Summary
CD9 is a member of the tetraspanin protein family, which has been widely studied in inflammation and cancer, but not in pathological cardiac hypertrophy. In this study, we found that the expression of CD9 was increased in transaortic constriction (TAC) myocardial tissue. Knockdown of CD9 alleviated damage to cardiac function in the TAC model and reduced heart weight, cardiomyocyte size, and degree of fibrosis, and vice versa. Mechanistically, co-immunoprecipitation results showed that CD9 and GP130 can bind to each other in cardiomyocytes, and knockdown of CD9 can reduce the protein level of GP130 and the phosphorylation of STAT3 in vivo and in vitro, and vice versa. GP130 knockdown reversed the aggravating effects of CD9 on pathological cardiac hypertrophy. Therefore, we conclude that CD9 exacerbates pathological cardiac hypertrophy by regulating the GP130/STAT3 signaling pathway and may serve as a therapeutic target for pathological cardiac hypertrophy.
Subject areas: Cardiovascular medicine, Pathophysiology, Molecular physiology
Graphical abstract

Highlights
-
•
The expression of CD9 increased in TAC myocardial tissue
-
•
Knockdown of CD9 alleviated TAC-induced pathological cardiac hypertrophy, and vice versa
-
•
CD9 exacerbates pathological cardiac hypertrophy through regulating GP130/STAT3
Cardiovascular medicine; Pathophysiology; Molecular physiology
Introduction
The increase in cardiomyocyte size in response to an increased workload is called cardiac hypertrophy. Hypertrophy initially develops as an adaptive response to physiological and pathological stimuli. However, pathological cardiac hypertrophy, which is associated with increased interstitial fibrosis, cell death, and cardiac dysfunction, is a key risk factor for heart failure.1 Due to the poor overall prognosis of heart failure, it is important to identify new therapeutic targets to prevent or reverse cardiac hypertrophy and heart failure.
The pathogenesis of pathological cardiac hypertrophy is complex, and the entire process involves abnormal protein synthesis, participation of various cytokines, excessive activation of signal transduction pathways, and re-expression of cardiac embryonic bases. Many signaling cascades and proteins have been implicated in mediating pathological cardiac hypertrophy. GP130/STAT3 signaling plays a key role in pathological cardiac hypertrophy.2,3 In addition to its cytoprotective action, GP130-dependent signaling induces cardiomyocyte hypertrophy.4 Ancey et al. have demonstrated that GP130 activation in human cardiac cells leads to cardiomyocyte hypertrophy.5 GP130 receptor-mediated signaling promotes cardiomyocyte survival, induces hypertrophy, and modulates cardiac extracellular matrix and cardiac function.6 GP130 is critical in pressure overload-induced cardiac hypertrophy, possibly through the STAT3 pathway.7
CD9 is a transmembrane glycoprotein that belongs to the tetraspanin family.8 Depending on the cell type and associated molecules, CD9 has various biological activities such as cell adhesion, cell migration, cellular signaling, inflammation, and tumor-related processes.9,10,11,12 Shi et al. have found that CD9 stabilizes GP130 by preventing ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells.13 Kim et al. found that CD9 expression may play an important role in vascular senescence and the pathogenesis of atherosclerosis.14 Cho et al. have found that CD9 induces cellular senescence and aggravates atherosclerotic plaque formation.15
Therefore, we speculated that CD9 may be a therapeutic target for pathological cardiac hypertrophy. In this study, we investigated whether CD9 plays a role in the pathology of myocardial hypertrophy.
Results
The expression of CD9 increased in TAC myocardial tissue
We searched and analyzed three public mRNA-seq datasets in the GEO database (GSE101977 and GSE180720 from mice and GSE89714 from humans). The results showed increased expression of CD9 in a mouse model of stress-induced cardiac hypertrophy and in the heart tissues of patients with hypertrophic cardiomyopathy (p < 0.05 for CD9, as shown in Figure 1A). We established a mouse model of pathological cardiac hypertrophy using TAC, and the expression of CD9 and ANP in the cardiac tissue four weeks after TAC surgery was detected by western blotting and RT-PCR. The results showed that, compared with the sham group, the expression of CD9 and ANP increased in cardiac tissue after TAC, both at the protein and mRNA levels (Figures 1B and 1C). To determine whether the CD9 protein was expressed in cardiomyocytes, we isolated cardiomyocytes and fibroblasts from neonatal rats and found that the CD9 content in cardiomyocytes was significantly higher than that in fibroblasts (Figure 1D). We used Ang II to stimulate neonatal rat cardiomyocytes (NRCMs) to establish a pathological myocardial hypertrophy model at the cellular level. The results showed that the expression of CD9 and ANP increased in cardiomyocytes stimulated by Ang II at both the protein and mRNA levels (Figures 1E and 1F). Immunofluorescence of CD9 co-stained with the cardiomyocyte marker cTNT in the Sham and TAC groups verified the elevated cardiomyocyte expression of CD9 after TAC surgery (Figure 1G), IgG was used as a negative control (Figure S1A).
Figure 1.
The Expression of CD9 in TAC Myocardial Tissue
(A) The mRNA-seq data (GSE101977, GSE180720, and GSE89714) displayed increased CD9 expression in a stress-induced cardiac hypertrophy mouse model and hypertrophic cardiomyopathy patients' heart tissues (p < 0.05 for CD9).
(B) Western blot analysis of CD9 protein expression in heart tissue from mice 4 weeks post-TAC surgery and the Sham group, with quantification in the right panel (n = 6).
(C) RT-PCR detected CD9 mRNA expression in heart tissue from mice 4 weeks post-TAC surgery and Sham group (n = 6).
(D) CD9 protein expression in cardiomyocytes and fibroblasts analyzed by Western blot, with quantification below (n = 3).
(E) CD9 protein expression in cardiomyocytes under Ang II stimulation examined by Western blot, with quantification below (n = 3).
(F) CD9 mRNA expression in cardiomyocytes under Ang II stimulation detected through RT-PCR.
(G) Representative immunofluorescence image depicting CD9 (Green) co-stained with cTNT (Red) and DAPI in Sham and TAC groups, Scale bar: 100 μm, with quantification in the right panel (n = 3). See also Figure S1A.
(H) Western blot analysis of HIF1α and CD9 protein expression in cardiomyocytes, with quantification in the right panel (n = 3).
(I) The luciferase reporter results showed the CD9 promoter activity (n = 5). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
It has been reported that hypoxia Inducible Factor 1α (HIF1α), the master transcription factor involved in hypoxia response, bind directly CD9 promoter and induce CD9 transcription in leukemic cells.16 We stimulated Ang II after knocking down HIF1α in NRCMs, and found that knocking down HIF1α could abolish the promoting effect of Ang II on CD9 expression (Figure 1H). In addition, we constructed a CD9 promoter reporter plasmid with hypoxia responsive elements (HRE) mutated. The luciferase reporter results showed that Ang II could promote CD9 promoter activity, but had a poor effect on HRE MUT promoter activity in NRCMs (Figure 1I). These results suggest that Ang II may promote CD9 expression through HIF1α.
Knockdown of CD9 ameliorated TAC-induced pathological cardiac hypertrophy
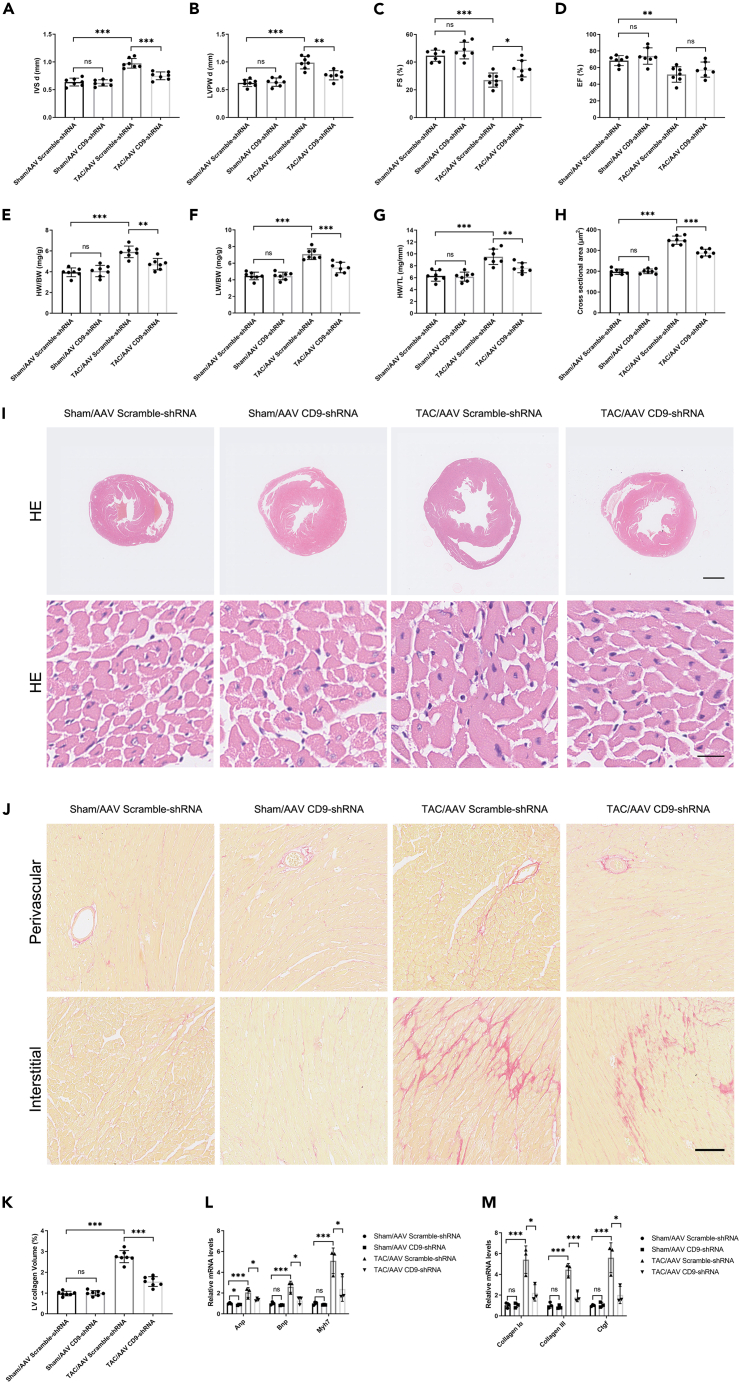
To verify the effect of CD9 on pressure-stressed myocardial hypertrophy in vivo, we injected C57BL/6J mice with AAV CD9-shRNA and AAV Scramble-shRNA virus through the tail vein and performed TAC surgery on the mice 2 weeks later. The cell specificity of AAV9 was determined by immunofluorescence. The GFP signal of the control group injected with the AAV GFP virus was co-stained with ACTA1 (Alexa Fluor 594-conjugated secondary antibody) and DAPI. The specificity of the Alexa Fluor 594-conjugated secondary antibody was determined in the absence of an ACTA1 antibody. The negative control GFP signal was detected in the group injected with AAV Scramble-shRNA (Figure S1B). The knockout efficacy of CD9 is shown by western blotting in Figure S1C. The cardiac function of the mice was detected by ultrasound, and the representative ultrasound images are shown in Figure S1D. Compared with the sham group, the degree of cardiac hypertrophy index IVSd and LVPWd was increased, and the myocardial systolic function EF% and FS% decreased at 4 weeks after TAC. Meanwhile, compared with the TAC/AAV Scramble-shRNA group, the IVSd and LVPWd decreased, and the myocardial systolic function EF% and FS% increased in the TAC/AAV CD9-shRNA group (although the EF% was not statistically different, there was an increasing trend, probably because the sample size was small). (Figures 2A–2D), suggesting that CD9 knockdown can alleviate cardiac function damage after TAC. The mice were sacrificed four weeks after surgery to detect relevant indicators to clarify the effect of CD9 knockdown on pressure-stressed cardiac hypertrophy. As with the ultrasound results, HW/BW, LW/BW, and HW/TL increased significantly at 4 weeks after TAC, and the ratio decreased after CD9 knockdown, indicating that the knockdown of CD9 reduced the heart weight of mice (Figures 2E–2G).
Figure 2.
Knockdown of CD9 Ameliorates TAC-Induced Pathological Cardiac Hypertrophy
C57 mice injected with AAV CD9-shRNA two weeks pre-TAC surgery, sacrificed 4 weeks post-surgery.
(A and B) Ultrasound assessed cardiac hypertrophy degree pre-sacrifice (n = 7).
(C and D) Ultrasound evaluated cardiac function pre-sacrifice (n = 7).
(E–G) Stats on heart weight/body weight, lung weight/body weight, and heart weight/tibia length (n = 7).
(H and I) HE staining of cardiac tissue, with cardiomyocyte cross-sectional area statistics (n = 7). Scale bar: 1 mm in upper panel, 20 μm in lower panel.
(J and K) Sirius Red staining and cardiac tissue statistics (n = 7). Scale bar: 100 μm.
(L) RT-PCR measured Anp, Bnp, and Myh7 expression in cardiac tissue (n = 3).
(M) RT-PCR assessed cardiac tissue fibrosis marker expression (n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figures S1B–S1F.
To further verify the effect of CD9 knockdown on cardiomyocyte size after TAC, HE staining was performed on cardiac tissue sections from mice in each group. The cross-sectional area increased significantly four weeks after TAC and decreased after CD9 knockdown (Figures 2H and 2I). The mouse heart tissue was stained with Sirius Red to detect the effect of CD9 knockdown on the degree of myocardial fibrosis in mice after TAC. The results showed that the degree of interstitial fibrosis increased significantly at 4 weeks after TAC, and the degree of interstitial fibrosis was significantly decreased in CD9 knockdown mice (Figures 2J and 2K). RT-PCR showed that the expressions of Anp, Bnp, Myh7, and Collagen Iα, Collagen III, Ctgf were increased in cardiac tissue after surgery, and knockdown of CD9 could reduce the levels of Anp, Bnp, Myh7, and Collagen Iα, Collagen III, Ctgf (Figures 2L and 2M). Inflammation plays an important role in the development of cardiac hypertrophy. ELISA assays were applied to detect the levels of TNFα and IL6 in serum. The results showed that TAC increased the levels of cytokines in the serum, but the knockdown of CD9 had no effect (Figure S1E). We detected mRNA expression in the heart tissue of these mice and found that TAC increased the mRNA levels of cytokines in heart tissue, and knockdown of CD9 decreased the mRNA expression of cytokines in heart tissue (Figure S1F).
Overexpression of CD9 aggravates TAC-induced pathological cardiac hypertrophy
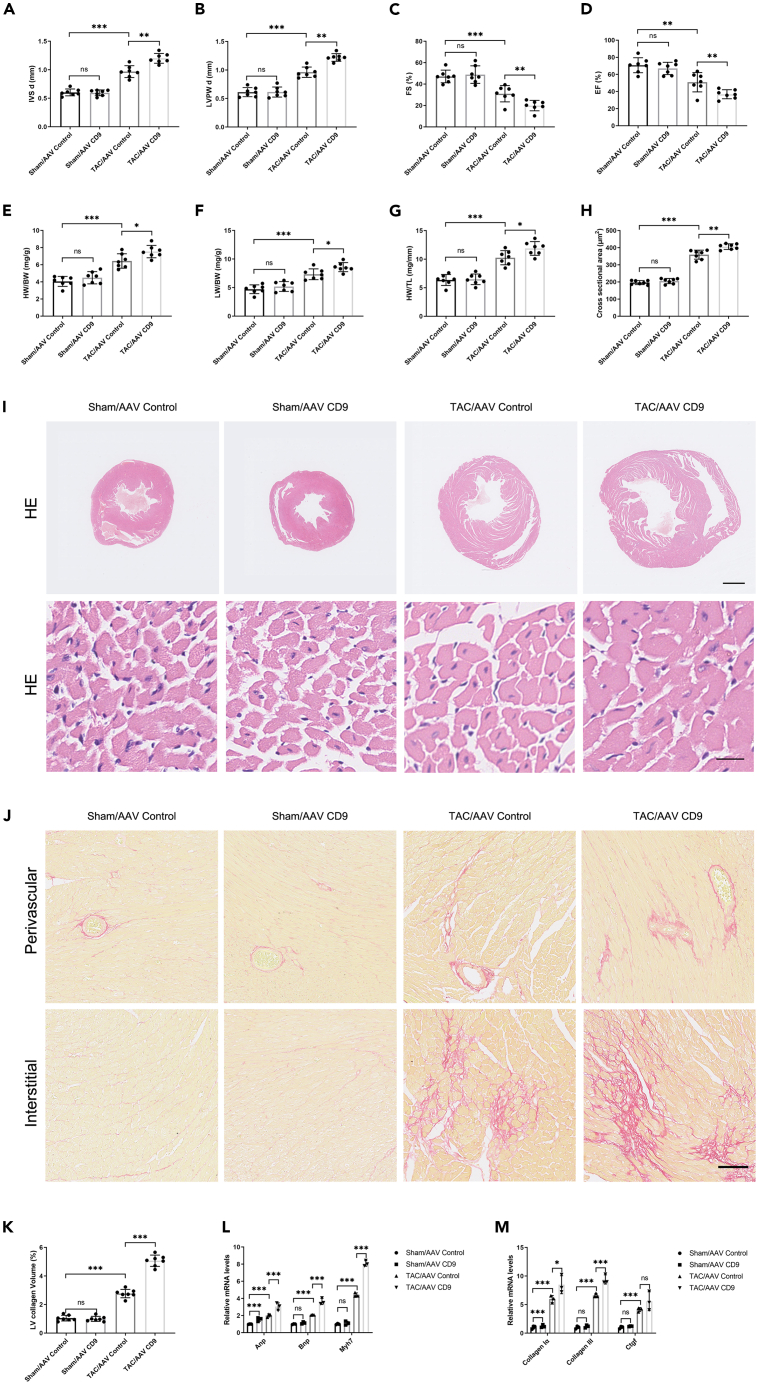
We investigated the effect of CD9 on TAC-induced pathological cardiac hypertrophy. We injected C57BL/6J mice with AAV CD9 and AAV Control virus through the tail vein, and the overexpression efficacy of CD9 was shown by Western blot in Figure S2A; representative ultrasound images are shown in Figure S2B, and the results showed that compared with the TAC/AAV Control group, the degree of cardiac hypertrophy index IVSd and LVPWd were increased, and the myocardial systolic function EF% and FS% were decreased in the TAC/AAV CD9 group (Figures 3A–3D), suggesting that CD9 can exacerbate cardiac function damage after TAC. And the HW/BW, LW/BW, and HW/TL increased after CD9 overexpression (Figures 3E–3G). HE staining was performed on cardiac tissue sections of the mice in each group. The cross-sectional area increased after CD9 overexpression (Figures 3H and 3I). Sirius red staining showed that the degree of interstitial fibrosis was significantly increased in CD9 overexpression mice (Figures 3J and 3K). RT-PCR showed that overexpression of CD9 could increase the mRNA levels of Anp, Bnp, Myh7, Collagen Iα, Collagen III, and Ctgf (Figures 3L and 3M). However, overexpression of CD9 did not affect the levels of cytokines in the serum (Figure S2C), but overexpression of CD9 could increase the mRNA expression of cytokines in the heart tissue (Figure S2D).
Figure 3.
Overexpression of CD9 Aggravates TAC-Induced Pathological Cardiac Hypertrophy
C57 mice injected with AAV CD9 two weeks pre-TAC surgery, sacrificed 4 weeks post-surgery.
(A and B) Ultrasound assessed cardiac hypertrophy degree pre-sacrifice (n = 7).
(C and D) Ultrasound evaluated cardiac function pre-sacrifice (n = 7).
(E–G) Stats on heart weight/body weight, lung weight/body weight, and heart weight/tibia length (n = 7).
(H and I) HE staining of cardiac tissue, with cardiomyocyte cross-sectional area statistics (n = 7). Scale bar: 1 mm in upper panel, 20 μm in lower panel.
(J and K) Sirius Red staining and cardiac tissue statistics (n = 7). Scale bar: 100 μm.
(L) RT-PCR measured Anp, Bnp, and Myh7 expression in cardiac tissue (n = 3).
(M) RT-PCR assessed cardiac tissue fibrosis marker expression (n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S2.
CD9 promotes cardiac hypertrophy by regulating GP130 to activate STAT3
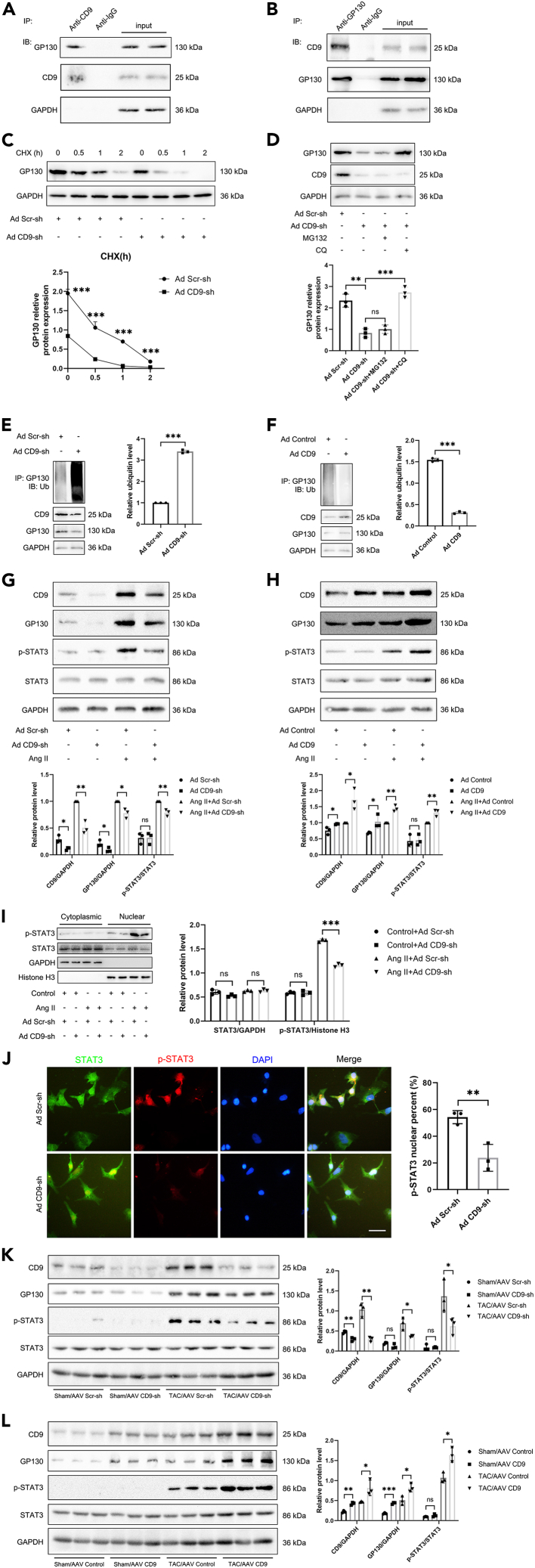
The above gain- and loss-of-function experiments prove that CD9 overexpression can aggravate pressure overload-induced myocardial hypertrophy severity. It has been reported that CD9 can activate STAT3 by stabilizing GP130 and that STAT3 may participate in pathological myocardial hypertrophy. GP130/STAT3 inhibition alleviates TAC-induced cardiac remodeling. Therefore, we aimed to confirm whether CD9 functions via GP130 signaling in cardiac hypertrophy. Co-IP experiments showed that CD9 and GP130 bind to each other in cardiomyocytes (Figures 4A and 4B). Next, we investigated whether CD9 regulated the translation or degradation of GP130. Cardiomyocytes transfected with Ad CD9-shRNA or Ad Scr-shRNA were treated with cycloheximide (CHX), the protein synthesis inhibitor CHX. The abundance of GP130 protein at serial time points (0, 0.5, 1, and 2 h) was analyzed by western blotting. As expected, the results showed that when protein translation was inhibited by CHX, the half-life of the synthesized GP130 was shortened by CD9-shRNA treatment, suggesting that the knockdown of CD9 could promote GP130 degradation (Figure 4C). Ubiquitination-dependent protein degradation is generally divided into two pathways: lysosomal and proteasomal. We treated cardiomyocytes with the lysosome inhibitor CQ and the proteasome inhibitor MG132. Figure 4D shows that the lysosome pathway inhibitor, CQ, slowed down the degradation of GP130 in cardiomyocytes transfected with Ad CD9-shRNA, suggesting that knockdown of CD9 may accelerate the lysosome-mediated degradation of GP130. To prove this, cardiomyocytes were transfected with Ad CD9-shRNA or Ad Scr-shRNA and then subjected to co-IP with GP130, followed by immunoblotting of the precipitates for ubiquitin, showing that knockdown of CD9 increased the ubiquitination of GP130 (Figure 4E). Correspondingly, CD9 decreased the ubiquitination of GP130 (Figure 4F). Furthermore, the expression of GP130 and the phosphorylation of STAT3 decreased when CD9 was knocked down in cardiomyocytes under Ang II stimulation (Figure 4G). Overexpression of CD9 increased the expression of GP130 and the phosphorylation of STAT3 under Ang II stimulation (Figure 4H).
Figure 4.
CD9 Promotes Cardiac Hypertrophy by Regulating GP130 to Activate STAT3
(A and B) Immunoprecipitation and Western blot assays used anti-CD9 and anti-GP130 antibodies to detect GP130 binding with CD9 in cardiomyocytes, with IgG as control (n = 1).
(C) Cardiomyocytes transfected with Ad CD9-shRNA or Ad Scr-shRNA underwent cycloheximide treatment (CHX). GP130 protein abundance analyzed through Western blot over time (0, 0.5, 1, and 2 h), quantified below (n = 3).
(D) Effects of lysosome inhibitor CQ and proteasome inhibitor MG132 on GP130 expression in cardiomyocytes transfected with Ad CD9-shRNA or Ad Scr-shRNA examined (n = 3).
(E) Cardiomyocytes transfected with Ad CD9-shRNA or Ad Scr-shRNA underwent co-IP of GP130, followed by immunoblotting of precipitates for ubiquitin (n = 3).
(F) Cardiomyocytes transfected with Ad CD9 or Ad Control co-IP’d for GP130, followed by immunoblotting of precipitates for ubiquitin (n = 3).
(G and H) Western blot assays performed on cardiomyocytes with CD9 knockdown or overexpression under Ang II stimulation, assessing CD9, GP130, p-STAT3, and STAT3 expression. GAPDH was loading control, with quantification below (n = 3).
(I) Western blot nuclear plasma separation analysis of p-STAT3 nuclear translocation in cardiomyocytes transfected with Ad CD9-shRNA or Ad Scr-shRNA with or without Ang II stimulation (n = 3).
(J) Representative immunofluorescence image showed p-STAT3 nuclear translocation in cardiomyocytes transfected with Ad CD9-shRNA or Ad Scr-shRNA with Ang II stimulation. Scale bar: 50 μm.
(K and L) C57 mice injected with AAV CD9-shRNA or AAV CD9 two weeks pre-TAC surgery, sacrificed 4 weeks post-surgery. Western blot assays on cardiac tissues assessed CD9, GP130, p-STAT3, and STAT3 expression. GAPDH was the loading control, with quantification in the right panel (n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Western blot nuclear plasma separation analysis was performed to investigate the nuclear translocation of p-STAT3 in cardiomyocytes transfected with Ad CD9-shRNA or Ad Scr-shRNA, with or without Ang II stimulation. The results showed that the knockdown of CD9 decreased the nuclear translocation of p-STAT3 in cardiomyocytes following Ang II stimulation (Figure 4I). Immunofluorescence also showed that the nuclear translocation of p-STAT3 decreased in cardiomyocytes transfected with Ad CD9-shRNA with Ang II stimulation. (Figure 4J). C57 mice were injected with AAV CD9-shRNA or AAV CD9 two weeks before TAC surgery, and the mice were sacrificed 4 weeks after surgery. Western blot assays were performed on cardiac tissues to detect the expression of CD9, GP130, p-STAT3, and STAT3. The results showed that GP130 and STAT3 phosphorylation levels decreased when CD9 was knocked down in the TAC model (Figure 4K), and GP130 level and STAT3 phosphorylation levels increased when CD9 was overexpressed in the TAC model (Figure 4L).
Knockdown of GP130 reverses CD9-induced pathological cardiac hypertrophy
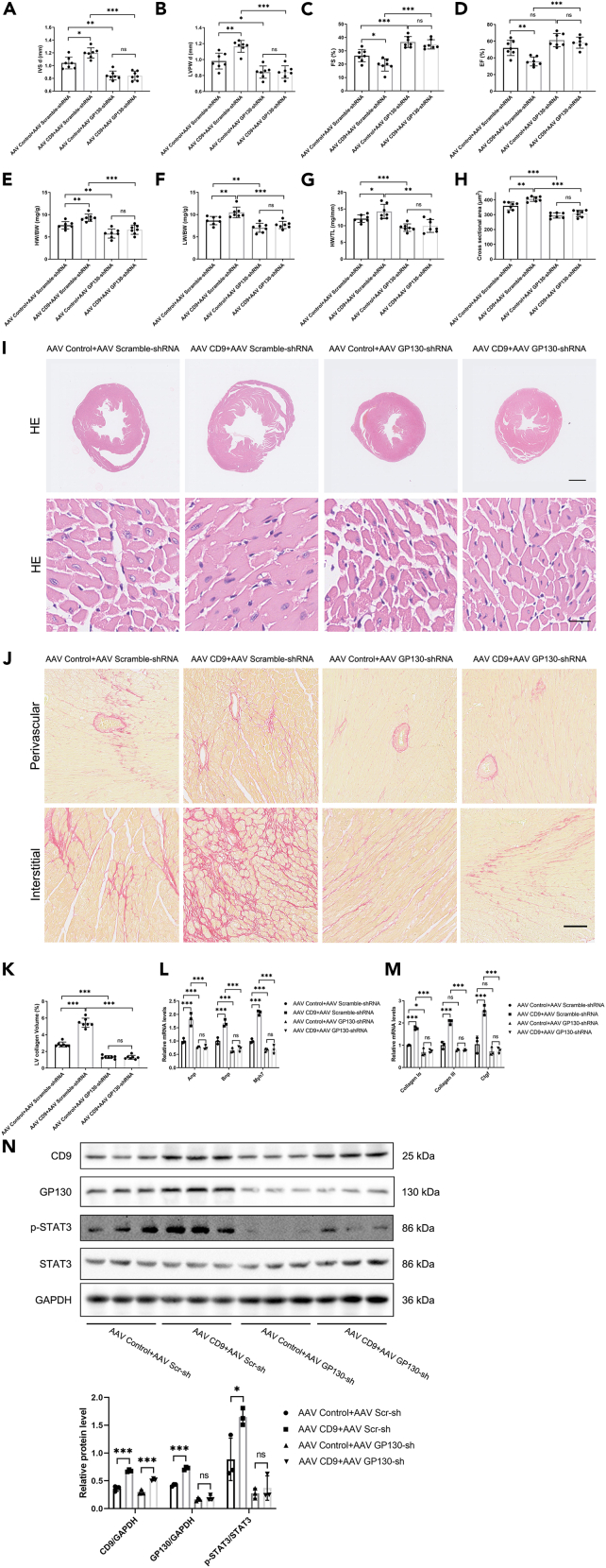
These results indicated that CD9 may be involved in pathological cardiac hypertrophy via GP130/STAT3 signaling. To confirm this, we knocked down GP130 based on overexpression of CD9 in a TAC model. The knockout efficacy of GP130 is shown by western blotting in Figure S3A, and representative ultrasound images are shown in Figure S3B. The results showed that the knockdown of GP130 could reverse the increase in IVSd and LVPWd caused by overexpression of CD9 and increase myocardial contractile function EF% and FS% (Figures 5A–5D). The increase in HW/BW, LW/BW, and HW/TL by overexpressing CD9 was also reversed by the knockdown of GP130 (Figures 5E–5G). Correspondingly, the increased cardiomyocyte cross-sectional area and cardiac fibrosis in CD9-overexpressing mice were reversed by GP130 (Figures 5H–5K). At the same time, the elevated levels of Anp, Bnp, Myh7, Collagen Iα, Collagen III, and Ctgf in CD9-overexpressing mice were reversed by knockdown of GP130 (Figures 5L and 5M). Furthermore, the phosphorylation of STAT3 in CD9-overexpressing mice was reversed by GP130 knockdown (Figure 5N). Therefore, the effect of CD9 on the aggravation of pathological cardiac hypertrophy was reversed by the GP130 knockdown.
Figure 5.
Knockdown of GP130 Reverses CD9-Induced Pathological Cardiac Hypertrophy
C57 mice injected with AAV CD9 and AAV GP130-shRNA two weeks pre-TAC surgery, sacrificed 4 weeks post-surgery.
(A and B) Ultrasound assessed cardiac hypertrophy degree pre-sacrifice (n = 7).
(C and D) Ultrasound evaluated cardiac function pre-sacrifice (n = 7).
(E–G) Stats on heart weight/body weight, lung weight/body weight, and heart weight/tibia length (n = 7).
(H–I) HE staining of cardiac tissue, with cardiomyocyte cross-sectional area statistics (n = 7). Scale bar: 1 mm in upper panel, 20 μm in lower panel.
(J and K) Sirius Red staining and cardiac tissue statistics (n = 7). Scale bar: 100 μm.
(L) RT-PCR measured Anp, Bnp, and Myh7 expression in cardiac tissue (n = 3).
(M) RT-PCR assessed cardiac tissue fibrosis marker expression (n = 3).
(N) Western blot detected the phosphorylation level of STAT3, with quantification below (n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S3.
Discussion
In this study, our results showed that the expression of CD9 increased in TAC myocardial tissue. In addition, knockdown CD9 ameliorated TAC-induced pathological cardiac hypertrophy, whereas CD9 overexpression aggravated it. Mechanistically, our results showed that CD9 partially promotes cardiac hypertrophy by regulating GP130 to activate STAT3.
When the cardiac pressure load increases, early myocardial cell hypertrophy, called compensatory myocardial hypertrophy, ensures cardiac ejection. The number of organelles in hypertrophic cells increases, and their functions are enhanced. However, the compensatory effects of cell hypertrophy are limited. As the pressure load continues, when cardiomyocytes become excessively hypertrophic, the blood supply to the cardiomyocytes becomes relatively deficient, eventually leading to the myocardium’s overloading and a reduction in contractility. When the normal ejection fraction is not met, insufficiency is induced, that is, decompensated pathological myocardial hypertrophy. Pathological myocardial hypertrophy progresses, leading to heart failure.1
The pathophysiological relevance of the GP130 signaling pathway in the development of heart failure was clearly demonstrated using different genetic approaches. For example, Uozumi et al. showed that transgenic mice expressing a dominant negative mutant of GP130 in cardiomyocytes appeared to protect against pressure overload-induced hypertrophy and suggested that GP130 plays a critical role in pressure overload-induced cardiac hypertrophy, possibly through the STAT3 pathway,7 Our results showed that CD9 promotes cardiac hypertrophy partially by regulating GP130 to activate STAT3 in cardiomyocytes, which is consistent with the results of Uozumi et al. In contrast to GP130 deficiency, continuous activation of GP130 in the heart, achieved by the combined overexpression of IL-6 and IL-6 receptors, leads to ventricular hypertrophy.17 Kunisada et al. showed that cardiac-specific transgenic mice overexpressing STAT3 displayed cardiac hypertrophy that was protective against doxorubicin-induced cardiomyopathy.18
These experimental data clearly demonstrate that GP130 receptor signaling is required for compensatory hypertrophy and cardioprotection during episodes of pressure overload. However, other data suggest that the GP130/JAK/STAT pathway plays a protective role in the heart.6,19,20 Therefore, the role of the GP130/STAT3 pathway in pressure overload-induced hypertrophy remains controversial. Recently, Huo et al. showed that IL-6/gp130/STAT3 inhibition by raloxifene alleviates TAC-induced myocardial inflammation, cardiac remodeling, and dysfunction.21 We speculate that the GP130/STAT3 pathway is protective in the compensatory phase of hypertrophy, but has side effects in pathological cardiac hypertrophy and even heart failure.
Using single-cell sequencing of adult human heart tissue, Wang et al.22 found that the expression of CD9 was higher in endothelial cells, smooth muscle cells, and immune cells. However, in our study, by separating rat cardiomyocytes and fibroblasts, we found that CD9 protein was highly expressed in cardiomyocytes, which was inconsistent with the results of single-cell sequencing, which showed that CD9 was expressed more in fibroblasts than in cardiomyocytes. This difference may be due to post-translational modifications that result in inconsistent protein and mRNA expression. In the present study, Ang II stimulation could up-regulate the expression of CD9 in cardiomyocytes. Moreover, it has been reported that CD9 can activate STAT3 by stabilizing GP13013 and that STAT3 may participate in pathological myocardial hypertrophy.7 Our results also showed that knockdown of CD9 may accelerate the ubiquitination and degradation of GP130, and overexpression of CD9 decreases the ubiquitination and degradation of GP130. Furthermore, the knockdown of CD9 decreased the nuclear translocation of p-STAT3 in cardiomyocytes following Ang II stimulation. The effect of CD9 intervention on the GP130/STAT3 pathway in cardiomyocytes indicated that CD9 has an effect, at least in cardiomyocytes.
Specifically, CD9 promotes endothelial-leukocyte adhesion by co-localizing with the adhesion molecules ICAM-1 and VCAM-1 at the endothelial cell surface, reinforcing the adhesion of leukocytes to the endothelium and facilitating the process of extravasation.23 Furthermore, the knockdown of CD9 reduced ICAM-1 and VCAM-1 surface expression, thus reducing leukocyte adhesion and transendothelial migration.24 These results suggest that CD9 is a key regulator of leukocyte recruitment during the inflammatory cascade.11
We detected the levels of inflammatory factors TNFα and IL6 in the serum of mice, and their expression had no change under CD9 intervention. RNA was extracted from the myocardium of mice, and RT-PCR was performed to detect the mRNA expression of Tnfα and Il6. The results showed that overexpression of CD9 promoted the mRNA expression of myocardial inflammatory factors, whereas CD9 knockdown inhibited the mRNA expression of myocardial inflammatory factors.
The ligand binding to the GP130 and LIF receptor complex triggers dimerization, activating Janus kinase (JAK). Activated JAKs rapidly phosphorylate the tyrosine residues of these receptors, which subsequently recruit various signaling molecules, including signal transducer and activator of transcription 3 (STAT3), to the receptor complex. Activated STAT3 dimers translocate to the nucleus, activating the transcription of downstream target genes.25 JAK-mediated activation of the GP130 signaling pathway during pressure overload has been well described, and the cessation of activation of this pathway to prevent GP130 cytokine overstimulation may have independent pathological effects on cardiac function.4 Our study shows that CD9 may serve as a therapeutic target for pathological cardiac hypertrophy.
To our knowledge, the present study demonstrated for the first time that CD9 exacerbates pathological cardiac hypertrophy by regulating the GP130/STAT3 signaling pathway. These findings indicated that CD9 is a potentially important therapeutic target for treating pathological cardiac hypertrophy. CD9 exacerbates pathological cardiac hypertrophy by regulating the GP130/STAT3 signaling pathway.
Limitations of the study
This study was based on a mouse model and may not be applicable to humans. Without conditional knockout genetic engineering mice, cell specificity cannot be guaranteed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Polyclonal anti-CD9 | Affinity Biosciences | Cat# AF5139; RRID:AB_2837625 |
| Rabbit Polyclonal anti-NPPA | Proteintech | Cat# 27426-1-AP; RRID:AB_2880868 |
| Rabbit Polyclonal anti-HIF1A | Affinity Biosciences | Cat# AF1009; RRID:AB_2835328 |
| Mouse Monoclonal anti-GAPDH | Proteintech | Cat# 60004-1-Ig; RRID:AB_2107436 |
| Rabbit Polyclonal anti-CD130/gp130 | Affinity Biosciences | Cat# AF6291; RRID:AB_2835141 |
| Rabbit Polyclonal anti-STAT3 | Proteintech | Cat# 10253-2-AP; RRID:AB_2302876 |
| Phospho-Stat3 (Tyr705) (D3A7) XP Rabbit mAb | Cell Signaling Technology | Cat# 9145; RRID:AB_2491009 |
| Mouse monoclonal anti cTnT | Santa Cruz | Cat# sc-20025; RRID:AB_628403 |
| Rabbit monoclonal anti-actin | Abcam | Cat# ab179467; RRID:AB_2737344 |
| Goat anti-rabbit IgG (H+L), HRP conjugate | Proteintech | Cat# SA00001-2; RRID:AB_2722564 |
| Rabbit polyclonal ubiquitin anti-ub | Proteintech | Cat# 10201-2-AP; RRID:AB_671515 |
| Mouse Monoclonal anti-STAT3 | Proteintech | Cat# 60199-1-Ig; RRID: AB_10913811 |
| Mouse Monoclonal anti-CD9 | Proteintech | Cat# 60232-1-Ig; RRID: AB_11232215 |
| Bacterial and virus strains | ||
| pAAV:cTNT:CD9 3xflag | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| CHX | MCE, MedChemExpress | HY-12320 |
| CQ | MCE, MedChemExpress | HY-17589 |
| MG132 | MCE, MedChemExpress | HY-13259 |
| Ang II | R&D systems | Cat# 1158 |
| Deposited data | ||
| GSE101977 | National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101977 |
| GSE180720 | National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE180720 |
| GSE89714 | National Institutes of Health (NIH) | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE89714 |
| Experimental models: Organisms/strains | ||
| Eight-week-old male C57BL/6J mice | Beijing HFK Bioscience Co. Ltd | N/A |
| Oligonucleotides | ||
| See Table S4 for the primers sequences | This paper | N/A |
| CCGG GGAGAGCAAGTGCTATCAAAT CTCGAG ATTTGATAGCACTTGCTCTCC TTTTT | mice gp130 shRNA | N/A |
| CCGG GGATGAGGTGATTAAAGAACT CTCGAG AGTTCTTTAATCACCTCATCC TTTTT | Mice cd9 shRNA | N/A |
| CCGG GGGAAACACTCAAAGCCATCC CTCGAG GGATGGCTTTGAGTGTTTCCC TTTTT | Rat cd9 shRNA | N/A |
| Recombinant DNA | ||
| pAAV:cTNT:CD9 3xflag | This paper | N/A |
| Software and algorithms | ||
| ImageJ | National Institutes of Health (NIH) | https://imagej.net/ij/ |
| GraphPad Software | San Diego, CA | https://www.graphpad.com |
| Other | ||
| HE Staining Kit | Solarbio | Cat#G1120 |
| Sirius Red Staining Kit | Polysciences | Cat#24901 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chao Zhang (zhangchao1@hust.edu.cn).
Materials availability
This study did not generate new unique reagents.
Method details
Experimental model and study participant details
Eight-week-old male C57BL/6J mice provided by HFK Bioscience (Beijing, China) were housed in a specific pathogen-free (SPF) animal room with a 12-hour light/12-hour dark cycle and free access to food and water. Primary myocardial cells were isolated from SD rats within 1–2 days of birth. All experimental procedures were approved by the Animal Use Subcommittee of Tongji Medical College of Huazhong University of Science and Technology.
Isolation and culture of primary myocardial cells
According to previous reports,26 Neonatal rat cardiomyocytes (NRCMs) and cardiac fibroblasts (NRCFs) were isolated from newborn Sprague-Dawley rats within 48 h of birth using 0.04% collagen II (Worthington) and 0.03% trypsin (Gibco). The two cell populations were separated based on their distinct adhesion times. NRCMs were then resuspended in DMEM with 10% FBS and seeded at a density of 2ⅹ105 /mL into culture dishes or flasks, and the corresponding intervention and treatment were administered to the cells 24 h later. Stable cells were infected with Adenovirus Control (Ad Control), Adenovirus-CD9 (Ad CD9), adenovirus scrambled shRNA (Ad-Scr-sh), and Adenovirus CD9-shRNA (Ad CD9-sh), respectively. After 24 h of infection, the cardiomyocytes were treated with Ang II (50 μM) for 24 h. The cells were then collected for further experiments.
Recombinant adenovirus and adeno-associated virus production
Recombinant adenoviruses containing CD9, CD9 shRNA (a short hairpin RNA targeting CD9), green fluorescent protein (GFP, as a negative control), or scrambled shRNA (as a nonspecific control) were used to infect primary cardiomyocytes in vitro. Recombinant adeno-associated virus serotype 9 (rAAV9; Obio Technology) was used to overexpress and knockdown CD9 in vivo. The recombinant adeno-associated virus (AAV) system (type 9), which contains control and CD9 with a heart-specific promoter (TNT promoter) or scrambled shRNA and CD9 shRNA, was employed to manipulate the expression of CD9 in vivo via tail vein injection. Mice were injected via the tail vein with 100 μl of virus containing 2×1011 VG of the AAV9 vector genomes. The recombinant adeno-associated virus (AAV) system (type 9) containing scrambled shRNA and GP130 shRNA was used to manipulate the expression of GP130 in vivo via tail vein injection. Mice were injected via the tail vein with 100 μl of virus containing 2×1011 VG of the AAV9 vector genomes. All adenoviruses and adeno-associated viruses were provided by Obio Technology Corp., Ltd. (Shanghai, China).
Transaortic constriction (TAC) model
Male C57BL/6J mice aged 8-10 weeks were randomly divided into four groups. Subsequently, 0.5% pentobarbital was injected into the abdominal cavity for anesthesia, fixed on the surgical plate, and depilated the anterior area. The skin was cut from the upper limbs of the mice to separate soft tissues on both sides. A 7-0 medical suture was inserted into the adipose tissue at the upper margin of the aorta and out of the triangular area of the adipose soft tissue at the lower margin of the aorta with ophthalmic tweezers. The suture was pulled out, and the original route was withdrawn from the threading device. The 27G needle was placed at the aortic arch, and the 7-0 suture was tied with a surgical knot. After the two knots were tightened, the excess suture was cut, and the 27G needle was withdrawn. 4-0 medical sutures were used to suture the left and right pectoralis major muscles. The skin was sutured and disinfected with alcohol cotton balls, and the mice were placed on a heating pad at 37°C until they recovered from anesthesia. The Sham group was sutured after aortic arch exposure. The heart function of the mice was assessed using ultrasound 4 weeks after TAC and before surgery, and the mice's body, heart, and lung weights were measured.
Luciferase assays
Rat CD9 promoters was amplified using PCR with rat genomic DNA and cloned into the pGL3.0-Basic vector (Promega, Madison, WI) using the One Step Cloning Kit (C112-02, Vazyme Biotech Ltd., Nanjing, China). The putative binding site of HIF1α of the CD9 promoter was constructed by site-directed mutagenesis using a QuikChange II Kit (Stratagene, La Jolla, CA). Luciferase reporter constructs (wild-type or mutated) were co-transfected with an internal control plasmid pRL-TK (Renilla luciferase reporter plasmid, Promega) into NRCMs. The luciferase activity was determined with Dual-Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s instructions.
Echocardiography and electrocardiogram
After the mice were anesthetized with isoflurane, hair in the anterior chest area was removed after the mice were in a stable state. Furthermore, they were fixed to the ultrasound examination table in the supine position, and then the ultrasound coupling agent was applied to the anterior chest area; the mice's anal temperature was 37±0.4°C, and the ambient temperature of the ultrasound room was 25°C. Long-axis and short-axis B-mode ultrasound images of the mouse heart were collected through a Vevo2100 small animal ultrasound, and the cardiac ejection fraction (EF%), fractional shortening (FS%), and left ventricular end-diastolic phase were measured and calculated to analyze the cardiac function of mice. The interventricular septal (IVS) and left ventricular posterior wall diastolic thicknesses (LVPWd) were used to compare the degree of cardiac hypertrophy. The detailed parameters of echocardiography were provided in the Tables S1–S3.
Histological assessments
Tissue sections were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin by the Bios Biological Company. Serial 5 mm-thick transverse sections were stained in batches with hematoxylin and eosin (H&E). Digital images were obtained using an inverted microscope (OLYMPUS IX73, Tokyo, Japan). ImageJ software was used for the morphometric analyses.
Sirius red staining
Tissue sections were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin by the Bios Biological Company. Serial 5 mm-thick transverse sections were stained in batches with Lapis lazuli blue dye for 5-10 min, washed with distilled water three times, and stained in batches with Sirius red saturated picric acid dye for 15-30 min. Anhydrous ethanol was used for direct differentiation, dehydration, and neutral gum sealing. Digital images were obtained using an inverted microscope (OLYMPUS IX73, Tokyo, Japan). ImageJ software was used for the morphometric analyses. The percentage of the positive area of Sirius red staining relative to the total area of the left ventricle was calculated.
Quantitative real-time RT-PCR (qRT-PCR)
Total RNA was extracted from mouse cells and heart tissues using TRIzol reagent (D9108A; TaKaRa Bio). Reverse transcription of isolated RNA into complementary DNA (cDNA) was performed using the RR047A PrimeScript™ RT reagent Kit (Perfect Real Time) (TaKaRa), which removes genomic DNA. SYBR Green (Vazyme) was used to quantify the amplification products using a PRISM 7900 Sequence Detector System (Applied Biosystems, Foster City). All genes were quantified using β-actin as the internal control. The primer sequences were listed in the Table S4.
Co-immunoprecipitation (Co-IP) and western blot
Cells were lysed in ice-cold IP buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 0.5 mM β-glycerophosphate disodium salt hydrate and 1 mM phenylmethylsulfonyl) containing complete protease inhibitor (no. 04693132001, Roche) and centrifuged at 12,000 g for 15 min. The cell lysate was collected and incubated with protein G Sepharose beads (no. 11719416001, Roche) with the indicated antibody overnight at 4°C and then washed in an immunoprecipitation buffer. The immune complexes were collected and immunized with the indicated antibodies. Whole-cell lysates were isolated from tissues or cells at the indicated times using RIPA lysis buffer with a PhosSTOP phosphatase inhibitor (Roche Diagnostics, Barcelona, Spain) and a complete protease inhibitor cocktail (Roche). A BCA protein assay kit (Thermo Scientific, Waltham, MA, USA) was used to determine protein concentrations. Equal amounts of proteins were subjected to PAGE and transferred onto a PVDF membrane (Millipore, Billerica, MA, USA). After incubation in non-fat milk in TBST to block the membranes, they were incubated with the following primary antibodies: anti-CD9 (Affinity, AF5139, 1:1000), anti-ANP (Proteintech, 27426-1-AP, 1:1000), anti-HIF1α (Affinity, AF1009, 1:1000), anti-GAPDH (Proteintech, 60004-1-Ig, 1:100000), anti-GP130 (Affinity, AF6291, 1:1000), anti-STAT3 (Proteintech, 10253-2-AP, 1:2000), anti-p-STAT3 (Tyr705) (Cell Signaling Technology, #9145, 1:1000) at 4°C overnight. Protein STAT3 was blotted on different blots with GAPDH using the same sample, and all other proteins were blotted on the same blot with GAPDH. The samples were then incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Chemiluminescent signals were detected using the ChemiDoc XRS+ imaging system (Bio-Rad, Hercules, CA, USA).
Immunofluorescence
To analyze the colocalization of STAT3 and p-STAT3 in cardiomyocytes, NRCMs were seeded onto coverslips in 24-well plates. The cells were fixed in 10% formalin and blocked with 2.5% goat serum. Mouse STAT3 (Proteintech; 60199-1-Ig) and rabbit p-STAT3 (Tyr705) (Cell Signaling Technology; 9145) were used as primary antibodies. Anti-mouse IgG (H+L), F(ab’)2 fragment (Alexa Fluor® 488 Conjugate) (Cell Signaling Technology; 4408), and Anti-rabbit IgG (H+L), F(ab’)2 fragment (Alexa Fluor® 594 Conjugate) (Cell Signaling Technology; 8889) were used as corresponding secondary antibodies. CD9 expression and localization in the heart sections of mice were investigated using CD9 and cTnT (a marker of cardiomyocytes) co-staining. Mouse cTnT-specific antibody (Santa Cruz, sc-20025) and rabbit CD9 antibody (Affinity Biosciences; AF5139) were used as primary antibodies. Anti-mouse IgG (H+L), F (ab’) 2 fragment (Alexa Fluor® 488 Conjugate) (Cell Signaling Technology; 4408), and Anti-rabbit IgG (H+L), F(ab’)2 Fragment (Alexa Fluor® 594 Conjugate) (Cell Signaling Technology; 8889) antibodies were applied as the corresponding secondary antibodies. Cell nuclei were stained with DAPI. Immunofluorescent images were obtained using a fluorescence microscope (Olympus). The software ImageJ was used to measure the average fluorescence intensity of CD9 in heart sections, the Mean gray value=Integrated Density/Area ( Arbitrary Units A.U.). p-STAT3 nuclear translocation in cardiomyocytes was qualified using the ImageJ software by calculating the ratio of the number of p-stat3 positive cells and the number of stat3 positive cells.
Quantification and statistical analysis
Data are presented as mean ± SD values. Data were tested for normality using the Shapiro–Wilk and Kolmogorov–Smirnov normality tests. All normally distributed data were analyzed using Student’s t-test and one-way ANOVA, followed by Tukey’s post-hoc analysis. Statistical analyses were performed using the non-parametric Kruskal–Wallis test for data that were not normally distributed. All experiments were performed at least in triplicate and representative data are shown. All analyses were performed using the Prism software (GraphPad Software, San Diego, CA, USA). p < 0.05 was defined as statistically significant.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (No: 82000050, No: 82170239 and No: 81900257). The Medical Science and Technology Program of Henan Province, LHGJ20220286. Postdoctoral funding for the First Affiliated Hospital of Zhengzhou University (71983 and 366733).
Author contributions
K.H., C.Z.., and Z.L. conceived and designed the research; Y.L., S.F., and L.K. performed the experiments; Y.L., S.F., L.K., Z.H., Y.Z., J.S., L.G., M.W., Y.K., and X.L. contributed to analysis the data; K.H., C.Z., and Z.L. wrote and revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare that they have no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: September 28, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108070.
Contributor Information
Kun Huang, Email: 446372485@qq.com.
Chao Zhang, Email: zhangchao1@hust.edu.cn.
Zhibo Liu, Email: lzl1041100271@163.com.
Supplemental information
Data and code availability
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Nakamura M., Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018;15:387–407. doi: 10.1038/s41569-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 2.Bernardo B.C., Weeks K.L., Pretorius L., McMullen J.R. Molecular distinction between physiological and pathological cardiac hypertrophy: experimental findings and therapeutic strategies. Pharmacol. Ther. 2010;128:191–227. doi: 10.1016/j.pharmthera.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Molkentin J.D., Dorn G.W., II Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu. Rev. Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 4.Yasukawa H., Hoshijima M., Gu Y., Nakamura T., Pradervand S., Hanada T., Hanakawa Y., Yoshimura A., Ross J., Chien K.R. Suppressor of cytokine signaling-3 is a biomechanical stress–inducible gene that suppresses gp130-mediated cardiac myocyte hypertrophy and survival pathways. J. Clin. Invest. 2001;108:1459–1467. doi: 10.1172/jci200113939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ancey C., Menet E., Corbi P., Fredj S., Garcia M., Rücker-Martin C., Bescond J., Morel F., Wijdenes J., Lecron J.C., Potreau D. Human cardiomyocyte hypertrophy induced in vitro by gp130 stimulation. Cardiovasc. Res. 2003;59:78–85. doi: 10.1016/s0008-6363(03)00346-8. [DOI] [PubMed] [Google Scholar]
- 6.Fischer P., Hilfiker-Kleiner D. Survival pathways in hypertrophy and heart failure: the gp130-STAT3 axis. Basic Res. Cardiol. 2007;102:279–297. doi: 10.1007/s00395-007-0658-z. [DOI] [PubMed] [Google Scholar]
- 7.Uozumi H., Hiroi Y., Zou Y., Takimoto E., Toko H., Niu P., Shimoyama M., Yazaki Y., Nagai R., Komuro I. gp130 plays a critical role in pressure overload-induced cardiac hypertrophy. J. Biol. Chem. 2001;276:23115–23119. doi: 10.1074/jbc.M100814200. [DOI] [PubMed] [Google Scholar]
- 8.Umeda R., Satouh Y., Takemoto M., Nakada-Nakura Y., Liu K., Yokoyama T., Shirouzu M., Iwata S., Nomura N., Sato K., et al. Structural insights into tetraspanin CD9 function. Nat. Commun. 2020;11:1606. doi: 10.1038/s41467-020-15459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes R., Cardeñes B., Machado-Pineda Y., Cabañas C. Tetraspanin CD9: A Key Regulator of Cell Adhesion in the Immune System. Front. Immunol. 2018;9:863. doi: 10.3389/fimmu.2018.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powner D., Kopp P.M., Monkley S.J., Critchley D.R., Berditchevski F. Tetraspanin CD9 in cell migration. Biochem. Soc. Trans. 2011;39:563–567. doi: 10.1042/BST0390563. [DOI] [PubMed] [Google Scholar]
- 11.Brosseau C., Colas L., Magnan A., Brouard S. CD9 Tetraspanin: A New Pathway for the Regulation of Inflammation? Front. Immunol. 2018;9:2316. doi: 10.3389/fimmu.2018.02316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorico A., Lorico-Rappa M., Karbanová J., Corbeil D., Pizzorno G. CD9, a tetraspanin target for cancer therapy? Exp. Biol. Med. 2021;246:1121–1138. doi: 10.1177/1535370220981855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y., Zhou W., Cheng L., Chen C., Huang Z., Fang X., Wu Q., He Z., Xu S., Lathia J.D., et al. Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells. Cell Death Differ. 2017;24:167–180. doi: 10.1038/cdd.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J.R., Choi J.H. CD9 expression in vascular aging and atherosclerosis. Histol. Histopathol. 2020;35:1449–1454. doi: 10.14670/hh-18-268. [DOI] [PubMed] [Google Scholar]
- 15.Cho J.H., Kim E.C., Son Y., Lee D.W., Park Y.S., Choi J.H., Cho K.H., Kwon K.S., Kim J.R. CD9 induces cellular senescence and aggravates atherosclerotic plaque formation. Cell Death Differ. 2020;27:2681–2696. doi: 10.1038/s41418-020-0537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouger-Gaudichon J., Cousin E., Jakobczyk H., Debaize L., Rio A.G., Forestier A., Arnaud M.P., Villacreces A., Praloran V., Jacamo R., et al. Hypoxia regulates CD9 expression and dissemination of B lymphoblasts. Leuk. Res. 2022;123:106964. doi: 10.1016/j.leukres.2022.106964. [DOI] [PubMed] [Google Scholar]
- 17.Hirota H., Yoshida K., Kishimoto T., Taga T. Continuous activation of gp130, a signal-transducing receptor component for interleukin 6-related cytokines, causes myocardial hypertrophy in mice. Proc. Natl. Acad. Sci. USA. 1995;92:4862–4866. doi: 10.1073/pnas.92.11.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunisada K., Negoro S., Tone E., Funamoto M., Osugi T., Yamada S., Okabe M., Kishimoto T., Yamauchi-Takihara K. Signal transducer and activator of transcription 3 in the heart transduces not only a hypertrophic signal but a protective signal against doxorubicin-induced cardiomyopathy. Proc. Natl. Acad. Sci. USA. 2000;97:315–319. doi: 10.1073/pnas.97.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer P., Hilfiker-Kleiner D. Role of gp130-mediated signalling pathways in the heart and its impact on potential therapeutic aspects. Br. J. Pharmacol. 2008;153:S414–S427. doi: 10.1038/bjp.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boengler K., Hilfiker-Kleiner D., Drexler H., Heusch G., Schulz R. The myocardial JAK/STAT pathway: from protection to failure. Pharmacol. Ther. 2008;120:172–185. doi: 10.1016/j.pharmthera.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Huo S., Shi W., Ma H., Yan D., Luo P., Guo J., Li C., Lin J., Zhang C., Li S., et al. Alleviation of Inflammation and Oxidative Stress in Pressure Overload-Induced Cardiac Remodeling and Heart Failure via IL-6/STAT3 Inhibition by Raloxifene. Oxid. Med. Cell. Longev. 2021;2021:6699054. doi: 10.1155/2021/6699054. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Wang L., Yu P., Zhou B., Song J., Li Z., Zhang M., Guo G., Wang Y., Chen X., Han L., Hu S. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat. Cell Biol. 2020;22:108–119. doi: 10.1038/s41556-019-0446-7. [DOI] [PubMed] [Google Scholar]
- 23.Bailey R.L., Herbert J.M., Khan K., Heath V.L., Bicknell R., Tomlinson M.G. The emerging role of tetraspanin microdomains on endothelial cells. Biochem. Soc. Trans. 2011;39:1667–1673. doi: 10.1042/BST20110745. [DOI] [PubMed] [Google Scholar]
- 24.Barreiro O., Yáñez-Mó M., Sala-Valdés M., Gutiérrez-López M.D., Ovalle S., Higginbottom A., Monk P.N., Cabañas C., Sánchez-Madrid F. Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood. 2005;105:2852–2861. doi: 10.1182/blood-2004-09-3606. [DOI] [PubMed] [Google Scholar]
- 25.Yamauchi-Takihara K., Kishimoto T. A Novel Role for STAT3 in Cardiac Remodeling. Trends Cardiovasc. Med. 2000;10:298–303. doi: 10.1016/s1050-1738(01)00066-4. [DOI] [PubMed] [Google Scholar]
- 26.Yuan L., Bu S., Du M., Wang Y., Ju C., Huang D., Xu W., Tan X., Liang M., Deng S., et al. RNF207 exacerbates pathological cardiac hypertrophy via post-translational modification of TAB1. Cardiovasc. Res. 2023;119:183–194. doi: 10.1093/cvr/cvac039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.