Abstract
Long viewed as an intermediary in protein translation, there is a growing awareness that tRNAs are capable of myriad other biological functions linked to human health and disease. These emerging roles could be tapped to leverage tRNAs as diagnostic biomarkers, therapeutic targets, or even as novel medicines. Furthermore, the growing array of tRNA-derived fragments, which modulate an increasingly broad spectrum of cellular pathways, is expanding this opportunity. Together, these molecules offer drug developers the chance to modulate the impact of mutations and to alter cell homeostasis. Moreover, because a single therapeutic tRNA can facilitate readthrough of a genetic mutation shared across multiple genes, such medicines afford the opportunity to define patient populations not based on their clinical presentation or mutated gene but rather on the mutation itself. This approach could potentially transform the treatment of patients with rare and ultrarare diseases. In this review, we explore the diverse biology of tRNA and its fragments, examining the past and present challenges to provide a comprehensive understanding of the molecules and their therapeutic potential.
Keywords: tRNA, tRNA fragments, genetic disease, RNA modification, transcription, translation, machine learning, drug design, programmable RNA, tRNA therapeutics, tRNA medicines
Since their initial description in 1958, tRNAs have largely been viewed as intermediaries in the translation of the genetic code into proteins. Recent research, however, continues to reveal that although protein translation is their predominant role, tRNAs perform many more functions within the cell, dictating both health and disease. The growing understanding of the origins and relevance of small noncoding RNAs that result from tRNA fragments has further expanded our understanding of the diversity of tRNA biology (1, 2, 3). These studies revealed that tRNA-derived fragments (tRFs) were not simply degradation intermediates of tRNA turnover but were instead processed precisely with distinct expression patterns, suggesting possible functions in cell biology. As detailed later (see also review by Qi Chen and Tong Zhou in this issue), the array of cellular functions and correlations with human disease have only grown, expanding the therapeutic potential of tRFs and by extension, tRNAs.
The central role of tRNA in so many biological processes, including protein translation, makes it an attractive molecule for therapeutic development, offering researchers an opportunity to control an array of functions with a single molecule. The mechanisms by which tRNAs function also offer an opportunity to intervene in human health in ways unlike other DNA- and RNA-based therapies that aim to replace or modulate the activity of specific genes (Table 1).
Table 1.
Unique features of tRNA medicines as compared with other RNA modalities
| Therapeutic mechanism & application | siRNA | mRNA | tRNA | tRF |
| Mechanism of action | Targets specific RNAs for degradation | Provides a functional mRNA | Functionally corrects/recodes endogenous mutation | Generally regulates biological pathways |
| Control of target gene expression | Fully leverages endogenous control pathways | Encodes in the exogenous mRNA | Fully leverages endogenous control pathways | Fully leverages endogenous control pathways |
| Addressable patient population for each medicine | Each disease target requires full development | Each disease target requires full development | Addresses entire class of mutations independent of nature of gene/protein | Depends on biological application |
| Specifications | siRNA | mRNA | tRNA | tRF |
| Drug substance/RNA optimization | Full scope of combinatorial sequence and modification exploration optimizes drug properties∗ | Primary sequence and higher order structure design restricts encoded protein sequence and translationally competent modifications | Full scope of combinatorial sequence and modification exploration optimizes drug properties | Full scope of combinatorial sequence and modification exploration optimizes drug properties∗ |
| Size | Small (<25 nucleotides for single strand) | Hundreds to thousands of nucleotides | Relatively small (<100 nucleotides) | Small (<40 nucleotides) |
| Synthesis | Chemical | Enzymatic | Chemical Enzymatic |
Chemical Enzymatic |
| Delivery | Nanoparticle Viral Conjugation | Nanoparticle Viral | Nanoparticle Viral Conjugation |
Nanoparticle Viral Conjugation |
A comparison of the molecular mechanisms of action, addressable patient population, and modality design and delivery parameters across RNA modalities, namely siRNA, mRNA, tRNA, and tRFs.
With restrictions based on complementarity to target.
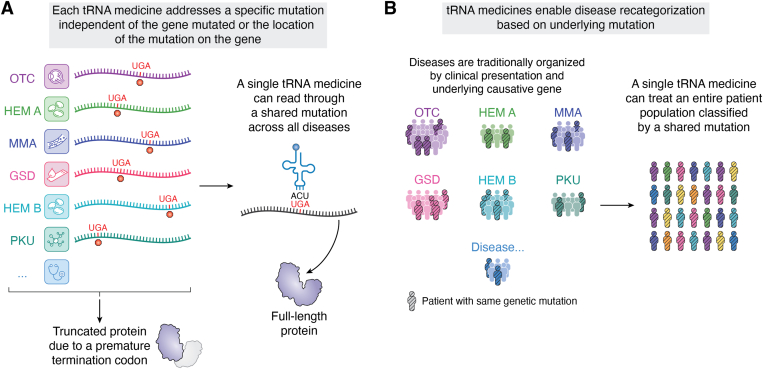
The codon–anticodon pairing between tRNAs and mRNA transcripts means that a single tRNA medicine could potentially modulate the impact of a shared genetic mutation independent of the gene in which that mutation resides or where within the gene it is located (Fig. 1A). From a clinical perspective, using a single tRNA medicine to treat multiple and varied disorders would be transformative, enabling the novel grouping of patients based on their underlying mutation, rather than their clinical presentation or the identity of the mutated gene (Fig. 1B). This approach would impact clinical care and patient lives by enabling more tenable and commercially viable drug development for unaddressed rare and ultrarare diseases that currently have no or few treatment options.
Figure 1.
Transformative clinical potential of tRNA medicines.A, a single tRNA medicine can readthrough a shared genetic mutation, such as a premature termination codon, across different genes independent of the mutation location along the transcript. B, the clinical impact of this modality feature enables a novel grouping of patients, not based on their clinical presentation or on the identity of the gene mutated as is traditionally done, but based on the underlying mutation patients carry. GSD, glycogen storage disease; HEM A, hemophilia A; HEM B, hemophilia B; MMA, methylmalonic aciduria; OTC, ornithine transcarbamylase; PKU, phenylketonuria.
Therapeutic potential of tRNAs
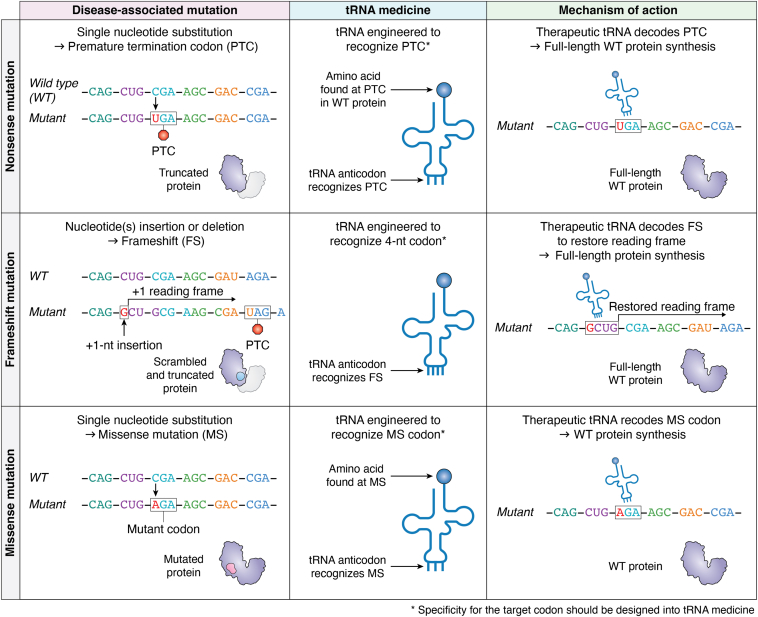
The therapeutic potential of tRNAs is already being actively explored, building on decades of research insights. Mutations within protein coding sequences can disrupt translation by altering the sequence from an amino acid codon to a stop codon (premature termination codon or PTC mutations), deleting or inserting nucleotides to shift the translational reading frame (frameshift or FS mutations), or changing a codon to trigger incorporation of a different amino acid (missense or MS mutation) (Fig. 2). These mutations lead to the production of sequence- and/or length-altered proteins, changes that impact the protein’s expression, stability, and function and that potentially translate to disease pathology. Engineered tRNAs could be a universal all-in-one tool to address each of these mutations during translation by introducing the appropriate amino acid at the mutation site to read through the PTCs, adjust the reading frame disrupted by the FS mutation, or correct the MS mutation, allowing the restoration of the wildtype polypeptide (Fig. 2).
Figure 2.
Design and molecular mechanism of therapeutic tRNAs for genetic mutation correction. Examples of disease-inducing mutations (first column) that can be treated with unique tRNA medicines (second column) and the mechanism of action (third column) by which this occurs.
Stop codon tRNA medicines
In attempting to overcome PTC mutations, one also has to consider the influence of these genetic alterations on a natural mRNA quality control mechanism known as nonsense-mediated mRNA decay (NMD) (reviewed in Refs. (4, 5, 6)). This translation-dependent surveillance mechanism accelerates the destruction of some PTC-encoding mRNAs to limit the cellular impact of the expression of truncated and potentially toxic proteins. A PTC-reading tRNA therapeutic could potentially act at two levels—enabling the synthesis of full-length proteins and restoring native mRNA levels for continued translation through inhibition of the NMD pathway.
Also of concern is the realization that both PTCs and natural termination codons (NTCs) rely on the same triplet sequences—UAA (ochre), UAG (amber), and UGA (opal)—to terminate translation. This limitation raises the possibility that any effort to overcome a given PTC might increase the likelihood of also inducing readthrough of corresponding NTCs, whether in the same transcript or in other transcripts (Fig. 2). Fortunately, several studies have shown that a number of environmental factors influence termination efficiency and thereby the “strength” of the termination codon, allowing differentiation of PTCs and NTCs ((7); reviewed in Ref. (8)). These factors include translation factors that interact with the mRNA and/or the ribosome, the sequence context around the termination codon, the presence of splicing junctions, and the general vicinity of the 3′ terminus of the mRNA. Efforts to develop tRNA medicines that directly address PTCs while limiting their impact on NTCs will require a systematic prerescue and postrescue assessment of cellular proteomic inventories, which should be programmed into the tRNA medicine design.
The high incidence of PTC-induced diseases—responsible for 5 to 15% of mutations linked to inherited diseases (www.clinicalgenome.org/data-sharing/clinvar/)—has led to concerted efforts to discover mechanisms to induce stop-codon readthrough. To date, much of this effort has centered on small molecules, such as the aminoglycoside antibiotics like gentamicin and G418 and the oxadiazole ataluren (reviewed in Ref. (9)), which alter the translational fidelity at the ribosome’s decoding center. These molecules permit the binding of near- and noncognate aminoacylated tRNAs (aa-tRNAs), reading through the PTC, and promoting elongation of the growing polypeptide chain. A drawback with this approach is that by engaging any aa-tRNA, the amino acid incorporated into the protein at the site of mutation does not necessarily reflect the wildtype residue. The resulting pool of proteins will be a heterogenous mix of sequences, and the mutated forms may be structurally and/or functionally altered. Additional challenges relate to the potential impact on readthrough of NTCs and the overall integrity of the proteome because of the possibility of miscoding, making it difficult to balance drug efficacy and safety. Several of these compounds induced ototoxicity (reviewed in Ref. (10)) and nephrotoxicity, limiting their practical use (reviewed in Ref. (11)).
A better therapeutic would recognize the PTC itself and then insert the correct amino acid at that site in the growing peptide, thereby producing a full-length wildtype functional protein. This precision medicine approach is enabled by the natural role and function of tRNAs. Building on decades of research into tRNA-based PTC readthrough, several groups have engineered tRNAs to suppress PTC mutations. Because there are 19 amino-acid stop mutation combinations resulting from a single nucleotide change that transforms a sense codon into a stop codon, developing tRNA medicines to treat diseases caused by stop codons has the potential to reclassify thousands of genetic disorders into 19 amino-acid-stop codon diseases.
Lueck et al. (12) developed a high-throughput cell-based assay to explore anticodon-engineered (ACE) tRNAs capable of suppressing PTCs both in vitro and in vivo. When transiently expressed in cells, several ACE-tRNA candidates promoted PTC readthrough better than gentamicin or G418, and ribosome profiling suggested that ACE-tRNAs selectively suppressed PTCs over NTCs. Ko et al. (13) later showed that ACE-tRNAs were able to suppress PTCs in the cystic fibrosis transmembrane regulator (CFTR) gene and inhibit NMD, restoring functional CFTR to levels that could produce a therapeutic effect. Both studies showed that ACE-tRNAs could be effectively delivered as complementary DNA (cDNA) or RNA, expanding the potential delivery options. Furthermore, stable genomic integration of ACE-tRNA expression cassettes suppressed PTCs in a reporter construct without perturbing cell morphology or growth rates, suggesting that persistent ACE-tRNA expression was not toxic. Chen et al. (14) also showed that a PTC-suppressing tRNA construct could integrate into mouse genomes and efficiently transmit the new gene to subsequent generations with no obvious impact on animal health.
Albers et al. (15, 16) took a more systematic approach to designing suppressor tRNAs, computationally selecting sequences likely to adopt optimal secondary and tertiary structures. They found that changing the anticodon loop did not improve the efficiency of PTC suppression nearly as much as changes in the TψC-stem, which interacts with translational elongation factors, expanding our understanding of the role of structural elements in PTC readthrough. Administering the suppressor tRNAs in mice restored production of functional proteins from genes bearing nonsense codons. Expression and function of CFTR in cell systems and patient-derived nasal epithelia was also restored.
This preclinical research has led a handful of companies to explore the therapeutic potential of tRNAs that read through stop codons. ReCode Therapeutics (https://recodetx.com/science/) reported in vitro studies showing that its PTC-suppressing tRNA (RCT223) was able to restore CFTR function in bronchial epithelial cells derived from cystic fibrosis patients and was generally well tolerated in cell culture even after repeated dosing (https://recodetx.com/recode-therapeutics-presents-preclinical-data-using-sort-lnptm-and-rna-platforms-to-rescue-cftr-function-at-the-44th-european-cystic-fibrosis-conference-ecfs/). Shape Therapeutics (https://shapetx.com/) has developed its RNAskip platform for PTC suppression, targeting stop codon mutations in MECP2 that cause Rett Syndrome. Tevard Biosciences and hC Bioscience are also leveraging tRNA technology to overcome stop codon mutations, targeting Dravet syndrome (the SCN1A gene) and liver cancer, respectively.
Working with Arcturus Therapeutics (https://arcturusrx.com/rna-mrna-proprietary-technologies/), Ignatova et al. (patent publication no.: WO 2019/175316) extended the tRNA anticodon loop to accommodate two anticodon triplets, the PTC and an adjacent codon, in the target mRNA. The engineered aa-tRNA inserts the amino acid reflective of the PTC-adjacent codon, resulting in a protein one residue shorter than the wildtype analog. The double anticodon should offer greater specificity for its target than a tRNA only targeting a PTC, reducing the risk of off-target binding. It would, however, limit the use of the tRNA to situations involving a specific hexanucleotide sequence, increasing the number of therapeutics required to address patient mutations.
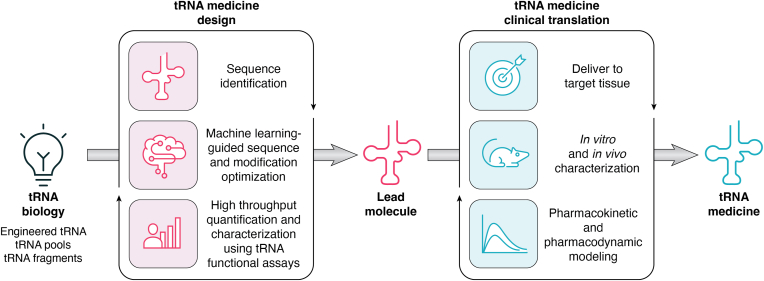
Alltrna's platform (www.alltrna.com/#science; Fig. 3) utilizes an automated and high-throughput screening infrastructure and machine learning to program tRNAs. Starting with a basic tRNA scaffold, the tRNA sequence is optimized and chemically modified to achieve desired therapeutic features, such as high activity, specificity and stability, lower immunogenicity, and targeted biodistribution. A large dataset produced from automated and high-throughput assays is used to train and retrain machine learning models. These iterative design cycles precisely generate high-performing tRNAs as medicines. Focusing initially on PTC-reading tRNA medicines to restore full-length wildtype proteins, Alltrna’s highly programmable platform can be adapted further to the design of tRNAs for correcting FS and MS mutations, modulating tRNA pools, or developing tRF-based medicines.
Figure 3.
Taking a tRNA medicine from concept to programmable medicine. Iterative cycles of tRNA design and validation through clinical translation inform the rules of programmable therapeutic features to create RNA medicines from a tRNA-based drug concept (such as engineered tRNAs, tRNA pools, and tRNA fragments).
Frameshift and Missense tRNA medicines
Expansion of the anticodon loop is one strategy to overcome FS mutations (17). The first sequenced FS-reading tRNA inserted a cytosine immediately 5′ of its CCC anticodon, creating (18). Sako et al. (19) tested the ability of several quadruplet-anticodon tRNAs to correct an FS mutation caused by a 26-nucleotide deletion in the COL α2 (VI) gene, a mutation linked to Ullrich disease. Transfecting patient fibroblasts with different constructs of FS increased mRNA levels and restored expression of collagen VI α2 protein. Rodriguez et al. (20) used nonsense and FS tRNAs to incorporate unnatural amino acids into a neuroreceptor expressed in Xenopus oocytes. Although the overall efficiency of the FS tRNAs— and —was lower than that of the tRNA that reads through stop codons, their efficiency increased nonlinearly with the amount of injected tRNA, suggesting competition with endogenous triplet-recognizing tRNA.
Recently, DeBenedictis et al. (17, 21) engineered quadruplet Escherichia coli (E. coli) tRNAs and then used phage-assisted continuous evolution to improve translation efficiencies by upward of 80-fold over the initial candidates. Many of the “evolved” tRNAs were aminoacylated by their cognate aminoacyl tRNA synthetases and were able to translate adjacent quadruplet codons. The group also showed that changes outside the anticodon may be necessary to increase the efficiency of translation by FS tRNAs.
Other models to overcome FS mutations have also been suggested, including a wobble model where a triplet anticodon covers a quadruplet codon, and a slippage model where a triplet anticodon shifts by one nucleotide along the mRNA as the tRNA transitions from the ribosomal A site to the P site (22). Fagan et al. (23) suggested that the activity of FS tRNAs may be due to structural changes during ribosome engagement rather than codon–anticodon interactions.
As cognate tRNAs for MS-mutated codons already exist in cells, treatment with tRNAs engineered to correct the MS codon by inserting the amino acid found in the wildtype protein may not be as straightforward as with PTCs. Competing with the endogenous cognate tRNA risks interference with the translation, function, and stability of other proteins using that codon. Thus, this approach may only be useful under specific circumstances, such as when the MS mutation creates a rare codon or when even low levels of wildtype protein restoration can ameliorate disease symptoms or pathology.
In conditions like hemoglobinopathies, Hatfield et al. (24) suggested it may be possible to introduce MS-suppressing tRNAs to red blood cell precursors rather than systemically to minimize the risks involved. Alternatively, post-transcriptional modifications of the anticodon could impact how the introduced tRNA competes for codon binding.
Future design efforts would require a much more thorough understanding of the impact of sequence and post-transcriptional modifications on tRNA structure and function, building on the vast natural libraries of tRNAs that already exist. Similarly, an expanded understanding of the sequence determinants for tRNA–mRNA interactions on the ribosome would facilitate the engineering of molecules with greater specificity for the transcripts of interest and thereby lessen the risks of off-target effects.
tRNA pools
The diversity of tRNA molecules and genes plays a significant regulatory role in cell proliferation, differentiation, and response to environmental cues, as the pool of any given tRNA fluctuates under certain conditions to modulate the translation of specific proteins (25). It is a supply-and-demand scenario, where tRNA composition and abundance typically reflect the codon usage of key mRNA transcripts. Several studies have shown how the interplay between tRNA and codon abundance directly impacts cellular function and can alter cell fate.
Dittmar et al. (26) noted that expression levels of individual tRNAs can vary up to 10-fold between cell types and observed a correlation between abundance and codon usage in highly expressed tissue-specific genes. These tissue-specific patterns can also be impacted by outside factors. Huang et al. (27) found that the microbiome reprogrammed tRNA expression patterns in four different human tissues and altered the post-transcriptional modification profiles of the tRNAs.
Such changes can directly impact cellular function and therefore health. Alterations in tRNA pools impact the transition of a cell from proliferation to differentiation states with upregulated and downregulated tRNAs reflecting the codon usage demands of the cell (28). Rak et al. (29) noted significant changes in tRNA abundance and modifications during the process of T-cell activation. Similarly, Goodarzi et al. (30) showed that upregulation of specific tRNAs in cell culture drives a proteomic shift toward a prometastatic state. In addition, Santos et al. (31) showed that upregulating levels in normal bronchial cells increased proliferation and the transforming potential of the cells in vitro. These cells also generated tumors when injected into mice. Pavon-Eternod et al. (32) noted similar tumorigenic results with breast tissue.
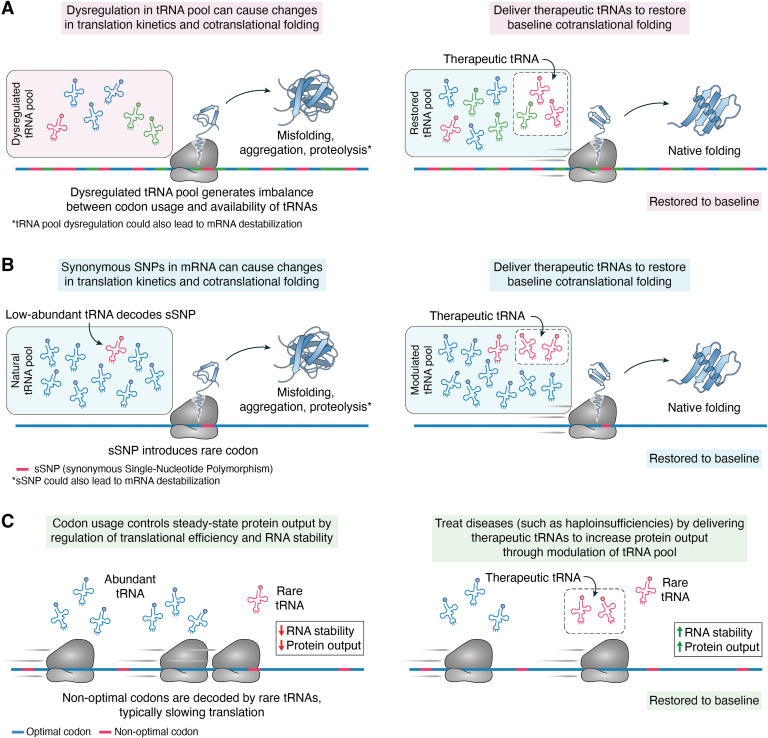
Beyond cancer (33), several groups have shown that cells alter their tRNA expression in response to stress and that these changes correlate with altered expression of proteins and codon content. Pang et al. (34), Chan et al. (35) and Torrent et al. (36) found that the levels of different tRNAs and/or their modification states responded dynamically when different forms of stress were applied to yeast cells. The codon profiles of the altered tRNA pools correlated with codon usage by genes involved in stress response pathways. Collectively, these studies highlight the breadth of impacts altered tRNA pools play in various tissues, both in terms of natural physiology and in the onset of disease (Fig. 4A).
Figure 4.
Examples of translation dynamic control through tRNA pool modulation.A, dysregulation in tRNA pools or (B) synonymous SNPs encoding rare codons alter translation kinetics and cotranslational folding, leading to changes in protein expression and increased protein misfolding, aggregation, and proteolysis. C, codon usage impacts steady-state protein output by regulation of translation efficiency and mRNA stability. By restoring tRNA pool imbalance with a tRNA medicine, a baseline steady state can be restored.
An imbalance in this supply-and-demand relationship could lead to significant problems in protein translation, causing disease. Transcripts that include rare codons may be translated more slowly as the ribosomes await delivery of the appropriate aa-tRNA. Such changes in translation kinetics can impact folding of the nascent polypeptide chain (reviewed in Ref. (37)) or influence transcript folding, splicing, or stability (reviewed in Refs. (38, 39)) (Fig. 4B). It may be possible to design tRNAs or tRNA pools that modulate molecular pathways by altering the expression of proteins arising from rare-codon-bearing transcripts or overcome the effects of mutations that generate rare codons (Fig. 4C). As noted by Liu (37), codon usage influences translation elongation and efficiency and can potentially lead to ribosome stalling, even resulting in premature termination.
The impact of these rare codons has been well characterized in yeast and bacteria. Arias et al. (40) showed that the stability of the yeast cell-cycle protein Cdc13 may depend in part on two nonoptimal glycine codons. Although overexpression of the cognate tRNAs did not seem to impact the amount of protein produced, up to half of the resulting protein was in the aggregated fraction. Similar influences have been seen in E. coli (41) and in higher eukaryotes, including human cells, although sometimes with the opposite outcome (38).
Transcripts using rare codons can benefit from an increase in global tRNA levels rather than in individual tRNAs. Guimaraes et al. (42) reported selectively increased translation in proproliferative rare-codon-laden transcripts over transcripts carrying more common codons when total tRNA levels increased. These results, however, also highlight a potential safety risk with any efforts to modulate tRNA pools given the distinct codon usage bias of genes involved in cell proliferation and metastasis.
Codon usage can also significantly impact transcript stability. In yeast, Hanson et al. (43) documented that transcripts with more optimal codons were translated more quickly and were more stable. Similar findings linking mRNA stability, codon optimality, tRNA levels, and translation efficiency have been reported for Xenopus and zebrafish (44) as well as humans (45). This impact may extend beyond a simple codon–anticodon pairing, as Gamble et al. (46) showed that translation efficiency in yeast is modulated by the sequences of adjacent codon pairs.
Any effort to develop tRNA medicines will require broader efforts to map codon usage across the transcriptome as it relates to tRNA cellular levels, particularly in genes related to disease phenotypes. It may be possible to design tRNAs to target specific rare-codon-bearing genes, whether based on the sequence context of the rare or nonoptimal codon or the cells in which the transcript is most relevant.
While changes in tRNA pools often cause a cell to transition from a healthy state to a diseased state, a deeper understanding of the mechanisms by which these transitions occur may facilitate identification of ways in which these transitions can be reversed. It may be possible, for example, to introduce therapeutic tRNAs that support expression of genes involved in nonproliferative or antiproliferative pathways or that restore homeostasis to perturbed tissues.
Altering tRNA pools could also facilitate the rescue of haploinsufficiency, where the mutation or deletion of a single copy of a gene reduces production of its protein product (Fig. 4C). This idea is being evaluated by Tevard Biosciences, which is using a viral vector to overexpress a cocktail of tRNAs in targeted cells to increase production of functional SCN1A protein by increasing the stability of its mRNA. Alltrna is leveraging its platform (i) to learn how the rules of protein synthesis, including translation dynamics, transcript stability, or cotranslational folding of proteins, are impacted by varying pools of tRNAs across cell types and cell states and (ii) to precisely manipulate cellular tRNA pools to address disease. Although much work remains to be done to fully understand and harness tRNA pools, the impact of these efforts would be a new approach to controlling protein expression and disease pathology.
tRNA fragments
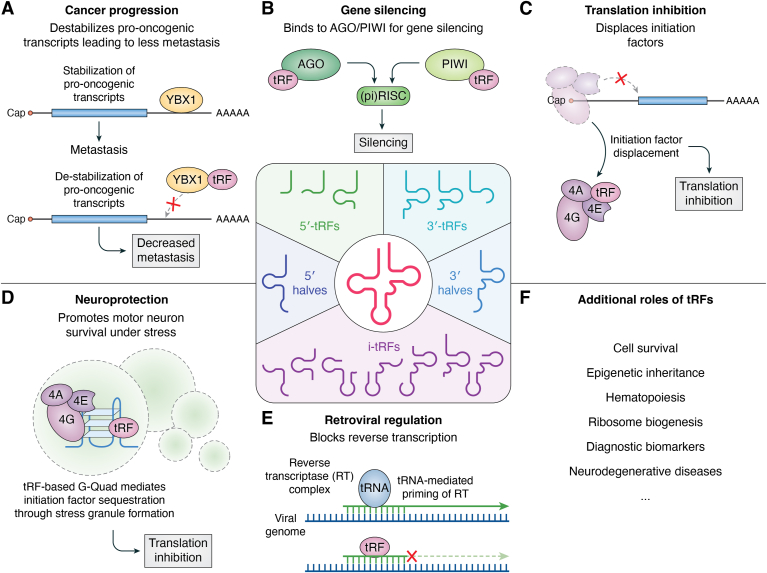
More than simply byproducts of tRNA degradation, tRFs are a family of precisely defined molecules arising from endonucleolytic cleavage of parent tRNA molecules. The number and variety of tRFs continues to grow as researchers expand their search. Aligned with that expanding diversity is an understanding of their myriad functions as regulatory mechanisms in cells and throughout the body (Fig. 5). Goodarzi et al. (47) showed that tRFs could limit the metastatic potential of breast cancer cells by selectively sequestering YBX1, an RNA-binding protein that stabilizes oncogenic transcripts. Other studies have shown that tRFs can impact the growth and differentiation of embryonic (48) and hematopoietic (49) stem cells.
Figure 5.
Examples of proposed functions of tRNA fragments. Role of tRFs in regulating metastasis (A), gene silencing (B), translation (C), stress response (D); retroviral replication (E), and other functions (F). tRF, tRNA-derived fragment.
Most studies of tRFs have reflected their potential as prognostic biomarkers or diagnostic biomarkers. Zhang et al. (50) identified differentially regulated tRFs in Alzheimer’s and Parkinson’s disease models that targeted genes involved in synapse formation and the synaptic vesicle cycle pathway. Green et al. (51) similarly noted altered tRF expression profiles in osteoarthritis cartilage, including a tRF that repressed Janus kinase 3 expression. Gao et al. (52) analyzed tRFs in blood samples and identified 11 differentially expressed molecules with prognostic value for non–small cell lung cancer. Other studies expanded the potential of tRF-based diagnostics in liver (53), gastric (54) and colorectal cancers (55), as well as in disorders as diverse as autoimmune nephritis (56), Parkinson’s disease (57), endometriosis (58), and osteoporosis (59).
Although tRFs have been less thoroughly studied as a therapeutic, several studies highlight their potential (Fig. 5). Fan et al. (60) showed that the tRF AS-tDR-007872 was preferentially downregulated in tissues and plasma samples from non–small cell lung cancer patients compared with healthy subjects, and they indicated that they could reverse this oncogenic phenotype in vitro by overexpressing the tRF, decreasing cancer cell viability, migration, and invasion. Xu et al. (61) showed similar capacity for tRF-Glu-TTC-027 in gastric carcinoma, where transfection into cells reduced cell proliferation, migration, and invasion in vitro. The tRF also reduced tumor growth when injected subcutaneously in a mouse xenograft with no significant difference in bodyweight from controls, a measure of toxicity.
Other studies have shown that the tRF does not have to be endogenous to the tissues being studied. Cao et al. (62) identified tRFs from nonpathogenic E. coli strains that were able to inhibit colorectal cancer in vitro. They also noted that modification of one tRF mimic, EC83, with 2-O-methylguanosine improved efficacy, offering IC50 values 700-fold lower than 5-fluorouracil. The 2-O-methylguanosine-modified EC83 was also more structurally stable than the parent molecule or two other modified variants.
The same group (63) also identified plant-derived tRFs with in vitro cytotoxicity against ovarian cancer. In a mouse xenograft model, tRF-11 reduced tumor growth, achieving comparable efficacy to the well-known chemotherapy drug taxol but at a 16-fold lower dose. Treatment also appeared safe, showing no significant histological changes in major organs or altered bodyweight when compared with the control group.
Screening ginseng extracts for cardioprotective activity in ischemia/reperfusion injury, Hu et al. (64) found that tRF HC83 was 1000-fold more potent than ginsenoside Rg1 in increasing cell viability in vitro. Furthermore, this protection extended to mitochondrial function as HC83 suppressed both injury-induced intracellular and mitochondrial generation of reactive oxygen species. Nanoparticulate HC83 also significantly reduced ischemia/reperfusion-induced myocardial infarctions in mice, showing comparable efficacy to metoprolol but at a 500-fold lower dose.
Significantly more foundational work needs to be done to better understand the breadth of tRF functions on human health and disease and to develop novel methodologies to study tRFs at a scale that allows us to learn the rules of design to transform them into a novel class of medicines.
Translating tRNAs to medicines
For all their promising medicinal potential, until recently, there has not been a systematic effort to program tRNA into medicines. The reason for this delay largely centered on challenges common to RNA-based therapeutics in general, including a lack of tools and methodologies to synthesize or express and characterize these molecules. Without these resources, it was much more difficult to fully comprehend the correlations between sequence, modification, and structure with biological function and the properties required to be effective drugs.
Although we could isolate existing molecules, for example, it was not possible to synthesize modified oligoribonucleotides of tRNA size to construct drug-candidate libraries for functional characterization or test design rules for sequence and modification optimization. Furthermore, existing analytical methods did not permit accurate molecular quantitation or chemical characterization of naturally occurring tRNAs. And even if a tRNA or a tRF could be developed as a medicine, it was only recently that systems have been developed to deliver those molecules to target tissues.
These limitations are systematically being addressed through recent and continuing advances that can be broken into six categories: (i) improvements in existing or development of enabling technologies; (ii) exploration and optimization of the sequence space; (iii) understanding and utilization of the full chemical modification space; (iv) optimization of drug-like properties; (v) leveraging machine learning; and (vi) delivery of tRNAs to patients.
Need for enabling technologies
Efforts to fully understand the breadth of tRNA and tRF biology and chemistry have been hampered by inherent limitations within current technologies that do not address the unique characteristics (sequence, structure, and epitranscriptomic modification) of these molecules. Existing methods will need to evolve, and new technologies be developed to realize the full potential of tRNA- and tRF-based medicines (Table 1).
Synthesis
The technical challenges and cost of synthesizing RNAs with modified bases has made it difficult to develop experimental systems that allow full characterization of molecules and systematic testing of biological hypotheses. Chemical synthetic methods that easily generate DNAs 100 nt or longer become less efficient when producing RNAs because of the presence of the 2′-hydroxyl group of the ribose moiety (65). Ohtsuka et al. (66) reported total synthesis of E. coli tRNAfMet in 1981, but the protocol required the ligation of oligoribonucleotides 3 to 10 nt in length using RNA ligase, and the resulting molecule only possessed 4 to 6% of the aminoacylation activity of native tRNAfMet. Ogilvie et al. (67) later performed total synthesis of a 77-nt E. coli tRNAfMet that also incorporated modified bases, but this molecule still only possessed 11% aminoacylation activity. Although researchers in neither study could explain why the aminoacylation activities were so low, they speculated that these lower rates may be due to the requirement of a particular nucleotide modification for direct interaction with the aminoacyl-tRNA synthetase. Another possibility was that the synthetic sample was contaminated with nondeprotected tRNA species that could not be aminoacylated.
Despite these achievements, the complexities of solid-phase synthesis of RNA, where protected and possibly modified ribonucleosides are sequentially added to extend the oligoribonucleotide chain attached to a solid support, have historically limited the generation of molecules larger than approximately 50 nt, making it challenging to fully synthesize a tRNA of 70 to 80 nt. Although tRFs fit well within this size limit, their synthesis and that of tRNAs has been hampered by the limited availability of chemical intermediates reflective of epitranscriptomic modification (68).
Furthermore, methods like solid-phase synthesis also suffer from the generation of incomplete strands as byproducts (65). To address this problem, He et al. (69) developed a solid-phase synthesis protocol that includes bio-orthogonal tagging to separate full-sized molecules from failed sequences, synthesizing and purifying a 76-nucleotide tRNA without the need for chromatography. Other groups have combined solid-phase synthesis with chemoenzymatic methods to introduce modifications at specific locations within tRNAs (70).
For tRNA- and tRF-based medicines to be clinically and commercially viable, the incremental improvements at the experimental level must be scalable at the manufacturing level. To be cost-effective, the efficiency of each synthetic coupling reaction must be as high as possible (reviewed in Ref. (71)). To produce a 25-nt oligonucleotide, 99% coupling efficiency results in 79% overall yield, whereas a relatively small drop to 97% coupling efficiency reduces the overall yield to 48%. The ligation of two shorter RNA strands to produce a larger molecule further diminishes yields.
The extra costs of new synthetic intermediates heighten the need for highly efficient reactions that require as little postsynthesis purification as possible and offer optimal quality to increase the potential for regulatory approval. In addition, chemical synthesis involves extensive use of solvents, which when used at-scale, produce significant environmental concerns.
Continued progress and optimization in the development of synthetic methods and generation of chemically modified synthetic intermediates will be imperative as drug developers look to expand their libraries of therapeutic candidates from tens of molecules to hundreds or thousands. Such methods will also need to be scalable, producing high-quality molecules in quantities sufficient for everything from initial high-throughput screening to large-scale drug manufacture.
Sequencing
The advent and evolution of next-generation sequencing changed the landscape for high-throughput and large-scale sequencing of RNAs, yet these technologies have historically struggled to cope with the compact structure of tRNAs and post-transcriptional modifications (72, 73). Stem–loop structures can impede RNA sequencing library development by limiting first-strand synthesis by reverse transcriptase. Many reverse transcriptases also struggle to elongate past the many modified nucleotides found in tRNA, terminating cDNA synthesis reactions when the enzyme reaches these residues. This challenge makes it difficult to quantify and characterize individual Trna molecules in any meaningful way. Several groups have tried to circumvent these issues by incorporating enzymes like demethylases to remove certain epitranscriptomic modifications (e.g., demethylase-thermostable group II intron reverse transcription sequencing or DM-TGIRT-Seq; (74)), whereas others incorporated the distinct footprints left by different modifications during the reverse transcriptase step in their sequencing and analysis protocols (e.g., tRNA mutational profiling or tRNA-MaP; (75)). Alternatively, some protocols include fragmentation of tRNA molecules to facilitate the priming of shorter RNA molecules for cDNA synthesis (e.g., quantitative mature- or QuantM-tRNA-Seq; (72)).
Rather than go through the cDNA intermediary required by next-generation sequencing, other groups have attempted the direct sequencing of full-length tRNAs using a nanopore-based platform (76). This method directly reads the sequence of linearized tRNAs and tRFs based on ion current changes as each nucleotide passes through the nanopore; the positions of modifications can also be inferred based on alterations in the ion current signal when compared with corresponding synthetic controls. Ramasamy et al. (77) combined nanopore-based direct RNA sequencing with a chemical probe to identify pseudouridine modifications, keeping other modifications intact for further analysis. Abebe et al. (78) leveraged advances in bioinformatics and neural networks to compare direct RNA sequencing datasets and identify sequencing errors caused by modifications. Thomas et al. (76) introduced custom adapters to their nanopore protocol to improve end-to-end base calling for full-length tRNA and were able to align base miscalls with known post-transcriptional modifications. One limitation of nanopore sequencing is the low basecall rate at the 5' end because of accelerated RNA translocation as the molecule exits the helicase (79). Using adaptors that leverage the 3' CCA overhang found in tRNAs, Lucas et al. (79) combined nanopore sequencing with a computational framework to increase tRNA reads 10-fold over other methods, more accurately quantifying tRNA abundance. Their method (Nano-tRNAseq) also allowed them to capture modifications and to monitor changes in tRNA populations as cells responded to oxidative stress.
Another challenge to distinguishing very similar tRNAs is that epitranscriptomic modifications are not identified directly using sequencing methods but are inferred from the data. Several groups have therefore adapted liquid chromatography–mass spectrometry to map RNA modifications, in part relying on the specificity of RNases such as guanosine-specific RNase T1. Thakur et al. (80) leveraged the uridine-specific RNase MC1, which cannot cleave thiouracil, and cytidine-specific cusativin, which cannot cleave methylcytidine or acetylcytidine, to expand coverage of tRNA modifications. Adjacent cytosines (CpC) also inhibit cleavage by cusativin (81). Suzuki et al. (82) also used mass spectrometry to characterize modifications on all human mitochondrial tRNAs (mt-tRNAs). These methods are intrinsically low throughput, limiting their applicability to efforts to screen large candidate libraries, but they can be useful to validate the characterization of promising candidates.
Such advances may also help bring clarity to another challenge, the identification of active tRFs versus the short RNAs generated incidentally by RNA degradation processes (reviewed in Ref. (83)) or by premature termination during reverse transcription of full-length tRNAs. The growing awareness of the biological relevance of tRNA fragmentation has led to a more sustained effort to identify and characterize these molecules, resulting in searchable databases such as tRFdb, which contains more than 12,000 tRFs from organisms as diverse as yeast, worms, zebrafish, mice, and humans ((84); reviewed in Ref. (85)).
Improvements in synthesis and sequencing will subsequently increase the demand for development of new screening methodologies as candidate libraries rapidly expand. Experiments that could once be easily handled manually will increasingly require technologies that facilitate high-throughput and high-sensitivity characterization of both biological function and drug-like properties. Because tRNA quantitation remains challenging at the single-cell level, Gao et al. (86) validated the use of single-cell sequencing assay for transposase-accessible chromatin data as a proxy for tRNA gene usage and compared that to scRNA-Seq-derived codon usage and amino acid demand to resolve translation efficiency differences across multiple tissues. Similarly, Isakova et al. (87) developed Smart-Seq-total to provide absolute quantitation of all coding/noncoding RNA transcripts in a single cell.
Several groups are also developing methods to scan libraries for parameters such as molecular stability or target specificity. Biedenbänder et al. (88) used NMR spectroscopy to show how post-transcriptional modifications can stabilize tRNA structure while at the same time facilitate conformational flexibility for tRNA function. Leamy et al. (89) used small-angle X-ray scattering to evaluate the impact of single nucleotide changes on tRNA folding and stability, whereas Gremminger et al. (90) used microscale thermophoresis to monitor interactions between HIV-1 viral RNA and tRNALys3.
Optimizing the sequence space
Despite the sequence diversity of tRNA genes in the human genome, targeted nucleotide changes may yet afford opportunities to enhance molecular function. Central to this design approach will be a better understanding how tRNAs and tRFs interact with and modulate their targets as well as how sequence influences the structure, stability, and potential therapeutic efficacy and safety of these RNAs.
Sequence optimization
Protein translation centers on the recognition of triplet sequences or codons within mRNAs by the anticodons of cognate aa-tRNAs in order to add specific amino acids to the growing polypeptide chain. Of the 64 (43) possible codons, 61 are amino acid coding, whereas the other three are translation termination or stop codons. Thus, protein translation should only require 61 tRNAs, one for each amino acid–coding codon, but several factors disrupt this one-to-one correlation. The wobble position of the tRNA anticodon loop allows non-Watson–Crick base pairing to occur, resulting in many tRNA species having a one-to-many relationship with mRNA codons (91). For example, decodes the codons UUC and UUU.
Yet, despite the potential to use fewer tRNAs, organisms across the spectrum encode multiple copies of different tRNA genes throughout their genomes. E. coli only requires 47 different tRNA species and yet has 86 tRNA genes. The human genome encodes more than 270 tRNA sequences from approximately 450 unique and duplicated tRNA genes (26), whereas the zebrafish genome encodes 12,292 tRNA genes (92). Gene numbers can even vary significantly across highly related species, as shown in Saccharomyces spp., which encode from 171 to 322 tRNA genes (93).
In the almost 60 years since the first tRNA molecule was sequenced (94), much has been learned about the diversity of sequences that fold into the cloverleaf and L-shape secondary and tertiary structures, respectively. The tRNAdb, for example, includes almost 13,000 tRNA sequences from almost 700 species, whereas mitotRNAdb contains more than 30,000 mt-tRNA sequences across more than 1500 species (http://trnadb.bioinf.uni-leipzig.de/). Furthermore, the genome tRNA database, compiled from tRNAscan-SE profiling of complete or nearly complete genomes, predicts more than 400,000 tRNA genes across bacteria, archaea, and eukaryotes (http://gtrnadb.ucsc.edu/).
These sequence data are fundamentally descriptive and offer us little to no insight into how tRNA sequences influence their structure, activity, or stability or what factors influence their processing into tRFs. Understanding those parameters requires a more thorough analysis of tRNA sequences, starting with native species and then experimenting with tRNA design.
A key influence of sequence variation is its impact on tRNA structure, a characteristic critical to therapeutics development whether in terms of biological stability and function or shelf-life. Leamy et al. (95) sought to determine whether tRNA secondary structure or tertiary structure was more important to its thermostability. They found that thermostability of tRNAPhe increased when mutations strengthened base pairing, suggesting the importance of the stem structures. This theme was consistent among tRNAPhe sequences of species living across a range of temperatures. Leveraging such findings, drug developers could optimize sequences to design therapeutic molecules that offer superior stability.
Another factor that can be influenced by tRNA sequence is the degree to which it is aminoacylated, which can determine its translational functions or extratranslational functions. To facilitate such analysis, Evans et al. (96) modified their high-throughput sequencing method DM-tRNA-Seq to determine the charging levels of different tRNAs in mammalian cells at single-nucleotide resolution. They found that while most tRNAs were charged at levels >80%, a couple of tRNAs showed distinctly lower levels. Such analysis could be critical to comparing cell types and states and to ensure designed tRNAs either restore endogenous aminoacylation rates or perhaps outcompete endogenous tRNAs for activity, such as in the development of MS-correcting tRNAs.
The use of tRNA-based medicines could also be challenged by the processing of the intended medicine into tRFs, potentially causing unanticipated secondary effects. Thus, drug development requires a deeper understanding of the parameters, like sequence motifs, that influence endonucleolytic processing of tRNAs. Complicating this effort is the challenge that tRF biogenesis involves a variety of RNases, only some of which are known, including angiogenin (reviewed in Ref. (97)), dicer (98), and ELAC2 (99). Understanding how sequence governs tRNA processing will be critical to learning how to design molecules resistant to cleavage while maintaining optimal biological function.
To optimize the biological half-life of tRNAs and tRFs, strategies to protect them from nuclease digestion will have to be developed (100). One challenge is to generate molecules that optimize protection without impeding function.
Given the diversity of known and as yet undiscovered influences of sequence on structure and function, there is an impetus to apply computational tools such as machine learning to scan the myriad sequences and screening data. As explained later, identifying correlations in these datasets will help elucidate the design rules for producing optimal tRNA medicines. This demand will be further increased with the growing awareness of the multiple roles played by post-transcriptional modifications in tRNAs and tRFs.
Optimizing the chemical space
Aside from being the most abundant RNA molecule in the cell, tRNAs are also the most heavily modified, each molecule bearing an average of 13 modifications (reviewed in Ref. (101)), which can include methylation, acetylation, pseudouridylation, adenosine-to-inosine conversion, and highly complex chemical alterations. This epitranscriptomic diversity and ubiquity are both a challenge and an opportunity as these modifications are critical to activities and characteristics, such as post-transcriptional processing, structural stability, cellular transport, and biological function and yet offer layer of diversity in the design of therapeutic candidate libraries.
Epitranscriptomic alteration at multiple nucleotide positions within the RNA as well as in multiple positions within an individual nucleotide (e.g., sugar, backbone, base) significantly expand the explorable universe of tRNA and tRF design (reviewed in Ref. (102)). The 2021 update of the MODOMICS RNA modifications database added 172 new modified residues to its collection, currently totaling 180 nucleotides, 152 nucleosides, and 3 bases ((103); https://genesilico.pl/modomics/statistics/).
Different modifications within an individual molecule will likely provide unforeseen effects that can either be harnessed to improve the molecule’s drug-like characteristics, from stability to efficacy to safety, or that must be overcome. Joshi et al. (104) and Hoffer et al. (105) demonstrated that alterations of tRNA modifications can significantly impact the fidelity of codon–anticodon interactions. Changes in modification at one position within a tRNA impact modifications at other positions within the same molecule, reflective of a hierarchy of modification events (reviewed in Ref. (106)). Thus, it is not only whether a modification exists but also its type, location, and the presence of other modifications that determine its impact on tRNA features.
Modifications also influence the sensitivity of tRNAs to endonucleolytic cleavage to form tRFs (107, 108). Cleavage-sensitive modification status will therefore likely be critical when designing molecules to serve as tRNAs or to be processed into tRFs, helping to optimize function while limiting potential side effects from undesirable processing or degradation.
The limitations of current qualitative and quantitative technologies to accommodate post-transcriptional modifications have slowed efforts to develop these molecules as medicines. The combination of sequencing methodology and computational analysis, as described earlier, is helping to confirm the identity of specific modifications within a tRNA or a tRF, but until this work has been performed on a vast library of natural and synthetic molecules, it will be difficult to fully understand what combination and location of modifications will help in the design of the most impactful therapies. As with efforts to correlate sequence with molecular characteristics and biological activity, machine learning tools will provide pivotal insights to research efforts to optimize epitranscriptomic modifications in candidate tRNA and tRF medicines.
Drug-like properties
The success of translating a therapeutic candidate from the laboratory to the clinic does not rest purely within its biological activity. Instead, a drug candidate—whether small molecule, protein, or nucleic acid—must be optimized for several drug-like properties, such as solubility; target selectivity; immunogenicity; absorption, distribution, metabolism, excretion; and toxicity (109). Efforts to design novel tRNAs and tRFs will require a thorough examination of the impact of sequence or modification changes on aspects such as molecular structure and function, including interactions with functional partners such as RNA-binding proteins and ribosomes. Barraud et al. (110) used time-resolved NMR spectroscopy to follow the maturation and sequential modification of yeast tRNAPhe, noting that modification events occur in a defined sequential order dictated by crosstalk between modifications. Szameit et al. (111) used small-angle X-ray scattering and mass spectrometry to explore the interaction of a full and truncated RNA aptamer with its protein partner. They observed that the aptamers folded into a number of distinct structures and that dimerization was likely required for protein binding.
Pharmacokinetics (PK) and pharmacodynamics of drugs, including RNA medicines, are critical to many therapeutic considerations, such as dosing. In the absence of tRNA-specific data, experiences with other noncoding RNAs offer insights. Godinho et al. (112) developed what they termed PK-modifying anchors for siRNAs that, in some cases, reduced renal clearance by 23-fold and improved tissue exposure by 26-fold. McDougall et al. (113) similarly examined the PK/pharmacodynamics and absorption, distribution, metabolism, excretion properties of siRNA conjugated to GalNAc in both animal models and humans, finding differences between intravenous and subcutaneous dosing. How applicable such mechanisms are to tRNAs or tRFs will need to be explored.
Immunogenicity also represents a major concern for RNA-based medicines given that innate immune systems constantly monitor their environment for the dsRNA signature of invading viruses (reviewed in Ref. (114)). Epitranscriptomic modification may help to reduce immune activation (114, 115). Freund et al. (116) noted that loss of guanosine 2′-O-methylation of tRNA increased immunostimulation of peripheral blood mononuclear cells, suggesting that modifications suppress the immune response. Keller et al. (117) observed a similar impact for 2′-O-methylthymidine in tRNALys3-treated peripheral blood mononuclear cells. Nallagatla et al. (118) found that altering tRNAs with a variety of epitranscriptomic modifications abrogated their ability to activate PKR, a protein kinase involved in innate immunity.
Immunological considerations will also be important in the development of tRF-based medicines as several fragments have been shown to interact with Toll-like receptors to induce Th1-mediated (119) and interferon-mediated immune responses (reviewed in Ref. (120)).
Given the novelty of developing tRNAs and tRFs as therapeutic interventions, little is known about the design challenges of ensuring that these molecules can be safely delivered to and enter target tissues to perform the desired functions before being metabolized and eliminated, hopefully without detrimental effects. The expansion and diversification of candidate libraries and the continued evolution of high-throughput screens to explore these parameters will be a critical first step to building that reservoir of understanding.
Leveraging machine learning
The large volume and great variety of data arising from high-throughput automated characterization methods provide both opportunities and challenges to the interpretation and integration of data. This could make it more difficult to identify correlations between experimental changes and biological outcomes within and across existing experiments or to develop testable in silico models. To achieve these ends, researchers will need to use and evolve artificial intelligence approaches and machine learning tools.
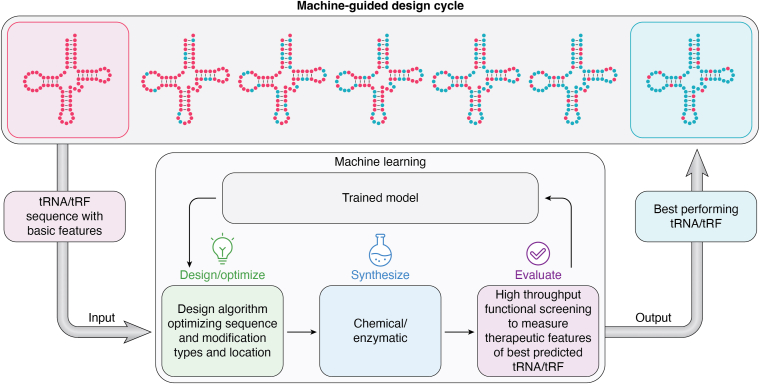
Machine learning models are of particular interest for drug design as they enable the development process to shift from largely empirical approaches to leverage powerful predictive tools, thus augmenting and accelerating discovery and optimization processes (Fig. 6). Although the use of machine learning tools for tRNA-related applications is still in its early stages, ongoing work seeks to align these tools with automated large-scale screening methods, which in turn strive to identify specific patterns and elucidate parameters influencing RNA structure, stability, efficacy, and delivery, as well as toxicity and immunogenicity.
Figure 6.
Machine learning application to program tRNAs into medicines. Optimizing the vast sequence and modification space of tRNA–tRF molecules is enabled through high-throughput and automated screening tools paired with the power of predictive machine learning models that can be trained and retrained on large datasets to generate high-performing tRNA–tRF medicines with specific therapeutic features. tRF, tRNA-derived fragment.
A deep learning approach, tRNA-DL, was applied to improve one of the most commonly available tRNA annotation tools, tRNAscan-SE to reduce false-positive outputs (121). Panwar and Raghava (122) published a webserver, tRNAmod, to predict post-transcriptional modifications in tRNAs based on the MODOMICS datasets. Sonney et al. (123) also evaluated the possible impact of all single-base variants and deletions across mitochondrial tRNAs. The Mitochondrial tRNA Informatics Predictor provided good sensitivity and specificity in predicting pathogenicity and highlighted potential inconsistencies in the database sources where specific variants may have been incorrectly associated with disease outcomes.
Machine learning is a critical component of Alltrna's platform to develop therapeutic tRNAs. Using an iterative, high-throughput, and automated loop of design–build–test–learn, the company seeks to increase the predictive capacity of its in silico models, leading to tRNA and tRF medicines with optimized therapeutic features. Learning from in silico models may also suggest new avenues of exploration or highlight unproductive lines of inquiry that can be abandoned. It will be important to find ways to improve the quality, comprehensiveness, and scale of the data arising from high-throughput experimentation and model systems, as well as identify ways to better integrate these data sources to generate stronger predictions of therapeutic parameters like product stability, efficacy, safety, and manufacturability.
Delivering tRNAs to patients
Given the inherent sensitivity of all RNAs to degradation and their immunogenic properties, as well as the need for effective targeting of RNA-based medicines to the appropriate tissues, such medicines are likely to be administered within a delivery vehicle, tRNAs and tRFs included. Following the lead of gene therapy and gene editing as well as RNA-based medicines, such as the severe acute respiratory syndrome coronavirus 2 vaccines, a broad array of viral and nonviral delivery methods has been developed over the years that modulate cellular targeting, uptake and intracellular release, as well as mitigate the challenges of therapeutic administration of RNA. The unique structural and chemical composition of tRNAs and tRFs combined with their compact size opens prospects for testing established delivery tools and designing novel vehicles (Table 1).
Viral delivery
Because of their efficiency in delivering genetic material to cells, viral vectors such as adenovirus and adeno-associated virus (AAV) have been extensively studied for therapeutic application of RNAs. Although cargo capacity limits prevent these viruses from delivering large genes, the small size of tRNAs and tRFs means that viral constructs can carry single or multiple copies of a given sequence.
Buvoli et al. (124) created plasmid constructs with up to 16 copies of an ochre-suppressing tRNASer to modulate expression of a reporter gene in mouse heart and skeletal muscle. Recently, recombinant AAV delivery of a suppressor tRNA safely and efficiently rescued mice carrying a PTC-mutated alpha-l-iduronidase (IDUA) gene (125). Mechanistic studies suggested that the construct induced PTC readthrough and inhibited NMD. Adjusting the AAV capsid and routes of administration, therapeutic efficacy of the engineered tRNA was achieved in multiple target tissues with a single treatment lasting through 6 months. It should also be possible to combine different tRNA or tRF sequences into a single construct, thereby expressing multiple molecules and potentially modulating different regulatory mechanisms or molecular pathways simultaneously.
To overcome issues related to immunogenicity and genomic integration efficiency, which have slowed the clinical application of viral vectors (reviewed in the study by Ghosh et al. (126)), Havlik et al. (127) evolved a humanized AAV8 capsid with altered surface loop structures. The resulting virus displayed tropism for human hepatocytes and improved gene transfer efficiency, while at the same time evading neutralizing antibodies. The results were consistent with Ogden et al. (128), who designed and verified a diverse in vivo-targeted capsid library using systematic mutagenesis and machine-guided engineering. Given the impact of transcriptional processing and post-transcriptional modification on tRNA and tRF function and stability, one drawback of delivering DNA rather than RNA is that cellular processing and modification are limited to the natural make-up of the respective target cells. To leverage the full scope of drug design and programmability of tRNA and tRF molecules, it may be more desirable to deliver appropriately modified RNA rather than DNA.
Lipid nanoparticles
The development of lipid nanoparticle (LNP)-based RNA medicines has gained momentum because of its recent use in vaccines, for treatment of various genetic diseases and cancers, and in gene editing. Kawamura et al. (129) used a liposomal formulation known as MITO-Porter to deliver wildtype pre-tRNAPhe into cells from patients with a mutation in mitochondrial tRNAPhe. In vitro, the encapsulated pre-tRNA was taken up by cells, where it was transported to mitochondria, improving mitochondrial function. ReCode Therapeutics similarly used an LNP formulation to restore CFTR function in patient-derived bronchial epithelial cells (https://recodetx.com/recode-therapeutics-presents-preclinical-data-using-sort-lnptm-and-rna-platforms-to-rescue-cftr-function-at-the-44th-european-cystic-fibrosis-conference-ecfs/). The growing experience with and successful use of LNPs in RNA delivery, such as with the severe acute respiratory syndrome coronavirus 2 vaccines and the siRNA-based patisiran, has led to proven processes for LNP manufacturing, formulation, and storage. There is also a growing awareness of the factors that influence product biocompatibility, bioavailability, and safety (reviewed in Ref. (130)).
One of the limitations of the original LNP formulations was their preferential distribution to the liver (reviewed in Ref. (131)). Although this may not present a problem for some clinical applications, it potentially limits efficacy for extrahepatic targets and may trigger hepatoxicity. Modifying LNPs to target specific cell types, tissues, or organs is an area of active research. Dilliard et al. (132) altered LNPs with selective organ targeting molecules that interact with distinct serum proteins that in turn recognize specific receptors in targeted tissues (e.g., liver, lung, spleen). Other groups are modifying LNPs with ligands that directly interact with receptors on target tissues or that help the LNPs transit biological barriers (reviewed in Ref. (133)).
Several studies have shown that cationic and ionizable lipids can stimulate the secretion of proinflammatory cytokines (reviewed in Ref. (134)), which can interfere with drug efficacy. Lokugamage et al. (135) observed that LNP-mediated Toll-like receptor 4 activation blocked mRNA translation in all the cell types they tested, and yet, it did not disrupt LNP uptake by cells. Others have noticed that the attachment of PEG to enhance LNP performance may also induce hypersensitivity reactions, accelerating blood clearance of pegylated LNPs and thereby reducing their efficacy.
The transient nature of RNA therapeutics is both a distinct advantage of the modality and a potential drawback as it impacts dosage and dosing frequency. Many disease states will require repeat dosing of the RNA, making the overall tolerability of LNP-encapsulated RNA an absolute necessity. This need is driving drug developers to find the ideal lipid composition for efficient targeting, reduced hepatotoxicity and immunogenicity, and fast biodegradability (reviewed in Ref. (136)), without compromising the required potency or stability.
Other nanoparticles
Other nonlipid nanoparticles are comprised of natural (e.g., chitosan, hyaluronic acid) or synthetic polymers (e.g., PEG, polyethyleneimine derivatives) that are biocompatible, bioavailable, and can be functionalized to improve delivery efficiency and targeting. Although there is no reported use of these delivery vehicles with tRNAs or tRFs, preclinical results with other RNA species offer promise. Chan et al. (137) created PEG–polyamine nanocomplexes with luciferase and green fluorescent reporter mRNAs for injection into the brains of mice. Whereas naked mRNA produced little luminescence, the nanocomplex exhibited strong and lasting signal in mouse brains with lower cytotoxicity and immunogenicity than the naked mRNA.
Mbatha et al. (138) modified gold nanoparticles with polyamidoamine to deliver mRNA to cells in vitro. Compared with mRNA complexed with polyamidoamine alone, the addition of the gold nanoparticles improved cell viability and was more efficient in transfecting mRNA into multiple cell lines. Selenium- and zinc-based nanoparticles were used to target cancer cells in vitro with mRNA (139) and siRNA (140) or miRNA (141), respectively. It remains to be seen if these nanoparticles can support tRNA or tRF delivery.
Ligand conjugation
Avoiding the complexities of nanoparticles or viruses, smaller RNA molecules can also be conjugated directly to specific molecules/moieties, such as ligands of cell receptors for tissue-specific delivery. By homing treatment to the organ of interest, such ligands could conceivably lead to lower amounts and less frequent drug dosing for patients and reduce the risk of off-target effects in other tissues. This approach of “self-deliverable” RNA has been promising for small RNA species, including tRNAs. Wang et al. (142) appended a fragment of H1 RNA, a component of human RNase P, to in vitro synthesized mt-tRNA precursors and found that not only were the molecules transported into isolated mitochondria but also they were processed into mature tRNAs that partially rescued the translation defect in mitochondria from patients with myoclonic epilepsy with ragged red fibers and mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MERRF and MELAS, respectively).
Other ligands have been more thoroughly tested with siRNAs. Chernikov et al. (143) developed a nuclease-resistant anti-MDR1 cholesterol–siRNA conjugate that significantly reduced expression of P-glycoprotein in mice carrying a human xenograft overexpressing MDR1. Rider et al. (144) showed that conjugation of GalNAc to antilipoprotein (a) (LPA) siRNA significantly reduced LPA expression in human and cynomolgus hepatocytes in vitro and reduced LPA expression and serum LPA levels in vivo in cynomolgus monkeys.
GalNAc conjugates may also influence therapeutic dosing and duration of response, as shown by Brown et al. (145), who delivered an identical siRNA to mice using an LNP formulation (intravenous) and a GalNAc conjugate (subcutaneous). Whereas the LNP formulation produced a rapid onset but short duration of activity, the GalNAc formulation had a much slower onset but sustained duration of activity. If these differences translate from siRNA to tRNAs, they too could significantly influence the choice of delivery vehicle for tRNA medicines as researchers try to optimize pharmacological factors, such as dosing, speed of onset, and duration of activity.
Extracellular vesicles
Within organisms, both tRNAs and tRFs function within the cells in which they are transcribed and processed and readily move between cells, often encapsulated within extracellular vesicles (EVs)—lipid-bound vesicles released by cells as a natural component of intercellular signaling—or in ribonucleoprotein complexes (reviewed in Ref. (146)). As their molecular cargo reflects the cell of origin, EVs have become attractive targets as possible disease biomarkers that can be acquired noninvasively from the body fluids in which they circulate. They may also provide a mechanism to actively transport engineered molecules for therapeutic purposes.
Gámbaro et al. (147) packaged synthetic tRNA halves into EVs by first transfecting the molecules into donor cells and then used purified donor EVs to transfer the RNA molecules to acceptor cells. They found that the level of tRNA halves in acceptor cells reflected levels in the donor cells and that the molecules were biologically active, altering the transcriptional profile of acceptor cells. Although EVs can be harvested from multiple sources that make them biocompatible and could help with cellular targeting, their biological origin presents challenges such as the possible lack of size or composition uniformity that is demanded for other delivery vehicles (reviewed in Refs. (148, 149)). Work is ongoing to develop clinic-grade bioproduction and purification processes to ensure consistent quality and efficacy. EVs endogenous to the patient receiving treatment could reduce immunogenicity risk, but even off-the-shelf vehicles can be adjusted to facilitate therapy without immunogenicity. Methods for the large-scale manufacturing of quality-assured, economically viable EVs are still in development.
Clinical impact of tRNA medicines
Despite the relatively nascent efforts to translate tRNAs and tRFs into medicines, the potential transformative impact of these medicines on the treatment of a range of diseases is clear. It may no longer be necessary to treat many conditions based on their pathology or to tackle genetic disorders one gene or one protein at a time. Rather, treatments may become agnostic to the specific gene, focusing instead on the genetic mutation itself (Fig. 1). Thus, one tRNA medicine may be able to treat multiple diseases caused by a shared mutation. Given its long experimental history, the first developed tRNA-based therapies may be PTC-readthrough molecules to treat “stop codon disease,” disorders that arise from the mutation of a codon to a PTC.
The universal approach of using tRNA medicines to treat shared genetic mutations would enable clinicians to target diseases that currently have no treatment options and would facilitate the treatment of disorders so rare that their small patient populations lessen commercial interest for therapy development. Expanding the potential patient population for a mutation-specific treatment would allow clinicians and companies to design clinical trials as basket trials, as is the case with many oncology studies. Furthermore, a larger patient population under the umbrella of stop codon disease could enable viable drug development programs that were previously financially unfeasible. This approach would be a paradigm shift in the treatment of patients with rare and ultrarare diseases.
Summary
While the promise of RNA medicines is well documented, until recently, tRNAs have not been included in this well of potential. Instead, tRNAs have been largely viewed simply as mediators of protein translation, as their broader roles in biology remained poorly understood. Fortunately, this perspective is changing—reflected by the numerous companies exploring tRNAs as therapeutics, including Shape Therapeutics, hC Bioscience, and Tevard Biosciences, as well as by companies like Alltrna building entire platforms around this molecule.
Bringing tRNA-based medicines to fruition will require a concerted effort to fill the large gaps that remain in our understanding of tRNA and tRF biology and chemistry. Existing automated and high-throughput screening technologies will need to be aligned and augmented with new methodologies for tRNA characterization. Using artificial intelligence and machine learning tools, the resulting data will need to be transformed into actionable information that can be used to correlate molecular sequence and modification with biological structure and function. These correlations will form the basis of a systematic design–build–test–learn cycle that will enable the programmability of tRNAs into potent, selective, and safe medicines.
The ever-expanding understanding of the factors influencing tRNA and tRF biology, structure, and function will facilitate efforts to tackle human diseases differently—leveraging, rather than circumventing or overcoming, intrinsic biological mechanisms. By shifting the therapeutic focus from disorder pathology to disease genetics and thereby making treatment both clinically practical and commercially viable, this approach offers hope to patients with rare and ultrarare diseases.
By moving away from conventional treatment approaches that address each disease or gene individually, drug developers can directly tackle the once insurmountable challenge of developing treatments for the more than 6000 genetic disorders that affect over 300 million people worldwide. A concerted effort to address yet unanswered questions will help to progress this novel tRNA modality and usher in a reality in which far more than today’s 5% of rare genetic diseases have Food and Drug Administration–approved medicines, helping to provide treatment for the millions of people with rare diseases but currently no disease-modifying therapy.
Conflict of interest
Dr Anastassiadis and Dr Köhrer are employees and shareholders of Alltrna. The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We extend special thanks to Randall C Willis and Elizabeth Pavone for draft copy editing support and Paula Adduci for graphic design of the figures. We also thank Rachel Green and Alltrna colleagues Lovisa Afzelius, Stephen Eichhorn, Samantha Prezioso, Siddharth Shukla, and Michelle Werner for the helpful comments and informative discussion that improved the article. The work was funded by Alltrna, Inc.
Author contributions
T. A. and C. K. writing–original draft; T. A. and C. K. visualization.
Reviewed by members of the JBC Editorial Board. Edited by Karin Musier-Forsyth
References
- 1.Kawaji H., Nakamura M., Takahashi Y., Sandelin A., Katayama S., Fukuda S., et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole C., Sobala A., Lu C., Thatcher S.R., Bowman A., Brown J.W.S., et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y.S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hug N., Longman D., Cáceres J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurosaki T., Maquat L.E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 2016;129:461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nickless A., Bailis J.M., You Z. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci. 2017;7:26. doi: 10.1186/s13578-017-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schilff M., Sargsyan Y., Hofhuis J., Thoms S. Stop codon context-specific induction of translational readthrough. Biomolecules. 2021;11:1006. doi: 10.3390/biom11071006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dabrowski M., Bukowy-Bieryllo Z., Zietkiewicz E. Translational readthrough potential of natural termination codons in eucaryotes--The impact of RNA sequence. RNA Biol. 2015;12:950–958. doi: 10.1080/15476286.2015.1068497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeling K.M., Xue X., Gunn G., Bedwell D.M. Therapeutics based on stop codon readthrough. Annu. Rev. Genomics Hum. Genet. 2014;15:371–394. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selimoglu E. Aminoglycoside-induced ototoxicity. Curr. Pharm. Des. 2007;13:119–126. doi: 10.2174/138161207779313731. [DOI] [PubMed] [Google Scholar]
- 11.Lopez-Novoa J.M., Quiros Y., Vicente L., Morales A.I., Lopez-Hernandez F.J. New insights into the mechanism of aminoglycoside nephrotoxicity: an integrative point of view. Kidney Int. 2011;79:33–45. doi: 10.1038/ki.2010.337. [DOI] [PubMed] [Google Scholar]
- 12.Lueck J.D., Yoon J.S., Perales-Puchalt A., Mackey A.L., Infield D.T., Behlke M.A., et al. Engineered transfer RNAs for suppression of premature termination codons. Nat. Commun. 2019;10:822. doi: 10.1038/s41467-019-08329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko W., Porter J.J., Sipple M.T., Edwards K.M., Lueck J.D. Efficient suppression of endogenous CFTR nonsense mutations using anticodon-engineered transfer RNAs. Mol. Ther. Nucleic Acids. 2022;28:685–701. doi: 10.1016/j.omtn.2022.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Ma J., Lu W., Tian M., Thauvin M., Yuan C., et al. Heritable expansion of the genetic code in mouse and zebrafish. Cell Res. 2017;27:294–297. doi: 10.1038/cr.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albers S., Beckert B., Matthies M.C., Mandava C.S., Schuster R., Seuring C., et al. Repurposing tRNAs for nonsense suppression. Nat. Commun. 2021;12:3850. doi: 10.1038/s41467-021-24076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albers S., Allen E.C., Bharti N., Davyt M., Joshi D., Perez-Garcia C.G., et al. Engineered tRNAs suppress nonsense mutations in cells and in vivo. Nature. 2023;618:842–848. doi: 10.1038/s41586-023-06133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeBenedictis E.A., Söll D., Esvelt K.M. Measuring the tolerance of the genetic code to altered codon size. Elife. 2022 doi: 10.7554/elife.76941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riddle D.L., Carbon J. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nat. New Biol. 1973;242:230–234. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- 19.Sako Y., Usuki F., Suga H. A novel therapeutic approach for genetic diseases by introduction of suppressor tRNA. Nucleic Acids Symp. Ser. (Oxf). 2006;50:239–240. doi: 10.1093/nass/nrl119. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez E.A., Lester H.A., Dougherty D.A. In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8650–8655. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeBenedictis E.A., Carver G.D., Chung C.Z., Söll D., Badran A.H. Multiplex suppression of four quadruplet codons via tRNA directed evolution. Nat. Commun. 2021;12:5706. doi: 10.1038/s41467-021-25948-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian Q., Li J.N., Zhao H., Hagervall T.G., Farabaugh P.J., Björk G.R. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell. 1998;1:471–482. doi: 10.1016/s1097-2765(00)80048-9. [DOI] [PubMed] [Google Scholar]
- 23.Fagan C.E., Maehigashi T., Dunkle J.A., Miles S.J., Dunham C.M. Structural insights into translational recoding by frameshift suppressor tRNASufJ. RNA. 2014;20:1944–1954. doi: 10.1261/rna.046953.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatfield D., Lee B.J., Smith D.W.E., Oroszlan S. Progress in Molecular and Subcelluar Biology. Springer Berlin Heidelberg; Berlin, Heidelberg: 1990. Role of nonsense, frameshift, and missense suppressor tRNAs in mammalian cells. [Google Scholar]
- 25.Aharon-Hefetz N., Frumkin I., Mayshar Y., Dahan O., Pilpel Y., Rak R. Manipulation of the human tRNA pool reveals distinct tRNA sets that act in cellular proliferation or cell cycle arrest. Elife. 2020;9 doi: 10.7554/eLife.58461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dittmar K.A., Goodenbour J.M., Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J., Chen W., Zhou F., Pang Z., Wang L., Pan T., et al. Tissue-specific reprogramming of host tRNA transcriptome by the microbiome. Genome Res. 2021;31:947–957. doi: 10.1101/gr.272153.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gingold H., Tehler D., Christoffersen N.R., Nielsen M.M., Asmar F., Kooistra S.M., et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 29.Rak R., Polonsky M., Eizenberg-Magar I., Mo Y., Sakaguchi Y., Mizrahi O., et al. Dynamic changes in tRNA modifications and abundance during T cell activation. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2106556118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodarzi H., Nguyen H.C.B., Zhang S., Dill B.D., Molina H., Tavazoie S.F. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165:1416–1427. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos M., Fidalgo A., Varanda A.S., Soares A.R., Almeida G.M., Martins D., et al. Upregulation of tRNA-Ser-AGA-2-1 promotes malignant behavior in normal bronchial cells. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.809985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavon-Eternod M., Gomes S., Geslain R., Dai Q., Rosner M.R., Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dedon P.C., Begley T.J. Dysfunctional tRNA reprogramming and codon-biased translation in cancer. Trends Mol. Med. 2022;28:964–978. doi: 10.1016/j.molmed.2022.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pang Y.L.J., Abo R., Levine S.S., Dedon P.C. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res. 2014;42:e170. doi: 10.1093/nar/gku945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan C.T.Y., Deng W., Li F., DeMott M.S., Babu I.R., Begley T.J., et al. Highly predictive reprogramming of tRNA modifications is linked to selective expression of codon-biased genes. Chem. Res. Toxicol. 2015;28:978–988. doi: 10.1021/acs.chemrestox.5b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torrent M., Chalancon G., de Groot N.S., Wuster A., Babu M.M. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 2018 doi: 10.1126/scisignal.aat6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y. A code within the genetic code: codon usage regulates co-translational protein folding. Cell Commun. Signal. 2020;18:145. doi: 10.1186/s12964-020-00642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng Z., Bromberg Y. Predicting functional effects of synonymous variants: a systematic review and perspectives. Front. Genet. 2019;10:914. doi: 10.3389/fgene.2019.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Presnyak V., Alhusaini N., Chen Y.-H., Martin S., Morris N., Kline N., et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arias L., Martínez F., González D., Flores-Ríos R., Katz A., Tello M., et al. Modification of transfer RNA levels affects cyclin aggregation and the correct duplication of yeast cells. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.607693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosano G.L., Ceccarelli E.A. Rare codon content affects the solubility of recombinant proteins in a codon bias-adjusted Escherichia coli strain. Microb. Cell Fact. 2009;8:41. doi: 10.1186/1475-2859-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guimaraes J.C., Mittal N., Gnann A., Jedlinski D., Riba A., Buczak K., et al. A rare codon-based translational program of cell proliferation. Genome Biol. 2020;21:44. doi: 10.1186/s13059-020-1943-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanson G., Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bazzini A.A., Del Viso F., Moreno-Mateos M.A., Johnstone T.G., Vejnar C.E., Qin Y., et al. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 2016;35:2087–2103. doi: 10.15252/embj.201694699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Q., Medina S.G., Kushawah G., DeVore M.L., Castellano L.A., Hand J.M., et al. Translation affects mRNA stability in a codon-dependent manner in human cells. Elife. 2019;8 doi: 10.7554/eLife.45396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamble C.E., Brule C.E., Dean K.M., Fields S., Grayhack E.J. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell. 2016;166:679–690. doi: 10.1016/j.cell.2016.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodarzi H., Liu X., Nguyen H.C.B., Zhang S., Fish L., Tavazoie S.F. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guzzi N., Cieśla M., Ngoc P.C.T., Lang S., Arora S., Dimitriou M., et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173:1204–1216. doi: 10.1016/j.cell.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 49.Goncalves K.A., Silberstein L., Li S., Severe N., Hu M.G., Yang H., et al. Angiogenin promotes hematopoietic regeneration by dichotomously regulating quiescence of stem and progenitor cells. Cell. 2016;166:894–906. doi: 10.1016/j.cell.2016.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S., Li H., Zheng L., Li H., Feng C., Zhang W. Identification of functional tRNA-derived fragments in senescence-accelerated mouse prone 8 brain. Aging (Albany NY) 2019;11:10485–10498. doi: 10.18632/aging.102471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Green J.A., Ansari M.Y., Ball H.C., Haqqi T.M. tRNA-derived fragments (tRFs) regulate post-transcriptional gene expression via AGO-dependent mechanism in IL-1β stimulated chondrocytes. Osteoarthritis Cartilage. 2020;28:1102–1110. doi: 10.1016/j.joca.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Z., Jijiwa M., Nasu M., Borgard H., Gong T., Xu J., et al. Comprehensive landscape of tRNA-derived fragments in lung cancer. Mol. Ther. Oncolytics. 2022;26:207–225. doi: 10.1016/j.omto.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhan S., Yang P., Zhou S., Xu Y., Xu R., Liang G., et al. Serum mitochondrial tsRNA serves as a novel biomarker for hepatocarcinoma diagnosis. Front. Med. 2022;16:216–226. doi: 10.1007/s11684-022-0920-7. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y., Gu X., Qin X., Huang Y., Ju S. Evaluation of serum tRF-23-Q99P9P9NDD as a potential biomarker for the clinical diagnosis of gastric cancer. Mol. Med. 2022;28:63. doi: 10.1186/s10020-022-00491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen H., Xu Z., Cai H., Peng Y., Yang L., Wang Z. Identifying differentially expressed tRNA-derived small fragments as a biomarker for the progression and metastasis of colorectal cancer. Dis. Markers. 2022;2022 doi: 10.1155/2022/2646173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang P., Zhang X., Chen S., Tao Y., Ning M., Zhu Y., et al. A novel serum tsRNA for diagnosis and prediction of nephritis in SLE. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Magee R., Londin E., Rigoutsos I. tRNA-derived fragments as sex-dependent circulating candidate biomarkers for Parkinson’s disease. Parkinsonism Relat. Disord. 2019;65:203–209. doi: 10.1016/j.parkreldis.2019.05.035. [DOI] [PubMed] [Google Scholar]
- 58.Li Y., Cui S., Xu Z., Zhang Y., Wu T., Zhang J., et al. Exosomal tRF-Leu-AAG-001 derived from mast cell as a potential non-invasive diagnostic biomarker for endometriosis. BMC Womens Health. 2022;22:253. doi: 10.1186/s12905-022-01827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y., Cai F., Liu J., Chang H., Liu L., Yang A., et al. Transfer RNA-derived fragments as potential exosome tRNA-derived fragment biomarkers for osteoporosis. Int. J. Rheum. Dis. 2018;21:1659–1669. doi: 10.1111/1756-185X.13346. [DOI] [PubMed] [Google Scholar]
- 60.Fan H., Liu H., Lv Y., Song Y. AS-tDR-007872: a novel tRNA-derived small RNA acts an important role in non-small-cell lung cancer. Comput. Math. Methods Med. 2022;2022 doi: 10.1155/2022/3475955. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Xu W., Zhou B., Wang J., Tang L., Hu Q., Wang J., et al. tRNA-derived fragment tRF-Glu-TTC-027 regulates the progression of gastric carcinoma via MAPK signaling pathway. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.733763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao K.-Y., Pan Y., Yan T.-M., Tao P., Xiao Y., Jiang Z.-H. Antitumor activities of tRNA-derived fragments and tRNA halves from non-pathogenic Escherichia coli strains on colorectal cancer and their structure-activity relationship. mSystems. 2022;7 doi: 10.1128/msystems.00164-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao K.-Y., Yan T.-M., Zhang J.-Z., Chan T.-F., Li J., Li C., et al. A tRNA-derived fragment from Chinese yew suppresses ovarian cancer growth via targeting TRPA1. Mol. Ther. Nucleic Acids. 2022;27:718–732. doi: 10.1016/j.omtn.2021.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu K., Yan T.-M., Cao K.-Y., Li F., Ma X.-R., Lai Q., et al. A tRNA-derived fragment of ginseng protects heart against ischemia/reperfusion injury via targeting the lncRNA MIAT/VEGFA pathway. Mol. Ther. Nucleic Acids. 2022;29:672–688. doi: 10.1016/j.omtn.2022.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.El-Sagheer A.H., Brown T. New strategy for the synthesis of chemically modified RNA constructs exemplified by hairpin and hammerhead ribozymes. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15329–15334. doi: 10.1073/pnas.1006447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohtsuka E., Tanaka S., Tanaka T., Miyake T., Markham A.F., Nakagawa E., et al. Total synthesis of a RNA molecule with sequence identical to that of Escherichia coli formylmethionine tRNA. Proc. Natl. Acad. Sci. U. S. A. 1981;78:5493–5497. doi: 10.1073/pnas.78.9.5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogilvie K.K., Usman N., Nicoghosian K., Cedergren R.J. Total chemical synthesis of a 77-nucleotide-long RNA sequence having methionine-acceptance activity. Proc. Natl. Acad. Sci. U. S. A. 1988;85:5764–5768. doi: 10.1073/pnas.85.16.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanpure A.A., Balasubramanian S. Synthesis and multiple incorporations of 2’-O-methyl-5-hydroxymethylcytidine, 5-hydroxymethylcytidine and 5-formylcytidine monomers into RNA oligonucleotides. Chembiochem. 2017;18:2236–2241. doi: 10.1002/cbic.201700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He M., Wu X., Mao S., Haruehanroengra P., Khan I., Sheng J., et al. Bio-orthogonal chemistry enables solid-phase synthesis and HPLC and gel-free purification of long RNA oligonucleotides. Chem. Commun. (Camb). 2021;57:4263–4266. doi: 10.1039/d1cc00096a. [DOI] [PubMed] [Google Scholar]
- 70.Blümler A., Schwalbe H., Heckel A. Solid-phase-supported chemoenzymatic synthesis of a light-activatable tRNA derivative. Angew. Chem. Int. Ed. Engl. 2022;61 doi: 10.1002/anie.202111613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ryczek M., Pluta M., Błaszczyk L., Kiliszek A. Overview of methods for large-scale RNA synthesis. Appl. Sci. 2022;12:1543. [Google Scholar]
- 72.Pinkard O., McFarland S., Sweet T., Coller J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 2020;11:4104. doi: 10.1038/s41467-020-17879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Behrens A., Rodschinka G., Nedialkova D.D. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol. Cell. 2021;81:1802–1815. doi: 10.1016/j.molcel.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng G., Qin Y., Clark W.C., Dai Q., Yi C., He C., et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamagami R., Hori H. Application of mutational profiling: new functional analyses reveal the tRNA recognition mechanism of tRNA m1A22 methyltransferase. J. Biol. Chem. 2023;299 doi: 10.1016/j.jbc.2022.102759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thomas N.K., Poodari V.C., Jain M., Olsen H.E., Akeson M., Abu-Shumays R.L. Direct nanopore sequencing of individual full length tRNA strands. ACS Nano. 2021;15:16642–16653. doi: 10.1021/acsnano.1c06488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ramasamy S., Mishra S., Sharma S., Parimalam S.S., Vaijayanthi T., Fujita Y., et al. An informatics approach to distinguish RNA modifications in nanopore direct RNA sequencing. Genomics. 2022;114 doi: 10.1016/j.ygeno.2022.110372. [DOI] [PubMed] [Google Scholar]
- 78.Abebe J.S., Price A.M., Hayer K.E., Mohr I., Weitzman M.D., Wilson A.C., et al. DRUMMER--rapid detection of RNA modifications through comparative nanopore sequencing. Bioinformatics. 2022;38:3113–3115. doi: 10.1093/bioinformatics/btac274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lucas M.C., Pryszcz L.P., Medina R., Milenkovic I., Camacho N., Marchand V., et al. Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nat. Biotechnol. 2023 doi: 10.1038/s41587-023-01743-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thakur P., Estevez M., Lobue P.A., Limbach P.A., Addepalli B. Improved RNA modification mapping of cellular non-coding RNAs using C- and U-specific RNases. Analyst. 2020;145:816–827. doi: 10.1039/c9an02111f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Addepalli B., Venus S., Thakur P., Limbach P.A. Novel ribonuclease activity of cusativin from Cucumis sativus for mapping nucleoside modifications in RNA. Anal. Bioanal. Chem. 2017;409:5645–5654. doi: 10.1007/s00216-017-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suzuki T., Yashiro Y., Kikuchi I., Ishigami Y., Saito H., Matsuzawa I., et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020;11:4269. doi: 10.1038/s41467-020-18068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar P., Kuscu C., Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends Biochem. Sci. 2016;41:679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar P., Mudunuri S.B., Anaya J., Dutta A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43:D141–D145. doi: 10.1093/nar/gku1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie Y., Yao L., Yu X., Ruan Y., Li Z., Guo J. Action mechanisms and research methods of tRNA-derived small RNAs. Signal. Transduct. Target Ther. 2020;5:109. doi: 10.1038/s41392-020-00217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao W., Gallardo-Dodd C.J., Kutter C. Cell type-specific analysis by single-cell profiling identifies a stable mammalian tRNA-mRNA interface and increased translation efficiency in neurons. Genome Res. 2022;32:97–110. doi: 10.1101/gr.275944.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Isakova A., Neff N., Quake S.R. Single-cell quantification of a broad RNA spectrum reveals unique noncoding patterns associated with cell types and states. Proc. Natl. Acad. Sci. U. S. A. 2021;118:e211356811. doi: 10.1073/pnas.2113568118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Biedenbänder T., de Jesus V., Schmidt-Dengler M., Helm M., Corzilius B., Fürtig B. RNA modifications stabilize the tertiary structure of tRNAfMet by locally increasing conformational dynamics. Nucleic Acids Res. 2022;50:2334–2349. doi: 10.1093/nar/gkac040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leamy K.A., Yamagami R., Yennawar N.H., Bevilacqua P.C. Single-nucleotide control of tRNA folding cooperativity under near-cellular conditions. Proc. Natl. Acad. Sci. U. S. A. 2019;116:23075–23082. doi: 10.1073/pnas.1913418116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gremminger T., Song Z., Ji J., Foster A., Weng K., Heng X. Extended interactions between HIV-1 viral RNA and tRNALys3 are important to maintain viral RNA integrity. Int. J. Mol. Sci. 2020;22:58. doi: 10.3390/ijms22010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Crick F.H. Codon--anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 92.Soares A.R., Fernandes N., Reverendo M., Araújo H.R., Oliveira J.L., Moura G.M.R., et al. Conserved and highly expressed tRNA-derived fragments in zebrafish. BMC Mol. Biol. 2015;16:22. doi: 10.1186/s12867-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iben J.R., Maraia R.J. tRNAomics: tRNA gene copy number variation and codon use provide bioinformatic evidence of a new anticodon:codon wobble pair in a eukaryote. RNA. 2012;18:1358–1372. doi: 10.1261/rna.032151.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Holley R.W., Apgar J., Everett G.A., Madison J.T., Marquisee M., Merrill S.H., et al. Structure of a ribonucleic acid. Science. 1965;147:1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 95.Leamy K.A., Yennawar N.H., Bevilacqua P.C. Molecular mechanism for folding cooperativity of functional RNAs in living organisms. Biochemistry. 2018;57:2994–3002. doi: 10.1021/acs.biochem.8b00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Evans M.E., Clark W.C., Zheng G., Pan T. Determination of tRNA aminoacylation levels by high-throughput sequencing. Nucleic Acids Res. 2017;45:e133. doi: 10.1093/nar/gkx514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wen J., Huang Z., Li Q., Chen X., Qin H., Zhao Y. Research progress on the tsRNA classification, function, and application in gynecological malignant tumors. Cell Death Discov. 2021;7:388. doi: 10.1038/s41420-021-00789-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kazimierczyk M., Wojnicka M., Biała E., Żydowicz-Machtel P., Imiołczyk B., Ostrowski T., et al. Characteristics of transfer RNA-derived fragments expressed during human renal cell development: the role of dicer in tRF biogenesis. Int. J. Mol. Sci. 2022;23:3644. doi: 10.3390/ijms23073644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choi E.-J., Wu W., Zhang K., Lee I., Kim I.-H., Lee Y.S., et al. ELAC2, an enzyme for tRNA maturation, plays a role in the cleavage of a mature tRNA to produce a tRNA-derived RNA fragment during respiratory syncytial virus infection. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.609732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tosar J.P., Gámbaro F., Darré L., Pantano S., Westhof E., Cayota A. Dimerization confers increased stability to nucleases in 5’ halves from glycine and glutamic acid tRNAs. Nucleic Acids Res. 2018;46:9081–9093. doi: 10.1093/nar/gky495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28:395–404. doi: 10.1038/s41422-018-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Väre V.Y.P., Eruysal E.R., Narendran A., Sarachan K.L., Agris P.F. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules. 2017;7:29. doi: 10.3390/biom7010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Boccaletto P., Stefaniak F., Ray A., Cappannini A., Mukherjee S., Purta E., et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022;50:D231–D235. doi: 10.1093/nar/gkab1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Joshi K., Bhatt M.J., Farabaugh P.J. Codon-specific effects of tRNA anticodon loop modifications on translational misreading errors in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2018;46:10331–10339. doi: 10.1093/nar/gky664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoffer E.D., Hong S., Sunita S., Maehigashi T., Gonzalez R.L., Jr., Whitford P.C., et al. Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon:anticodon pairing. Elife. 2020;9 doi: 10.7554/eLife.51898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Motorin Y., Helm M. RNA nucleotide methylation: 2021 update. Wiley Interdiscip Rev RNA. 2022;13 doi: 10.1002/wrna.1691. [DOI] [PubMed] [Google Scholar]
- 107.Blanco S., Dietmann S., Flores J.V., Hussain S., Kutter C., Humphreys P., et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schaefer M., Pollex T., Hanna K., Tuorto F., Meusburger M., Helm M., et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Di L., Kerns E., Carter G.T. Drug-like property concepts in pharmaceutical design. Curr. Pharm. Des. 2009;15:2184–2194. doi: 10.2174/138161209788682479. [DOI] [PubMed] [Google Scholar]
- 110.Barraud P., Gato A., Heiss M., Catala M., Kellner S., Tisné C. Time-resolved NMR monitoring of tRNA maturation. Nat. Commun. 2019;10:3373. doi: 10.1038/s41467-019-11356-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Szameit K., Berg K., Kruspe S., Valentini E., Magbanua E., Kwiatkowski M., et al. Structure and target interaction of a G-quadruplex RNA-aptamer. RNA Biol. 2016;13:973–987. doi: 10.1080/15476286.2016.1212151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Godinho B.M.D.C., Knox E.G., Hildebrand S., Gilbert J.W., Echeverria D., Kennedy Z., et al. PK-modifying anchors significantly alter clearance kinetics, tissue distribution, and efficacy of therapeutics siRNAs. Mol. Ther. Nucleic Acids. 2022;29:116–132. doi: 10.1016/j.omtn.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McDougall R., Ramsden D., Agarwal S., Agarwal S., Aluri K., Arciprete M., et al. The nonclinical disposition and pharmacokinetic/pharmacodynamic properties of N-acetylgalactosamine-conjugated small interfering RNA are highly predictable and build confidence in translation to human. Drug Metab. Dispos. 2022;50:781–797. doi: 10.1124/dmd.121.000428. [DOI] [PubMed] [Google Scholar]
- 114.Mu X., Hur S. Immunogenicity of in vitro-transcribed RNA. Acc. Chem. Res. 2021;54:4012–4023. doi: 10.1021/acs.accounts.1c00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nelson J., Sorensen E.W., Mintri S., Rabideau A.E., Zheng W., Besin G., et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Freund I., Buhl D.K., Boutin S., Kotter A., Pichot F., Marchand V., et al. 2′-O-methylation within prokaryotic and eukaryotic tRNA inhibits innate immune activation by endosomal Toll-like receptors but does not affect recognition of whole organisms. RNA. 2019;25:869–880. doi: 10.1261/rna.070243.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Keller P., Freund I., Marchand V., Bec G., Huang R., Motorin Y., et al. Double methylation of tRNA-U54 to 2′-O-methylthymidine (Tm) synergistically decreases immune response by Toll-like receptor 7. Nucleic Acids Res. 2018;46:9764–9775. doi: 10.1093/nar/gky644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nallagatla S.R., Jones C.N., Ghosh S.K.B., Sharma S.D., Cameron C.E., Spremulli L.L., et al. Native tertiary structure and nucleoside modifications suppress tRNA’s intrinsic ability to activate the innate immune sensor PKR. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Z., Xiang L., Shao J., Yuan Z. The 3’ CCACCA sequence of tRNAAla(UGC) is the motif that is important in inducing Th1-like immune response, and this motif can be recognized by Toll-like receptor 3. Clin. Vaccin. Immunol. 2006;13:733–739. doi: 10.1128/CVI.00019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pandey K.K., Madhry D., Kumar Y.S.R., Malvankar S., Sapra L., Srivastava R.K., et al. Regulatory roles of tRNA-derived RNA fragments in human pathophysiology. Mol. Ther. Nucleic Acids. 2021;26:161–173. doi: 10.1016/j.omtn.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gao X., Wei Z., Hakonarson H. tRNA-DL: a deep learning approach to improve tRNAscan-SE prediction results. Hum. Hered. 2018;83:163–172. doi: 10.1159/000493215. [DOI] [PubMed] [Google Scholar]
- 122.Panwar B., Raghava G.P. Prediction of uridine modifications in tRNA sequences. BMC Bioinformatics. 2014;15:326. doi: 10.1186/1471-2105-15-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sonney S., Leipzig J., Lott M.T., Zhang S., Procaccio V., Wallace D.C., Sondheimer N. Predicting the pathogenicity of novel variants in mitochondrial tRNA with MitoTIP. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Buvoli M., Buvoli A., Leinwand L.A. Suppression of nonsense mutations in cell culture and mice by multimerized suppressor tRNA genes. Mol. Cell Biol. 2000;20:3116–3124. doi: 10.1128/mcb.20.9.3116-3124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang J., Zhang Y., Mendonca C.A., Yukselen O., Muneeruddin K., Ren L., et al. AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice. Nature. 2022;604:343–348. doi: 10.1038/s41586-022-04533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ghosh S., Brown A.M., Jenkins C., Campbell K. Viral vector systems for gene therapy: a comprehensive literature review of progress and biosafety challenges. Appl. Biosaf. 2020;25:7–18. doi: 10.1177/1535676019899502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Havlik L.P., Simon K.E., Smith J.K., Klinc K.A., Tse L.V., Oh D.K., et al. Coevolution of adeno-associated virus capsid antigenicity and tropism through a structure-guided approach. J. Virol. 2020;94 doi: 10.1128/JVI.00976-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ogden P.J., Kelsic E.D., Sinai S., Church G.M. Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science. 2019;366:1139–1143. doi: 10.1126/science.aaw2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kawamura E., Maruyama M., Abe J., Sudo A., Takeda A., Takada S., et al. Validation of gene therapy for mutant mitochondria by delivering mitochondrial RNA using a MITO-Porter. Mol. Ther. Nucleic Acids. 2020;20:687–698. doi: 10.1016/j.omtn.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Han X., Mitchell M.J., Nie G. Nanomaterials for therapeutic RNA delivery. Matter. 2020;3:1948–1975. [Google Scholar]
- 131.Mashima R., Takada S. Lipid nanoparticles: a novel gene delivery technique for clinical application. Curr. Issues Mol. Biol. 2022;44:5013–5027. doi: 10.3390/cimb44100341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dilliard S.A., Cheng Q., Siegwart D.J. On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2021 doi: 10.1073/pnas.2109256118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Montoto S.S., Muraca G., Ruiz M.E. Solid lipid nanoparticles for drug delivery: pharmacological and biopharmaceutical aspects. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.587997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lokugamage M.P., Gan Z., Zurla C., Levin J., Islam F.Z., Kalathoor S., et al. Mild innate immune activation overrides efficient nanoparticle-mediated RNA delivery. Adv. Mater. 2020;32 doi: 10.1002/adma.201904905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kulkarni J.A., Cullis P.R., van der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 2018;28:146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- 137.Chan L.Y., Khung Y.L., Lin C.-Y. Preparation of messenger RNA nanomicelles via non-cytotoxic PEG-polyamine nanocomplex for intracerebroventicular delivery: a proof-of-concept study in mouse models. Nanomaterials (Basel) 2019 doi: 10.3390/nano9010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mbatha L.S., Maiyo F., Daniels A., Singh M. Dendrimer-coated gold nanoparticles for efficient folate-targeted mRNA delivery in vitro. Pharmaceutics. 2021 doi: 10.3390/pharmaceutics13060900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Singh D., Singh M. Hepatocellular-targeted mRNA delivery using functionalized selenium nanoparticles in vitro. Pharmaceutics. 2021 doi: 10.3390/pharmaceutics13030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.He C., Lu K., Liu D., Lin W. Nanoscale metal-organic frameworks for the co-delivery of cisplatin and pooled siRNAs to enhance therapeutic efficacy in drug-resistant ovarian cancer cells. J. Am. Chem. Soc. 2014;136:5181–5184. doi: 10.1021/ja4098862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao H., Li T., Yao C., Gu Z., Liu C., Li J., et al. Dual roles of metal-organic frameworks as nanocarriers for miRNA delivery and adjuvants for chemodynamic therapy. ACS Appl. Mater. Inter. 2021;13:6034–6042. doi: 10.1021/acsami.0c21006. [DOI] [PubMed] [Google Scholar]
- 142.Wang G., Shimada E., Zhang J., Hong J.S., Smith G.M., Teitell M.A., et al. Correcting human mitochondrial mutations with targeted RNA import. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4840–4845. doi: 10.1073/pnas.1116792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chernikov I.V., Gladkikh D.V., Meschaninova M.I., Ven’yaminova A.G., Zenkova M.A., Vlassov V.V., et al. Cholesterol-containing nuclease-resistant siRNA accumulates in tumors in a carrier-free mode and silences MDR1 gene. Mol. Ther. Nucleic Acids. 2017;6:209–220. doi: 10.1016/j.omtn.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rider D.A., Eisermann M., Löffler K., Aleku M., Swerdlow D.I., Dames S., et al. Pre-clinical assessment of SLN360, a novel siRNA targeting LPA, developed to address elevated lipoprotein (a) in cardiovascular disease. Atherosclerosis. 2022;349:240–247. doi: 10.1016/j.atherosclerosis.2022.03.029. [DOI] [PubMed] [Google Scholar]
- 145.Brown C.R., Gupta S., Qin J., Racie T., He G., Lentini S., et al. Investigating the pharmacodynamic durability of GalNAc–siRNA conjugates. Nucleic Acids Res. 2020;48:11827–11844. doi: 10.1093/nar/gkaa670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tosar J.P., Cayota A. Extracellular tRNAs and tRNA-derived fragments. RNA Biol. 2020;17:1149–1167. doi: 10.1080/15476286.2020.1729584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gámbaro F., Calzi M.L., Fagúndez P., Costa B., Greif G., Mallick E., et al. Stable tRNA halves can be sorted into extracellular vesicles and delivered to recipient cells in a concentration-dependent manner. RNA Biol. 2020;17:1168–1182. doi: 10.1080/15476286.2019.1708548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ahn S.-H., Ryu S.-W., Choi H., You S., Park J., Choi C. Manufacturing therapeutic exosomes: from bench to industry. Mol. Cells. 2022;45:284–290. doi: 10.14348/molcells.2022.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Claridge B., Lozano J., Poh Q.H., Greening D.W. Development of extracellular vesicle therapeutics: challenges, considerations, and opportunities. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.734720. [DOI] [PMC free article] [PubMed] [Google Scholar]