA, processing can involve alternative splicing, RNA editing, and post-transcriptional modifications. Furthermore, both lncRNA and mRNA can be involved in gene expression regulation: while mRNA carries the genetic information required to synthesize proteins, lncRNA plays a role in the regulation of gene expression by various mechanisms, including transcriptional, post-transcriptional, and epigenetic regulation. Our findings indicate that particular cancer types exhibit transcriptional strand bias for specific types of mutations, typically linked to the cancer's etiology. For instance, skin cancer predominantly features C>T mutations owing to exposure to ultra violet light, while lung cancer is often characterized by C>A mutations attributed to tobacco smoking [18].
Mutation density fluctuations manifest in numerous cancer types across diverse genomic features. Notably, mutation density peaks encircle mRNA gene transcription start sites (TSS) [3], [19], while mutational density dips manifest in the vicinity of retrotransposons [3]. These distinctive mutation density patterns typically arise from specific underlying biological mechanisms. For instance, the mutation density peak is attributed to Transcription-coupled repair processes. Perera et al. [19] proposed that in highly transcribed promoters, the transcription pre-initiation complex hinders the recognition of DNA damage by repair machinery, including Xeroderma Pigmentosum C (XPC), thereby resulting in the frequently observed mutation peaks coinciding with TSS. The mutation density dip observed in retrotransposons implies the existence of an unidentified mechanism responsible for the diminished mutation rate within these elements, warranting further investigation. The above examples highlight the importance of mutation density, and how it uncover hidden biological mechanism. The aforementioned instances underscore the significance of mutation density and its capacity to unveil concealed biological mechanisms.
LncRNAs are transcribed from DNA using largely the same machinery as mRNAs. Thus, if assuming mutations occur indiscriminately across the genome, observing similar mutation density patterns around TSS between mRNA and lncRNA is within reasonable expectation. Our analyses confirm this hypothesis: 55 cancer cohorts presented TSS-coincident mutation peaks in all six mutation types. While TSS-coincident mutation peaks tend to appear frequently in many cancer types for both mRNA and lncRNA, the contrary phenomenon of TSS-coincident mutation dip is rare. In screening 81 cancer cohorts on six mutation categories, we identified six statistically significant lncRNA-TSS mutation dips, including the cases for melanoma (MELA-AU) on T > A and liver cancer (LIRI-JP) on T > C. The two mutation dips were also observed at mRNA TCC vicinity. Previously, a C>T mutation dip was found for enhancer regions in the same Australian melanoma cohort [3]. Future studies are necessary to investigate and elucidate these intriguing mutation peaks and dips associated with TSS in various scenarios.
Our results show that specific cancer type shows transcriptional strand bias for specific mutation types. This is usually related to the etiology of the cancer. For example, skin cancer is dominated by C>T mutations due to UV light exposure; lung cancer is usually dominated by C>A mutation due to tobacco smoking. Thus, it is not surprising that the mutation density patterns we observe are often associated with certain types of mutation.
The transcription strand bias is a direct consequence of Transcription-coupled repair mechanisms. Strong Transcription-coupled repair capability may result in more severe bias. Based on this fact, we hypothesized that transcription strand bias may negatively affect the response rate of chemotherapy. We quantified transcription strand bias at the sample level and conducted survival analysis. Thirteen prognostic significant results were identified, which support our hypothesis. A majority of the 13 scenarios did not show overall transcription strand bias at the cohort level, meaning that a cancer type without a coherent bias may still show a considerable range of transcription strand bias across patients and the individual-level bias strength can have prognostic value. Four of the 13 scenarios showed overall transcription strand bias, and three scenarios were consistent with our hypothesis in directional prognosis: the stronger the bias, the poorer the survival. Skin cancer, lymphoma, gastric cancer, lung cancer, and bladder cancer each possessed at least one mutation category as plausible patient-level prognostic marker.
Our research findings underscore the inherent association between transcriptional strand bias and survival outcomes, largely driven by its potential impact on the efficacy of chemotherapy. However, to conclusively validate and integrate transcriptional strand bias as a clinical parameter, the acquisition of precise drug treatment data and the development of drug-specific predictive models are imperative prerequisites. Moreover, our study showcases the potential significance of exploring the role of lncRNA mutations in the context of cancer. This highlights the broader benefits of mutational density investigations, thereby encouraging further research into mutational density patterns across other RNA types, such as enhancer RNAs.
CRediT authorship contribution statement
All authors contributed to the manuscript. Guo and Bai supervised the project, Guo secured funding and designed the study. Zhang and Yu conducted formal analysis and data curation. Zhang, Yu, Bai and Guo contributed to writing of the manuscript.
Acknowledgments
This study was supported by the Biostatistics and Bioinformatics Shared Resource at Sylvester Comprehensive Cancer Center, University of Miami. YG and HY were supported by the cancer center support grant P30CA240139 and grant R01ES030993 from the National Cancer Institute, USA.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.09.027.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Sung H., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Croce C.M. Oncogenes and cancer. N Engl J Med. 2008;358(5):502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 3.Yu H., et al. Surveying mutation density patterns around specific genomic features. Genome Res. 2022;32(10):1930–1940. doi: 10.1101/gr.276770.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sebastian-delaCruz M., et al. The Role of lncRNAs in Gene Expression Regulation through mRNA Stabilization. Non-Coding RNA. 2021;7(1) doi: 10.3390/ncrna7010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y., Sun H., Wang H.T. Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci. 2016;6 doi: 10.1186/s13578-016-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nie W., et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 8.Atianand M.K., Fitzgerald K.A. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med. 2014;20(11):623–631. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volders P.J., et al. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019;47(D1):D135–D139. doi: 10.1093/nar/gky1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haradhvala N.J., et al. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell. 2016;164(3):538–549. doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheng Q., et al. AnnoGen: annotating genome-wide pragmatic features. Bioinformatics. 2020;36(9):2899–2901. doi: 10.1093/bioinformatics/btaa027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degasperi A., et al. Substitution mutational signatures in whole-genome-sequenced cancers in the UK population. Science. 2022;376(6591) doi: 10.1126/science.abl9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkle M., El-Daly S.M., Fabbri M., Calin G.A. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20(8):629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arun G., et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30(1):34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson N., et al. A disease-linked lncRNA mutation in RNase MRP inhibits ribosome synthesis. Nat Commun. 2022;13(1) doi: 10.1038/s41467-022-28295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh B., et al. Genome sequencing and RNA-Motif analysis reveal novel damaging noncoding mutations in human tumors. Mol Cancer Res. 2018;16(7):1112–1124. doi: 10.1158/1541-7786.MCR-17-0601. [DOI] [PubMed] [Google Scholar]
- 17.Qin T., Li J., Zhang K.Q. Structure, regulation, and function of linear and circular long non-coding RNAs. Front Genet. 2020;11 doi: 10.3389/fgene.2020.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell P.J., et al. Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82. doi: 10.1038/s41586-020-1969-6. (-+) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perera D., et al. Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature. 2016;532(7598):259. doi: 10.1038/nature17437. (-+) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material