Abstract
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variants has led to concerns that ancestral SARS-CoV-2-based vaccines may not be effective against newly emerging Omicron subvariants. The concept of “imprinted immunity” suggests that individuals vaccinated with ancestral virus-based vaccines may not develop effective immunity against newly emerging Omicron subvariants, such as BQ.1.1 and XBB.1. In this study, we investigated this possibility using hamsters. Although natural infection induced effective antiviral immunity, breakthrough infections in hamsters with BQ.1.1 and XBB.1 Omicron subvariants after receiving the 3-dose mRNA-lipid nanoparticle vaccine resulted in only faintly induced humoral immunity, supporting the possibility of imprinted immunity.
Keywords: imprinted immunity, mRNA vaccine, omicron, SARS-CoV-2
The emergence of SARS-CoV-2 Omicron XBB subvariants raises vaccine effectiveness concerns. Our hamster model demonstrated that ancestral virus-based vaccines failed to produce effective neutralizing antibodies against Omicron subvariants (eg, BQ.1.1, XBB.1) during breakthrough infections, supporting the concept of imprinted immunity.
The coronavirus disease 2019 (COVID-19) pandemic has led to a global vaccination effort to protect against severe COVID-19 and further to possibly prevent SARS-CoV-2 infections. First-generation COVID-19 vaccines were developed based on the spike protein of the now-ancestral Wuhan-Hu-1 strain of SARS-CoV-2 (ie, monovalent vaccines) [1]. As of June 13, 2023, 70.1% of the world population has received at least 1 dose of COVID-19 vaccine [2].
During the COVID-19 pandemic, a variety of SARS-CoV-2 variants with multiple mutations, particularly in the spike gene, have emerged. The B.1.1 strain, which harbors the D614G substitution in the spike protein, predominantly spread worldwide in 2020 [3]. Subsequently, variants such as Alpha, Beta, Gamma, Delta, and Omicron (B.1.529 and BA lineages) emerged and spread around the world [4]. The Omicron variant has greatly diversified, and various Omicron subvariants, such as BA.2, BA.5, BQ.1.1, XBB.1, and XBB.1.5, have emerged one after another. More importantly, these subvariants have acquired greater immune escape ability than the ancestral B.1.1 strain and former variants [5–8]. The emergence of highly immune-evasive Omicron variants led to the development of bivalent vaccines (ie, a combination of ancestral virus-based and Omicron BA.1 or BA.4/5 based vaccines) to boost the immunity against newly emerging Omicron subvariants [9]. Nevertheless, booster shots with these bivalent vaccines tend to induce neutralizing antibodies (NAbs) against the ancestral SARS-CoV-2 strain rather than those against newer Omicron subvariants of concern [10]. Similarly, breakthrough infection (ie, natural infection after vaccination) of Omicron subvariants more strongly induces antiviral humoral immunity against the ancestral SARS-CoV-2 strain than the Omicron subvariant naturally infected [8]. These observations raise a possible concept called “imprinted immunity”—prior exposure to a primary antigen (eg, ancestral SARS-CoV-2 in this case) can attenuate the induction magnitude of immunity against certain secondary antigens (eg, Omicron subvariants) [9, 11–13]. However, to date, the immunological background of humans against SARS-CoV-2 has been highly diversified, because the vaccination status, history of natural infection, the SARS-CoV-2 variant infected, and the time after vaccination/natural infection in individuals vary significantly. Moreover, because most people have been vaccinated, the comparison of immune status between vaccinated and unvaccinated individuals after natural infection of certain SARS-CoV-2 variants of concern is challenging. Therefore, directly addressing the possibility of imprinting immunity by vaccination using human samples is technically difficult. In this study, we use an experimental hamster model to address the possibility of imprinting immunity by vaccination.
METHODS
Ethics Statement
All experiments with hamsters were performed in accordance with the Science Council of Japan's Guidelines for the Proper Conduct of Animal Experiments. The protocols were approved by the Institutional Animal Care and Use Committee of National University Corporation Hokkaido University (approval ID: 20-0060).
Cell Culture
HOS-ACE2/TMPRSS2 cells (HOS cells stably expressing human ACE2 and TMPRSS2) were maintained in Dulbecco’s modified Eagle’s medium ([DMEM] high glucose) (Sigma-Aldrich, Catalog Number 6429-500ML) containing 10% fetal bovine serum ([FBS] Sigma-Aldrich, Catalog Number 172012-500ML) and 1% penicillin–streptomycin ([PS] Sigma-Aldrich, Catalog Number P4333-100ML). VeroE6/TMPRSS2 cells (VeroE6 cells stably expressing human TMPRSS2; JCRB Cell Bank, JCRB1819) were maintained in DMEM (low glucose) (Wako, Catalog Number 041-29775) containing 10% FBS, G418 (1 mg/mL; Nacalai Tesque, Catalog Number G8168-10ML), and 1% PS.
mRNA Vaccination in Hamsters
Syrian hamsters (male, 4 weeks old) were purchased from Japan SLC Inc. (Shizuoka, Japan). Spikevax (0.2 mg/mL; Moderna) was thawed at room temperature in darkness and diluted in saline to vaccinate a hamster at 5 µg/50 µL per hamster intramuscularly. The vaccination was performed 3 times at a 28-day interval starting from 5 weeks old. All hamsters were monitored daily and weighed once a week. They did not show any clinical manifestations during the vaccination period.
Severe Acute Respiratory Syndrome Coronavirus 2 Infection in Hamsters
Animal experiments were performed as previously described, and sera for the unvaccinated hamster group were obtained from a previous study [5–7]. Syrian hamsters (male, 4 weeks old) were purchased from Japan SLC Inc. for the unvaccinated group. For the virus infection experiments, hamsters were anaesthetized by intramuscular injection of a mixture of 0.15 mg/kg medetomidine hydrochloride, 2.0 mg/kg alphaxaone (Alfaxanâ, Jurox), and 2.5 mg/kg butorphanol. The clinical isolates of SARS-CoV-2 B.1.1 (strain TKYE610670; GISAID ID: EPI_ISL_479681) [5], Omicron BA.2 (strain TY40-385; GISAID ID: EPI_ISL_9595859) [5], Omicron BQ.1.1 (strain TY41-796-P1; GISAID ID: EPI_ISL_15579783) [6], or Omicron XBB.1 (strain TY41-795; GISAID ID: EPI_ISL_15669344) [7] (10 000 50% tissue culture infective dose in 100 µL), or saline (100 µL) were intranasally inoculated under anesthesia. Sera of infected hamsters were collected at 16 days postinfection (d.p.i.). using cardiac puncture under anesthesia with isoflurane and used for neutralization assay.
Reverse-Transcription Quantitative Polymerase Chain Reaction
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR) (Figure 1B) was performed as previously described [5]. Briefly, 5-μL culture supernatant was mixed with 5 μl of 2× ribonucleic acid (RNA) lysis buffer [2% Triton X-100; Nacalai Tesque, Catalog Number 35501-15], 50 mM KCl, 100 mM Tris-HCl [pH 7.4], 40% glycerol, and 0.8 U/μL recombinant RNase inhibitor [Takara, Catalog Number 2313B]) and incubated at room temperature for 10 minutes. RNase-free water (90 μL) was added, and then a diluted sample (2.5 μL) was taken as the template for real-time RT-PCR (targeting nucleoprotein-coding plus-strand RNA) performed according to the manufacturer's protocol using One Step TB Green PrimeScript PLUS RT-PCR kit (Takara, Catalog Number RR096A) and the following primers: Forward N, 5′-AGC CTC TTC TCG TTC CTC ATC AC-3′; and Reverse N, 5′-CCG CCA TTG CCA GCC ATT C-3′. The viral RNA copy number was standardized with a SARS-CoV-2 direct detection RT-qPCR kit (Takara, Catalog Number RC300A). Fluorescent signals were acquired using a CFX Connect Real-Time PCR Detection system (Bio-Rad).
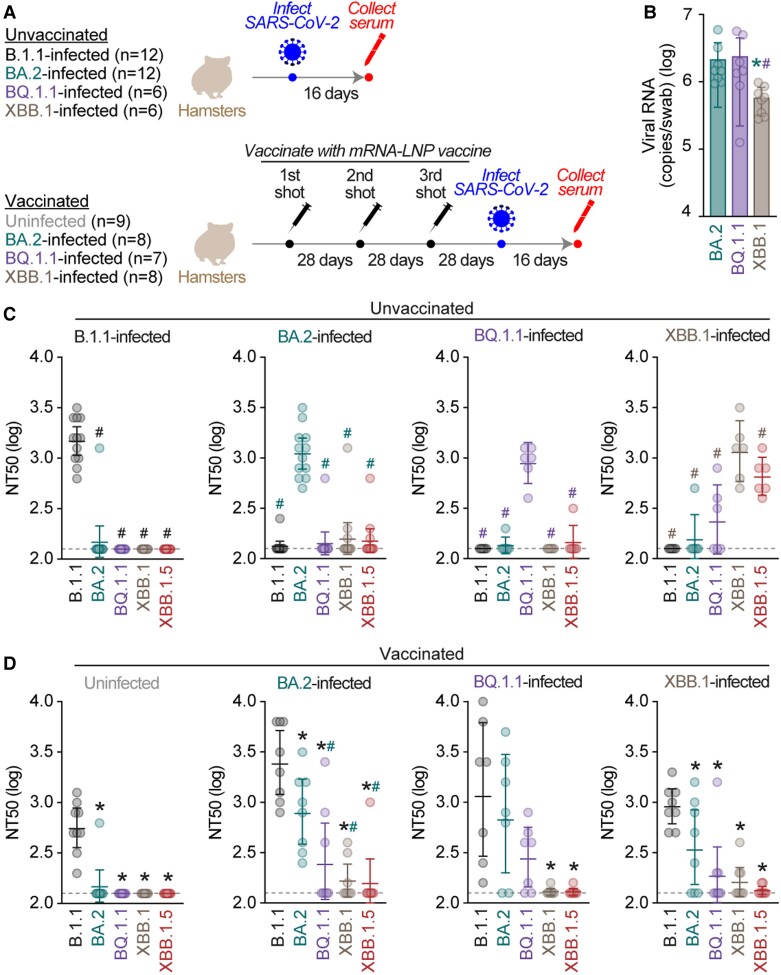
Figure 1.
Impact of immune imprinting by vaccination. (A) Schematic time course of animal experiment in this study. The number of hamsters in each group (N) is indicated in parenthesis. (B) Viral ribonucleic acid (RNA) loads in the oral swab. The copy numbers of viral RNA in the oral swab of breakthrough infection hamsters at 2 days postinfection were quantified by reverse-transcription quantitative polymerase chain reaction. The presented data are expressed as the average ± standard error of the mean, and each dot indicates the result of an individual hamster. Statistically significant differences versus BA.2 (*, P < .05) or BQ.1.1 (#, P < .05) were determined by two-sided Mann-Whitney U tests. (C and D) Neutralization assay. The assay was performed with pseudoviruses harboring the S proteins of B.1.1 (the D614G-bearing ancestral virus), BA.2, BQ.1.1, XBB.1, and XBB.1.5. The following sera were used: (C) the sera obtained from hamsters infected with B.1.1 (12 hamsters), BA.2 (12 hamsters), BQ.1.1 (6 hamsters), and XBB.1 (6 hamsters) without vaccination; (D) and the sera obtained from hamsters receiving 3-dose mRNA vaccination before severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, uninfected (ie, only vaccinated; 9 hamsters), and breakthrough-infected with BA.2 (8 hamsters), BQ.1.1 (7 hamsters), and XBB.1 (8 hamsters). Assays for each serum sample were performed in triplicate to determine the 50% neutralization titer (NT50). Each dot represents 1 NT50 value, and the geometric mean and 95% confidential interval are shown. Statistically significant differences versus B.1.1 (*P < .05) or infected variant (#P < .05) were determined by two-sided Wilcoxon signed-rank tests. The color of the asterisks indicates the variant compared. The horizontal dashed line indicates the detection limit (120-fold).
Neutralization Assay
Pseudoviruses were prepared as previously described [5]. Briefly, lentivirus (human immunodeficiency virus [HIV]-1)-based, luciferase-expressing reporter viruses were pseudotyped with SARS-CoV-2 S proteins. HEK293T cells (1 000 000 cells) were cotransfected with 1 μg psPAX2-IN/HiBiT, 1 μg pWPI-Luc2, and 500 ng of plasmids expressing parental S or its derivatives using PEI Max (Polysciences, Catalog Number 24765-1) according to the manufacturer's protocol. Two days posttransfection, the culture supernatants were harvested and centrifuged. The pseudoviruses were stored at −80°C until use.
The neutralization assay was prepared as previously described [5–8]. Briefly, the SARS-CoV-2 S pseudoviruses (counting ∼20 000 relative light units) were incubated with serially diluted (120-fold to 87 480-fold dilution at the final concentration) heat-inactivated sera at 37°C for 1 hour. Pseudoviruses without sera were included as controls. Then, a 40-μL mixture of pseudovirus and serum/antibody was added to HOS-ACE2/TMPRSS2 cells (10 000 cells/50 μL) in a 96-well white plate. At 2 d.p.i., the infected cells were lysed with a Bright-Glo luciferase assay system (Promega, Catalog Number E2650), and the luminescent signal was measured using a GloMax explorer multimode microplate reader 3500 (Promega).
RESULTS
Preparation of Hamster Antisera by Severe Acute Respiratory Syndrome Coronavirus 2 Infection With and Without Prior Vaccination
To investigate the effect of vaccination on the induction of NAbs after breakthrough infection, we prepared (1) 4 groups of unvaccinated Syrian hamsters (Figure 1A, top) and (2) 4 groups of hamsters that intramuscularly received a 3-dose vaccination with monovalent mRNA-LNP (Spikevax, Moderna) at a 28-day interval (Figure 1A, bottom). Then, hamsters were challenged with SARS-CoV-2 ancestral strain B.1.1, Omicron BA.2, Omicron BQ.1.1, or Omicron XBB.1. To verify successful breakthrough infection in vaccinated hamsters, the viral RNA copy number via oral swabs at 2 d.p.i. was measured by RT-qPCR. As shown in Figure 1B, the viral RNA loads in the oral swabs of the BA.2- and BQ.1.1-infected hamsters were comparable, whereas that of XBB.1-infected hamsters was significantly lower than that of other groups. These RNA loads indicate breakthrough SARS-CoV-2 infection was established in the vaccinated hamsters.
Evaluation of Antiviral Humoral Immunity Induced by Severe Acute Respiratory Syndrome Coronavirus 2 Infection With and Without Prior Vaccination
At 16 d.p.i., the sera were collected from infected hamsters (Figure 1A). We then performed neutralization assays using the sera and HIV-1-based pseudoviruses harboring the spike protein of B.1.1, BA.2, BQ.1.1, XBB.1, and XBB.1.5. The antisera obtained from unvaccinated hamsters exhibited neutralization activity against the variant infected, whereas antisera from hamsters infected with XBB.1 also cross-reacted with XBB.1.5 (Figure 1C). Consistent with our previous findings [5–7], natural SARS-CoV-2 infection efficiently induced antiviral humoral immunity against the variant infected.
In the case of hamster sera with a 3-dose vaccination (without SARS-CoV-2 infection), neutralization activity against the ancestral B.1.1 variant was observed (Figure 1D). However, these antisera did not exhibit neutralizing activity against other Omicron subvariants such as BA.2, BQ.1.1, XBB.1, and XBB.1.5 (Figure 1D). These observations mirror findings in humans [8, 12]. We then assessed whether the breakthrough infections of 3 Omicron subvariants (BA.2, BQ.1.1, and XBB.1) induced the neutralization activity against the variant infected, as observed in unvaccinated hamsters above. However, although anti-B.1.1 (ancestral SARS-CoV-2) neutralization activity was observed, the sera obtained from breakthrough infection hamsters did not exhibit prominent antiviral effects against the variant infected (Figure 1D). Although the 50% neutralization titer (NT50) of the sera obtained from BA.2 breakthrough infection hamsters against BA.2 was significantly lower than that against B.1.1 (4.3-fold), all sera obtained from 8 breakthrough infection hamsters with BA.2 exhibited anti-BA.2 activity (Figure 1D). However, the NT50 values of the sera obtained from BQ.1.1 and XBB.1 breakthrough infection hamsters against the variant infected were profoundly lower than that against B.1.1 (Figure 1D). Moreover, the NT50 values of the sera of 3 and 5 hamsters’ breakthrough infection with BQ.1.1 and XBB.1, respectively, were below detection limit (Figure 1D). These observations suggest that the induction of antiviral humoral immunity against a SARS-CoV-2 variant of infection is attenuated by comparatively ancestral vaccine inoculation before infection.
DISCUSSION
In this study, we investigated the impact of vaccination on the induction of humoral immunity against the SARS-CoV-2 variants infected using an experimental animal model and addressed the possibility of immune imprinting by vaccination. We showed that the hamsters infected with a variety of SARS-CoV-2 Omicron subvariants exhibited specific humoral immunity against the SARS-CoV-2 variant infected. On the other hand, the infection of SARS-CoV-2 Omicron subvariants, particularly BQ.1.1 and XBB.1, in the hamsters that received 3 doses of monovalent mRNA vaccine did not induce specific humoral immunity against the SARS-CoV-2 variant infected. Consistent with findings in human samples [8, 12], these results suggest that breakthrough infections tend to boost the humoral immunity against the vaccine strain (ie, ancestral SARS-CoV-2) rather than the variant infected after vaccination. In particular, although the immunogenicity of XBB.1 was comparable to those of the other variants (Figure 1C), vaccination before XBB.1 infection failed to effectively induce antiviral humoral immunity against XBB.1 (Figure 1D). Because the antigenicity of XBB.1 is prominently different from that of ancestral SARS-CoV-2 [7, 8, 12], imprinted immunity can be observed when variants of SARS-CoV-2 breakthrough infections have different immunogenicity from the vaccine strain.
In this study, we showed experimental data suggesting the existence of imprinted immunity by vaccination in hamsters. However, it should be noted that our results do not necessarily suggest the ineffectiveness of anti-SARS-CoV-2 mRNA vaccines to date. First, these findings are brought from experiments using an animal model, and the antibody repertoire induced by SARS-CoV-2 infection should differ between hamsters and humans. Therefore, it is unclear to what extent the results shown in this study can be extrapolated to humans. Second, we only analyzed monovalent vaccine effects, which is based on ancestral SARS-CoV-2, and therefore the impact of BA.1 or BA.5 bivalent vaccines remains unclear. Third, we only evaluated the possibility of immune imprinting on humoral immunity. In studies using human samples, it is evident that monovalent mRNA vaccines can induce efficient cellular immunity against multiple SARS-CoV-2 subvariants [14]. Therefore, mRNA vaccines can contribute to prevent COVID-19 severity. Fourth, although the time interval in this study is fixed (28-day interval for vaccination, 28-day interval for infection after the third dose of vaccine, and the serum collection at 16 d.p.i.), previous studies using human samples suggest the time interval between vaccination and breakthrough infection can impact the magnitude of humoral immunity [15, 16]. Therefore, the time interval between vaccinations and breakthrough infection after the last vaccination may modulate the magnitude of immune imprinting by vaccination.
CONCLUSIONS
In our study, experimental data using hamsters indicates the possibility of imprinted immunity as an effect of vaccination. With this in mind, the potential impact of imprinted immunity in humans requires extensive investigation.
Contributor Information
Shigeru Fujita, Division of Systems Virology, Department of Microbiology and Immunology, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan; Graduate School of Medicine, The University of Tokyo, Tokyo, Japan.
Keiya Uriu, Division of Systems Virology, Department of Microbiology and Immunology, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan; Graduate School of Medicine, The University of Tokyo, Tokyo, Japan.
Lin Pan, Division of Systems Virology, Department of Microbiology and Immunology, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan; Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Japan.
Naganori Nao, Division of International Research Promotion, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Japan; One Health Research Center, Hokkaido University, Sapporo, Japan.
Koshiro Tabata, Division of Molecular Pathobiology, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Japan.
Mai Kishimoto, Division of Molecular Pathobiology, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Japan.
Yukari Itakura, Division of Molecular Pathobiology, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Japan.
Hirofumi Sawa, Division of International Research Promotion, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Japan; One Health Research Center, Hokkaido University, Sapporo, Japan; Division of Molecular Pathobiology, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Japan; Institute for Vaccine Research and Development: HU-IVReD, Hokkaido University, Sapporo, Japan; International Collaboration Unit, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Hokkaido, Japan.
Izumi Kida, Division of Risk Analysis and Management, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Japan.
Tomokazu Tamura, Department of Microbiology and Immunology, Faculty of Medicine, Hokkaido University, Sapporo, Japan.
Takasuke Fukuhara, Department of Microbiology and Immunology, Faculty of Medicine, Hokkaido University, Sapporo, Japan; AMED-CREST, Japan Agency for Medical Research and Development (AMED), Tokyo, Japan; Laboratory of Virus Control, Research Institute for Microbial Diseases, Osaka University, Suita, Japan.
Jumpei Ito, Division of Systems Virology, Department of Microbiology and Immunology, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan.
Keita Matsuno, One Health Research Center, Hokkaido University, Sapporo, Japan; Institute for Vaccine Research and Development: HU-IVReD, Hokkaido University, Sapporo, Japan; International Collaboration Unit, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Hokkaido, Japan; Division of Risk Analysis and Management, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Japan.
Kei Sato, Division of Systems Virology, Department of Microbiology and Immunology, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan; Graduate School of Medicine, The University of Tokyo, Tokyo, Japan; Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Japan; International Research Center for Infectious Diseases, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan; International Vaccine Design Center, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan; Collaboration Unit for Infection, Joint Research Center for Human Retrovirus infection, Kumamoto University, Kumamoto, Japan; CREST, Japan Science and Technology Agency, Kawaguchi, Japan.
The Genotype to Phenotype Japan (G2P-Japan) Consortium:
Yu Kaku, Naoko Misawa, Arnon Plianchaisuk, Ziyi Guo, Alfredo A Hinay, Jr, Jarel Elgin M Tolentino, Luo Chen, Mai Suganami, Mika Chiba, Ryo Yoshimura, Kyoko Yasuda, Keiko Iida, Naomi Ohsumi, Adam P Strange, Shiho Tanaka, Rigel Suzuki, Saori Suzuki, Hayato Ito, Shinya Tanaka, Masumi Tsuda, Lei Wang, Yoshikata Oda, Zannatul Ferdous, Kenji Shishido, Kenji Sadamasu, Kazuhisa Yoshimura, Hiroyuki Asakura, Isao Yoshida, Mami Nagashima, So Nakagawa, Kotaro Shirakawa, Akifumi Takaori-Kondo, Kayoko Nagata, Ryosuke Nomura, Yoshihito Horisawa, Yusuke Tashiro, Yugo Kawai, Kazuo Takayama, Rina Hashimoto, Sayaka Deguchi, Yukio Watanabe, Ayaka Sakamoto, Naoko Yasuhara, Takao Hashiguchi, Tateki Suzuki, Kanako Kimura, Jiei Sasaki, Yukari Nakajima, Hisano Yajima, Takashi Irie, Ryoko Kawabata, Kaori Tabata, Terumasa Ikeda, Hesham Nasser, Ryo Shimizu, MST Monira Begum, Michael Jonathan, Yuka Mugita, Otowa Takahashi, Kimiko Ichihara, Takamasa Ueno, Chihiro Motozono, Mako Toyoda, Akatsuki Saito, Maya Shofa, Yuki Shibatani, and Tomoko Nishiuchi
Notes
Acknowledgments. We thank all members belonging to The Genotype to Phenotype Japan (G2P-Japan) Consortium.
Financial support. This study was supported in part by AMED SCARDA Japan Initiative for World-leading Vaccine Research and Development Centers “UTOPIA” (Grant JP223fa627001; to KS), AMED SCARDA Program on research and development of new-generation vaccine including new modality application (Grant JP223fa727002; to KS); AMED SCARDA World-leading Institutes for Vaccine Research and Development Hokkaido Synergy Campus (Grant JP223fa627005; to HS, TF, and KM); AMED Research Program on Emerging and Re-emerging Infectious Diseases (Grants JP22fk0108146 [to KS], JP21fk0108493 and JP22fk0108617 [to TF], JP21fk0108494 [to G2P-Japan Consortium, TF, and KS], JP21fk0108425 [to KS], and JP21fk0108432 [to TF and KS]); AMED Research Program on HIV/AIDS (Grant JP22fk0410039; to KS); AMED CREST (Grant JP22gm1610008; to TF); JST PRESTO (Grant JPMJPR22R1; to JI); AMED Japan Program for Infectious Diseases Research and Infrastructure (Grant JP22wm0125008; to HS); JST CREST (Grant JPMJCR20H4; to KS); JSPS KAKENHI Grant-in-Aid for Scientific Research B (Grant 21H02736; to TF); JSPS KAKENHI Grant-in-Aid for Early-Career Scientists (Grant 20K15767; to JI); JSPS Core-to-Core Program (A. Advanced Research Networks) (Grant JPJSCCA20190008; to KS); JSPS Research Fellow DC2 (Grant 22J11578; to KU); JST SPRING (Grant JPMJSP2108; to SF); World-leading Innovative and Smart Education (WISE) Program 1801 from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to NN).
CONSORTIA
The Genotype to Phenotype Japan (G2P-Japan) Consortium
The Institute of Medical Science, The University of Tokyo, Japan
Yu Kaku, Naoko Misawa, Arnon Plianchaisuk, Ziyi Guo, Alfredo A. Hinay Jr., Jarel Elgin M. Tolentino, Luo Chen, Mai Suganami, Mika Chiba, Ryo Yoshimura, Kyoko Yasuda, Keiko Iida, Naomi Ohsumi, Adam P. Strange, Shiho Tanaka
Hokkaido University, Japan
Rigel Suzuki, Saori Suzuki, Hayato Ito, Shinya Tanaka, Masumi Tsuda, Lei Wang, Yoshikata Oda, Zannatul Ferdous, Kenji Shishido
Tokyo Metropolitan Institute of Public Health, Japan
Kenji Sadamasu, Kazuhisa Yoshimura, Hiroyuki Asakura, Isao Yoshida, Mami Nagashima
Tokai University, Japan
So Nakagawa
Kyoto University, Japan
Kotaro Shirakawa, Akifumi Takaori-Kondo, Kayoko Nagata, Ryosuke Nomura, Yoshihito Horisawa, Yusuke Tashiro, Yugo Kawai, Kazuo Takayama, Rina Hashimoto, Sayaka Deguchi, Yukio Watanabe, Ayaka Sakamoto, Naoko Yasuhara, Takao Hashiguchi, Tateki Suzuki, Kanako Kimura, Jiei Sasaki, Yukari Nakajima, Hisano Yajima
Hiroshima University, Japan
Takashi Irie, Ryoko Kawabata
Kyushu University, Japan
Kaori Tabata
Kumamoto University, Japan
Terumasa Ikeda, Hesham Nasser, Ryo Shimizu, MST Monira Begum, Michael Jonathan, Yuka Mugita, Otowa Takahashi, Kimiko Ichihara, Takamasa Ueno, Chihiro Motozono, Mako Toyoda
University of Miyazaki, Japan
Akatsuki Saito, Maya Shofa, Yuki Shibatani, Tomoko Nishiuchi
References
- 1. Krammer F. SARS-CoV-2 vaccines in development. Nature 2020; 586:516–27. [DOI] [PubMed] [Google Scholar]
- 2. Our world in data. “Coronavirus (COVID-19) vaccinations June 13, 2023).” Available at: https://ourworldindata.org/covid-vaccinations. Accessed 13 June 2023.
- 3. Hou YJ, Chiba S, Halfmann P, et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020; 370:1464–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization . “Tracking SARS-CoV-2 variants (March 30, 2023)”. Available at: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants. Accessed 30 March 2023.
- 5. Kimura I, Yamasoba D, Tamura T, et al. Virological characteristics of the novel SARS-CoV-2 omicron variants including BA.4 and BA.5. Cell 2022; 185:3992–4007.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ito J, Suzuki R, Uriu K, et al. Convergent evolution of the SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant. Nat Commun 2023; 14:2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tamura T, Ito J, Uriu K, et al. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat Commun 2023; 14:2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uriu K, Ito J, Zahradnik J, et al. Enhanced transmissibility, infectivity, and immune resistance of the SARS-CoV-2 omicron XBB.1.5 variant. Lancet Infect Dis 2023; 23:280–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Offit PA. Bivalent Covid-19 vaccines—a cautionary tale. N Engl J Med 2023; 388:481–3. [DOI] [PubMed] [Google Scholar]
- 10. Wang Q, Bowen A, Valdez R, et al. Antibody response to Omicron BA.4-BA.5 bivalent booster. N Engl J Med 2023; 388:567–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reynolds CJ, Pade C, Gibbons JM, et al. Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure. Science 2022; 377:eabq1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao Y, Jian F, Wang J, et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 2023; 614:521–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park YJ, Pinto D, Walls AC, et al. Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. Science 2022; 378:619–27. [DOI] [PubMed] [Google Scholar]
- 14. Liu J, Chandrashekar A, Sellers D, et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature 2022; 603:493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levin EG, Lustig Y, Cohen C, et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N Engl J Med 2021; 385:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaku CI, Starr TN, Zhou P, et al. Evolution of antibody immunity following Omicron BA.1 breakthrough infection. Nat Commun 2023; 14:2751. [DOI] [PMC free article] [PubMed] [Google Scholar]