Abstract
Objective
Osteoarthritis (OA) is a degenerative joint disease associated with excessive mechanical loading. The aim here was to elucidate whether different subpopulations of chondrocytes exhibit distinct phenotypes in response to variations in loading conditions. Furthermore, we seek to investigate the transcriptional switches and cell crosstalk among these chondrocytes subsets.
Methods
Proteomic analysis was performed on cartilage tissues isolated from weight-bearing and non-weight-bearing regions. Additionally, single-cell RNA sequencing was employed to identify different subsets of chondrocytes. For disease-specific cells, in vitro differentiation induction was performed, and their presence was confirmed in human cartilage tissue sections using immunofluorescence. The molecular mechanisms underlying transcriptional changes in these cells were analysed through whole-transcriptome sequencing.
Results
In the weight-bearing regions of OA cartilage tissue, a subpopulation of chondrocytes called OA hypertrophic chondrocytes (OAHCs) expressing the marker genes SLC39A14 and COL10A1 are present. These cells exhibit unique characteristics of active cellular interactions mediated by the TGFβ signalling pathway and express OA phenotypes, distinct from hypertrophic chondrocytes in healthy cartilage. OAHCs are mainly distributed in the superficial region of damaged cartilage in human OA tissue, and on TGFβ stimulation, exhibit activation of transcriptional expression of iron metabolism-related genes, along with enrichment of associated pathways.
Conclusion
This study identified and validated the existence of a subset of OAHCs in the weight-bearing area of OA cartilage tissue. Our findings provide a theoretical basis for targeting OAHCs to slow down the progression of OA and facilitate the repair of cartilage injuries.
Keywords: osteoarthritis; chondrocytes; osteoarthritis, knee; inflammation; Magnetic Resonance Imaging
WHAT IS ALREADY KNOWN ON THIS TOPIC.
The multifaceted and intricate phenomenon of the impact of mechanical stimulation under different loading states on osteoarthritis (OA) remains enigmatic. It is unclear whether the hypertrophic chondrocytes generated in this pathological state of OA are synonymous with those in the normal physiological state.
WHAT THIS STUDY ADDS
This study introduces the effects of weight-bearing conditions and mechanical stimulation on patients with OA, using MRI-based grading of cartilage damage.
This study examined the dissimilarities in chondrocytes of patients with OA under divergent loading states at the levels of proteomics, transcriptomics and single-cell resolution, and validated the findings through in vitro cellular induction and clinical specimens.
SLC39A14+/COL10A1+ osteoarthritis hypertrophic chondrocytes (OAHCs) were identified in OA cartilage tissue stimulated by excessive mechanical loading. These OAHCs exhibited a sensitive response to the TGFβ signalling pathway.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
These findings have not only established a fundamental basis for targeted therapies for chondrocytes but have also provided novel insights into alleviating cartilage degeneration under mechanical loading.
Introduction
Osteoarthritis (OA) represents failed repair of joint damage resulting from stresses initiated by any joint or periarticular tissue abnormality, usually causing joint pain and impairment of function.1 OA can involve any joint in the body and the most commonly affected joint is the knee, which affects more than 240 million people worldwide.2 Many patients present to orthopaedic surgeons with a painful knee attributed to OA of the medial compartment, with the lateral compartment and patella-femoral joint being relatively spared.3 Up to 90% of unicompartment knee OA had exclusively medial involvement,4 which is closely associated with the loading of forces on the medial side during walking, the most common site of knee degeneration, due to the adduction moment of weight-acceptance phase.5 The knees are subject to continuous mechanical load as a weight-bearing joints, and cartilage is the only tissue on the articular surface that cushions this load. At the same time, chondrocytes are the only cell type in articular cartilage tissue.6 The transition mechanism of chondrocytes under mechanical stimuli during joint loading has an important impact on cartilage homoeostasis and function.
Chondrocytes are surrounded by a highly specialised extracellular matrix (ECM) that does not have a homogeneous structure and composition, with type II collagen forming the network and receiving high concentrations of proteoglycans (such as aggrecan, lubricin and perlecan) and glycosaminoglycans (such as hyaluronan), as well as type VI collagen and type IX collagen, which together form the pericellular matrix.7 8 Mechanical loading brings not only ‘wear and tear’ but actually a driving force that promotes chondrocyte phenotypic drift and OA development by activating mechanoresponsive cell signalling (mechanosignalling) and the resulting proinflammatory mediators and catabolic enzymes.9–11
Previous studies have demonstrated that poor alignment is a strong predictor of disease progression in patients with knee OA.12 In addition, single-cell RNA sequencing (scRNA-seq) has demonstrated the presence of different subpopulations of chondrocytes that differentially drive inflammation in patients with OA.13 Therefore, the purpose of this study was to determine whether patient-reported sites of excessive stress loading exhibit cartilage tissue with a different transcriptomic phenotype containing different subpopulations of chondrocytes, compared with sites of normal mechanical loading.
Methods
Patient recruitment, pain scoring and tissue collection
On obtaining ethical review approval, 44 patients diagnosed with knee joint pain and receiving treatment at the Third Affiliated Hospital of Sun Yat-sen University were enrolled as study subjects. Among them, 25 individuals underwent joint replacement surgery, while the remaining patients underwent arthroscopic examination or received non-surgical treatment. Comprehensive clinical data were collected for all participants. For patients undergoing total knee arthroplasty (TKA), cartilage tissue samples were extracted from the medial and lateral tibial plateaus. The specific methodology employed is as follows: subsequent to the tibial plateau osteotomy, full-thickness cartilage (~1.5 g) was resected from identical locations at the central region (central lateral articular cartilage and central medial articular cartilage). In addition, we also collected cartilage tissue samples from the weight-bearing and non-weight-bearing regions of facet joints in three patients diagnosed with scoliosis, who underwent osteotomy corrective surgery at the Third Affiliated Hospital of Sun Yat-sen University. These collected cartilage tissues were used for protein and RNA extraction, as well as for tissue section staining.
OA scoring using MRIs
The presurgical MRI images were evaluated using the MRI Osteoarthritis Knee Score (MOAKS), which is a semiquantitative assessment tool derived from the Whole Organ MRI Score and the Boston Leeds Osteoarthritis Knee Score scoring system. Articular cartilage and bone marrow lesions were scored in 15 distinct subregions, which include the medial and lateral patella, medial and lateral trochlea, medial and lateral central femur, medial and lateral posterior femur, anterior medial articular cartilage, central medial articular cartilage, posterior medial articular cartilage, anterior lateral articular cartilage, central lateral articular cartilage, posterior lateral articular cartilage and tibial spines.14 The scoring process was conducted by a consultant radiologist and research fellow.
Protein mass spectrometry
For the four TKA patients in the study population with varus knees in whom there was a significant difference between the medial and lateral weight-bearing zones of the tibial plateau in the preoperative MOAKS score (table 1), we performed protein mass spectrometry of their cartilage tissues. The sterile cartilage tissue was cleansed with phosphate buffered saline (PBS) and then pulverised into a fine powder using a mortar and pestle in liquid nitrogen. The resulting powder was combined with trichloroacetic acid (TCA)/acetone (1:9) in a fivefold volume and the mixture was subsequently dried and resuspended in sodium dodecyl sulfate-dithiothreitol-tris (SDT) lysis buffer at a mass-to-volume ratio of 1:30. After ultrasonication, the samples were centrifuged at 14 000g for 15 min and the supernatant was filtered through a 0.22 µm membrane. Protein concentration was assessed using the bicinchoninic acid (BCA) assay. For each group, 200 µg of protein was digested with filter aided sample preparation (FASP), and 100 µg of peptides were labelled using the Thermo Tandem Mass Tag (TMT) labelling kit according to the manufacturer’s instructions. The resulting labelled peptides from each group were mixed and fractionated using an Agilent 1260 Infinity II HPLC system. The samples were subsequently separated by nanoflow chromatography and analysed by mass spectrometry using a Q Exactive Plus mass spectrometer. Mascot 2.6 and Proteome Discoverer 2.1 software were employed for library search identification and quantitative analysis. A total of 5177 unique peptides were detected in 10 cartilage tissues, and 967 proteins were used for further bioinformatics analysis.
Table 1.
Tandem Mass Tag Patients MRI Osteoarthritis Knee Score (MOAKS) information
| Side | Left | Right | Right | Right | Right | |
| MOAKS | Medial patella | 0 | 1 | 0 | 1 | 0 |
| Lateral patella | 1 | 1 | 0 | 0 | 0 | |
| Medial trochlea | 2 | 4 | 1 | 1 | 0 | |
| Lateral trochlea | 0 | 0 | 0 | 0 | 0 | |
| Medial central femur | 1 | 3 | 1 | 0 | 0 | |
| Lateral central femur | 0 | 0 | 0 | 0 | 0 | |
| Medial posterior femur | 0 | 1 | 0 | 0 | 0 | |
| Lateral posterior femur | 0 | 0 | 0 | 0 | 0 | |
| Anterior medial articular cartilage | 6 | 8 | 2 | 4 | 4 | |
| Central medial articular cartilage | 7 | 11 | 9 | 11 | 6 | |
| Posterior medial articular cartilage | 9 | 6 | 6 | 6 | 8 | |
| Anterior lateral articular cartilage | 0 | 0 | 0 | 0 | 0 | |
| Central lateral articular cartilage | 0 | 0 | 0 | 0 | 0 | |
| Posterior lateral articular cartilage | 0 | 0 | 0 | 0 | 0 | |
| Tibial spines | 0 | 0 | 0 | 0 | 0 | |
| Total score | 26 | 35 | 19 | 23 | 18 | |
Single-cell sequencing analysis
Using the search terms osteoarthritis and single cell in the GEO database (https://www.ncbi.nlm.nih.gov/geo/), the dataset GSE152805 was obtained (GSE152805 was uploaded by Kraus in 2020).15 16 The data sets collected a total of 26 228 chondrocytes from 3 patients undergoing knee arthroplasty due to OA. The dataset was single-cell high-throughput sequencing using the Illumina HiSeq 4000 (Homo sapiens) platform. The sequencing data were analysed using Seurat v4.0.5 (https://satijalab.org/Seurat/) for the purpose of data processing.17 Following quality control standardisation and log transformation, highly variable genes were identified and used for performing principal component analysis (PCA). Harmony was employed to further integrate the data. Uniform Manifold Approximation and Projection (UMAP) was performed using the top 30 PCs to cluster cells, with a resolution of 0.5.
Monocle v2.22.0 was used to analyse the pseudotime trajectory (http://Monocle-bio.sourceforge.net/).18 We established a ‘CellDataSet object’ derived from Seurat object. Subsequently, we identified differentially expressed highly variable genes that were expressed in a minimum of 10 cells with a q value<0.01. The dimensionality of the data was reduced using the DDRTree method, and cells were pseudotime sorted.
CellChat v1.6.0 was used to quantitatively characterise and compare the inferred cell-cell communication networks.19 We first established a ‘CellChat object’ derived from Seurat objects, encompassing cell type identity for each cell. The comprehensive collection of human ligand–receptor interactions documented in the CellChatDB was employed for the examination of cell–cell communication. Subsequently, we identified the ligands and receptors that were overexpressed within each cell type, and determined the overexpressed interactions between these ligands and receptors. The probabilities of cell–cell communication for each ligand–receptor interaction were deduced using permutation tests. Importance of each cell cluster in the signalling pathway of interest was also computed and represented.
miloR V.1.9.1 was employed for the identification of disease-specific cell populations by performing differential abundance analysis.20 We generated a ‘Milo object’ from Seurat objects, in which PCA and UMAP dimensionality reduction had been executed. Subsequently, representative cell neighbourhoods were established by randomly selecting 10% of all cells within the object, setting d and k values at 40. The cell count within each neighbourhood was then computed, followed by the execution of differential abundance analysis between conditions (‘iMT’ vs ‘oLT’).
Cell culture
Following the protocol of Haseeb and Lefebvre,21 the tibial plateau and femoral head of 3-day-old C57BL/6 mice were dissected open. Initially, the tissues were digested with 5 mL of 0.25% Trypsin-EDTA (Gibco, USA) at 37℃ in a 5% CO2 incubator for 1 hour, followed by 5 mL of 0.2% collagenase type II (Biofroxx, Germany) digestion for 5 hours at 37℃ to collect and culture primary mouse chondrocytes. Chondrocytes and ATDC5 (Sigma-Aldrich) were cultured in DF12 (Gibco, USA) supplemented with 10% fetal bovine serum (Nobimpex) and 1% penicillin-streptomycin (Gibco) at 37℃ and 5% CO2. The culture medium was replenished every 72 hours. ATDC5 or chondrocytes were treated with 1% ITS (#41400045, Gibco), 250 µM L-Ascorbic acid (#4055, Tocris), or 20 ng/mL TGF-β (#7666-MB, R&D) as indicated.
RNA sequencing analysis
Total RNA was isolated from primary chondrocytes (n=6) in the presence or absence of TGF-β using Trizol Reagent (#15596018, Invitrogen), followed by RNA library preparation using the QIAseq RNA Library Kit (QIAGEN, Germany). Subsequently, the Illumina high-throughput sequencing technology platform was used to obtain sequencing data which were then normalised for differential gene expression analysis after alignment with BWA (v0.7.12-r1039). We used the DEGSeq algorithm to selectively isolate differentially expressed microRNA and circular RNA genes. Moreover, we employed the DESeq2 algorithm to identify and filter differentially expressed mRNA and long non-coding RNA (lncRNA) genes. These algorithms conducted correction based on false discovery rate (FDR) control method in multiple tests to reduce the probability of Class I errors, and determined differentially expressed genes with significant differences based on criteria of >1.5 fold change and FDR<0.05.
Pathway analysis
The proteins that showed significant differences (>1.2 fold change, p<0.05) were subjected to analysis using the Ingenuity Pathway Analysis (IPA; Qiagen, UK). This tool incorporates a database that integrates machine learning and manual curation by researchers, encompassing approximately 8 million relevant pieces of information on humans, mice and rats from biomedical literature and over 30 authoritative databases. In addition, the differential genes or marker genes were subjected to enrichment analysis of gene ontology, Kyoto Encyclopedia of Genes and Genomes, and Reactome pathways using David (https://david.ncifcrf.gov/home.jsp).22 This allowed us to determine the interactions and regulatory network relationships of the differential genes.
Quantitative real-time PCR (qPCR)
mRNA isolation and qPCR were performed following previously described methods.23 Total RNA was extracted and mRNA was purified using the E.Z.N.A. Total RNA Kit I (Omega, USA). mRNA was then reverse transcribed into cDNA using Evo M-MLV RT Premix (AG11706, China). The primer sequences used are as follows: SLC39A14,F:5’AAGGCCCTACTCAACCACCT3’,R:5’CGACTGCTCGCTGAAATTGTG3’;Slc39a14,F:5’GTGTCTCACTGATTAACCTGGC3’,R:5’AGAGCAGCGTTCCAATGGAC3’;COL10A1,F:5’ATGCTGCCACAAATACCCTTT3’,R:5’GGTAGTGGGCCTTTTATGCCT3’;Col10a1,F:5’GGTGTGAATGGGCGGAAAG3’,R:5’GCTTCCCAATACCTTCTCGTC3’;Col2a1,F:5’CACACTGGTAAGTGGGGCAAGACCG3’,R:5’GGATTGTGTTGTTTCAGGGTTCGGG3’;Mmp13,F:5’TGTTTGCAGAGCACTACTTGAA3’,R:5’CAGTCACCTCTAAGCCAAAGAAA3’.
Gene expression was detected using the LightCycler 480 system (Roche, USA) and amplified using SYBR-Green Premix (AG11701, China) according to the user manual. mRNA expression values were normalised to Beta-Actin and relative calculations were performed using the 2−ΔΔCt method.
Immunofluorescence and immunohistochemical staining
The histological slides of human cartilage were deparaffinised and hydrated. They were then subjected to blocking using 5% bovine serum albumin for 1 hour at room temperature, followed by overnight incubation with primary antibodies against COL10A1 (orb312179, Biobyt). A secondary antibody labelled with horseradish peroxidase (HRP) corresponding to the primary antibody species was added to the tissue. After CY3-tyramide signal amplification, the primary antibody against SLC39A14 (PA5-21077, Thermo) was added along with the corresponding fluorescein isothiocyanate (FITC)-fluorescent secondary antibody. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The samples were imaged using both fluorescence microscope (Nikon, Germany) and confocal laser scanning microscope (Leica, Germany). Under blinded conditions, two observers quantified the proportion of positive cells. For safranin o-fast green counterstaining, the sections were first stained with plant safranin and plant o-fast, followed by rapid differentiation with ethanol. To quench endogenous peroxidase activity, 3% (v/v) H2O2 was used for immunohistochemical staining. FTH1 (PA5-27500, Invitrogen) was used as the primary antibody, and incubated overnight. A goat anti-rabbit immunoglobulin G conjugated with HRP (BL003A, BioSharp, Shanghai, China) was used as the secondary antibody. The immunohistochemical staining was observed under an inverted microscope (Nikon, Germany). Quantification of positively cells in immunofluorescence images, encompassing regions located within 700 µm from the articular surface were included in the analysis.
GSH detection in primary mouse chondrocytes
The quantification of GSH content in primary mouse chondrocytes was conducted by thoroughly mixing the extracted cell protein solution with the GSH detection reagent (A006-2-1, Jiancheng), as protocol. The resultant supernatant was collected for analysis. GSH content was assessed through colorimetric analysis at 405 nm, with reference to a standard curve.
Statistical analysis
Data were analysed using Graphpad Prism V.9. Specific tests performed are detailed within the figure legends, with statistical significance determined to be p<0.05. To avoid bias the investigators were blinded during data analysis.
Results
Differences in arthritis severity, radiographic assessments and proteomic profiles among patients with knee OA different weight-bearing states
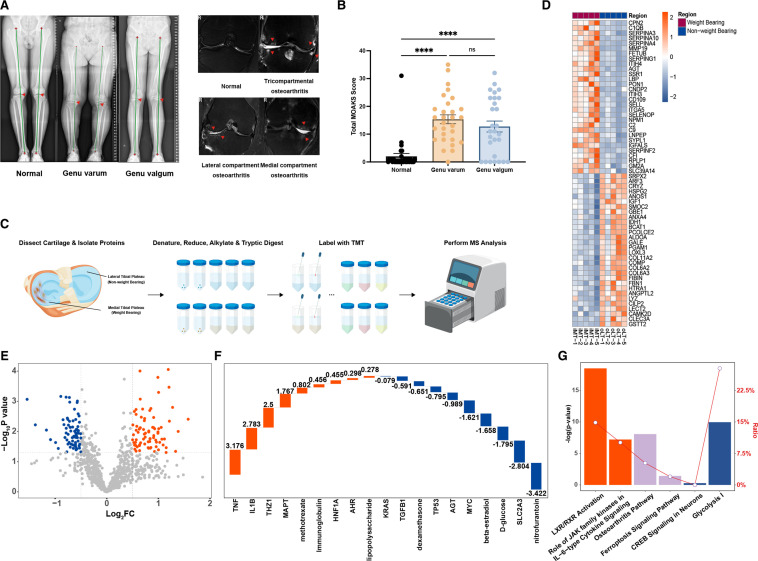
Based on preoperative imaging findings, most patients with OA exhibited genu varum (figure 1A). Compared with knee joints with a normal force line (median OKS=48), patients with OA with altered force line due to genu varum or genu valgum (genu varum median OKS=18, genu valgum median OKS=24) exhibit significantly (p<0.05) more severe joint arthritis(table 2). Naturally, patients with genu varum or genu valgum also had a larger hip-knee-ankle angle than normal patients(table 2).
Figure 1.
Clinical radiographic alterations in patients with knee osteoarthritis (OA) under different weight-bearing types and Tandem Mass Tag (TMT) analysis of cartilage tissues. (A) Representative imaging of the knee joint in patients with OA, with X-ray (full-length standing anteroposterior scan of both lower limbs) showing different weight-bearing types (indicated by red triangles) and MRI (coronal scan) showing cartilage damage and bone marrow oedema in different anatomical locations (indicated by red triangles). (B)Total MRI Osteoarthritis Knee Score score for normal knees (n=31), varus knees (n=29), and valgus knees (n=28). ****P<0.0001 (one-way analysis of variance (ANOVA)), with significant differences observed in both valgus and varus knees compared with normal knees. (C) Schematic workflow illustrating the experimental strategy, outlining the sequence of steps involved in TMT. (D) Heatmap of top 20 differentially expressed proteins in cartilage tissue from non-weight-bearing (oLT), compared with weight-bearing (iMT) regions in patients with OA (5 samples/group). Colour of tile indicates z-scored expression. (E) Volcano plot of differentially expressed proteins (|FC|>1.2) in cartilage tissue from non-weight-bearing (oLT), compared with weight-bearing (iMT) regions in patients with OA (5 samples/group). (F) Z-scores of predicted upstream regulators in the proteomes of non-weight-bearing (oLT) and weight-bearing (iMT) cartilage tissues. Positive Z-scores (orange bars) represent predicted ‘activated’ upstream regulators, while negative Z-scores (blue bars) represent predicted ‘inhibited’ upstream regulators. The numerical values indicate the Z-scores. (G) Significant enrichment of differential proteins in classic pathways as analysed by Ingenuity Pathway Analysis. Orange denotes predicted ‘activation’ of a pathway, blue denotes predicted ‘inhibition’ and purple indicates a lack of activation or inhibition prediction for the pathway. The y-axis on the left represents the significance level of enrichment (-Log P-value), while the ratio shown by the line indicates the proportion of differential proteins in the pathway compared with all proteins in that pathway.
Table 2.
Patients information
| All patients | Genu varum | Genu valgum | Normal | ||
| Age | 59.48±1.28 | 64.83±1.12* | 60.43±2.77* | 53.61±1.99 | |
| Gender (male:female) | 11:33 | 2:14 | 2:14 | 9:10 | |
| Height (cm) | 166.25±0.74 | 165.24±1.26 | 164.71±1.11 | 168.58±1.37 | |
| Weight (kg) | 65.10±1.07 | 63.90±1.70 | 62.52±1.72* | 68.56±1.96 | |
| BMI | 23.56±0.37 | 23.42±0.60 | 23.11±0.68 | 24.10±0.63 | |
| Oxford Knee Score (0–48) | Median | 31.5 (8–48) | 18 (8–41)* | 24(8-48)* | 48(17-48) |
| Oxford Knee Score Category | Severe (0–19) | 33% (29) | 59% (17) | 39.2% (11) | 3% (1) |
| Moderate (20–29) | 15% (13) | 17% (5) | 28.6% (8) | 0% (0) | |
| Mild (30–39) | 8% (7) | 21% (6) | 3.6% (1) | 0% (0) | |
| Normal (40–48) | 44% (39) | 3% (1) | 28.6% (8) | 97% (30) | |
| Hip-Knee-Ankle Angle (°) | 4.60±0.46 | 7.21±0.73* | 6.4±0.74* | 0.53±0.06 | |
| Osteotomy (TKA:UKA:None) | 15:10:63 | 8:7:14 | 6:3:19 | 1:0:30 |
Values represent mean±SEM for all patients (n=44), genu varum (n=29), genu valgum (n=28) and normal knee joints (n=31).
Values represent median (IQR).
Values represent % (number) of patients in each category.
*A statistically significant comparison with normal patients.
BMI, body mass index; TKA, total knee arthroplasty; UKA, unicompartmental knee arthroplasty.
The total MOAKS scores, which were calculated by combining scores from MRI analysis at 15 anatomical locations, were significantly higher in patients with OA with genu varum (15.34±1.60) and genu valgum (12.75±1.97) when compared with patients with a normal force line (2±1.02) (figure 1B). On comparing MOAKS cartilage grades in different anatomical locations, it was observed that most cartilage injuries in patients with OA with genu varum and genu valgum were concentrated in the weight-bearing area of the tibial plateau. The cartilage injury grade in the non-weight-bearing area of the contralateral tibial plateau was found to be significantly lower (p<0.05) (online supplemental figure S1A).
rmdopen-2023-003255supp002.xlsx (328.7KB, xlsx)
Considering the differential response of cartilage tissue to mechanical stimuli under varying weight-bearing states, we conducted TMT-based proteomic analysis on tibial plateau cartilage tissue from matched weight-bearing and non-weight-bearing sites in patients with OA (5 samples/group) (table 1, figure 1C). Comparing weight-bearing and non-weight-bearing cartilage tissue in patients with OA, a total of 224 proteins were differentially expressed (> 1.2 fold change, p<0.05) of which 101 were upregulated and 123 were downregulated proteins in weight-bearing-associated cartilage (figure 1D,E).
To further investigate these differentially expressed proteins, we then performed an upstream analysis to identify any activated or inhibited upstream regulators of the weight-bearing-associated proteomes. TNF, IL1B and THZ1 were identified as activated upstream regulators, while KRAS, TGFB1 and SLC2A3 were identified as inhibited upstream regulators (figure 1F). It is noteworthy that the expression of TGFB1, an inhibited upstream regulator, is significantly downregulated in weight-bearing cartilage tissue. IPA analysis also revealed that the activated pathways included ‘LXR/RXR Activation’ and ‘Role of JAK family kinases in IL-6-type Cytokine Signalling’, while the inhibited pathways included ‘CREB Signalling in Neurons’ and ‘Glycolysis I’. OA and Ferroptosis Signalling pathways were also enriched, but without direct prediction of activation or inhibition (figure 1G).
Single-cell RNA sequencing identifies differences in chondrocyte subpopulations among different weight-bearing types of knee OA
After identifying differences in the proteomes of weight-bearing and non-weight-bearing cartilage tissues, we proceeded to investigate whether chondrocytes from weight-bearing-associated cartilage tissue exhibited distinct cellular subpopulations. To this end, an analysis was performed on publicly available scRNA-seq data (GSE152805) sourced from outer lateral (non-weight-bearing) and inner medial (weight-bearing) tibial articular cartilage (n=3).15 16
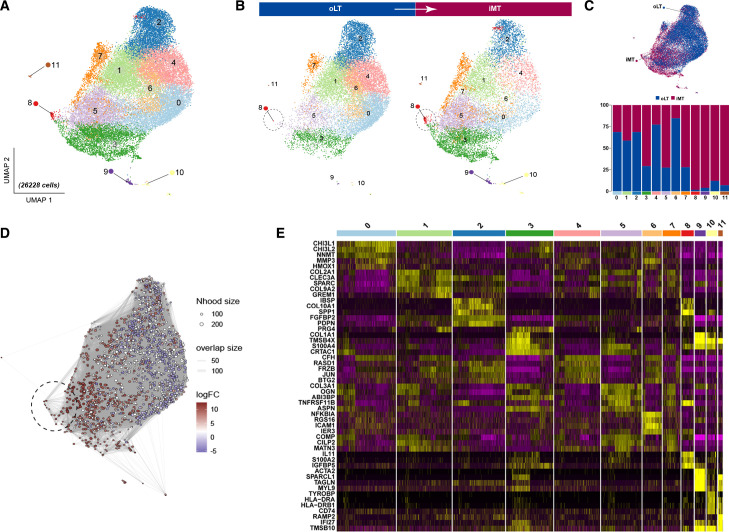
UMAP analysis revealed 12 discrete subsets of chondrocytes, designated as clusters 0–11(figure 2A), exhibiting evident distribution disparities between weight-bearing and non-weight-bearing cartilage sites (figure 2B). Chondrocytes isolated from weight-bearing sites were mainly observed in clusters 3, 5, 7, 8, 9, 10 and 11. In contrast, chondrocytes obtained from non-weight-bearing regions were predominantly observed in clusters 0, 1, 2, 4 and 6 (figure 2C). In particular, cluster 8 (black circle) appears to emerge in the iMT region as a consequence of changes in weight-bearing conditions. Through Milo analysis,20 we further discerned that in comparison to the non-weight-bearing region represented by oLT, there is a significant abundance alteration within the region of cluster 8(figure 2D). This suggests that cluster 8 represents a population of disease-specific cells under weight-bearing conditions. Considering that each cluster exhibited distinct gene expression patterns and marker genes(figure 2E). Based on an extensive review of high-quality literature (online supplemental table 1),13 24–68 we have summarised the widely accepted and recognised types of chondrocyte subpopulations and their corresponding marker genes: hypertrophic chondrocytes (HCs) (COL10A1, RUNX2, SPP1), endothelial cells (ECs) (CD93, CDH5), fibrochondrocytes (FCs) (COL1A1, COL3A1, COL6A1), stable chondrocytes (SCs) (COL2A1, COL6A1, COL9A1) and chondrogenic progenitor cells (CPCs) (CDK1, BIRC5).
Figure 2.
Single-cell sequencing of human knee articular cartilage tissues from weight-bearing and non-weight-bearing regions. (A) The single-cell RNA sequencing (scRNAseq) of chondrocytes was visualised using Uniform Manifold Approximation and Projection (UMAP) plots, which demonstrated 12 distinct chondrocyte subsets. A total of n=3 patients with OA were included in the study, and chondrocytes from both weight-bearing (iMT) and non-weight-bearing (oLT) cartilage patient-matched sites were subjected to scRNAseq analysis. (B) Identification of a disparate population (black circle) in OA cartilage undergoing transitions from non-weight-bearing to weight-bearing states, visualised through UMAP plot split by weight-bearing conditions (oLT vs iMT). (C) UMAP plot illustrates the distribution of chondrocyte subsets under different weight-bearing conditions (upper panel). Percentage distribution of the 12 different subsets according to either weight-bearing (iMT) or non-weight-bearing (oLT) sites (lower panel). (D) Milo analysis reveals a significant upregulation in the abundance of cell neighbourhoods in the disparate subset (black circle) compared with other subpopulations. (E) Heatmap showing the z-score average gene signature expression of the top 5 most differentially expressed genes within each of the 12 chondrocytes clusters.
rmdopen-2023-003255supp001.xlsx (14.5KB, xlsx)
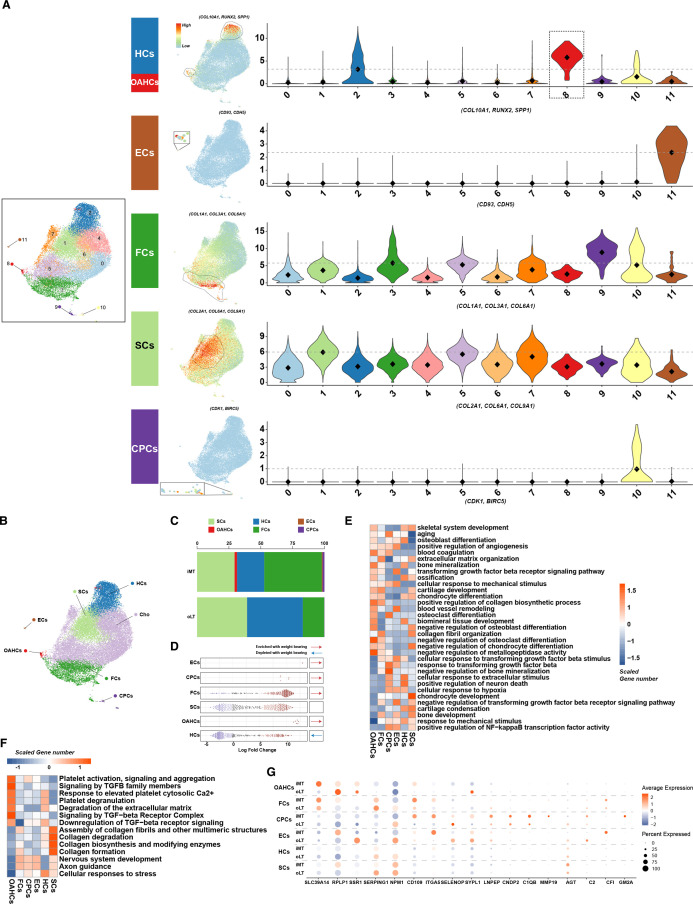
On visualising the expression levels of these marker genes using UMAP and violin plots (figure 3A), we observed the following: both clusters 2 and 8 exhibit high expression level of HC markers. Considering the Milo analysis that suggested cluster 8 as a disease-specific cell population, we have identified cluster 8 as ‘osteoarthritis hypertrophic chondrocytes’ (OAHCs) and cluster 2 as HCs. Furthermore, clusters 10 and 11 can be distinctly identified as CPCs and ECs, respectively. In light of the high expression levels of marker genes within clusters 1, 3 and 9, coupled with the concentrated distribution of these highly expressing cells, we have, respectively, labelled them as SCs or FCs. Due to the lack of distinct features exhibited by clusters 0, 4, 5, 6 and 7 during the visualisation of marker genes, we have provisionally labelled these cells as ‘Cho’ to facilitate streamlined analysis. After identifying and annotating the chondrocyte subtypes (figure 3B), we compared the differences in the proportion of chondrocyte subtypes between weight-bearing and non-weight-bearing cartilage tissues. It was observed that the proportion of SCs and HCs decreased, while that of FCs increased in weight-bearing cartilage tissues. In addition, OAHCs appeared, and the proportion of CPCs increased after activation. ECs also emerged in weight-bearing cartilage tissues (figure 3C). Additional Milo analysis further reveals that the alterations in abundance of these chondrocyte subpopulations during the transition to weight-bearing states align consistently with changes in their proportions (figure 3D).
Figure 3.
Single-cell RNA sequencing identifies distinct chondrocyte subsets in the cartilage from weight-bearing (iMT) and non-weight-bearing (oLT) sites in patients with osteoarthritis (OA). (A) Visualisation of chondrocytes subsets by Uniform Manifold Approximation and Projection (UMAP) plot (left panel). Feature plots show the expression distribution of subset-specific markers for each of the seven chondrocyte subsets (clusters 1–3, 8–11) on the UMAP plot (middle panel). Violin plots showing the expression levels of these markers, the grey dashed line highlights a comparatively higher average expression level relative to other subsets (right panel). (B) Visualisation of the spatial distribution of annotated chondrocyte subsets on the UMAP plot. (C) The percentage distribution of stable chondrocytes, hypertrophic chondrocytes, endothelial cells, osteoarthritis hypertrophic chondrocytes (OAHCs), fibrochondrocytes, and chondrogenic progenitor cells in cartilage tissues under different weight-bearing conditions. (D) Milo analysis reveals a pronounced enriched associated with weight-bearing conditions in the abundance of cell neighbourhoods in the OAHCs compared with other chondrocytes. (E) Heatmap of biological process significantly associated with the transcriptome of each of the six annotated chondrocytes subsets as identified using DAVID functional annotation tool, and coloured by scaled gene number. (F) Heatmap of significant pathways for each of the six chondrocytes subsets as identified using Reactome Pathway Database, and coloured by scaled gene number. (G) DotPlot depicts the expression pattern of differentially expressed proteins identified through Tandem Mass Tag proteomics analysis in each of the annotated chondrocyte subsets. The size of the circles represents the proportion of protein expression in the corresponding subset, while the colour indicates the average expression level.
Apart from these alterations, we assessed the functional enrichment of characteristic genes in each subtype. Remarkably, despite the relatively small number of OAHCs, they displayed more characteristic genes enriched in biological processes like ‘skeletal system development’, ‘cartilage development’ and ‘chondrocyte differentiation’, which were also enriched in the proteomics analysis of weight-bearing and non-weight-bearing cartilage tissue (figure 3E). These findings all suggest that OAHCs may modulate protein-level biological functions under mechanical stimulation from weight bearing. Through pathway enrichment analysis of each chondrocyte subpopulation, we found that OAHCs have more characteristic genes responsive to the TGF-β signalling pathway compared with other chondrocyte subpopulations (figure 3F). This suggests that despite the prediction from the proteomics analysis that TGFB1, an upstream regulatory factor, is inhibited and downregulated, OAHCs are still able to exhibit a more sensitive response to TGF-β-related signalling pathways compared with other chondrocyte subpopulations. In addition, by comparing the transcription levels of RNAs corresponding to the differentially expressed proteins identified in the proteomics analysis across each cell subtype, we found that OAHCs better reproduced the differences between weight-bearing and non-weight-bearing cartilage tissue compared with the other cell subtypes (figure 3G).
Functional characteristics of OAHCs populations
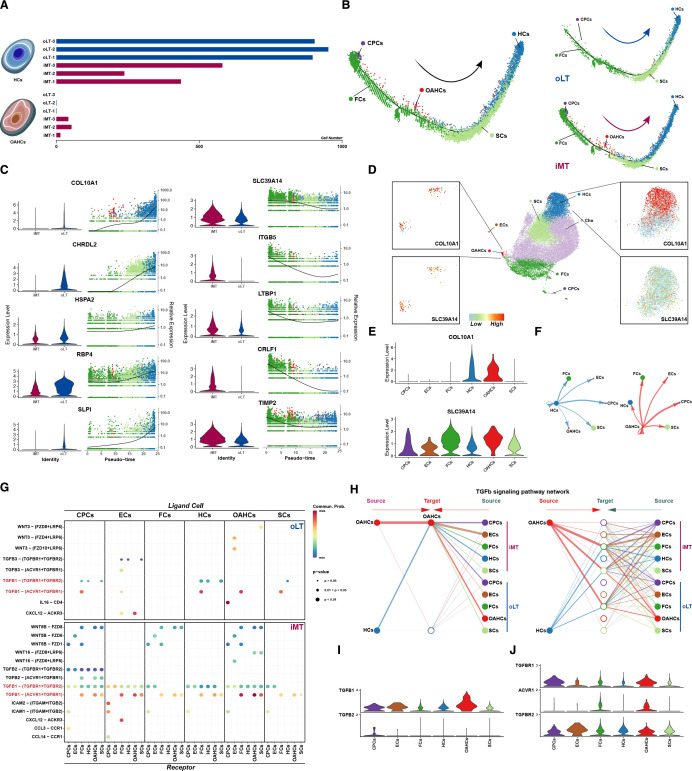
To further investigate the differences between OAHCs and HCs, we calculated the proportion of these two subtypes in different samples. OAHCs were present in all three weight-bearing cartilage tissue samples, while the number of HCs in weight-bearing cartilage tissue was lower than that in their corresponding non-weight-bearing cartilage tissue (figure 4A). Subsequently, we employed Monocle to construct a pseudotime trajectory to investigate the transcriptional alterations of chondrocytes during their maturation and development, as well as throughout the progression of OA. We discovered that in the physiological state, the maturation trajectory of chondrocytes proceeds from CPCs to FCs, SCs and finally HCs, which is consistent with previous studies. However, in weight-bearing cartilage tissue subjected to more intense mechanical stress, the proportion of CPCs and FCs increased while the ratio of SCs and HCs decreased. Notably, a subset of cells exhibited a sudden differentiation pattern similar to that of ‘tumour cells’, deviating from the normal maturation trajectory, and these cells, namely OAHCs, may be a critical factor in the development and progression of OA (figure 4B). Through the analysis of pseudotemporal expression dynamics, we have identified COL10A1 and SLC39A14, which are marker genes of OAHCs, as potential key regulatory genes involved in inducing the fate of differentiation in chondrocytes. When both COL10A1 and SLC39A14 are highly expressed, OAHCs are present. In contrast, when SLC39A14 expression is reduced and COL10A1 expression is further increased, normal chondrocyte hypertrophy occurs (figure 4C). Based on the aforementioned results, as well as the expression levels of COL10A1 and SLC39A14 as displayed in the UMAP and violin plots (figure 4D,E), we conclude that chondrocytes expressing both COL10A1 and SLC39A14 can be accurately identified as OAHCs.
Figure 4.
Transcriptional switch and ligand–receptor analysis of hypertrophic phenotypes associated with osteoarthritis (OA) disease progression to evaluate the crosstalk between hypertrophic chondrocytes and other chondrocyte types after weight bearing. (A) Comparison of the number of two hypertrophic chondrocyte subsets under different weight-bearing conditions. (B) Monocle pseudotime trajectory of the transcriptome phenotype transition of chondrocytes (left panel). The trajectory is divided into different weight-bearing conditions (right panel). Cells are ordered in pseudotime based on differentially expressed genes (q-value<0.01), coloured according to the chondrocyte types. (C) Expression dynamics of upregulated and downregulated genes, which serve as markers of osteoarthritis hypertrophic chondrocytes (OAHCs), were analysed over the progression of chondrocyte phenotype towards pseudotime, and were overlaid with cell distribution. Violin plots and feature plots showing the expression levels (y-axis) of the upregulated/downregulated genes for different weight-bearing conditions (x-axis). (D) Feature plots illustrate the expression of markers specific to OAHCs on the Uniform Manifold Approximation and Projection (UMAP) (identified by performing differential expression analysis on the non-parametric Wilcoxon rank sum test). (E) Violin plots below the UMAP display the expression levels (y-axis) of these markers across different chondrocyte cohorts (x-axis). (F Quantification of intercellular communication intensity between two subtypes of hypertrophic chondrocytes and other chondrocytes using Cellchat (thicker line segments representing higher intensity). (G) Dot plot visualise the interaction scores between chondrocyte subtypes, where each subtype was either the ligand-expressing cell (top axis) or the receptor-expressing cell (bottom axis). The specific ligand–receptor pairs were listed along the left axis. Size of the dot indicates -Log p value. Colour of the dot indicated the interaction score, with dark red dots representing stronger predicted interactions. (H) Hierarchical plot display the activation of the TGFβ signalling pathway in chondrocyte subtypes, with the left portion indicating the extent of signalling received by two hypertrophic chondrocyte subtypes and the right part indicating the signalling received by the other chondrocyte groups under different weight-bearing conditions. (I) Violin plots show the expression levels of TGFβ signalling pathway ligands TGFB1 and TGFB2 in each chondrocyte subsets. (J) Violin plots depict the distribution of expression levels of TGFβ signalling pathway receptors TGFBR1, AVCR1 and TGFBR2 in each annotated chondrocyte subset. The y-axis represents the expression values, and the x-axis represents the different chondrocyte subsets (same in (I)).
To gain deeper insights into the cellular interaction underlying the generation of OAHCs in response to mechanical stimulation in load-bearing cartilage, we used CellChat to predict the differential strength of cellular crosstalk and signalling between cell types in different load-bearing states. We found that compared with HCs, OAHCs showed higher interaction weights with all chondrocyte subpopulations, including themselves (figure 4F), indicating that even though OAHCs are present in low numbers, they may be regulating the state of various cell subpopulations in load-bearing cartilage through strong cellular crosstalk. On comparing the signal transduction interactions among diverse cell subpopulations, acting as ligand and receptor cells, we have discovered that the proportion of TGFB1-related signalling pathways is significantly escalated in the weight-bearing cartilage (figure 4G). Further examination of the communication strength of TGFβ signal transduction pathways in different cell subpopulations exposed to different weight-bearing conditions has demonstrated that OAHCs hold critical importance as both targets and sources of the TGFβ signal, particularly when compared with HCs (figure 4H). Additionally, the analysis of ligand and receptor transcription levels within the TGFβ signalling pathway has revealed that OAHCs demonstrate higher expression levels of TGFB1, TGFBR1, and ACVR1 than other cell subpopulations (figure 4I,J). Collectively, the data we have obtained offer a rational and temporal framework for how OAHCs respond to excessive mechanical stimulation under weight-bearing conditions by interacting with various cell types through the TGFβ pathway, emphasising the autocrine and paracrine functions of OAHCs in this process.
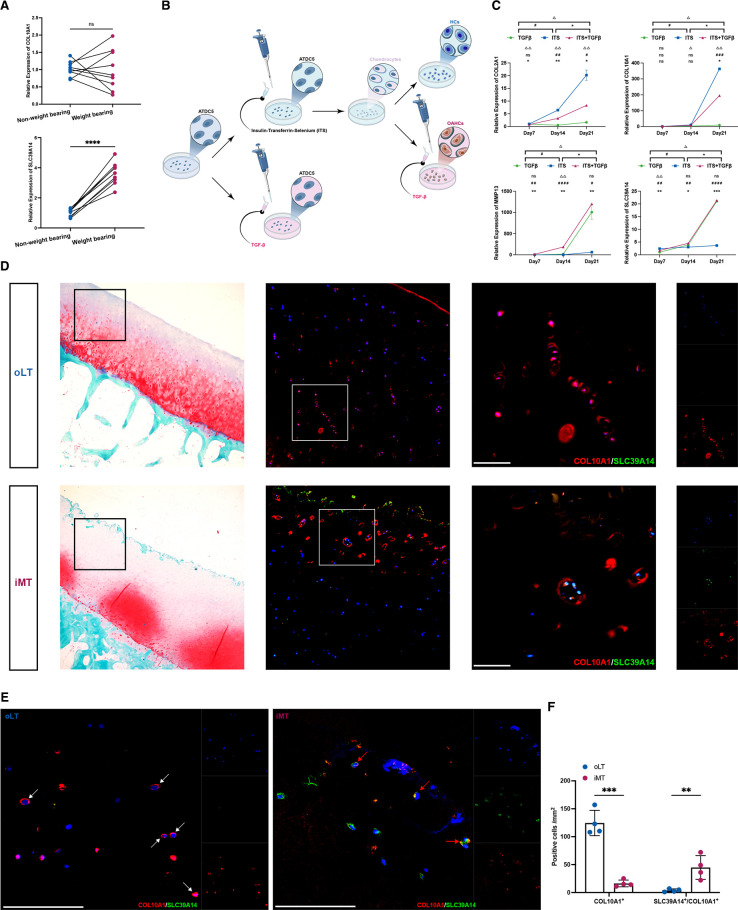
Induction of OAHCs in vitro and histological validation
To verify the distinctiveness of OAHCs from normal HCs, we initially compared the transcript levels of OAHCs marker genes, SLC39A14 and COL10A1, in the cartilage of the non-weight-bearing and the weight-bearing regions of the tibial plateau. Our findings were in line with the scRNAseq data and proteomic analysis, revealing an elevated expression level of SLC39A14 in the weight-bearing region (figure 5A). Subsequently, we used ATDC5, a chondrogenic cell line that undergoes a sequential process analogous to chondrocyte differentiation, to simulate the overloading-induced OAHCs production process via TGFβ stimulation. We monitored the transcript-level changes of ATDC5 cells at various time points during this process (figure 5B).
Figure 5.
Osteoarthritis hypertrophic chondrocytes (OAHCs) induced to differentiate by TGFβ stimulation, and these specific OAHCs, unlike typical hypertrophic chondrocytes, exist in the weight-bearing cartilage surface of joints. (A) Relative expression levels of OAHCs marker genes were assessed in either weight-bearing or non-weight-bearing cartilage tissues (nine knee patients with osteoarthritis (OA) with total knee arthroplasty (TKA)). ns=no significant difference, **** = p < 0.0001 (paired t-test). (B) The schematic diagram of the pattern for inducing the differentiation of OAHCs using the ATDC5 chondrogenesis cell line in vitro experimental conditions through insulin, transferrin, selenium (ITS) and TGFβ stimulation. (C) Relative expression levels of OA-related genes and OAHCs marker genes were assessed in ATDC5 cells differentiated with TGFβ, ITS, or ITS and TGFβ stimulation for 7, 14 or 21 days. Significant differences between OAHCs and HCs differentiated by ATDC5 were determined by repeated measures analysis of variance (ANOVA) followed by Sidak test, with significance levels denoted as follows: ns=no significant difference, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001. (D) Histological validation of the tibial plateau cartilage from patients with OA was performed. Non-weight-bearing (oLT) and weight-bearing (iMT) tibial plateau cartilage sections were stained with safranin o-fast green and immunohistochemically labelled for SLC39A14 (green) to identify OAHCs and for COL10A1 (red) to label HCs that overlie the hypertrophic site. Captured using fluorescence microscopy, scale bars, 100 µm. (E) Immunohistological (COL10A1 in red, SLC39A14 in green) validation of HCs and OAHCs in response to weight bearing. White arrowheads denote COL10A1+ HCs, and red arrowheads denote SLC39A14+/COL10A1+ OAHCs. DAPI is in blue labelling cell nuclei. Captured using confocal laser scanning microscope, scale bars, 100 µm. (F) Quantification of positive hypertrophic chondrocytes numbers within the cartilage tissues of oLT and iMT (four patients with knee OA with TKA), ** = p < 0.01, ∗∗∗p<0.001 (two-way ANOVA with Bonferroni’s multiple comparisons test).
Initially, under standard induction by insulin-transferrin-selenium, ATDC5 cells exhibited high expression of COL2A1, a chondrocyte-specific marker, at day 14. As time elapsed, the expression levels of COL2A1 steadily escalated to 21 days. Notably, concomitant with this temporal progression, the expression of COL10A1, indicative of HCs, also exhibited a substantial increase, signifying the advancing differentiation of cells into the hypertrophic phenotype. On stimulation with TGF-β cytokine on this basis, at day 14, phenotypes related to OA appeared, such as a decrease in COL2A1 expression and an increase in MMP13 expression. At day 21, the expression level of SLC39A14 was elevated, indicating the emergence of the OAHCs. Notably, the application of TGF-β alone was insufficient to induce the differentiation of ATDC5 cells into chondrocytes, as demonstrated by our control experiments (figure 5C).
To validate the presence of OAHCs subtypes in vivo, we employed immunohistochemistry with antibodies specific targeting to SLC39A14 and COL10A1, enabling us to spatially validate our findings. We observed a considerable number of HCs in the deeper layers of non-weight-bearing articular cartilage, consistent with previous research. However, in regions of weight-bearing cartilage adjacent to the injury site, we identified the presence of OAHCs positive for both SLC39A14 and COL10A1 (figure 5D). Given the limitations of fluorescent microscopy in precisely localising the distribution of both marker proteins, we employed laser confocal microscopy for a more comprehensive examination. The results revealed that, unlike normal HCs where COL10A1 uniformly envelops the cell, in OAHCs, COL10A1 and SLC39A14 are dispersed around the cell periphery (figure 5E). We also quantified the number of positive cells within cartilage tissues under different weight-bearing conditions, confirming the augmentation of SLC39A14+/COL10A1+ OAHCs in weight-bearing cartilage tissues (figure 5F). Hence, both the in vitro induction and the immunohistochemical data obtained from human articular cartilage provide compelling evidence for the molecular identification of OAHC subtypes and their temporal progression following excessive mechanical loading stimuli on the weight-bearing surface. In summary, the analysis of chondrocytes has uncovered a novel and pivotal subtype of OAHCs that arises during the progression of OA due to excessive mechanical loading.
Transcriptional conversion mechanism of OAHCs phenotype under weight-bearing conditions
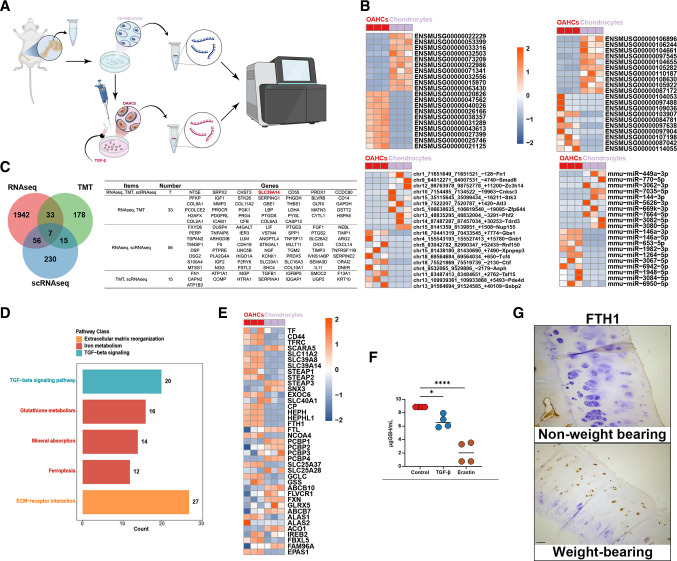
To explore the precise mechanism underlying transcriptional conversion in OAHCs under weight-bearing conditions, we used TGFβ to directly stimulate primary mouse chondrocytes and mimic the chondrocytes receiving excessive mechanical stimuli under weight-bearing conditions. We then performed and analysed whole-transcriptome sequencing on the stimulated cells and compared them with untreated primary chondrocytes (figure 6A). A total of 2222 genes were found to be differentially expressed (> 1.5 fold change, p<0.05) in OAHCs, of which 1252 were upregulated and 970 were downregulated. These genes comprised of 1186 protein coding genes, 66 lncRNAs, 752 circular RNAs, and 83 micro RNAs. The top 10 most significant upregulated and downregulated genes are presented in the heatmap (figure 6B). Seven genes were identified to have significant differential expression in both transcriptome sequencing and single-cell sequencing results, which were also validated at the protein level through TMT analysis. Among these genes is the marker gene SLC39A14 for the identified OAHCs, and its expression trend is completely consistent (figure 6C).
Figure 6.
Functional characteristics of TGFβ-induced osteoarthritis hypertrophic chondrocytes (OAHCs) contributes to the progression of osteoarthritis (OA). (A) Schematic workflow illustrat the experimental strategy outlines the sequential steps involved in RNA sequencing of TGFβ-induced differentiated OAHCs from primary mouse chondrocytes. (B) Heatmap showing the top 10 upregulated/downregulated RNAs, long non-coding RNAs, circRNAs, and mircroRNAs that are differentially expressed in OAHCs and chondrocytes. Colour of tile indicates z-scored expression. (C) Venn diagram display the significantly different genes identified through RNA sequencing between OAHCs and chondrocytes, the significantly different proteins identified in non-weight-bearing and weight-bearing cartilage tissues through Tandem Mass Tag, as well as the overlap and total number of characteristic marker genes of OAHCs identified through single-cell RNA sequencing. Genes are listed in the left-hand table. (D) Significant pathways enriched by differential genes, categorised by colour, with bar graphs representing the number of enriched proteins in each pathway. (E) Heatmap representing the differential expression of all genes related to iron metabolism pathway. Colour of tile indicates z-scored expression. (F) The levels of total glutathione in primary mouse chondrocytes stimulated with TGFβ or erastin (ferroptosis inducer) (n=4). * = p < 0.05, **** = p < 0.0001 (one-way analysis of variance). (G) Immunohistochemical staining on cartilage tissue sections of non-weight-bearing and weight-bearing facet joints from scoliosis patients to investigate the expression of FTH1, a major iron storage protein, in chondrocytes. Scale bars, 100 µm.
The functional pathway enrichment analysis of differentially expressed genes encoding proteins demonstrated the significant association of the ‘TGF-beta signalling pathway’ as expected. Additionally, we observed that pathways associated with iron metabolism, such as ‘Glutathione metabolism’, ‘Mineral absorption’ and ‘Ferroptosis’ also displayed significant association (figure 6D). Given that SLC39A14 has been widely recognised as an important iron ion transport protein related to iron metabolism, we thoroughly examined all genes related to iron metabolism (including iron uptake, transport, storage and export) and found that the majority of them displayed significant differences (figure 6E). Consequently, we validated the changes in glutathione levels following TGFβ stimulation of chondrocytes. Correspondingly to erastin-induced chondrocyte ferroptosis, we observed a decrease in the level of glutathione in chondrocytes (figure 6F). Furthermore, in order to corroborate the presence of abnormal iron metabolism in weight-bearing cartilage tissues beyond the knee joint, we employed immunohistochemical sections of facet joint cartilage from weight-bearing and non-weight-bearing joints of patients with scoliosis to scrutinise the expression of FTH1, the sole protein that stores iron ions in chondrocytes. The findings indicated a significant increase in FTH1 content in weight-bearing joints, particularly in the superficial layer of cartilage tissue (figure 6G). In summary, our analysis emphasises the utility of our whole transcriptome sequencing data in identifying signalling mechanisms underlying the presumed changes in the transcriptional profile of weight-bearing cartilage tissue. Iron metabolism can be hypothesised as an important mechanistic factor mediating the production of OAHCs in weight-bearing cartilage tissue and the progression of OA.
Discussion
This study represents the first identification and validation of a novel subset of OAHCs in weight-bearing OA cartilage tissues, which are distinguished from normal HCs. The study further investigates the transcriptional conversion and potential mechanisms associated with this subset. Additionally, this is the first to report on the phenotypic changes and intercellular communication of chondrocytes in OA cartilage tissue subjected to mechanical stimulation, specifically between weight-bearing and non-weight-bearing regions.
There are numerous risk factors for OA, such as age, female gender, obesity, genetic and environmental factors, and trauma. However, the underlying cause of irreversible cartilage tissue damage remains controversial.69 Our clinical data results indicate that patients with OA with altered force lines, particularly those clearly categorised as genu varum or genu valgum, exhibit more severe arthritis than patients with normal force lines. This is reflected in higher MOAKS scores and cartilage damage that is concentrated in the weight-bearing region. We have also noted the relatively higher number of female patients with OA included in our study. We speculate that factors contributing to their susceptibility include a higher prevalence of genu varum, greater peak joint power in females and a higher-paced walking pattern compared with males.70–73 These factors collectively result in increased mechanical loading on female joint cartilage. These results also highlight the significance of weight bearing and the overload mechanical stimulation it causes when assessing the severity of OA in patients. Our analysis of cartilage tissue on weight-bearing and non-weight-bearing surfaces in patients with OA provides the first evidence linking proteomic differences in cartilage tissue to cartilage damage on the weight-bearing surface of the knee. The findings indicate that various upstream regulators and pathways, including TNF, IL1B, TGFB1 and ferroptosis, are significantly enriched in the cartilage tissue of weight-bearing surfaces in patients.
Through the analysis of single-cell sequencing data, we delved deeper into the cellular-level disparities between weight-bearing and non-weight-bearing cartilage tissue. As single-cell technology continues to advance, new sequencing data and subpopulation classifications are constantly emerging. In our preceding investigation,23 we annotated chondrocyte subpopulations based on the enriched biological processes and pathways. However, a uniform and rigorous standard has yet to be established. To tackle this issue, we thoroughly reviewed a multitude of high-quality literature and identified the most meticulous marker genes for each subpopulation of chondrocytes. For subpopulations of chondrocytes without well-defined marker genes, additional research is needed to clarify their identities. Overall, our study unveiled the existence of six unique subsets of chondrocytes, each displaying conspicuous differences in their predominance under differing weight-bearing conditions. Moreover, we ascertained that the proteomic differences observed in weight-bearing cartilage tissue of knee OA extend to the cellular level of OA chondrocytes.
The destruction of cartilage is the primary outcome of OA, and chondrocytes that have been injured endeavour to mend themselves through a process that resembles their growth and maturation. As previous studies have revealed, initially, CPCs situated in the superficial layer become activated and proliferate to fill the area with FCs.74 75 However, only a small fraction of FCs have the ability to differentiate into SCs and migrate towards the deeper layers of cartilage tissue.76 As the SCs mature, they undergo a transformation into HCs that are typically found at the tidemark of the cartilage tissue.77 The ultimate fate of these HCs is to undergo gradual calcification and deposition.78 Our pseudotime analysis aligns and accurately reproduces this process. Nonetheless, an important issue that has long vexed the field is whether the normal HCs and the HCs involved in cartilage repair after OA are the same cell type.79–82 By performing further analysis of the pseudotime trajectories of chondrocytes under various weight-bearing conditions, we have discovered that the OAHCs, a type of HCs that only manifests in weight-bearing OA cartilage tissue, appear directly in the early phases of chondrocyte development and do not follow the normal maturation path. Combined with the proteomics differences that are most representative of weight-bearing cartilage tissue, as well as the expression of a greater number of genes that respond to the TGFβ signalling pathway, we believe that the OAHCs that appear in the cartilage repair process following OA are indeed a novel subset of cells that differ from normal HCs. Based on these findings, we have ascertained marker genes that can effectively label OAHCs: COL10A1 and SLC39A14 are both highly expressed. This has been validated through both in vitro and in vivo experiments, demonstrating successful identification of this subset of cells.
It is noteworthy that in our study, TGFB1 was predicted as an upstream regulator that is inhibited at the protein level. However, cell communication analysis of OAHCs revealed that the TGFβ signalling pathway is highly important and active. Extensive research has been conducted on the pathophysiology of TGFβ in OA.83 In healthy cartilage, TGFβ stimulates ECM protein synthesis as a metabolic factor and plays a protective role in mechanically loaded cartilage tissue in healthy joints.84 Active TGFβ1 encoded by TGFB1 and subsequent downstream signalling can prevent chondrocyte hypertrophy.85 86 The expression of TGFβ decreases with age, which indeed exposes chondrocytes to ECM degradation by matrix metalloproteinases.87 88 However, there is also evidence that high concentrations of TGFβ combined with altered TGFβ receptor expression lead to chondrocyte hypertrophy.88 89 Facing this pleiotropic cytokine, TGFβ, we have found that OAHCs effectively explain this dramatically different role. TGFβ can indeed prevent chondrocyte hypertrophy and differentiation in normal chondrocytes, but due to the sensitive response of OAHCs to the TGFβ signalling pathway in weight-bearing cartilage tissue, even minimal stimulation by active TGFβ induces their hypertrophic differentiation and the development of the OA phenotype. Specifically, when abnormal mechanical loading in weight-bearing cartilage tissue results in the release of TGFβ ligands, although it is an effective protective signal for normal chondrocytes and a large number of binding sites make it inactive,88 it still results in the appearance of OAHCs, which further mediate destructive processes in weight-bearing cartilage tissue. This represents a groundbreaking advancement in our understanding of the complex role of the TGFβ signalling pathway in OA.
Regarding the role of OAHCs in OA disease progression, we have observed that abnormal iron metabolism may be a significant contributing factor. Elevated expression of iron uptake-related genes, including the OAHCs marker gene SLC39A14, suggests that this group of chondrocytes may be susceptible to ferroptosis, as indicated by reduced glutathione content and increased ferritin levels in loaded cartilage tissue. Recent important studies have also highlighted the critical role of ferroptosis in OA pathogenesis.90 91 It is conceivable that the mechanical overload caused by the invasion of vascularised tissues, such as synovium and bone, into the avascular cartilage tissue may create an abnormal iron concentration environment. This is also one of the important pathological driving factors of known haemophilic arthropathy.92 Therefore, these findings suggest that abnormal iron metabolism may mediate OA disease progression in OAHCs. Further mechanistic studies are required to validate the role of ferroptosis in OAHCs in the future.
In conclusion, we identified and validated the existence of a subset of OAHCs in the weight-bearing area of OA cartilage tissue, which are distinct from HCs in normal cartilage. The presence of these cells explains the bidirectional effect of TGFβ signalling pathway in both normal and diseased cartilage tissue. Therefore, targeting OAHCs could provide a theoretical basis for delaying and treating OA under abnormal weight-bearing conditions.
rmdopen-2023-003255supp003.pdf (59.9KB, pdf)
Footnotes
Twitter: @DiJarvan
Contributors: JDi and ZC performed the research. JDi, ZC, TH, DW and LM performed the experiments and bioinformatics analysis. JDi, ZW, CW and JDeng collected and processed the samples. JDi and ZC wrote the manuscript. LR, LH and KW provided the several suggestions for manuscript revision. All authors read and approved the final manuscript. LR acts as the guarantor of the study.
Funding: This study was supported by the National Natural Science Foundation of China (Grant Number: 82102576), Basic and Applied Basic Research Foundation of Guangzhou City, China (Grant Number: 202201011001; 202201011036), Medical Science and Technology Research Foundation of Guangdong Province, China (Grant Number: A2021330), Basic and Applied Basic Research Foundation of Guangdong Province, China (Grant Number: 2021A1515111181).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information. Data are available in a public, open access repository. The single-cell RNA-seq data are available at the NCBI’s Gene Expression Omnibus (GEO) data repository with the accession ID GSE152805. All the reagents and experimental protocols detailed in this study are accessible upon request that is deemed reasonable.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by the Third Affiliated Hospital of Sun Yat-sen University (SL-RG2023-090-01). Participants gave informed consent to participate in the study before taking part.
References
- 1.Sharma L. Osteoarthritis of the knee. N Engl J Med 2021;384:51–9. 10.1056/NEJMcp1903768 [DOI] [PubMed] [Google Scholar]
- 2.Yue L, Berman J. What is osteoarthritis JAMA 2022;327:1300. 10.1001/jama.2022.1980 [DOI] [PubMed] [Google Scholar]
- 3.McCormack DJ, Puttock D, Godsiff SP. Medial compartment osteoarthritis of the knee: a review of surgical options. EFORT Open Rev 2021;6:113–7. 10.1302/2058-5241.6.200102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T-R, Wang H-D, Chen W, et al. Proximal fibular osteotomy alleviates medial compartment knee osteoarthritis in a mouse model. Int Orthop 2020;44:1107–13. 10.1007/s00264-020-04497-x [DOI] [PubMed] [Google Scholar]
- 5.Amis AA. Biomechanics of high tibial osteotomy. Knee Surg Sports Traumatol Arthrosc 2013;21:197–205. 10.1007/s00167-012-2122-3 [DOI] [PubMed] [Google Scholar]
- 6.Vincent TL, Wann AKT. Mechanoadaptation: articular cartilage through thick and thin. J Physiol 2019;597:1271–81. 10.1113/JP275451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgkinson T, Kelly DC, Curtin CM, et al. Mechanosignalling in cartilage: an emerging target for the treatment of osteoarthritis. Nat Rev Rheumatol 2022;18:67–84. 10.1038/s41584-021-00724-w [DOI] [PubMed] [Google Scholar]
- 8.Schätti OR, Marková M, Torzilli PA, et al. Mechanical loading of cartilage explants with compression and sliding motion modulates gene expression of lubricin and Catabolic enzymes. Cartilage 2015;6:185–93. 10.1177/1947603515581680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert SJ, Blain EJ. Chapter 4 - cartilage Mechanobiology: how Chondrocytes respond to mechanical load. In: Verbruggen SW, ed. Mechanobiology in health and disease. Academic Press, 2018: 99–126. [Google Scholar]
- 10.Glasson SS, Askew R, Sheppard B, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 2005;434:644–8. 10.1038/nature03369 [DOI] [PubMed] [Google Scholar]
- 11.Ismail HM, Yamamoto K, Vincent TL, et al. Interleukin-1 acts via the JNK-2 signaling pathway to induce aggrecan degradation by human chondrocytes. Arthritis Rheumatol 2015;67:1826–36. 10.1002/art.39099 [DOI] [PubMed] [Google Scholar]
- 12.Hunter DJ, Sharma L, Skaife T. Alignment and osteoarthritis of the knee. J Bone Joint Surg Am 2009;91 Suppl 1:85–9. 10.2106/JBJS.H.01409 [DOI] [PubMed] [Google Scholar]
- 13.Ji Q, Zheng Y, Zhang G, et al. Single-cell RNA-Seq analysis reveals the progression of human osteoarthritis. Ann Rheum Dis 2019;78:100–10. 10.1136/annrheumdis-2017-212863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter DJ, Guermazi A, Lo GH, et al. Evolution of semi-quantitative whole joint assessment of knee OA: MOAKS (MRI osteoarthritis knee score). Osteoarthritis Cartilage 2011;19:990–1002. 10.1016/j.joca.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou C-H, Jain V, Gibson J, et al. Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. Sci Rep 2020;10:10868. 10.1038/s41598-020-67730-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraus VB. Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. NCBI GEO Database 2020. Available: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu X, Hill A, Packer J, et al. Single-cell mRNA quantification and differential analysis with census. Nat Methods 2017;14:309–15. 10.1038/nmeth.4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021;12:1088. 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dann E, Henderson NC, Teichmann SA, et al. Differential abundance testing on single-cell data using K-nearest neighbor graphs. Nat Biotechnol 2022;40:245–53. 10.1038/s41587-021-01033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haseeb A, Lefebvre V. Isolation of Mouse growth Plategrowth plates and Articular Chondrocytesarticular Chondrocytes for primary cultures. In: Haqqi TM, Lefebvre V, eds. Chondrocytes: methods and protocols. New York, NY: Springer US, 2021: 39–51. 10.1007/978-1-0716-1119-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 2022;50:W216–21. 10.1093/nar/gkac194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao H, Di J, Yin M, et al. Identification of chondrocyte subpopulations in osteoarthritis using single-cell sequencing analysis. Gene 2023;852. 10.1016/j.gene.2022.147063 [DOI] [PubMed] [Google Scholar]
- 24.Oláh T, Reinhard J, Gao L, et al. Topographic modeling of early human osteoarthritis in sheep. Sci Transl Med 2019;11:eaax6775. 10.1126/scitranslmed.aax6775 [DOI] [PubMed] [Google Scholar]
- 25.Rim YA, Nam Y, Ju JH. The role of chondrocyte hypertrophy and senescence in osteoarthritis initiation and progression. Int J Mol Sci 2020;21:2358. 10.3390/ijms21072358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu C-L, Dicks A, Steward N, et al. Single cell transcriptomic analysis of human pluripotent stem cell chondrogenesis. Nat Commun 2021;12:362. 10.1038/s41467-020-20598-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He X, Ohba S, Hojo H, et al. AP-1 family members act with Sox9 to promote chondrocyte hypertrophy. Development 2016;143:3012–23. 10.1242/dev.134502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Z, Lin J, Yan R, et al. Atlas of musculoskeletal stem cells with the soft and hard tissue Diff Erentiation architecture. Adv Sci (Weinh) 2020;7:2000938. 10.1002/advs.202000938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baryawno N, Przybylski D, Kowalczyk MS, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell 2019;177:1915–32. 10.1016/j.cell.2019.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samvelyan HJ, Madi K, Törnqvist AE, et al. Characterisation of growth plate dynamics in murine models of osteoart Hritis. Front Endocrinol (Lausanne) 2021;12:734988. 10.3389/fendo.2021.734988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dreier R, Opolka A, Grifka J, et al. Collagen IX-deficiency seriously compromises growth cartilage developm ent in mice. Matrix Biol 2008;27:319–29. 10.1016/j.matbio.2008.01.006 [DOI] [PubMed] [Google Scholar]
- 32.Cescon M, Gattazzo F, Chen P, et al. Collagen VI at a glance. J Cell Sci 2015;128:3525–31. 10.1242/jcs.169748 [DOI] [PubMed] [Google Scholar]
- 33.Di Martino A, Cescon M, D’Agostino C, et al. Collagen VI in the musculoskeletal system. Int J Mol Sci 2023;24:5095. 10.3390/ijms24065095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brachvogel B, Zaucke F, Dave K, et al. Comparative proteomic analysis of normal and collagen IX null mouse cartilage reveals altered extracellular matrix composition and novel components of the collagen IX Interactome. J Biol Chem 2013;288:13481–92. 10.1074/jbc.M112.444810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Ning Y, Zhang P, et al. Comparison of the major cell populations among osteoarthritis, Kashin-Beck disease and healthy chondrocytes by single-cell RNA-Seq analysis. Cell Death Dis 2021;12:551. 10.1038/s41419-021-03832-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexopoulos LG, Youn I, Bonaldo P, et al. Developmental and osteoarthritic changes in Col6A1-knockout mice: biomechanics of type VI collagen in the cartilage pericellular matrix. Arthritis Rheum 2009;60:771–9. 10.1002/art.24293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbera S, Raucci L, Tassone G, et al. Dimerization of the C-type lectin-like receptor Cd93 promotes its binding to multimerin-2 in endothelial cells. Int J Biol Macromol 2023;224:453–64. 10.1016/j.ijbiomac.2022.10.136 [DOI] [PubMed] [Google Scholar]
- 38.Horikawa O, Nakajima H, Kikuchi T, et al. Distribution of type VI collagen in chondrocyte microenvironment: study of chondrons isolated from human normal and degenerative articular cartilage and cultured chondrocytes. J Orthop Sci 2004;9:29–36. 10.1007/s00776-003-0737-4 [DOI] [PubMed] [Google Scholar]
- 39.Ono K, Hata K, Nakamura E, et al. DMRT2 promotes transition of endochondral bone formation by linking Sox9 and Runx2. Commun Biol 2021;4:326. 10.1038/s42003-021-01848-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chavkin NW, Genet G, Poulet M, et al. Endothelial cell cycle state determines propensity for arterial-venous fate. Nat Commun 2022;13:5891. 10.1038/s41467-022-33324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JB, Zhao Q, Nguyen T, et al. Environment-sensing aryl hydrocarbon receptor inhibits the chondrogenic fate of modulated smooth muscle cells in atherosclerotic lesions. Circulation 2020;142:575–90. 10.1161/CIRCULATIONAHA.120.045981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potluri T, Taylor MJ, Stulberg JJ, et al. An estrogen-sensitive fibroblast population drives abdominal muscle fibrosis in an inguinal hernia mouse model. JCI Insight 2022;7:e152011. 10.1172/jci.insight.152011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Upadhyay U, Kolla S, Chelluri LK. Extracellular matrix composition analysis of human articular cartilage for the development of organ-on-a-chip. Biochem Biophys Res Commun 2023;667:81–8. 10.1016/j.bbrc.2023.04.117 [DOI] [PubMed] [Google Scholar]
- 44.Shi Y, Liao X, Long JY, et al. Gli1+ progenitors mediate bone anabolic function of teriparatide via Hh and Igf signaling. Cell Rep 2021;36:109542. 10.1016/j.celrep.2021.109542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xi H, Langerman J, Sabri S, et al. A human skeletal muscle atlas identifies the trajectories of stem and progenitor cells across development and from human pluripotent stem cells. Cell Stem Cell 2020;27:181–5. 10.1016/j.stem.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu J, Lu Y, Li F, et al. Identification and characterization of the novel Col10A1 regulatory mechanism during Chondrocyte hypertrophic differentiation. Cell Death Dis 2014;5:e1469. 10.1038/cddis.2014.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellayr IH, Marklein RA, Lo Surdo JL, et al. Identification of predictive gene markers for multipotent stromal cell proliferation. Stem Cells Dev 2016;25:861–73. 10.1089/scd.2015.0374 [DOI] [PubMed] [Google Scholar]
- 48.Guo A, Zhang J, Tian Y, et al. Identify the immune characteristics and immunotherapy value of CD93 in the pan-cancer based on the public data SETS. Front Immunol 2022;13:907182. 10.3389/fimmu.2022.907182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito M, Mulati M, Talib SZA, et al. The indispensable role of Cyclin-dependent kinase 1 in skeletal development. Sci Rep 2016;6:20622. 10.1038/srep20622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang F, Rummukainen P, Pehkonen M, et al. Mesenchymal cell-derived WNT1 signaling regulates subchondral bone remodeling but has no effects on the development of growth plate or articular cartilage in mice. Bone 2022;163. 10.1016/j.bone.2022.116497 [DOI] [PubMed] [Google Scholar]
- 51.Melnik S, Gabler J, Dreher SI, et al. Mir-218 affects hypertrophic differentiation of human mesenchymal Stromal cells during Chondrogenesis via targeting RUNX2, MEF2C, and COL10A1. Stem Cell Res Ther 2020;11. 10.1186/s13287-020-02026-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomez-Camarillo MA, Kouri JB. Ontogeny of rat chondrocyte proliferation: studies in embryo, adult and osteoarthritic (OA) cartilage. Cell Res 2005;15:99–104. 10.1038/sj.cr.7290273 [DOI] [PubMed] [Google Scholar]
- 53.Pullig O, Weseloh G, Gauer S, et al. Osteopontin is expressed by adult human osteoarthritic chondrocytes: protein and mRNA analysis of normal and osteoarthritic cartilage. Matrix Biol 2000;19:245–55. 10.1016/s0945-053x(00)00068-8 [DOI] [PubMed] [Google Scholar]
- 54.Chang C-J, Lai Y-J, Tung Y-C, et al. Osteopontin mediation of disturbed flow-induced endothelial mesenchymal transition through CD44 is a novel mechanism of Neointimal hyperplasia in arteriovenous fistulae for hemodialysis access. Kidney Int 2023;103:702–18. 10.1016/j.kint.2022.12.022 [DOI] [PubMed] [Google Scholar]
- 55.Zhang P, Jimenez SA, Stokes DG. Regulation of human COL9A1 gene expression. activation of the proximal promoter region by Sox9. J Biol Chem 2003;278:117–23. 10.1074/jbc.M208049200 [DOI] [PubMed] [Google Scholar]
- 56.Chabronova A, van den Akker GGH, Housmans BAC, et al. Ribosomal RNA-based epitranscriptomic regulation of chondrocyte translation and proteome in osteoarthritis. Osteoarthritis and Cartilage 2023;31:374–85. 10.1016/j.joca.2022.12.010 [DOI] [PubMed] [Google Scholar]
- 57.Tossetta G, Piani F, Borghi C, et al. Role of CD93 in health and disease. Cells 2023;12:1778. 10.3390/cells12131778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin X, Jiang Q, Nagano K, et al. Runx2 is essential for the transdifferentiation of chondrocytes into osteoblasts. PLoS Genet 2020;16:e1009169. 10.1371/journal.pgen.1009169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoon DS, Kim E-J, Cho S, et al. Runx2 stabilization by long non-coding RNAs contributes to hypertrophi C changes in human chondrocytes. Int J Biol Sci 2023;19:13–33. 10.7150/ijbs.74895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guerrero-Juarez CF, Dedhia PH, Jin S, et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat Commun 2019;10:650. 10.1038/s41467-018-08247-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z, Ye D, Dai L, et al. Single-cell RNA sequencing reveals the difference in human normal and degenerative nucleus pulposus tissue profiles and cellular interactions. Front Cell Dev Biol 2022;10:910626. 10.3389/fcell.2022.910626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun H, Wen X, Li H, et al. Single-cell RNA-Seq analysis identifies meniscus progenitors and reveals the progression of meniscus degeneration. Ann Rheum Dis 2020;79:408–17. 10.1136/annrheumdis-2019-215926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dy P, Wang W, Bhattaram P, et al. Sox9 directs hypertrophic maturation and blocks Osteoblast differentiation of growth plate Chondrocytes. Dev Cell 2012;22:597–609. 10.1016/j.devcel.2011.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hattori T, Müller C, Gebhard S, et al. SOX9 is a major negative regulator of cartilage Vascularization, bone marrow formation and Endochondral ossification. Development 2010;137:901–11. 10.1242/dev.045203 [DOI] [PubMed] [Google Scholar]
- 65.Yuan Y, Loh Y-H, Han X, et al. Spatiotemporal cellular movement and fate decisions during first pharyngeal arch morphogenesis. Sci Adv 2020;6:eabb0119. 10.1126/sciadv.abb0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang C, Brisson BK, Terajima M, et al. Type III collagen is a key regulator of the collagen fibrillar structure and biomechanics of articular cartilage and meniscus. Matrix Biol 2020;85–86:47–67. 10.1016/j.matbio.2019.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Komori T. Whole aspect of Runx2 functions in skeletal development. Int J Mol Sci 2022;23:5776. 10.3390/ijms23105776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Houben A, Kostanova-Poliakova D, Weissenböck M, et al. Β-Catenin activity in late hypertrophic chondrocytes locally orchestra Tes osteoblastogenesis and osteoclastogenesis. Development 2016;143:3826–38. 10.1242/dev.137489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao Q, Wu X, Tao C, et al. Osteoarthritis: pathogenic signaling pathways and therapeutic targets. Signal Transduct Target Ther 2023;8:56. 10.1038/s41392-023-01330-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tschon M, Contartese D, Pagani S, et al. Gender and sex are key determinants in osteoarthritis not only confounding variables. A systematic review of clinical data. J Clin Med 2021;10:3178. 10.3390/jcm10143178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kerrigan DC, Todd MK, Della Croce U. Gender differences in joint biomechanics during walking: normative study in young adults. Am J Phys Med Rehabil 1998;77:2–7. 10.1097/00002060-199801000-00002 [DOI] [PubMed] [Google Scholar]
- 72.Peshkova M, Lychagin A, Lipina M, et al. Gender-related aspects in osteoarthritis development and progression: a review. Int J Mol Sci 2022;23:2767. 10.3390/ijms23052767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan Y-S, Qu Z, Yu D-Q, et al. Sex steroids and osteoarthritis: a Mendelian randomization study. Front Endocrinol 2021;12:683226. 10.3389/fendo.2021.683226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu W, Feng M, Xu P. From regeneration to osteoarthritis in the knee joint: the role shift of cartilage-derived progenitor cells. Front Cell Dev Biol 2022;10:1010818. 10.3389/fcell.2022.1010818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y-X, Zhao Z-D, Wang Q, et al. Biological potential alterations of migratory chondrogenic progenitor cells during knee osteoarthritic progression. Arthritis Res Ther 2020;22:62. 10.1186/s13075-020-2144-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Varela-Eirin M, Loureiro J, Fonseca E, et al. Cartilage regeneration and ageing: targeting cellular plasticity in osteoarthritis. Ageing Res Rev 2018;42:56–71. 10.1016/j.arr.2017.12.006 [DOI] [PubMed] [Google Scholar]
- 77.Madry H, van Dijk CN, Mueller-Gerbl M. The basic science of the subchondral bone. Knee Surg Sports Traumatol Arthrosc 2010;18:419–33. 10.1007/s00167-010-1054-z [DOI] [PubMed] [Google Scholar]
- 78.Salhotra A, Shah HN, Levi B, et al. Mechanisms of bone development and repair. Nat Rev Mol Cell Biol 2020;21:696–711. 10.1038/s41580-020-00279-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Du X, Cai L, Xie J, et al. The role of TGF-Beta3 in cartilage development and osteoarthritis. Bone Res 2023;11:2. 10.1038/s41413-022-00239-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li M, Yin H, Yan Z, et al. The immune microenvironment in cartilage injury and repair. Acta Biomaterialia 2022;140:23–42. 10.1016/j.actbio.2021.12.006 [DOI] [PubMed] [Google Scholar]
- 81.Lepage SIM, Robson N, Gilmore H, et al. Beyond cartilage repair: the role of the osteochondral unit in joint health and disease. Tissue Eng Part B Rev 2019;25:114–25. 10.1089/ten.TEB.2018.0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simkin PA. A biography of the chondrocyte. Ann Rheum Dis 2008;67:1064–8. 10.1136/ard.2007.084574 [DOI] [PubMed] [Google Scholar]
- 83.van der Kraan PM, van den Berg WB. Chondrocyte hypertrophy and osteoarthritis: role in initiation and progression of cartilage degeneration. Osteoarthritis Cartilage 2012;20:223–32. 10.1016/j.joca.2011.12.003 [DOI] [PubMed] [Google Scholar]
- 84.Zhen G, Guo Q, Li Y, et al. Mechanical stress determines the configuration of TGFβ activation in articular cartilage. Nat Commun 2021;12:1706. 10.1038/s41467-021-21948-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang C, Shen J, Ying J, et al. Foxo1 is a crucial mediator of TGF-Β/TAK1 signaling and protects against osteoarthritis by maintaining articular cartilage homeostasis. Proc Natl Acad Sci USA 2020;117:30488–97. 10.1073/pnas.2017056117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang X, Chen L, Xu X, et al. TGF-beta/SMAD3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol 2001;153:35–46. 10.1083/jcb.153.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pujol J-P, Chadjichristos C, Legendre F, et al. Interleukin-1 and transforming growth factor-beta 1 as crucial factors in osteoarthritic cartilage metabolism. Connect Tissue Res 2008;49:293–7. 10.1080/03008200802148355 [DOI] [PubMed] [Google Scholar]
- 88.van der Kraan PM. The changing role of TGFβ in healthy, ageing and osteoarthritic joints. Nat Rev Rheumatol 2017;13:155–63. 10.1038/nrrheum.2016.219 [DOI] [PubMed] [Google Scholar]
- 89.Thielen NGM, Neefjes M, Vitters EL, et al. Identification of transcription factors responsible for a transforming growth factor-Β-driven hypertrophy-like phenotype in human osteoarthritic chondrocytes. Cells 2022;11:1232. 10.3390/cells11071232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lv Z, Han J, Li J, et al. Single cell RNA-Seq analysis identifies ferroptotic chondrocyte cluster and reveals TRPV1 as an anti-ferroptotic target in osteoarthritis. EBioMedicine 2022;84:104258. 10.1016/j.ebiom.2022.104258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miao Y, Chen Y, Xue F, et al. Contribution of ferroptosis and GPX4’s dual functions to osteoarthritis progression. EBioMedicine 2022;76:103847. 10.1016/j.ebiom.2022.103847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun K, Guo Z, Hou L, et al. Iron homeostasis in arthropathies: from pathogenesis to therapeutic potential. Ageing Res Rev 2021;72:101481. 10.1016/j.arr.2021.101481 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
rmdopen-2023-003255supp002.xlsx (328.7KB, xlsx)
rmdopen-2023-003255supp001.xlsx (14.5KB, xlsx)
rmdopen-2023-003255supp003.pdf (59.9KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information. Data are available in a public, open access repository. The single-cell RNA-seq data are available at the NCBI’s Gene Expression Omnibus (GEO) data repository with the accession ID GSE152805. All the reagents and experimental protocols detailed in this study are accessible upon request that is deemed reasonable.